Abstract
Signal transduction by Toll-like receptor 2 (TLR2) and TLR4 requires the adaptors MyD88 and Mal (MyD88 adaptor-like) and serine/threonine kinases, interleukin-1 receptor-associated kinases IRAK1 and IRAK4. We have found that both IRAK1 and IRAK4 can directly phosphorylate Mal. In addition, co-expression of Mal with either IRAK resulted in depletion of Mal from cell lysates. This is likely to be due to Mal phosphorylation by the IRAKs because kinase-inactive forms of either IRAK had no effect. Furthermore, lipopolysaccharide stimulation resulted in ubiquitination and degradation of Mal, which was inhibited using an IRAK1/4 inhibitor or by knocking down expression of IRAK1 and IRAK4. MyD88 is not a substrate for either IRAK and did not undergo degradation. We therefore conclude that Mal is a substrate for IRAK1 and IRAK4 with phosphorylation promoting ubiquitination and degradation of Mal. This process may serve to negatively regulate signaling by TLR2 and TLR4.
Keywords: Death Domain, Interleukin Receptor-associated Kinase (IRAK), Mal TIRAP, Phosphorylation Enzymes, Ubiquitination
Introduction
Activation of Toll-like receptor (TLR)4 signaling pathways by microbial products initiates a cascade of events starting at the receptor level and leading eventually to the induction of an array of genes that encode for immune and inflammatory proteins. Ligand binding typically induces receptor dimerization, the recruitment of adaptor molecules, and the activation of a series of kinase cascades. In the case of the lipopolysaccharide (LPS) receptor TLR4 or the lipoprotein receptor TLR2, the adaptor molecules MyD88 (myeloid differentiation factor 88) and Mal (MyD88 adaptor-like) are recruited to activate NF-κB and induce proinflammatory cytokines, such as interleukin-1, tumor necrosis factor α, and interleukin-6 (1–3). Two other adaptors, TRIF (Toll/interleukin-1 receptor domain-containing inducing interferon-β) and TRAM (TRIF adaptor molecule), also participate in TLR4 signaling. Recruitment of these proteins leads to the activation of the transcription factor, IRF3 (interferon regulatory factor-3), and the induction of the type I interferons (4–6).
Mal and MyD88 both contain a Toll/interleukin-1 receptor (TIR) domain and function together to optimally transduce signals from TLR2 as well as TLR4. MyD88 differs from Mal in that it possesses an N-terminal death domain responsible for mediating interactions between MyD88 and members of the interleukin-1 receptor-associated kinase (IRAK) family. Four members of this family have been described, and the pathways leading to their activation have recently been dissected (7). In resting cells, IRAK1 can be found in a receptor complex containing MyD88 and Tollip. Following stimulation with LPS, IRAK1 is recruited to the TLR4 receptor complex, where it undergoes phosphorylation by IRAK4 on key threonine residues. This in turn promotes the autophosphorylating activity of IRAK1, its dissociation from the receptor complex, and subsequent interaction with TRAF6 (tumor necrosis factor receptor-associated factor) and a TAK1-TAB1-TAB2 kinase complex leading to the activation of NFκB and mitogen-activated protein kinases. IRAK1 undergoes degradation within 1 h of activation, after which time IRAK2 functions to maintain sustained TLR responses (8). IRAKM, unlike the other IRAK family members, lacks a kinase activity and functions as a negative regulator of TLR signaling (9). In addition to IRAK1, Pellino 2 has also been described as an IRAK4 substrate, whereas STAT3, histone H3, and IRF7 have been described as substrates for IRAK1 (10–13). Pellino 3 has been shown to possess ligase activity and is also an IRAK1 and IRAK4 substrate. Only kinase-active forms of these enzymes promote the ubiquitination and subsequent degradation of Pellino3 (14).
In this paper, we demonstrate that the TIR domain of Mal interacts with both IRAK1 and IRAK4. We also show that overexpression of Mal with active IRAK4 and IRAK1 but not the kinase-dead forms leads to Mal degradation. An interaction can only be observed between Mal and the inactive forms of these enzymes presumably because of degradation of Mal by the kinase-active forms. It is known that phosphorylation is often a prelude to ubiquitination. We also found that both enzymes could phosphorylate this adaptor molecule in vitro. Finally, we show that Mal undergoes ubiquitination prior to degradation following LPS stimulation and that this can be inhibited by an IRAK1/4 inhibitor or by knockdown of IRAK1 and IRAK4. These features do not apply to MyD88 and may represent an important negative feedback process in TLR2 and TLR4 signaling.
EXPERIMENTAL PROCEDURES
Biological Reagents and Cell Culture
HEK293 cells were obtained from the Centre for Applied Microbiology and Research (Porton Down, Salisbury, Wiltshire, UK) and were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 100 units/ml gentamycin, and 2 mm l-glutamine and maintained at 37 °C in a humidified atmosphere of 5% CO2. HEK293 TLR4/MD2/CD14 cells (which will now be referred to as 293MTC cells) were obtained from Invivogen. HA-tagged Mal in the pCDNA3 expression vector has been described previously (15) and was used for mammalian cell-based transfections. FLAG-tagged IRAK4 and IRAK4K213A (kinase-inactive mutant) was obtained previously from Amgen. Myc-tagged IRAK2 was a kind gift from Martha Muzio. Recombinant Mal, IRAK1, and IRAK4 were prepared as described previously (10, 15). Anti-FLAG monoclonal antibody cross-linked to Sepharose beads (M2 beads) was purchased from Sigma. Monoclonal antibodies against the epitope tags FLAG (12CA5) and Myc (9E10) were obtained from Sigma. The polyclonal antibody against the HA epitope tag (Y-11) was obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Anti-IRAK1 antibody (sc-5288) was obtained from Santa Cruz Biotechnology, Inc. Anti-IRAK4 (catalog no. 4363) antibody was purchased from Cell Signaling. Lys48-specific antibody was purchased from Millipore (clone Apu2). Protein A/G beads were obtained from Santa Cruz Biotechnology, Inc. All other reagents were obtained from Sigma unless otherwise stated.
Co-immunoprecipitation and Co-expression Studies
HA-tagged Mal was co-expressed in HEK293 cells with various FLAG-tagged constructs in 10 cm. Cells were seeded at 105 cells/ml 24 h prior to transfection with GeneJuice according to the manufacturer's instructions (Novagen). Cells were washed in ice-cold phosphate-buffered saline and lysed in 500 μl of high stringency lysis buffer (50 mm HEPES, pH 7.5, 100 mm NaCl, 1 mm EDTA, 10% glycerol, 1% Nonidet P-40). Supernatants were removed and added to the relevant precoupled antibody. 50 μl of each lysate was retained to confirm expression of Mal and IRAK1 or IRAK4. Samples were incubated overnight at 4 °C. Following incubation, the immune complexes were washed twice with 1 ml of lysis buffer and once with ice-cold phosphate-buffered saline. All supernatants were removed, and beads were resuspended in 30 μl of 5× sample buffer. The samples were boiled for 5 min and subjected to SDS-PAGE analysis and Western blotting with the appropriate epitope tag antibodies.
Degradation Studies
HA-tagged Mal was transiently transfected into 293MTC in 6-well dishes. Cells were seeded at 105 cells/ml 24 h prior to transfection with GeneJuice according to the manufacturer's instructions (Novagen). Cells were stimulated with 100 ng/ml LPS for the times indicated in the figure legends. Cells were pretreated for 3 h prior to stimulation with 5 μm IRAK1/4 inhibitor (Calbiochem) or with 5 μm MG132 (Calbiochem). Cells were washed in ice-cold phosphate-buffered saline and lysed in 150 μl of high stringency lysis buffer (50 mm HEPES, pH 7.5, 100 mm NaCl, 1 mm EDTA, 10% glycerol, 1% Nonidet P-40). Samples were immunoblotted with HA and also with β-actin to ensure equal protein loading.
THP1 cells were seeded at 106 cells/ml in 10-cm dishes 24 h prior to treatment with 500 nm IRAK1/4 inhibitor (Sigma) for 3 h. Cells were then stimulated for the indicated time points with 100 ng/ml LPS. Samples were lysed directly into sample buffer, sonicated, and boiled. Samples were diluted 1:2 and probed with anti-Mal antibody (16). Samples were diluted 1:5 and probed with β-actin to ensure equal protein loading.
In Vitro Kinase Assay and Mass Spectrometry Analysis
Recombinant Mal, IRAK1, and IRAK4 were purified as described previously (10, 15). FLAG-tagged IRAK4 was overexpressed in HEK293 cells and immunoprecipitated with anti-FLAG M2-agarose beads. The immune complex was washed twice in lysis buffer and an additional three times in kinase buffer (20 mm HEPES, pH 7.5, 2 mm dithiothreitol, 10 mm MgCl2, 50 mm NaCl, 100 μm sodium orthovanadate, 20 mm β-glycerophosphate, 1 mm aprotinin, 1 mm phenylmethylsulfonyl fluoride). The wash buffer was removed, and the immune complex was resuspended in 30 μl of kinase buffer with 2 μCi of [γ-32P]ATP, 0.6 mm ATP, and 100 ng of recombinant Mal. The reaction was left for 20 min at 30 °C. For direct kinase assays, 100 ng of recombinant IRAK1 or IRAK4 was added to 100 ng of recombinant Mal in 30 μl of kinase buffer. The reactions were stopped by boiling in 20 μl of 2× SDS sample buffer. Proteins were separated via SDS-PAGE, and gels were dried and exposed to x-ray film. For mass spectrometry analysis, kinase assays were performed with cold ATP with larger sample volumes, and protein bands were resolved in 14 × 24-cm SDS-polyacrylamide gels before being excised. Digests were prepared in 0.1 ml of 1% formic acid and analyzed for 20 μl by liquid chromatography-mass spectrometry with precursor ion scanning.
Ubiquitination Studies
HA-tagged Mal was either co-expressed in HEK293MTC cells with various FLAG-tagged constructs as well as Myc-tagged ubiquitin in 6-well dishes or transfected with Mal and Myc-tagged ubiquitin, followed by stimulation with 100 ng/ml LPS for the indicated times. Cells were seeded at 105 cells/ml 24 h prior to transfection with GeneJuice according to the manufacturer's instructions (Novagen). The indicated samples were pretreated with 5 μm IRAK1/4 inhibitor for 3 h prior to stimulation with LPS. Cells were washed in ice-cold phosphate-buffered saline and lysed in 150 μl of high stringency lysis buffer (50 mm HEPES, pH 7.5, 100 mm NaCl, 1 mm EDTA, 10% glycerol, 1% Nonidet P-40). Supernatants were removed and added to HA-precoupled antibody. 50 μl of each lysate was retained to confirm expression of Mal and IRAK1 or IRAK4. Samples were incubated overnight at 4 °C. Following incubation, the immune complexes were washed twice with 1 ml of lysis buffer and once with ice-cold phosphate-buffered saline. All supernatants were removed, and beads were resuspended in 30 μl of 5× sample buffer. The samples were boiled for 5 min, and SDS-PAGE analysis was performed. Samples were probed with Myc antibody.
U373 cells were seeded at 105 cells/ml. After 24 h, cells were transfected with siRNA oligonucleotides to IRAK1 and IRAK4 or a negative control. After 72 h, samples were transfected with 200 ng of HA-tagged Mal. After 6 h, cells were treated with the proteosomal inhibitor lactacystin (Biotrend) for 24 h. Cells were treated with 100 ng/ml LPS for the indicated times and harvested as described previously for the 293 cells.
Small Interfering RNA
Oligonucleotides for IRAK1 (Hs_IRAK1_5) and IRAK4 (Hs_IRAK4_5) were purchased from Qiagen. A non-targeting oligonucleotide (catalog no. 1027310) was used as a negative control. siRNAs were used at 50 nm. U373 or 293MTC cells were seeded at 105 cells/ml in 6-well plates. After 24 h, oligonucleotides were transfected using Lipofectamine RNAiMAX (Invitrogen) in serum-free medium. After 72 h, cells were stimulated with 100 ng/ml LPS for the indicated times. Cells were then harvested and used for further analysis.
RESULTS
Mal Associates with the IRAK1 and IRAK4 through Its TIR Domain
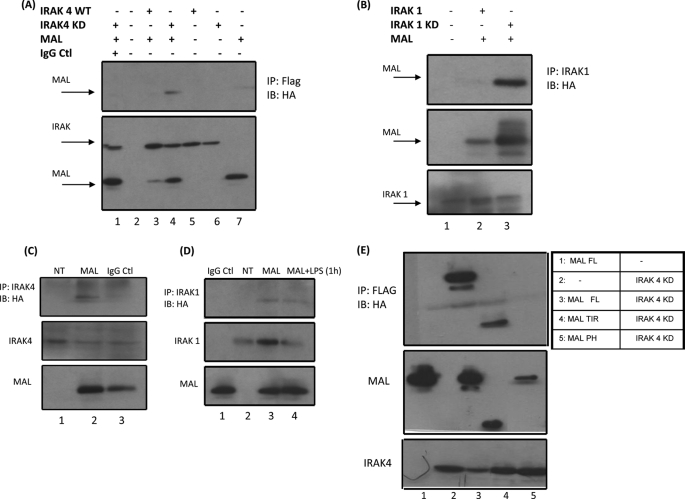
We have previously reported that Mal is capable of interacting with IRAK2 (12). We therefore sought to determine if Mal could interact with other members of the IRAK family. When Mal was overexpressed with the active form of IRAK4, no interaction was observed (Fig. 1A, top, lane 3). Importantly, there was a notable depletion of Mal in the corresponding cell lysates (Fig. 1A, bottom, lane 3), suggesting that Mal may have been undergoing degradation when co-expressed with IRAK4. It is known that IRAK4 undergoes autoactivation when overexpressed. We therefore carried out co-immunoprecipitation assays with Mal and kinase-dead IRAK4. As shown in Fig. 1A (bottom), Mal was not depleted in these lysates (compare lane 4 with lane 3). Furthermore, we were able to detect an interaction between the two proteins (Fig. 1A, top, lane 4). This was also the case for IRAK1, with Mal interacting only with the kinase-dead version of this protein as opposed to overexpressed active IRAK1 (Fig. 1B, top, lane 3). As was the case for IRAK4, Mal expression levels were decreased when overexpressed with wild-type IRAK1 (Fig. 1B, middle, lane 2). Furthermore, Mal was capable of interacting with endogenous IRAK4 and IRAK1, suggesting that the two proteins may be associated in resting cells (Fig. 1, C (lane 2) and D (lane 3)). It is known that IRAK1 itself undergoes degradation following LPS stimulation, and this may explain the reduced interaction we observed between Mal and IRAK1 in stimulated cells (Fig. 1D, top, lane 4).
FIGURE 1.
Mal interacts with IRAK1 and IRAK4. A, HEK293 cells were transiently transfected with 1 μg of a plasmid encoding HA-Mal (lanes 1, 3, 4, and 7), FLAG-IRAK4 (lanes 3 and 5), or FLAG-IRAK4KD (lanes 1, 4, and 6) or mock-transfected (lane 2). Cell lysates were prepared and immunoprecipitated (IP) with FLAG antibody (lanes 2–7) or IgG control (Ctl) (lane 1) and immunoblotted (IB) with HA antibody. Sample lysates were immunoblotted with FLAG and HA antibody for IRAK and Mal expression (bottom). B, HEK293 cells were mock-transfected (lane 1) or transiently transfected with 1 μg of plasmids encoding HA-Mal with either FLAG-IRAK1 (lane 2) or FLAG-IRAK1KD (lane 3). Samples were immunoprecipitated with an IRAK1-specific antibody and probed with anti-HA (top). Lysates were probed with IRAK antibody for IRAK1 and IRAK1KD expression (bottom) and HA for Mal expression (middle). C, HEK293 cells were mock-transfected (top, lane 1) or transiently transfected with plasmids encoding HA-Mal (top, lanes 2 and 3). Samples were immunoprecipitated with IRAK4 antibody or with IgG control (lane 3) and probed with HA antibody. Lysates were immunoblotted with IRAK4 antibody for IRAK4 expression (middle) or with HA antibody for Mal expression (bottom). D, HEK293 MTC cells were mock-transfected or transfected with plasmid encoding HA-Mal. After 24 h, cells were stimulated for 1 h with 100 ng/ml LPS. Samples were immunoprecipitated with IRAK1 antibody (lanes 2–4) or with IgG control (lane 1) and probed with HA antibody. Samples were immunoblotted with IRAK1 antibody for IRAK1 expression (middle). Lysates were probed with HA antibody for Mal expression (bottom). HEK293 cells were transfected with 1 μg of IRAK4KD and 1 μg of either full-length Mal, TIR domain of Mal, or proline to histidine (P125H) mutant. Cell lysates were prepared and immunoprecipitated with FLAG antibody and immunoblotted with HA antibody. Sample lysates were immunoblotted with FLAG and HA antibody for IRAK4 and Mal expression (E, bottom panels). Results are representative of three separate experiments.
We next carried out co-immunoprecipitation assays in order to identify the region of Mal that was interacting with IRAK4. As shown in Fig. 1E, Mal interacts with IRAK4KD via its TIR domain (lane 4). Mutation of the conserved proline residue within the BB loop of Mal abolished this interaction (Fig. 1E, lane 5). We did not observe any interaction between the N-terminal region of Mal and IRAK4KD (data not shown). This finding suggests that Mal interacts with the IRAKs via a TIR domain.
Co-expression of Mal with IRAK4 and IRAK1 Leads to Mal Degradation
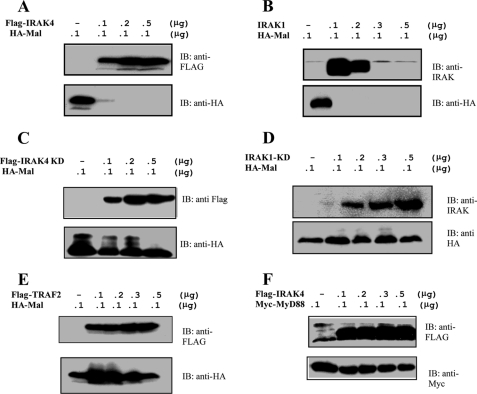
As mentioned above, we observed on several occasions that overexpression of Mal with the IRAKs led to a dramatic reduction in Mal protein levels as observed by Western blotting. In order to address whether IRAK expression results in Mal degradation, we co-expressed Mal with increasing concentrations of plasmids expressing either IRAK1 or IRAK4 and analyzed the cell lysates by Western blotting. As shown in Fig. 2, A and B (bottom panels), Mal was completely depleted from cell lysates in the presence of both enzymes. Importantly, co-expression of Mal with kinase-inactive forms of the IRAKs did not cause any degradation (Fig. 2, C and D, bottom panels). The unrelated protein TRAF2 also had no effect on Mal stability (Fig. 2E, bottom) further attesting to the specificity of the effect of the IRAKs. In addition, MyD88 expression levels remained unchanged when co-expressed with the IRAKs (as shown for IRAK4 in Fig. 2F (bottom)). Taken together, our results suggest that Mal undergoes IRAK-dependent degradation.
FIGURE 2.
Mal is degraded in the presence of IRAK1 and IRAK4. A–E, HEK293 cells were transiently transfected with 100 ng of a plasmid encoding HA-Mal and increasing concentrations of plasmids encoding active IRAK4 (A), active IRAK1 (B), inactive IRAK4 (C), inactive IRAK1 (D), and TRAF2 (E). F, HEK293 cells were transiently transfected with 100 ng of a plasmid encoding Myc-MyD88 and increasing concentrations of plasmid encoding active IRAK4. Cell lysates were prepared, and samples were analyzed by SDS-PAGE and immunoblotting. Results shown are representative of three individual experiments. IB, immunoblot.
Mal Undergoes IRAK1/4-dependent Phosphorylation
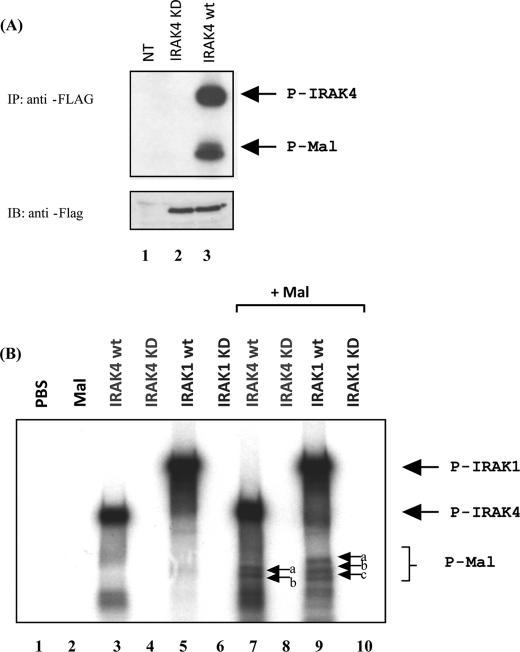
We have previously shown that overexpression of Mal in HEK293 cells results in the appearance of higher molecular weight forms of the protein that are absent upon pretreatment with calf intestinal alkaline phosphatase (17). In vitro kinase assays and mutagenesis studies implicated Bruton's tyrosine kinase as the enzyme responsible for phosphorylating Mal on key tyrosine residues, an event required for Mal activity. Calf intestinal alkaline phosphatase is actually a serine/threonine phosphatase; however, high concentrations will dephosphorylate tyrosine residues. Given that Mal only undergoes degradation with the active forms of the IRAKs, we next sought to determine if Mal was being phosphorylated by these kinases. Cells were transfected with FLAG-tagged IRAK4, and after 24 h, the enzyme was immunoprecipitated, and the resulting immune complex was incubated with purified recombinant Mal in kinase buffer. As shown in Fig. 3A, Mal was clearly phosphorylated by IRAK4 (lane 3). Autophosphorylation of IRAK4 can also be observed (upper band). A kinase-dead version of the IRAK4 (IRAK4KD) failed to phosphorylate Mal (lane 2), suggesting that the kinase activity of IRAK4 is required for this effect. A similar result was obtained with immunoprecipitated IRAK1 (not shown).
FIGURE 3.
Mal is phosphorylated in vitro by both IRAK1 and IRAK4. A, top, HEK293 cells were mock-transfected (NT; lane 1) or transiently transfected with plasmids encoding either FLAG-IRAK4KD (lane 2) or FLAG-IRAK4 (lane 3), cell lysates were prepared, and FLAG tagged proteins were immunoprecipitated with a FLAG antibody. Immunoprecipitates were washed twice with kinase buffer and incubated with 100 ng of recombinant Mal. Samples were then incubated with [γ-32P]ATP and a kinase buffer for 30 min at 37 °C. Samples were separated by SDS-PAGE and visualized by autoradiography. Bottom, cell lysates were immunoblotted for IRAK4 and IRAK4KD expression. B, phosphate-buffered saline (PBS), recombinant Mal, recombinant IRAK4, recombinant IRAK4KD, recombinant IRAK1 wild type (wt), and recombinant IRAK1KD were all incubated alone (lanes 1–6) or with recombinant Mal (lanes 7–10). Samples were incubated with [γ-32P]ATP and a kinase buffer for 30 min at 37 °C. Samples were then separated by SDS-PAGE and visualized by autoradiography.
We next performed a direct in vitro kinase assay with recombinant Mal and purified recombinant forms of the IRAKs in a cell-free system. As shown in Fig. 3B, Mal was directly phosphorylated by IRAK4 and IRAK1. Both enzymes underwent autophosphorylation (lanes 3 and 5), which was not evident with the kinase-inactive mutants (lanes 4 and 6). Incubation of IRAK4 (lane 7) and IRAK1 (lane 9) with Mal led to Mal phosphorylation, again not evident with the kinase-inactive forms (lanes 8 and 10). For IRAK4, two phosphobands are evident (lane 7, arrows b and c), whereas with IRAK1, an additional phosphoband appears (arrow a). This indicates that Mal is differentially phosphorylated by both IRAKs. MyD88 was not phosphorylated by either enzyme (data not shown).
We next sought to identify the site(s) on Mal undergoing phosphorylation. A non-radioactive in vitro kinase assay was carried out with purified recombinant Mal and either IRAK1 or IRAK4. Samples were run on a 14 × 16-cm SDS gel, and the slower migrating form of Mal that was visible on staining was excised from the gel and subject to trypsin digestion followed by liquid chromatography-mass spectrometry with precursor ion scanning. As shown in Table 1, threonine 28 was identified as the phospho-accepting residue in both cases. Mutation of this residue to arginine (T28A) had no affect on Mal-mediated NFκB activity in luciferase reporter assays (data not shown). Furthermore, this mutant was degraded to the same extent as wild type Mal when overexpressed with IRAK1 or IRAK4, suggesting that more than one phosphorylation site is required for this effect (data not shown). We therefore generated point mutants of 23 serine residues that could possibly undergo phosphorylation within the protein; however, no single point mutation showed any variation in comparison with wild type Mal. Given that we observed multiple phosphobands in the in vitro kinase assay, it is likely that Mal is phosphorylated on more than one site by the IRAKs and that multiple phosphorylations are required for Mal degradation.
TABLE 1.
Mal is phosphorylated on threonine 28 by IRAK1 and IRAK4
Purified recombinant Mal was incubated with either recombinant IRAK1 or IRAK4 in kinase buffer containing ATP and subjected to SDS-PAGE. Upper bands that were visible in lanes containing ATP were excised from the gel and subjected to liquid chromatography-mass spectrometry.
| Phosphopeptide ion | Sequence |
|---|---|
| 744.86 (2+) | MADWFRQpTLLK |
| 753.39 (2+) | MADWFRQpTLLK |
| 1006.48 (2+) | KPLGKMADWFRQpTLLK |
| 665.38 (1+) | ZpTLLKa |
| 710.39 (1+) | QpTLLK + formic acid |
a Z, pyro-Glu.
Mal Undergoes LPS-induced Ubiquitination and Degradation
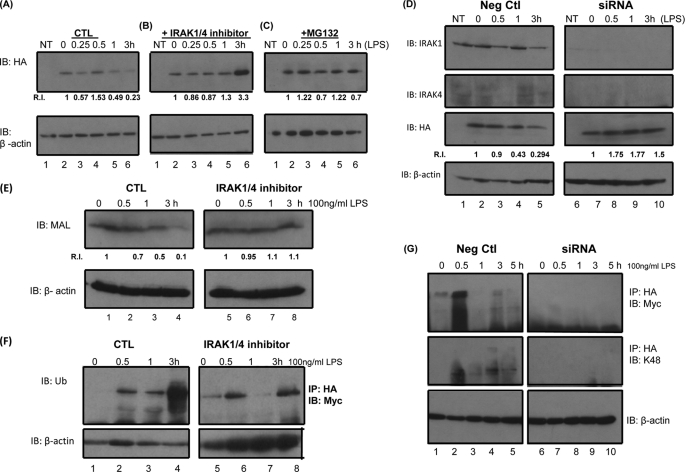
It has previously been shown that Mal undergoes LPS-induced degradation (18). In order to examine if the IRAKs play a role in this event, we pretreated cells with an IRAK1/4 inhibitor for 3 h prior to stimulation with LPS. LPS induced the degradation of Mal in cells after treatment times of 60 min and 3 h (Fig. 4A, lanes 5 and 6). The inhibitor prevented the degradation of Mal and also appeared to stabilize the protein at later time points (Fig. 4B, lanes 5 and 6). Pretreatment of cells with the proteosome inhibitor MG132 also prevented LPS-induced degradation of Mal, as shown in Fig. 4C (lanes 5 and 6).
FIGURE 4.
Mal undergoes LPS induced ubiquitination and degradation. A–C, HEK293 MTC cells were mock-transfected (NT) or transiently transfected with 100 ng of HA-Mal. Samples were stimulated with 100 ng/ml LPS for the indicated times (A). Samples were pretreated with 5 μm IRAK1/4 inhibitor (B) or with 5 μm MG132 (C). Samples were probed with HA antibody for Mal expression or with β-actin antibody to ensure equal protein loading in each sample. Densitometric analyses of band intensities were determined using ImageJ. Relative intensity (R.I.) values were calculated relative to time 0 for each experiment. D, 293 MTC cells were transfected with siRNA oligonucleotides specific to IRAK1 and IRAK4 (50 nm of each) or with an equivalent concentration of a negative control. After 72 h, cells were transfected with 100 ng of Mal. After 24 h, samples were stimulated with 100 ng/ml LPS for the indicated times. Samples were probed with HA antibody for Mal expression or with β-actin antibody to ensure equal protein loading in each sample. Densitometric analyses of band intensities were determined using ImageJ. Relative intensity values were calculated relative to time 0 for each experiment. E, THP1 cells were pretreated with 500 nm IRAK1/4 inhibitor followed by LPS stimulation (100 ng/ml) for the indicated times. Samples were probed with anti-Mal antibody or with β-actin antibody to ensure equal protein loading in each sample. Densitometric analyses of band intensities were determined using ImageJ. Relative intensity values were calculated relative to time 0 for each experiment. F, HEK293 MTC cells were transfected with the 100 ng of Mal and 100 ng of Myc-ubiquitin. After 24 h, samples were pretreated with 5 μm IRAK1/4 inhibitor followed by LPS stimulation (100 ng/ml) for the indicated times. Samples were immunoprecipitated with HA antibody and probed with Myc antibody (top). Lysates were probed with β-actin as a loading control (bottom). G, U373 cells were transfected with siRNA oligonucleotides specific to IRAK1 and IRAK4 (50 nm) or with an equivalent concentration of a negative control. After 72 h, cells were transfected with 100 ng of Mal and 100 ng of Myc-ubiquitin, and after 6 h, samples were treated with 1 μm lactacystin. After 24 h, samples were stimulated with 100 ng/ml LPS for the indicated times. Samples were probed with HA antibody for Mal expression or with β-actin antibody to ensure equal protein loading. Results shown are representative of two or three individual experiments.
In order to further confirm a role for the IRAKs in LPS-induced Mal degradation, we carried out siRNA experiments to knockdown IRAK1 and IRAK4 in 293MTC cells. Both proteins were successfully knocked down, as shown in Fig. 4D (top panel and second panel, respectively). Knockdown of IRAK1 and IRAK4 reduced LPS-induced degradation of Mal as shown in Fig. 4D (panel 3; compare lanes 9 and 10 with lanes 4 and 5).
To further confirm the involvement of IRAKs in LPS-induced Mal degradation at a physiological level, we pretreated the human monocytic cell line, THP1, with an IRAK1/4 inhibitor for 3 h prior to stimulation with LPS. In agreement with previous results, LPS induced degradation of endogenous Mal maximally at 3 h. Pretreatment of cells with the IRAK1/4 inhibitor attenuated this effect (Fig. 4E, compare lanes 1–4 with lanes 5–8).
It is well established that ubiquitination of proteins occurs prior to degradation. In order to determine if Mal undergoes this form of modification, we overexpressed Mal together with Myc-tagged ubiquitin. As shown in Fig. 4F, stimulation of cells with LPS promoted Mal ubiquitination in a time-dependent manner. The IRAK1/4 inhibitor blocked LPS-induced Mal ubiquitination particularly at the later time point of 3 h (Fig. 4F, compare lanes 4 and 8). An additional IRAK1/4 inhibitor also blocked LPS-induced Mal ubiquitination at a lower concentration (100 nm) (data not shown). We also examined Mal ubiquitination in an additional LPS-responsive astrocytoma cell line (U373). In this case, ubiquitination of Mal occurred at an earlier time point (Fig. 4G, top left, lane 2). However, Mal ubiquitination was completely abolished when both IRAK1 and IRAK4 were knocked down, as shown in Fig. 4G (top right). Finally, we confirmed that Mal is subject to Lys48-linked ubiquitination that is indicative of degradation, using a Lys48-specific antibody as shown in Fig. 4G (middle).
DISCUSSION
In this paper, we demonstrate that Mal can interact with IRAK1 and IRAK4 and acts as a substrate for both enzymes in vitro. Furthermore, we show that Mal undergoes IRAK-dependent ubiquitination and degradation, which may serve to negatively regulate TLR signaling. Mal therefore joins Pellino2, Pellino3, IRAK1, IRF7, STAT3, and histone H3 as an IRAK substrate. Of these, IRAK1 is the best studied in terms of phosphorylation. Kollewe et al. (17) have used a series of IRAK1 peptides and deletion mutants in order to analyze the sequence of events surrounding IRAK1 phosphorylation. It is thought that IRAK1 is initially phosphorylated on Thr209, which lies in the ProST region of the protein. In vitro, this reaction is catalyzed by IRAK1 itself; however, phosphorylation of a peptide containing this residue was also observed with IRAK4. The addition of a phosphate group results in a conformational change in IRAK1 and effectively opens the kinase domain to expose an activation loop, which is then phosphorylated on Thr387 and results in a full enzymatic activity. At this point, IRAK1 undergoes extensive hyperphosphorylation in the ProST region, leading to its eventual dissociation from MyD88 and Tollip and engagement with downstream signaling proteins. Hyperphosphorylation of IRAK1 also serves to limit the availability of the protein, which undergoes phosphorylation-dependent degradation.
In this study, we observed that co-expression of Mal with both IRAK4 and IRAK1 leads to depletion of Mal from cell lysates. This effect is dependent on the kinase activity of both enzymes because catalytically inactive forms did not cause degradation. It should be noted that Mal degradation by IRAK4 may also involve IRAK1 because IRAK1 is known to be activated upon overexpression of IRAK4. Other examples of degradation of components of the TLR system have emerged. TLR3, TLR5, and TLR9 have all been shown to be depleted from cell lysates when overexpressed with Triad3A, an E3 ubiquitin ligase that functions to modulate TLR signaling (19). Similarly, MyD88 has been shown to undergo ubiquitin-mediated proteolysis following stimulation of cells with TGF-β (20). A recent paper has investigated the effect of pharmacological inhibition of IRAK1 and IRAK4 on proinflammatory cytokine production in human cells (21). The most profound effects were observed with a dual inhibitor of both IRAKs, suggesting redundancy in the activities of the two enzymes. It has also been demonstrated that IRAK1/4 inhibitors result in a decrease in the production of interleukin-8 following LPS stimulation (22). We found that LPS-induced Mal degradation is also prevented upon inhibition of IRAK1/4. In addition, we show that knocking down both IRAK1 and IRAK4 blocks LPS-induced degradation of Mal.
Both IRAK1 and IRAK4 are capable of phosphorylating Mal in a cell-free system. IRAK4 itself also undergoes phosphorylation when overexpressed in cells; however, it is not clear at present if this is an autophosphorylation event or if indeed another kinase is involved. Mass spectrometric analysis identified threonine 28 as a novel phosphorylation site on Mal. Single point mutants of threonine and possible serine phosphorylation sites were not enough to prevent activation or degradation of Mal, further emphasizing the complex nature of the phosphorylation of Mal by IRAK1 and -4. This also suggests that it is more than a single site that is important. We are currently generating double and triple mutants to try to determine the key phosphorylation sites on Mal. We were unable to detect phosphorylation of endogenous Mal using phosphopeptide antibodies due to the low level of Mal expression and ensuing detection problems. MyD88 was not phosphorylated by the IRAKs; nor did it undergo degradation. It is possible that this process may serve to dampen TLR2 and TLR4 responses via specific targeting of Mal.
Protein phosphorylation is often a prelude to ubiquitination. This may also be the case for Mal because pretreatment with IRAK1/4 inhibitors or depletion of the kinases from cells prevented LPS-induced Mal ubiquitination. Furthermore, we confirmed that Mal undergoes Lys48-linked ubiquitination, which serves as a marker for proteosomal degradation. It has previously been shown that SOCS1 (suppressor of cytokines 1) is involved in Mal polyubiquitination and degradation following tyrosine phosphorylation by Bruton's tyrosine kinase (16). More recently, it has been reported that wild-type but not kinase-inactive forms of IRAK1 and IRAK4 promote the polyubiquitination and degradation of Pellino family members, whereas IRAK4 has been shown to induce the degradation of IRAK1 in a proteosome-independent manner (23, 24).
In conclusion, Mal is a substrate of both IRAK1 and IRAK4 in vitro; the consequence of Mal phosphorylation is ubiquitination followed by degradation. This is in contrast to phosphorylation of Mal by Bruton's tyrosine kinase, which is required for Mal activation (18). We have therefore revealed an additional substrate for IRAK1 and IRAK4 and uncovered what is likely to be an important control mechanism in TLR2 and TLR4 signal transduction.
This work was supported by grants from the Science Foundation of Ireland and the Health Research Board.
- TLR
- Toll-like receptor
- LPS
- lipopolysaccharide
- IRAK
- interleukin-1 receptor-associated kinase
- TIR
- Toll/interleukin-1 receptor
- HA
- hemagglutinin
- siRNA
- small interfering RNA.
REFERENCES
- 1.Wesche H., Henzel W. J., Shillinglaw W., Li S., Cao Z. (1997) Immunity 7, 837–847 [DOI] [PubMed] [Google Scholar]
- 2.Fitzgerald K. A., Palsson-McDermott E. M., Bowie A. G., Jefferies C. A., Mansell A. S., Brady G., Brint E., Dunne A., Gray P., Harte M. T., McMurray D., Smith D. E., Sims J. E., Bird T. A., O'Neill L. A. (2001) Nature 413, 78–83 [DOI] [PubMed] [Google Scholar]
- 3.Horng T., Barton G. M., Medzhitov R. (2001) Nat. Immunol. 2, 835–841 [DOI] [PubMed] [Google Scholar]
- 4.Yamamoto M., Sato S., Mori K., Hoshino K., Takeuchi O., Takeda K., Akira S. (2002) J. Immunol. 169, 6668–6672 [DOI] [PubMed] [Google Scholar]
- 5.Oshiumi H., Sasai M., Shida K., Fujita T., Matsumoto M., Seya T. (2003) J. Biol. Chem. 278, 49751–49762 [DOI] [PubMed] [Google Scholar]
- 6.Fitzgerald K. A., Rowe D. C., Barnes B. J., Caffrey D. R., Visintin A., Latz E., Monks B., Pitha P. M., Golenbock D. T. (2003) J. Exp. Med. 198, 1043–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gottipati S., Rao N. L., Fung-Leung W. P. (2008) Cell. Signal. 20, 269–276 [DOI] [PubMed] [Google Scholar]
- 8.Meylan E., Tschopp J. (2008) Nat. Immunol. 9, 581–582 [DOI] [PubMed] [Google Scholar]
- 9.Kobayashi K., Hernandez L. D., Galán J. E., Janeway C. A., Jr., Medzhitov R., Flavell R. A. (2002) Cell 110, 191–202 [DOI] [PubMed] [Google Scholar]
- 10.Strelow A., Kollewe C., Wesche H. (2003) FEBS Lett. 547, 157–161 [DOI] [PubMed] [Google Scholar]
- 11.Huang Y., Li T., Sane D. C., Li L. (2004) J. Biol. Chem. 279, 51697–51703 [DOI] [PubMed] [Google Scholar]
- 12.Liu G., Park Y. J., Abraham E. (2008) FASEB J. 22, 2285–2296 [DOI] [PubMed] [Google Scholar]
- 13.Uematsu S., Sato S., Yamamoto M., Hirotani T., Kato H., Takeshita F., Matsuda M., Coban C., Ishii K. J., Kawai T., Takeuchi O., Akira S. (2005) J. Exp. Med. 201, 915–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Butler M. P., Hanly J. A., Moynagh P. N. (2007) J. Biol. Chem. 282, 29729–29737 [DOI] [PubMed] [Google Scholar]
- 15.Dunne A., Ejdeback M., Ludidi P. L., O'Neill L. A., Gay N. J. (2003) J. Biol. Chem. 278, 41443–41451 [DOI] [PubMed] [Google Scholar]
- 16.Mansell A., Smith R., Doyle S. L., Gray P., Fenner J. E., Crack P. J., Nicholson S. E., Hilton D. J., O'Neill L. A., Hertzog P. J. (2006) Nat. Immunol. 7, 148–155 [DOI] [PubMed] [Google Scholar]
- 17.Kollewe C., Mackensen A. C., Neumann D., Knop J., Cao P., Li S., Wesche H., Martin M. U. (2004) J. Biol. Chem. 279, 5227–5236 [DOI] [PubMed] [Google Scholar]
- 18.Gray P., Dunne A., Brikos C., Jefferies C. A., Doyle S. L., O'Neill L. A. (2006) J. Biol. Chem. 281, 10489–10495 [DOI] [PubMed] [Google Scholar]
- 19.Chuang T. H., Ulevitch R. J. (2004) Nat. Immunol. 5, 495–502 [DOI] [PubMed] [Google Scholar]
- 20.Naiki Y., Michelsen K. S., Zhang W., Chen S., Doherty T. M., Arditi M. (2005) J. Biol. Chem. 280, 5491–5495 [DOI] [PubMed] [Google Scholar]
- 21.Song K. W., Talamas F. X., Suttmann R. T., Olson P. S., Barnett J. W., Lee S. W., Thompson K. D., Jin S., Hekmat-Nejad M., Cai T. Z., Manning A. M., Hill R. J., Wong B. R. (2009) Mol. Immunol. 46, 1458–1466 [DOI] [PubMed] [Google Scholar]
- 22.Bhattacharyya S., Borthakur A., Pant N., Dudeja P. K., Tobacman J. K. (2007) Am. J. Physiol. Gastrointest. Liver Physiol. 293, G429–G437 [DOI] [PubMed] [Google Scholar]
- 23.Butler M. P., Hanly J. A., Moynagh P. N. (2007) J. Biol. Chem. 282, 29729–29737 [DOI] [PubMed] [Google Scholar]
- 24.Kubo-Murai M., Hazeki K., Nigorikawa K., Omoto T., Inoue N., Hazeki O. (2008) J. Biochem. 143, 295–302 [DOI] [PubMed] [Google Scholar]