Abstract
Human mitochondrial transcription is driven by a single subunit RNA polymerase and a set of basal transcription factors. The development of a recombinant in vitro transcription system has allowed for a detailed molecular characterization of the individual components and their contribution to transcription initiation. We found that TFAM and TFB2M act synergistically and increase transcription efficiency 100–200-fold as compared with RNA polymerase alone. Both the light-strand promoter (LSP) and the HSP1 promoters displayed maximal levels of in vitro transcription when TFAM was present in an amount equimolar to the DNA template. Importantly, we did not detect any significant transcription activity in the presence of the TFB2M paralog, TFB1M, or when templates containing the putative HSP2 promoter were used. These data confirm previous observations that TFB1M does not function as a bona fide transcription factor and raise questions as to whether HSP2 serves as a functional promoter in vivo. In addition, we did not detect transcription stimulation by the ribosomal protein MRPL12. Thus, only two essential initiation factors, TFAM and TFB2M, and two promoters, LSP and HSP1, are required to drive transcription of the mitochondrial genome.
Keywords: DNA-Protein Interaction, Mitochondria, RNA Polymerase, Transcription, Transcription Promoter
Introduction
Transcription of the human mitochondrial genome is governed by a nuclear-encoded single-subunit RNA polymerase (POLRMT) that is assisted by two transcription initiation factors, TFAM and TFB2M (see Refs. 1 and 2 and references therein). POLRMT possesses promoter recognition functions but depends on TFAM and TFB2M for promoter melting (3). TFAM, a high mobility group class protein, binds to mitochondrial DNA, protects a region 14–35 bp upstream of the light-strand promoter (LSP)4 transcription start site, and assists in assembly of the initiation complex by attracting POLRMT-TFB2M and/or by causing initial melting of the promoter (4). The primary role of TFB2M is to melt the promoter and to stabilize the open promoter complex by simultaneous binding of the priming substrate and the templating DNA base (5). Although the basic requirements for mitochondrial transcription have been established, a number of existing controversial observations preclude a comprehensive view of gene transcription and its regulation in mitochondria. For example, in addition to TFB2M, human mitochondria also contain a homologous factor TFB1M that was reported to stimulate transcription initiation in vitro with 10–100-fold lower efficiency (6, 7). However, in vivo studies demonstrated that although TFB1M is an essential methyltransferase required to methylate 12 S ribosomal RNA, it plays no role in transcription (8).
Another paradox in the field of mitochondrial transcription concerns the existence of an additional promoter in the heavy strand of mtDNA. Transcription initiated at the LSP results in synthesis of a single mRNA that encodes subunit 6 of the NADH dehydrogenase and eight tRNAs (9). The heavy strand of mtDNA encodes 12 polypeptides, two rRNAs, and the rest of the tRNAs; transcription of this strand appears to involve two promoters (10–13). The transcription start site of the HSP1 promoter was shown to be at position 561, whereas HSP2-initiated transcripts were found to originate within tRNAPhe at position 646, 2 nt upstream of the 12 S rRNA 5′-end (10). However, although in vitro transcription using HSP1 and LSP promoters is well documented, no experimental data for HSP2-driven transcription have been reported. Mapping experiments using guanylyltransferase, an enzyme that has specificity toward 5′-end tri- or diphosphates of the RNA, suggest that the identified labeled RNA products are the RNA transcripts and not the products of RNA processing or degradation (10, 12). Notwithstanding these data, in this work, we demonstrate that HSP2 cannot be transcribed in vitro using purified components of the transcription system and is not recognized by POLRMT in a factor-independent assay. We also demonstrate that TFB1M has no transcription activity in vitro, supporting reports that it lacks in vivo activity. Our data suggest that transcription of mitochondrial genes involves only two promoters and depends upon only two absolutely essential factors, TFAM and TFB2M. Our findings underline the importance of corroborating data obtained in vivo by in vitro studies and vice versa.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification of the Components of Mitochondrial Transcription
Human POLRMT, TFB2M, and TFAM were expressed and purified from Escherichia coli (5) or insect cells (2) as described previously. Human mitochondrial TFB1M having a C-terminal His6 tag was cloned and purified as described previously (6). TFB1M lacking 27 N-terminal amino acids (presumptive mitochondrial localization signal) was cloned as an N-terminal intein fusion using the pTYB11 vector (New England Biolabs) and purified using the TFB2M protocol described in Ref. 5 (supplemental Fig. S1).
The coding sequence of human MRPL12 without the mitochondrial targeting signal, residues 46–198 (14, 15), was amplified by PCR from human heart cells cDNA (Panomics) and cloned using the pTYB11 vector as an N-terminal intein fusion. MRPL12 was expressed in BL21 RIPL (DE3) cells (Stratagene) using the TFB2M protocol (5). MRPL12 was purified using chitin-agarose (New England Biolabs) followed by anion exchange chromatography on a MonoQ column (GE Healthcare) in 50–400 mm NaCl gradient (supplemental Fig. S3).
Cloning and purification of yeast (Saccharomyces cerevisiae) Rpo41 and Mtf1 was described previously (16). The budding yeast analog of MRPL12, Mnp1 (residues 35–194), was cloned into the pTrcHisC expression vector (Invitrogen) using NcoI and XhoI restriction sites and expressed and purified as described previously (17).
Transcription Assays
Standard transcription reactions were carried out as described (5). The HSP1 templates contained 83 bp of wild-type human mitochondrial DNA (positions 501–583), 62 bp of LSP DNA (positions 386–447), and 90 bp of HSP2 DNA (positions 605–694). Reactions were carried out at 35 °C using an ApA primer and NTP mixtures, as indicated in the figure legends, for 30 min and stopped by the addition of an equal volume of 95% formamide, 0.05 m EDTA. The products were resolved by 20% PAGE in the presence of 6 m urea and visualized by PhosphorImagerTM (GE Healthcare). In vitro transcription using recombinant proteins and plasmid DNA fragments was performed as reported previously (3). Transcription activity in mitochondrial lysates was monitored as described in Ref. 18. Transcription assays involving yeast mitochondrial proteins were performed as in Ref. 19.
Cross-linking of the Priming Nucleotide by Catalytic Autolabeling
Crosslinking using 2-hydroxybenzaldehyde AMP was performed as described previously (5). The cross-linking was activated by the addition of 0.1 mm NaBH4 for 15 min at room temperature. Autolabeling was carried out by the addition of 1 μl of [α-32P]ATP (800 Ci/mmol) followed by incubation of the reaction for 45 min at 35 °C. The reaction was stopped by the addition of SDS loading buffer, and the products were resolved using a 4–12% Bis-Tris NuPAGE gel (Invitrogen) and visualized by PhosphorImager (GE Healthcare).
RESULTS AND DISCUSSION
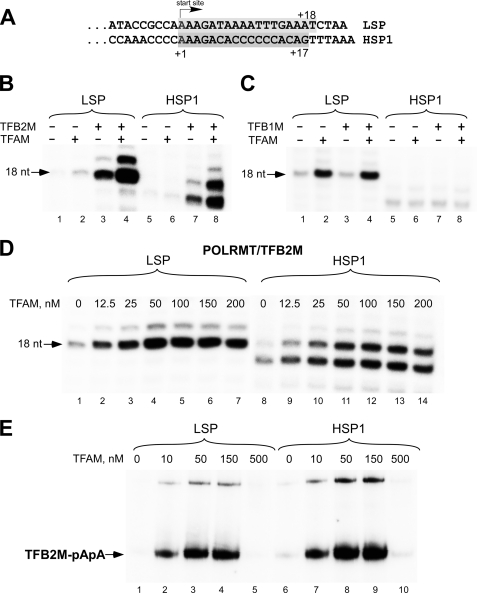
To monitor transcription initiation, we used synthetic templates containing LSP and HSP1 promoters that allow synthesis of 17–18-mer transcripts when one of the NTPs is omitted from the transcription reaction (Fig. 1A). Consistent with a previous report (3), we found that efficient transcription initiation from LSP and HSP1 required both TFAM and TFB2M (Fig. 1B). A faint RNA product was observed when only POLRMT was present in the transcription reaction (Fig. 1B, lane 1, and 1C, lane 1); however, the efficiency of transcription was ∼0.01–1% of that in the presence of both TFB2M and TFAM and varied from one template preparation to another, possibly reflecting the presence of a small amount of a single-stranded template strand. Nevertheless, the addition of TFAM to POLRMT caused a 3-fold stimulation of RNA synthesis, whereas the addition of TFB2M resulted in a 6–10-fold stimulation of transcription of both LSP and HSP1. In either case, transcription was still well below the levels seen when POLRMT, TFAM, and TFB2M were used in combination. The synergistic action of TFB2M and TFAM added together resulted in more than 100–200-fold stimulation of transcription from LSP and HSP1. At the same time, no stimulation of transcription activity on either LSP or HSP1 was observed when TFB1M or TFB1M together with TFAM were added to POLRMT, suggesting that this protein lacks transcription stimulation activity (Fig. 1C). Similar results were also obtained in abortive initiation assays (supplemental Fig. S2A) or when a longer DNA template and the full-length TFB1M (containing the N-terminal, mitochondrial-targeting sequence) were used (supplemental Fig. S2B). We speculate that the previously observed in vitro transcription activity in the presence of TFB1M was due to stimulation of the transcription on the LSP promoter by TFAM. Alternatively, it is possible that co-purification of TFB1M with RNAP resulted in stabilization of RNAP, thus causing the observed differences in activity between RNAP and the RNAP-TFB1M complex (6). Our findings, together with the data of in vivo experiments (8, 20), demonstrate that TFB1M plays no role in mitochondrial transcription. Unlike TFB2M, TFB1M is a functional methyltransferase that dimethylates a conserved region of the rRNA of the small mitochondrial ribosomal subunit in most eukaryotic organisms (8, 21, 22). A notable exception of this conserved function is yeast cells, in which mitochondrial rRNA is not methylated at this position. Accordingly, yeast mitochondrial transcription relies on a single TFB2M-like factor (Mtf1) (19, 23). Although Mtf1 is also related to the family of methyltransferases, its other function (besides transcription initiation) is not known. It is therefore likely that transcription and methyltransferase functions in human and yeast mitochondria are performed by strictly dedicated enzymes and not by the bifunctional proteins.
FIGURE 1.
TFB2M but not TFB1M is required to stimulate human mitochondrial transcription initiation. A, sequences of LSP and HSP1 aligned at their transcription start sites. Shaded boxes indicate transcripts obtained in reaction having limited sets of the substrate NTPs. B, transcription of HSP1 and LSP in the presence of POLRMT, TFAM, and TFB2M. The reactions were carried out as described under “Experimental Procedures” using ApA primer and limited NTP mixtures, and the products were resolved by 20% PAGE containing 6 m urea. C, transcription of HSP1 and LSP in the presence of POLRMT, TFAM, and TFB1M. The reactions were carried out as described above. Note that the image is overexposed (6-fold) as compared with the experiment shown in panel B to demonstrate TFAM effect on transcription by POLRMT. D, transcription of LSP and HSP1 depends on TFAM stimulation. The reactions were carried out as described above using POLRMT, TFB2M (all 50 nm), and TFAM concentrations as indicated (0–200 nm). E, single-round transcription using catalytic autolabeling. Reactions containing POLRMT (150 nm), TFB2M (150 nm), and TFAM (0–500 nm) were performed as described under “Experimental Procedures” and resolved using 4–12% Bis-Tris NuPAGE.
We next investigated the response of transcription initiation to increasing TFAM concentrations (Fig. 1D). The highest level of transcription initiation by POLRMT-TFB2M on either promoter was observed at around 50 nm TFAM (i.e. at a 1:1 template:TFAM ratio), which, considering TFB2M binding to POLRMT, suggests equimolar stoichiometry for the initiation complex (1:1:1:1 template:TFAM:POLRMT:TFB2M). Increased concentrations of TFAM had an inhibitory effect on transcription, probably due to nonspecific binding of this factor to the DNA template. These data indicate that HSP1 and LSP respond to TFAM in a similar fashion. Interestingly, when the two promoters are present on the same template, as is the case in vivo, there is a distinct difference in the TFAM dependence, with LSP being active at much lower TFAM concentrations (2). This difference could be due to a number of factors that include differences in the length and sequence of LSP and HSP1 transcripts, rates of POLRMT release at the end of the template (“turnover” rates), and interference between two closely located promoters, etc. To clarify this issue, we performed a single-round transcription assay based on catalytic autolabeling (5) and monitored formation of the first phosphodiester bond between the cross-linked AMP derivative and [α-32P]ATP (Fig. 1E). The resulting 32P-labeled dinucleotide product (pApA) can be resolved by SDS-PAGE because it is covalently attached to TFB2M. As in the experiment described above, we observed similar response of LSP and HSP1 promoters to TFAM stimulation, suggesting identical mechanisms of transcription initiation at these promoters.
Changes in TFAM concentration seem to be important for regulating rates of mitochondrial transcription in vitro and in vivo, as demonstrated by studies on transcription in organello and cell lines (24, 25). However, besides being a transcription factor, TFAM is also an important component of mitochondrial nucleoids and is essential for the maintenance of mtDNA (26–28). This suggests that a situation where TFAM is absent from mitochondria is not physiologically relevant, and therefore transcription of the mitochondrial genes by the POLRMT-TFB2M complex does not play any significant role. However, it is possible that when the TFB2M concentration in mitochondria is low, transcription by RNAP-TFAM may contribute to the synthesis of the RNA primers necessary for mtDNA replication (29). In this scenario, although mRNA synthesis on both LSP and HSP1 would be significantly decreased, RNA primers generated from LSP (i.e. oriH) and oriL could be at a level sufficient to support replication. Indeed, it has been recently demonstrated that primer generation at oriL is TFB2M-independent (29).
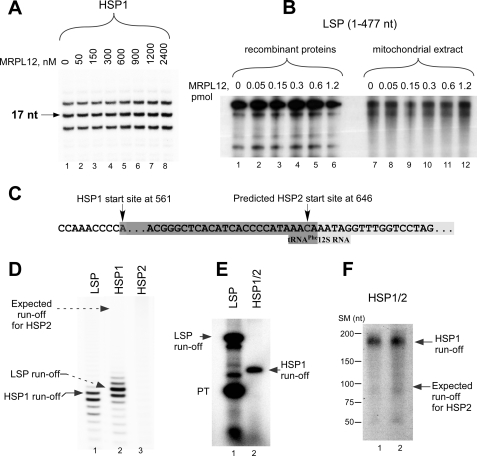
Ribosomal protein MRPL12 was recently shown to stimulate mitochondrial transcription (14). We failed to observe any stimulatory effect of MRPL12 during transcription initiation in vitro using the LSP and HSP1 promoters (Fig. 2A). MRPL12 had no effect on transcription elongation when tested in the factor-independent assay performed on “bubble” promoter template (supplemental Fig. S4A) or in an abortive initiation assay (supplemental Fig. S4B). Identical results were obtained when the experiments were repeated using recombinant proteins purified from another source (insect cells) and using a longer DNA template (about 400 nt) (Fig. 2B). Furthermore, we could not observe any stimulatory effect of MRPL12 on transcription activity in mitochondrial extracts (Fig. 2B), arguing against the existence of an additional factor required for the stimulatory activity of MRPL12. We also cloned and isolated the yeast homolog of MRPL12, Mnp1. Similarly to MRPL12, this protein had no effect on transcription initiation in the yeast mitochondrial system (supplemental Fig. S4C). This is consistent with a previous study that did not detect Mnp1 among yeast proteins that interact with Rpo41 (16). Both Mnp1 and MRPL12 proteins failed to stimulate transcription in heterological systems (data not shown and supplemental Fig. S4C). We conclude that MRPL12 does not stimulate mitochondrial transcription in vitro.
FIGURE 2.
Basal transcription system requires only TFAM, TFB2M, and two promoters, LSP and HSP1. A, MRPL12 does not stimulate mitochondrial transcription in vitro in the presence of the purified recombinant proteins. Reactions containing synthetic HSP1 template were performed as in Fig. 1B except that ATP, GTP, and CTP (all 0.3 mm) were used. B, MRPL12 does not stimulate transcription in the presence of mitochondrial extracts. For experiments with recombinant proteins (lanes 1–6), the individual transcription reaction mixtures contained POLRMT (400 fmol), TFB2M (400 fmol), TFAM (2.5 pmol), and the indicated mtDNA template (85 fmol). An S-100 mitochondrial lysate was used for transcription with the extracts (lanes 7–12). C, sequence of the HSP2 region. Shaded boxes indicate the 3′-terminal end of the tRNAPhe gene and the 5′-end of 12 S RNA gene. D, run-off transcription assay using promoter templates containing LSP1, HSP1, and HSP2. Transcription was carried out in the presence of TFAM, TFB2M, and POLRMT for 40 min at 35 °C using 0.5 mm substrate NTPs. E, run-off transcription assay using plasmid templates containing LSP and HSP1/HSP2. Transcription reactions were performed as indicated in panel B. HSP1/HSP2 template was obtained by linearization of a human mtDNA fragment at position 741. Transcription of HSP1 transcription generated a 181-nt product, but no 96-nt product expected for initiation at HSP2 was observed. LSP transcription using an mtDNA fragment corresponding to positions 1–477 generated run-off and prematurely terminated (PT) products. F, transcription is initiated from HSP1, but not from HSP2, in the presence of an S-100 mitochondrial lysate. A transcription reaction using HSP1/HSP2 template (lanes 1 and 2) described above was performed in the presence of the mitochondrial extracts supplemented with TFAM (1 pmol) and TFB2M (0.25 pmol). The positions of size markers (SM) are given to the left.
The results of the above experiments are in contrast to the data reported by Shadel and colleagues (14), who observed stimulation of LSP transcription in an in vitro system using recombinant TFAM, MRPL12, and partially purified POLRMT-containing mitochondrial extracts. It should be noted that the transcription conditions used in these experiments were not optimal as both LSP and HSP1 transcription showed no response to TFAM, and the TFB2M concentration in partially purified mitochondrial extracts could not be determined. Although overexpression of MRPL12 results in 1.5–2-fold stimulation of mRNA synthesis (14), additional experiments will be required to clarify the effect of this protein on transcription in vivo.
Transcription using the HSP2 promoter has never been reported. Although efficient run-off transcription was observed on LSP and HSP1, we observed no specific HSP2 transcripts, indicating that it is not an active promoter in vitro (Fig. 2, C and D). We also repeated the experiments using a longer template containing both HSP1 and HSP2. On this template, we could only observe HSP1 transcription initiation (Fig. 2E). Similar results were obtained in an abortive initiation assay (supplemental Fig. S5A), in which RNA synthesis was observed only using LSP- and HSP1-containing templates.
The absence of specific transcription from the HSP2 promoter could hypothetically be explained by the existence of other, currently unknown, transcription initiation factor(s) that are used exclusively to activate this otherwise dormant promoter. This would be reminiscent of the situation where transcription of different promoters by a bacterial core RNAP requires different specificity subunits: σ factors. However, transcription using mitochondrial extracts (Fig. 2, B and F) produced RNA transcripts only from LSP and HSP1, and we observed no distinct HSP2 transcript. To test this further, we took advantage of a factor-independent assay based on the ability of POLRMT to transcribe premelted promoter templates in the absence of TFAM and TFB2M (supplemental Fig. S5, B and C) (5). As in the experiments shown above, specific RNA synthesis was observed only for templates containing the LSP and HSP1 promoters, suggesting that HSP2 does not contain specific recognition determinants required for promoter function (supplemental Fig. S5C).
The start site of HSP2 is located only 63 bp downstream of HSP1, and thus, the HSP2 region was included in a number of the template constructs used for in vitro transcription (30, 31); however, no data are available that demonstrate synthesis of HSP2-specific transcripts. Moreover, the identified HSP2 start site is located in the immediate proximity of the main processing RNA point, between tRNAPhe and 12 S rRNA. Transcription initiation at this site would result in synthesis of 12 S rRNA with two additional nt at its 5′-end as opposed to the transcript generated from the HSP1 promoter and processed with RNase Z (32). It should be feasible to address experimentally the question of whether 12 S rRNA is heterogeneous at its 5′-end.
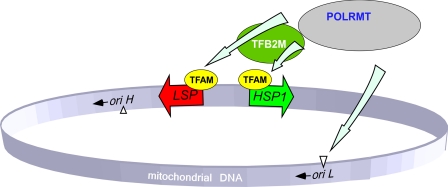
To conclude, the biochemical data obtained in in vitro transcription experiments using highly purified recombinant proteins allow us to simplify the current model of mitochondrial transcription initiation in humans (Fig. 3). Transcription of all mitochondrial genes appears to require only two promoters, LSP and HSP1. Efficient initiation on these promoters requires only TFAM and TFB2M; however, generation of a primer at oriL can be performed by POLRMT on its own. Other recently identified transcription factors such as MTERFs 2 and 3 were shown to affect transcription, but unlike TFAM and TFB2M, they are not essential for initiation (18, 33). Although it is likely that there are other, as yet unidentified, transcription factors in human mitochondria, we favor the view that the basal transcription machinery essential for initiation of transcription has been already defined (6, 21).
FIGURE 3.
Schematics of the basal transcription initiation machinery in human mitochondria. Human mitochondrial genome contains two promoters located in the opposing DNA strands. The HSP1 promoter is responsible for synthesis of most mitochondrial genes and is activated by POLRMT-TFAM-TFB2M complex. During replication, when DNA region near oriL becomes single-stranded and forms stem-loop structure, transcription by POLRMT generates short RNA primers. This initiation event is TFAM- and TFB2M-independent. Transcription from the LSP promoter generates primers for replication at oriH as well as the rest of the tRNAs and mRNA. This initiation event, similar to HSP1, requires cooperative action of POLRMT, TFAM, and TFB2M for efficient transcription and replication.
Supplementary Material
Acknowledgments
We thank Katrina Stehle for assistance in cloning of human MRPL12. We thank Dr. Walter Rossmanith for the productive discussion. Dr. William T. McAllister is acknowledged for fruitful discussion and critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM38147 (to Dr. William McAllister). This work was also supported in part by a University of Medicine and Dentistry of New Jersey (UMDNJ) Research Foundation grant (to D. T.) and by grants from the Swedish Research Council, the Swedish Cancer Society, the Goran Gustafsson Foundation, and the Foundation for Strategic Research (to C. M. G. and M. F.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- LSP
- light-strand promoter
- RNAP
- RNA polymerase
- nt
- nucleotide(s)
- Bis-Tris
- 2-(bis(2-hydroxyethyl)amino)-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1.Asin-Cayuela J., Gustafsson C. M. (2007) Trends Biochem. Sci. 32, 111–117 [DOI] [PubMed] [Google Scholar]
- 2.Bonawitz N. D., Clayton D. A., Shadel G. S. (2006) Mol. Cell 24, 813–825 [DOI] [PubMed] [Google Scholar]
- 3.Gaspari M., Falkenberg M., Larsson N. G., Gustafsson C. M. (2004) EMBO J. 23, 4606–4614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dairaghi D. J., Shadel G. S., Clayton D. A. (1995) Biochim. Biophys. Acta. 1271, 127–134 [DOI] [PubMed] [Google Scholar]
- 5.Sologub M., Litonin D., Anikin M., Mustaev A., Temiakov D. (2009) Cell 139, 934–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Falkenberg M., Gaspari M., Rantanen A., Trifunovic A., Larsson N. G., Gustafsson C. M. (2002) Nat. Genet. 31, 289–294 [DOI] [PubMed] [Google Scholar]
- 7.McCulloch V., Shadel G. S. (2003) Mol. Cell. Biol. 23, 5816–5824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Metodiev M. D., Lesko N., Park C. B., Cámara Y., Shi Y., Wibom R., Hultenby K., Gustafsson C. M., Larsson N. G. (2009) Cell Metab. 9, 386–397 [DOI] [PubMed] [Google Scholar]
- 9.Fisher R. P., Topper J. N., Clayton D. A. (1987) Cell 50, 247–258 [DOI] [PubMed] [Google Scholar]
- 10.Martin M., Cho J., Cesare A. J., Griffith J. D., Attardi G. (2005) Cell 123, 1227–1240 [DOI] [PubMed] [Google Scholar]
- 11.Montoya J., Christianson T., Levens D., Rabinowitz M., Attardi G. (1982) Proc. Natl. Acad. Sci. U.S.A. 79, 7195–7199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoza B. K., Bogenhagen D. F. (1984) J. Biol. Chem. 259, 3909–3915 [PubMed] [Google Scholar]
- 13.Montoya J., Gaines G. L., Attardi G. (1983) Cell 34, 151–159 [DOI] [PubMed] [Google Scholar]
- 14.Wang Z., Cotney J., Shadel G. S. (2007) J. Biol. Chem. 282, 12610–12618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marty L., Fort P. (1996) J. Biol. Chem. 271, 11468–11476 [DOI] [PubMed] [Google Scholar]
- 16.Markov D. A., Savkina M., Anikin M., Del Campo M., Ecker K., Lambowitz A. M., De Gnore J. P., McAllister W. T. (2009) Yeast 26, 423–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sato H., Miyakawa I. (2004) Protoplasma 223, 175–182 [DOI] [PubMed] [Google Scholar]
- 18.Park C. B., Asin-Cayuela J., Cámara Y., Shi Y., Pellegrini M., Gaspari M., Wibom R., Hultenby K., Erdjument-Bromage H., Tempst P., Falkenberg M., Gustafsson C. M., Larsson N. G. (2007) Cell 130, 273–285 [DOI] [PubMed] [Google Scholar]
- 19.Savkina M., Temiakov D., McAllister W. T., Anikin M. (2010) J. Biol. Chem. 285, 3957–3964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsushima Y., Adán C., Garesse R., Kaguni L. S. (2005) J. Biol. Chem. 280, 16815–16820 [DOI] [PubMed] [Google Scholar]
- 21.Cotney J., Wang Z., Shadel G. S. (2007) Nucleic Acids Res. 35, 4042–4054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seidel-Rogol B. L., McCulloch V., Shadel G. S. (2003) Nat. Genet. 33, 23–24 [DOI] [PubMed] [Google Scholar]
- 23.Matsunaga M., Jaehning J. A. (2004) J. Biol. Chem. 279, 44239–44242 [DOI] [PubMed] [Google Scholar]
- 24.Maniura-Weber K., Helm M., Engemann K., Eckertz S., Möllers M., Schauen M., Hayrapetyan A., von Kleist-Retzow J. C., Lightowlers R. N., Bindoff L. A., Wiesner R. J. (2006) Nucleic Acids Res. 34, 6404–6415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maniura-Weber K., Goffart S., Garstka H. L., Montoya J., Wiesner R. J. (2004) Nucleic Acids Res. 32, 6015–6027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ekstrand M. I., Falkenberg M., Rantanen A., Park C. B., Gaspari M., Hultenby K., Rustin P., Gustafsson C. M., Larsson N. G. (2004) Hum. Mol. Genet. 13, 935–944 [DOI] [PubMed] [Google Scholar]
- 27.Antoshechkin I., Bogenhagen D. F. (1995) Mol. Cell. Biol. 15, 7032–7042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bogenhagen D. F., Rousseau D., Burke S. (2008) J. Biol. Chem. 283, 3665–3675 [DOI] [PubMed] [Google Scholar]
- 29.Wanrooij S., Fusté J. M., Farge G., Shi Y., Gustafsson C. M., Falkenberg M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 11122–11127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hixson J. E., Clayton D. A. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 2660–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Christianson T. W., Clayton D. A. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 6277–6281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dubrovsky E. B., Dubrovskaya V. A., Levinger L., Schiffer S., Marchfelder A. (2004) Nucleic. Acids. Res. 32, 255–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wenz T., Luca C., Torraco A., Moraes C. T. (2009) Cell Metab. 9, 499–511 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.