Abstract
The serine/threonine protein kinase LKB1 is a tumor suppressor gene mutated in Peutz-Jeghers syndrome patients. The mutations are found also in several types of sporadic cancer. Although LKB1 is implicated in suppression of cell growth and metastasis, the detailed mechanisms have not yet been elucidated. In this study, we investigated the effect of LKB1 on cell motility, whose acquisition occurs in early metastasis. The knockdown of LKB1 enhanced cell migration and PAK1 activity in human colon cancer HCT116 cells, whereas forced expression of LKB1 in Lkb1-null mouse embryonic fibroblasts suppressed PAK1 activity and PAK1-mediated cell migration simultaneously. Notably, LKB1 directly phosphorylated PAK1 at Thr109 in the p21-binding domain in vitro. The phosphomimetic T109E mutant showed significantly lower protein kinase activity than wild-type PAK1, suggesting that the phosphorylation at Thr109 by LKB1 was responsible for suppression of PAK1. Consistently, the nonphosphorylatable T109A mutant was resistant to suppression by LKB1. Furthermore, we found that PAK1 was activated in the hepatocellular carcinomas and the precancerous liver lesions of Lkb1(+/−) mice. Taken together, these results suggest that PAK1 is a direct downstream target of LKB1 and plays an essential role in LKB1-induced suppression of cell migration.
Keywords: Cancer/Colon, Cancer/Liver, Cell/Migration, Diseases/Cancer, Enzymes/Kinase, Signal Transduction/Protein Kinases/Serine/Threonine
Introduction
LKB1 is a serine/threonine kinase whose mutations have been found not only in Peutz-Jeghers syndrome patients (1–3) but also in various types of sporadic cancer (4–6). These results suggest that LKB1 is a tumor suppressor gene. In addition, we and others (7–9) have previously shown that the heterozygous Lkb1 mutations in mice cause gastrointestinal hamartomas after 20 weeks of age and cause hepatocellular carcinomas (HCCs)2 after 50 weeks (10). Notably, the Lkb1 gene in all HCC and hepatic precancerous lesions shows loss of heterozygosity (10, 11). These phenotypes in Lkb1(+/−) mice further indicate that LKB1 is a tumor suppressor gene.
In mammalian cells, LKB1 forms a complex with STE20-related adaptor pseudokinase (STRAD) and scaffolding protein MO25, both of which are required for LKB1 enzymatic activity (12, 13). It can phosphorylate and activate at least 14 kinases, including AMP-activated protein kinase (AMPK) and microtubule-associated protein/microtubule affinity-regulating kinases (MARKs) (5). Activation of AMPK by LKB1 leads to inactivation of mammalian target of rapamycin complex 1 via phosphorylation of the tuberous sclerosis complex 1/2, and this pathway has been implicated in tumor suppressor functions of LKB1. In addition to growth control, LKB1 also plays important roles in establishing cell polarity in mammalian cells (14). LKB1 regulates tight junction assembly and cell polarity through AMPK in mammalian cells (15, 16), and we have shown that LKB1 suppresses tubulin polymerization by activating MARK microtubule-associated protein signaling (17). We have also reported that HCCs in Lkb1(+/−) mice metastasize to the lungs (10). However, the mechanisms by which LKB1 suppresses cancer metastasis have not yet been explored. In this study, we have investigated the effect of LKB1 on cell motility whose acquisition occurs in early metastasis (18).
The serine/threonine kinase PAK1 is known as a key regulator of cell motility, proliferation, differentiation, and survival (19). It can activate diverse signaling pathways, including LIM motif-containing protein kinase 1, mitogen-activated protein kinase (MAPK), and NF-κB signaling (20). PAK1 interacts with the GTP-bound forms of Cdc42 or Rac, which cause PAK1 activation (21). In addition to activation by Cdc42/Rac, its activity is positively regulated also by other signaling molecules, including phosphoinositide 3-kinase (22), 3-phosphoinositide-dependent kinase-1 (23), and AKT (24).
PAK1 is overexpressed in various types of cancer, including breast (25, 26), colon (27), and liver (28) cancers. A high level of PAK1 expression is correlated with more aggressive tumor behaviors such as metastatic phenotype and advanced tumor stages in hepatocellular carcinomas (28) as well as in colorectal cancer (27). Consistently, overexpression of constitutively active mutant PAK1 (T423E) helps develop malignant mammary tumors in a transgenic mouse model (29). These results indicate that PAK1 plays a key role in carcinogenesis and cancer metastasis. Here, we present evidence that LKB1 inhibits cell motility and PAK1-mediated signaling through direct phosphorylation of PAK1 at Thr109 in the p21-binding domain.
EXPERIMENTAL PROCEDURES
Cell Culture
HCT116 cells and HEK293T cells were maintained in low glucose Dulbecco's modified Eagle's medium (Nacalai Tesque, Kyoto, Japan) supplemented with 10% fetal bovine serum (BioWest, Nuaillé, France), and 100 units of penicillin/streptomycin.
Mice
Construction of Lkb1 knock-out mice has been described previously (10). We only used males due to the low incidence of nodular foci and HCCs in female Lkb1(+/−) mice, as reported previously. All animal experiments were approved by the Animal Care and Use Committee of Kyoto University.
Reagents
A mouse monoclonal anti-LKB1 (clone Ley37D/G6), anti-Myc (clone 9E10) and rabbit polyclonal anti-PAK1 antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit polyclonal anti-phospho-PAK1 (Ser144)/PAK2 (Ser141), anti-phospho-PAK1 (Ser199/204)/PAK2 (Ser201), anti-PAK1 and anti-phospho-Vasodilator-stimulated phosphoprotein (VASP) (Ser239) antibodies were from Cell Signaling Technology (Danvers, MA), and rabbit polyclonal anti-VASP antibody was from EMD Chemicals (Gibbstown, NJ). A mouse monoclonal anti-β-actin antibody was from Sigma- Aldrich (St. Louis, MO), and a mouse monoclonal anti-Hemagglutinin antibody was from Invivogen (San Diego, CA). Two different small interfering RNAs against human LKB1, and scramble siRNA pools were purchased from Dharmacon, Inc. (Lafayette, CO).
cDNA
The following cDNAs were isolated by standard PCR-based cloning techniques: human LKB1, NCBI accession no. NM_000455 and human STRAD, respectively, NCBI accession no. NM_001003787. HA and FLAG tags were attached to the N termini of LKB1 and STRAD, as described previously (17). LKB1 (kinase-dead, a catalytically inactive version of LKB1 with a D194A mutation (30)), was generated with QuikChange II site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's protocol. Wild-type of PAK1, constitutively active mutant T423E, and a dominant-negative mutant H83LH86LK299R were kindly gifted by Dr. Jonathan Chernoff (Fox Chase Cancer Center). Appropriate PCR primers were used to generate the point mutants PAK1-T109E and PAK1-T109A with the QuikChange II site-directed mutagenesis kit. These BamHI/EcoRI fragments were subcloned into pCMV-Tag 3B (Stratagene), which has an N-terminal Myc tag and a cytomegalovirus promoter.
PAK1-K299R Lentivirus
The NotI/SalI fragment of pCMV-Tag3B-PAK1-K299R was subcloned into pLEX vector (Open Biosystems, Huntsville, AL). Recombinant lentivirus encoding K299R was prepared according to the manufacturer's protocol.
Establishment of an Lkb1-null Mouse Embryonic Fibroblast (MEF) Cell Line
Lkb1(+/−) mice (C57BL/6N background) were crossed with outbred ICR mice, and offspring were intercrossed. Lkb1-null embryos at 9.5 days post-coitum were minced, trypsinized briefly, and placed on 24-well plates. These cells were cultured in RPMI 1640 (Sigma) with 10% (v/v) fetal bovine serum and 50% (v/v) conditioned medium from the MEFs that were derived from wild-type embryos at 12.5 days post-coitum. We obtained a spontaneously immortalized cell line (MEF3) by continuous passages (>3 months) of the Lkb1-null MEFs and further established the subclone MEF3-2 cell line. Finally, we cultured MEF3-2 in Dulbecco's modified Eagle's medium (Sigma) with 10% (v/v) fetal bovine serum.
Recombinant Adenoviruses
Recombinant adenoviruses were constructed by Adeno-X expression system 1 (Clontech Laboratories, Mountain View, CA), according to the manufacturer's protocol. The titers of the recombinant adenoviruses were determined by the method previously reported by others (31).
Wound-healing Assay
Cells were infected with recombinant adenovirus; after 24 h of infection, wounds were incised by scratching the cell monolayer using 10-μl pipette tips. Photographs were taken at 0 and 20 h after the wound was made. Cell migration was normalized so that 100% represents the migration distance of control cells.
Transfection
HEK293T cells were transfected with plasmid DNA of the indicated PAK1 and LKB1 expression vectors using Lipofectamine 2000 (Invitrogen). After 24 h of transfection, cell lysates were prepared for Western blotting or in vitro kinase assay. Lipofectamine 2000 RNAiMAX (Invitrogen) was used for small interfering RNA (siRNA) transfection according to manufacturer's protocol.
Recombinant PAK1 Proteins Produced in Escherichia coli
PAK1-PBD was isolated by standard PCR-based cloning techniques. The PAK1-PBD-T109A mutant was generated with the QuikChange II site-directed mutagenesis kit. To attach GST protein to PAK1-PBD, PAK1-PBD-T109A, PAK1-K299R, or PAK1-T109A/K299R was inserted in the BamHI-double digested pGEX-6P-1 vector (GE Healthcare). pGEX-2TK-VASP-(158–277) was generated previously (32). 200 ml of 2×YT medium (1.6% tryptone, 1% yeast extract, and 0.5% NaCl) medium was inoculated with E. coli (BL21 strain) containing the recombinant pGEX-2TK plasmid that encodes GST-VASP-(158–277), or the recombinant pGEX-6P-1 plasmid that encodes GST-PAK1-PBD, GST-PAK1-PBD-T109A, GST-PAK1-K299R, or GST-PAK1-T109A/K299R and was incubated at 37 °C in the presence of 100 μg/ml ampicillin until A600 reached 0.8. To induce expression of the GST-tagged proteins, isopropyl-d-thiogalactopyranoside was added at 100 μm. After culture for 24 additional hours at 25 °C, cells were harvested, and recombinant proteins were purified as described previously (17).
Western Blotting
Cells were lysed in the lysis buffer (50 mm Tris-HCl (pH 7.4), 150 mm NaCl, 5 mm Na4P2O7, 10 mm β-glycerophosphate, 25 mm NaF, 1 mm EDTA, 1 mm EGTA, 1 mm Na3VO4, and 1% (v/v) Triton X-100) containing Complete mini protease inhibitor mixture (Roche Applied Science). Cell lysates were separated by SDS-PAGE and transferred to Immobilon-P membrane (Millipore, Billerica, MA). After blocking with Blocking One or Blocking One-P (Nacalai Tesque, Kyoto, Japan), the membranes were probed with the indicated antibodies. The signals were visualized by Immobilon western detection system (Millipore) or ECL Western blotting detection reagents (GE Healthcare).
In Vitro Phosphorylation Assay
Phosphorylation reactions were performed at 30 °C in the phosphorylation reaction buffer (50 mm Tris-HCl (pH 7.0), 10 mm MgCl2, 2 mm MnCl2, 5 mm β-glycerophosphate, 100 μm Na3VO4, 1 mm dithiothreitol, and 200 μm ATP). As shown in Figs. 1C and 2B, the immunoprecipitates obtained from cell lysates using anti-PAK1 antibody were incubated with GST-VASP-(158–277) for 20 min. The reactions were stopped by the addition of 3× sample buffer (150 mm Tris-HCl (pH 6.8), 6% (w/v) SDS, 0.03% (w/v) bromphenol blue, 30% (v/v) glycerol, and 15% (v/v) 2-mercaptoethanol). The anti-Myc immunoprecipitates from HEK293T cell lysates were incubated with 5 μg of GST-VASP-(158–277) for 20 min with [γ-32P]-ATP (see Figs. 3A and 5). The reactions were then terminated by the addition of 3× sample buffer. The samples were electrophoresed on 5–20% SDS-PAGE gels. The blots were exposed to a phosphor imaging plate (Fujifilm, Tokyo, Japan). The signals were detected by using BAS-5000 Bio-imaging Analyzer (Fujifilm). All assays using [γ-32P]ATP were done at the Radioisotope Research Center of Kyoto University. Fold induction was determined using NIH Image (version 1.62). Recombinant LKB1/STRAD/MO25 (Millipore) was preincubated with recombinant PAK1 (EMD Chemicals) in the presence of ATP for 20 min, and then the reaction mixtures were incubated with 5 μg of GST-VASP-(158–277) for 20 min (see Fig. 3B). The cellular levels of phosphorylated VASP were detected using an anti-phospho-VASP (Ser239) antibody.
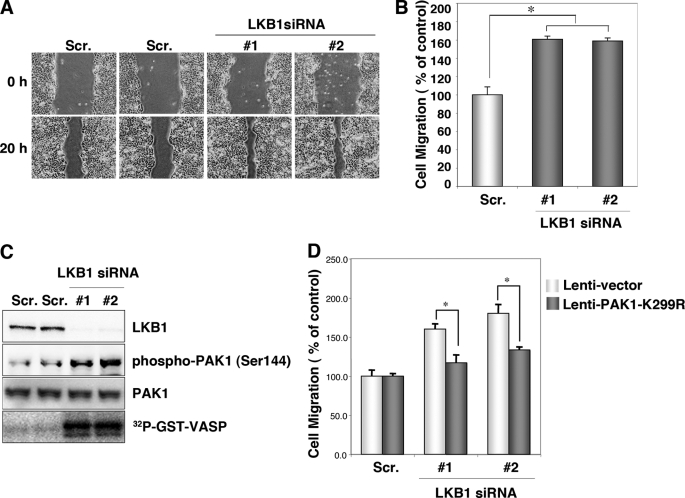
FIGURE 1.
Knockdown of endogenous LKB1 increases cell migration and PAK1 activity in HCT116 cells. A, knockdown of LKB1 increases cell migration. HCT116 cells were transfected with two different specific siRNAs against human LKB1. Control cells were transfected with a scrambled (Scr.) siRNA pool. After 24 h of plating, scratches were made with 10-μl pipette tips. Photographs were taken at 0 and 20 h after the wound was made. B, cell migration was normalized so that 100% represented migration distance of control cells. Error bars indicate S.D. The asterisk indicates significant increases compared with control cells (p < 0.001). C, cellular levels of phospho-PAK1 (Ser144), total PAK1 and in vitro PAK1 activity in LKB1 knockdown HCT116 cells. Lysates were prepared from HCT116 cells transfected with the indicated siRNAs. The levels of phospho-PAK1 (Ser144) and PAK1 were determined by Western blotting with the indicated antibodies. Lysates were prepared from HCT116 cells transfected with the indicated siRNAs and immunoprecipitated using an anti-PAK1 antibody. The precipitates were used for in vitro kinase assays using GST-VASP-(158–277) as a substrate. The phosphorylation of GST-VASP was visualized using BAS-5000 Bio-imaging Analyzer. D, induction of PAK1-K299R suppressed the increase in migration in LKB1 knockdown cells. The cells transfected with the indicated siRNAs were then infected with lentivirus lenti-PAK1-K299R or lenti-vector. At 24 h post-infection, cultures were scratches were made with 10-μl pipette tips. Photographs were taken at 0 and 20 h thereafter. Cell migration was normalized so that 100% represented the migration distance of control cells. Error bars indicate S.D. The asterisk represents significant increases compared with control cells (p < 0.001).
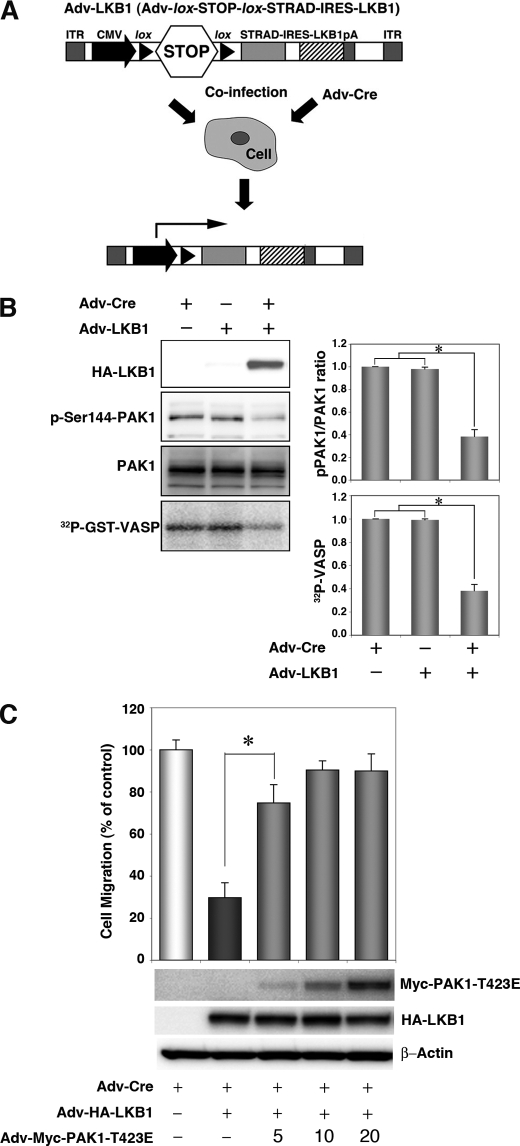
FIGURE 2.
Expression of LKB1 in the Lkb1-null MEF cell line (MEF3-2) inhibits cell migration and PAK1 activation. A, schematic structure of the adenoviral construct, adapted from Ref. 17. ITR, inverted terminal repeat; CMV, human cytomegalovirus immediate early promoter; lox, loxP site sequence; STOP; transcriptional termination cassette; IRES, internal ribosome entry site; pA, polyadenylation signal. Co-infection with Adv-Cre removed the STOP cassette from Adv-LKB1. B, cellular levels of PAK1 autophosphorylation, total PAK1, and in vitro PAK1 activity in LKB1-expressing MEF3-2 cells. MEF 3-2 cells were co-infected with recombinant adenoviruses Adv-Cre and Adv-LKB1. Control cells were infected with Adv-Cre alone. The levels of phospho-PAK1 and PAK1 were determined by Western blotting with anti-phospho-PAK1 (Ser144)/PAK2 (Ser141) and anti-PAK1 antibodies, respectively. Lysates were prepared from MEF3-2 cells infected with the indicated recombinant adenoviruses and immunoprecipitated using an anti-PAK1 antibody. The precipitates were used for in vitro kinase assays with GST-VASP-(158–277) as a substrate. The phosphorylation of GST-VASP was visualized and quantified using BAS-5000 Bio-imaging Analyzer. Error bars indicate S.D. The asterisk represents significant decreases compared with control cells (p < 0.001). C, the constitutively active mutant of PAK1 rescues LKB1-induced suppression of cell migration in MEF3-2 cells. MEF 3-2 cells were co-infected with recombinant adenoviruses Adv-Cre, Adv-LKB1, and Adv-PAK1-T423E at a multiplicity of infection of 5, 10, or 20. Control cells were infected with Adv-Cre alone. Cell migration assays were performed as Fig. 1. Error bars indicate S.D. The asterisk represents significant changes in Adv-Cre/Adv-LKB1/Adv-PAK1-T423E (5) infected cells compared with Adv-Cre/Adv-LKB1-infected cells (p < 0.001). Similar results were obtained in three independent experiments.
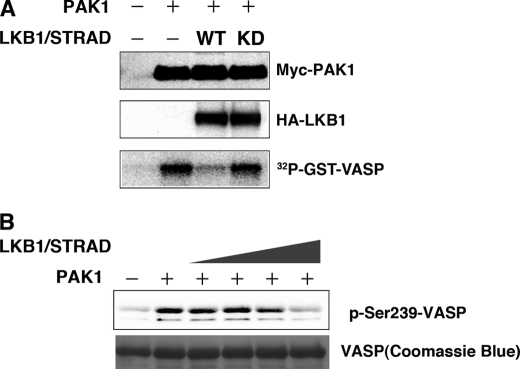
FIGURE 3.
LKB1 activity is necessary for suppression of PAK1 activity. A, the wt LKB1, but not the kinase-dead mutant (KD), inhibits PAK1 activity. HEK293T cells were co-transfected with the indicated constructs. At 24 h post-transfection, immunoprecipitates were prepared with an anti-Myc antibody. The precipitates were used for in vitro kinase assays using GST-VASP-(158–277) as a substrate. The kinase activities were visualized using BAS-5000 Bio-imaging Analyzer. Cellular levels of Myc-PAK1 and HA-LKB1 were determined by Western blotting with anti-Myc or anti-HA antibody. B, recombinant LKB1 complex (LKB1-STRAD-MO25) directly inhibits PAK1 activity in vitro in a dose-dependent manner. The LKB1 complex (LKB1-STRAD-MO25) was preincubated with recombinant PAK1 and ATP in the absence of any PAK1 substrates, followed by addition of GST-VASP. The phosphorylated VASP was detected using an anti-phospho-VASP (Ser239) antibody. Coomassie Brilliant Blue was used for detection of GST-VASP-(158–277). Similar results were obtained in three independent experiments.
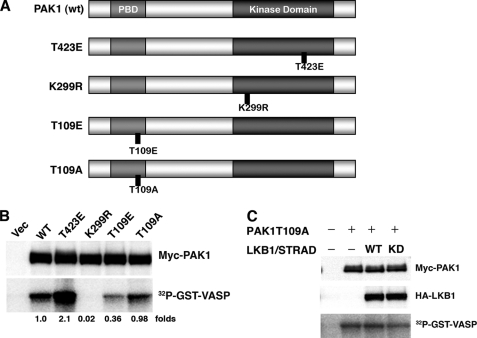
FIGURE 5.
Phosphorylation of PAK1 at Thr109 reduces its activity. A, structures of PAK1 constructs. wt PAK1 contains the p21-binding domain (light gray) and the kinase domain (dark gray). The noncatalytic domains are in white. Also displayed are the constitutively active mutant (T423E), dominant-negative mutant (K299R), phosphorylation-mimicked mutant for Thr109 (T109E), and the nonphosphorylatable mutant for Thr109 (T109A). B, the kinase activities of PAK1 mutants. The HEK293T cells were co-transfected with the indicated constructs. At 24 h post-transfection, the immunoprecipitates were prepared with an anti-Myc antibody. The precipitates were used for in vitro kinase assays using GST-VASP-(158–277) as a substrate. Cellular levels of Myc-PAK1 were determined by Western blotting with anti-Myc antibody. C, LKB1 (wt or kinase-dead (KD)) failed to inhibit the activity of the PAK1-T109A mutant. Cultured HEK293T cells were co-transfected with the indicated constructs. The kinase activities were determined as described under “Experimental Procedures.” Cellular levels of Myc-PAK1 and HA-LKB1 were determined by Western blotting with anti-Myc or anti-HA antibody. Similar results were obtained in three independent experiments. Vec, vector.
Statistical Analysis
Results of the experimental studies were reported as mean ± S.D. Differences were analyzed by Student's t test. A value of p < 0.05 was regarded as significant.
RESULTS
Knockdown of Endogenous LKB1 Enhances Cell Migration and PAK1 Activity in HCT116 Cells
We have reported previously that HCCs in Lkb1(+/−) mice metastasize to the lungs (10). Consistently, LKB1 also appears to suppress lung cancer metastasis (33). To investigate the mechanism(s) by which LKB1 suppressed metastasis of cancer cells, we determined the effect of LKB1 knockdown on cell migration using a wound-healing assay. As test cells, we used a human colon cancer cell line HCT116 that expressed a significant level of LKB1 endogenously. HCT116 cells transfected with LKB1 siRNA (1 and 2) migrated 1.5× faster than those with scrambled siRNA pool (Fig. 1, A and B, Scr.). Because PAK1 is an important regulator of cell migration (34), we investigated a possible role of PAK1 in cell migration enhanced by LKB1 knockdown. To determine the PAK1 activity in LKB1 knockdown cells, we then analyzed the level of phospho-PAK1 (Ser144), its active form (35). Knockdown of LKB1 increased the cellular level of the phosphorylated PAK1, without affecting that of total PAK1 (Fig. 1C). These results suggest that knockdown of LKB1 increases the PAK1 activity. To confirm this interpretation, we determined the effect of LKB1 knockdown on in vitro PAK1 activity. Because histone H4, a widely used substrate for PAK1, was phosphorylated by LKB1 (data not shown), we screened in silico candidate substrate sequences for PAK1 using the consensus phosphorylation motif ((K/R)(R/X)(X)(pS/pT)) (34) as a query. As a candidate, we identified VASP-(158–277), which is a major substrate for both PKA and PKG (32, 36). We first verified that recombinant PAK1 efficiently phosphorylated GST-VASP-(158–277) (data not shown). We found that PAK1 activity was significantly higher in LKB1 knockdown cells than in the control cells (Fig. 1C). To further investigate whether enhanced PAK1 activity contributed to the cell migration increase in LKB1 knockdown cells, we tested the effect of a dominant-negative mutant PAK1-K299R in LKB1 knockdown cells. Notably, expression of PAK1-K299R suppressed the increase in migration of these cells (Fig. 1D). These results suggest that LKB1 inhibits PAK1 activity, which can suppress cell migration.
Expression of LKB1 in Lkb1-null MEFs Inhibits PAK1 Activity and Cell Migration
To investigate the role of LKB1 in suppression of PAK1-mediated signaling, we further evaluated the effect of forced LKB1 expression on PAK1 activation in mouse embryonic fibroblasts that lack wild-type LKB1 (MEF3-2). To this end, we used the recombinant adenovirus encoding LKB1/STRAD (Adv-LKB1) that contained a transcription termination cassette (STOP sequence) flanked by the loxP sequences and expressed LKB1/STRAD by co-infection of MEF3-2 with Adv-Cre, which removed the STOP sequence from Adv-LKB1. The floxed inducible system was employed because LKB1 had some cytotoxic effects on host HEK293 cells and hampered production of the LKB1-expressing recombinant adenovirus. Such cytotoxicity was not observed with MEF3-2 cells up to 48 h post-induction. (Fig. 2A) (17). The cells infected with Adv-LKB1 alone showed similar activity of PAK1 to those infected with Adv-Cre alone (control cells) (Fig. 2B). Notably, forced expression of LKB1/STRAD by co-infection of Adv-LKB1 and Adv-Cre decreased the cellular level of the phosphorylated PAK1 but not that of total PAK1 and inhibited the PAK1 activity by ∼60% as compared with that in the control cells infected with Adv-Cre or Adv-LKB1 alone (Fig. 2B). Furthermore, forced expression of LKB1/STRAD reduced cell migration in MEF3-2 cells (to 35% of Adv-Cre alone) (Fig. 2C). To directly test whether LKB1 reduces cell migration through inhibition of PAK1, we infected MEF3-2 cells with various multiplicity of infection of the adenovirus encoding the constitutively active mutant of PAK1 (Adv-PAK1-T423E) together with Adv-Cre/Adv-LKB1. Co-infection with Adv-PAK1-T423E has recovered the LKB1-induced reduction of cell migration in a dose-dependent manner (Fig. 2C). These results suggest that LKB1 can contribute to the suppression of cell migration through inhibition of PAK1.
LKB1 Activity Is Necessary for Suppression of PAK1
PAK1 can be activated by recruitment of the active forms of Cdc42/Rac and phosphorylation by 3-phosphoinositide-dependent kinase-1 (19). Thus, we investigated whether the protein kinase activity of LKB1 was necessary to suppress the PAK1 activation. HEK293T cells were transiently transfected with PAK1 and/or LKB1/STRAD expression vector to determine PAK1 activity. The wild-type LKB1 (wt) inhibited the activity significantly, whereas the kinase-dead mutant of LKB1 (kinase-dead) did not affect activity (Fig. 3A). These results indicated that LKB1 activity was necessary for PAK1 inhibition. Accordingly, we next hypothesized that LKB1-mediated phosphorylation of PAK1 reduced its kinase activity. To test the possibility, we preincubated recombinant PAK1 with various amounts of LKB1 in the presence of ATP and then added the substrate of PAK1 to determine its kinase activity. LKB1 directly inhibited the PAK1 activity in vitro in a dose-dependent manner (Fig. 3B). These results indicate that LKB1-mediated PAK1 phosphorylation decreases its kinase activity.
LKB1 Can Phosphorylate PAK1 at Thr109
To study PAK1 phosphorylation by LKB1 further, we searched the LKB1 substrate amino acid sequence motif “LXT” (5, 37) in full-length PAK1. We found an LQT sequence (107–109) within the p21-binding domain (PBD) in the regulatory region located near the N terminus of PAK1 (Fig. 4A). To determine whether Thr109 of PAK1 could be phosphorylated by LKB1, we constructed a GST-fused PBD recombinant protein (GST-PBD) and its mutated form in PBD, GST-PBD-T109A (Fig. 4A). Recombinant LKB1 failed to phosphorylate GST-PBD-T109A in vitro, although it phosphorylated GST-PBD efficiently (Fig. 4B). Next, we tested whether LKB1 phosphorylated the full-length PAK1 at Thr109. To this end, we used a full-length dominant-negative PAK1 recombinant protein (GST-PAK1-K299R) as the substrate to minimize the effect of autophosphorylation and introduced alanine substitution at Thr109, GST-PAK1-K299R/T109A (Fig. 4A). The full-length PAK1 (GST-PAK1-K299R) was phosphorylated by LKB1 efficiently, whereas the GST-PAK1-K299R/T109A was at a much reduced level (Fig. 4C). These results suggest that full-length PAK1 can be a substrate for LKB1 and that Thr109 is a likely target of phosphorylation by LKB1, although it is possible that LKB1 phosphorylates other residues of PAK1 in addition to Thr109. To determine whether LKB1 phosphorylates PAK1 at Thr109 in vivo, we performed a series of mass spectrometric analyses of PAK1 purified from HEK293T cells transiently co-transfected with plasmids encoding PAK1 and LKB1/STRAD (supplemental Fig. S1). The protonated molecular ions of peptide LLQTSNITK-(106–114) obtained from tryptic digestion of wt-PAK1 were detected in its nonphosphorylated form ([M + H]+ at m/z 1017.55) and in its monophosphorylated form ([M + H]+ at m/z 1097.54). To further confirm the specific phosphorylation site of the peptide, we used HEK293T cells co-transfected with plasmids encoding PAK1-T109A and LKB1/STRAD for another set of mass spectrometric analyses. The molecular ion of LLQASNITK appeared at m/z 987.59 corresponding to the substitution of threonine at residue 109 with alanine. Notably, the corresponding monophosphopeptide was not detected in PAK1-T109A, excluding phosphorylation at the other Thr or Ser residue of the peptide. These results are consistent with our interpretation that LKB1 phosphorylates PAK1 at Thr109 in vivo.
FIGURE 4.
LKB1 phosphorylates PAK1 in vitro at Thr109. A, schematic presentation of PAK1 and its mutant forms. wt PAK1 contains the p21-binding domain (light gray) and the kinase domain (dark gray). The noncatalytic domains are in white. Thr109 is a possible phosphorylation site for LKB1. B, LKB1 phosphorylates GST-PAK1-PBD at Thr109 in vitro. The recombinant LKB1 complex (LKB1-STRAD-MO25) was incubated with GST-PBD or GST-PBD-T109A. C, LKB1 phosphorylates full-length GST-PAK1-K299R and GST-PAK1-T109A/K299R to a lesser extent. The in vitro kinase assays were performed using the recombinant LKB1 complex with GST-PAK1-K299R or GST-PAK1-T109A/K299R. The phosphorylation of the indicated recombinant proteins was visualized using BAS-5000 Bio-imaging Analyzer. These experiments were repeated at least twice with similar results.
Phosphorylation of PAK1 at Thr109 by LKB1 Reduces Its Activity
We have demonstrated that the kinase activity of LKB1 is necessary to inhibit that of PAK1 (Fig. 3A). Therefore, we next investigated whether phosphorylation of Thr109 by LKB1 regulated PAK1 activity. Namely, we constructed a PAK1-T109E mutant whose amino acid residue 109 was converted to glutamic acid from threonine. This substitution mimics the phosphorylation of threonine with the acidic moiety of glutamic acid. As an inactive (negative) control, we also constructed the alanine substitution PAK1-T109A (Fig. 5A). We transiently transfected HEK293T cells with plasmids encoding these constructs of PAK1: wt, constitutively-active (T423E), dominant-negative (K299R), T109E, and T109A, respectively (Fig. 5A). We then determined the in vitro PAK1 activities in the cell lysates using GST-VASP-(158–277) as substrate. The PAK1 mutant T423E showed the highest activity (2.1×), whereas mutant K299R with defective kinase (kinase-dead) had only 2% of the wt-PAK1 activity. Notably, T109E mutant showed significantly reduced PAK1 activity (36% of wt), whereas T109A mutant had a similar activity to the wt (98%) (Fig. 5B). These results suggest that phosphorylation of PAK1 at Thr109 decreases its kinase activity. To confirm the role of phosphorylation at Thr109 in suppression of PAK1 activity by LKB1, we next tested nonphosphorylatable PAK1-T109A in HEK293T cells. Co-expression of LKB1 failed to suppress the activity of PAK1-T109A (Fig. 5C), in sharp contrast to the effect on wt-PAK1 (Fig. 3A). These results strongly suggest that LKB1 suppresses the PAK1 activity by phosphorylating it at Thr109.
Activation of PAK1 in Lkb1(+/−) Mouse HCCs and Lkb1(−/−) MEFs
We have previously demonstrated that Lkb1(+/−) mice develop HCC after 50 weeks of age and that HCCs and nodular foci of Lkb1(+/−) liver show loss of the Lkb1 heterozygosity (10, 11). To test the involvement of LKB1 in PAK1-mediated signaling in vivo, we next determined the level of phosphorylated form of PAK1 in HCCs of Lkb1(+/−) mice, using two different types of phospho-specific PAK1 antibodies that recognized the active form of PAK1. Notably, we found that the level of phosphorylated PAK1 was increased in HCCs (H1 and H2) and in precancerous lesions (nodular foci, F1 and F2), compared with the normal liver of Lkb1(+/−) or Lkb1(+/+) mice, while the levels of total PAK1 in nodular foci and HCCs were similar to those in normal liver (Fig. 6A). These results are therefore consistent with the interpretation that loss of LKB1 induces PAK1 activation. We further investigated the status of PAK1 activation in Lkb1(−/−) MEFs (MEF3-2) and wt MEFs. Both the level of phosphorylated PAK1 and in vitro PAK1 activity were significantly higher in Lkb1(−/−) MEFs than those in wt MEFs (Fig. 6B). These data together suggest that loss of LKB1 contributes to the activation of PAK1.
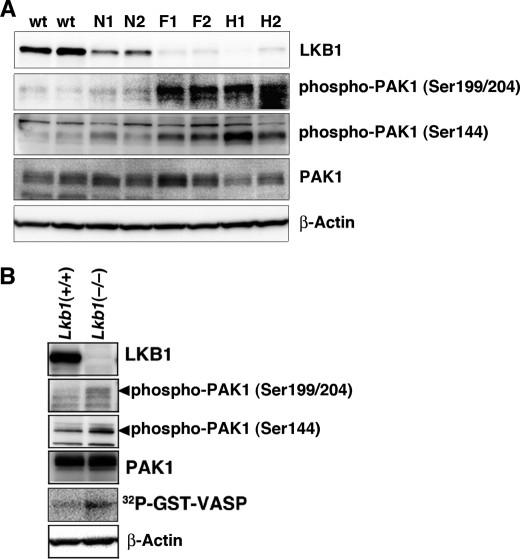
FIGURE 6.
Activation of PAK1 in Lkb1(+/−) mouse HCCs and Lkb1(−/−) MEFs. A, activation of PAK1 in Lkb1(+/−) mice HCCs. Cellular levels of phospho-PAK1, PAK1, and β-actin in tissue lysates were determined by Western blotting with anti-phospho-PAK1 (Ser144), anti-phospho-PAK1 (Ser199/204), anti-PAK1, and anti-β-actin. Two wild-type livers (W1 and W2), pairs of tumor (H1 and H2), precancerous lesions (F1 and F2) and adjacent normal tissue (N1 and N2) from two Lkb1(+/−) mice were analyzed. B, increased PAK1 activity in Lkb1(−/−) MEFs. Cellular levels of phospho-PAK1, PAK1, and β-actin in tissue lysates were determined by Western blotting with the indicated antibodies. For in vitro kinase assay, cell lysates were prepared from MEF cells and immunoprecipitated using an anti-PAK1 antibody. The precipitates were used for in vitro kinase assays using GST-VASP-(158–277) as a substrate. The phosphorylation of GST-VASP was visualized using BAS-5000 Bio-imaging Analyzer. Similar results were obtained in three independent experiments.
DISCUSSION
In this study, we have provided evidence that LKB1 inhibits PAK1 activation by direct phosphorylation of PAK1 at Thr109. We first showed that knockdown of LKB1 increased cell migration and PAK1 activity in HCT116 cells (Fig. 1), whereas introduction of LKB1 in Lkb1-null MEFs reduced cell migration and PAK1 activity simultaneously (Fig. 2). Notably, a constitutively active mutant of PAK1 (PAK1-T423E) recovered the LKB1-mediated reduction of cell migration in a dose-dependent manner (Fig. 2C). Because HCCs in Lkb1(+/−) mice can metastasize to the lung (10), it is conceivable that inhibition of PAK1 by LKB1 contributes to suppression of cancer metastasis. We also demonstrated that LKB1 directly phosphorylated PAK1 at Thr109 both in vitro (Fig. 4) and in vivo (supplemental Fig. S1). Importantly, the phosphomimetic PAK1-T109E mutation reduced its activity (Fig. 5B), whereas the activity of a nonphosphorylatable mutant PAK1-T109A was not suppressed by LKB1 (Fig. 5C). These results indicate that phosphorylation of Thr109 is both necessary and sufficient for inhibition of PAK1 activity by LKB1, although it is possible that LKB1 phosphorylates other residues of PAK1 in addition to Thr109 (Fig. 4C). Consistently, we found that active forms of PAK1 were increased in the nodular foci and HCCs in Lkb1(+/−) mice, compared with the normal liver of Lkb1(+/−) and Lkb1(+/+) mice (Fig. 6A).
Our present results indicate that PAK1 is a novel LKB1 substrate. So far, 14 kinases have been identified as LKB1 substrates, including AMPKα1, AMPKα2, MARK1, and MARK2. LKB1 phosphorylates these kinases at the LXT motif in the activation loop (5, 37), and such phosphorylations can induce kinase activation. In contrast, LKB1-induced phosphorylation of PAK1 at Thr109 in the LXT motif of PBD causes a decrease in the kinase activity (Fig. 5B). Because this motif is well conserved among group I PAKs (PAK1, PAK2, and PAK3), it is likely that regulation of the PAK1 activity by LKB1 applies to PAK2 and PAK3 as well. According to the structural analysis of PAK1, Thr109 is located at the helix loop Iα in an inhibitory switch (38) and exposed to the outer space. In addition, the L107F mutation of PAK1 prevents the interaction between the autoinhibitory region and the C-terminal catalytic region, making the constitutively active kinase (39). Because this mutant lacks the LXT motif, the constitutive activation is likely due to the loss of negative regulation by LKB1. We therefore speculate that the phosphorylation of this Thr residue enhances the interaction between the autoinhibitory (N-terminal regulatory) and kinase domains, leading to inhibition of the PAK1 activity. Further experiments are required to elucidate the precise role of Thr109 phosphorylation in PAK1 conformation.
It has been reported recently that LKB1 maintains cancer cell polarity by recruiting Cdc42 to the leading edges and that inhibition of LKB1 reduces PAK1 autophosphorylation (40). In contrast, we have found that LKB1 suppresses PAK1 activity by direct phosphorylation. The reason for the discrepancy is not clear but may be related to the methods used to overexpress LKB1 (transient transfection of LKB1 expression vector alone versus co-infection of LKB1 with STRAD using adenovirus vector) or the cell context, genetic constitution such as p53 mutation status (non-small cell lung cancer cells (p53 mutant) versus HEK293T or Lkb1-null MEFs (wt-p53)). Notably, LKB1 plays key roles in growth arrest or apoptosis through a p53-dependent mechanism (41, 42). Thus, it is conceivable that LKB1-mediated PAK1 inhibition may be dependent on the p53 status.
Numerous PAK1 substrates and/or interacting proteins have been identified that activate diverse signaling processes involved in cell motility, morphology, growth, survival, and epithelial-mesenchymal transition (19). For example, PAK1 stabilizes microtubules through stathmin/Op18 phosphorylation (43, 44). Therefore, LKB1 also may contribute to microtubule destabilization by regulating PAK1, whereas we have reported previously that LKB1 inhibits tubulin polymerization through activation of MARK2 (17). Further investigation is needed to address the role of LKB1 in various downstream processes of PAK1 other than cell motility. In conclusion, our present results suggest that PAK1 is a novel downstream target of LKB1, and that LKB1-mediated PAK1 phosphorylation may play important roles in tumor suppression in the liver.
Supplementary Material
Acknowledgment
We thank Dr. J. Chernoff for PAK1 wild-type, T423E, H83LH86LK299R, and 86L-K299R constructs.
This work was supported by a grant-in-aid for scientific research (to M. M. T.), a grant-in-aid for young scientist (B) from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and Littlefield-American Association for Cancer Research grants in metastatic colon cancer research (to M. M. T.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures” and Fig. S1.
- HCC
- hepatocellular carcinoma
- PAK1
- p21-activated kinase-1
- PBD
- p21-binding domain
- Adv
- adenovirus
- AMPK
- AMP-activated protein kinase
- STRAD
- STE20-related adaptor pseudo-kinase
- MARK
- microtubule-associated protein/microtubule affinity-regulating kinase
- siRNA
- small interfering RNA
- wt
- wild-type
- GST
- glutathione S-transferase
- HA
- hemagglutinin
- VASP
- vasodilator-stimulated phosphoprotein.
REFERENCES
- 1.Giardiello F. M., Welsh S. B., Hamilton S. R., Offerhaus G. J., Gittelsohn A. M., Booker S. V., Krush A. J., Yardley J. H., Luk G. D. (1987) N. Engl. J. Med. 316, 1511–1514 [DOI] [PubMed] [Google Scholar]
- 2.Hemminki A., Markie D., Tomlinson I., Avizienyte E., Roth S., Loukola A., Bignell G., Warren W., Aminoff M., Höglund P., Järvinen H., Kristo P., Pelin K., Ridanpää M., Salovaara R., Toro T., Bodmer W., Olschwang S., Olsen A. S., Stratton M. R., de la Chapelle A., Aaltonen L. A. (1998) Nature 391, 184–187 [DOI] [PubMed] [Google Scholar]
- 3.Jenne D. E., Reimann H., Nezu J., Friedel W., Loff S., Jeschke R., Müller O., Back W., Zimmer M. (1998) Nat. Genet. 18, 38–43 [DOI] [PubMed] [Google Scholar]
- 4.Launonen V. (2005) Hum. Mutat. 26, 291–297 [DOI] [PubMed] [Google Scholar]
- 5.Alessi D. R., Sakamoto K., Bayascas J. R. (2006) Annu. Rev. Biochem. 75, 137–163 [DOI] [PubMed] [Google Scholar]
- 6.Katajisto P., Vallenius T., Vaahtomeri K., Ekman N., Udd L., Tiainen M., Mäkelä T. P. (2007) Biochim. Biophys. Acta 1775, 63–75 [DOI] [PubMed] [Google Scholar]
- 7.Miyoshi H., Nakau M., Ishikawa T. O., Seldin M. F., Oshima M., Taketo M. M. (2002) Cancer Res. 62, 2261–2266 [PubMed] [Google Scholar]
- 8.Rossi D. J., Ylikorkala A., Korsisaari N., Salovaara R., Luukko K., Launonen V., Henkemeyer M., Ristimaki A., Aaltonen L. A., Makela T. P. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 12327–12332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bardeesy N., Sinha M., Hezel A. F., Signoretti S., Hathaway N. A., Sharpless N. E., Loda M., Carrasco D. R., DePinho R. A. (2002) Nature 419, 162–167 [DOI] [PubMed] [Google Scholar]
- 10.Nakau M., Miyoshi H., Seldin M. F., Imamura M., Oshima M., Taketo M. M. (2002) Cancer Res. 62, 4549–4553 [PubMed] [Google Scholar]
- 11.Miyoshi H., Deguchi A., Nakau M., Kojima Y., Mori A., Oshima M., Aoki M., Taketo M. M. (2009) Cancer Sci. 100, 2046–2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baas A. F., Boudeau J., Sapkota G. P., Smit L., Medema R., Morrice N. A., Alessi D. R., Clevers H. C. (2003) EMBO J. 22, 3062–3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boudeau J., Baas A. F., Deak M., Morrice N. A., Kieloch A., Schutkowski M., Prescott A. R., Clevers H. C., Alessi D. R. (2003) EMBO J. 22, 5102–5114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baas A. F., Smit L., Clevers H. (2004) Trends. Cell Biol. 14, 312–319 [DOI] [PubMed] [Google Scholar]
- 15.Zhang L., Li J., Young L. H., Caplan M. J. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 17272–17277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng B., Cantley L. C. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 819–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kojima Y., Miyoshi H., Clevers H. C., Oshima M., Aoki M., Taketo M. M. (2007) J. Biol. Chem. 282, 23532–23540 [DOI] [PubMed] [Google Scholar]
- 18.Partin A. W., Isaacs J. T., Treiger B., Coffey D. S. (1988) Cancer Res. 48, 6050–6053 [PubMed] [Google Scholar]
- 19.Bokoch G. M. (2003) Annu. Rev. Biochem. 72, 743–781 [DOI] [PubMed] [Google Scholar]
- 20.Kumar R., Gururaj A. E., Barnes C. J. (2006) Nat. Rev. Cancer 6, 459–471 [DOI] [PubMed] [Google Scholar]
- 21.Manser E., Leung T., Salihuddin H., Zhao Z. S., Lim L. (1994) Nature 367, 40–46 [DOI] [PubMed] [Google Scholar]
- 22.Tsakiridis T., Taha C., Grinstein S., Klip A. (1996) J. Biol. Chem. 271, 19664–19667 [DOI] [PubMed] [Google Scholar]
- 23.King C. C., Gardiner E. M., Zenke F. T., Bohl B. P., Newton A. C., Hemmings B. A., Bokoch G. M. (2000) J. Biol. Chem. 275, 41201–41209 [DOI] [PubMed] [Google Scholar]
- 24.Tang Y., Zhou H., Chen A., Pittman R. N., Field J. (2000) J. Biol. Chem. 275, 9106–9109 [DOI] [PubMed] [Google Scholar]
- 25.Balasenthil S., Sahin A. A., Barnes C. J., Wang R. A., Pestell R. G., Vadlamudi R. K., Kumar R. (2004) J. Biol. Chem. 279, 1422–1428 [DOI] [PubMed] [Google Scholar]
- 26.Holm C., Rayala S., Jirström K., Stål O., Kumar R., Landberg G. (2006) J. Natl. Cancer Inst. 98, 671–680 [DOI] [PubMed] [Google Scholar]
- 27.Carter J. H., Douglass L. E., Deddens J. A., Colligan B. M., Bhatt T. R., Pemberton J. O., Konicek S., Hom J., Marshall M., Graff J. R. (2004) Clin. Cancer Res. 10, 3448–3456 [DOI] [PubMed] [Google Scholar]
- 28.Ching Y. P., Leong V. Y., Lee M. F., Xu H. T., Jin D. Y., Ng I. O. (2007) Cancer Res. 67, 3601–3608 [DOI] [PubMed] [Google Scholar]
- 29.Wang R. A., Zhang H., Balasenthil S., Medina D., Kumar R. (2006) Oncogene 25, 2931–2936 [DOI] [PubMed] [Google Scholar]
- 30.Sapkota G. P., Kieloch A., Lizcano J. M., Lain S., Arthur J. S., Williams M. R., Morrice N., Deak M., Alessi D. R. (2001) J. Biol. Chem. 276, 19469–19482 [DOI] [PubMed] [Google Scholar]
- 31.Sandig V., Youil R., Bett A. J., Franlin L. L., Oshima M., Maione D., Wang F., Metzker M. L., Savino R., Caskey C. T. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 1002–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deguchi A., Soh J. W., Li H., Pamukcu R., Thompson W. J., Weinstein I. B. (2002) Mol. Cancer. Ther. 1, 803–809 [PubMed] [Google Scholar]
- 33.Ji H., Ramsey M. R., Hayes D. N., Fan C., McNamara K., Kozlowski P., Torrice C., Wu M. C., Shimamura T., Perera S. A., Liang M. C., Cai D., Naumov G. N., Bao L., Contreras C. M., Li D., Chen L., Krishnamurthy J., Koivunen J., Chirieac L. R., Padera R. F., Bronson R. T., Lindeman N. I., Christiani D. C., Lin X., Shapiro G. I., Jänne P. A., Johnson B. E., Meyerson M., Kwiatkowski D. J., Castrillon D. H., Bardeesy N., Sharpless N. E., Wong K. K. (2007) Nature 448, 807–810 [DOI] [PubMed] [Google Scholar]
- 34.Kumar R., Vadlamudi R. K. (2002) J. Cell. Physiol. 193, 133–144 [DOI] [PubMed] [Google Scholar]
- 35.Chong C., Tan L., Lim L., Manser E. (2001) J. Biol. Chem. 276, 17347–17353 [DOI] [PubMed] [Google Scholar]
- 36.Butt E., Abel K., Krieger M., Palm D., Hoppe V., Hoppe J., Walter U. (1994) J. Biol. Chem. 269, 14509–14517 [PubMed] [Google Scholar]
- 37.Lizcano J. M., Göransson O., Toth R., Deak M., Morrice N. A., Boudeau J., Hawley S. A., Udd L., Mäkelä T. P., Hardie D. G., Alessi D. R. (2004) EMBO J. 23, 833–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lei M., Lu W., Meng W., Parrini M. C., Eck M. J., Mayer B. J., Harrison S. C. (2000) Cell 102, 387–397 [DOI] [PubMed] [Google Scholar]
- 39.Brown J. L., Stowers L., Baer M., Trejo J., Coughlin S., Chant J. (1996) Curr. Biol. 6, 598–605 [DOI] [PubMed] [Google Scholar]
- 40.Zhang S., Schafer-Hales K., Khuri F. R., Zhou W., Vertino P. M., Marcus A. I. (2008) Cancer Res. 68, 740–748 [DOI] [PubMed] [Google Scholar]
- 41.Karuman P., Gozani O., Odze R. D., Zhou X. C., Zhu H., Shaw R., Brien T. P., Bozzuto C. D., Ooi D., Cantley L. C., Yuan J. (2001) Mol. Cell 7, 1307–1319 [DOI] [PubMed] [Google Scholar]
- 42.Tiainen M., Ylikorkala A., Mäkelä T. P. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 9248–9251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Belmont L. D., Mitchison T. J. (1996) Cell 84, 623–631 [DOI] [PubMed] [Google Scholar]
- 44.Larsson N., Marklund U., Gradin H. M., Brattsand G., Gullberg M. (1997) Mol. Cell. Biol. 17, 5530–5539 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.