Abstract
Rotaviruses perform the remarkable tasks of transcribing and replicating 11 distinct double-stranded RNA genome segments within the confines of a subviral particle. Multiple viral polymerases are tethered to the interior of a particle, each dedicated to a solitary genome segment but acting in synchrony to synthesize RNA. Although the rotavirus polymerase specifically recognizes RNA templates in the absence of other proteins, its enzymatic activity is contingent upon interaction with the viral capsid. This intraparticle strategy of RNA synthesis helps orchestrate the concerted packaging and replication of the viral genome. Here, we review our current understanding of rotavirus RNA synthetic mechanisms.
Keywords: Double-stranded RNA Viruses, Reovirus, Transcription, Viral Polymerase, Viral Replication, RNA Synthesis, RNA-dependent RNA Polymerase, Rotavirus
Introduction
A distinguishing feature of viruses with segmented double-stranded RNA (dsRNA)2 genomes is that they perform all stages of RNA synthesis inside icosahedral subviral particles (1–4). Each genome segment is dedicated to a single polymerase complex (PC) anchored to the interior of the particle. Particle-associated PCs act in synchrony to transcribe and replicate the viral dsRNA segments. Thus, subviral particles of dsRNA viruses can be viewed as nanoscale factories containing multiple highly coordinated PC machines. During transcription, each genome segment serves as a template for the generation of plus-strand RNAs (+RNAs). In addition to directing viral protein expression, +RNAs also serve as templates for minus-strand synthesis to recreate dsRNA duplexes. The intraparticle mechanism of RNA synthesis not only protects the genome from recognition by antiviral dsRNA sensors but also provides a mechanism for coordinating +RNA packaging with genome replication (4).
As the primary cause of life-threatening gastroenteritis in young children, rotaviruses (RVs) have long been the subject of basic research (5). Specifically, biochemical studies have probed the functions of viral proteins involved in RNA synthesis and have elucidated critical features of viral RNA templates. Moreover, the recent determination of a high resolution structure of the RV RNA-dependent RNA polymerase (RdRp) in complex with RNA has greatly enhanced our understanding of intraparticle RNA synthesis (6).
Triple-layered Structure of RV Particles
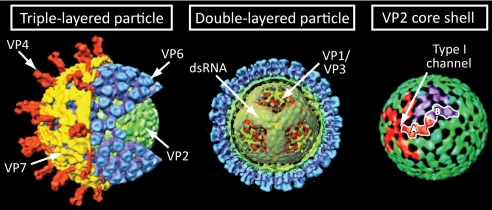
Infectious RVs are non-enveloped, icosahedral, triple-layered particles (TLPs) that encapsidate genomes consisting of 11 segments of dsRNA (7–10). The outer protein layer of the TLP has T = 13 symmetry and is composed of 60 trimers of the viral attachment protein (VP4) and 260 trimers of the viral glycoprotein (VP7) (Fig. 1) (8–12). The intermediate layer also has T = 13 symmetry and is formed by 260 trimers of VP6 (Fig. 1) (8, 9, 13). The innermost layer of the TLP, referred to as the T = 1 core shell, is made up of 120 copies of VP2 organized as 12 decamers (Fig. 1) (13, 14). Each decamer is composed of five VP2-A monomers and five structurally quasi-equivalent but chemically identical VP2-B monomers. VP2-A monomers tightly encircle the 5-fold vertex of the decamer, whereas VP2-B monomers are situated farther back and intercalate VP2-A (13). An aqueous channel traversing the VP6 and VP2 layers at each 5-fold vertex (type I channel) serves as a conduit for the exit of +RNAs, and additional channels permit the entry of nucleotide triphosphates (NTPs) and divalent cations (9, 15, 16). Tethered to the inner surface of the VP2 shell near each 5-fold vertex is a single PC, consisting of the viral RdRp (VP1) and the viral RNA-capping enzyme (VP3) (13, 17–20). Within the core shell, the genome is organized in symmetrical layers, with each segment spooled around one PC (13, 19). Because there are 12 5-fold vertices in the icosahedral core particle and only 11 segments of dsRNA in the RV genome, one vertex may lack an associated RNA molecule or PC. Together, the 11 RV genome segments encode six viral structural proteins (VP1–VP4, VP6, and VP7) and six nonstructural proteins (NSP1–NSP6) (7).
FIGURE 1.
Structural organization of the RV virion as determined by cryoelectron microscopy and image reconstruction. Left, cutaway of the TLP, identifying the outer capsid attachment protein VP4, glycoprotein VP7, and the internal VP6 and VP2 protein layers. Middle, the DLP, which has been colored based on radial distance from the center of the particle. Part of the VP2 and VP6 layers has been computationally removed to reveal the tubular organization of dsRNA spooled around internal projections formed at 5-fold vertices by VP2 N termini, VP1, and VP3. Right, the VP2 core shell, which is shown primarily in green, with VP2-A monomers encircling one 5-fold vertex shown in red and VP2-B monomers encircling one 3-fold vertex shown in purple. A single asymmetrical VP2 dimer formed by the interaction of a VP2-A monomer of one vertex and the VP2-B monomer of another vertex is outlined in white. The location of a type I channel is indicated. Images were kindly provided by B. V. V. Prasad (Baylor College of Medicine).
RNA Synthesis during the RV Life Cycle
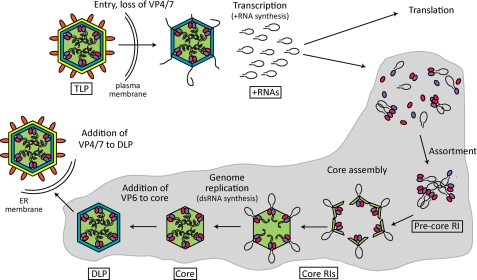
During entry into a host cell, the VP4–VP7 outer capsid layer of the TLP is lost, yielding a double-layered particle (DLP) (Fig. 2) (7, 21). In the cytoplasm, the DLP becomes transcriptionally active; its interior PCs act in concert to produce the 11 species of viral +RNA (1, 15, 22–24). Transcription is conservative, with the parental plus and minus strands of dsRNA segments retained within the DLP. Multiple rounds of transcription result in the accumulation of numerous copies of each +RNA (1, 24). Following synthesis of the first few nucleotides of nascent transcripts, the VP3 component of the PC adds a 5′-terminal m7G cap (17, 25). Thereafter, +RNA molecules are extruded out of the DLP and into the cytosol through type I channels located above the PCs (1, 15, 26). These +RNAs not only direct protein synthesis but also accumulate in electron-dense cytoplasmic inclusions termed viroplasms.
FIGURE 2.
Model of RNA synthesis during the RV life cycle. During entry into target cells, VP4 (orange) and VP7 (yellow) are lost from TLPs. In the resultant DLPs, PCs composed of VP1 (purple) and VP3 (pink) catalyze synthesis of multiple copies of each of the 11 species of RV +RNAs (black ellipses), which anneal in cis to form panhandle structures, with the 3′CS+ remaining largely unbase-paired. The +RNAs serve as templates for translation or are packaged into assembling particles. As viral proteins and +RNAs accumulate in the viroplasm (gray), VP1, VP3, and +RNAs associate to form precore RIs, followed by addition of VP2 (green) to form catalytically active core RIs. Genome replication results in synthesis of the 11 genomic dsRNA segments. VP6 (blue) is added to mature cores, forming DLPs. Finally, DLPs move into the endoplasmic reticulum (ER) where, by budding, they acquire the outer VP4 and VP7 layer.
Viroplasms represent viral factories that are nucleated by the interaction of two multifunctional nonstructural proteins (NSP2 and NSP5) (27–29). The NSP2 component is a homo-octamer with strong affinity for single-stranded RNA and helix unwinding and NTPase activities (28, 30–33). Its counterpart, NSP5, is a serine/threonine-rich phosphoprotein that forms dimers, which bridge NSP2 octamers (28, 29, 34–36). NSP2 and NSP5 are presumed to be responsible not only for recruiting unassembled core proteins to viroplasms but also for retaining +RNAs made by DLPs in these inclusions. The interaction of NSP2 and NSP5 with core proteins and +RNA may be vital for regulating the complex sequence of events required to assemble progeny cores (24, 28, 29). Indeed, the NSP2 octamer undergoes ATP-dependent conformational shifts, consistent with the possibility that it serves as a packaging motor that introduces +RNAs into cores (30).
The characterization of replication intermediates (RIs) isolated from infected cells has provided important clues into the process by which core assembly is coordinated with +RNA packaging and dsRNA synthesis (Fig. 2) (37–40). These studies indicate that the PC proteins, VP1 and VP3, interact with +RNAs in the viroplasm to form replication-incompetent precore RIs (28, 37, 40). It is during the formation of precore RIs that RNA-RNA interactions are thought to drive assortment, selection of the 11 different viral +RNAs that will be packaged and replicated. Interactions of VP2 with precore RIs mediate the formation of replication-competent core RIs, structures that contain the full complement of viral +RNAs (37, 40). The PC proteins of core RIs synthesize 11 viral dsRNAs from the packaged +RNAs. Each PC directs a single round of minus-strand synthesis, and the dsRNA product is retained within the core. Interestingly, core RIs decrease in size and become RNase-resistant as genome replication proceeds, yet genomic dsRNA is protected from dsRNA-specific RNase (40). These observations suggest that, initially, +RNAs are incompletely encapsidated by the VP2 shell of the core RI but are completely protected by the time the VP2 shell has fully matured and genome replication is complete. Following replication, VP6 trimers interact with core RIs, generating progeny DLPs. Some newly formed DLPs may become transcriptionally active, allowing them to contribute to the pool of +RNAs in the viroplasm that will be encapsidated and replicated within assembling cores (41). In the final stages of virion assembly, DLPs transit to the endoplasmic reticulum, where they acquire VP4 and VP7 prior to lytic release from the cell (7, 42).
Protein Requirements for RV RNA Synthesis
Although VP3 is a component of RV PCs, VP1 does not require the viral capping enzyme for dsRNA synthesis in vitro (43). Notably, this enzyme does have an important role in genome replication in vivo, as small interfering RNA-mediated knockdown of VP3 expression in infected cells yields a reduction in progeny particles containing dsRNA (41, 44). In contrast, both in vitro and in vivo experiments have shown that dsRNA synthesis by VP1 is strictly dependent upon the presence of the core shell protein VP2 (41, 45–47). It is not clear how VP1 is regulated by VP2, but prevailing evidence suggests that an interaction between these two proteins during core assembly triggers the RdRp to begin minus-strand synthesis (48, 49). Recombinant VP2 is capable of self-assembling into empty core-like particles and putative assembly intermediates (i.e. decamers), making it an ideal scaffold (50, 51). The N termini of VP2 molecules cluster to form an internal hub at each 5-fold vertex that is in proximity to engage VP1 (Fig. 1) (13). Removal of VP2 N termini not only abrogates VP1 and VP3 encapsidation into core-like particles but also severely diminishes VP1-mediated in vitro genome replication (47, 51–53). This result suggests that the VP2 N-terminal structure plays an important role in genome replication either by directly triggering VP1 to begin RNA synthesis or by facilitating interactions between the RdRp and the T = 1 shell. The molar ratio of VP1 to VP2 for optimal in vitro genome replication is 1:10, which reflects the stoichiometry of the RV core (49). Thus, an attractive model for RV assembly and genome replication is that VP2 decamers form around individual +RNA-bound PCs, making complete 5-fold vertices. It is likely that VP1 within the PC remains inactive until all 12 vertices interact to form an icosahedron.
Unlike genome replication, RV transcription has not been reconstituted in vitro using recombinant proteins. Consequently, the minimal components required for +RNA synthesis are not known. DLPs isolated from RV-infected cells are transcriptionally active in vitro, whereas single-layered core particles are not, suggesting that VP6 is required (1). Addition of recombinant VP6 to isolated RV core particles can restore their transcriptional activity (54–56). Still, mere recoating is not sufficient, as some forms of VP6 are capable of binding cores but not inducing transcription (54–56). These results suggest that VP6 induces precise but as of yet unidentified structural changes in the VP2 layer, which influence the activity of the internally bound PCs. In addition to VP6, the outer VP7 layer is also an important regulator of transcription. Specifically, structural studies have shown that removal of VP7 exposes channels in the VP6 and VP2 layers, presumably allowing NTPs and divalent cations to flow into the DLP interior (57). Moreover, in the absence of VP7, the diameter of the type I channels is increased, facilitating exit of newly made +RNAs (57). During assembly of TLPs, addition of VP7 may serve to inhibit the transcriptional activity of newly formed DLPs.
Critical Features of RV RNA Templates
RV RNA molecules contain important sequence and structural elements that promote their use as templates for replication and transcription. The +RNA templates for dsRNA synthesis contain 5′-m7G caps but lack 3′-poly(A) tails (58). RV +RNAs typically contain a single open reading frame flanked by 5′- and 3′-untranslated regions, which are of variable length. There is little sequence identity among the +RNAs, except for short consensus sequences (CSs) located at the extreme termini (59). The 3′CS of RV +RNA (3′CS+) has the sequence 5′-UGUGACC-3′. This element has been functionally defined, using in vitro replication assays, as the minimal essential promoter for dsRNA synthesis (43, 49, 60, 61). The 5′- and 3′-untranslated regions contain complementary sequences that mediate folding of +RNAs in cis into structures with extended panhandles. In these structures, the 3′CS+ is either not base-paired or only partially so, leaving the 3′-end of the +RNAs in a single-stranded conformation accessible to the RdRp (62, 63). Cell-free replication assays have established that the 3′-terminal CC residues of the 3′CS+ are required for initiation of minus-strand synthesis; on the other hand, electrophoretic mobility assays have shown that the UGUG residues of the 3′CS+ are important for specific recognition of +RNAs by VP1 (46, 49, 60, 61, 64). Thus, the 3′CS+ is a multifunctional element that contributes not only to template recognition but also to RNA catalysis.
VP1 recognition of the highly conserved 3′CS+ likely represents a critical step in packaging of +RNAs into viral cores, providing a mechanism by which viral RNAs can be distinguished from nonviral RNAs. Stem-loops are predicted to extend from the 5′–3′-panhandles of folded RV +RNAs (63, 65, 66). Because the precise location and length of the putative stem-loops differ among the 11 RV +RNAs, these structures are hypothesized to serve as assortment signals that mediate RNA-specific packaging of the 11 viral RNAs during core formation (65).
The minus strands of dsRNA genome segments serve as the templates for transcription. Unlike the 5′-end of the plus strand, the minus strand lacks a 5′-cap (3). The 3′CS of the minus strand (3′CS−) is less conserved than the 3′CS+ of +RNAs, usually consisting of the sequence 5′-(A/U)7GCC-3′ (3). Our understanding of the critical residues and elements in the minus strand that promote +RNA synthesis is limited by the lack of an efficient in vitro transcription system that can utilize exogenous dsRNA templates. Nonetheless, the conservation of the terminal CC residues in both the 3′CS− and 3′CS+ suggest that this dinucleotide plays a critical role in initiating both +RNA and minus-strand RNA (−RNA) synthesis.
Structure of VP1 and Its Interactions with RNA
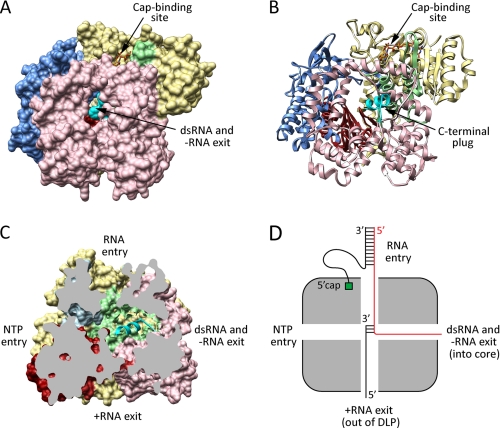
The catalytic polypeptide of the RV PC is VP1, a hollow enzyme composed of three domains: a globular N-terminal domain, a central polymerase domain, and a C-terminal bracelet domain (Fig. 3) (6). The polymerase domain is similar to those of other RdRps, containing fingers, palm, and thumb subdomains as well as the six canonical motifs (A–F) (Figs. 3 and 4) (67). The large N- and C-terminal domains of VP1 sandwich the polymerase domain, enclosing its catalytic center and forming four tunnels between the interior and exterior of the enzyme (Fig. 3, C and D). Based on a comparison with the reovirus RdRp (λ3), the functions of the VP1 tunnels have been identified as (i) template RNA entry, (ii) NTP entry and pyrophosphate exit, (iii) dsRNA and −RNA exit, and (iv) +RNA exit (68). Although other viral RdRps typically contain only three tunnels, the unique four-tunnel arrangement of reovirus and RV RdRps permits the exit of transcription and replication products through different conduits (48). Specifically, during transcription, nascent +RNAs are directed out of VP1 by a route that permits capping via VP3 and exit from the DLP through type I channels. In contrast, during genome replication, nascent dsRNAs are directed out of a separate tunnel in VP1 and placed into the interior of the core. In the solved structure of VP1, the extreme C terminus forms an α-helix that resides within the dsRNA/−RNA exit tunnel (Fig. 3, B and C) (6). The diameter of this C-terminal “plug” is sufficient to block passage of dsRNA, suggesting that it must be removed during genome replication. A putative cap-binding site resides adjacent to the template entry tunnel on the exterior of VP1 (Fig. 3, A and B). Although its contribution to RV RNA synthesis has not yet been formally tested, the cap-binding site may tether the 5′-end of RNA templates and indirectly recruit the 3′ termini toward the entry tunnel via base pairing (48). This feature would be of particular significance during transcription, whereupon VP1 must continuously re-engage the minus strand of dsRNA genome segments.
FIGURE 3.
Structural organization of the RV RdRp. A and B, surface and ribbon representations of VP1 (Protein Data Bank code 2R7R). A m7G cap (orange sticks) is shown in the putative cap-binding site. C, cutaway of VP1 (rotated 90° to the right relative to the image in A) showing the four-tunnel architecture of the polymerase. Putative functions of the tunnels are indicated. For A–C, the N-terminal domain of VP1 is colored yellow; the C-terminal domain is colored pink; and the fingers, palm, and thumb subdomains of the polymerase domain are colored blue, red, and green, respectively. The C-terminal plug is colored cyan. D, schematic depicting the four tunnels of VP1, oriented as in C, during transcription. +RNA is colored black, and −RNA is colored red. The 5′-cap (green) of the plus strand of a dsRNA template undergoing transcription is shown anchored into the cap-binding site.
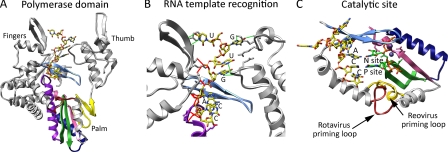
FIGURE 4.
VP1 RNA recognition and polymerization. Shown are ribbon drawings of the polymerase domain (A), template recognition region (B), and catalytic site (C) of VP1 (Protein Data Bank code 2R7R). In all images, a bound oligonucleotide (3′CS+, 5′-UGUGACC-3′; yellow sticks) is shown, and RdRp motifs are colored as follows: motif A (pink), motif B (purple), motif C (green), motif D (navy), motif E (yellow), and motif F (light blue). The remaining portions of VP1 are colored gray. In B, nucleotide bases of the 3′CS+ are labeled, and side chains of VP1 residues that interact with the RNA are shown. Hydrogen bonds formed with the RNA bases and ribose phosphate backbone are shown with green and red lines, respectively. In A and C, the RV priming loop is colored red. In C, nucleotide bases of the 3′-terminal ACC stack are labeled, and the catalytic aspartates of motifs A and C are shown. NTPs (green sticks), divalent cations (slate-blue spheres), and the priming loop (gold) from the structure of reovirus λ3 RdRp (Protein Data Bank code 1N1H) have been overlaid to highlight the overshot position of the RNA and the differences in priming loop orientation of the two polymerases.
Structures derived from soaks of VP1 crystals with RNA oligonucleotides representing the 3′CS+ and 3′CS− confirmed the results of biochemical studies and provided additional insight into mechanisms of RV template recognition (6). VP1 binds the 3′CS+ within the template entry tunnel, with the 3′ terminus of the oligonucleotide extending into the catalytic center of the RdRp (Fig. 4). The 3′CS+ is bound via both nonspecific and sequence-specific interactions (Fig. 4B). In particular, residues in the VP1 template entry tunnel form seven hydrogen bonds with the UGUG bases as well as 11 hydrogen bonds with the phosphate backbone of the 3′CS+. VP2 and VP3 are also capable of binding RNA, but VP1 is the sole component of the RV core that exhibits specific recognition of +RNAs (6, 46, 49, 69). Thus, VP1 is likely to play an important role in selecting RV +RNAs during packaging, and interactions with the UGUG bases of the 3′CS+ may contribute to this process. Within the center of VP1, the bases of the ACC nucleotides of the 3′CS+ form a stack that sits adjacent to the catalytic site (Fig. 4C) (6). These nonspecific stacking interactions might help align templates with the priming site (P site) and incoming nucleotide site (N site) NTPs for catalysis. Interestingly, specific recognition of the 3′CS+ by residues in the VP1 template entry tunnel aligns the +RNA template such that it has overshot the correct register for initiation by a single nucleotide (6). As a consequence, the AC residues of the terminal ACC of the 3′CS+ sit adjacent to the N and P sites. This overshot alignment results in the formation of an autoinhibited VP1/+RNA complex that would need to be corrected for RNA synthesis to commence with the first residue of the template strand.
During transcription, VP1 interacts with the 3′ terminus of the minus strand of dsRNA. In contrast to the base-specific recognition observed for the 3′CS+, VP1 recognizes the 3′CS− solely through nonspecific interactions (6). The manner in which VP1 engages the 3′CS− is nearly identical to the nonspecific contacts between VP1 and the 3′CS+. Thus, VP1 interactions with the 3′CS− likely represent a conserved mechanism of nonspecific RNA recognition that permits VP1 to guide RNA templates of differing sequence into its catalytic center. Specific recognition of the 3′CS− is not as important as specific recognition of the 3′CS+ because dsRNA templates for transcription have already been encapsidated within the DLP.
De Novo Initiation of RV RNA Synthesis and VP1 Activation
Generation of a stable initiation complex is the first step in RV RNA synthesis and involves (i) in-register alignment of the 3′-end of the RNA template; (ii) occupation of the P and N sites by NTPs; and (iii) formation of the first phosphodiester bond, creating an initiating dinucleotide. Unlike RdRps that utilize a protein or RNA primer, VP1 uses a de novo mechanism of initiation for RNA synthesis. Therefore, VP1 must be able to support the priming NTP yet permit elongation of the nascent strand. A loop located between two α-helices of the palm subdomain, close to the P site, is an ideal candidate structure to serve as a platform for initiation complex formation (Fig. 4C) (6). In the enzymatically inactive form of VP1, this priming loop is bent away from the P site such that it is incapable of NTP binding. However, in the structure of enzymatically active reovirus RdRp, the homologous loop is bent toward the P site, supporting the priming NTP by binding its triphosphates (68). This result suggests that the priming loop of VP1 must be repositioned for RNA catalysis to occur.
Although VP1 can bind viral +RNA templates alone, initiation of genome replication requires the core shell protein VP2 (6, 45–47). Thus, in addition to its role as a scaffold, VP2 appears to activate VP1, likely by inducing multiple conformational changes in the RdRp. These changes are anticipated to include (i) realignment of overshot +RNA templates, (ii) movement of the priming loop, and (iii) removal of a C-terminal α-helical plug that obstructs dsRNA exit (6, 48). As mentioned above, VP1 recognizes the 3′CS+ sequence in a manner that causes the terminus of the template to overshoot the initiation register by a single nucleotide (Fig. 4C) (6). It is possible that interactions with VP2 shift the equilibrium toward a properly aligned +RNA template. Oligonucleotides representing a 3′CS+ with a deleted A residue (5′-UGUGCC-3′) align in register for initiation (6), yet templates with this 3′-sequence still cannot be replicated by VP1 in the absence of VP2 (6). Thus, proper alignment of the template may be important, but it is insufficient to render VP1 replication-competent. In addition to the overshot +RNA templates, the position of the priming loop might contribute to the incapacity of VP1 to replicate +RNA in the absence of VP2 (Fig. 4C) (6). Interactions with VP2 are expected to move the flexible VP1 priming loop toward the P site, permitting initiation complex formation. Moreover, a C-terminal α-helical plug resides in the VP1 dsRNA/−RNA exit tunnel (Fig. 3, B and C) (6). In the presence of the plug, the tunnel is too narrow to permit passage of dsRNA. However, movement of the plug, perhaps as a result of interaction with VP2, is anticipated to widen the tunnel sufficiently for exit of nascent genome segments. VP1 proteins in which the C-terminal plug has been deleted are equivalently active to wild-type VP1 in vitro (6). However, they do not gain the capacity to replicate RNA in the absence of VP2. Thus, similar to template alignment, removal of the C-terminal plug is insufficient to render VP1 replication-competent on its own. Taken together, current findings suggest that at least these three conformational changes are required to activate VP1. Since initiation complex formation for transcription entails the same steps as for genome replication, it is likely that some of these conformational changes are also required for VP1 activation during +RNA synthesis.
Future Directions and Concluding Remarks
Although biochemical and structural studies have shed light on the molecular mechanism of RV RNA synthesis, there are several key gaps in knowledge that warrant future investigation. One of the most interesting unresolved events of RV biology is assortment, the efficient packaging of 11 different +RNA segments. Since VP1 is the only viral protein known to bind +RNAs specifically, and it does so via sequences that are conserved across all 11 genome segments, it is likely that RNA-RNA interactions primarily mediate assortment (6). Sequences differ substantially among the 11 RV genome segments and may contain segment-specific packaging information (3, 59). RNA secondary structural elements, exemplified by predicted stem-loop structures (mentioned above), likely play a prominent role in gene-specific packaging (63, 65, 66). Assortment based on RNA-RNA interactions is a strategy that is not unique to RVs. Mounting evidence suggests that assortment for influenza viruses (which encapsidate eight different segments of negative-sense RNA) involves a segment-specific arrangement of RNA mediated by RNA-RNA interactions (70–75). Further insight into RV assortment may be gained from the development of a tractable packaging assay or reverse genetics system.
Aspects of the structural features and interactions between proteins involved in RV intraparticle RNA synthesis also remain incompletely understood. In particular, little is known about how the core shell protein VP2 activates the RV RdRp VP1. The precise interactive domains between these proteins have not yet been mapped, and the changes that occur in VP1 following VP2 binding remain speculative. Although there is evidence that VP1 and VP3 reside near 5-fold vertices in the interior of the core, the precise location of these enzymes remains unclear (19). The nearly immediate capping of nascent +RNAs suggests that VP3 abuts the +RNA exit tunnel of VP1 (25). However, a formal interaction between VP1 and VP3 has not been demonstrated, and structural information for VP3 is currently unavailable. Although addition of VP6 to RV cores results in a switch in VP1 function from replication to transcription, the mechanism of this switch and the components through which these signals are mediated (VP1, VP2, VP3, or RNA) remain unknown (54–56). Future structural and biochemical studies may clarify these unresolved issues.
Intraparticle RNA synthesis is a strategy that is shared with other segmented dsRNA viruses such as reovirus and bluetongue virus. Although the specific arrangement of proteins and number of layers composing virions differ among these viruses, each forms a T = 1 core structure that houses the viral RdRps and capping machinery (2, 10, 14, 19, 25, 76–79). Surprisingly, unlike VP1, the RdRps of reovirus and bluetongue virus are active in vitro in the absence of other viral proteins (68, 80). An independently active RdRp could synthesize dsRNA prior to core assembly, a process that would expose the genome to the cellular contents and that has not been observed previously in infected cells. It remains to be seen whether these differences in RdRp activity are artifacts of in vitro assays and whether we can exploit them to engineer VP1 proteins whose activity is independent of VP2 for future studies of RV RNA synthesis.
Supplementary Material
This work was supported, in whole or in part, by the National Institutes of Health Intramural Research Program, NIAID. This minireview will be reprinted in the 2010 Minireview Compendium, which will be available in January, 2011.
- dsRNA
- double-stranded RNA
- PC
- polymerase complex
- +RNA
- plus-strand RNA
- −RNA
- minus-strand RNA
- RV
- rotavirus
- RdRp
- RNA-dependent RNA polymerase
- TLP
- triple-layered particle
- NTP
- nucleotide triphosphate
- DLP
- double-layered particle
- RI
- replication intermediate
- CS
- consensus sequence
- 3′CS+
- CS at the 3′ terminus of RV +RNA
- 3′CS−
- CS at the 3′ terminus of RV −RNA
- P site
- priming site
- N site
- incoming nucleotide site.
REFERENCES
- 1.Lawton J. A., Estes M. K., Prasad B. V. (2000) Adv. Virus Res. 55, 185–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mertens P. P., Diprose J. (2004) Virus Res. 101, 29–43 [DOI] [PubMed] [Google Scholar]
- 3.Patton J. T., Vasquez-Del Carpio R., Tortorici M. A., Taraporewala Z. F. (2007) Adv. Virus Res. 69, 167–201 [DOI] [PubMed] [Google Scholar]
- 4.McDonald S. M., Patton J. T. (2009) in Viral Genome Replication ( Cameron C. E., Gotte M., Raney K. D. eds) pp.201– 224, Springer Science+Business Media, New York [Google Scholar]
- 5.Parashar U. D., Gibson C. J., Bresse J. S., Glass R. I. (2006) Emerg. Infect. Dis. 12, 304–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu X., McDonald S. M., Tortorici M. A., Tao Y. J., Vasquez-Del Carpio R., Nibert M. L., Patton J. T., Harrison S. C. (2008) Structure 16, 1678–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Estes M. K., Kapikian A. Z. (2007) in Fields Virology ( Knipe D. M., Howley P. M. eds)5th Ed., pp. 1917– 1974, Lippincott Williams & Wilkins, Philadelphia [Google Scholar]
- 8.Li Z., Baker M. L., Jiang W., Estes M. K., Prasad B. V. (2009) J. Virol. 83, 1754–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prasad B. V., Wang G. J., Clerx J. P., Chiu W. (1988) J. Mol. Biol. 199, 269–275 [DOI] [PubMed] [Google Scholar]
- 10.Yeager M., Dryden K. A., Olson N. H., Greenberg H. B., Baker T. S. (1990) J. Cell Biol. 110, 2133–2144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dormitzer P. R., Nason E. B., Prasad B. V., Harrison S. C. (2004) Nature 430, 1053–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prasad B. V., Burns J. W., Marietta E., Estes M. K., Chiu W. (1990) Nature 343, 476–479 [DOI] [PubMed] [Google Scholar]
- 13.McClain B., Settembre E., Temple B. R., Bellamy A. R., Harrison S. C. (2010) J. Mol. Biol. 397, 587–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lawton J. A., Zeng C. Q., Mukherjee S. K., Cohen J., Estes M. K., Prasad B. V. (1997) J. Virol. 71, 7353–7360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lawton J. A., Estes M. K., Prasad B. V. (1997) Nat. Struct. Biol. 4, 118–121 [DOI] [PubMed] [Google Scholar]
- 16.Lawton J. A., Estes M. K., Prasad B. V. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 5428–5433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen D., Luongo C. L., Nibert M. L., Patton J. T. (1999) Virology 265, 120–130 [DOI] [PubMed] [Google Scholar]
- 18.Liu M., Mattion N. M., Estes M. K. (1992) Virology 188, 77–84 [DOI] [PubMed] [Google Scholar]
- 19.Prasad B. V., Rothnagel R., Zeng C. Q., Jakana J., Lawton J. A., Chiu W., Estes M. K. (1996) Nature 382, 471–473 [DOI] [PubMed] [Google Scholar]
- 20.Valenzuela S., Pizarro J., Sandino A. M., Vásquez M., Fernández J., Hernández O., Patton J., Spencer E. (1991) J. Virol. 65, 3964–3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tihova M., Dryden K. A., Bellamy A. R., Greenberg H. B., Yeager M. (2001) J. Mol. Biol. 314, 985–992 [DOI] [PubMed] [Google Scholar]
- 22.Cohen J., Dobos P. (1979) Biochem. Biophys. Res. Commun. 88, 791–796 [DOI] [PubMed] [Google Scholar]
- 23.Cohen J. (1977) J. Gen. Virol. 36, 395–402 [DOI] [PubMed] [Google Scholar]
- 24.Jayaram H., Estes M. K., Prasad B. V. (2004) Virus Res. 101, 67–81 [DOI] [PubMed] [Google Scholar]
- 25.Lawton J. A., Estes M. K., Prasad B. V. (2001) J. Virol. 75, 1632–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Libersou S., Siebert X., Ouldali M., Estrozi L. F., Navaza J., Charpilienne A., Garnier P., Poncet D., Lepault J. (2008) J. Virol. 82, 2844–2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fabbretti E., Afrikanova I., Vascotto F., Burrone O. R. (1999) J. Gen. Virol. 80, 333–339 [DOI] [PubMed] [Google Scholar]
- 28.Taraporewala Z. F., Patton J. T. (2004) Virus Res. 101, 57–66 [DOI] [PubMed] [Google Scholar]
- 29.Patton J. T., Silvestri L. S., Tortorici M. A., Vasquez-Del Carpio R., Taraporewala Z. F. (2006) Curr. Top. Microbiol. Immunol. 309, 169–187 [DOI] [PubMed] [Google Scholar]
- 30.Jayaram H., Taraporewala Z., Patton J. T., Prasad B. V. (2002) Nature 417, 311–315 [DOI] [PubMed] [Google Scholar]
- 31.Taraporewala Z. F., Patton J. T. (2001) J. Virol. 75, 4519–4527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schuck P., Taraporewala Z., McPhie P., Patton J. T. (2001) J. Biol. Chem. 276, 9679–9687 [DOI] [PubMed] [Google Scholar]
- 33.Vasquez-Del Carpio R., Gonzalez-Nilo F. D., Riadi G., Taraporewala Z. F., Patton J. T. (2006) J. Mol. Biol. 362, 539–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eichwald C., Vascotto F., Fabbretti E., Burrone O. R. (2002) J. Virol. 76, 3461–3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vende P., Taraporewala Z. F., Patton J. T. (2002) J. Virol. 76, 5291–5299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poncet D., Lindenbaum P., L'Haridon R., Cohen J. (1997) J. Virol. 71, 34–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gallegos C. O., Patton J. T. (1989) Virology 172, 616–627 [DOI] [PubMed] [Google Scholar]
- 38.Helmberger-Jones M., Patton J. T. (1986) Virology 155, 655–665 [DOI] [PubMed] [Google Scholar]
- 39.Patton J. T., Gallegos C. O. (1988) Virology 166, 358–365 [DOI] [PubMed] [Google Scholar]
- 40.Patton J. T., Gallegos C. O. (1990) J. Gen. Virol. 71, 1087–1094 [DOI] [PubMed] [Google Scholar]
- 41.Ayala-Breton C., Arias M., Espinosa R., Romero P., Arias C. F., López S. (2009) J. Virol. 83, 8819–8831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pesavento J. B., Crawford S. E., Estes M. K., Prasad B. V. (2006) Curr. Top. Microbiol. Immunol. 309, 189–219 [DOI] [PubMed] [Google Scholar]
- 43.Wentz M. J., Zeng C. Q., Patton J. T., Estes M. K., Ramig R. F. (1996) Arch. Virol. 12, 59–67 [DOI] [PubMed] [Google Scholar]
- 44.Vásquez M., Sandino A. M., Pizarro J. M., Fernández J., Valenzuela S., Spencer E. (1993) J. Gen. Virol. 74, 937–941 [DOI] [PubMed] [Google Scholar]
- 45.Mansell E. A., Patton J. T. (1990) J. Virol. 64, 4988–4996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patton J. T. (1996) J. Virol. 70, 7940–7947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patton J. T., Jones M. T., Kalbach A. N., He Y. W., Xiaobo J. (1997) J. Virol. 71, 9618–9626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McDonald S. M., Tao Y. J., Patton J. T. (2009) Curr. Opin. Struct. Biol. 19, 775–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tortorici M. A., Broering T. J., Nibert M. L., Patton J. T. (2003) J. Biol. Chem. 278, 32673–32682 [DOI] [PubMed] [Google Scholar]
- 50.Crawford S. E., Labbé M., Cohen J., Burroughs M. H., Zhou Y. J., Estes M. K. (1994) J. Virol. 68, 5945–5952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Labbé M., Charpilienne A., Crawford S. E., Estes M. K., Cohen J. (1991) J. Virol. 65, 2946–2952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Labbé M., Baudoux P., Charpilienne A., Poncet D., Cohen J. (1994) J. Gen. Virol. 75, 3423–3430 [DOI] [PubMed] [Google Scholar]
- 53.Zeng C. Q., Estes M. K., Charpilienne A., Cohen J. (1998) J. Virol. 72, 201–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Charpilienne A., Lepault J., Rey F., Cohen J. (2002) J. Virol. 76, 7822–7831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kohli E., Pothier P., Tosser G., Cohen J., Sandino A. M., Spencer E. (1993) Arch. Virol. 133, 451–458 [DOI] [PubMed] [Google Scholar]
- 56.Kohli E., Pothier P., Tosser G., Cohen J., Sandino A. M., Spencer E. (1994) Arch. Virol. 135, 193–200 [DOI] [PubMed] [Google Scholar]
- 57.Chen J. Z., Settembre E. C., Aoki S. T., Zhang X., Bellamy A. R., Dormitzer P. R., Harrison S. C., Grigorieff N. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 10644–10648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Imai M., Akatani K., Ikegami N., Furuichi Y. (1983) J. Virol. 47, 125–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mitchell D. B., Both G. W. (1990) Virology 177, 324–331 [DOI] [PubMed] [Google Scholar]
- 60.Patton J. T., Wentz M., Xiaobo J., Ramig R. F. (1996) J. Virol. 70, 3961–3971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wentz M. J., Patton J. T., Ramig R. F. (1996) J. Virol. 70, 7833–7841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barro M., Mandiola P., Chen D., Patton J. T., Spencer E. (2001) Virology 288, 71–80 [DOI] [PubMed] [Google Scholar]
- 63.Chen D., Patton J. T. (1998) J. Virol. 72, 7387–7396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen D., Barros M., Spencer E., Patton J. T. (2001) Virology 282, 221–229 [DOI] [PubMed] [Google Scholar]
- 65.Patton J. T., Spencer E. (2000) Virology 277, 217–225 [DOI] [PubMed] [Google Scholar]
- 66.Tortorici M. A., Shapiro B. A., Patton J. T. (2006) RNA 12, 133–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ferrer-Orta C., Arias A., Escarmís C., Verdaguer N. (2006) Curr. Opin. Struct. Biol. 16, 27–34 [DOI] [PubMed] [Google Scholar]
- 68.Tao Y., Farsetta D. L., Nibert M. L., Harrison S. C. (2002) Cell 111, 733–745 [DOI] [PubMed] [Google Scholar]
- 69.Patton J. T., Chen D. (1999) J. Virol. 73, 1382–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fujii K., Fujii Y., Noda T., Muramoto Y., Watanabe T., Takada A., Goto H., Horimoto T., Kawaoka Y. (2005) J. Virol. 79, 3766–3774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fujii K., Ozawa M., Iwatsuki-Horimoto K., Horimoto T., Kawaoka Y. (2009) J. Gen. Virol. 90, 1734–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fujii Y., Goto H., Watanabe T., Yoshida T., Kawaoka Y. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 2002–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Noda T., Sagara H., Yen A., Takada A., Kida H., Cheng R. H., Kawaoka Y. (2006) Nature 439, 490–492 [DOI] [PubMed] [Google Scholar]
- 74.Ozawa M., Maeda J., Iwatsuki-Horimoto K., Watanabe S., Goto H., Horimoto T., Kawaoka Y. (2009) J. Virol. 83, 3384–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Watanabe T., Watanabe S., Noda T., Fujii Y., Kawaoka Y. (2003) J. Virol. 77, 10575–10583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Grimes J. M., Burroughs J. N., Gouet P., Diprose J. M., Malby R., Ziéntara S., Mertens P. P., Stuart D. I. (1998) Nature 395, 470–478 [DOI] [PubMed] [Google Scholar]
- 77.Hewat E. A., Booth T. F., Loudon P. T., Roy P. (1992) Virology 189, 10–20 [DOI] [PubMed] [Google Scholar]
- 78.Reinisch K. M., Nibert M. L., Harrison S. C. (2000) Nature 404, 960–967 [DOI] [PubMed] [Google Scholar]
- 79.Zhang X., Tang J., Walker S. B., O'Hara D., Nibert M. L., Duncan R., Baker T. S. (2005) Virology 343, 25–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Boyce M., Wehrfritz J., Noad R., Roy P. (2004) J. Virol. 78, 3994–4002 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.