Abstract
The mammalian B-cell receptor-associated proteins of 29 and 31 kDa (BAP29 and BAP31) are conserved integral membrane proteins that have reported roles in endoplasmic reticulum (ER) quality control, ER export of secretory cargo, and programmed cell death. In this study we investigated the yeast homologs of BAP29 and BAP31, known as Yet1p and Yet3p, to gain insight on cellular function. We found that Yet1p forms a complex with Yet3p (Yet complex) and that complex assembly was important for subunit stability and proper ER localization. The Yet complex was not efficiently packaged into ER-derived COPII vesicles and therefore does not appear to act as an ER export receptor. Instead, a fraction of the Yet complex was detected in association with the ER translocation apparatus (Sec complex). Specific mutations in the Sec complex or Yet complex influenced these interactions. Moreover, associations between the Yet complex and Sec complex were increased by ER stress and diminished when protein translocation substrates were depleted. Surprisingly, yet1Δ and yet3Δ mutant strains displayed inositol starvation-related growth defects. In accord with the biochemical data, these growth defects were exacerbated by a combination of certain mutations in the Sec complex with yet1Δ or yet3Δ mutations. We propose a model for the Yet-Sec complex interaction that places Yet1p and Yet3p at the translocation pore to manage biogenesis of specific transmembrane secretory proteins.
Keywords: Cell/trafficking, Membrane/Biogenesis, Membrane/Function, Membrane/Proteins, Membrane/Trafficking, Protein/Sorting, Protein/Translocation, Subcellular Organelles/Endoplasmic Reticulum
Introduction
Nascent polypeptides enter the secretory pathway via the endoplasmic reticulum (ER)2-localized Sec61 protein translocation channel. Translocation into the ER provides access to a number of enzymes that facilitate secretory protein biogenesis including signal peptidase, luminal chaperones, core glycosylation machinery, and thiol oxidases. Several proteins associate with the Sec61 translocon, apparently coordinating aspects of protein biogenesis with translocation (1, 2). The core translocation apparatus (Sec61 complex) in the yeast, Saccharomyces cerevisiae, consists of Sec61p, Sss1p, and Sbh1p (3–5). The Sec63 complex, comprised of Sec63p, Sec62p, Sec71p, and Sec72p, associates with the Sec61 complex to form the Sec complex, and together with the luminal chaperone Kar2p has a role in post-translational translocation (6). Sec63p and Kar2p also appear to be important for co-translational translocation (7, 8).
The fidelity of secretory protein biogenesis is essential to ensure that only properly folded/assembled protein molecules exit the ER in COPII-coated vesicles. Accordingly, incorrectly folded or assembled proteins are retained in the ER, and are either fully folded and exported in COPII vesicles or are retrotranslocated to the cytoplasm for proteasomal degradation in a process known as ER-associated degradation (ERAD) (reviewed in Ref. 9). Establishing a role for the Sec61 translocon in ERAD has been confounded by anterograde translocation defects associated with most sec61 alleles, although data supporting such a role for Sec61p continues to emerge (10–13).
The mammalian protein BAP31 and its paralog BAP29 are ubiquitously expressed residents of the early secretory pathway originally identified in association with membrane-bound immunoglobulin D molecules (mIgD) (14). In the past decade and a half, numerous studies have found BAP31 associated with various transmembrane proteins, with reported effects on secretory protein biogenesis including ER export (e.g. cellubrevin and major histocompatibility complex I) (15–17), ER retention (e.g. mIgD) (18), and degradation (e.g. CFTRΔF508) (19, 20). BAP31 and BAP29 appear to be conserved across eukaryotic species, suggesting preserved function for these proteins. However, the underlying mechanism by which BAP31 proteins act in ER secretory protein biogenesis remains unclear. S. cerevisiae possess three sequence homologs of BAP31 known as Yet1p, Yet2p, and Yet3p. Like BAP31, these proteins are predicted to have three transmembrane segments with a cytoplasmic, coiled-coil C-terminal domain (21). Deletion of the YET genes does not affect yeast cell viability under standard laboratory conditions and little is known about Yet protein function, although YET3 has been reported to be important for invertase secretion (21).
In this study, we used biochemical and genetic approaches to investigate the function of Yet1p and Yet3p. Our results indicate that Yet1p forms a complex with Yet3p and that a fraction of this Yet1p-Yet3p complex is associated with the Sec complex. Moreover, we show that the level of Yet-Sec complex association is influenced by ER stress (induced by DTT, inositol starvation, and IRE1 deletion), by the availability of translocation substrates, and by mutations in either the Yet or Sec complexes. Our data supports a model that places the Yet1p-Yet3p complex at the ER translocon to interact with translocation substrates. Moreover, we find that Yet1p and Yet3p are required for robust growth in the absence of inositol, suggesting a role for Yet1p and Yet3p in the biogenesis or regulation of specific components involved in inositol synthesis.
EXPERIMENTAL PROCEDURES
Yeast Strains and Media
Yeast strains used in this study are listed in supplemental Table S1. All C-terminal epitope tagging and deletion of YET1 (CBY2613) was achieved using the described methods (22). The functionality of chromosomally tagged Yet1p and Yet3p (HA and GFP) was tested by growth in the absence of inositol and found to be similar to untagged control. To construct CBY0310, YET1-MYC/pRS306 was integrated at ura3. CBY2791 (yet3Δ with natMX4 marker) was generated using p4339 as described (23). For Yet3pΔCT-HA (CBY2815), the cassette containing the HA epitope was integrated 207 nucleotides upstream of the YET3 stop codon removing the C-terminal 69 amino acids (six amino acids after last predicted transmembrane domain). To generate CBY2614 (sec61-2 in FY834) and CBY2708 (sec63-1 in BY4741) the “PCR-mediated integration of conditional allele” method was used (24). Briefly, the sec61-2 open reading frame (ORF) and 299 nucleotides downstream of the stop codon were amplified from RSY533 (25). In a separate reaction, the natMX4 cassette was amplified from p4339 (23). Primers were designed with 5′ sequences to direct homologous recombination so that the sec61-2 ORF with 299 3′ nucleotides directly followed by the natMX4 cassette would replace the wild-type SEC61 ORF. The resulting PCR products were transformed into FY834 and transformants were selected for nourseothricin (clonNAT, Werner BioAgents, Jena, Germany) resistance and temperature sensitivity (37 °C) characteristic of cells harboring the sec61-2 allele. Sequencing was used to confirm the presence of the sec61-2 allele (G213D). For construction of sec63-1 (RSY151) in BY4741, the sec63-1 ORF and 458 nucleotides downstream of the stop codon were amplified from RSY151 (26). Otherwise, the method was comparable with that used for the sec61-2. Sequencing was used to confirm the presence of the sec63-1 allele (A179T). Yeast transformations were performed using the lithium acetate technique (27).
Yeast were grown at 30 °C in 1% yeast extract, 1% peptone, 2% dextrose (YPD) medium unless otherwise noted. For plasmid selection, yeast were grown in 0.67% yeast nitrogen base without amino acids, 2% dextrose, and appropriate dropout supplements (YMD). For inositol starvation growth assays (see Fig. 7 and supplemental Fig. S4), strains were grown overnight in YMD (see Fig. 7A with plasmid selection). After washing with sterile water, strains were plated on YMD with or without 75 μm inositol (no plasmid selection) and grown at the indicated temperature. Cells used in supplemental Fig. S3B were grown to early log phase in 0.67% yeast nitrogen base (without inositol) with complete supplement (Bio 101, Inc.), 2% dextrose, and 75 μm inositol (CSMD). Cells were washed extensively in sterile deionized water and then grown for another 3 h in either CSMD without inositol or CSMD with 75 μm inositol before harvesting. All defined growth media lacked choline.
FIGURE 7.
Influence of inositol starvation on the growth of yet1Δ or yet3Δ mutants and synthetic growth defects of yet sec double mutants. A, cells were grown overnight in minimal medium containing inositol (YMD) with plasmid selection. After washing in water, cells were plated in 10-fold serial dilutions on YMD (without plasmid selection) either with (75 μm) or without inositol (INO) and grown at the indicated temperature for 4 days. Strains are CBY2913 (yet1Δ + pRS426), CBY2986 (yet1Δ + YET1/pRS423), CBY2987 (yet1Δ + YET3/pRS426), CBY2905 (yet3Δ + pRS426), CBY2992 (yet3Δ + YET1/pRS423), and CBY2991 (yet3Δ + YET3/pRS426). B and C, cells were grown overnight in minimal medium containing the required supplements and inositol (75 μm). Cells were then washed and plated as in A and grown for 5 days at 30 °C. Strains in B are wild-type (BY4742), sec63-1 (CBY2708), yet1Δ (CBY2198), yet1Δ sec63-1 (CBY3004), yet3Δ (CBY2200), and yet3Δ sec63-1 (CBY3005); C, wild-type (CBY2651), sec61-2 (CBY2650), yet1Δ (CBY2649), and yet1Δ sec61-2 (CBY2648).
Plasmid Construction
For synthesis of plasmid inserts, genomic DNA from BY4742 was used as the template. For the construction of YET1/pRS423, YET1 including 354 nucleotides upstream of the ORF and 348 nucleotides downstream of the ORF was amplified and cloned into SpeI and EcoRI sites in the polylinker of pRS423 (28). YET3/pRS426 was constructed using yeast-based homologous recombination (29). YET3 ORF plus 300 nucleotides upstream and 317 nucleotides downstream of the ORF and the appropriate homologous sequence was amplified and transformed into yeast with pRS426 linearized with HindIII. To generate YET1-MYC/pRS306 for CBY0310, a fragment consisting of the YET1 sequence including 210 nucleotides upstream to 223 nucleotides downstream of the ORF with a single MYC epitope inserted 60 nucleotides upstream of the stop codon was placed in pRS306 (30). For the construction of SEC71/pRS425, SEC71 including 508 nucleotides upstream and 349 nucleotides downstream of the ORF was amplified and cloned into SpeI and BamHI polylinker sites of pRS425. For SEC63/pRS425, SEC63 including 501 nucleotides upstream and 458 nucleotides downstream of the ORF was amplified and cloned into NotI and SacII sites of pRS425.
Antibodies
Antiserum directed against Yet1p (21), Sec63p (31), Sec61p (32), Sec71p (33), Erv41p (34), Kar2p (35), Sec22p (36), and Sec12p (37) have been described previously. The sheep anti-mouse and donkey anti-rabbit secondary ECL horseradish peroxidase-linked antibodies (GE Healthcare) and monoclonal antibodies against c-myc (9E10, Covance) and HA (HA.11, Covance) are available commercially. Yet3p antibodies were raised against a recombinant fusion protein consisting of the C-terminal 65 amino acids of Yet3p fused to glutathione S-transferase in pGEX-2T (GE Healthcare). The glutathione S-transferase-Yet3p fusion protein was expressed and purified using an Escherichia coli-based system according to the manufacturer's instructions (GE Healthcare) and used to raise polyclonal antiserum in rabbits (Covance).
Yeast Cell Lysates and Immunoblotting
Lysates were derived from ∼1 A600 equivalent of yeast cells and prepared using the glass bead-beater method in JR lysis buffer (20 mm HEPES, pH 7.4, 0.1 m sorbitol, 50 mm KOAc, 2 mm EDTA, 1 mm phenylmethylsulfonyl fluoride). Lysates were then cleared by centrifugation for 3 min at 1,000 × g at 4 °C to create low speed supernatants. For the lysates shown in Fig. 1A, this low speed supernatant was mixed 2:1 with 5× reducing sample buffer, heated for 6 min at 75 °C, and resolved by SDS-PAGE. For the lysates shown in Fig. 2A, low speed supernatants were further centrifuged for 10 min at 16,000 × g at 4 °C. The resulting pellets were dissolved in 5× reducing sample buffer, heated for 6 min at 75 °C, and resolved by SDS-PAGE. Immunoblots were developed with SuperSignal chemiluminescent substrate (Pierce), imaged with a CCD camera (UVP BioImaging) and quantified with LabWorks software (UVP BioImaging). Statistical analyses (Figs. 5 and 6 and supplemental Fig. S3) were performed with Excel (Microsoft).
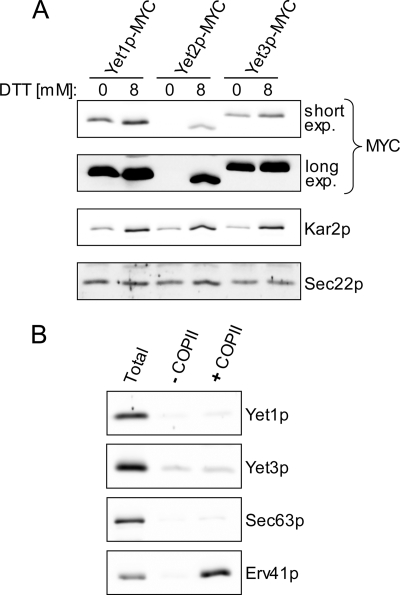
FIGURE 1.
Yet protein expression and COPII vesicle incorporation. A, log phase cells with chromosomally MYC-tagged versions of Yet1p (CBY2291), Yet2p (CBY2615), and Yet3p (CBY2661) were treated with or without 8 mm DTT for 2 h before harvesting. Whole cell lysates were generated and immunoblotted. Longer exposures (long exp.) indicate the identical immunoblot captured on a CCD camera after an increased exposure time. B, wild-type (BY4742) microsomes were incubated in the presence (+COPII) or absence (−COPII) of purified coat proteins and the vesicle fraction analyzed by immunoblot. The Total lane represents one-tenth of the reaction material from which the COPII vesicles were isolated.
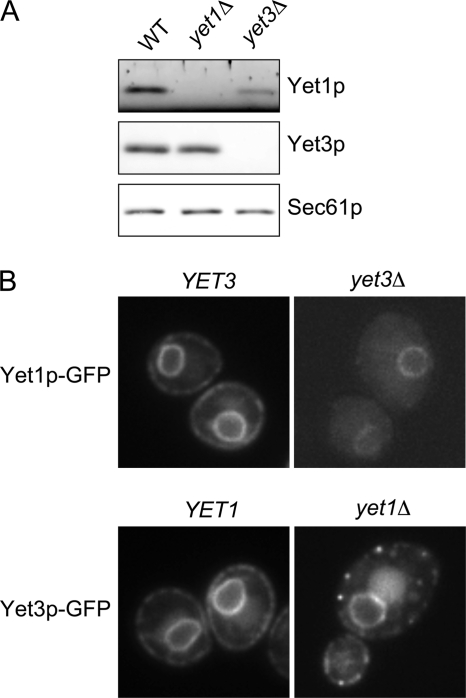
FIGURE 2.
Influence of yet1Δ and yet3Δ mutations on stability and localization of Yet3p and Yet1p, respectively. A, wild-type (BY4742), yet1Δ (CBY2198), and yet3Δ (CBY2200) strains were harvested in log phase. Whole cell lysates from these strains were centrifuged at 16,000 × g for 10 min and the resulting pellets were analyzed by immunoblot. B, live cell GFP fluorescence imaging was performed on overnight cultures expressing Yet1p-GFP (wild-type (WT), CBY2727; yet3Δ CBY2799) or Yet3p-GFP (wild-type, CBY2728; yet1Δ, CBY2800) strains. For clarity, image intensity levels of both deletion strains were increased to be comparable with wild-type controls. Image boxes are 10 × 10 μm.
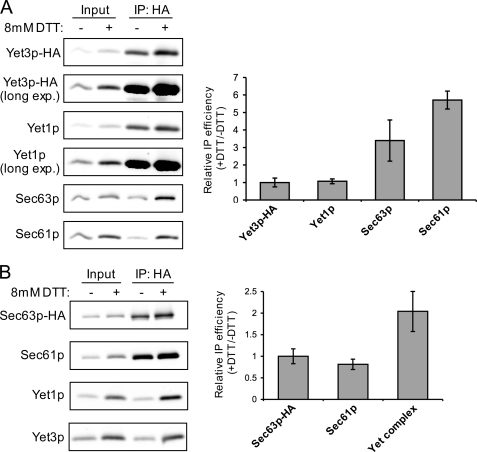
FIGURE 5.
Yet-Sec complex interaction is enhanced by DTT-induced ER stress. A, semi-intact cells from untagged (CBY2615) or Yet3p-HA (CBY2647) strains were produced from log phase cells treated with or without 8 mm DTT for 2 h prior to harvest. HA immunoprecipitations were performed and analyzed from these semi-intact cells as described in the legend to Fig. 3B. B, semi-intact cells from untagged (BY4742) and Sec63p-HA (CBY2733) strains were generated and used for HA immunoprecipitations as in A. For both A and B, immunoblots were quantified and immunoprecipitation efficiency of each protein was calculated by dividing the amount in the IP lane by the amount in the respective Input lane. Data are plotted as the mean of the ratio of +DTT IP efficiency over −DTT IP efficiency and are normalized to an IP efficiency of unity for Yet3p-HA (A) or Sec63p-HA (B). Error bars represent the S.D. with n = 3 except n = 5 for Yet complex (n = 3 for Yet1p and n = 2 for Yet3p) in B.
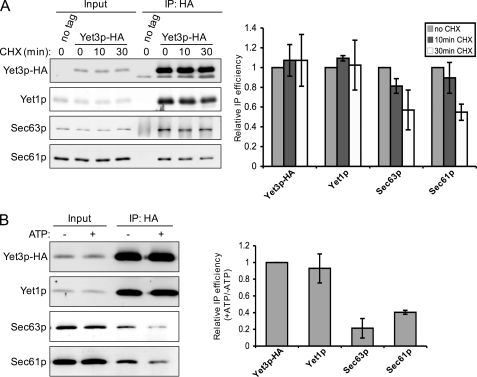
FIGURE 6.
Yet-Sec complex interaction is diminished by treatments that purge translocation substrates. A, semi-intact cells were produced from untagged (CBY2615) or Yet3p-HA (CBY2647) strains, which were treated with or without 0.1 mg/ml of cycloheximide (CHX) 10 or 30 min prior to harvesting. HA immunoprecipitations were performed and analyzed as described in the legend to Fig. 3B. Quantification data from immunoblots are plotted as the mean of the ratio of immunoprecipitation over input, normalized to the no CHX condition (relative IP efficiency). B, semi-intact cells from untagged (CBY740) or Yet3p-HA (CBY2647) strains were washed and incubated with or without an ATP regeneration system as described under “Experimental Procedures” prior to HA immunoprecipitation as in A. Immunoblot data are plotted as the mean of the ratio of +ATP IP efficiency over −ATP IP efficiency (as in Fig. 5), normalized to an IP efficiency of unity for Yet3p-HA. Error bars in both A and B represent the standard deviation with n = 3. For A, p values of <0.05 were achieved between Yet1p and Sec63p, and Yet1p and Sec61p at the 30-min time point (Student's t test, two tailed, paired).
Immunoprecipitation and Mass Spectrometry
For immunoprecipitation experiments, ∼7.5 A280 units of semi-intact cell membranes (38) were warmed to room temperature with 1 volume (200 μl) of B88 (20 mm HEPES, pH 7.0, 0.25 m sorbitol, 0.15 m KOAc, 5 mm MgOAc) and then solubilized with 2 volumes (0.8 ml) of B88-8 (B88, pH 8.0) containing 1% digitonin (Calbiochem) and 1 mm phenylmethylsulfonyl fluoride for 15 min at room temperature. Digitonin-insoluble material was cleared by centrifugation at 16,000 × g for 5 min at room temperature. The resulting supernatant (1 ml), i.e. “Input,” was used for immunoprecipitation with 1 μl of anti-HA (Covance 11) or 1 μl of anti-myc (Covance 9E10) monoclonal antibodies and 30 μl of 20% protein A-Sepharose (GE Healthcare). After immunoprecipitation reactions were incubated at 4 °C with rotation for 2 h, beads were washed 4 times with 1 ml of ice-cold B88-8 containing 0.05% digitonin. One volume of 5× reducing sample buffer (20 μl) was added, samples were heated for 6 min at 75 °C, and subjected to SDS-PAGE and immunoblot analysis. Input lanes represent 2% of the material used for immunoprecipitation. For immunoprecipitations involving ATP treatment, 16 A280 units of semi-intact cell membranes were washed twice with 1 ml of ice-cold B88. After the second wash, membranes were resuspended in 200 μl of B88 and evenly distributed into two tubes. For ATP treatment of the washed membranes, 80 μl of B88 and 20 μl of a 10× ATP regenerating system (38) containing 1 mm GTP was added. For the minus ATP control, 100 μl of B88 was added. Membranes were incubated at 12 °C for 15 min to stimulate translocation, 0.8 ml of B88-8 with 1% digitonin was then added and immunoprecipitations were performed as described above. The immunoprecipitations used for mass spectrometry analysis were performed from 2 A280 units (∼0.4 mg of protein) of microsomes (39) and were otherwise carried out as described for semi-intact cells. Samples were resolved on a 10% Novex Tris glycine gel and stained with a Colloidal Blue Staining Kit (Invitrogen) or silver stained (40). Bands of interest were excised from the Colloidal Blue-stained gel (not shown) and submitted to ProtTech (Norristown, PA) for reversed phase chromatography and peptide sequence analysis using a quadrupole ion trap mass spectrometer (Thermo, Palo Alto, CA).
In Vitro Vesicle Budding
Experiments to assess the COPII-dependent packaging of Yet1p and Yet3p were performed using microsomes (39) in 250-μl reactions essentially as described (34, 41).
Microscopy
Live cell imaging of Yet1p-GFP and Yet3p-GFP was carried out using an Olympus BX51 microscope with a 100-watt mercury arc lamp, a ×60, 1.4 numerical aperture Olympus Plan Apochromat objective, and a Cooke Sensicam QE CCD camera. Images were captured using IPLab software (BD Biosciences) and whole image level adjustments were made with Photoshop Elements (Adobe).
RESULTS
Expression of Yet Proteins Is Influenced by Reductive ER Stress
Exposure of yeast cells to the reducing agent DTT is known to induce the unfolded protein response (UPR), which up-regulates nearly 400 genes (42). The corresponding proteins are involved in a diverse set of processes, including protein folding, trafficking, and turnover, which are thought to collectively contribute to the clearance of misfolded proteins from the ER. According to microarray data, the YET mRNAs, especially YET2 mRNA, are up-regulated by exposure to DTT (42, 43). It is unknown how changes in YET mRNA levels relate to changes in protein expression. To test the influence of DTT on Yet protein expression, log phase cells expressing MYC epitope-tagged versions of YET1, YET2, or YET3 were treated with 8 mm DTT for 2 h before harvest and immunoblot analysis. Yet2p-MYC protein was undetectable in the absence of DTT but clearly present after DTT exposure, thus documenting a large increase in Yet2p expression upon DTT exposure, consistent with microarray observations (Fig. 1A). Yet1p-MYC and Yet3p-MYC were also up-regulated after DTT treatment, although the magnitude (∼2-fold) was less (also see Fig. 5). Kar2p and Sec22p are included as positive and negative controls, respectively, for the expression effects of DTT (42). In contrast to Yet2p-MYC, Yet1p-MYC and Yet3p-MYC are expressed constitutively at similar levels, which exceed the level of Yet2p-MYC expression even at 8 mm DTT. These observations led us to focus on Yet1p and Yet3p as the major cellular species in subsequent experiments. DTT up-regulation of the Yet proteins suggested a link to protein quality control.
The Yet Proteins Are Not Efficiently Packaged into COPII Vesicles in Vitro
BAP31 appears to localize to the ER, but has also been observed in vesicular elements proposed to be ER-Golgi vesicles or an intermediate compartment, indicative of transport between the ER and Golgi (15). Several studies have suggested that BAP31 influences early (ER-Golgi) trafficking events of transmembrane proteins including cellubrevin and major histocompatibility complex I (15–17). These observations led to the proposal that BAP31 functions as an ER export receptor (15, 44). A salient feature of characterized ER export receptors is efficient incorporation into in vitro generated COPII vesicles (37, 45–47). We reasoned that if the yeast BAP31 homologs acted as ER export receptors they should be efficiently packaged into COPII vesicles. As shown in Fig. 1B, Yet1p and Yet3p were not efficiently packaged into ER vesicles generated with purified COPII proteins. The ER vesicle protein Erv41p and the ER resident protein Sec63p serve as positive and negative controls, respectively. Even when a crude cytosol was used to support budding, Yet1p was not enriched in ER-derived vesicles (data not shown). These observations suggest that the Yet proteins are not actively cycling between the ER and Golgi as reported for other ER export receptors (37, 48). In addition, previous studies have reported a typical ER localization pattern for Yet1p and Yet3p (21, 49), in accord with our COPII budding results and microscopy (Fig. 2B). Taken together, these data suggest that the yeast homologs of BAP31 do not function as ER export receptors, at least not as expected for cycling receptors that traffic in COPII vesicles (reviewed in Ref. 50).
Interdependence of Yet1p and Yet3p for Expression and Localization
We next asked whether removal of YET1 or YET3 would influence the expression and/or localization of the remaining Yet protein. Protein levels were observed by immunoblot of lysates generated from log phase wild-type, yet1Δ, or yet3Δ strains. As shown in Fig. 2A, Yet3p expression was not detectably altered in a yet1Δ mutant although some decrease was observed when Yet3p-HA was expressed in a yet1Δ mutant (Fig. 3C, input lanes). In contrast, Yet1p was strongly destabilized in the yet3Δ strain. We next examined the localization of Yet1p-GFP and Yet3p-GFP in the context of yet3Δ and yet1Δ, respectively. As previously observed (49), both Yet1p-GFP and Yet3p-GFP displayed a typical ER fluorescence pattern (Fig. 2B). However, deletion of either YET3 or YET1 significantly influenced the localization of the remaining GFP-tagged Yet protein. In addition to reduced expression, Yet1p-GFP was absent from cortical regions in the yet3Δ strain. In contrast, the yet1Δ mutation caused the cortical pool of Yet3p-GFP to shift into a punctate pattern. Sec63p-RPF localization in yet1Δ and yet3Δ mutants was indistinguishable from wild-type cells indicating that the Yet1p-GFP and Yet3p-GFP localization defects were not the result of general alterations in ER morphology (data not shown). Together, these results document an interdependence between Yet1p and Yet3p for normal stability and localization and are indicative of co-assembly into a functional complex.
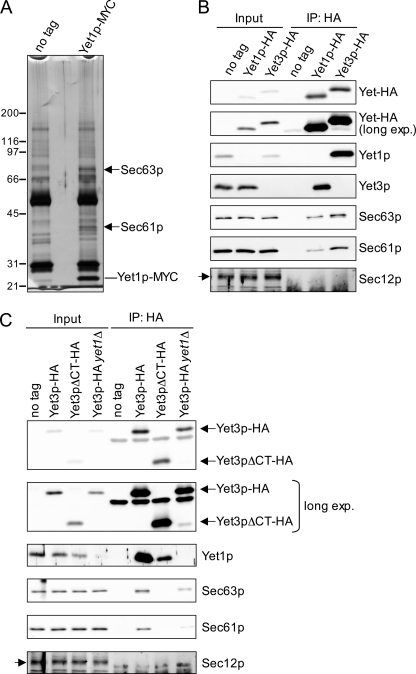
FIGURE 3.
Yet1p and Yet3p form a complex and interact with Sec63p and Sec61p. A, MYC immunoprecipitations were performed on digitonin-solubilized microsomes from untagged (YPH500) and Yet1p-MYC (CBY0310) strains. Bound material was resolved by SDS-PAGE and silver stained. Arrows indicate bands that were excised (from a colloidal Coomassie-stained gel run in parallel, not shown) and identified by mass spectrometry. The line indicates a prominent band presumed to be Yet1p-MYC. B, HA immunoprecipitations were carried out on digitonin-solubilized semi-intact cells from untagged (BY4742), Yet1p-HA (CBY2795), and Yet3p-HA (CBY2805) tagged strains and analyzed by immunoblot. C, HA immunoprecipitations performed and analyzed as in B from untagged (BY4742), Yet3p-HA (CBY2805), Yet3pΔCT-HA (CBY2815), and Yet3p-HA yet1Δ (CBY2810). Immunoblots for the ER resident protein Sec12p are included as a negative control for immunoprecipitations and is indicated by an arrow.
Yet Proteins Interact and Form a Complex with the Translocation Machinery
To explore function of the Yet proteins, we performed a series of immunoprecipitation experiments using C-terminal epitope-tagged versions of Yet1p and Yet3p. Immunoprecipitation of Yet1p-MYC from digitonin-solubilized microsomes consistently yielded protein staining species at ∼40 and 75 kDa (Fig. 3A). These proteins were identified by mass spectrometry as Sec61p and Sec63p, respectively, and confirmed by immunoblot (Fig. 3B). In addition to Sec61p and Sec63p, Yet3p was also efficiently recovered in the Yet1p-HA immunoprecipitation (Fig. 3B). Yet3p was not visible by protein staining of the Yet1p-MYC immunoprecipitation (Fig. 3A) because of a similar mobility to Yet1p-MYC. Immunoprecipitation of Yet3p-HA also yielded Sec61p and Sec63p, as well as an abundance of Yet1p (Fig. 3B). Notably, the Sec61p homolog, Ssh1p, was not detected in complex with the Yet proteins (data not shown). In a reciprocal set of immunoprecipitation experiments we observed that Sec63p-HA and Sec61p-HA specifically recovered Yet1p-Yet3p complex, although recovery of this complex was 2–3-fold greater when Sec63p-HA was the target compared with Sec61p-HA (supplemental Fig. S1). Collectively, these results indicate that Yet1p and Yet3p form a complex that interacts with Sec61p and Sec63p.
To examine whether a heteromeric Yet1p-Yet3p complex is required for interaction with Sec61p and Sec63p, we immunoprecipitated Yet3p-HA from yet1Δ cells (Fig. 3C). The amount of Sec63p and Sec61p that co-immunoprecipitated with Yet3p-HA was significantly reduced by the yet1Δ mutation indicating that Yet1p-Yet3p complex formation is required for efficient association with Sec complex components. It has been reported that the C-terminal cytoplasmic domain of BAP31 is dispensable for interaction with cellubrevin but important for ER export (15). This domain is proposed to join BAP29 and BAP31 to form an oligomer (51). Thus, we tested the importance of this domain by deleting most of the cytoplasmic C-terminal segment (terminal 69 amino acids) of Yet3p (Yet3pΔCT-HA) and examined interactions with Yet1p and the Sec complex. Strikingly, this truncation substantially reduced the amount of co-immunoprecipitated Yet1p and reduced the association with Sec61p and Sec63p to undetectable levels (Fig. 3C). These data show that a heteromeric Yet1p-Yet3p complex (Yet complex) is necessary for maximal association with the Sec complex. Although we cannot exclude the possibility that this C-terminal truncation influences Yet3p folding, these results suggest that the cytoplasmic C-terminal domain of Yet3p mediates interactions with Sec61p, Sec63p, and Yet1p.
Yet Protein Interactions with the Sec61 and Sec63 Complexes Appear to Be Separable
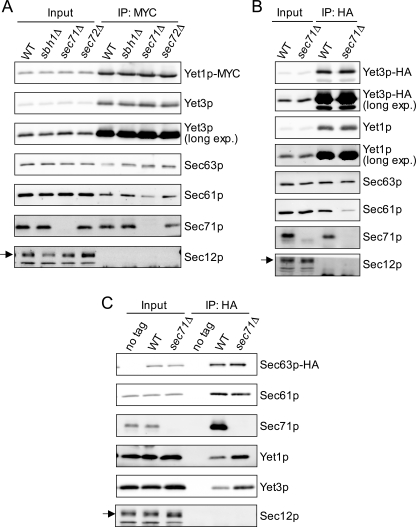
We reasoned that the interaction between the Yet1p-Yet3p and Sec61p-Sec63p complexes might be disrupted if Sec complex subunits important for the interaction were removed. Of the seven subunits that comprise the Sec complex, three (sbh1Δ, sec71Δ, and sec72Δ) can be individually deleted without causing inviability. To examine the influence of these mutations on the Yet-Sec complex interaction, we performed Yet1p-MYC immunoprecipitation experiments in these Sec complex deletion mutants. As shown in Fig. 4A, Yet1p-MYC immunoprecipitated from sec71Δ cells had notably less co-associated Sec61p. In contrast, the amount of co-immunoprecipitated Sec63p was not significantly reduced in the absence of SEC71. Consistent with this observation, Yet3p-HA immunoprecipitated from sec71Δ membranes also had considerably less associated Sec61p. (Fig. 4B). The reduced amount of Yet1p/Yet3p-associated Sec61p in sec71Δ membranes could represent a direct disruption of this interaction or a disruption between the Sec63 and Sec61 subcomplexes. Thus, to assess Sec63p-Sec61p and Sec63p-Yet complex interactions, we immunoprecipitated Sec63p-HA from SEC71 and sec71Δ digitonin-solubilized semi-intact cells and examined the level of co-associated Sec61p, Yet1p, and Yet3p. As shown in Fig. 4C, the association of Sec61p with Sec63p-HA was not detectably altered in sec71Δ membranes. Furthermore, immunoprecipitation of Sec63p-HA in the absence of Sec71p did not diminish the amount of associated Yet1p or Yet3p. In fact, the levels of Yet1p and Yet3p recovered with Sec63p were increased by sec71Δ mutation. In summary, these results suggest that Sec71p is important for association of the Yet complex with Sec61p, but not for Yet complex association with Sec63p. Thus, Yet1p-Yet3p interaction with the Sec63 complex appears distinct from interaction with the Sec61 complex.
FIGURE 4.
Influence of sec71Δ mutation on Yet-Sec complex interaction. MYC and HA immunoprecipitations were performed and analyzed as described in the legend to Fig. 3B from: A, Yet1p-MYC (CBY2291), Yet1p-MYC sbh1Δ (CBY2292), Yet1p-MYC sec71Δ (CBY2293) and Yet1p-MYC sec72Δ (CBY2363); B, Yet3p-HA (CBY2805) and Yet3p-HA sec71Δ (CBY2813); C, no tag (BY4742), Sec63p-HA (CBY2733), and Sec63p-HA sec71Δ (CBY2814). Immunoblots for the ER resident protein Sec12p are included as a negative control and noted with an arrow.
Influence of DTT Treatment on Yet and Sec Complex Associations
Because the Yet proteins are up-regulated by DTT treatment, we reasoned that accumulation of unfolded proteins might also facilitate Yet complex interactions important during ER stress. Therefore, we tested whether DTT treatment altered the association of the Yet complex with the Sec complex. To address this possibility, we treated mid-log phase cultures with 8 mm DTT for 2 h prior to harvest. As shown in Fig. 5A (input lanes), Yet1p, Yet3p-HA, and Sec61p were up-regulated by DTT treatment. Immunoprecipitation of Yet3p-HA from these digitonin-solubilized membranes revealed a substantial increase in the amount of co-associated Sec63p and Sec61p, even after accounting for elevated protein expression (Fig. 5A). The increase in Yet3p-HA-associated Sec63p was ∼3-fold in DTT-treated samples compared with untreated control. Remarkably, ∼6-fold more Sec61p was associated with Yet3p-HA after DTT treatment, whereas the amount of Yet1p associated with Yet3p-HA was constant. Examination of these digitonin-solubilized protein complexes by velocity sedimentation on sucrose gradients revealed co-migration of the Yet3p and Sec61p containing complexes. However, the sedimentation properties were not detectably altered after DTT treatment of cells (supplemental Fig. S2). Yet3p-HA immunoprecipitation experiments performed with membranes derived from ire1Δ cells treated with DTT also showed increased levels of Sec61p and Sec63p with Yet3p-HA (supplemental Fig. S3A). Interestingly, we observed an overall decrease in the amount of Sec complex associated with Yet3p-HA in ire1Δ cells under both DTT and untreated conditions. Thus, Ire1p influences Yet-Sec complex interactions, but DTT-induced ER stress enhances Yet-Sec complex associations independent of Ire1p and the UPR. Furthermore, the differential increases in Sec61p and Sec63p with Yet3p-HA are consistent with the distinct interactions observed between the Yet complex and these translocon subcomplexes (Fig. 4).
We next examined the influence of DTT-induced ER stress on Sec61-Sec63 complex association by performing immunoprecipitation experiments as described in Fig. 5A, but using membranes from cells expressing Sec63p-HA. In line with the results obtained from Yet3p-HA immunoprecipitations, the amount of Yet1p and Yet3p (shown in graph as Yet complex) associated with Sec63p-HA was increased ∼2-fold in DTT-treated samples. In contrast, the amount Sec61p associated with Sec63p-HA did not increase, confirming that the observed increases in association were between the Yet and Sec complexes. These results show that ER stress, at least when caused by DTT, promotes Yet-Sec complex association and that the Yet-Sec61 complex association is differentially enhanced compared with the Yet-Sec63 complex association.
Yet-Sec Complex Interaction Is Diminished by Translocation Substrate Depletion
We next asked whether the Yet-Sec complex association was sensitive to the availability of translocation substrates. To address this idea, Yet3p-HA was immunoprecipitated from digitonin-solubilized membranes derived from cells that had been treated with the protein synthesis inhibitor cycloheximide for 10 or 30 min prior to harvest. Interestingly, the amount of Yet3p-HA associated Sec63p and Sec61p decreased after exposure to cycloheximide, which was especially apparent after the 30-min treatment, whereas the level of associated Yet1p was relatively unchanged (Fig. 6A). To further study the influence of translocation substrates, we examined Yet-Sec complex association after stimulating translocation in vitro. We reasoned that stimulating translocation in the absence of translation would deplete the ER of translocation substrates. Thus, if Yet-Sec complex associations are increased by active translocation, conditions that deplete the ER of translocation substrates should reduce the level of Yet-Sec complex interaction. Indeed, as shown in Fig. 6B, incubation of washed membranes with an ATP/GTP system (to stimulate translocation) prior to immunoprecipitation of Yet3p-HA significantly reduced the amount of associated Sec63p and Sec61p. In contrast, Yet1p association with Yet3p-HA was not detectably altered. Together, these results suggest that Yet-Sec complex association is influenced by nascent polypeptide availability and/or translocation status of the Sec complex with the Yet complex preferentially associated with Sec complexes engaged in translocation.
Cells Lacking YET1 or YET3 Are Sensitive to Inositol Starvation
The increase in Yet protein expression and Yet-Sec complex association observed after DTT treatment suggests that the Yet proteins may have an important role during ER stress. However, cells lacking the YET genes did not exhibit detectable growth defects in the presence of DTT or tunicamycin (data not shown). Interestingly, a recent chemical genomics study indicated co-fitness correlations between yet1Δ and yet3Δ mutants and several deletion mutants with known inositol auxotrophies (52). This observation suggested to us that yet1Δ and yet3Δ mutants might also be sensitive to inositol starvation. Indeed, as shown in Fig. 7A, yet1Δ and yet3Δ cells grow very slowly in the absence of inositol. These growth defects were apparent at room temperature (not shown) or 30 °C and exacerbated at 37 °C. Importantly, the growth defect exhibited by either mutant could be rescued by episomal expression of the gene deleted. However, 2-μm overexpression of YET3 did not improve the growth of the yet1Δ mutant in the absence of inositol. Likewise, overexpression of YET1 did not rescue the growth phenotype of yet3Δ. These results establish a requirement for YET1 and YET3 in inositol prototrophy and support the proposal that Yet1p and Yet3p function as a hetero- oligomer. Notably, the top co-fitness deletion mutants that correlated with yet2Δ did not include established inositol auxotrophs (52). Consistent with this result, yet2Δ cells did not display a growth defect in the absence of inositol (supplemental Fig. S4A). Importantly, double yet1Δ yet3Δ and triple yet1Δ yet2Δ yet3Δ mutants display inositol starvation growth deficiencies similar to single yet1Δ and yet3Δ mutants (data not shown), ruling out the possibility that this phenotype is the result of a dominant-negative effect of unpartnered Yet1p or Yet3p.
The yet1Δ and yet3Δ Mutants Display Synthetic Growth Defects When Combined with Certain Translocation Mutants
The observation that deletion of YET1 or YET3 reduces yeast cell growth in the absence of inositol, together with the observed interactions between the Yet proteins and the Sec complex, prompted us to examine whether cells harboring mutations in both complexes would display synthetic growth defects in the absence of inositol. Indeed, when yet1Δ or yet3Δ mutants were combined with the sec63-1 translocation mutant (26), clear synthetic growth defects were observed in the absence of inositol (Fig. 7B). Importantly, cells with only the sec63-1 mutation were not sensitive to inositol starvation. Similar results were noted when yet1Δ was combined with sec61-2 (25) in a distinct genetic background (Fig. 7C). We also found that yet1Δ sec71Δ and yet3Δ sec71Δ double mutants showed synthetic growth defects in the absence of inositol (supplemental Fig. S4B), an effect not seen when a yet3Δ mutant was combined with sbh1Δ or sec72Δ mutants (supplemental Fig. S4C). Together, these data provide the first genetic evidence linking the Yet complex to the ER translocation apparatus.
DISCUSSION
In this study, we report several findings on the yeast BAP31 homologs, Yet1p and Yet3p, indicating a functional role in ER homeostasis. Assembly of Yet1p and Yet3p into a heteromeric complex was required for normal localization, expression, and function. Yet1p and Yet3p were not efficiently incorporated into COPII vesicles and were not detected in post-ER compartments, inconsistent with an export receptor function as suggested for BAP31 (15, 44). Consistent with a functional role at the ER, both Yet1p and Yet3p co-immunoprecipitated the Sec complex, and this interaction was responsive to the availability of translocation substrates. Yet1p and Yet3p levels were elevated during ER stress, as was their interaction with the Sec complex. Additionally, cells with yet1Δ or yet3Δ mutations displayed attenuated growth in the absence of inositol, a defect that was exacerbated when combined with certain mutations in the Sec complex. Taken together, our data indicate a functional link between the Yet and Sec complex.
The interaction we observed between the Yet proteins and Sec61p appears to be conserved as recently reported in mammalian cells (20). In addition to members of the Sec61 complex (Sec61α, Sec61β, and TRAM) found in association with BAP31 by Wang et al. (20), we discovered the Yet proteins associated with members of the Sec63 complex (Sec63p and Sec71p). Interestingly, Wang et al. (20) showed that translocon-locked CFTRΔF508 also interacts with BAP31, suggesting that BAP31 can associate with its substrates as they emerge from the translocation apparatus. Our observation that the association between the Yet and Sec complex was enhanced during ER stress suggests that this interaction is especially important under such conditions. We propose that a subset of nascent secretory proteins engaged by Yet1p-Yet3p at the translocon may be up-regulated during ER stress. Alternatively, the folding stress caused by DTT could in turn slow translocation and thus prolong interactions between the Yet and Sec complex.
We observed a decrease in the Yet-Sec complex interaction when translocation substrates were depleted, suggesting that the Yet complex preferentially associates with Sec complexes engaged in nascent polypeptide translocation. However, it seems unlikely that Yet-Sec complex interaction is important for translocation in general because we found that cells lacking the yet genes (yet1Δ yet2Δ yet3Δ) were fully competent for translocation of CPY and α-factor as assessed by pulse-chase and in vitro translocation experiments, respectively (data not shown). Furthermore, we found that whereas 2-μm overexpression of SEC63 was able to partially suppress thermosensitive growth of a sec71Δ strain, 2-μm overexpression of YET1 and YET3 had no detectable effect on the growth of sec71Δ (supplemental Fig. S5), sec61-2, or sec63-1 mutants (data not shown).
Several observations suggest that the association between the Yet and Sec complexes involves multiple interaction sites. We found that immunoprecipitation of epitope-tagged Yet1p or Yet3p in the absence of Sec71p (sec71Δ) resulted in decreased co-immunoprecipitation of Sec61p, whereas the association with Sec63p appeared unchanged. In contrast, when Sec63p-HA was immunoprecipitated from membranes derived from sec71Δ cells, the amount of associated Yet1p and Yet3p was not decreased, but instead appeared to be elevated. We also found that the association between Sec61p and Sec63p was not diminished in sec71Δ cells, suggesting that the decrease in Sec61p seen in the Yet protein immunoprecipitations resulted from a defect in Yet-Sec61p interaction. One explanation for these results is that the Yet complex first associates with the Sec63 complex and then, perhaps facilitated by Sec71p, is “handed off” to the Sec61p complex. Notably, Sec72p is unstable in cells lacking Sec71p (53) suggesting that Sec63p, or potentially Sec62p, is the primary point of association for the Yet complex. Our experiments examining the effect of DTT on the Yet-Sec complex interaction are consistent with the idea that the Yet proteins form distinct interactions with the Sec63 and Sec61 complexes. When Yet3p-HA was immunoprecipitated from membranes isolated from DTT-treated cells, we observed an ∼3-fold increase in associated Sec63p, whereas the increase in Sec61p was ∼6-fold. Immunoprecipitation of Sec63p-HA under identical conditions demonstrated that the amount of associated Sec61p was unaltered and that, similar to the Yet3p-HA immunoprecipitation results, an ∼2-fold increase in recovery of Yet1p-Yet3p complex was observed. Inositol starvation also enhanced Yet-Sec complex association, although the level of enhancement was similar for both Sec63p and Sec61p (supplemental Fig. S3B). Together, these results show that Yet-Sec complex association is modulated by ER stress and suggest that the Yet complex is capable of distinct associations with the Sec63 and Sec61 complexes.
A number of studies propose a role for BAP31 in ER protein quality control (19, 20, 54, 55). Consistent with these reports, we observed that Yet protein expression was elevated by UPR-inducing conditions (DTT treatment). The observation that Yet2p expression was highly influenced by DTT suggested to us that it may perform a specialized role in ER stress-related Yet complex function. However, Yet2p-MYC immunoprecipitations revealed that only minor amounts were co-associated with Yet1p and the Sec complex (supplemental Fig. S6A). Although Yet2p-MYC was detected in association with Yet3p-HA, deletion of YET2 did not influence Yet3p-HA co-immunoprecipitation of Yet1p or the Sec complex (supplemental Fig. S6B). Furthermore, removal of all three YET genes in the yet1Δ yet2Δ yet3Δ triple mutant (yetΔΔΔ) did not cause growth defects in the presence of DTT or tunicamycin (data not shown), suggesting that these genes are not essential components of the ER stress response induced by these compounds. Several studies have shown that certain ER retained molecules including CFTRΔF508 (19, 20) and cytochrome P450 C2C (55) are stabilized by the down-regulation of BAP31, thus pointing to a role for BAP31 in ERAD. We examined the turnover rate of two model ERAD substrates, CPY* and Ste6* (56, 57), in yetΔΔΔ cells and, consistent with a previous study that reported a wild-type degradation rate for CPY* in yet2Δ mutant cells (58), found no obvious differences compared with wild-type cells (data not shown). We also investigated the turnover rate of CFTR-HA (59) in a yet3Δ mutant and found that it was similar to wild-type cells (data not shown). Additionally, yetΔΔΔ mutant cells did not exhibit a constitutive UPR that often accompanies defects in ERAD (data not shown) (42). Thus, we conclude that if the yeast homologs of BAP31 are involved in ER quality control, they act on a specific subset of ERAD substrates that have not been characterized.
The observation that cells lacking YET1 or YET3 exhibit growth defects in the absence of inositol is intriguing because cells with null mutations in IRE1 or HAC1, genes central to UPR activation, are also defective for growth in the absence of inositol (60–62). Another connection between the UPR and the inositol starvation response is the elevation of INO1 mRNA levels when the UPR is activated by tunicamycin treatment (63). However, we found that yetΔΔΔ mutant cells were able to mount a UPR similar to wild-type cells (data not shown). Our favored explanation for the inositol growth defect in yet1Δ or yet3Δ mutants is that deletion of these genes impairs the translocation/biogenesis of protein(s) important for the response to inositol starvation. It is important to note that none of the single Sec complex mutants examined exhibited inositol starvation-specific growth defects, signifying that the translocation of such a Yet-dependent substrate is not significantly compromised in these mutants. Additionally, this observation suggests that the ER stress attributable to the translocation defects in the Sec complex mutants is not additive or synergistic with the stress induced by inositol starvation. Consistent with this idea, an erv25Δ strain, known to have a constitutive UPR (64, 65), did not exhibit an inositol starvation growth defect and a yet3Δ erv25Δ mutant grew similarly to a yet3Δ mutant in the absence of inositol (data not shown).
Our data suggest that interactions between Yet1p and Yet3p and the Sec complex rely on the C-terminal cytoplasmic domain of Yet3p. In addition to abrogating Yet and Sec complex interactions, deletion of this domain resulted in inositol auxotrophy that was indistinguishable from a yet3Δ mutant (supplemental Fig. S4A). Consistent with the notion that Yet1p stability requires interaction with Yet3p, the analogous truncation in Yet1p was destabilizing (data not shown) presumably because of reduced Yet3p association. Together these observations suggest that the C-terminal cytoplasmic domains of Yet1p and Yet3p function as essential interaction modules.
In summary, our results show that Yet1p and Yet3p form a heteromeric complex probably through association of coiled-coil motifs within their cytoplasmic C-terminal domains, as proposed for BAP31/29 (51). We envisage this heteromeric coiled-coil structure to form an interaction domain that mediates the Yet-Sec complex association. We speculate that the Yet complex interacts via their coiled-coil domains with the signal sequence recognition domains of the Sec63 complex. This may occur in the absence of Sec63 complex-dependent signal sequences, thus facilitating access of the Yet complex to Sec61p for co-translational translocation. This model is consistent with reported BAP31 substrates, which are transmembrane proteins and presumably follow the co-translational translocation pathway. The Sec63 complex, which has an established role in the post-translational translocation pathway, may serve to guide Yet1p-Yet3p to the Sec61 complex. Notably, a heteromeric BAP31/29 complex appears to be important for interaction with mIgD molecules (18), via transmembrane domain interactions (51). Based on these interactions, we propose a model for Yet1p and Yet3p in which their cytoplasmic C-terminal domains facilitate association with the Sec complex, leaving their transmembrane domains available to manage biogenesis of specific translocation substrates.
Supplementary Material
Acknowledgments
We thank C. Bentivoglio for generating the Yet3p antiserum and J. Powers for construction of CBY0310. We also thank R. Schekman, M. Makarow, and C. Stirling for yeast strains and antisera. We are grateful to J. Merritt, J. Scarcelli, A. Lorente, P. Shindiapina, S. Thompson, S. Bakhoum, C. Hodge, W. Wickner, L. Myers, J. Brickner, and C. Cole for helpful suggestions, J. Scarcelli for technical assistance, and W. Wickner for use of his microscope.
This work was supported, in whole or in part, by National Institutes of Health Grant GM052549.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6 and Table S1.
- ER
- endoplasmic reticulum
- ERAD
- ER-associated degradation
- mIgD
- membrane-bound immunoglobulin D molecules
- DTT
- dithiothreitol
- HA
- hemagglutinin
- GFP
- green fluorescent protein
- ORF
- open reading frame
- UPR
- unfolded protein response.
REFERENCES
- 1.Kalies K. U., Rapoport T. A., Hartmann E. (1998) J. Cell Biol. 141, 887–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scheper W., Thaminy S., Kais S., Stagljar I., Römisch K. (2003) J. Biol. Chem. 278, 37998–38003 [DOI] [PubMed] [Google Scholar]
- 3.Hartmann E., Sommer T., Prehn S., Görlich D., Jentsch S., Rapoport T. A. (1994) Nature 367, 654–657 [DOI] [PubMed] [Google Scholar]
- 4.Esnault Y., Feldheim D., Blondel M. O., Schekman R., Képès F. (1994) J. Biol. Chem. 269, 27478–27485 [PubMed] [Google Scholar]
- 5.Finke K., Plath K., Panzner S., Prehn S., Rapoport T. A., Hartmann E., Sommer T. (1996) EMBO J. 15, 1482–1494 [PMC free article] [PubMed] [Google Scholar]
- 6.Panzner S., Dreier L., Hartmann E., Kostka S., Rapoport T. A. (1995) Cell 81, 561–570 [DOI] [PubMed] [Google Scholar]
- 7.Brodsky J. L., Goeckeler J., Schekman R. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 9643–9646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Young B. P., Craven R. A., Reid P. J., Willer M., Stirling C. J. (2001) EMBO J. 20, 262–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vembar S. S., Brodsky J. L. (2008) Nat. Rev. Mol. Cell Biol. 9, 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalies K. U., Allan S., Sergeyenko T., Kröger H., Römisch K. (2005) EMBO J. 24, 2284–2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oyadomari S., Yun C., Fisher E. A., Kreglinger N., Kreibich G., Oyadomari M., Harding H. P., Goodman A. G., Harant H., Garrison J. L., Taunton J., Katze M. G., Ron D. (2006) Cell 126, 727–739 [DOI] [PubMed] [Google Scholar]
- 12.Scott D. C., Schekman R. (2008) J. Cell Biol. 181, 1095–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Willer M., Forte G. M., Stirling C. J. (2008) J. Biol. Chem. 283, 33883–33888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim K. M., Adachi T., Nielsen P. J., Terashima M., Lamers M. C., Köhler G., Reth M. (1994) EMBO J. 13, 3793–3800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Annaert W. G., Becker B., Kistner U., Reth M., Jahn R. (1997) J. Cell Biol. 139, 1397–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paquet M. E., Cohen-Doyle M., Shore G. C., Williams D. B. (2004) J. Immunol. 172, 7548–7555 [DOI] [PubMed] [Google Scholar]
- 17.Ladasky J. J., Boyle S., Seth M., Li H., Pentcheva T., Abe F., Steinberg S. J., Edidin M. (2006) J. Immunol. 177, 6172–6181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schamel W. W., Kuppig S., Becker B., Gimborn K., Hauri H. P., Reth M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 9861–9866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lambert G., Becker B., Schreiber R., Boucherot A., Reth M., Kunzelmann K. (2001) J. Biol. Chem. 276, 20340–20345 [DOI] [PubMed] [Google Scholar]
- 20.Wang B., Heath-Engel H., Zhang D., Nguyen N., Thomas D. Y., Hanrahan J. W., Shore G. C. (2008) Cell 133, 1080–1092 [DOI] [PubMed] [Google Scholar]
- 21.Toikkanen J. H., Fatal N., Hildén P., Makarow M., Kuismanen E. (2006) J. Biol. Sci. 6, 446–456 [Google Scholar]
- 22.Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998) Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 23.Tong A. H., Evangelista M., Parsons A. B., Xu H., Bader G. D., Pagé N., Robinson M., Raghibizadeh S., Hogue C. W., Bussey H., Andrews B., Tyers M., Boone C. (2001) Science 294, 2364–2368 [DOI] [PubMed] [Google Scholar]
- 24.Tong A. H., Boone C. (2006) Methods Mol. Biol. 313, 171–192 [DOI] [PubMed] [Google Scholar]
- 25.Deshaies R. J., Schekman R. (1987) J. Cell Biol. 105, 633–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rothblatt J. A., Deshaies R. J., Sanders S. L., Daum G., Schekman R. (1989) J. Cell Biol. 109, 2641–2652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ito H., Fukuda Y., Murata K., Kimura A. (1983) J. Bacteriol. 153, 163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christianson T. W., Sikorski R. S., Dante M., Shero J. H., Hieter P. (1992) Gene 110, 119–122 [DOI] [PubMed] [Google Scholar]
- 29.Oldenburg K. R., Vo K. T., Michaelis S., Paddon C. (1997) Nucleic Acids Res. 25, 451–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sikorski R. S., Hieter P. (1989) Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feldheim D., Rothblatt J., Schekman R. (1992) Mol. Cell. Biol. 12, 3288–3296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stirling C. J., Rothblatt J., Hosobuchi M., Deshaies R., Schekman R. (1992) Mol. Biol. Cell 3, 129–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feldheim D., Yoshimura K., Admon A., Schekman R. (1993) Mol. Biol. Cell 4, 931–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Otte S., Belden W. J., Heidtman M., Liu J., Jensen O. N., Barlowe C. (2001) J. Cell Biol. 152, 503–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brodsky J. L., Hamamoto S., Feldheim D., Schekman R. (1993) J. Cell Biol. 120, 95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Y., Barlowe C. (2002) Mol. Biol. Cell 13, 3314–3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Powers J., Barlowe C. (1998) J. Cell Biol. 142, 1209–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baker D., Hicke L., Rexach M., Schleyer M., Schekman R. (1988) Cell 54, 335–344 [DOI] [PubMed] [Google Scholar]
- 39.Wuestehube L. J., Schekman R. W. (1992) Methods Enzymol. 219, 124–136 [DOI] [PubMed] [Google Scholar]
- 40.Blum H., Beier H., Gross H. J. (1987) Electrophoresis 8, 93–99 [Google Scholar]
- 41.Barlowe C., Orci L., Yeung T., Hosobuchi M., Hamamoto S., Salama N., Rexach M. F., Ravazzola M., Amherdt M., Schekman R. (1994) Cell 77, 895–907 [DOI] [PubMed] [Google Scholar]
- 42.Travers K. J., Patil C. K., Wodicka L., Lockhart D. J., Weissman J. S., Walter P. (2000) Cell 101, 249–258 [DOI] [PubMed] [Google Scholar]
- 43.Gasch A. P., Spellman P. T., Kao C. M., Carmel-Harel O., Eisen M. B., Storz G., Botstein D., Brown P. O. (2000) Mol. Biol. Cell 11, 4241–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spiliotis E. T., Manley H., Osorio M., Zúñiga M. C., Edidin M. (2000) Immunity 13, 841–851 [DOI] [PubMed] [Google Scholar]
- 45.Belden W. J., Barlowe C. (2001) Science 294, 1528–1531 [DOI] [PubMed] [Google Scholar]
- 46.Kim J., Hamamoto S., Ravazzola M., Orci L., Schekman R. (2005) J. Biol. Chem. 280, 7758–7768 [DOI] [PubMed] [Google Scholar]
- 47.Powers J., Barlowe C. (2002) Mol. Biol. Cell 13, 880–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kappeler F., Klopfenstein D. R., Foguet M., Paccaud J. P., Hauri H. P. (1997) J. Biol. Chem. 272, 31801–31808 [DOI] [PubMed] [Google Scholar]
- 49.Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. (2003) Nature 425, 686–691 [DOI] [PubMed] [Google Scholar]
- 50.Lee M. C., Miller E. A., Goldberg J., Orci L., Schekman R. (2004) Annu. Rev. Cell Dev. Biol. 20, 87–123 [DOI] [PubMed] [Google Scholar]
- 51.Adachi T., Schamel W. W., Kim K. M., Watanabe T., Becker B., Nielsen P. J., Reth M. (1996) EMBO J. 15, 1534–1541 [PMC free article] [PubMed] [Google Scholar]
- 52.Hillenmeyer M. E., Fung E., Wildenhain J., Pierce S. E., Hoon S., Lee W., Proctor M., St. Onge R. P., Tyers M., Koller D., Altman R. B., Davis R. W., Nislow C., Giaever G. (2008) Science 320, 362–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feldheim D., Schekman R. (1994) J. Cell Biol. 126, 935–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang B., Pelletier J., Massaad M. J., Herscovics A., Shore G. C. (2004) Mol. Cell. Biol. 24, 2767–2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Szczesna-Skorupa E., Kemper B. (2006) J. Biol. Chem. 281, 4142–4148 [DOI] [PubMed] [Google Scholar]
- 56.Wolf D. H., Fink G. R. (1975) J. Bacteriol. 123, 1150–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Loayza D., Tam A., Schmidt W. K., Michaelis S. (1998) Mol. Biol. Cell 9, 2767–2784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Caldwell S. R., Hill K. J., Cooper A. A. (2001) J. Biol. Chem. 276, 23296–23303 [DOI] [PubMed] [Google Scholar]
- 59.Zhang Y., Michaelis S., Brodsky J. L. (2002) Methods Mol. Med. 70, 257–265 [DOI] [PubMed] [Google Scholar]
- 60.Nikawa J., Yamashita S. (1992) Mol. Microbiol. 6, 1441–1446 [DOI] [PubMed] [Google Scholar]
- 61.Cox J. S., Shamu C. E., Walter P. (1993) Cell 73, 1197–1206 [DOI] [PubMed] [Google Scholar]
- 62.Nikawa J. (1994) Gene 149, 367–372 [DOI] [PubMed] [Google Scholar]
- 63.Cox J. S., Chapman R. E., Walter P. (1997) Mol. Biol. Cell 8, 1805–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Belden W. J., Barlowe C. (2001) Mol. Biol. Cell 12, 957–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wilson J. D., Liu Y., Bentivoglio C. M., Barlowe C. (2006) Traffic 7, 1213–1223 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.