Abstract
The sulfhydryl oxidase Ero1 oxidizes protein disulfide isomerase (PDI), which in turn catalyzes disulfide formation in proteins folding in the endoplasmic reticulum (ER). The extent to which other members of the PDI family are oxidized by Ero1 and thus contribute to net disulfide formation in the ER has been an open question. The yeast ER contains four PDI family proteins with at least one potential redox-active cysteine pair. We monitored the direct oxidation of each redox-active site in these proteins by yeast Ero1p in vitro. In this study, we found that the Pdi1p amino-terminal domain was oxidized most rapidly compared with the other oxidoreductase active sites tested, including the Pdi1p carboxyl-terminal domain. This observation is consistent with experiments conducted in yeast cells. In particular, the amino-terminal domain of Pdi1p preferentially formed mixed disulfides with Ero1p in vivo, and we observed synthetic lethality between a temperature-sensitive Ero1p variant and mutant Pdi1p lacking the amino-terminal active-site disulfide. Thus, the amino-terminal domain of yeast Pdi1p is on a preferred pathway for oxidizing the ER thiol pool. Overall, our results provide a rank order for the tendency of yeast ER oxidoreductases to acquire disulfides from Ero1p.
Keywords: Electron Transfer, Enzymes/Flavin, Enzymes/Oxidase, Glutathione, Organisms/Yeast, Subcellular Organelles/Endoplasmic Reticulum, Sulfhydryls/Disulfide
Introduction
The sulfhydryl oxidase Ero1p supports oxidative protein folding in the yeast endoplasmic reticulum (ER)3 by catalyzing oxidation of thiol pairs to disulfides (1, 2). Ero1p directly oxidizes protein disulfide isomerase (PDI; Pdi1p in yeast), which then oxidizes newly synthesized substrate proteins (3). The balance between reduced and oxidized glutathione in the ER may also be altered as a consequence of Ero1p activity, because glutathione contributes to the reduction of mis-paired disulfides, either directly or via reduction of Pdi1p (4, 5). Pdi1p is composed of four thioredoxin fold (trx) domains, of which the amino-terminal (a domain) and carboxyl-terminal (a′ domain) contain redox-active Cys-X-X-Cys motifs (6, 7) (Fig. 1). These two redox-active domains have been proposed to play complementary roles in catalysis of proper disulfide formation in downstream substrate proteins (8, 9).
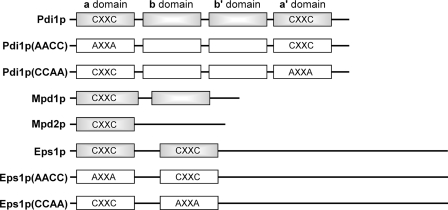
FIGURE 1.
Domain organization of yeast PDI family proteins and location of Cys-X-X-Cys motifs. Standard nomenclature for Pdi1p domains (a, b, b′, and a′ domains) is indicated. Boxes correspond to trx domains. Gray boxes distinguish wild-type proteins from mutants used in this study, which are shown with white boxes.
Three other ER proteins related to Pdi1p and containing Cys-X-X-Cys motifs are found in yeast, namely Mdp1p, Mpd2p, and Eps1p (Fig. 1). Mpd1p and Mpd2p each contain a single redox-active trx domain, whereas Eps1p has two predicted trx domains that contain the Cys-X-X-Cys motif. In principle, these other PDI family proteins could, like Pdi1p, be targets of Ero1p oxidase activity. Mpd1p, Mpd2p, and Eps1p are encoded by non-essential genes in yeast, whereas the gene for Pdi1p is essential (10). Furthermore, Pdi1p is expressed at higher levels than the other family members (10, 11). Nevertheless, biochemical and genetic experiments suggest that other members of the family can indeed interact with, and perhaps function downstream of, the Ero1p sulfhydryl oxidase. First, Mpd2p was trapped in a mixed disulfide with Ero1p (3), suggesting the possibility of electron transfer between these two proteins. Second, overexpression of Mpd1p rescued cells bearing the otherwise lethal deletion of the genes encoding Pdi1p and its ER homologs (10, 12). This finding indicates that, when present at higher than normal levels, Mpd1p can perform the essential functions of Pdi1p, one of which may be to transfer disulfides from Ero1p to substrate proteins folding in the ER. However, the extent to which Ero1p discriminates between Pdi1p and these other ER oxidoreductases is unknown.
The ability of Ero1p to oxidize a PDI family oxidoreductase is likely to influence the extent to which the latter oxidizes downstream substrates in the ER. However, re-reduction of the oxidoreductase by glutathione after oxidation by Ero1p would produce a futile cycle (13). Therefore, both a sufficient rate of oxidation by Ero1p and an appropriate reduction potential relative to the reduced and oxidized glutathione couple are important for an oxidoreductase to function on the protein thiol oxidation pathway of the ER. We compared the suitability of each yeast PDI-family redox-active Cys-X-X-Cys motif to serve downstream of Ero1p by measuring its equilibrium reduction potential and rate of oxidation by Ero1p. Importantly, in vitro results were supported by physical and genetic interactions in vivo. We provide a new perspective on the two active sites of Pdi1p by showing that yeast Ero1p preferentially oxidizes the a domain of Pdi1p, which should consequently function as the primary oxidant of protein thiols in the yeast ER. Furthermore, our study provides a broad view of the interactions between Ero1p and PDI family proteins and better defines the physiological pathways for disulfide bond formation in yeast.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The sequence coding for Saccharomyces cerevisiae Mpd1p (residues 23–310) and a His6 tag at the carboxyl terminus was inserted into the pET-15b vector (Novagen) between the NcoI and BamHI restriction sites. The coding sequence for S. cerevisiae Eps1p (residues 28–645) was inserted into pET-15b as described (14), and the Eps1p(CCAA) and Eps1p(AACC) mutants were produced by introducing the site-specific mutations C200A/C203A or C60A/C63A, respectively, using the QuikChange kit (Stratagene). The Pdi1p(CCAA) and Pdi1p(AACC) mutants were constructed by site-directed mutagenesis of the pVW828 plasmid (15). Thioredoxin mutants were generated by site-directed mutagenesis, and protein was prepared as described for wild type (16).
Mpd1p, Pdi1p, and their variants were produced in the E. coli strain BL21 (DE3) plysS (Novagen). Eps1p and its variants were produced in Origami B(DE3) pLysS (Novagen) cells. In all cases, cells were grown to an A595 nm of 0.4–0.6 at 37 °C, at which point isopropyl-1-thio-β-d-galactopyranoside was added to a final concentration of 1 mm. Mpd1p and Eps1p wild-type and mutant cultures were grown for a further 12 h at 25 °C before harvesting, whereas Pdi1p and its mutants were grown for 3 h at 30 °C. The supernatants of the cell lysates were applied to an Ni-NTA-Sepharose column, and proteins were eluted with a step gradient of imidazole. Eps1p and variants were dialyzed against 300 mm NaCl, 10 mm Tris, pH 8. The N-terminal His6 tag of Eps1p was cleaved using thrombin (Sigma) and removed using Ni-NTA-Sepharose (except for protein used in reduction potential measurements, for which the His6 tag was not removed). Thrombin activity was inhibited by phenylmethanesulfonyl fluoride.
A glutathione S-transferase-tagged version of Ero1p (residues 10–424) and the corresponding mutant Ero1p(C150A/C295A) were produced from the pGEX-4T1 vector (Amersham Biosciences) using similar bacterial growth and induction conditions as previously described (17). The fusion proteins were loaded onto a glutathione-Sepharose column and subsequently cleaved by thrombin on the column as described (17). Enzyme released from the column by cleavage was further purified using size-exclusion chromatography.
Mpd2p produced in bacteria gave low yields and unstable protein. Mpd2p was instead produced by transient transfection of human HEK293T cells. The Mpd2p coding sequence (residues 20–272) was inserted into the pHLsec plasmid between a signal sequence and a His6 tag (18), and the resulting pHLsec-Mpd2p plasmid was purified using the Qiagen Plasmid Mega Kit. HEK293T cells were split onto 30 tissue culture plates of diameter 15 cm. The following measures are given per plate. After 24 h, when cells were at ∼80% confluence, 40 μg of plasmid DNA was added to 4.5 ml of serum-free medium, and mixing was followed by the addition of 90 μl of 1 mg/ml polyethyleneimine (25-kDa branched) and brief vortexing. The solution was incubated for 15 min at room temperature to allow DNA-polyethyleneimine complex formation. Meanwhile, cells were washed with serum-free medium, and 14 ml of fresh medium was added to each plate. DNA-polyethyleneimine complex was then added. After 5 h, cells were washed once, and 22 ml of serum-free medium was added to each plate. Cell culture supernatants were collected after 3 days, filtered, dialyzed against 20 mm Tris pH 8, 200 mm NaCl, 10 mm imidazole, and applied to an Ni-NTA column. Eluted fractions containing Mpd2p were pooled and concentrated. The protein appeared less prone to degradation than the bacterially expressed material, but yields were still low (∼1 mg/liter of culture medium).
Protein Reduction
Mpd1p, Eps1p, and Pdi1p(AACC) at ∼500 μm were reduced by incubation with 10 mm reduced glutathione (GSH) for 1 h at room temperature, whereas Pdi1p and Pdi1p(CCAA) were incubated with 20 mm GSH. These conditions do not reduce the CX6C disulfide present in the a domain of Pdi1p. Mpd2p was reduced at a concentration of ∼50 μm with 23 mm GSH for 1 h at room temperature. Thioredoxin and its mutants at ∼2 mm were reduced with 50 mm dithiothreitol (DTT) for 1 h at room temperature. GSH or DTT was removed using a PD-10 desalting column equilibrated in 50 mm phosphate buffer, pH 7.5, 300 mm NaCl, 0.5 mm EDTA (buffer A).
For the experiment in Fig. 2B, the Eps1p(AACC) mutant at ∼25 μm was incubated for approximately 0.5 h either with 100 mm DTT or with 1 mm DTT in the presence or absence of 2% SDS. After incubation, reactions with 1 mm DTT were mixed with an equal volume of 5 mm PEG-maleimide MW 5000 (mal-PEG5K, Nektar Therapeutics, San Carlos, CA) in 2% SDS, 50 mm Tris, pH 6.8, 0.1% bromphenol blue, 20% glycerol (loading buffer). The reaction with 100 mm DTT was applied to a PD-10 desalting column equilibrated in 300 mm NaCl, 10 mm Tris, pH 8, concentrated in <20 min back to ∼25 μm using a Vivaspin centrifugal concentrator (Vivascience), and then mixed with mal-PEG5K in loading buffer. All samples were then treated with 10 mm DTT and applied to a 10% polyacrylamide gel. After separation, proteins were visualized by Coomassie Blue staining.
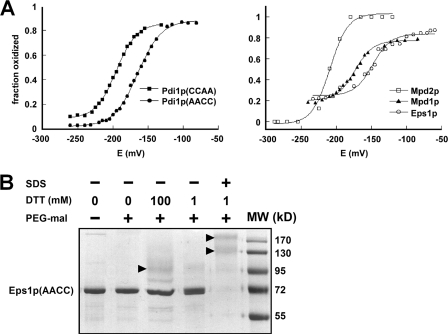
FIGURE 2.
Redox characteristics of yeast PDI family Cys-X-X-Cys motifs. A, redox potential of the samples was adjusted by varying the GSH/GSSG ratio at pH 7.0. After equilibration in redox buffer, the proteins were precipitated and reacted with mal-PEG5K, separated by SDS-PAGE, and visualized using fluorescent stain. The intensities of the bands were quantified and plotted as fraction oxidized. Data for the five Cys-X-X-Cys motifs are shown on two graphs for clarity. Each titration was fitted to the Nernst equation for a two-electron transfer to give reduction potential values of Pdi1p(CCAA): −194 mV, Pdi1p(AACC): −164 mV, Mpd1p: −174 mV, and the Eps1p amino-terminal domain: −149 mV. The Mpd2p reduction potential is estimated to be −210 mV. B, the Cys-X-X-Cys of the second Eps1p trx domain is not fully reduced by high concentrations of DTT in the folded state. Alkylation of free thiols with mal-PEG5K was used to distinguish oxidized from reduced fractions after the indicated treatments. The Eps1p(AACC) mutant was incubated with low (1 mm) or high (100 mm) DTT concentrations in the folded state or with 1 mm DTT after denaturing the protein with detergent. In the folded state, the protein was highly resistant to reduction. Only in the denatured state did substantial reduction occur, as indicated by the disappearance of the unmodified protein (labeled Eps1p(AACC) to the side of the gel) and the appearance of species of higher apparent molecular weights (indicated by arrowheads). Eps1p(AACC) contains six cysteines, so the various higher molecular weight species may have different subsets of these cysteines reduced and modified by mal-PEG5K.
Oxidation Assays
For gel analysis of Ero1p oxidative activity, reactions were performed in buffer A. Each experiment, with the exception of Mpd2p oxidation, contained 150 μm thiols from the PDI family protein and 2 μm Ero1p or Ero1p(C150A/C295A). Due to the limited material obtained from transient transfection of mammalian cells, the starting Mpd2p thiol concentration was 50 μm. The somatostatin peptide, when present, was at a concentration of 150 μm. At each time point, 10 μl were removed and mixed with 10 μl 5 mm mal-PEG5K in loading buffer. After 20 min, remaining mal-PEG5K was quenched by addition of DTT, and samples were subjected to SDS-PAGE on a 12% acrylamide gel. Proteins were visualized with Coomassie Blue stain. Band intensities were determined using the ImageQuant 5.0 program. Fraction reduced was calculated as follows: the intensity of the band corresponding to the oxidized, unmodified material in each lane was divided by the intensity of a band corresponding to an oxidized sample not treated with mal-PEG5K run on the same gel, and this value was subtracted from 1.
Oxygen consumption was measured in 0.8 ml of buffer A using an Oxygraph Clark-type oxygen electrode (Hansatech Instruments). For PDI family protein substrates, assays were performed in the presence of 10 mm GSH. Each experiment initially contained 100 μm protein thiols as determined using Ellman's assay (19), which corresponded to ∼50 μm protein as determined by absorbance at 280 nm in guanidine solution. The only exception was Pdi1p(CCAA), in which thiols (100 μm) were obtained from 70 μm protein, indicating incomplete reduction. More vigorous reduction protocols reduced the Pdi1p CX6C disulfide and thus were not used. Oxygen levels in the substrate solution were monitored until a linear baseline was established, at which time the reaction was initiated by injection of 20 μl of 40 μm Ero1p or Ero1p(C150A/C295A) in buffer A to achieve a final enzyme concentration of 1 μm. To confirm that re-reduction of protein substrate by GSH was not rate-limiting, oxygen consumption was measured in solutions containing Pdi1p(CCAA), GSH, and either 1 or 2 μm Ero1p(C150A/C295A). After halving the time axis of the reaction at 2 μm enzyme concentration, the two curves were superposable (data not shown). This observation indicates that, even at double the concentration of the fastest Ero1p mutant and the best substrate used in the experiments, the rate of oxidation was limited by the quantity of enzyme and not by substrate re-reduction. For thioredoxin variants, oxygen consumption experiments were done at a starting protein thiol content of 200 μm and an enzyme concentration of 1 μm, without GSH.
Reduction Potential Measurements
Proteins were diluted to ∼9 μm into 300 mm NaCl, 100 mm sodium phosphate buffer, pH 7.0, 1 mm EDTA containing different ratios of reduced to oxidized glutathione (GSH/GSSG) and incubated at room temperature for 1 h. The stock solutions of GSH and GSSG were titrated to pH 7 using 10 n NaOH. The GSH/GSSG redox potential was calculated according to the Nernst equation (Eh = E0 + 2.3*RT/nF*log([GSSG]/[GSH]2), where E0 = −240 mV, pH 7.0 (20), and n = 2 for the two-electron oxidation. After the 1-h incubation, protein was precipitated by addition of ice-cold trichloroacetic acid to a final concentration of 25%, and the pellet was washed with ice-cold acetone. The protein was resuspended in a solution containing 5 mm mal-PEG5K in loading buffer. Samples were resolved by SDS-PAGE on a 10% gel and visualized using InVisionTM His tag In-gel Stain (Invitrogen). Band intensities were determined using the ImageQuant 5.0 program. We assumed that the His tag stain labeled the oxidized and mal-PEG5K modified proteins equally, so fraction oxidized is given as the ratio of the intensity of the unmodified band to the sum of the intensities of the mal-PEG5K modified and unmodified bands.
S. cerevisiae Strains and Plasmids
PDI1 plasmids for expression in yeast contain the PDI1 coding region and ∼875 and ∼150 bp of the 5′- and 3′-untranslated regions, respectively. To allow for purification of Pdi1p from yeast, DNA sequence encoding a tandem FLAG-His6 (FLAG-H6) tag was inserted at a NheI site introduced after the signal peptide coding region. To add a thrombin recognition site within Pdi1p, the coding sequence for amino acids 370–373 (Ile-Val-Arg-Ser) was replaced with the sequence Leu-Val-Pro-Arg-Gly-Ser. An LEU2-marked, CEN plasmid encoding MPD1 was created by PCR amplification of the MPD1 coding region plus 1 kb and 200 bp of the 5′- and 3′-untranslated regions, respectively. To create pCS205 (MPD1-myc), a myc tag was added by ligation of a triple-myc epitope fragment into a NotI site created by site-directed mutagenesis prior to residue 300. Two copies of the triple-myc fragment inserted during the ligation yielded six tandem copies of the myc tag. Cysteine amino acid replacement alleles of Pdi1p and Mpd1p (pCS619 [mpd1-C59A-C62A-myc]) were created using QuikChange or by subcloning from previously constructed plasmids using standard techniques. ERO1 coding URA3 CEN plasmids pCS483 (ero1-C100A-C105A-C150A-C295A-myc), pCS452 (PGAL1-ERO1-C150A-C295A-myc), and pCS504 (PGAL1-ero1-C100A-C105A-C150A-C295A-myc) have been described previously (21). Plasmid pCS456 (ero1-C150A-C295A-myc) is the URA3-marked equivalent of the LEU2 plasmid pCS524 described in (21).
To generate a chromosomal PDI1 deletion strain, the PDI1 coding sequence was replaced with KanMX in a homozygous GAL2 ura3–52 leu2–3,112 diploid background. The resultant heterozygous diploid was transformed with pCS213 (URA3 CEN PDI1), and viable MATa and MATα URA+ KanMX+ segregants were recovered after sporulation (CKY1044 and CKY1045, respectively). The viability of CKY1044 and CKY1045 depends on the episomal PDI1 allele, as demonstrated by the failure of these strains to grow on medium containing 5-fluoroorotic acid (Toronto Research Laboratories, Downsview, Ontario, Canada). Strains CKY1046–1056 were generated by transformation of CKY1044 with the CEN LEU2-marked plasmids pCS463 (PDI1), pCS465 (pdi1-C61S-C64S), pCS464 (pdi1-C406S-C409S), pCS462 (pdi1-C64S-C409S), pSK45 (pdi1-C64S), pSK46 (pdi1-C409S), pSK32 (FLAG-H6-PDI1), pSK49 (FLAG-H6-pdi1-C406S-C409S), pSK50 (FLAG-H6-pdi1-C61S-C64S), pSK42 (FLAG-H6-pdi1-C64S-thrombin-C409S), or pSK47 (FLAG-H6-pdi1-C64S-C409S), respectively, followed by selection against pCS213 by plating on synthetic minimal medium (SMM) with 5-fluoroorotic acid. To allow for purification of a cysteine-less Pdi1p, CKY1044 was transformed with pSK36 (FLAG-H6-pdi1-C61S-C64S-thrombin-C406S-C409S), and the wild-type PDI1 plasmid was maintained (strain CKY1057). To create the pdi1Δ ero1–1 double mutant (CKY1058), the yeast strains CKY1045 (pdi1Δ) and CKY598 (ero1–1) were mated, and a MATa temperature-sensitive URA+ KanMX+ segregant was selected after sporulation. The mpd1Δ yeast strain (CKY1059) was made by one-step gene replacement of the coding region of MPD1 with KanMX in a MATa GAL2 ura3–52 leu2–3,112 background.
CPY Radiolabeling and Immunoprecipitation
CKY1046, CKY1047, and CKY1048 cells were grown to exponential phase, and cells were harvested and suspended at 5 A600 units/ml in SMM lacking methionine. Cells were labeled at 30 °C for 7 min with [35S]methionine and [35S]cysteine (EXPRESS, PerkinElmer Life Sciences) and chased for 0–15 min with cold methionine and cysteine. Cells lysis and immunoprecipitation of carboxypeptidase Y (CPY) were completed as described before (22).
In Vivo Oxidation State of Pdi1p
To analyze the redox state of Pdi1p, cells encoding FLAG-H6-Pdi1p or FLAG-H6-Pdi1p mutants (CKY1052, CKY1053, CKY1054, and CKY1057) were grown to exponential phase in SMM and lysed by agitation with glass beads in 10% ice-cold trichloroacetic acid at 4 °C. Proteins were collected by centrifugation, washed once with chilled acetone, and resuspended in 0.2 m Tris-HCl, 2% SDS, pH 7.8, with 50 mm N-ethylmaleimide (NEM). After 30-min incubation at room temperature, NEM-modified cell lysates were clarified by centrifugation. SDS was removed by treating with SDS-out (Pierce) and replaced with 1% Triton X-100. Lysates were diluted into binding buffer (50 mm phosphate buffer, pH 7.8, 500 mm NaCl, 1% Triton X-100) and incubated with Ni-NTA-agarose beads (Qiagen) for 2 h at room temperature. Beads were washed with binding buffer, and Pdi1p was eluted with 250 mm imidazole containing 1% Triton X-100. Eluted proteins were reduced by boiling for 5 min in the presence of 50 mm DTT and 1% SDS. DTT was removed by trichloroacetic acid precipitation of the protein, and proteins were treated with 5 mm PEG-maleimide MW 2000 (mal-PEG2K, NOF Corp., Japan) in modified loading buffer (0.2 m Tris-HCl, pH 8, 3% SDS, 10% glycerol, 0.1% bromphenol blue) for 30 min at 30 °C. Free mal-PEG2K was quenched with DTT. To prepare oxidized and reduced Pdi1p controls, cells were lysed and incubated in phosphate-buffered saline with 10 mm diamide or 50 mm DTT, respectively, prior to the initial trichloroacetic acid-NEM treatment. All samples were deglycosylated with endoglycosidase Hf (New England Biolabs) and analyzed by SDS-PAGE and Western blotting with anti-Pdi1p antibody.
In Vivo Oxidation State of Mpd1p
To analyze the redox state of Mpd1p-Myc, cells were grown to exponential phase in SMM lacking leucine. Cells (2 A600 units) were harvested by centrifugation, resuspended in 50 μl of 20% (w/v) trichloroacetic acid, and disrupted by agitation with glass beads. Proteins were collected by centrifugation, washed with cold acetone, and resuspended in 40 μl of modified loading buffer with 5 mm mal-PEG2K and protease inhibitors. Samples were incubated for 30 min at 30 °C and quenched with 10 μl of 0.25 m DTT in loading buffer. Insoluble material was removed by centrifugation, samples were resolved by 10% SDS-PAGE and transferred to nitrocellulose, and Mpd1p was detected with anti-Myc antibody (9E10). For reduced and oxidized control samples, cells were suspended in modified loading buffer containing protease inhibitors and 10 mm DTT or 10 mm diamide, lysed by agitation with glass beads, and incubated for 30 min at 30 °C. Proteins were precipitated with trichloroacetic acid and subsequently treated with mal-PEG2K.
Detection of Mixed-disulfide Intermediate between Pdi1p and Ero1p in Vivo
To analyze the requirements for capture of the Pdi1p-Ero1p mixed-disulfide intermediate in yeast, the Ero1p plasmids pCS452 and pCS504 were transformed into the Pdi1p mutant strains CKY1049, CKY1050, and CKY1051. Cells were grown in SMM lacking leucine and uracil with 1% galactose and 1.5% raffinose. Cell lysates were prepared and modified by NEM as described above for the in vivo oxidation studies with Pdi1p. NEM-treated lysates were subsequently deglycosylated with endoglycosidase Hf, and Pdi1p and Ero1p-Myc were detected by immunoblotting with anti-Pdi1p or anti-Myc antibody.
For purification and subsequent thrombin cleavage of the mixed-disulfide intermediate between Ero1p and Pdi1p variants, the Ero1p plasmid pCS456 was transformed into the Pdi1p(CSCS) mutant strains CKY1055 and CKY1056. Cell lysates were prepared, modified with NEM, and treated with SDS-out using the methods described under “Experimental Procedures” subheading “In Vivo Oxidation State of Pdi1p.” Cell lysates were diluted into TBS/Triton (50 mm Tris, pH 7.6, 150 mm NaCl, 1% Triton X-100) and incubated with anti-FLAG affinity beads (Sigma) for 4 h at room temperature. Beads were washed with TBS/Triton, and Pdi1p was eluted with 0.2 mg/ml FLAG peptide (Sigma) in TBS/Triton. Eluants were incubated with 0.5 or 3 units of thrombin (Sigma) for 3 h at 37 °C, and proteins were detected by immunoblotting with anti-Ero1p and anti-FLAG antibody.
RESULTS
Reduction Potentials of Yeast PDI Family Cys-X-X-Cys Motifs
We first measured the equilibrium reduction potentials of the yeast PDI family proteins Mpd1p, Mpd2p, and Eps1p, as well as of the two redox-active domains of Pdi1p (Fig. 2A). The values we obtained for the a and a′ domains of Pdi1p were −194 and −164 mV, respectively, compared with previously published values of −188 and −154 mV (23, 24). Mpd1p gave a reduction potential of −174 mV, and Eps1p gave a value of −149 mV. Eps1p has two Cys-X-X-Cys motifs. The lack of reduction of the carboxyl-terminal trx domain even after incubation with 100 mm DTT (Fig. 2B) suggests that the measured reduction potential for Eps1p corresponds to the amino-terminal trx domain. Although the Mdp2p measurements were not performed as rigorously as the others due to insufficient material, the approximate reduction potential obtained for this protein was −210 mV. Thus, the order of yeast ER Cys-X-X-Cys motifs from most to least oxidizing is: Eps1p first domain (−149 mV), Pdi1p a′ domain (−164 mV), Mpd1p (−174 mV), Pdi1p a domain (−194 mV), Mpd2p (−210 mV), followed presumably by the second domain of Eps1p (not determined).
Oxidation Rates of ER Oxidoreductases by Ero1p in Vitro
In the first set of kinetics experiments, enzymatic reactions were performed in a simple system monitoring direct transfer of electrons from reduced Cys-X-X-Cys motifs of PDI family proteins to Ero1p under multiple turnover conditions. Rates of oxidation were measured by mixing the reduced PDI family member with recombinant Ero1p and blocking the reaction after various times using mal-PEG5K (9, 23). The remaining reduced substrate, containing mal-PEG5K at each unpaired cysteine residue, was separated by SDS-PAGE from the oxidized fraction, in which cysteines are protected from reaction with mal-PEG5K by participating in disulfide bonds. It should be noted that kinetic parameters such as maximum rates and Michaelis constants may not be obtainable for the oxidation of PDI family proteins by Ero1p. Ero1p activation and deactivation processes result in lag phase kinetics (16, 21) and other complexities that interfere with measurement of initial rates or mathematical modeling of enzyme progress curves. Furthermore, we were not able to produce PDI family protein substrates with high enough yields or to concentrate them sufficiently to achieve the maximal oxidation rate. Therefore, qualitative comparisons were made by performing oxidation reactions at the same initial reduced substrate concentration.
Comparing the direct oxidation rates of the Cys-X-X-Cys motifs in the four proteins under study revealed that the amino-terminal domain (a domain) of Pdi1p is oxidized most rapidly by recombinant yeast Ero1p (Fig. 3A). Mdp2p was also oxidized relatively rapidly (Fig. 3B), but the results cannot be compared quantitatively with the other proteins due to the lower starting Mpd2p concentration in the reaction. In comparison to the a domain, the carboxyl-terminal domain (a′ domain) of Pdi1p was oxidized at a slower but measurable rate (Fig. 3A). The rate of oxidation of Mpd1p was similar or slightly greater than that observed for the Pdi1p a′ domain. Oxidation of the amino-terminal domain of Eps1p was virtually undetectable. In all cases, a mutant version of Ero1p, Ero1p(C150A/C295A), which was previously shown to have enhanced activity in oxidation of the non-natural substrate thioredoxin (21), also oxidized the set of potentially native ER substrates more rapidly than did wild-type Ero1p (Fig. 3A). For example, the Ero1p(C150A/C295A) mutant at 2 μm concentration oxidized 75 μm Pdi1p(CCAA) completely within the first minute of the reaction, whereas wild-type Ero1p oxidized half the Pdi1p(CCAA) in approximately 5 min. In general, the order of substrate preference seen for wild-type Ero1p was maintained by the hyperactive Ero1p(C150A/C295A) mutant. As observed previously using thioredoxin as a substrate (21), the Ero1p(C90A/C349A) mutant of Ero1p oxidized ER substrates slightly faster than did wild-type Ero1p, and again the order of substrate preference was maintained (data not shown).
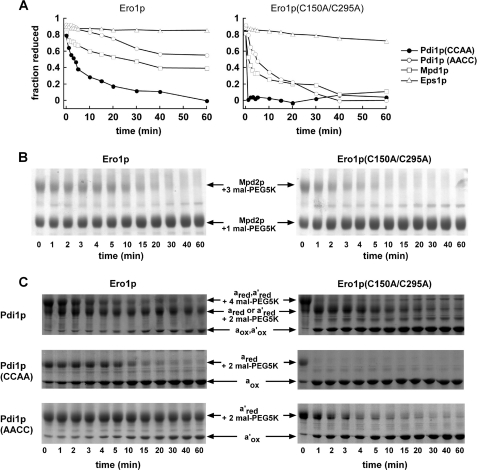
FIGURE 3.
Direct oxidation rates of PDI family proteins by Ero1p monitored by gel assays. A, reduced PDI family proteins (150 μm thiols) were mixed with 2 μm Ero1p or Ero1p(C150A/C295A). Loss of reactivity with mal-PEG5K upon disulfide bond formation was monitored by quantification of the accumulating unmodified protein after separation by SDS-PAGE. Lag phases are less evident in this experiment than in Fig. 4, because of the poor time resolution of gel-based assays. Furthermore, due to the relatively low substrate/enzyme ratio in this experiment (37.5:1), a significant fraction (estimated 5–10%) of substrate is oxidized by dithiol/disulfide exchange with the Ero1p shuttle and regulatory disulfides prior to oxygen consumption and enzyme turnover. B, except for a lower Mpd2p concentration, the experiment was conducted as in A, but the gels, rather than results of quantification, are shown. Mpd2p was partially reduced by incubation with 23 mm GSH for 1 h. Following removal of GSH, Mpd2p (50 μm thiols) was mixed with 2 μm Ero1p or Ero1p(C150A/C295A). The Mpd2p protein lacking its signal sequence has seven cysteine residues. One unpaired cysteine reacts with mal-PEG5K regardless of the redox status of the Cys-X-X-Cys motif. C, the two Pdi1p active sites appear to function independently during oxidation by Ero1p, because the two oxidation events seen for wild-type Pdi1p (top panel) occur at similar rates as the oxidation of the Pdi1p(CCAA) (middle panel) and Pdi1p(AACC) (bottom panel) mutants. This observation holds for both wild-type Ero1p and the C150A/C295A mutant. Bands are labeled according to the state of each Cys-X-X-Cys motif present (subscript “red” indicates reduced and “ox” oxidized) and the number of mal-PEG5K additions present. The gels shown here, which are similar to those used to obtain the data in A, further demonstrate that only a single disulfide was reduced initially in Pdi1p(AACC) and Pdi1p(CCAA), and two disulfides in wild-type Pdi1p, indicating that the Cys-X6-Cys motif remained oxidized as intended.
In addition to monitoring oxidation of each Cys-X-X-Cys alone, we tested whether the simultaneous presence of the two redox-active centers of Pdi1p affects the reactivity of either site. The disappearance of the wild-type Pdi1p species containing four reduced and modified cysteines (Fig. 3C, top band in top gels) correlates with the relatively rapid disappearance of reduced Pdi1p(CCAA) (Fig. 3C, upper band in middle gels). Full oxidation of Pdi1p then correlates with the slow disappearance of reduced Pdi1p(AACC) (Fig. 3C, upper band of bottom gels). In short, the rates of the two oxidation events observed in wild-type Pdi1p are consistent with the rates observed for Pdi1p(CCAA) and Pdi1p(AACC) separately. We also tested whether the presence of a potential Pdi1p-binding peptide influenced the absolute or relative reactivity of the two domains of Pdi1p with Ero1p; a somatostatin-derived peptide lacking cysteine residues (25) did not affect the rate of oxidation of Pdi1p(CCAA) or Pdi1p(AACC) (data not shown).
In the second set of experiments, oxidation rates of PDI family proteins by Ero1p were monitored by oxygen consumption in the presence of 10 mm GSH (Fig. 4A). These conditions mimic an overly reducing ER, enabling us to test whether Ero1p substrate selectivity changes or expands under these conditions. Oxidized protein substrate generated by Ero1p activity in these reactions is re-reduced by GSH, providing more substrate for oxidation. The reaction continues until the oxygen in the chamber is consumed. These experiments showed a similar order of substrate preference as observed in the SDS-PAGE assays conducted without GSH, with the a domain of Pdi1p remaining the most highly favored substrate for Ero1p. Mpd1p and the a′ domain of Pdi1p showed similar rates of reaction to one another, when oxidized either by wild-type Ero1p or by the Ero1p(C150A/C295A) mutant. Addition of glutathione did not improve the relative reactivity of Eps1p.
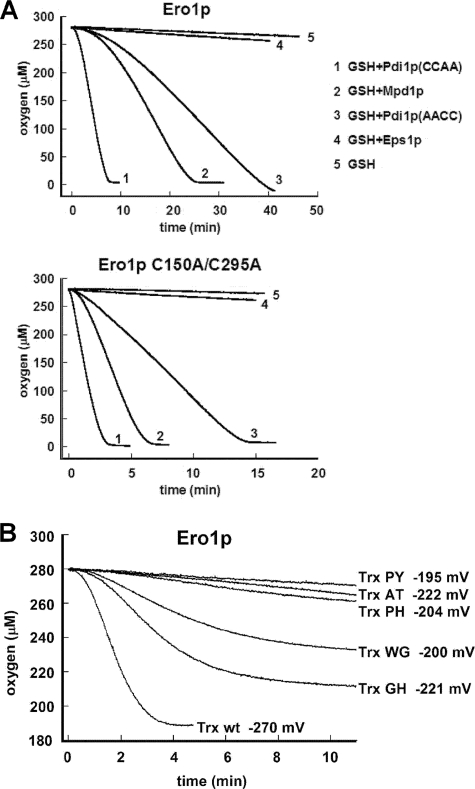
FIGURE 4.
Oxidation rates measured by oxygen consumption. A, reduced Pdi1p(CCAA), Pdi1p(AACC), Mpd1p, or Eps1p at an initial thiol concentration of 100 μm were mixed with 10 mm GSH. At time zero, Ero1p or Ero1p(C150A/C295A) was added to a concentration of 1 μm. Representative progress curves are shown, but in some experiments, oxidation of Mpd1p was slightly slower. Lag phases are evident in this experiment. B, oxidation of E. coli thioredoxin and mutant series. Reduced thioredoxin or active-site mutants (the two letters indicate the identity of the X-X amino acids in the Cys-X-X-Cys motif) at 200 μm thiol concentration were oxidized by 1 μm Ero1p.
The rough correlation observed between rate of oxidation by Ero1p and reduction potential for the Cys-X-X-Cys motifs in yeast PDI family proteins prompted us to test whether changes in reduction potential affect reactivity with Ero1p. To do this, we measured Ero1p oxidation rates on a series of mutants derived from the non-native substrate E. coli thioredoxin (Fig. 4B). Changing the two residues between the cysteines in the Cys-X-X-Cys motif in thioredoxin was previously reported to affect the reduction potential of the di-cysteine motif (26, 27). Although all the thioredoxin mutants tested had reduction potentials below that of the Pdi1p a domain, suggesting that they should be good electron donors, most of them were not oxidized by Ero1p at significant rates. Exceptions were wild-type thioredoxin and a mutant containing a Cys-Gly-His-Cys motif, the same motif as found in both active sites of Pdi1p and in Mpd1p. Interestingly, thioredoxin containing a Cys-Trp-Gly-Cys motif was also oxidized by Ero1p, despite having a higher reduction potential than some of the mutants that were not appreciably oxidized. This mutant was previously shown to functionally replace Pdi1p in vivo to a limited extent (28). Our results indicate that amino acid changes that alter reduction potential can dramatically affect reactivity with Ero1p, but that there is no absolute correlation between reduction potential and rate of oxidation.
Redox States of ER Oxidoreductases in Vivo
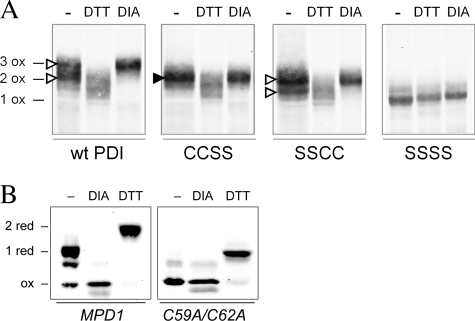
We next tested the hypothesis that rates of oxidation by Ero1p will determine the steady-state condition of the di-cysteine motifs of PDI family oxidoreductases in yeast cells. The redox states of the two active sites of Pdi1p were compared with each other and with Mpd1p. Pdi1p and its active-site mutants were purified from cell lysates after blocking reduced cysteines with NEM, and then oxidized Pdi1p disulfides were completely reduced and modified with mal-PEG2K (29). The number of mal-PEG2K modifications therefore indicates the number of cysteines that were oxidized in vivo. Wild-type Pdi1p appeared mostly in the fully oxidized form based on the co-migration of the majority of wild-type Pdi1p with Pdi1p fully oxidized by treatment with the small molecule oxidant diamide (Fig. 5A, top open arrowhead). However, a small but reproducible portion of wild-type Pdi1p migrated more quickly than the completely oxidized form and thus evidently contained a single reduced disulfide (Fig. 5A, bottom open arrowhead). The two Pdi1p active-site mutants allowed us to identify the reduced site; Pdi1p(CCSS) was observed in the fully oxidized form co-migrating with diamide-treated Pdi1p(CCSS) (Fig. 5A, filled arrowhead), whereas Pdi1p(SSCC), like wild-type Pdi1p, showed a mixture of oxidized and reduced forms, with ∼10–20% of its remaining active site in the reduced form (Fig. 5A, open arrowheads). Therefore, the Pdi1p a domain is fully oxidized, but the a′ domain is partially reduced in yeast cells.
FIGURE 5.
Redox states of Pdi1p and Mpd1p in vivo. A, wild-type Pdi1p and mutants Pdi1p(CCSS), Pdi1p(SSCC), and Pdi1p(SSSS) were purified from yeast cell lysates that had been reacted with NEM to block free thiols. Purified proteins were subsequently reduced with DTT and modified by mal-PEG2K. Pdi1p was visualized by Western blotting with anti-Pdi1p serum. With this protocol, the presence of a disulfide bond in the original cell lysate is reflected by mal-PEG2K modification and a corresponding decreased migration during SDS-PAGE. Fully oxidized Pdi1p controls (e.g. three disulfide bonds for wild-type Pdi1p (3 ox)) were prepared by lysis of cells in the presence of 10 mm diamide (DIA). Reduced migration controls were prepared by cell lysis in the presence of DTT; note that these lysis conditions did not result in reduction of the structural disulfide in Pdi1p as evident by the co-migration of the DTT and diamide-treated samples for Pdi1p(SSSS). The filled arrowhead indicates the single band representing a species with an oxidized a domain in the mutant lacking the active-site cysteines of the a′ domain. The open arrowheads indicate the two bands corresponding to the oxidized and reduced forms of the a′ domain in the mutant lacking the active-site cysteines of the a domain. B, yeast mpd1Δ cells containing plasmids encoding Myc-tagged Mpd1p or an Mpd1p active-site mutant, Mpd1p(C59A/C62A), were grown in SMM at 30 °C. The oxidation state of Mpd1p was assessed after resolving mal-PEG2K-modified cellular proteins by reducing SDS-PAGE and immunoblotting with anti-Myc. Using this protocol, free cysteine thiols within Mpd1p in the original cell lysate result in mal-PEG2K modification and a corresponding decrease in migration. Oxidized and reduced controls were prepared by treatment of cell lysates with diamide or DTT prior to mal-PEG2K.
For comparison, the redox state of Mpd1p was assessed in vivo. Here cell lysates were treated directly with mal-PEG2K, and thus the slower migrating forms of Mpd1p contain cysteines that were reduced in the cell. The majority of wild-type Mpd1p was observed to contain a single reduced cysteine pair (Fig. 5B). Mpd1p contains two pairs of cysteines: a Cys-X-X-Cys active site and a Cys-X8-Cys pair. An Mpd1p mutant lacking the active-site cysteines co-migrated with fully oxidized Mpd1p (Fig. 5B), indicating that the active-site cysteines are the ones reduced in vivo. In contrast to the current findings for Mpd1p, the active-site di-cysteine motif of Mpd2p was found previously to be oxidized in vivo (3). The in vivo redox states of these three proteins are thus roughly consistent with the preference of Ero1p in vitro for the a domain of Pdi1p and Mpd2p relative to Mpd1p and the a′ domain of Pdi1p.
Ero1p Interacts Preferentially with the N-terminal Domain of Pdi1p in Vivo
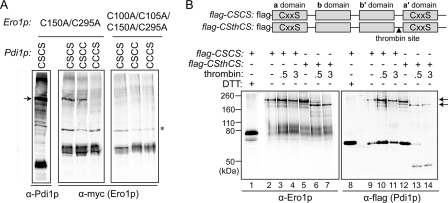
Because the a domain redox-active site of Pdi1p was more completely oxidized than the a′ domain in yeast cells, we investigated whether this difference was caused by Ero1p preference in vivo. As mixed disulfides are obligatory intermediates in dithiol-disulfide transfer events, trapping of transient complexes between Ero1p and ER oxidoreductases suggests preferred disulfide transfer pathways. Trichloroacetic acid-treated, NEM-modified lysates of cells expressing the Ero1p mutant Ero1p(C150A/C295A) and either Pdi1p(CSCS), Pdi1p(CSCC), or Pdi1p(CCCS) were examined by Western blot. The presence of a serine in place of the second cysteine in either Pdi1p Cys-X-X-Cys motif prevents the formation of an active-site disulfide bond and thus stabilizes the mixed-disulfide complex formed by the corresponding N-terminal interchange cysteine. Importantly, mixed disulfides were observed for the Pdi1p(CSCS) and Pdi1p(CSCC) variants, but not for Pdi1p(CCCS) (Fig. 6A), indicating that the first cysteine of the a domain Cys-X-X-Cys motif is used more frequently as the interchange thiol with Ero1p than is the first cysteine of the a′ domain di-cysteine motif. Absolute recovery of mixed disulfides was greater with the Ero1p(C150A/C295A) mutant, although the relative preference for the Pdi1p a and a′ domains was the same when wild-type Ero1p was used (data not shown). To exclude the possibility that the Pdi1p-Ero1p mixed disulfides are formed during other processes, such as catalysis of Ero1p folding by Pdi1p, catalytically inactive Ero1p(C100A/C105A/C150A/C295A) was examined as a negative control. Ero1p(C100A/C105A/C150A/C295A) did not form a mixed-disulfide intermediate, suggesting that the mixed disulfide between Ero1p(C150A/C295A) and Pdi1p(CSCS) was formed by attack of a Pdi1p cysteine on the catalytically relevant Ero1p disulfide, namely C100-C105.
FIGURE 6.
Ero1p preferentially forms a mixed-disulfide intermediate with the a domain of Pdi1p. A, formation of mixed-disulfide intermediates between Ero1p(C150A/C295A) (with a Myc epitope tag) and Pdi1p mutants Pdi1p(CSCS), Pdi1p(CSCC), and Pdi1p(CCCS). NEM-modified yeast cell lysates were resolved by non-reducing SDS-PAGE, and mixed-disulfide intermediates between Ero1p and Pdi1p were identified by Western blotting with either anti-Pdi1p or anti-Myc. Catalytically inactive Ero1p(C100A/C105A/C150A/C295A) was used as a negative control. An arrow denotes the mixed-disulfide complexes between mutant Ero1p and a subset of Pdi1p variants. Other bands in the leftmost (α-Pdi1p) lane are mixed-disulfide complexes between Pdi1p(CSCS) and other proteins (high molecular weight bands) or free Pdi1p(CSCS) (intense lower band). The asterisk indicates a nonspecific background band that does not disappear upon reduction (not shown). B, thrombin digestion of the Pdi1p(CSCS)-Ero1p(C150A/C295A) mixed-disulfide intermediate. To enable dissection of Pdi1p(CSCS), a thrombin site was introduced into the segment linking the b′ and a′ domains to generate the construct Pdi1p(CSthCS). Mixed-disulfide intermediates were isolated under non-reducing conditions from cells expressing a FLAG-epitope-tagged Pdi1p by affinity to anti-FLAG beads. Purified Pdi1p(CSCS)- and Pdi1p(CSthCS)-Ero1p(C150A/C295A) mixed-disulfide complexes were digested by thrombin, and association of the Pdi1p fragments with Ero1p was analyzed by immunoblotting with either anti-Ero1p or anti-FLAG. Arrows highlight the major Pdi1p-Ero1p mixed-disulfide complexes observed before (top) and after (bottom) thrombin cleavage.
To support these results, a thrombin cleavage site was introduced into Pdi1p(CSCS), allowing the two Pdi1p active sites to be separated by thrombin digestion after formation of the trapped complex between Ero1p(C150A/C295A) and Pdi1p(CSCS). The Pdi1p domain responsible for covalent association with Ero1p could then be identified by the size of the mixed-disulfide complex remaining after proteolysis and its immunoreactivity, because the thrombin cleavage site was closer to the carboxyl terminus of Pdi1p(CSCS), and a FLAG tag was present near the amino terminus (Fig. 6B). As for Fig. 6A, the Ero1p(C150A/C295A) mutant was used to enhance recovery. Pdi1p(CSCS), with or without a thrombin cleavage site, was purified from trichloroacetic acid- and NEM-treated yeast lysates using anti-FLAG affinity beads. Purified samples were treated with thrombin, and complexes of the Pdi1p and Ero1p variants were analyzed under non-reducing conditions by Western blot. The presence of a thrombin site within Pdi1p(CSCS) did not affect the recovery of complex with the Ero1p mutant; cleavable Pdi1p(CSCS) showed the same amount of complex with Ero1p(C150A/C295A) as Pdi1p(CSCS) without a thrombin site (Fig. 6B). After thrombin addition, the complex behaved as a single faster-migrating species, suggesting that Ero1p was associated with a single fragment of Pdi1p (Fig. 6B, lane 7). The major thrombin-cleaved Pdi1p(CSCS) fragment bound to Ero1p(C150A/C295A) reacted with the anti-FLAG antibody, which recognizes only amino-terminal fragments of the Pdi1p construct, suggesting a complex between Ero1p and the Pdi1p a domain (Fig. 6B, lane 14). Furthermore, the Ero1p-associated thrombin-cleaved fragment was only slightly smaller than the uncleaved complex, consistent with the presence of the larger abb′ fragment and with the loss of the smaller a′ domain from the complex after cleavage. Together these data suggest that the a domain rather than the a′ domain was most often responsible for the complex between Ero1p(C150A/C295A) and thrombin-cleavable Pdi1p(CSCS).
Phenotypic Effects of Disrupting the Interaction between Ero1p and the Pdi1p a Domain
It was previously shown that yeast cells containing a Pdi1p mutant lacking a functional a domain are more sensitive to DTT treatment than cells expressing Pdi1p lacking the a′ domain cysteines (30). We observed a similar sensitivity to DTT treatment with our Pdi1p mutant strains; Pdi1p(SSCC) yeast strains showed a more pronounced growth delay in the presence of 1 mm DTT than wild-type or Pdi1p(CCSS) containing yeast (Fig. 7A, first column).
FIGURE 7.
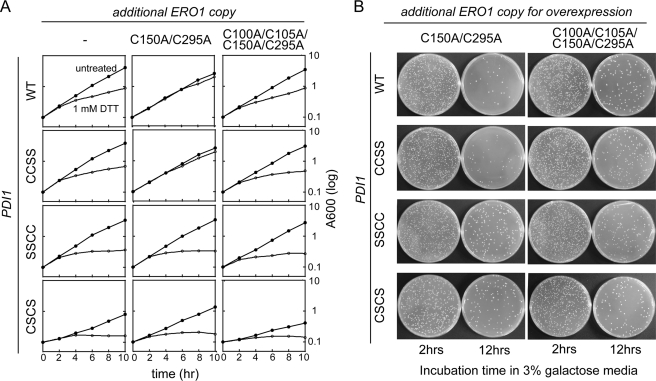
Disulfide relay system between Ero1p and the a domain of Pdi1p. A, the de-regulated Ero1p(C150A/C295A) mutant requires a functional Pdi1p a domain to suppress growth defects observed upon addition of 1 mm DTT. Yeast strains CKY1046–1049 were transformed with Ero1p(C150A/C295A) (pCS456) or the catalytically inactive Ero1p(C100A/C105A/C150A/C295A) (pCS483). Yeast strains were cultured in SMM at 30 °C, and growth was followed by light scattering (at 600 nm). At 2 h, DTT was added to half the cultures to a final concentration of 1 mm. B, cellular toxicity by overexpressed Ero1p(C150A/C295A) is alleviated by disruption of the Pdi1p a domain. CKY1046–1049 were transformed with a plasmid encoding Ero1p(C150A/C295A) (pCS452) or catalytically inactive Ero1p(C100A/C105A/C150A/C295A) (pCS504) under the control of the GAL promoter. After 2 and 12 h of growth in SMM with 3% galactose, 10−4 A600 unit of cells were spread onto SMM plates and incubated for 2 days at 30 °C. Cell viability was calculated by comparing colony forming units between cells expressing Ero1p(C150A/C295A) and cells expressing Ero1p(C100A/C105A/C150A/C295A) 12 h after induction.
We next examined the importance of the Pdi1p a and a′ domains in other tests of a functional Ero1p-Pdi1p oxidation pathway. Ectopic expression of hyperactive Ero1p(C150A/C295A) confers resistance to DTT and alleviates the growth defect observed when wild-type yeast are grown in the presence of DTT (Fig. 7A, top row). DTT resistance conferred by Ero1p(C150A/C295A) requires Ero1p catalytic activity as shown by the lack of resistance with ectopic expression of the Ero1p(C100A/C105A/C150A/C295A) catalytically inactive mutant (Fig. 7A). We have extended these observations by demonstrating that the resistance to DTT treatment conferred by hyperactive Ero1p(C150A/C295A) requires a functional Pdi1p a domain, but not a Pdi1p a′ domain; expression of Ero1p(C150A/C295A) confers resistance to DTT in a Pdi1p(CCSS) strain, but not in a Pdi1p(SSCC) strain (Fig. 7A).
Overexpressed Ero1p(C150A/C295A) is toxic to yeast cells (21). Here we show that the decrease in yeast cell viability observed after 12 h of Ero1p(C150A/C295A) overexpression can be alleviated by mutation of the Pdi1p a domain redox-active cysteines (Fig. 7B). This observation supports the idea that the a domain of Pdi1p is the primary domain that Ero1p(C150A/C295A) uses to hyperoxidize the ER. A slight loss in viability was observed in the Pdi1p(SSCC) background upon overexpression of Ero1p(C150A/C295A), but this toxicity did not appear to depend on Ero1p catalytic activity, because the same toxicity was observed upon overexpression of catalytically inactive Ero1p(C100A/C105A/C150A/C295A) (Fig. 7B). Toxicity from overexpression of catalytically inactive Ero1p may reflect an increased burden on the ER folding machinery due to increased protein expression levels. Despite the ability of Ero1p to form a mixed-disulfide intermediate with a Pdi1p(CSCC) mutant (Fig. 6A), expression of Ero1p(C150A/C295A) in a Pdi1p(CSCC) mutant strain did not confer resistance to DTT treatment, nor did overexpression of Ero1p(C150A/C295A) in a Pdi1p(CSCC) mutant strain result in a loss of cell viability (data not shown). Thus, mixed disulfide formation alone is not sufficient to confer these Ero1p-mediated phenotypes. Rather, a functional Pdi1p a domain active site is necessary for Ero1p-mediated hyperoxidation of the ER. These results suggest that the a domain is the major conduit of oxidizing equivalents from Ero1p to the ER.
Phenotypes for Pdi1p a Domain Mutant in S. cerevisiae Mimic Loss of Ero1p Function
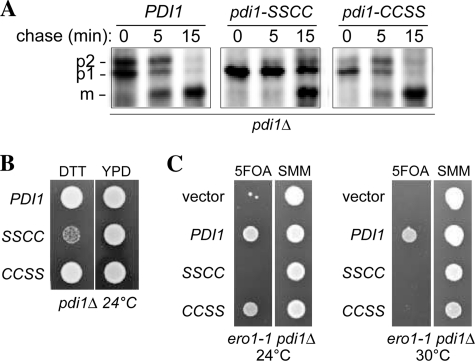
Decreased Ero1p function in vivo leads to a defect in the processing of CPY and to increased sensitivity of yeast cells to DTT (1). Consistent with preferential electron flow between Ero1p and the a domain of Pdi1p, similar phenotypes to those seen with an Ero1p loss-of-function mutant are observed in cells bearing a domain mutants of Pdi1p (Fig. 8, A and B). In addition, a Pdi1p mutant with a non-functional a domain redox center shows synthetic lethality with Ero1p (Fig. 8C), consistent with disruption of electron flow via a pathway containing Ero1p and the a domain of Pdi1p. Loss of Ero1p function and Pdi1p a′ domain function also showed some synthetic defects, lowering the temperature sensitivity of the ero1-1 strain to 30 °C, suggesting that there is some cooperation between Ero1p and the a′ domain as well.
FIGURE 8.
Pdi1p a domain mutant shows diminished oxidative folding capacity. A, processing of CPY in pdi1Δ cells containing PDI1 plasmids. Cells were pulse labeled for 7 min and chased at 30 °C, and CPY was immunoprecipitated and resolved by SDS-PAGE. ER (p1), Golgi (p2), and vacuolar (m) forms of CPY are indicated. B, yeast strains from panel A were grown for 12 h in SMM without leucine and spotted onto YPD plates with or without 10 mm DTT. Strains were grown for 2 days at 24 °C. C, the pdi1Δ strain CKY1058 covered by an URA3-marked PDI1 plasmid was transformed with LEU2-marked plasmids encoding Pdi1p (pCS463), Pdi1p(SSCC) (pCS465), Pdi1p(CCSS) (pCS464), or empty vector. These strains were spotted onto SMM plates or SMM plates containing 5-fluoroorotic acid (to select for yeast able to grow in the absence of the URA3 plasmid). Plates were incubated at 24 °C, 30 °C, or 37 °C for 3 days. All four strains were inviable at 37 °C (not shown), consistent with the previously characterized lethality of the ero1-1 mutant at 37 °C (1).
DISCUSSION
Oxidoreductases in the ER are responsible for net oxidation of protein cysteines with proper connectivity. To achieve correct cysteine pairing, disulfide isomerization or cycles of reduction and reoxidation may be necessary (31, 32). Furthermore, to maintain the quality of proteins oxidized and transported through the secretory pathway, the ER performs reduction and retrotranslocation of misfolded proteins (33). A major outstanding question related to biosynthetic disulfide bond formation in the ER is how the physiological functions of oxidation, rearrangement, and reduction of disulfide bonds are distributed among PDI family proteins. If the different oxidoreductases exist to carry out different functions, on what structural or biochemical basis do they partition into those that physiologically oxidize, reduce, or rearrange cysteines in their target proteins?
For comparison, elegant models have been developed to explain how a balance between oxidation and reduction is maintained to allow for native disulfide formation the E. coli periplasm. The proteins DsbB and DsbA cooperate to catalyze oxidation of substrate thiols, whereas DsbD and DsbC accomplish reduction and isomerization (reviewed in Ref. 34). These counteracting pathways are kept separate by kinetic barriers that prevent mis-reduction of functional oxidants (i.e. DsbA) (35) or mis-oxidation of reductants (i.e. DsbC) (36). For example, the kinetic barrier preventing oxidation of DsbC by DsbB arises from dimerization of DsbC (36, 37), which sterically limits access of DsbB to the DsbC active site (36, 38). Our results suggest that, analogous to DsbB, Ero1p selectivity toward ER oxidoreductases allows the ER to maintain both oxidation and reduction pathways. However, the mechanism by which selectivity is achieved in the ER is distinct from in the periplasm, because many ER oxidoreductases (e.g. Pdi1p, Mpd1p, and Eps1p) are predominantly monomeric (see however Ref. 39), and the redox-active domains are structurally very similar (40).
Here we show that the yeast ER oxidoreductases are oxidized at dramatically different rates by Ero1p. Within a single protein, different domains also showed differential reactivity. For example, the two trx domains of Eps1p were the most different from one another. The amino-terminal Eps1p domain remained in the reduced form even upon prolonged incubation with Ero1p, whereas the second Eps1p trx domain remained in oxidized form even upon incubation with high concentrations of the strong reducing agent DTT and thus could not be tested as an Ero1p substrate. The two redox-active domains of Pdi1p also showed clear differences in behavior; the a domain was oxidized by Ero1p more readily than the a′ domain.
In a previous study, the opposite preference was reported for the two redox-active domains of Pdi1p (8). The reason for the contradictory findings is unclear, but we note a few differences between our study and the previous one. First, in Kulp et al. (23), Pdi1p was reduced under conditions (10 mm DTT) known to reduce a Cys-X6-Cys disulfide in the a domain as well as the Cys-X-X-Cys motifs, whereas the Cys-X6-Cys disulfide remained oxidized in our experiments (Fig. 3C). Second, the previous study compared the a and a′ reactivities at much lower starting Pdi1p concentrations than the experiments reported here, which were done closer (but not yet achieving) the estimated near millimolar physiological Pdi1p concentration in the ER (11, 41). Activation of Ero1p may be less efficient at low Pdi1p concentrations. Another study that reported greater Ero1p oxidation of the Pdi1p a′ domain compared with the a domain also used high concentrations of DTT to prepare reduced Pdi1p and used a very low starting Pdi1p concentration (42); the vast excess of Ero1p disulfides compared with starting Pdi1p thiols meant that Ero1p turnover was not required to observe oxidation of the Pdi1p mutants. In any event, in vivo data reported here and elsewhere support the ranking we observe. Mutation of the Pdi1p a domain cysteines lowers resistance to DTT treatment to a greater extent than mutation of the Pdi1p a′ domain cysteines (30) (Fig. 7A), suggesting that Ero1p predominantly oxidizes the a domain of Pdi1p to counteract excess thiol species in the ER. In addition, a number of genetic and biochemical experiments (Figs. 6–8) demonstrate that oxidizing equivalents flow from Ero1p primarily through the a domain of Pdi1p.
Although we did not observe the same results on which the model was based, there is logical appeal to the idea that Ero1p might oxidize the domain of Pdi1p with the higher reduction potential (i.e. the a′ domain), as has been proposed (8). This model would involve rapid oxidation of a good oxidant by Ero1p and relatively slow oxidation of a good reductant. This system would be useful for catalysis of oxidative protein folding in the ER, because it would lead to formation of a pool of strong oxidant to promote disulfide bonding in substrate proteins, while at the same time a pool of good reductant would be available for rearrangement or reduction of erroneously paired disulfides. One potential problem with this model, however, is that oxidized Pdi1p a′ domain, precisely because it is a better oxidant, would be re-reduced more readily than oxidized a domain by ambient glutathione. Thus, preferential oxidation of the Pdi1p a′ domain by Ero1p would seemingly lead to futile cycles of oxidation and reduction. In contrast, an oxidized Pdi1p a domain would be less likely to be reduced by ambient glutathione and would be more specific for structural disulfides in substrate proteins, which presumably have lower reduction potentials than the overall ER environment.
In vitro kinetic assays with isolated components revealed a clear hierarchy in the interaction of ER oxidoreductases with Ero1p. However, each of the events we studied in isolation is potentially coupled in vivo to a number of other redox processes. For example, the redox states of PDI family oxidoreductases in cells will be determined from the relative rates of reduction by substrate and oxidation by Ero1p. Furthermore, ER oxidoreductases may react with other redox-active proteins in the compartment or with glutathione. Finally, complexes of ER oxidoreductases with other ER factors may modulate their redox properties. For example, the association of Mpd1p with the chaperone Cne1p (14) may affect the rate of Mpd1p oxidation by Ero1p. Along these lines, the extremely low reduction potential of the second trx domain of Eps1p is particularly striking and raises the question of whether this di-cysteine motif serves a structural role or whether it becomes redox-active in the ER as a result of quaternary structural interactions. The studies presented herein can serve as benchmarks for exploring the effect on disulfide bond formation pathways of higher order associations among redox-active ER proteins and their partners. It is important to note, however, that Ero1p substrate preferences determined by enzyme assays performed in vitro and reported here correlate well with preferred pathways for oxidation in the ER assessed in vivo using biochemical and genetic approaches.
Acknowledgment
We thank Konstantin Kogan for assistance in reduction potential measurements.
This work was supported in part by National Institutes of Health Grant RO1 GM046941 (to C. A. K.) from NIGMS. This work was also supported by a grant from the U.S.-Israel Binational Science Foundation (to D. F. and C. A. K.) and by the Kimmelman Center for Macromolecular Assemblies (to D. F.).
- ER
- endoplasmic reticulum
- trx
- thioredoxin fold
- PDI
- protein disulfide isomerase
- mal-PEG
- maleimide polyethylene glycol
- DTT
- dithiothreitol
- GSSG
- oxidized glutathione
- NEM
- N-ethylmaleimide
- SMM
- synthetic minimal medium
- Ni-NTA
- nickel-nitrilotriacetic acid
- CPY
- carboxypeptidase Y.
REFERENCES
- 1.Frand A. R., Kaiser C. A. (1998) Mol. Cell 1, 161–170 [DOI] [PubMed] [Google Scholar]
- 2.Pollard M. G., Travers K. J., Weissman J. S. (1998) Mol. Cell 1, 171–182 [DOI] [PubMed] [Google Scholar]
- 3.Frand A. R., Kaiser C. A. (1999) Mol. Cell 4, 469–477 [DOI] [PubMed] [Google Scholar]
- 4.Jessop C. E., Bulleid N. J. (2004) J. Biol. Chem. 279, 55341–55347 [DOI] [PubMed] [Google Scholar]
- 5.Molteni S. N., Fassio A., Ciriolo M. R., Filomeni G., Pasqualetto E., Fagioli C., Sitia R. (2004) J. Biol. Chem. 279, 32667–32673 [DOI] [PubMed] [Google Scholar]
- 6.Edman J. C., Ellis L., Blacher R. W., Roth R. A., Rutter W. J. (1985) Nature 317, 267–270 [DOI] [PubMed] [Google Scholar]
- 7.Darby N. J., Kemmink J., Creighton T. E. (1996) Biochemistry 35, 10517–10528 [DOI] [PubMed] [Google Scholar]
- 8.Kulp M. S., Frickel E. M., Ellgaard L., Weissman J. S. (2006) J. Biol. Chem. 281, 876–884 [DOI] [PubMed] [Google Scholar]
- 9.Lyles M. M., Gilbert H. F. (1994) J. Biol. Chem. 269, 30946–30952 [PubMed] [Google Scholar]
- 10.Nørgaard P., Westphal V., Tachibana C., Alsøe L., Holst B., Winther J. R. (2001) J. Cell Biol. 152, 553–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marquardt T., Hebert D. N., Helenius A. (1993) J. Biol. Chem. 268, 19618–19625 [PubMed] [Google Scholar]
- 12.Tachikawa H., Takeuchi Y., Funahashi W., Miura T., Gao X. D., Fujimoto D., Mizunaga T., Onodera K. (1995) FEBS Lett. 369, 212–216 [DOI] [PubMed] [Google Scholar]
- 13.Thorpe C., Kodali V. K. (2010) Antioxid. Redox. Signal., Epub ahead of print [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kimura T., Hosoda Y., Sato Y., Kitamura Y., Ikeda T., Horibe T., Kikuchi M. (2005) J. Biol. Chem. 280, 31438–31441 [DOI] [PubMed] [Google Scholar]
- 15.Westphal V., Darby N. J., Winther J. R. (1999) J. Mol. Biol. 286, 1229–1239 [DOI] [PubMed] [Google Scholar]
- 16.Gross E., Sevier C. S., Heldman N., Vitu E., Bentzur M., Kaiser C. A., Thorpe C., Fass D. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 299–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gross E., Kastner D. B., Kaiser C. A., Fass D. (2004) Cell 117, 601–610 [DOI] [PubMed] [Google Scholar]
- 18.Aricescu A. R., Lu W., Jones E. Y. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 1243–1250 [DOI] [PubMed] [Google Scholar]
- 19.Ellman G. L. (1959) Arch. Biochem. Biophys. 82, 70–77 [DOI] [PubMed] [Google Scholar]
- 20.Schafer F. Q., Buettner G. R. (2001) Free Radic. Biol. Med. 30, 1191–1212 [DOI] [PubMed] [Google Scholar]
- 21.Sevier C. S., Qu H., Heldman N., Gross E., Fass D., Kaiser C. A. (2007) Cell 129, 333–344 [DOI] [PubMed] [Google Scholar]
- 22.Sevier C. S., Kaiser C. A. (2006) Mol. Biol. Cell 17, 2256–2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilkinson B., Xiao R., Gilbert H. F. (2005) J. Biol. Chem. 280, 11483–11487 [DOI] [PubMed] [Google Scholar]
- 24.Tian G., Xiang S., Noiva R., Lennarz W. J., Schindelin H. (2006) Cell 124, 61–73 [DOI] [PubMed] [Google Scholar]
- 25.Klappa P., Ruddock L. W., Darby N. J., Freedman R. B. (1998) EMBO J. 17, 927–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krause G., Lundström J., Barea J. L., Pueyo de la Cuesta C., Holmgren A. (1991) J. Biol. Chem. 266, 9494–9500 [PubMed] [Google Scholar]
- 27.Mössner E., Huber-Wunderlich M., Glockshuber R. (1998) Protein Sci. 7, 1233–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chivers P. T., Laboissière M. C., Raines R. T. (1996) EMBO J. 15, 2659–2662 [PMC free article] [PubMed] [Google Scholar]
- 29.Appenzeller-Herzog C., Ellgaard L. (2008) Antioxid. Redox. Signal 10, 55–64 [DOI] [PubMed] [Google Scholar]
- 30.Holst B., Tachibana C., Winther J. R. (1997) J. Cell Biol. 138, 1229–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwaller M., Wilkinson B., Gilbert H. F. (2003) J. Biol. Chem. 278, 7154–7159 [DOI] [PubMed] [Google Scholar]
- 32.Laboissiere M. C., Sturley S. L., Raines R. T. (1995) J. Biol. Chem. 270, 28006–28009 [DOI] [PubMed] [Google Scholar]
- 33.Fagioli C., Mezghrani A., Sitia R. (2001) J. Biol. Chem. 276, 40962–40967 [DOI] [PubMed] [Google Scholar]
- 34.Kadokura H., Katzen F., Beckwith J. (2003) Ann. Rev. Biochem. 72, 111–135 [DOI] [PubMed] [Google Scholar]
- 35.Rozhkova A., Stirnimann C. U., Frei P., Grauschopf U., Brunisholz R., Grütter M. G., Capitani G., Glockshuber R. (2004) EMBO J. 23, 1709–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bader M. W., Hiniker A., Regeimbal J., Goldstone D., Haebel P. W., Riemer J., Metcalf P., Bardwell J. C. (2001) EMBO J. 20, 1555–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCarthy A. A., Haebel P. W., Törrönen A., Rybin V., Baker E. N., Metcalf P. (2000) Nat. Struct. Biol. 7, 196–199 [DOI] [PubMed] [Google Scholar]
- 38.Inaba K., Murakami S., Suzuki M., Nakagawa A., Yamashita E., Okada K., Ito K. (2006) Cell 127, 789–801 [DOI] [PubMed] [Google Scholar]
- 39.Tian G., Kober F. X., Lewandrowski U., Sickmann A., Lennarz W. J., Schindelin H. (2008) J. Biol. Chem. 283, 33630–33640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vitu E., Gross E., Greenblatt H. M., Sevier C. S., Kaiser C. A., Fass D. (2008) J. Mol. Biol. 384, 631–640 [DOI] [PubMed] [Google Scholar]
- 41.Lyles M. M., Gilbert H. F. (1991) Biochemistry 30, 613–619 [DOI] [PubMed] [Google Scholar]
- 42.Tsai B., Rapoport T. A. (2002) J. Cell Biol. 159, 207–216 [DOI] [PMC free article] [PubMed] [Google Scholar]