Abstract
Cytolethal distending toxins (CDTs) are tripartite protein exotoxins produced by a diverse group of pathogenic Gram-negative bacteria. Based on their ability to induce DNA damage, cell cycle arrest, and apoptosis of cultured cells, CDTs are proposed to enhance virulence by blocking cellular division and/or directly killing epithelial and immune cells. Despite the widespread distribution of CDTs among several important human pathogens, our understanding of how these toxins interact with host cells is limited. Here we demonstrate that CDTs from Haemophilus ducreyi, Aggregatibacter actinomycetemcomitans, Escherichia coli, and Campylobacter jejuni differ in their abilities to intoxicate host cells with defined defects in host factors previously implicated in CDT binding, including glycoproteins, and glycosphingolipids. The absence of cell surface sialic acid sensitized cells to intoxication by three of the four CDTs tested. Surprisingly, fucosylated N-linked glycans and glycolipids, previously implicated in CDT-host interactions, were not required for intoxication by any of the CDTs tested. Finally, altering host-cellular cholesterol, also previously implicated in CDT binding, affected intoxication by only a subset of CDTs tested. The findings presented here provide insight into the molecular and cellular basis of CDT-host interactions.
Keywords: Bacterial Toxins, Cholesterol, DNA Damage, Glycolipids, Glycoprotein, Aggregatibacter actinomycetemcomitans, Campylobacter jejuni, Cytolethal Distending Toxin, Escherichia coli, Haemophilus ducreyi
Introduction
Cytolethal distending toxins (CDTs)6 are members of a group of bacterial toxins and effectors called “cyclomodulins” that interfere with the eukaryotic cell cycle rather than inducing overt cytotoxicity (1, 2). Inhibiting cell cycle disrupts many of the normal functions of rapidly dividing eukaryotic cells, including lymphocytes and epithelial cells, which provide immunity and physical barriers to microbial pathogens (3–5). Thus, it is not surprising that cdt genes are found in a diverse group of Gram-negative pathogens that colonize different niches within the host. Although a growing body of evidence supports the importance of CDTs in bacterial virulence and host-pathogen interactions (6), the manner in which individual CDTs interact with and intoxicate host cells remains poorly understood.
CDTs are AB2 toxins, consisting of a hetero-trimeric complex of three proteins (CdtA, CdtB, and CdtC) at a 1:1:1 molar ratio (5, 7, 8). The current model is that CdtA and CdtC are the binding “B” moieties that collaborate to facilitate binding and entry of the catalytic “A” subunit, CdtB, into mammalian cells. CdtB shares a common tertiary structure with DNase I and phosphatidylinositol 3,4,5-triphosphate phosphatase enzymes and displays both activities in cell-free systems (9–13). It is not currently known which activity is of greater importance, and this may depend on the specific toxin and/or the host target cell type (12, 14). CdtB enzymatic activity induces cell cycle arrest predominantly at the G2/M transition, resulting in cellular distension and ultimately cell death (5, 15, 16).
Consistent with their proposed roles as binding subunits, CdtA and/or CdtC increase the ability of CdtB to associate with host cells and greatly enhance intoxication (7, 17–25). The identification of ricin-like lectin domains in CdtA and CdtC from structural and biochemical data first suggested that these subunits may interact with carbohydrates on the cell surface (13, 26, 27). Consistent with this hypothesis, CDT produced by Escherichia coli (Ec-CDT) was reported to require N-linked glycoproteins for binding and subsequent intoxication of HeLa cells (23). Moreover, Ec-CDT bound fucose in vitro, and fucose-specific lectins blocked Ec-CDT-mediated cell cycle arrest, presumably by preventing binding of toxin to its receptor. These findings suggested that fucose might serve as a binding determinant for Ec-CDT. Similarly, host glycans were reported to support Aggregatibacter actinomycetemcomitans (Aa-CDT) intoxication. Specifically, Aa-CDT bound three glycosphingolipids, GM1, GM2, and GM3, and intoxication of human monocytic U937 cells was blocked by preincubation of toxin with liposomes that contained GM3 (24). In addition, the CdtA subunit of Aa-CDT bound to the glycoprotein thyroglobulin (19). However, the functional significance of this binding is unknown, because mutants that failed to bind thyroglobulin retained near wild-type activity in intoxication assays.
In addition to a proposed role for host glycans in CDT binding, Aa-CdtC was recently demonstrated to possess a functional cholesterol recognition/interaction amino acid consensus (CRAC) motif important for binding of toxin to cholesterol-rich microdomains (28). However, it is not clear how CdtC binding to cholesterol relates to the previously proposed roles for glycolipids or glycoproteins. Further, it is not known whether CDTs from other bacteria possess a functional CRAC motif important for cellular binding and intoxication.
To determine if CDTs from various pathogens utilize similar host factors for intoxication, we set out to determine the role of several classes of cell surface biomolecules (i.e. glycoproteins, glycosphingolipids, cholesterol, and others) in intoxication by four CDTs. We chose to investigate two highly conserved CDTs, Aa-CDT and Haemophilus ducreyi CDT (Hd-CDT), which share 91.9% and 93.5% amino acid identity in their CdtA and CdtC subunits, respectively (5) (Fig. 1). In addition, we chose two CDTs, Ec-CDT and Campylobacter jejuni CDT (Cj-CDT), whose amino acid sequences are divergent (<30% amino acid identity between each other or with Aa/Hd-CDT). These four toxins were selected based on their relative sequence divergence as well as the host niche occupied by the respective CDT-producing pathogen. Specifically, CDTs from two enteric pathogens (E. coli and C. jejuni), an oral pathogen (A. actinomycetemcomitans), and a pathogen responsible for a sexually transmitted disease (H. ducreyi), were investigated. Our results reveal differences in the ability of each of these four CDTs to intoxicate host cells depending on the presence or absence of specific cell surface biomolecules. Furthermore, glycans previously implicated in CDT-host interactions were not required for intoxication by any of the toxins tested. Therefore, we propose that individual CDTs utilize distinct host factors to efficiently intoxicate target cells.
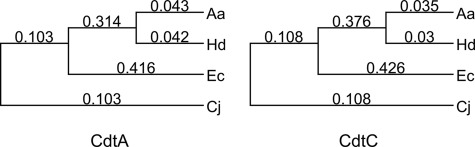
FIGURE 1.
Dendogram of CdtA and CdtC proteins. CdtA and -C protein sequences were aligned with ClustalW using BLOSUM 30 similarity matrix, and a midpoint rooted dendogram was constructed using MacVector version 7.2. Numbers indicate relative evolutionary distance.
EXPERIMENTAL PROCEDURES
Cell Culture
Pro−CHO (ProCHO), Pro−Lec8.D3 (Lec8), Pro−Lec23.11C (Lec23), Pro−Lec3.2.8 3B (Lec3.2.8), LdlD.Lec1, and Pro−Lec1.3C (Lec1) cells were provided by Pamela Stanley (Albert Einstein College of Medicine) (29–32). Pro−Lec13 (Lec13), Pro−Lec2 (Lec2), and CHO-K1 mutants defective in proteoglycan biosynthesis, PgsA745 and PgsD677 (51, 52), were provided by Jeff Esko (University of California at San Diego). Balb/3T3 clone A31 and CHO-K1 cells were a gift from John Young (Salk Institute). Y-1 cells were provided by Edward McCabe (UCLA). OT-1 cells were provided by Carrie Miceli (UCLA). HeLa, NIH/3T3, IC-21, and Raw 264.7 cells were obtained from ATCC. Unmutagenized CHO-K1 and ProCHO, glycosylation mutant CHO, and Y-1 cells were cultivated in F-12 media (Invitrogen) supplemented with 10% fetal bovine serum (FBS, Atlanta Biologicals), 100 units/ml penicillin, 100 μg/ml streptomycin, and 5 mm l-glutamine (PSG, Invitrogen). Lec13 and ldlD.Lec1 cells were cultured for 2 days prior to intoxication in F-12 containing 10% FBS dialyzed against phosphate-buffered saline (PBS). HeLa, NIH/3T3, Balb/3T3, and Raw 264.7 cells were cultured in Dulbecco's minimum essential medium (Invitrogen) supplemented with 10% FBS and PSG. IC-21 and OT-1 cells were cultivated in RPMI and Iscove's modified Dulbecco's medium, respectively, supplemented with 10% FBS and PSG. All cells were cultured at 37 °C in a humid atmosphere containing 5% CO2.
CDT Cloning, Expression, and Purification
Cloning and expression of CDTs was based on the method previously described (13). Cultures of A. actinomycetemcomitans (Y4) and E. coli (S5) were obtained from ATCC and C. jejuni (81–176) from Patricia Guerry (Naval Medical Research Center, Silver Spring, MD). Genomic DNA was purified from mid-log-phase cultures using the Wizard DNA Purification kit (Promega, Madison, WI). Genomic DNA of H. ducreyi (35000HP) was obtained from ATCC. cdtA, cdtB, and cdtC genes (Aa GI: 3786340, Hd GI: 2102681, Ec GI: 2218088, and Cj GI: 15791399) were PCR-amplified using primers corresponding to the 5′- and 3′-ends of each gene (Table 1). These primers were engineered such that 5′ NdeI and 3′ BamHI restriction sites (underlined in Table 1) were incorporated into the amplicon. Each PCR product was purified using the QIAquick PCR purification kit (Qiagen). The purified amplicons and pET15b vector were digested with NdeI and BamHI (New England Biolabs) to generate directional annealing sites. Digested vector and amplicons were ligated and electroporated into E. coli DH5α. The integrity of each gene was confirmed by DNA sequencing. E. coli BL21(DE3) strains transformed with cdt expression plasmids were grown in Luria-Bertani broth containing 100 μg/ml ampicillin (LB+amp) at 37 °C with continuous aeration. Starter cultures were diluted 1:400 into fresh LB+amp and grown until the optical density at 600 nm reached 0.4–0.6 at which point expression of Cdt was induced by addition of 0.25 mm isopropyl 1-thio-β-d-galactopyranoside (Fisher). Cultures were grown for an additional 3 h and then harvested by centrifugation at 5,000 × g for 10 min at 4 °C. The cell pellets were resuspended in 20 ml of PBS (8 g of NaCl, 0.2 g of KCl, 0.2 g of KH2PO4, and 1.15 g of Na2HPO4·7H2O in 1 liter) containing 8 m urea and disrupted by sonication (six 30-s cycles at 23 kHz and 30 watts) using Model 100 Sonic Dismembrator (Fisher). Cell lysates were then clarified by centrifugation at 16,000 × g for 30 min at 4 °C. Supernatants containing proteins were incubated with 3 ml of Talon Metal Affinity Resin (Clontech) overnight with gentle rotation at 4 °C. The resin was washed with buffer A (8 m urea, 20 mm HEPES, 200 mm NaCl, pH 7.5) and eluted with buffer A plus 100 mm EDTA. Protein concentrations were quantified using the Bradford Protein Assay and purity was estimated by SDS-PAGE. Proteins were stored in elution buffer at −20 °C. At time of use, proteins were diluted to 100 μg/ml with buffer A and dialyzed against buffer B (20 mm HEPES, 200 mm NaCl, 5% glycerol, 2.5 mm dithiothreitol, pH 7.5) using 6–8 kDa MWCO membranes (Spectrum Laboratories). Two final buffer changes were conducted in 3-liter volumes of PBS containing 5% glycerol and 2.5 mm dithiothreitol.
TABLE 1.
Primer sequences for PCR amplification of CDT
| Primer | Direction | Primer sequence (5′ → 3′) |
|---|---|---|
| Aa-CdtA | Forward | GGAGTTCCATATGAGTGACTATTCTCAGCCTG |
| Aa-CdtA | Reverse | CGGGATCCTTAATTAACCGCTGTTGC |
| Aa-CdtB | Forward | GGAGTTCCATATGAACTTGAGTGATTTCAAAGTAGC |
| Aa-CdtB | Reverse | CGGGATCCTTAGCGATCATGAACAAAACTAACAGG |
| Aa-CdtC | Forward | GGAGTTCCATATGGAATCAAATCCTGATCCG |
| Aa-CdtC | Reverse | CGGGATCCTTAGCTACCCTGATTTCTCC |
| Hd-CdtA | Forward | GGAGTTCCATATGAATGACTATTCTCAACCTGAATCTC |
| Hd-CdtA | Reverse | CGGGATCCTTAATTAACCGCTGTTGC |
| Hd-CdtB | Forward | GGAGTTCCATATGAACTTGAGTGACTTCAAAGTAG |
| Hd-CdtB | Reverse | CGGGATCCTTAGCGATCACGAACAAAACTAACAG |
| Hd-CdtC | Forward | GGAGTTCCATATGGAATCAAATCCTGATCCGAC |
| Hd-CdtC | Reverse | CGGGATCCTTAGCTACCCTGATTTCTTC |
| Ec-CdtA | Forward | GGAGTTCCATATGCATCTTGACCCCAAAG |
| Ec-CdtA | Reverse | CGGGATCCTCATTGTTCGCCTCCTG |
| Ec-CdtB | Forward | GGAGTTCCATATGGATTTAACTGATTTTCGCGTTG |
| Ec-CdtB | Reverse | CGGGATCCTTATCGTCTGGAAACG |
| Ec-CdtC | Forward | GGAGTTCCATATGGTCAATAATCAGATAGATGAGTTAG |
| Ec-CdtC | Reverse | CGGGATCCTTAAATAACAGGAGATTCTGTATTTAATG |
| Cj-CdtA | Forward | GGAGTTCCATATGTGTTCTTCTAAATTTGAAAATGTAAATCC |
| Cj-CdtA | Reverse | CGGGATCCTCATCGTACCTCTCC |
| Cj-CdtB | Forward | GGAGTTCCATATGAATTTAGAAAATTTTAATGTTGGCACTTGG |
| Cj-CdtB | Reverse | CGGGATCCCTAAAATTTTCTAAAATTTACTGGAAAATGATCTGAAAC |
| Cj-CdtC | Forward | GGAGTTCCATATGACTCCTACTGGAGATTTGAAAGATTTTACC |
| Cj-CdtC | Reverse | CGGGATCCTTATTCTAAAGGGGTAGCAGC |
Intoxication of Mammalian Cells with CDTs
Mammalian cells were trypsinized, counted, and allowed to adhere overnight in 6- or 384-well plates. The following day, medium was removed and toxin-containing medium was added for 10 min or 24 h as indicated in the figure legends. The concentration of toxin was selected to illustrate the presence or absence of a genetic or pharmacological effect on intoxication and differed depending on cell line and/or conditions used (Aa-CDT, 2–200 nm; Hd-CDT, 0.1–75 nm; Ec-CDT, 5–900 nm; and Cj-CDT, 50–1500 nm). Twenty-four hours after toxin addition, cells were analyzed for phosphorylation of histone 2AX (H2AX) or cell cycle arrest as described below. All results presented are representative of three or more independent experiments.
Histone H2AX Assay
Cells were intoxicated for 24 h in clear bottom 384-well plates then fixed with 2% formaldehyde, quenched with 100 mm glycine, and permeabilized with ice-cold methanol. The cells were subsequently blocked with 3% bovine serum albumin/0.3% Triton X-100 and incubated with rabbit anti-phospho-H2AX antibody (Cell Signal Technologies) overnight at 4 °C. After washing, cells were incubated with Alexa Fluor 488-labeled goat anti-rabbit antibody (Invitrogen) for 1 h at room temperature, washed, and counterstained with 1 μg/ml Hoechst 33342 (Invitrogen). Cytometric acquisition was performed on four 20× scan fields using an iCys laser scanning cytometer (CompuCyte) equipped with argon and violet diode lasers. Cytometric data analysis was conducted with iCys version 3.4 software (CompuCyte).
Cell Cycle Analysis
CHO glycosylation mutants and unmutagenized parental strains were intoxicated in 6-well plates for either 10 min or 24 h, then washed with Dulbecco's PBS (DPBS, Cellgro), detached with 0.05% trypsin/EDTA (Invitrogen), washed again with DPBS, and permeabilized with 60% ethanol for 30 min on ice. After washing again, cells were stained with a 50 μg/ml propidium iodide (PI) solution containing 1 mg/ml sodium citrate, 0.3% Nonidet P-40, and 20 μg/ml RNase-A. The fluorescence was quantified for 104 cells using a FACSCalibur flow cytometer (BD Biosciences) with CellQuest acquisition software (BD Biosciences). Flow cytometry data were subsequently analyzed using FloJo analysis software (Tree Star).
Fluorescent Lectin Binding
Cell surface glycan presentation was measured using the FITC-conjugated lectins (EY Laboratories) concanavalin A, Griffonia simplicifolia lectin, Lens culinaris agglutinin, Maclura pomifera agglutinin, Phaseolus vulgaris agglutinin, Pisum sativum agglutinin, Ulex europus agglutinin (UEA), and wheat germ agglutinin. Cells were detached as described above, mixed with an equal volume of complete media containing FBS, washed with DPBS, and resuspended in DPBS containing the manufacturer-recommended concentration of FITC-labeled lectin at room temperature for 15 min. Cells were washed with DPBS three times and resuspended in DPBS containing 1% formaldehyde (EMD Chemicals). Fluorescence was quantified as described above.
N-Linked Glycosylation Inhibitor Treatment and Cholesterol Loading
To inhibit N-linked glycosylation, CHO-K1 cells were cultured in the presence of 10 μg/ml tunicamycin (Sigma-Aldrich) for 2 days prior to intoxication. For cholesterol loading of CHO cells, 15 mg of cholesterol (Sigma-Aldrich) was solubilized by dissolving 15 mg in 0.4 ml of methanol:chloroform (2:1), then adding dropwise into 20 ml of PBS containing 0.37 g of methyl-β-cyclodextrin (Sigma-Aldrich) and stirred for 2 h at 80 °C. The solution was dried in a rotary-evaporator and resuspended in 6 ml of F-12 media (2.5 mg/ml cholesterol final). Cells were washed in serum-free F-12 media, incubated in solubilized cholesterol for 1 h, and washed in complete F-12 media three times. Following tunicamycin treatment, cholesterol loading, or both, the cells were intoxicated for 10 min, followed by a 24-h incubation, and analyzed for cell cycle arrest as described above.
Cholesterol was quantified by the use of Amplex Red reagent (Invitrogen). Cells were washed three times in PBS and detached using 1 mm EDTA in PBS. After centrifugation, cells were lysed by resuspension in 100 mm KH2PO4, 50 mm NaCl, 5 mm cholic acid, and 0.1% Triton X-100 and subjected to three freeze-thaw cycles. Lysates were added to an equal volume of the working Amplex Red assay buffer (prepared according to manufacturer's instructions). Fluorescence was measured with a Flexstation II microplate reader (Molecular Devices) using excitation at 560 nm and detection at 590 nm. Cholesterol levels were determined by comparing fluorescence values to a standard curve and normalizing to protein content as determined by Bio-Rad Protein Assay.
Fucosyltransferase Studies
Fucosyltransferase 1 (Fut1) cDNA was PCR-amplified from pCDM7 (33) (provided by John Lowe, Case Western Reserve University) using the forward primer, CCCCTCGAGATGTGGCTCCGGAGCCAT, and the reverse primer, CCCGAATTCTCAAGGCTTAGCCAATGTCC. The amplicon was digested with XhoI and EcoRI (engineered restriction sites are underlined in primer sequences), gel-purified, and ligated with T4 DNA ligase (New England Biolabs) into the retroviral vector pMSCVpuro (Clontech). A similar subcloning strategy was undertaken for Fut2–9. Plasmid DNA was purified and transfected into human 293 cells along with murine leukemia virus gag/pol and vesicular stomatitis virus G-spike protein expression plasmids as previously described (34). Forty-eight hours later, resulting retroviral particles were harvested, filter-sterilized, diluted 1:1 in fresh media with 8 μg/ml Polybrene (Sigma-Aldrich), and used to transduce cells in a 6-well plate. Cells were incubated with viral particles overnight. After 48 h, transduced cells were selected in medium containing 5 μg/ml puromycin.
Glycosphingolipid Studies
To deplete glycosylated sphingolipids, CHO-K1 cells were cultured in the presence of 5 μm 1-phenyl-2-palmitoylamino-3-morpholino-1-propanol (PPMP, Sigma-Aldrich) for 10 days then intoxicated for 24 h. Cell lines deficient in serine-palmitoyl transferase (LY-B) and the complimented counterpart LY-B/cLCB1 were obtained from the RIKEN cell bank. These cells were cultured in F-12 media containing PSG, 10 μm sodium oleate, 1× nutridoma (Roche Applied Science), and 0.1% fatty acid-free bovine serum albumin (Sigma-Aldrich) for 2 days prior to intoxication.
RESULTS
CDTs Display Differential Target Cell Preferences
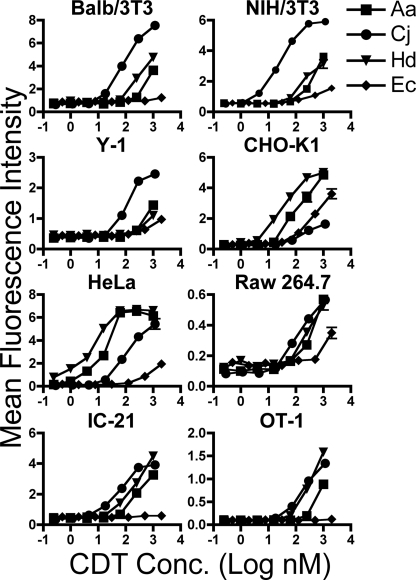
CDTs are able to intoxicate a wide variety of cell types derived from multiple species and progenitor tissues, indicating that each individual CDT utilizes receptor(s) and entry processes that are conserved among different hosts and target cells (5). To test whether host factors required for intoxication are shared between different CDTs, we set out to quantify the level of sensitivity to Ec-, Hd-, Aa-, and Cj-CDTs in a series of diverse cell lines. We intoxicated Chinese hamster ovary (CHO-K1) and HeLa cells, which represent epithelial cells commonly used for CDT studies, as well as T-cells (OT-1) and macrophage-like cells (RAW 264.7 and IC-21), which are proposed to be important targets for CDTs in vivo (35). In addition, we intoxicated 3T3 fibroblasts and Y-1 adrenal cells, which were reported to be resistant to Ec-, Hd-, and Cj-CDTs (16, 36–38).
Phosphorylation of the histone protein H2AX is a characteristic of double-stranded DNA breaks and has been used to monitor CDT intoxication (39). Therefore, CDT intoxication was assayed by immunofluorescence staining for phosphorylated H2AX (phospho-H2AX) followed by laser scanning cytometry. As predicted, 3T3 fibroblasts derived from NIH or BALB/c mice, as well as Y-1 cells, were highly resistant to Ec- and Hd-CDT intoxication (Fig. 2). This result was not due to an altered DNA damage response, because all cell lines tested induced phospho-H2AX in response to UV light-mediated DNA damage (data not shown). Consistent with its high sequence similarity with Hd-CDT, Aa-CDT was also ineffective at inducing H2AX phosphorylation in 3T3 or Y-1 cells (Figs. 1 and 2). Surprisingly, Cj-CDT efficiently intoxicated all three of these cell lines (Fig. 2), demonstrating for the first time that CDTs derived from different bacteria display variable target cell tropism. These data combined with the fact that the CdtB subunits from these four CDTs display similar enzymatic activities in vitro7 support a model whereby Cj-CDT utilizes a receptor and/or entry pathway that is distinct from the three other toxins (Aa-, Hd-, and Ec-CDTs). Further, Ec-CDT efficiently intoxicated CHO-K1 cells but displayed low to undetectable activity on all other cell types. This result was opposite the pattern seen with Cj-CDT, which efficiently intoxicated all cell types except CHO-K1 (Fig. 2). Although Aa- and Hd-CDTs were poorly active against 3T3 and Y-1 cells, these two toxins were more active than Ec- or Cj-CDTs on CHO-K1 and HeLa cells. Taken together, these data suggest that Aa- and Hd-CDT utilize a common set of host factors for intoxication that are distinct from those utilized by either Ec-CDT or Cj-CDT. Further, Ec-CDT appears to interact with host cells in manner that is distinct from Cj-, Aa-, and Hd-CDTs.
FIGURE 2.
Differential sensitivity of cell lines to divergent CDTs. Cells from indicated lines were seeded on 384-well plates, intoxicated for 24 h, fixed, permeabilized, and probed with anti-phospho H2AX antibodies followed by Alexa Fluor 488-labeled goat anti-rabbit antibodies. Nuclei were identified by staining with Hoechst. The relative level of activated H2AX per cell nucleus was determined by measuring Alexa Fluor 488 and Hoechst fluorescence intensity by laser scanning cytometry. Results are plotted as mean Alexa Fluor 488 fluorescence intensity per nucleus from triplicate samples ± S.E. Results are representative of three independent experiments.
Differential Roles for Cholesterol and Glycoproteins in CDT Intoxication
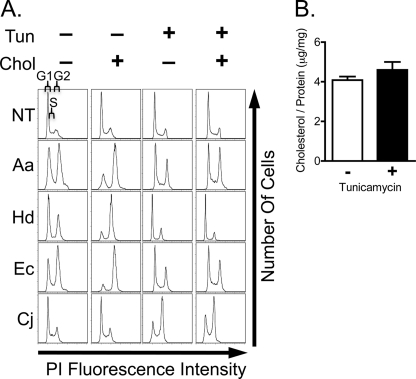
Cholesterol was recently reported as a direct binding determinant for Aa-CDT (28), and cholesterol-rich microdomains (e.g.“rafts”) were previously demonstrated to be required for binding and intoxication by Aa- and Hd-CDT (40–42). Thus, we sought to determine if cholesterol is important for intoxication of CHO-K1 cells by all CDTs. CDT-mediated cell cycle arrest in G2/M was measured by staining with the DNA-binding dye PI followed by flow cytometry to determine DNA content. Intoxication by Aa-, Hd-, and Ec-CDT was enhanced by cholesterol loading (Fig. 3A), indicating that these three toxins may bind directly to cholesterol and/or utilize lipid rafts for entry. Again, Cj-CDT behaved differently as cell cycle arrest with this toxin was not enhanced by cholesterol loading (Fig. 3A).
FIGURE 3.
CDT intoxication of tunicamycin-treated and cholesterol-loaded CHO-K1 cells. A, CHO-K1 cells were treated with 0.5 μg/ml tunicamycin for 2 days and/or incubated with 2 mg/ml methyl-β-cyclodextrin conjugated cholesterol for 30 min prior to intoxication. Cells were incubated with toxin for 10 min, then washed and incubated for a further 24 h. DNA content was determined by PI staining and flow cytometry. Histograms indicate the number of cells (y axis) at a given PI fluorescence intensity (x axis), with the left hand peak representing cells in G0/G1 phase of cell cycle, the peak on the right representing cells in G2/M, and the area between peaks representing cells in S phase as indicated in the top left histogram. B, total cellular cholesterol was quantified and normalized to total protein in CHO-K1 cells treated with tunicamycin. Average values calculated from triplicate samples ± S.E. are shown. A and B, results are representative of at least three independent experiments.
We next examined the role of host glycoproteins, which were previously implicated as important determinants for intoxication by Ec-CDT and Aa-CDT (19, 23). The N-linked glycans represent a common form of glycoprotein linkage and consist of carbohydrate covalently linked to an asparagine side chain. McSweeney and Dreyfus reported that treatment with tunicamycin resulted in diminished intoxication by Ec-CDT, leading them to conclude that N-linked glycans are required for this toxin (23). Tunicamycin blocks the addition of the dolichol pyrophosphate precursor (Glc3Man9GlcNAc2) to asparagine side chains in the endoplasmic reticulum, thereby inhibiting all N-linked glycosylation (43). Although tunicamycin is widely used to block the synthesis of N-linked glycans, there are several caveats to its use. Specifically, tunicamycin has been shown to block cholesterol biosynthesis by inhibiting the rate-limiting enzyme hydroxymethylglutaryl-CoA reductase. Furthermore, the absence of glycosylation induced by tunicamycin can result in misfolding of host-proteins and decreased presentation of these proteins on the cell surface (43, 44). To define the effects of tunicamycin on intoxication, we asked whether this drug inhibits intoxication by all four CDTs, and explored the mechanism by which this inhibition occurs.
CHO-K1 cells were pretreated with tunicamycin, then intoxicated with each CDT for 24 h. Inhibition of N-linked glycosylation by tunicamycin was confirmed using the FITC-labeled plant lectin, P. ativum agglutinin (data not shown). As expected, cells treated with tunicamycin were less sensitive to intoxication by Ec-CDT (Fig. 3A). In addition, tunicamycin-treated cells were less sensitive to Aa- and Hd-CDTs (Fig. 3A). Surprisingly, tunicamycin treatment enhanced intoxication by Cj-CDT (Fig. 3A), once more implicating this toxin as having distinct host cell requirements for intoxication.
Interestingly, tunicamycin inhibited host cell sensitivity to the same three CDTs that utilize cholesterol for intoxication (Fig. 3A). Therefore, we sought to determine if tunicamycin decreases sensitivity to Aa-, Hd-, and Ec-CDT through cholesterol depletion. Direct measurement of cellular cholesterol shows that levels of this lipid were unchanged by tunicamycin under the conditions tested here (Fig. 3B). Furthermore, cholesterol supplementation did not reverse resistance to intoxication observed in tunicamycin-treated cells (Fig. 3A). Together, these results suggest that the primary effect of tunicamycin in our experimental system is on inhibition of glycosylation. However, it is not clear from these data whether the block to CDT intoxication results from loss of specific glycans that are required for toxin interactions, or whether decreased glycosylation in the presence of tunicamycin leads to misfolding and/or destabilization of a protein that is required by CDTs.
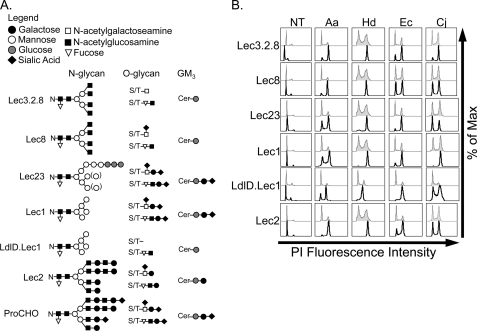
To test a direct role for specific glycans, we employed a panel of highly characterized glycan-deficient mutant CHO cells (45). Mutant cell lines with well defined deficiencies in N-, O-, or lipid-linked glycans (Fig. 4A) were challenged with each of the four CDTs for 24 h, stained with PI, and DNA content was measured by flow cytometry. As a control, the predicted glycan structures for each mutant (Fig. 4A) were confirmed using the appropriate glycan-binding FITC-labeled plant lectins (data not shown). Surprisingly, although several of the mutant cell lines have drastically truncated glycans (45), none displayed resistance to any CDT tested (Fig. 4B). Further, the mutant cell lines with the greatest defects in N-linked glycans (Lec1) or N- and O-linked glycans (ldlD.Lec1) demonstrated increased sensitivity to all four CDTs (Fig. 4B). Increased sensitivity to Aa-, Hd-, and Ec-CDT in cells lacking complex or hybrid N-glycans (e.g. Lec1 and Lec23) was confirmed by treating ProCHO cells with the α-mannosidase I inhibitor, kifunensine, which blocks trimming of mannose on N-linked glycans and therefore results in high mannose-containing N-linked glycans (data not shown). Taken together, these data indicate that the four CDTs tested do not require mature host cell N- or O-glycan structures for intoxication.
FIGURE 4.
CDT intoxication of CHO glycosylation mutants. A, predicted N- and O-linked glycan and glycolipid structures (adapted from Ref. 45). GM3 is the predominant glycolipid in CHO cells; therefore, the mutant structures are based on GM3. B, cells were intoxicated for 24 h, stained with PI, and subjected to flow cytometry. Unshaded histograms represent intoxicated loss of function mutants, and gray shaded histograms represent the intoxicated parental ProCHO controls. Results are representative of at least three independent experiments.
Of note, the Lec cell intoxication data support the previous claim that absence of sialic acid results in more efficient intoxication by CDT (23). It was previously reported that enzymatic removal of terminal sialic acid with neuraminidase resulted in increased sensitivity to Ec-CDT (23). However, the extent of sialic acid removal in this study was not quantified, and the acidic pH required for neuraminidase activity may lead to secondary effects on cell surface proteins and/or cell health. Here we used a genetic approach to determine if reduced sialylation results in increased sensitivity to Ec-CDT, and extended the study to test the remaining three CDTs. Lec2 cells lack the sialic acid transporter and are therefore unable to sialylate any of their glycans. Consistent with sialic acid blocking toxin interactions of some, but not all CDTs, Lec2 cells displayed increased sensitivity to Aa-, Hd-, and Ec-CDT. FITC-labeled wheat germ agglutinin was used to confirm the cell surface sialic acid deficiency in these cell lines (data not shown). An inhibitory role of sialic acid was further supported by the increased sensitivity of LdlD.Lec1 cells, which also lack terminal sialic acid due to the absence of sialotransferase substrates.
In addition to addressing a potential role for glycans, analysis of the mutant CHO intoxication data provided further evidence that Aa-, Hd-, Ec-, and Cj-CDTs utilize divergent host determinants. When compared with the parental ProCHO cells, all of the Lec mutants tested were hypersensitive to Aa- and Hd-CDT (Fig. 4B), supporting the hypothesis that these two CDTs have similar requirements for intoxication. In contrast, the intoxication profile for Cj-CDT on this panel of cells was markedly different, with Lec8, Lec23, and Lec2 cells being equally sensitive to this toxin when compared with parental ProCHO cells. These results further support the notion that Cj-CDT utilizes unique host determinants. Further, because Lec8, Lec23, and Lec2 cells were equally sensitive to Cj-CDT when compared with parental ProCHO cells, hypersensitivity to Aa- and Hd-CDT in these mutant cell lines is toxin-specific and does not derive from nonspecific effects in these mutant cells (e.g. altered growth rates, DNA damage response, etc.). The Ec-CDT intoxication profile was unique, being similar to Cj-CDT in that Lec8 showed no change in sensitivity, and similar to Aa- and Hd-CDT in that Lec3.2.8, Lec23, and Lec2 cells showed increased sensitivity.
Fucosylated Glycans Are Not Required for Intoxication by CDTs
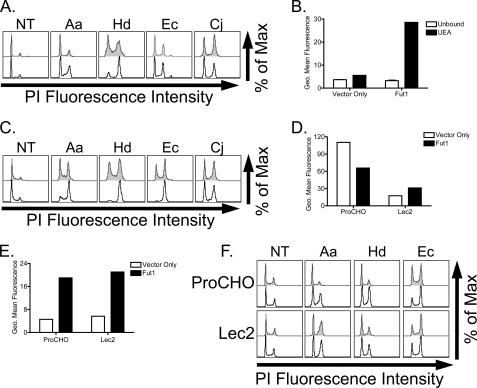
It was previously suggested that fucosylated glycans contribute to CDT binding based on findings that Ec-CDT bound directly to agarose-coupled fucose, and fucose-specific lectins blocked CdtA/C binding to HeLa cells, resulting in decreased CDT activity (23). Although data presented above indicate that complex N-linked glycans are not required for efficient CDT-host interactions (Fig. 4), they do not rule out a potential role for fucose-modified glycans in conferring sensitivity to this family of toxins. Specifically, CHO cells contain N-linked glycans with fucose attached to the first (reducing) N-acetylglucosamine in the core glycan structure (Fig. 4A), and this modification is maintained at reduced levels in Lec1 (46), but is absent following tunicamycin treatment. To test a role for fucosylated glycans in sensitivity to CDTs, we employed the mutant CHO cell line Lec13, which is deficient in the biosynthesis of fucose and therefore has reduced fucosylation of all glycans. Contrary to what was expected, Lec13 cells showed no alteration in sensitivity to any of the four CDTs tested (Fig. 5A). Therefore, fucosylated glycans are not required for CDT intoxication.
FIGURE 5.
Role of fucose in CDT intoxication. A, CDT intoxication of the fucose biosynthetic mutant (Lec13) cells. Cells were grown in media containing dialyzed serum for 2 days, intoxicated with CDT for 24 h, stained with PI, and subjected to flow cytometry. Unshaded histograms represent intoxicated Lec13 cells, and gray shaded histograms are intoxicated parental ProCHO cells. B, UEA binding of Fut1-expressing CHO-K1 cells. Cells transduced with Fut1-encoding or empty vectors were detached with EDTA, washed, and bound with FITC-conjugated UEA lectin for 15 min at room temperature. After three successive washes, the cells were resuspended in PBS plus 1% formaldehyde and subjected to flow cytometry. Data represent geometric mean fluorescence of 104 cells. C, CHO-K1 cells were transduced with a retroviral vector encoding Fut1 (unshaded histograms) or empty vector (gray histrograms), intoxicated for 24 h, stained with PI, and subjected to flow cytometry. D and E, ProCHO and Lec2 cells transduced with Fut1-encoding or empty vectors were bound with FITC-conjugated wheat germ agglutinin (D) or FITC-conjugated UEA (E), washed, and subjected to flow cytometry, as in B. F, CDT intoxication of parental (ProCHO) or sialic acid transporter mutant (Lec2) cells transduced with Fut1 encoding (unshaded histograms) or empty (gray histograms) retroviral vectors. Transduced cells were intoxicated for 24 h, stained with PI, and subjected to flow cytometry. Results are representative of at least three independent experiments.
The sensitivity of Lec13 cells to CDTs did not, however, address the question of whether terminally fucosylated glycans may serve to enhance intoxication. CHO-K1 cells express only two fucosyltransferase (Fut) genes, Fut8, which adds α1–6 fucose to the core of the N-linked glycan, and an O-fucosyltransferase that adds fucose directly to Ser/Thr residues on proteins. Many other cell types modify glycan structures by adding fucose to the terminal (non-reducing) end of N-, O-, and lipid-linked glycans through the activities of eight separate fucosyltransferases (FUT1–7 and FUT9) (47). To test whether fucosylated glycans can enhance CDT intoxication, cDNAs encoding Fut1–7 and Fut9 were subcloned into retroviral vectors that were used to transduce CHO-K1 cells either individually or pairwise. Expression of all Futs, except Fut2, gave rise to the expected fucose modification profiles as judged by binding to fluorescently labeled plant lectins and antibodies (Fig. 5B and data not shown) (48). All cell lines were incubated with each CDT and evaluated for G2/M arrest. Interestingly, overexpression of Fut1, but no other Fut, resulted in an increase in sensitivity to Aa-, Hd-, and Ec-CDTs (Fig. 5C). Furthermore, co-expression of any of the Fut genes (Fut3–7 and -9) in conjunction with Fut1 did not further sensitize CHO-K1 cells to CDT intoxication (data not shown).
The finding that Fut1 overexpression led to enhanced sensitivity to a subset of CDTs is consistent with a role for α1,2-fucosylated glycans in host-toxin interactions. However, an alternative explanation is that expression of Fut1 alters host glycan structures in ways other than addition of fucose. Indeed, it was previously demonstrated that expression of Fut1 reduces sialic acid on N-linked glycans (49). Consistent with this, expression of Fut1 in parental ProCHO cells resulted in decreased presentation of sialic acid (Fig. 5D). Because a reduction in sialic acid levels led to increased sensitivity to the same group of CDTs affected by Fut1 expression (compare Lec2 cells, Fig. 4), we questioned whether Fut1-dependent hypersensitivity to CDT derived from the addition of fucose or the absence of sialic acid. To test this, we transduced sialic acid transporter-deficient Lec2 cells with a retroviral vector encoding Fut1 and intoxicated with each CDT. Activity of Fut1 in transduced Lec2 cells and the transduced parental ProCHO cell line was confirmed using a fluorescently labeled fucose-specific plant lectin (Fig. 5E). Whereas ProCHO cells expressing Fut1 were hypersensitive to CDTs, Lec2 cells expressing Fut1 displayed no increase in sensitivity to intoxication (Fig. 5F). Therefore, increased sensitivity associated with Fut1 expression derives from loss of sialic acid and not from gain of fucosylation. Taken together, these data support a model where, contrary to previous reports, fucosylated glycans are not required for cellular intoxication by Ec-CDT or any of the other CDTs tested.
Glycolipid Deficiency Does Not Decrease Sensitivity to CDT
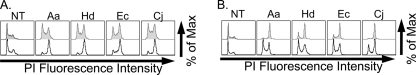
Next, we wished to address the proposed role of glycolipids in sensitivity to CDTs. An earlier study suggested that the glycosphingolipid GM3 functions as a receptor for Aa-CDT (24). Notably, GM3 is the predominant glycosphingolipid on CHO-K1 cells and consists of sialic acid (Neu5Ac), galactose (Gal), glucose (Glc), and ceramide (Cer) in the structure Neu5Acα2–3Galβ1–4Glcβ1–1Cer (Fig. 4A) (45). Therefore, mutant CHO cells defective in sialic acid (Lec2) or galactose (Lec8) modifications of glycoconjugates are predicted to lack GM3. Surprisingly, neither Lec2 nor Lec8 cells displayed resistance to Aa-CDT or any of the other three CDTs tested (Fig. 4B). In fact, Lec2 cells displayed increased sensitivity to Aa-, Hd-, and Ec-CDTs, and Lec8 cells were slightly more sensitive to Aa- and Hd-CDTs (Fig. 4B).
Although these data indicate that the terminal saccharides of GM3 are not essential for conferring cell sensitivity, it is possible that the glucosylceramide core of GM3 is important for intoxication. To evaluate this, we preincubated CHO-K1 cells with the glucosylceramide biosynthesis inhibitor PPMP, then challenged with each CDT. As with Lec2 and Lec8 cells (Fig. 4), CHO-K1 cells treated with PPMP displayed increased sensitivity to Ec-, Aa-, and Hd-CDT (Fig. 6A), a result opposite of that expected if GM3 were a receptor. Interestingly, PPMP also increased sensitivity of CHO-K1 cells to Cj-CDT, a result not predicted by the Lec mutants. Taken together, these results suggest that, contrary to previous reports, GM3 is not likely to function as a receptor for Aa-CDT, nor any of the three other CDTs tested. To further validate this result, a mutant CHO cell line deficient in synthesis of all sphingolipids (LY-B) (50) was incubated with each CDT. Consistent with the Lec cell and PPMP data, LY-B cells displayed increased sensitivity to all CDTs (Fig. 6B). These data suggest that GM3, or any other glycosphingolipid, is not required for conferring sensitivity to any of the CDTs studied here.
FIGURE 6.
Role of glycolipids in CDT intoxication. A, cells were treated with 5 μm PPMP for 10 days, intoxicated with CDT for 24 h, stained with PI, and subjected to flow cytometry. Unshaded histograms are intoxicated PPMP-treated cells, and gray shaded histograms are untreated controls. B, lipid glycosylation mutant cells (LY-B, gray shaded histograms) or the cDNA-complemented counterpart (Ly-BcLCB1, unshaded histograms) were intoxicated with CDT for 24 h, stained with PI, and subjected to flow cytometry. Results are representative of at least three independent experiments.
DISCUSSION
CDTs are expressed by several distantly related Gram-negative bacterial pathogens that occupy very different ecological niches and cause distinct diseases. Here we tested whether host cellular factors previously implicated in CDT-host interactions were required for cellular intoxication by CDTs derived from four bacterial pathogens that cause periodontal disease, gastroenteritis or chancroid. Surprisingly, we found that mature N-linked as well as O-, or lipid-linked glycans are not required for intoxication by any of the CDTs tested. Further, these four CDTs could be categorized into three groups based on their relative abilities to intoxicate a variety of wild-type and mutant host-cell types. We report that Aa- and Hd-CDTs behave similarly, whereas CDTs derived from E. coli and C. jejuni display distinct host cell preferences (Table 2). These findings are consistent with the degree of amino acid similarity and indicate that CDTs derived from different pathogens possess unique requirements to intoxicate host cells and may have different receptors.
TABLE 2.
Summary of differences between CDTs
+ represents increased sensitivity, − represents decreased sensitivity, and 0 represents no change.
| Toxin | Target cell preference | Tunicamycin treatment | Cholesterol addition | Sialic acid deficiency | Galactose transporter deficiency | Glycolipid deficiency |
|---|---|---|---|---|---|---|
| Aa | Highly active on most cell types | − | + | + | + | + |
| Hd | Highly active on most cell types | − | + | + | + | + |
| Ec | Most active on CHO-K1 cells | − | + | + | 0 | + |
| Cj | Active on 3T3 and Y-1 cells | + | 0 | 0 | 0 | + |
Prior studies with Ec- and Aa-CDTs supported a role for host carbohydrates in CDT interactions (23, 24). Here, we used a genetic approach to directly test the requirement of specific host glycans by employing cell lines with known glycan deficiencies. Surprisingly, mutant CHO cells that lack N-linked complex and hybrid carbohydrates (Fig. 4) or cells that lack glycosphingolipids (Fig. 6) were more sensitive to the CDTs tested. Hypersensitivity likely derives from the loss of sialic acid, a common feature of the Lec mutants tested here, and the only defect in Lec2 cells (Fig. 4). This result is in agreement with the previous finding that removal of sialic acid with neuraminidase leads to increased sensitivity to Ec-CDT (23). The negative charge associated with sialic acid may act to inhibit CDTs, a conclusion further supported by the finding that mutant CHO cells lacking negatively charged glycosaminoglycans (i.e. pgsA-745 and pgsD-677) (51, 52) are hypersensitive to all CDTs studied here (data not shown).
The difference in sensitivity of tunicamycin-treated and mutant Lec cell lines provides insight into toxin-host interactions and can be explained by one of two models. First, it is possible that the immature high mannose and/or core glycan structure present on Lec1, ldlD.Lec1, and Lec23 cells, but absent following tunicamycin treatment contributes to sensitivity to Aa-, Hd-, and Ec-CDTs. However, excess mannose or mannans did not block CDT intoxication (data not shown). Further, multiple attempts to identify Ec-CDT binding to over 300 individual glycan structures using glycan arrays revealed no consistent or detectable interactions.8 A second possibility is that the polypeptide component of glycoproteins, rather than glycans themselves, serve as host determinants for CDT sensitivity and that these proteins require glycan modification for proper folding and cell surface presentation (43). Indeed, Carette and colleagues recently identified a host membrane protein, TMEM181, that supports intoxication by Ec-CDT and may serve as a receptor for this toxin (53).
It is striking that our results appear to contradict previously reported roles for fucosylated glycans and GM3 (23, 24). The former may be explained by differences in toxins utilized. Specifically, E. coli can encode multiple distinct CDTs. Whereas McSweeney and Dreyfus studied type II Ec-CDT, results presented here are based on the closely related but distinct type III toxin. Discrepancies regarding PPMP-mediated inhibition of GM3 may be partly explained by the fact that inhibition of glycolipid synthesis in monocytes, the cell type utilized by Mise et al. (54), leads to altered trafficking following endocytosis and targeting of lipid-raft associated proteins to lysosomes. Trafficking of CDT to the lysosomes rather than the endoplasmic reticulum in PPMP-treated monocytes could have led to the previously reported results. However, Mise and colleagues also reported that glycolipid-deficient LY-B cells were resistant to Aa-CDT, in contrast to results presented here. Furthermore, Carette et al. recently reported that cells containing retroviral insertions in sphingomyelin synthase are resistant to Ec-CDT (53). LY-B cells used in our studies were originally isolated based on having deficiency in sphingomyelin content (50). Although the source of these discrepancies is still unclear, our conclusions are supported by multiple lines of evidence obtained with Lec2, Lec8, Lec3.2.8, LY-B, and PPMP-treated cells, all of which are deficient in GM3 and display increased sensitivity to one or more CDTs (Fig. 4B).
In summary, the results presented here show that CDTs derived from different pathogens utilize distinct host factors for intoxication. This tropism seems to correlate with the amino acid sequence of the CDT-binding subunits as opposed to bacterial niche. Although A. actinomycetemcomitans and H. ducreyi have different niches, the binding subunits of the CDTs they encode share high levels of amino acid identities, and they respond similarly to alterations in host factors such as glycans, cholesterol content, and cell lineage. Conversely, E. coli and C. jejuni have similar niches, but their binding subunits are quite different, leading to divergent target cell preferences. Future efforts to identify receptors for CDTs will provide insight into cellular and tissue tropism and may thus shed light on host interactions associated with CDT-producing pathogens.
Acknowledgments
We thank Dr. David Smith and members of the Consortium for Functional Glycomics Core H for helpful discussion and glycan array screening. We thank Dr. Pamela Stanley and Dr. Jeff Esko for glycan-deficient mutant cell lines and Dr. Esko and Dr. Linda Baum for expert consultation. Finally, we thank Dr. Baum for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants T32DE007296 (to A. E.), F31AI061837 (to F. J. M.-A.), and AI59095 (to S. R. B.).
A. Gargi, B. Powers, and S. R. Blanke, data not shown.
Data available on-line from the Consortium for Functional Glycomics.
- CDT
- cytolethal distending toxin
- Ec-CDT
- CDT produced by E. coli
- As-CDT
- CDT produced by A. actinomycetemcomitans
- Hd-CDT
- CDT produced by H. ducreyi
- Cj-CDT
- CDT produced by C. jejuni
- CHO
- Chinese hamster ovary
- FBS
- fetal bovine serum
- PBS
- phosphate-buffered saline
- DPBS
- Dulbecco's PBS
- PI
- propidium iodide
- FITC
- fluorescein isothiocyanate
- UEA
- U. europus agglutinin
- Fut1
- fucosyltransferase 1
- PPMP
- 1-phenyl-2-palmitoylamino-3-morpholino-1-propanol
- GM1
- Galβ1,3GalNAcβ1,4(NeuAcα2,3)-Galβ1,4Glc-ceramide
- GM2
- GalNAcβ1,4(NeuAcα2,3)-Galβ1,4Glc-ceramide
- GM3
- NeuAcα2,3Galβ1,4Glc-ceramide.
REFERENCES
- 1.Oswald E., Nougayrède J. P., Taieb F., Sugai M. (2005) Curr. Opin. Microbiol. 8, 83–91 [DOI] [PubMed] [Google Scholar]
- 2.Nougayrède J. P., Taieb F., De Rycke J., Oswald E. (2005) Trends. Microbiol. 13, 103–110 [DOI] [PubMed] [Google Scholar]
- 3.Shenker B. J., Hoffmaster R. H., Zekavat A., Yamaguchi N., Lally E. T., Demuth D. R. (2001) J. Immunol. 167, 435–441 [DOI] [PubMed] [Google Scholar]
- 4.Purdy D., Buswell C. M., Hodgson A. E., McAlpine K., Henderson I., Leach S. A. (2000) J. Med. Microbiol. 49, 473–479 [DOI] [PubMed] [Google Scholar]
- 5.Pickett C. L., Whitehouse C. A. (1999) Trends. Microbiol. 7, 292–297 [DOI] [PubMed] [Google Scholar]
- 6.Smith J. L., Bayles D. O. (2006) Crit. Rev. Microbiol. 32, 227–248 [DOI] [PubMed] [Google Scholar]
- 7.Frisk A., Lebens M., Johansson C., Ahmed H., Svensson L., Ahlman K., Lagergård T. (2001) Microb. Pathog. 30, 313–324 [DOI] [PubMed] [Google Scholar]
- 8.Pickett C. L., Pesci E. C., Cottle D. L., Russell G., Erdem A. N., Zeytin H. (1996) Infect. Immun. 64, 2070–2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elwell C., Chao K., Patel K., Dreyfus L. (2001) Infect. Immun. 69, 3418–3422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elwell C. A., Dreyfus L. A. (2000) Mol. Microbiol. 37, 952–963 [DOI] [PubMed] [Google Scholar]
- 11.Lara-Tejero M., Galán J. E. (2000) Science 290, 354–357 [DOI] [PubMed] [Google Scholar]
- 12.Shenker B. J., Dlakic M., Walker L. P., Besack D., Jaffe E., LaBelle E., Boesze-Battaglia K. (2007) J. Immunol. 178, 5099–5108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nesić D., Hsu Y., Stebbins C. E. (2004) Nature 429, 429–433 [DOI] [PubMed] [Google Scholar]
- 14.Rabin S. D., Flitton J. G., Demuth D. R. (2009) Infect. Immun. 77, 3161–3169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thelestam M., Frisan T. (2004) Rev. Physiol. Biochem. Pharmacol. 152, 111–133 [DOI] [PubMed] [Google Scholar]
- 16.Johnson W. M., Lior H. (1988) Microb. Pathog. 4, 115–126 [DOI] [PubMed] [Google Scholar]
- 17.Akifusa S., Heywood W., Nair S. P., Stenbeck G., Henderson B. (2005) Microbiology 151, 1395–1402 [DOI] [PubMed] [Google Scholar]
- 18.Akifusa S., Poole S., Lewthwaite J., Henderson B., Nair S. P. (2001) Infect. Immun. 69, 5925–5930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cao L., Volgina A., Huang C. M., Korostoff J., DiRienzo J. M. (2005) Mol. Microbiol. 58, 1303–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng K., Hansen E. J. (2003) Infect. Immun. 71, 6633–6640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee R. B., Hassane D. C., Cottle D. L., Pickett C. L. (2003) Infect. Immun. 71, 4883–4890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McSweeney L. A., Dreyfus L. A. (2004) Cell Microbiol. 6, 447–458 [DOI] [PubMed] [Google Scholar]
- 23.McSweeney L. A., Dreyfus L. A. (2005) Infect. Immun. 73, 2051–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mise K., Akifusa S., Watarai S., Ansai T., Nishihara T., Takehara T. (2005) Infect. Immun. 73, 4846–4852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nesic D., Stebbins C. E. (2005) PLoS. Pathog. 1, e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu X., Nesic D., Stebbins C. E. (2006) Proteins. 62, 421–434 [DOI] [PubMed] [Google Scholar]
- 27.Lara-Tejero M., Galán J. E. (2001) Infect. Immun. 69, 4358–4365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boesze-Battaglia K., Brown A., Walker L., Besack D., Zekavat A., Wrenn S., Krummenacher C., Shenker B. J. (2009) J. Biol. Chem. 284, 10650–10658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stanley P. (1989) Mol. Cell. Biol. 9, 377–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hong Y., Sundaram S., Shin D. J., Stanley P. (2004) J. Biol. Chem. 279, 49894–49901 [DOI] [PubMed] [Google Scholar]
- 31.Oelmann S., Stanley P., Gerardy-Schahn R. (2001) J. Biol. Chem. 276, 26291–26300 [DOI] [PubMed] [Google Scholar]
- 32.Chen W., Stanley P. (2003) Glycobiology 13, 43–50 [DOI] [PubMed] [Google Scholar]
- 33.Larsen R. D., Ernst L. K., Nair R. P., Lowe J. B. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 6674–6678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bradley K. A., Mogridge J., Jonah G., Rainey A., Batty S., Young J. A. (2003) J. Biol. Chem. 278, 49342–49347 [DOI] [PubMed] [Google Scholar]
- 35.Shenker B. J., Hoffmaster R. H., McKay T. L., Demuth D. R. (2000) J. Immunol. 165, 2612–2618 [DOI] [PubMed] [Google Scholar]
- 36.Cope L. D., Lumbley S., Latimer J. L., Klesney-Tait J., Stevens M. K., Johnson L. S., Purven M., Munson R. S., Jr., Lagergard T., Radolf J. D., Hansen E. J. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 4056–4061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cortes-Bratti X., Chaves-Olarte E., Lagergård T., Thelestam M. (1999) J. Clin. Invest. 103, 107–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson W. M., Lior H. (1988) Microb. Pathog. 4, 103–113 [DOI] [PubMed] [Google Scholar]
- 39.Li L., Sharipo A., Chaves-Olarte E., Masucci M. G., Levitsky V., Thelestam M., Frisan T. (2002) Cell Microbiol. 4, 87–99 [DOI] [PubMed] [Google Scholar]
- 40.Boesze-Battaglia K. (2006) Methods. Mol. Biol. 332, 169–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boesze-Battaglia K., Besack D., McKay T., Zekavat A., Otis L., Jordan-Sciutto K., Shenker B. J. (2006) Cell Microbiol. 8, 823–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guerra L., Teter K., Lilley B. N., Stenerlöw B., Holmes R. K., Ploegh H. L., Sandvig K., Thelestam M., Frisan T. (2005) Cell Microbiol. 7, 921–934 [DOI] [PubMed] [Google Scholar]
- 43.Zhou F., Xu W., Hong M., Pan Z., Sinko P. J., Ma J., You G. (2005) Mol. Pharmacol. 67, 868–876 [DOI] [PubMed] [Google Scholar]
- 44.Volpe J. J., Goldberg R. I. (1983) J. Biol. Chem. 258, 9220–9226 [PubMed] [Google Scholar]
- 45.Patnaik S. K., Stanley P. (2006) Methods Enzymol. 416, 159–182 [DOI] [PubMed] [Google Scholar]
- 46.Lin A. I., Philipsberg G. A., Haltiwanger R. S. (1994) Glycobiology 4, 895–901 [DOI] [PubMed] [Google Scholar]
- 47.Becker D. J., Lowe J. B. (2003) Glycobiology 13, 41R–53R [DOI] [PubMed] [Google Scholar]
- 48.Löfling J., Diswall M., Eriksson S., Borén T., Breimer M. E., Holgersson J. (2008) Glycobiology 18, 494–501 [DOI] [PubMed] [Google Scholar]
- 49.Mathieu S., Prorok M., Benoliel A. M., Uch R., Langlet C., Bongrand P., Gerolami R., El-Battari A. (2004) Am. J. Pathol. 164, 371–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hanada K., Hara T., Fukasawa M., Yamaji A., Umeda M., Nishijima M. (1998) J. Biol. Chem. 273, 33787–33794 [DOI] [PubMed] [Google Scholar]
- 51.Esko J. D., Stewart T. E., Taylor W. H. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 3197–3201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lidholt K., Weinke J. L., Kiser C. S., Lugemwa F. N., Bame K. J., Cheifetz S., Massagué J., Lindahl U., Esko J. D. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 2267–2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carette J. E., Guimaraes C. P., Varadarajan M., Park A. S., Wuethrich I., Godarova A., Kotecki M., Cochran B. H., Spooner E., Ploegh H. L., Brummelkamp T. R. (2009) Science 326, 1231–1235 [DOI] [PubMed] [Google Scholar]
- 54.Sillence D. J., Puri V., Marks D. L., Butters T. D., Dwek R. A., Pagano R. E., Platt F. M. (2002) J. Lipid. Res. 43, 1837–1845 [DOI] [PubMed] [Google Scholar]