Abstract
Histone deacetylases (HDACs) are best known as transcription corepressors through deacetylating histone tails. Here we show that HDAC2 is also involved in cap-dependent mRNA translation by promoting sumoylation of eukaryotic translation initiation factor 4E (eIF4E), which is independent of its deacetylase activity. By stimulating eIF4E sumoylation, HDAC2 induces the formation of the active eukaryotic initiation factor 4F (eIF4F) complex and induces the protein synthesis of a subset of eIF4E-responsive genes that are essential for cell proliferation and preventing apoptosis. These data demonstrate that HDAC2 has an unexpected sumoylation-promoting activity and regulates cap-dependent mRNA translation.
Keywords: Gene Expression, Histone Deacetylase, Post-translational Modification, Protein Synthesis, Sumoylation, HDAC2, cap-dependent mRNA Translation, eIF4E
Introduction
In eukaryotes, more than 95% of proteins are synthesized through cap-dependent mRNA translation. A rate-limiting step in cap-dependent translation is the formation of the eukaryotic initiation factor 4F (eIF4F)3 complex containing eIF4E (cap-binding protein), eIF4A (ATP-dependent mRNA helicase), and eIF4G (scaffold protein) (for review, see Refs. 1 and 2). eIF4E binding to the cap structure of mRNA is inhibited by a small family of eIF4E-binding proteins (4E-BPs). 4E-BP1, the most abundant member of the 4E-BP family, is phosphorylated at multiple sites, and several of these phosphorylations are necessary for the dissociation of 4E-BP1 from eIF4E, the subsequent formation of eIF4F complex, and the initiation of cap-dependent translation (3).
Recent studies indicate that eIF4E is modified by small ubiquitin-related modifier (SUMO-1) conjugation (4); we further demonstrate that sumoylation of eIF4E regulates its translation activity (5). SUMO is a reversible protein modifier that covalently attaches to its substrates. The human genome encodes four distinct SUMO proteins: SUMO-1, -2, -3, and -4. The sequence identity and expression of these four SUMO molecules are highly variable. Although SUMO-2 and -3 are 97% identical, they share only 50% identity with SUMO-1, and although SUMO-1, -2, and -3 are ubiquitously expressed, SUMO-4 seems to be expressed mainly in the kidney, lymph node, and spleen.
Protein sumoylation is mediated by activating (E1), conjugating (E2), and ligating (E3) enzymes (6). Ubc9 is the only identified SUMO E2 conjugating enzyme. However, several sequence-specific SUMO E3 ligases have been identified. These include the protein inhibitor of activated STAT (PIAS) family of proteins and vertebrate-specific nuclear pore protein RanBP2, polycomb group protein Pc2, and Mms21-Nse1 (for review, see Ref. 7). In addition, the class II histone deacetylases HDAC4 and HDAC7 promote sumoylation of transcription factor myocyte enhancer factor 2 (MEF2) and promyelocytic leukemia protein, respectively, through a deacetylase-independent mechanism (8–10). Human HDACs constitute a family of 18 enzymes that are divided into four classes according to their yeast homologues and sequence identity. Only HDAC4 and HDAC7 have been reported to possess SUMO E3 ligase activity.
Here we show that HDAC2, a well known transcription repressor, is present in the translation initiation complex and induces eIF4E sumoylation in a deacetylase-independent manner. By promoting eIF4E sumoylation-dependent formation of the eIF4F complex, HDAC2 activates protein synthesis of eIF4E-responsive genes including c-Myc, ornithine decarboxylase (ODC), and survivin. Taken together, our results have revealed a new feature and a novel function of HDAC2.
EXPERIMENTAL PROCEDURES
Most of the Experimental Procedures are described in the supplemental material.
Evaluation of in Vivo eIF4E Sumoylation
In vivo sumoylation of eIF4E was evaluated as described in our previous report (11). To reduce the interference from other sumoylated protein(s) that may be associated with eIF4E, 0.1% SDS was added to the lysis buffer for both immunoprecipitation and immunoblotting. Unless stated in the figure legends or labeled in the figures, 20 mm SUMO isopeptidase inhibitor N-ethylmaleimide (Sigma) was added to the lysis buffer for immunoprecipitation. For straight Western blotting, unless indicated, for the purpose of comparing the input or total amount of the protein, N-ethylmaleimide was not added to the lysis buffer. Sumoylation of total cellular eIF4E was evaluated by immunoprecipitation with anti-eIF4E and then subsequent immunoblotting with anti-SUMO-1 or immunoprecipitation with anti-eIF4E and subsequent immunoblotting with anti-eIF4E. Whole cell lysates equal to 10% of preimmunoprecipitation lysates were used for input control.
In Vitro Sumoylation Assay
The in vitro sumoylation assay was carried out using an in vitro sumoylation kit (LAE Biotech International, Rockville, MD). The kit provides necessary regents (including recombinant SUMO E1 enzymes SUMO-activating enzyme subunit I and SUMO-activating enzyme subunit II, SUMO E2 enzyme Ubc9, SUMO-1 or SUMO-2, and reaction buffer) for the assay except SUMO substrate. Substrate eIF4E was synthesized using a cell-free system. In vitro synthesis of human influenza hemagglutinin (HA)-tagged eIF4E (HA-eIF4E) or c-Myc-tagged NF-κB2/p100 (c-Myc-p100) was conducted using the TnT Quick Coupled Transcription/Translation Systems (Promega, Madison, WI) according to the protocol provided. Purified human recombinant protein HDAC2 was obtained from Biomol International (Plymouth Meeting, PA). The in vitro sumoylation was carried out according to the provided protocol. For each reaction, 1 μl of the in vitro synthesized HA-eIF4E or c-Myc-p100 along with 0–200 ng of HDAC2 was added to the reaction mixture. After in vitro sumoylation reaction, the reaction mixtures were subjected to immunoblotting. Non-sumoylated and sumoylated HA-eIF4E and c-Myc-p100 were detected by antibodies recognizing HA or c-Myc, respectively.
RESULTS AND DISCUSSION
HDCA2 Activates Luciferase mRNA Translation
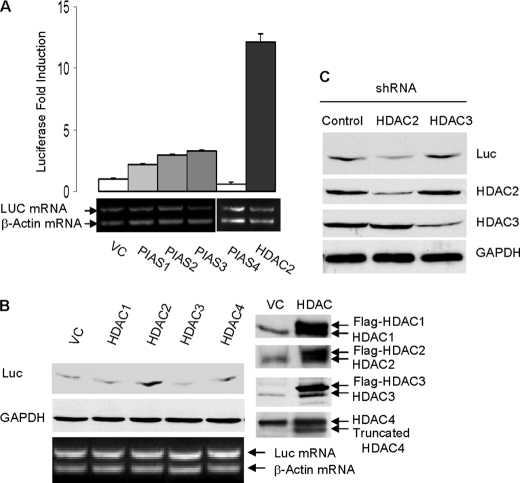
We have recently demonstrated that sumoylation activates the translation activity of eIF4E (5). We hypothesized that factors that stimulate eIF4E sumoylation activate cap-dependent translation. Therefore, to identify the SUMO E3 ligase for eIF4E, we compared the effects of overexpression of various SUMO E3 ligase candidates on cap-dependent translation of a luciferase reporter gene. The tested E3 candidates included the classic SUMO E3 ligase PIAS (PIAS1, -2, -3, and -4) as well as HDAC1, -2, -3, and -4. We found that luciferase protein expression was induced mostly by HDAC2 overexpression in various cell types including human colorectal cancer cell HCT-116 cells (Fig. 1, A and B) and human breast epithelial cell MCF-10A cells (supplemental Fig. 1). Accordingly, shRNA knockdown of HDAC2, but not HDAC3, down-regulated luciferase protein expression (Fig. 1C). HDAC2 did not affect the cytoplasmic mRNA level of the luciferase gene (Fig. 1, A and B), thus excluding the possibility that HDAC2 induces luciferase protein expression by activating gene transcription or mRNA transport of luciferase gene. Taken together, these findings point toward HDAC2 as a factor that is likely involved in eIF4E sumoylation.
FIGURE 1.
HDAC2 activates cap-dependent translation of luciferase gene. A, overexpression of PIAS and HDAC2 on Renilla luciferase mRNA (LUC mRNA) and protein expression. The HCT-116 cells were transfected with individual PIAS or HDAC2 along with luciferase cDNA reporter gene. The cells were lysed, and the luciferase activity was measured using a luciferase assay kit (Promega). The absolute relative light units values are in the 107-108 range. Reverse transcription-PCR was performed using cytosolic mRNAs. To minimize the interference resulting from unequal mRNA input, the primers for β-actin and the primers for luciferase were added to the same reaction tube. VC, vector control. Error bars indicate S.E. B, HDAC2 activates cap-dependent protein translation of luciferase protein in HCT-116 cells. Expressions of HDAC isoenzymes were verified by immunoblotting. C, knockdown of HDAC2 but not of HDAC3 inhibits luciferase expression in HCT-116 cells. Protein expression of luciferase was evaluated by immunoblotting with antibody against luciferase. shRNA knockdown of HDAC2 and HDAC3 were verified by immunoblotting. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) protein was used a loading control.
HDAC2 Promotes eIF4E Sumoylation
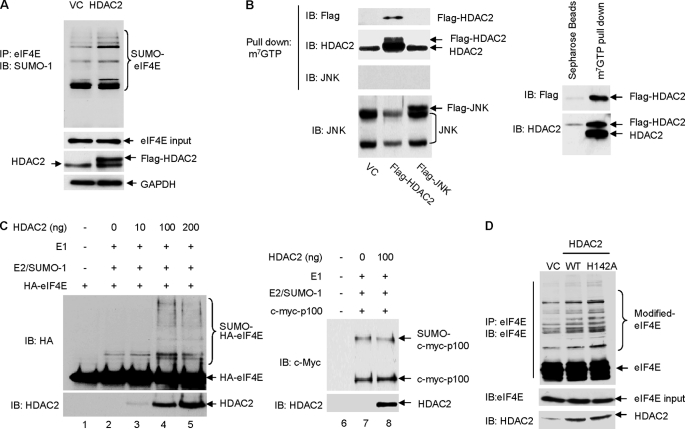
Using in vivo and in vitro assays, we directly examined the effect of HDAC2 on eIF4E sumoylation. Our prior study has revealed that transcription factor NF-κB2/p100 is modified by SUMO-1 conjugation (11); we therefore included NF-κB2/p100 as a control SUMO substrate. We found that without affecting overall cellular sumoylation and sumoylation of NF-κB2/p100 (supplemental Fig. 2, A and B), overexpression of HDAC2 enhanced sumoylation of eIF4E (Fig. 2A). Because overexpression of HDAC2 did not affect the protein expression level of Ubc9 or PIAS1–3 (supplemental Fig. 2C), HDAC2 may not promote eIF4E sumoylation through activating Ubc9 or the existing SUMO E3 ligases. Cell fractionation and immunofluorescence analysis indicated that HDAC2 localized in both cytoplasm and nucleus (supplemental Fig. 3, A and B), raising the possibility that HDAC2 directly stimulates sumoylation of eIF4E by acting as a SUMO E3 ligase-like factor. Although no direct interaction between HDAC2 and Ubc9 or between substrate eIF4E or eIF4A and eIF4G (supplemental Fig. 3C) was detected, by pulldown assay using 7-methyl-GTP-SepharoseTM 4B beads, we found that both endogenous and exogenous FLAG-tagged HDAC2, but not the control protein c-Jun N-terminal kinases (JNK, both endogenous and exogenous FLAG-tagged), were present in the cap-bound translation initiation complex (Fig. 2B, left panel), where it promotes eIF4E sumoylation. The specificity of the assay was further validated by the result that control Sepharose beads that are not conjugated with 7-methyl-GTP failed to precipitate HDAC2 (Fig. 2B, right panel).
FIGURE 2.
HDAC2 stimulates eIF4E sumoylation. A, overexpression of HDAC2 induces eIF4E sumoylation. The HCT-116 cells were transfected with FLAG-HDAC2 or control vectors (VC). Sumoylation of eIF4E was evaluated by immunoprecipitation (IP) of the whole cell lysates with anti-eIF4E followed by immunoblotting (IB) with anti-SUMO-1. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. B, HDAC2 but not JNK1 is associated with cap-bound translation initiation complex. The HCT-116 cells were transfected with empty vector or FLAG-HDAC2 or FLAG-JNK1. 48 h after transfection, the cells were harvested for the pulldown assays. Left panel, the cap-bound HDAC2 was eluted from the 7-methyl-GTP (m7GTP)-Sepharose resin using 7-methyl-GDP (m7GDP) and detected by immunoblotting with anti-FLAG or anti-HDAC2. Right panel, immunoprecipitation with Sepharose beads was included as the experimental control. C, the addition of purified recombinant HDAC2 protein directly stimulates in vitro sumoylation of eIF4E but not NF-κB2/p100. 1 μl of in vitro translated human influenza HA-tagged eIF4E or c-Myc-p100 was used as SUMO substrate. 0–200 ng of human recombinant HDAC2 protein (Biomol International) was added to the in vitro sumoylation reaction mixture as labeled in the figure. Sumoylated and non-sumoylated HA-eIF4E and c-Myc-p100 were detected by immunoblotting with anti-HA and anti-c-Myc, respectively. The addition of HDAC2 was verified by immunoblotting with anti-HDAC2. D, overexpression of HDAC2 and HDAC2 deacetylase-dead mutant (HDAC2-H142A) on eIF4E sumoylation in HCT-116 cells. WT, wild type.
In our earlier study, we found that the efficiency of eIF4E sumoylation in vitro is low (5). The low efficiency is partially attributed to the lack of prephosphorylation of in vitro synthesized eIF4E (5). It is also possible that sumoylation of eIF4E is SUMO E3 ligase-dependent and that lack of a SUMO E3 ligase contributes to the low efficiency of in vitro eIF4E sumoylation. To determine whether HDAC2 possesses a SUMO E3 ligase-like activity, we performed an in vitro eIF4E sumoylation assay using in vitro synthesized HA-eIF4E as the substrate and the purified human recombinant HDAC2 protein as a SUMO E3 ligase candidate. We also included the in vitro synthesized c-Myc-tagged NF-κB2/p100 as a control substrate. The same amount of HA-eIF4E or c-Myc-p100 was added to each assay; the only variable in each reaction was the quantity of the HDAC2 protein (0–200 ng). We found that the addition of HDAC2 protein dose-dependently stimulated the sumoylation of eIF4E (Fig. 2C, left panel) but not that of NF-κB2/p100 (Fig. 2C, right panel). Together, these findings indicate that HDAC2 substrate-specifically stimulates SUMO-1 conjugation to eIF4E both in cells and in an in vitro reconstituted sumoylation system.
Deacetylase activity is the best characterized feature of HDAC2. To determine whether the deacetylase activity of HDAC2 was required for the sumoylation of eIF4E, we overexpressed the enzymatically inert HDAC2-H142A (histidine 142 in HDAC2 was mutated to alanine) (12) and again examined eIF4E sumoylation. We found that loss of deacetylase activity did not inhibit HDAC2-induced eIF4E sumoylation (Fig. 2D). This result is consistent with prior findings that HDAC4 and HDAC7 regulate transcription factor MEF2 and promyelocytic leukemia protein, respectively, in a deacetylase-independent manner (8–10). Together, we have discovered a novel function of HDAC2; this protein promotes sumoylation of eIF4E, which is independent of its deacetylase activity.
Deacetylase-independent Activation of mRNA Translation by HDAC2
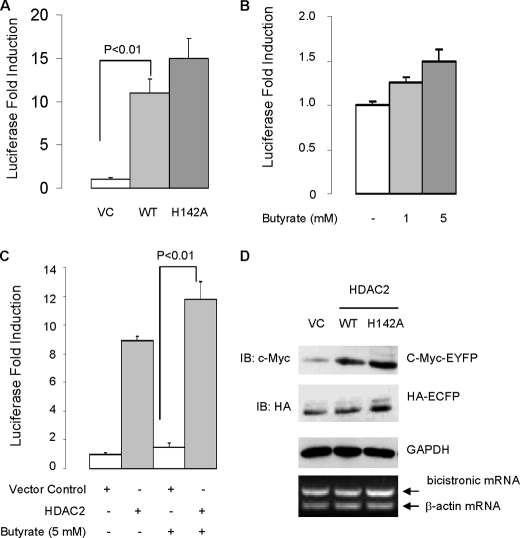
If deacetylase activity is not required for the sumoylation-promoting effect of HDAC2, one would expect that deacetylase is also dispensable for HDAC2-induced mRNA translation. As anticipated, we found that loss of deacetylase activity by point mutation (H142A) did not inhibit HDAC2-induced protein expression of luciferase gene (Fig. 3A). Consistent with this, the HDAC inhibitor butyrate induced basal luciferase protein and further enhanced HDAC2-mediated induction of luciferase protein (Fig. 3, B and C). These results thus suggest that HDAC regulates mRNA translation in a deacetylase-independent manner.
FIGURE 3.
Deacetylase activity is not required for HDAC2-induced mRNA translation of reporter genes. A, overexpression of wild-type HDAC2 and HDAC2-H142A mutant induces cap-dependent translation of luciferase reporter gene. The HCT-116 cells were transfected with empty vector (VC), wild-type (WT) HDAC2, or HDAC2-H142A mutant along with luciferase cDNA reporter. 48 h after transfection, the transfected cells were harvested for luciferase assay. The absolute luciferase relative light units values are in the 107–108 range. Statistics were performed by one-way analysis of variance followed by Tukey's multiple comparison test using the data obtained from three independent experiments. B, butyrate does not inhibit basal translation rate. The HCT-116 cells were transfected with luciferase cDNA reporter, and 24 h after transfection, the transfected cells were treated with butyrate at the indicated concentration for 24 h. C, butyrate treatment does not suppress HDAC2-medited induction of luciferase activity. Statistics were performed by one-way analysis of variance followed by Tukey's multiple comparison test using the data obtained from three independent experiments. Error bars in A–C indicate S.E. D, overexpression of HDAC2 on cap-dependent translation of c-Myc-tagged EYFP and IRES-dependent translation of HA-tagged ECFP. The HCT-116 cells were transfected with HDAC2 or empty vector along with bicistronic vector. Cap-dependent translation of c-Myc-EYFP protein was evaluated by immunoblotting with anti-c-Myc. IRES-dependent translation of HA-ECFP was evaluated by immunoblotting with anti-HA. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Proteins are synthesized through cap-dependent mRNA translation and internal ribosome entry site (IRES)-dependent mRNA translation. The IRES-dependent translation does not require eIF4E (for review, see Ref. 13). To determine whether HDAC2-mediated mRNA translation is eIF4E-dependent, we utilized a bicistronic fluorescent reporter construct (14). The bicistronic vector allows the expression of protein from two independent genes that are transcribed equally to be determined. Overexpression of HDAC2 and HDAC2-H142A induced cap-dependent translation of yellow fluorescent protein (EYFP), but not the IRES-dependent translation of cyan fluorescent protein (ECFP) (Fig. 3D). This finding thus indicated that HDAC2 is not involved in IRES-dependent mRNA translation, reinforcing the hypothesis that HDAC2 regulates mRNA translation through targeting eIF4E. Of note, consistent with the luciferase results (Fig. 3A), overexpression of HDAC2-H142A induced greater translation of EYFP and ECFP than wild-type HDAC2 (Fig. 3D). The increased translation of EYFP and ECFP in cells overexpressing the HDAC2-H142A mutant may be a result of derepression of gene transcription (Fig. 3D). Collectively, these results indicate that HDAC2 activates eIF4E-dependent mRNA translation in a deacetylase-independent manner.
HDAC2 Activates Protein Synthesis of eIF4E-responsive Genes
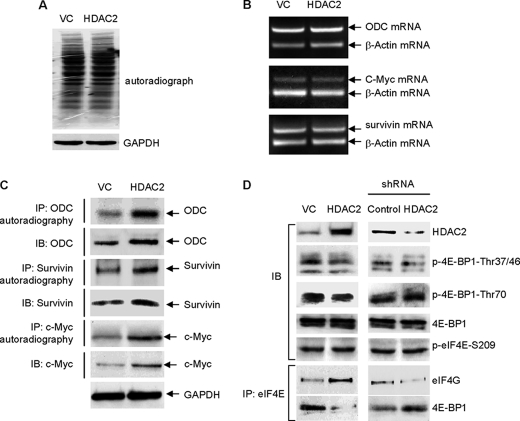
Next, we examined the effect of HDAC2 on the protein synthesis of some known eIF4E-responsive genes. Many of the eIF4E-regulated genes are growth-, proliferation-, and anti-apoptosis-related and include the oncogenes ODC, c-Myc, the Bcl family, and survivin (15–17). Our previous work has demonstrated that induction of mRNA translations of those genes by eIF4E is eIF4E sumoylation-dependent (5). The results from metabolic labeling using [35S]methionine showed that overexpression of HDAC2, without increasing global protein synthesis (Fig. 4A) and increasing the cytoplasmic mRNA levels of the genes (Fig. 4B), induced protein synthesis of ODC, c-Myc, and survivin, known eIF4E-responsive targets (Fig. 4C). These results thus indicate that HDAC2 gene-specifically activates protein synthesis of eIF4E-responsive genes.
FIGURE 4.
HDAC2 activates protein synthesis of eIF4E-responsive target genes. A, overexpression of HDAC2 does not change total protein synthesis rate in HCT-116 cells. The protein synthesis rate was assessed by [35S]methionine incorporation. VC, vector control. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. B, overexpression of HDAC2 does not induce cytoplasmic mRNA level of ODC, survivin, and c-Myc in HCT-116 cells. C, forced expression of HDAC2 induces protein synthesis rates of ODC, survivin, and c-Myc in HCT-116 cells. 35S-methioine-labled ODC, survivin, and c-Myc were detected by autoradiography. IP, immunoprecipitation; IB, immunoblotting. D, the effect of HDAC2 manipulation on 4E-BP1 phosphorylation and the formation of eIF4F complex in HCT-116 cells. The HCT-116 cells were transfected with HDAC2 or shRNA HDAC2 along with their individual control vector. 72 h after transfection, the cells were harvested, and the whole cell lysates were used for the immunoblotting and immunoprecipitation assays. Phosphorylation of 4E-BP1 at the threonines 37/46 or 70 was evaluated by immunoblotting using antibodies specifically recognizing phosphorylated (p) forms of 4E-BP1.
Our earlier studies have demonstrated that sumoylation of eIF4E activates cap-dependent mRNA translation by promoting the formation of the eIF4F complex (5). Because HDAC2 promotes sumoylation of eIF4E, we examined whether HDAC2 regulates the formation of eIF4F complex. We observed that overexpression of HDAC2, without changing the phosphorylation status of 4E-BP1, induced the binding between eIF4E and eIF4G and reduced the binding between eIF4E and 4E-BP1 (Fig. 4D). Further, shRNA knockdown of HDAC2 reduced the binding between eIF4E and eIF4G and increased the binding between eIF4E and 4E-BP1 (Fig. 4D). These results suggest that HDAC2 activates eIF4E-dependent mRNA translation by promoting the formation of the eIF4F complex.
Consistent with our finding that HDAC2 induces protein synthesis, experimental evidence from mouse studies has demonstrated that HDAC2 mutant mice (HDAC2 truncated after amino acid 288) are smaller and have a decreased number of cells in their intestines than their wild-type littermates (18). The HDAC2 mutant used in the study lacks deacetylase activity (18). However, because inhibition of deacetylase activity by either mutation (HDAC2-H142A) or deacetylase inhibitor butyrate does not prevent HDAC2-induced mRNA translation of luciferase gene, we do not believe that loss of HDAC2 deacetylase activity is responsible for the murine phenotype. We propose that lack of the sumoylation-promoting activity associated with HDAC2 is likely the cause of a reduced body size in the HDAC2 mutant mice.
Earlier findings link HDAC2 to many diseases including cancer. HDAC2 has been shown to be elevated in several human cancers (19, 20) and plays a rate-limiting role in mouse intestinal tumorigenesis (18, 21). Until now, the role of HDAC in biology and disease is largely attributed to its deacetylase activity-mediated transcriptional regulation. Here we provide the first evidence that 1) HDAC2 possesses deacetylase-independent SUMO E3 ligase-like activity and 2) HDAC2 gene-specifically activates protein synthesis of a subset of genes related to proliferation and apoptosis. Our findings indicate that HDAC2 regulates gene expression at both transcriptional and translational levels. HDAC2 is not only a transcription co-repressor but also a translation activator. Although dysregulated deacetylase activity of HDAC contributes to epigenetic silencing of tumor suppressor genes such as the cyclin-dependent kinase inhibitor p21 (22), elevated SUMO E3 ligase-like activity of HDAC2 may lead to induced protein synthesis of oncogenes that are related to antiapoptosis, cell growth or proliferation. Therefore, we speculate that both the deacetylase activity and the SUMO E3 ligase-like activity of HDAC2 contribute to human cancer.
In summary, we have identified HDAC2 as a factor that possesses a previously unidentified sumoylation-promoting activity. We believe that our findings offer new insights into how HDAC2 participates in physiological and pathological processes through its sumoylation-promoting activity.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant CA128681 through the United States Public Health Service (to J. H.). This work was also supported in part by grants from the Hillman Foundation fellowship and The Pittsburgh Foundation (to J. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures and Figs. 1–3.
- eIF4F
- eukaryotic initiation factor 4F
- eIF4E
- eukaryotic translation initiation factor 4E
- 4E-BP
- eIF4E-binding protein
- HDAC
- histone deacetylase
- SUMO
- small ubiquitin-like modifier
- PIAS
- protein inhibitor of activated STAT
- STAT
- signal transducers and activators of transcription
- HA
- hemagglutinin
- JNK
- c-Jun N-terminal kinase
- IRES
- internal ribosomal entry site
- EYFP
- enhanced yellow fluorescent protein
- ECFP
- enhanced cyan fluorescent protein
- shRNA
- short hairpin RNA.
REFERENCES
- 1.Sonenberg N., Hinnebusch A. G. (2009) Cell 136, 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gingras A. C., Raught B., Sonenberg N. (1999) Annu. Rev. Biochem. 68, 913–963 [DOI] [PubMed] [Google Scholar]
- 3.Gingras A. C., Raught B., Gygi S. P., Niedzwiecka A., Miron M., Burley S. K., Polakiewicz R. D., Wyslouch-Cieszynska A., Aebersold R., Sonenberg N. (2001) Genes Dev. 15, 2852–2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nie M., Xie Y., Loo J. A., Courey A. J. (2009) PLoS One 4, e5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu X., Vatsyayan J., Gao C., Bakkenist C. J., Hu J. (2010) EMBO Rep. 11, 299–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Di Bacco A., Gill G. (2006) Cell Cycle 5, 2310–2313 [DOI] [PubMed] [Google Scholar]
- 7.Geiss-Friedlander R., Melchior F. (2007) Nat. Rev. Mol. Cell Biol. 8, 947–956 [DOI] [PubMed] [Google Scholar]
- 8.Grégoire S., Yang X. J. (2005) Mol. Cell. Biol. 25, 2273–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao X., Sternsdorf T., Bolger T. A., Evans R. M., Yao T. P. (2005) Mol. Cell. Biol. 25, 8456–8464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao C., Ho C. C., Reineke E., Lam M., Cheng X., Stanya K. J., Liu Y., Chakraborty S., Shih H. M., Kao H. Y. (2008) Mol. Cell. Biol. 28, 5658–5667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vatsyayan J., Qing G., Xiao G., Hu J. (2008) EMBO Rep. 9, 885–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kee H. J., Eom G. H., Joung H., Shin S., Kim J. R., Cho Y. K., Choe N., Sim B. W., Jo D., Jeong M. H., Kim K. K., Seo J. S., Kook H. (2008) Circ. Res. 103, 1259–1269 [DOI] [PubMed] [Google Scholar]
- 13.Cullen B. R. (2009) Cell 136, 592–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nie M., Htun H. (2006) Nucleic Acids Res. 34, 5528–5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Benedetti A., Graff J. R. (2004) Oncogene 23, 3189–3199 [DOI] [PubMed] [Google Scholar]
- 16.Mamane Y., Petroulakis E., Martineau Y., Sato T. A., Larsson O., Rajasekhar V. K., Sonenberg N. (2007) PLoS ONE 2, e242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graff J. R., Konicek B. W., Vincent T. M., Lynch R. L., Monteith D., Weir S. N., Schwier P., Capen A., Goode R. L., Dowless M. S., Chen Y., Zhang H., Sissons S., Cox K., McNulty A. M., Parsons S. H., Wang T., Sams L., Geeganage S., Douglass L. E., Neubauer B. L., Dean N. M., Blanchard K., Shou J., Stancato L. F., Carter J. H., Marcusson E. G. (2007) J. Clin. Invest. 117, 2638–2648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zimmermann S., Kiefer F., Prudenziati M., Spiller C., Hansen J., Floss T., Wurst W., Minucci S., Göttlicher M. (2007) Cancer Res. 67, 9047–9054 [DOI] [PubMed] [Google Scholar]
- 19.Bolden J. E., Peart M. J., Johnstone R. W. (2006) Nat. Rev. Drug Discov. 5, 769–784 [DOI] [PubMed] [Google Scholar]
- 20.Weichert W., Röske A., Gekeler V., Beckers T., Stephan C., Jung K., Fritzsche F. R., Niesporek S., Denkert C., Dietel M., Kristiansen G. (2008) Br. J. Cancer 98, 604–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu P., Martin E., Mengwasser J., Schlag P., Janssen K. P., Göttlicher M. (2004) Cancer Cell 5, 455–463 [DOI] [PubMed] [Google Scholar]
- 22.Wilson A. J., Byun D. S., Popova N., Murray L. B., L'Italien K., Sowa Y., Arango D., Velcich A., Augenlicht L. H., Mariadason J. M. (2006) J. Biol. Chem. 281, 13548–13558 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.