Abstract
Arenavirus and bunyavirus RNA genomes are unusual in that they are found in circular nucleocapsids, presumably due to the annealing of their complementary terminal sequences. Moreover, arenavirus genome synthesis initiates with GTP at position +2 of the template rather than at the precise 3′ end (position +1). After formation of a dinucleotide, 5′ pppGpCOH is then realigned on the template before this primer is extended. The net result of this “prime and realign” mechanism of genome initiation is that 5′ pppG is found as an unpaired 5′ nucleotide when the complementary genome ends anneal to form a double-stranded (dsRNA) panhandle. Using 5′ pppRNA made in vitro and purified so that all dsRNA side products are absent, we have determined that both this 5′ nucleotide overhang, as well as mismatches within the dsRNA (as found in some arenavirus genomes), clearly reduce the ability of these model dsRNAs to induce interferon upon transfection into cells. The presence of this unpaired 5′ ppp-nucleotide is thus another way that some viruses appear to use to avoid detection by cytoplasmic pattern recognition receptors.
Keywords: Innate Immunity, Interferon, Negative-strand RNA Viruses, Pathogen-associated Molecular Pattern (PAMP), Viral Replication
Introduction
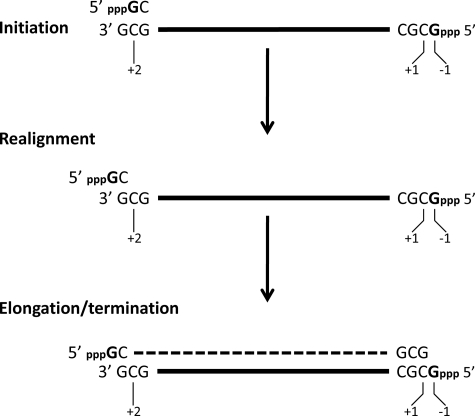
Most viral RNA-dependent RNA polymerases (RdRP)2 maintain genome length in a straightforward manner, by initiating genome (and antigenome) synthesis with an NTP opposite the precise 3′ end of their antigenome (and genome) templates, and terminating synthesis after incorporating the last nucleotide opposite their 5′ termini. Some segmented negative strand RNA viruses, however, have chosen a different approach. For example, when Tacaribe arenavirus (TCRV) genome and antigenome RNAs are annealed, this dsRNA contains a single unpaired 5′ nucleotide overhang (pppG) rather than flush or blunt ends (1). The initiating pppG could not have been coded for at the precise template/antigenome 3′ end, which is also G. Moreover, when TCRV RNA synthesis is reconstituted in vitro, certain di- and trinucleotides efficiently prime this synthesis, and GpC, the most efficient primer, led to transcripts whose 5′ ends were also at position −1 (see Fig. 1) (2). As the sequence 3′ OHGCG… is present at all arenavirus genome and antigenome 3′ ends, we proposed that TCRV genomes were initiated by 5′ pppGpCOH (an uncapped primer) formed at an internal position (in bold) of the 3′ OHGCG… sequence (Fig. 1). This pppGpCOH could then realign on the template such that its C residue is lined up opposite the G at the precise template 3′ end, and then extended for genome synthesis (Fig. 1). Support for this prime and realign mechanism was also obtained by studying Hantaan (bunya)virus (HTN), where both mRNA and genome synthesis are thought to involve priming and realigning (3) as well as mRNA synthesis of Germiston and Rift Valley fever virus, two other bunyaviruses (4). More recently, influenza A virus was also reported to use a prime and realign mechanism to initiate genome synthesis from antigenome templates (5). Thus all 3 families of segmented negative strand RNA viruses appear to use this unusual mechanism for initiating viral RNA synthesis to some extent.
FIGURE 1.
Prime and realign mechanism of initiating arenavirus genome synthesis. Antigenome synthesis is initiated by GTP opposite the template C at position +2 at the 3′ end of the genome (written 3′ to 5′; top level). After its extension to a dinucleotide, 5′ pppGpCOH is realigned such that its 3′ terminal COH is opposite the genome 3′ terminal G (middle level), and the realigned 5′ pppGpCOH then acts a primer for antigenome synthesis (dashed line; bottom level). The “pseudo-templated” 5′ pppG is somehow not copied during genome replication, as it is specifically cleaved from genome/antigenome dsRNA by single strand-specific RNase T1. An identical process occurs during genome synthesis.
In contrast to TCRV, when HTN genomes/antigenomes are aligned as dsRNA, the dsRNA ends are flush and contain 5′ pU rather than a 5′ triphosphate (3). As the HTN RdRP cap-dependent endonuclease strongly prefers to cleave host mRNAs after guanosines to generate capped fragments that prime viral mRNA synthesis, we originally proposed that HTN genome replication initiated with pppG opposite the C at position +3 (3′ OHUACUACUAC…), and following the addition of a few nucleotides, the uncapped primer, e.g. pppGAUGOH, realigns at the genome 3′ end creating an extra (pseudo-templated) overhanging 5′ pppG (in analogy to TCRV) (supplemental Fig. S1). The initiating pppG could then be removed by the G-specific RdRP endonuclease, leaving 5′ pU at position +1 (supplemental Fig. S1, right side) (3). It was thus assumed that this cap-dependent endonuclease can also work in a cap-independent fashion. With hindsight, it is now clear that once the 5′ end of the primer for genome synthesis has been removed, the primer can no longer be identified. This leads to a 2nd possible mechanism, in which the capped host-cell primer is used both for transcription and replication. For transcription, the endonuclease cleaves the host mRNA only once, to generate the capped primer. For replication the endonuclease cuts this capped primer a 2nd time, after it has been extended and realigned (supplemental Fig. S1, left side). In this case, the RdRP endonuclease works in a strictly cap-dependent fashion, as one would expect.
Independent of precisely how the 5′ pU HTN genome ends are formed, the advantage to an RNA virus of not having 5′ ppp-genome ends is also now more obvious. The innate immune system senses RNA virus infections primarily by two DEXD/H box helicases, RIG-I and mda-5 (6). These helicases act as pattern recognition receptors, which respond to two RNA PAMPs (pathogen-associated molecular patterns), namely dsRNA (e.g. poly(I)·poly(C), or poly(I/C) and 5′ pppRNA (7–9). These RNAs can act as PAMPs because their cytoplasmic presence is thought to be restricted to virus infection. RIG-I, which senses negative strand RNA virus infections (10), is a large protein composed of 3 domains; N-terminal tandem CARDs (caspase recruitment domains) (the effector domain), a DEXD/H box helicase whose ATPase powers the translocation of RIG-I on dsRNA (11), and a C-terminal regulatory domain that binds 5′ pppRNA (12, 13). When PAMPs bind to the RIG-I regulatory domain, the helicase ATPase is activated and the N-terminal CARDs are released for interacting with cardif/IPS-1, the central adaptor in the signaling pathway to IFNβ. In the earliest reports, 5′ pppRNA was thought to act as a PAMP whether present on ss- or dsRNA. More recently, good evidence has been presented that 5′ pppRNA without dsRNA character is not a PAMP (14, 15). In the latest descriptions of this signaling pathway, the regulatory domain is activated and IFN is induced only when RIG-I interacts with dsRNA, which also contains a 5′ ppp end, i.e. when both PAMPs are combined in a single molecule or complex.
Habjan et al. (16) have reported that Crimean Congo hemorrhagic fever virus (a bunyavirus of the Nairovirus genus) and Borna disease virus, as well as HTN, all generate 5′ monophosphate genome ends to avoid activation of the IFN system, e.g. their phenol-extracted virion RNAs do not bind to RIG-I or induce IFN upon transfection into cells. For HTN (and presumably Crimean Congo hemorrhagic fever virus), removal of the 5′ pppGOH or 7-MeGpppN10–20GOH from the genome end is a consequence of the prime and realign mechanism that generates these 5′ single-stranded overhangs during the initiation of genome synthesis. In the case of TCRV, however, the 5′ pppG is not removed and the single 5′ nucleotide overhang is maintained, because the additional or pseudo-templated 5′ pppG of the template is not copied during genome replication (2) (Fig. 1). This article examines whether the presence of this single 5′ nucleotide overhang is another possible way that some viruses avoid activating the IFN system.
EXPERIMENTAL PROCEDURES
Plasmids
pβ-IFN-fl-lucter containing the firefly luciferase gene under control of the human IFN-β promoter was described previously (17). pTK-rl-lucter, used as a transfection standard, contains the herpes simplex virus TK promoter region upstream of the Renilla luciferase gene (Promega). pEBS-E3L100–190 contains the dsRNA binding domain of the vaccinia virus E3L protein (18). pEBS-tom contains tomato, a red fluorescent protein.
Transfections
100,000 cells were plated into 6-well plates 20 h before transfection with 1.5 μg of pβ-IFN-fl-lucter, 0.5 μg of pTK-rl-lucter, 1 μg of plasmids expressing E3L or tomato (when indicated), and TransIT-LT1 transfection reagent (Mirus). At 24 h post-transfection, the cells were (or were not) transfected with the indicated RNAs using Trans-messenger transfection reagent (Qiagen)(a total of 3 μg of RNA was always transfected, the difference always being made up with tRNA). Twenty hours later, cells were harvested and assayed for firefly and Renilla luciferase activity (dual-luciferase reporter assay system; Promega). Relative expression levels were calculated by dividing the firefly luciferase values by those of Renilla luciferase.
In Vitro Synthesis of RNA and Purification
DNA for T7 RNA polymerase synthesis of model RNA1 was prepared by PCR using the partially complementary primers 5′-TAATACGACTCACTATAgggACACACCACAACCAACCCACAAC-3′ (forward; start site in lowercase type) and 5′-GAAAGAAAGGTGTGGTGTTGGTGTGGTTGTTGTGGGTTGGTTGTGG-3′ (reverse). Transcription was performed with 100 pmol of purified PCR product using T7 MEGAshortscript (Ambion) according to the manufacturer's instructions. The total T7 transcripts (unpurified ppp-ssRNA1) were digested with DNase I and then chromatographed on NucAway Spin columns (Ambion) to remove unincorporated nucleotides and DNA fragments. Slightly radiolabeled ([α-32P]CTP or ([α-32P]UTP, 0.37 MBq) T7 transcripts were further electrophoresed on 10% preparative denaturing gels. The major 54-nucleotide band was excised from the gel, and the RNA was eluted, followed by ethanol precipitation.
RNase Treatment
1 μg of ppp-RNA1 was digested with 1 unit of RNase III (Ambion) for 60 min at 37 °C in 40 μl of 50 mm NaCl, 10 mm Tris-HCl, 10 mm MgCl2, 1 mm dithiothreitol (pH 7.9), or 50 ng of RNase A in 40 μl of 10 mm Tris-HCl, 1 mm EDTA, and 0.4 m NaCl (pH 7.9). The digestion products were then phenol/chloroform-extracted, ethanol-precipitated with 10 μg of glycogen, and electrophoresed on a denaturing 10% polyacrylamide gel.
For further RNA purification, 300 μg of gel purified ppp-RNA1* was digested with 300 units of RNase III for 60 min at 37 °C in 300 μl of 50 mm NaCl, 10 mm Tris-HCl, 10 mm MgCl2, 1 mm dithiothreitol (pH 7.9). Then 60 μg of proteinase K was added for 15 min at 37 °C, followed by phenol-chloroform extraction and ethanol precipitation.
Annealed dsRNA
ppp-ssRNA1** (gel purified and RNase III treated) was mixed with the indicated synthetic complementary 5′ OH-oligoribonucleotides in a 1 to 3 molar ratio, in a final volume of 50 μl (300 mm NaCl, 50 mm Tris, pH 7.5, 1 mm EDTA), heated 1 min at 90 °C, and progressively cooled to room temperature. 200 ng (×1) or 1 μg of 54′-mer (×5) were then transfected into A549 cells, and their ability to activate the IFNβ promoter was determined (“Materials and Methods”). In all cases, 3 μg of total RNA was transfected, the difference being made up with tRNA. Transfection of tRNA by itself was neutral.
RESULTS
The Problem with in Vitro Synthesized 5′ ppp-ssRNA, and One Way to Fix It
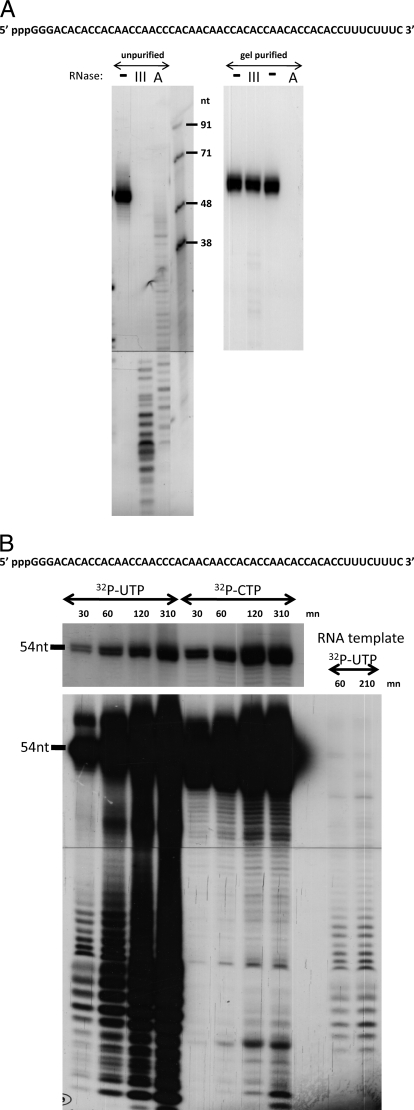
A model RNA1 (54 nt) was designed to contain little or no secondary structure. When synthesized in vitro with biotin-UTP, RNA1 (5′-pppGGGACACACCACAACCAACCCACAACAACCACACCAACACCACACCUUUCUUUCOH) can be stably bound to streptavidin beads, and used to study RIG-I/5′ ppp-ssRNA interactions in pulldown assays. Various non-biotinylated RNAs can then be used to compete with the binding of RIG-I to the RNA1 beads, to study the RNA determinants of this interaction. When [32P]CTP-labeled RNA1 was examined on sequencing gels, a single band of the expected mobility was seen (Fig. 2A, left panel, left lane), and so this product was bound to the beads without further purification. The binding of RIG-I to RNA1 beads appeared to depend on the presence of the 5′ triphosphate group, as several T7 transcripts competed efficiently, whereas the same chemically synthesized 5′ OHssRNAs were totally inactive. However, when we treated the competing in vitro transcripts with either phosphatase to remove the 5′ ppp, or RNase T1, which removes 5′ pppGGG (and this latter removal could be clearly verified by PAGE), we found that both 5′-dephosphorylated RNA1s had lost virtually none of their ability to compete for RIG-I binding to untreated RNA1. As the chemically synthesized 5′ OHssRNAs and the 5′-dephosphorylated T7 transcripts behaved so differently, the nature of these in vitro transcripts was further examined. When the in vitro synthesized transcripts were treated with RNase III (which specifically degrades dsRNA), or RNase A in 0.4 m NaCl (which specifically degrades ssRNA), these in vitro transcripts were clearly sensitive to both digestions (Fig. 2A, left panel).
FIGURE 2.
A, RNase III and RNase A sensitivity of 5′ ppp-RNA1 before and after PAGE purification. Left panel, [32P]CTP-labeled RNA1 was untreated, digested with RNase III, or digested with RNase A in 0.4 m NaCl, as indicated. After phenol extraction and ethanol precipitation, the samples were analyzed on a 15% sequencing gel. The right-hand lane shows RNA length markers. Right panel, [32P]CTP-labeled RNA1 was electrophoresed on a preparative denaturing 15% polyacrylamide gel and the 54-nt long band was recovered. PAGE-purified RNA1 (5′ ppp-RNA1*) was then digested (or not) with RNase III or RNase A. The sequence of RNA1 is shown above. B, kinetics of synthesis of [32P]UTP- or [32P]CTP-labeled RNA1. RNA1 was synthesized with T7 RNA polymerase directed by promoter-containing DNA, and the products were labeled with either [32P]CTP or [32P]UTP, as indicated. Samples were taken at various times and the products were examined by denaturing PAGE. A [32P]UTP-labeled reaction that was primed with PAGE-purified pppRNA1 in place of promoter-containing DNA was also carried out (RNA template). An underexposure of the 54-nt region of the gel is shown at the top. The sequence of RNA1 is shown above.
It appeared that our in vitro transcription reaction had also made RNA complementary to RNA1 (19) that would be poorly labeled with CTP. Upon annealing, this would result in dsRNA duplexes. We therefore examined the kinetics of RNA1 synthesis labeled with UTP as well as CTP. Full-length (54 nt) RNA1 was essentially the only product detected in the CTP-labeled reaction, even upon overexposure of the gel (best seen in 30-min time point). However, a series of shorter RNAs were seen, even in the 30-min UTP-labeled reaction. To examine whether these cRNA1 fragments were templated by the de novo RNA1 itself, gel-purified RNA1 (see below) was added to a reaction in place of promoter-containing DNA. As shown (Fig. 2B, right lanes), the same pattern of RNA fragments was detected in this reaction as in those containing DNA templates (no UTP incorporation was detected when neither nucleic acid template was added to the reaction). As the 54-nt ssRNA1 was the predominant reaction product (Fig. 2A), we purified this RNA from the mixture by recovering the 54-nt band from a preparative denaturing gel. The gel-purified in vitro transcript indeed appeared to consist essentially of ssRNA1, as the vast majority of this RNA was now resistant to RNase III digestion, and completely sensitive to digestion with RNase A in 0.4 m NaCl (Fig. 2A, right panel). The vast majority of the minimally purified RNA1 appears to have been partly ss and partly ds, and therefore sensitive to both RNases (Fig. 2A, left panel).
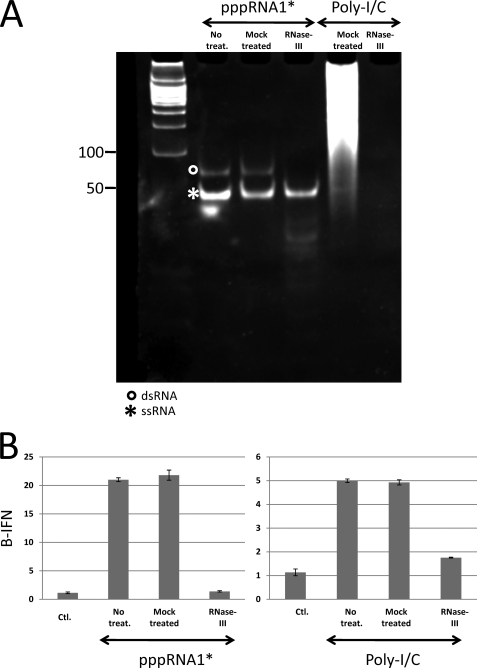
Gel-purified pppRNA1 (pppRNA1*) had also lost >95% of it ability to stimulate the ATPase activity of RIG-I (data not shown). Nevertheless, pppRNA1* retained a considerable ability to induce IFNβ upon transfection, and more importantly, this activity was essentially eliminated when the dsRNA binding domain of the vaccinia virus E3L protein was expressed in these cells (Fig. 3). Successive purification of 5′ ppp-RNA1 clearly showed that its ability to interact with RIG-I was due to dsRNA by-products of the T7 reaction, rather than the ssRNA1 (supplemental Fig. S3). If pppRNA1* still contained some dsRNA, it would have to be full-length dsRNA. We therefore examined pppRNA1* by non-denaturing PAGE (Fig. 4A). Ethidium bromide staining revealed that there was indeed some apparently full-length dsRNA present: ∼2% by Cerenkov counting of the excised bands (supplemental Fig. S2). When this minor dsRNA contamination was specifically removed by RNase III digestion (Fig. 4A), the doubly purified 5′ ppp- ssRNA1 (pppRNA1**) was found to be essentially inactive in inducing IFNβ (Fig. 4B), as recently reported by others using chemically synthesized 5′ ppp-ssRNA (14, 15). Moreover, annealing of a complementary RNA to pppRNA1** restored its ability to induce IFNβ (Figs. 5 and 6).
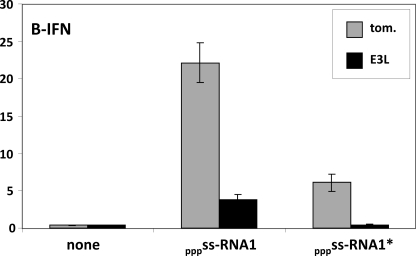
FIGURE 3.
The effect of E3L100–190 expression on 5′ pppRNA1-induced IFNβ activation. Parallel cultures of mouse embryo fibroblast were transfected with pIFNβ-(ff)luciferase and pTK-(ren)luciferase, plus and minus pEBS-E3L100–190. Twenty hours later, the cultures were further transfected with either unpurified or PAGE-purified 5′ pppRNA1. Cell extracts were prepared at 24 hours post transfection and their luciferase activities were determined. Tom refers to tomato, a red fluorescent protein, used as a control.
FIGURE 4.
RNase III treatment of PAGE-purified 5′ pppRNA1. A, PAGE-purified 5′ pppRNA1 and poly(I-C) were either mock-treated or digested with RNase III, as indicated. Samples were then separated by nondenaturing PAGE and the gel was stained with ethidium bromide. DNA restriction fragments were electrophoresed in the left-hand lane as size markers. B, samples were also transfected into A549 cells and their ability to activate the IFNβ promoter was determined.
FIGURE 5.
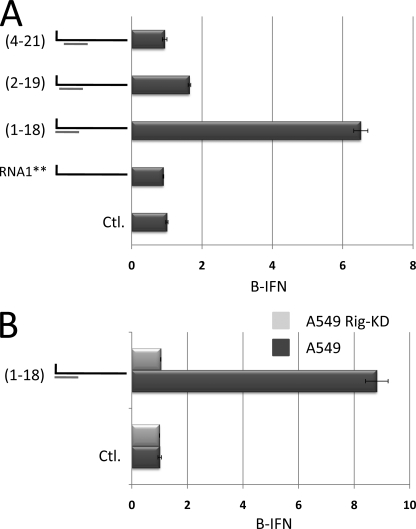
The importance of including the 5′ pppN within dsRNA. A, doubly purified 5′ ppp-ssRNA1** (54′mer) was annealed (or not) with 18′-mers complementary to positions 1–18, 2–19, and 4–21 (relative to the 5′ ppp-end), as indicated on the left. Samples were then transfected into A549 cells and their ability to activate the IFNβ promoter was determined. Transfection of the 18′-mers by themselves were inactive (supplemental Fig. S4). Line 1 (Ctl) shows the level of IFNβ activation in untransfected cells. B, doubly purified 5′ ppp-ssRNA1** (54′-mer) was annealed with an 18′-mer complementary to positions 1–18, as indicated on the left. Samples were then transfected into either wild-type A549 cells or A549 cells in which RIG-I was knocked down (KD) via miRNA expression (27), and their ability to activate the IFNβ promoter was determined. Ctl shows the level of IFNβ activation in untransfected cells.
FIGURE 6.
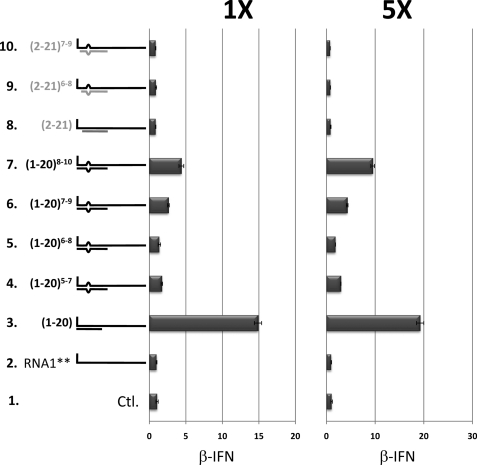
The effect of a 3-nt bulge plus the presence (or not) of the 5′ pppN within dsRNA of model RNA1 on PAMP function. 5′ ppp-ssRNA1 (54′-mer) was annealed (or not, line 2) with 20′-mer complementary to positions 1–20 or 2–21, which did or did not contain 2 mismatches at positions 5 and 7, 6 and 8, 7 and 9, or 8 and 10, as indicated on the left. 200 ng (×1) or 1 μg of 54′-mer (×5) were then transfected into A549 cells, and their ability to activate the IFNβ promoter was determined. 3 μg of total RNA was transfected in all cases, the difference being made up with tRNA (transfection of tRNA by itself was neutral). Line 1 (Ctl) shows the level of IFNβ activation in untransfected cells. Transfection of the 20′-mers by themselves were inactive (see supplemental Fig. S4).
Model Arenavirus dsRNA Panhandle Structures Are Poor PAMPs
As mentioned above, two groups have recently reported that chemically synthesized 5′ ppp-ssRNA by itself does not induce IFN via RIG-I, but becomes active when annealed to a complementary RNA. Both groups found that a stretch of dsRNA near the 5′ ppp end, as well as the 5′ ppp end itself, were essential for RIG-I to induce IFN. These reports, however, differed about the minimum length of the dsRNA region required, and more importantly for this study, whether the dsRNA region needed to include the nucleotide carrying the 5′ ppp. Schlee et al. (14) reported that a double strand spanning at least 19 nt and encompassing the 5′ pppN were essential; a 5′ ppp 24′-mer annealed to a 23′-mer, which left the 5′ pppN unpaired and lost most of its activity. Moreover, a 5′ ppp-24′-mer/antisense 24′-mer dsRNA, which contained a 3-nt bulge (starting at position 8 from the 5′ ppp-end) was also highly active, indicating that symmetrical bulges within the dsRNA region were well tolerated. In contrast, Schmidt et al. (15) reported that a double-strand encompassing the 5′ pppN as short as 10 bp was sufficient, and that whether this double-strand included the 5′ pppN was not critical; a 5′ ppp19′-mer annealed to a 18′-mer, which left the 5′ pppN unpaired and appeared to be fully active.
All arenavirus genomes (such as TCRV, lymphocytic choriomeninghitis virus, Junin virus and Lassa fever virus (LV)) contain at least 19 nt at their termini that are complementary, and many of their genome segments also contain 2 mismatches (e.g. at positions 7 and 9 from the unpaired 5′ end, or positions 6 and 8 from the base-paired 3′ end; Fig. 7) (20–26). These mismatches would create a symmetrical 3-nt bulge within the panhandle when the terminal sequences anneal to form dsRNA. This sequence complementarity and the mismatches presumably reflect the similarity and subtle differences, respectively, of the genomic and antigenomic promoters. We first examined the abilities of 5′ ppp-ssRNA1** (54′-mer) annealed with 18′-mers complementary to positions 1–18, 2–19, and 4–21 (relative to the 5′ ppp-end of RNA1) to activate the IFNβ promoter when transfected into A549 cells. As shown in Fig. 5A, displacement of the dsRNA region from the 5′ ppp end by even a single nucleotide was sufficient to reduce most of the activity, and displacement by 3 nt eliminated the remainder. The ability of 5′ ppp-ssRNA1**/1–18 to activate IFNβ appeared to depend on RIG-I, as this hybrid was inactive in A549 RIG-I knockdown cells (27)(Fig. 5B). 5′ ppp-ssRNA1** annealed with a 20′-mer complementary to positions 1–20 was 3-fold more active in activating the IFNβ promoter than that annealed with the analogous (1–18) 18′-mer (albeit in a separate experiment), whereas 5′ ppp-ssRNA1** annealed with a 20′-mer complementary to positions 2–21 showed no activity at all (Fig. 6, line 8 versus line 3). Thus, similar to the results of Schlee et al. (14), the dsRNA region needs to encompass the 5′ nt carrying the triphosphate to maximize its ability to activate RIG-I.
FIGURE 7.
The effect of a 3-nt bulge plus the presence (or not) of the 5′ pppN within dsRNA of modified Junin virus RNA on PAMP function. A, the sequence of the 5′ 61 nucleotides of the Junin virus S genome RNA is shown (5′ to 3′, top line), in which capital letters indicate uridines that were changed to the nucleotides shown to generate Jun −1/60mod RNA. The bottom line shows the sequence of the 3′ terminal 20 nucleotides of the S genome RNA (3′ to 5′). The mismatches in the dsRNA panhandle structure are indicated. B, 5′ ppp Jun −1/60mod RNA, RNase III treated (light gray) or not (dark gray), was annealed (or not, lane 7) with synthetic oligonucleotides representing positions −1 or +1 to 20 of the S genome panhandle as complementary RNA, maintaining the 3-nt bulge (lines 8 and 9, respectively), or oligonucleotides in which positions 5 and 7 were changed so that a perfect dsRNA panhandle would be formed to resemble to L segment (lines 10 and 11, respectively). The hybridized RNA (see supplemental Fig. S5B) were then transfected into A549 cells, and their ability to activate the IFNβ promoter was determined. 1 μg of 5′ ppp Jun −1/60mod RNA was transfected in all cases. Lane 1 (Ctl) shows the level of IFNβ activation in untransfected cells. Transfection of tRNA by itself is shown in lane 6. Transfection of the oligonucleotides by themselves is shown in lanes 2–5.
5′ ppp-ssRNA1 was also annealed with 20′-mers complementary to positions 1–20 or 2–21, which did or did not contain 2 mismatches at positions 5 and 7, 6 and 8, 7 and 9, or 8 and 10. As shown in Fig. 6, dsRNA regions that included the 5′ nt carrying the triphosphate and contained 2 mismatches progressively lost activity as the mismatches approached the 5′ ppp-end (Fig. 6, lines 4–7 versus line 3). Inclusion of 2 mismatches within dsRNAs that did not include the 5′ pppN did not further reduce the activity as without mismatches, which was already at background levels. Thus, an unbroken string of at least 6 base pairs including and following the 5′ ppp end appears to be required for 5′ ppp-dsRNA to activate the IFNβ promoter.
More Natural Arenavirus dsRNA Panhandle Structures Are Also Poor PAMPs
dsRNAs are known to differ in their ability to induce IFN. Poly(I)·poly(C), for example, is known to be a much stronger inducer of IFN than dsRNAs composed of other homopolymers (28). A dsRNA based on RNA1, containing mostly adenosines and cytosines on one strand, might also have unusual properties in its ability to interact with and activate RIG-I. It was therefore important to examine the ability of dsRNAs based on bona fide arenavirus panhandle structures to activate the IFNβ promoter.
As mentioned above, the terminal 19 nt of all arenavirus genome segments are highly conserved and complementary, such that the genome RNA termini form a dsRNA panhandle. Roughly half of all these genome segments contain a perfectly complementary dsRNA panhandle of around 19 bp in length, whereas the other half (including the Junin virus S segment) contain two mismatches that lead to a dsRNA with a symmetrical 3-nt bulge (20)(Fig. 7A). We first prepared a 61-nt RNA representing the 5′ end of the Junin virus S segment RNA (Jun −1/60), using T7 RNA polymerase as before. Jun −1/60 RNA, similar to model RNA1, continued to activate the IFNβ promoter upon transfection into cells, even after its isolation as full-length RNA on denaturing PAGE. However, in contrast to RNA1*, which was almost completely resistant to RNase III digestion after PAGE purification (Fig. 2A), >95% of Jun −1/60 RNA* was sensitive to RNase III digestion under similar conditions (not shown).
The reasons for this unexpected sensitivity to RNase III are unclear. Typical Escherichia coli RNase III substrates are cellular or viral RNAs that have two complementary segments that can fold back and form dsRNA regions. Although RNase III cleaves both strands of completely dsRNA in vitro, many natural substrates have ssRNA loops and bulges in addition to regions of dsRNA, and RNase III can also cleave the RNA within these ssRNA regions (29, 30). Model RNA1 is composed mostly of adenosines and cytosines so that it would contain little or no 2° structure and this presumably accounts for its resistance to RNase III. We therefore modified the sequence of Jun −1/60 RNA to minimize the possibilities for 2° structure. As there are only 2 uridines within the 19 5′ nt that form the dsRNA panhandle, these 2 uridines were changed to adenosines (to maintain dsRNA thermal stability), and the remaining 19 uridines were changed to adenosines or cytosines (Fig. 7A). Modified Jun −1/60 RNA (Jun −1/60mod RNA) cannot form A:U or G:U base pairs and so its 2° structures should be reduced. In addition, RNA synthesis in vitro can be carried out in the absence of UTP, which may eliminate the need to highly purify the RNA before transfection (14, 15).
In strong contrast to bona fide Jun −1/60 RNA, minimally purified Jun −1/60mod RNA was essentially resistant to RNase III digestion (supplemental Fig. S5A). Half of the preparation was nevertheless digested with RNase III as a control. Neither Jun −1/60mod RNA alone activated the IFNβ promoter upon transfection (Fig. 7B, lane 7), similar to tRNA used as a negative control (Fig. 7B, lane 6). In addition, transfection of oligonucleotides complementary to positions −1 to 20 of Jun −1/60mod RNA by themselves did not activate the IFNβ promoter (lanes 2–5). Both Jun −1/60mod RNAs, which were annealed to a 5′ OH RNA complementary to positions −1 to 20, except that it maintained the 2 mismatches present in the Junin S segment panhandle, induced IFNβ reporter gene activity 9–11-fold over background levels (lane 8), whereas both Jun −1/60mod RNAs annealed to an RNA exactly complementary to positions −1 to 20 induced IFNβ reporter gene activity 19–23-fold (lane 10). Most importantly, both Jun −1/60mod RNAs annealed to an RNA complementary to positions +1 to 20 that did or did not maintain the 2 mismatches present in the Junin S segment panhandle, did not induce IFNβ reporter gene activity over background levels (lanes 9 and 11, respectively). The formation of the dsRNAs between Jun −1/60mod RNAs and the complementary oligonucleotides was followed by nondenaturing PAGE, and found to be similar in all cases (supplemental Fig. S5B). Thus, similar to results obtained with model RNA1, dsRNAs representing the Junin S segment panhandle containing a 3-nt bulge, or the Junin L segment panhandle containing a perfect dsRNA panhandle (except for the 2 U:A bp inversions), are extremely poor inducers of IFNβ when the pseudo-templated 5′ pppG is not base paired, but naturally present as a single 5′-nt overhang. The 3-nt bulge present in many arenavirus genome segment panhandles also appears to weaken the ability of this structure to induce the activation of IFNβ.
DISCUSSION
Run-off transcription by bacteriophage T7 DNA-dependent RNA polymerase is often used to synthesize oligoribonucleotides (31). These reactions can also synthesize RNAs that are longer than intended when the run-off transcript displays self-complementarity at its 3′ end, and can therefore form intra- or intermolecular primed templates (19, 32). Two recent reports have pointed out that the ability of these in vitro transcripts to induce IFN is in fact due to their dsRNAs side products, e.g. the double-length hairpin structure that results from intramolecular self-priming (14, 15). Shorter than unit length RNAs complementary to the intended transcript have also been reported (14), presumably the result of RNA-coded de novo synthesis. Although these shorter complementary RNAs appeared to be minor side products in the previously reported reactions, they are by far the major side products of our reaction (Fig. 2B). There can be little doubt that our shorter complementary RNAs are the result of RNA1-coded de novo synthesis, as PAGE-purified RNA1 directs the synthesis of precisely the same pattern of predominantly 15–25′-mers present in the DNA-promoted reaction (Fig. 2B). The resemblance of this pattern of 15–25′-mers to a sequence ladder suggests that the T7 DdRP often started at the precise 3′ end of RNA1 (OHCUU….) as 5′ pppGAA is a preferred initiation sequence (31). This RNA1-coded reaction, however, was relatively non-processive. Only 1% of this complementary RNA appears to be full-length, accounting for the 2% of the PAGE-purified RNA1 found as dsRNA upon nondenaturing PAGE. We presume that this minor dsRNA contamination of PAGE-purified RNA1 is so active in inducing IFN (Fig. 3) because it contains two 5′ ppp blunt ends, and RIG-I translocates in both directions on dsRNA (11). T7 DdRP can be forced to uniquely synthesize ssRNA if this RNA contains only 3 bases and the 4th NTP is absent from the reaction (14, 15). Three NTP in vitro RNA synthesis efficiently eliminates synthesis of complementary RNAs, especially when the missing nucleotide is abundant at the 5′ end of the complementary RNA, as for Jun −1/60mod RNA (Fig. 7A). ssRNAs containing only 3 nucleotides also contain less 2° structure, are more resistant to RNase III degradation, and are more dependent on a complementary RNA annealed to the 5′ ppp-end of the in vitro transcript for IFN induction (Fig. 7B). This latter approach allowed us to test almost bona fide arenavirus panhandle structures for their ability to activate the IFNβ promoter.
Using doubly purified 5′ ppp- ssRNA1**, we examined the ability of 18 and 20 bp (perfectly paired) dsRNA regions that left either no or 1 (or in one case three) 5′ nucleotide unpaired to activate the IFNβ promoter. Similar to the results of Schlee et al. (14), we found that displacement of the dsRNA region from the 5′ ppp end by even a single nt clearly reduced this activity (Figs. 5 and 6). We also examined the effect of dsRNA regions, which contained 2 mismatches and included the nucleotide carrying the 5′ triphosphate. We found that the activity of dsRNA regions, which included the 5′ pppN but also contained 2 mismatches depended on the proximity of the mismatches to the 5′ ppp end. Mismatches that left only 4 or 5 bp following the 5′ ppp end were essentially inactive, but those containing 6 or 7 base pairs were increasingly active (Fig. 6). Regions of dsRNA 20 bp in length that did not include the 5′ ppp end were essentially inactive whether or not they contained the two mismatches that are found in some arenavirus genome segments. Similar results were obtained using Jun −1/60mod RNA, in which the dsRNAs formed to mimic arenavirus genome panhandles were almost exact copies of Junin virus, the etiological agent of Argentine hemorrhagic fever, and a member of the Tacaribe serogroup of arenaviruses. dsRNAs in which the 5′ ppp-nt was base paired were clearly active, and those containing a perfect dsRNA were clearly more active than those containing the 3-nt bulge. Moreover, dsRNAs in which the 5′ ppp-nt was present as a single nucleotide overhang were completely inactive, whether or not they contained the 3-nt bulge. Thus, the absence of a base-paired 5′ pppN end may be sufficient for arenavirus genomes to avoid being recognized as a PAMP by RIG-I, when these ends apparently form dsRNA panhandles within arenavirus nucleocapsids (see below).
The prime and realign manner of initiating viral genome replication is not unique to RNA viruses. For DNA viruses such as Φ29 (33), PRD1 (34), and adenovirus (35), a “jumping back” mechanism has been described in which the DNA primer is itself attached to a “terminal protein” primer, and where this mechanism is thought to restore small terminal deletions and errors. A somewhat similar mechanism is also used by picornaviruses, where a viral protein (VPg) is used as a primer for VPg-pUpUOH synthesis templated from an adenosine within a critical replication element, which can be located almost anywhere in the viral genome. VPg-pUpUOH is then realigned with the genome 3′ poly(A) tail, or the antigenome 3′ terminal OHAA… before the primer is extended for genome replication (36, 37). In all of these cases, as well as that proposed for influenza A virus genome synthesis (5), the oligonucleotide primer is realigned with the precise 3′ end of the genome or antigenome, thus maintaining genome length and the terminal sequences. However, in the case of TCRV (and presumably other arenaviruses), the primer is realigned on its template such that its 5′ pppN end is at position −1, and at the same time, this 5′ pppN end is not copied when it is part of a template (Fig. 1); presumably because these genome 3′ ends are determined by specific termination signals rather than run-off synthesis. This arrangement not only maintains genome length and the terminal sequences, as well as being able to restore small terminal deletions and errors, it leads to dsRNAs in which the 5′ pppN end is not blunt, but contains a single unpaired 5′ nt when the genome termini anneal.
Viruses counteract the cellular innate immune response by producing IFN antagonists and/or by limiting their PAMP exposure. The latter approach may be particularly important for arena- and bunyaviruses, because the complementary sequences at their genome ends apparently form dsRNA even within their helical RNP nucleocapsids. Unlike nonsegmented negative strand RNA viruses nucleocapsids, which are linear structures in the EM even when there are 109 perfectly complementary nt at each end (e.g. SeV DI-H4) (38), arena- and bunyaviruses nucleocapsids are circular, presumably due to the annealing of their complementary genome ends (39–41). For LaCrosse bunyavirus, the RNA within viral NCs can be cross-linked at high efficiency with psoralen, precisely at the positions expected if the genome RNA ends formed true dsRNA (42). Thus, we expect that the presence of a monophosphate rather than a triphosphate at the 5′ end of the HTN genome, as well as the presence of a single unpaired 5′ pppN of arenavirus genomes, helps these viruses limit their PAMP exposure. The presence of two mismatches within the terminal 19 nt of some arenavirus genome segments may also play a role in reducing their recognition as PAMPs. The bulge in the dsRNA panhandle of some arenavirus genome segments leaves only 5 bp between the bulge and the unpaired 5′ ppp end, and this can be expected to lower the stability of this short dsRNA region. Also, as RIG-I is thought to translocate on dsRNA so that (at least in part) its C-terminal regulatory domain can interact with a 5′ triphosphate group, a 3-nt bulge within the arenavirus dsRNA panhandle may interfere with this translocation. The precise position of the bulge within the dsRNA relative to the 5′ ppp end may be important in this respect as well.
Supplementary Material
Acknowledgments
We thank Daniel Pinschewer and Stephane Hausmann (Geneva) and Olke Uhlenbeck (Evanston) for useful discussions.
This work was supported by a grant from the Swiss National Science Fund.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- RdRP
- RNA-dependent RNA polymerase
- TCRV
- tacaribe arenavirus
- PAMP
- pathogen-associated molecular pattern
- dsRNA
- double-stranded RNA
- HTN
- hantaan (bunya)virus
- IFN
- interferon
- ssRNA
- single-stranded RNA
- nt
- nucleotide
- Jun −1/60mod RNA
- modified Jun −1/60 RNA.
REFERENCES
- 1.Garcin D., Kolakofsky D. (1990) J. Virol. 64, 6196–6203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garcin D., Kolakofsky D. (1992) J. Virol. 66, 1370–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garcin D., Lezzi M., Dobbs M., Elliott R. M., Schmaljohn C., Kang C. Y., Kolakofsky D. (1995) J. Virol. 69, 5754–5762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prehaud C., Lopez N., Blok M. J., Obry V., Bouloy M. (1997) Virology 227, 189–197 [DOI] [PubMed] [Google Scholar]
- 5.Deng T., Vreede F. T., Brownlee G. G. (2006) J. Virol. 80, 2337–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takeuchi O., Akira S. (2007) Immunol. Rev. 220, 214–224 [DOI] [PubMed] [Google Scholar]
- 7.Yoneyama M., Kikuchi M., Natsukawa T., Shinobu N., Imaizumi T., Miyagishi M., Taira K., Akira S., Fujita T. (2004) Nat. Immunol. 5, 730–737 [DOI] [PubMed] [Google Scholar]
- 8.Pichlmair A., Schulz O., Tan C. P., Näslund T. I., Liljeström P., Weber F., Reis e Sousa C. (2006) Science 314, 997–1001 [DOI] [PubMed] [Google Scholar]
- 9.Hornung V., Ellegast J., Kim S., Brzózka K., Jung A., Kato H., Poeck H., Akira S., Conzelmann K. K., Schlee M., Endres S., Hartmann G. (2006) Science 314, 994–997 [DOI] [PubMed] [Google Scholar]
- 10.Kato H., Takeuchi O., Sato S., Yoneyama M., Yamamoto M., Matsui K., Uematsu S., Jung A., Kawai T., Ishii K. J., Yamaguchi O., Otsu K., Tsujimura T., Koh C. S., Reis e Sousa C., Matsuura Y., Fujita T., Akira S. (2006) Nature 441, 101–105 [DOI] [PubMed] [Google Scholar]
- 11.Myong S., Cui S., Cornish P. V., Kirchhofer A., Gack M. U., Jung J. U., Hopfner K. P., Ha T. (2009) Science 323, 1070–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takahasi K., Yoneyama M., Nishihori T., Hirai R., Kumeta H., Narita R., Gale M., Jr., Inagaki F., Fujita T. (2008) Mol. Cell 29, 428–440 [DOI] [PubMed] [Google Scholar]
- 13.Cui S., Eisenächer K., Kirchhofer A., Brzózka K., Lammens A., Lammens K., Fujita T., Conzelmann K. K., Krug A., Hopfner K. P. (2008) Mol. Cell 29, 169–179 [DOI] [PubMed] [Google Scholar]
- 14.Schlee M., Roth A., Hornung V., Hagmann C. A., Wimmenauer V., Barchet W., Coch C., Janke M., Mihailovic A., Wardle G., Juranek S., Kato H., Kawai T., Poeck H., Fitzgerald K. A., Takeuchi O., Akira S., Tuschl T., Latz E., Ludwig J., Hartmann G. (2009) Immunity 31, 25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmidt A., Schwerd T., Hamm W., Hellmuth J. C., Cui S., Wenzel M., Hoffmann F. S., Michallet M. C., Besch R., Hopfner K. P., Endres S., Rothenfusser S. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 12067–12072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Habjan M., Andersson I., Klingström J., Schümann M., Martin A., Zimmermann P., Wagner V., Pichlmair A., Schneider U., Mühlberger E., Mirazimi A., Weber F. (2008) PLoS ONE 3, e2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.King P., Goodbourn S. (1994) J. Biol. Chem. 269, 30609–30615 [PubMed] [Google Scholar]
- 18.Strahle L., Marq J. B., Brini A., Hausmann S., Kolakofsky D., Garcin D. (2007) J. Virol. 81, 12227–12237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cazenave C., Uhlenbeck O. C. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 6972–6976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Albariño C. G., Bergeron E., Erickson B. R., Khristova M. L., Rollin P. E., Nichol S. T. (2009) J. Virol. 83, 5606–5614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iapalucci S., López N., Rey O., Zakin M. M., Cohen G. N., Franze-Fernández M. T. (1989) Virology 173, 357–361 [DOI] [PubMed] [Google Scholar]
- 22.Lukashevich I. S., Patterson J., Carrion R., Moshkoff D., Ticer A., Zapata J., Brasky K., Geiger R., Hubbard G. B., Bryant J., Salvato M. S. (2005) J. Virol. 79, 13934–13942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen M., Lan S., Ou R., Price G. E., Jiang H., de la Torre J. C., Moskophidis D. (2008) J. Gen. Virol. 89, 1421–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flatz L., Bergthaler A., de la Torre J. C., Pinschewer D. D. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 4663–4668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salvato M. S., Shimomaye E. M. (1989) Virology 173, 1–10 [DOI] [PubMed] [Google Scholar]
- 26.Salvato M., Shimomaye E., Southern P., Oldstone M. B. (1988) Virology 164, 517–522 [DOI] [PubMed] [Google Scholar]
- 27.Marq J. B., Hausmann S., Luban J., Kolakofsky D., Garcin D. (2009) J. Biol. Chem. 284, 25471–25478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Colby C., Chamberlin M. J. (1969) Proc. Natl. Acad. Sci. U.S.A. 63, 160–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robertson H. D. (1982) Cell 30, 669–672 [DOI] [PubMed] [Google Scholar]
- 30.MacRae I. J., Zhou K., Doudna J. A. (2007) Nat. Struct. Mol. Biol. 14, 934–940 [DOI] [PubMed] [Google Scholar]
- 31.Milligan J. F., Uhlenbeck O. C. (1989) Methods Enzymol. 180, 51–62 [DOI] [PubMed] [Google Scholar]
- 32.Triana-Alonso F. J., Dabrowski M., Wadzack J., Nierhaus K. H. (1995) J. Biol. Chem. 270, 6298–6307 [DOI] [PubMed] [Google Scholar]
- 33.Méndez J., Blanco L., Esteban J. A., Bernad A., Salas M. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 9579–9583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caldentey J., Blanco L., Bamford D. H., Salas M. (1993) Nucleic Acids Res. 21, 3725–3730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.King A. J., van der Vliet P. C. (1994) EMBO J. 13, 5786–5792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paul A. V., Yin J., Mugavero J., Rieder E., Liu Y., Wimmer E. (2003) J. Biol. Chem. 278, 43951–43960 [DOI] [PubMed] [Google Scholar]
- 37.Steil B. P., Barton D. J. (2009) Virus Res. 139, 240–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kolakofsky D. (1976) Cell 8, 547–555 [DOI] [PubMed] [Google Scholar]
- 39.Obijeski J. F., Bishop D. H., Palmer E. L., Murphy F. A. (1976) J. Virol. 20, 664–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palmer E. L., Obijeski J. F., Webb P. A., Johnson K. M. (1977) J. Gen. Virol. 36, 541–545 [DOI] [PubMed] [Google Scholar]
- 41.Pettersson R. F., von Bonsdorff C. H. (1975) J. Virol. 15, 386–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raju R., Kolakofsky D. (1989) J. Virol. 63, 122–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hausmann S., Marq J. B., Tapparel C., Kolakofsky D., Garcin D. (2008) PLoS ONE 3, e3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.