Abstract
Multiple endocrine neoplasia type 1 (MEN1) results from mutations in tumor suppressor gene Men1, which encodes nuclear protein menin. Menin up-regulates certain cyclin-dependent kinase inhibitors through increasing histone H3 lysine 4 (H3K4) methylation and inhibits G0/G1 to S phase transition. However, little is known as to whether menin controls G2/M-phase transition, another important cell cycle checkpoint. Here, we show that menin expression delays G2/M phase transition and reduces expression of Ccnb2 (encoding cyclin B2). Menin associates with the promoter of Ccnb2 and reduces histone H3 acetylation, a positive chromatin marker for gene transcription, at the Ccnb2 locus. Moreover, Men1 ablation leads to an increase in cyclin B2 expression, histone H3 acetylation at the Ccnb2 locus, and G2/M transition. In contrast, knockdown of cyclin B2 diminishes the number of cells at M phase and reduces cell proliferation. Furthermore, menin interferes with binding of certain positive transcriptional regulators, such as nuclear factor Y (NF-Y), E2 factors (E2Fs), and histone acetyltransferase CREB (cAMP-response element-binding protein)-binding protein (CBP) to the Ccnb2 locus. Notably, MEN1 disease-related mutations, A242V and L22R, abrogate the ability of menin to repress cyclin B2 expression and G2/M transition. Both of the mutants fail to reduce the acetylated level of the Ccnb2 locus. Together, these results suggest that menin-mediated repression of cyclin B2 is crucial for inhibiting G2/M transition and cell proliferation through a previously unrecognized molecular mechanism for menin-induced suppression of MEN1 tumorigenesis.
Keywords: Cancer Tumor Promoter, Cell Cycle, Epigenetics, Gene Transcription, Histone Modification, G2/M, Cyclin B2, Histone Deacetylation, Menin
Introduction
Multiple endocrine neoplasia type 1 (MEN1)2 is a dominantly inherited tumor syndrome characterized by development of certain endocrine tumors and non-endocrine tumors, such as collagenomas (1, 2). The tumor suppressor gene MEN1, whose mutation is responsible for MEN1 syndrome, was identified by positional cloning (3), and ∼1336 MEN1 mutations in MEN1 patients have been identified (4). Menin, the protein encoded by MEN1, is involved in regulation of gene transcription, cell cycle, and genome instability (5).
Menin plays an important role in regulating gene transcription. It represses transcriptional factor Jun D in a histone deacetylase (HDAC)-dependent manner, as shown by reporter assays in vitro, and mSin3A, a general co-repressor, bridging HDACs and menin (6–8). However, it is not yet clear whether menin-mediated and HDAC-dependent repression regulates any endogenous genes. Menin has also been shown to repress NF-κB-mediated transcription, which can at least partly account for a role of menin in tumor suppression (9). Our previous studies reveal that menin directly binds to double-stranded DNA (10) and represses expression of growth factor pleiotrophin (11).
Our previous study shows that excision of floxed MEN1 in mouse embryonic fibroblasts (MEFs) accelerates G0/G1 to S phase entry (12), at least partly by decreasing expression of cyclin-dependent kinase (CDK) inhibitors p18Ink4c and p27Kip1 and increasing the activity of CDK2 (12). This function of menin may partly account for its role in suppressing collagenomas from skin fibroblasts. However, little is known as to whether menin has any impact on G2/M transition, albeit we previously found more G2/M phase cells in menin-expressing MEFs than in menin-null cells (10, 12).
CDK1/cyclin B acts as the maturation-promoting factor (13–16), which is the checkpoint for G2/M transition (17, 18). The Ccnb2 promoter contains three CCAAT boxes, which bind to trimetric transcription factor NF-Y (19). Two elements, cell cycle-dependent element (CDE) and cell cycle gene homology region (CHR), in the Ccnb2 promoter, have been implicated in conferring cell cycle-regulated transcription of cyclin B2 (20). CDE/CHR is important for cell cycle-regulated expression of other cell cycle genes, such as cyclin A and cdc25 (21–24). Mutations in either CDE or CHR result in elevated transcriptional activity of the genes and consequential loss of cell cycle regulated repression (20, 23). These elements play an important role in transcription of cyclin B2 (20).
In our study we found that menin normally repressed G2/M transition through suppressing transcription of cyclin B2 via associating with the Ccnb2 promoter and reducing histone H3 acetylation at the locus. This function was disrupted by the tested MEN1 disease-related mutations, underscoring the importance of menin-mediated repression of cyclin B2 expression and G2/M transition in suppressing phenotypes of MEN1 syndrome.
EXPERIMENTAL PROCEDURES
Plasmid Construction
pMX-menin A242V and L22R point mutations of menin were constructed as previously described (25–27). PLK0.1-based constructs expressing cyclin B2 shRNAs were purchased from Open Biosystems (RMM4534, Huntsville, AL). Wild-type and mutants of cyclin B2 promoter luciferase constructs were kind gifts from Kurt Engeland (20): pGl3-cylcin B2-wt-Luc, pGl3-cylcin B2–3 CCAAT mut-Luc, pGl3-cylcin B2-CDE mut-Luc, and pGl3-cylcin B2-CHR mut-Luc.
Cell Culture
Cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and penicillin (100 units/ml). Menin-null MEFs complemented with control vector, menin, or its mutants, A242V and L22R, were established as previously described (28). Two independent batches of menin-null and menin-expressing MEFs were used. To monitor cell proliferation, on day 0 cells were seeded at 5 × 104 cells per well in 6-well plates and counted in a hemocytometer. Results were representative of two independent experiments with triplicates.
Chromatin Immunoprecipitation (ChIP)
ChIP assay was performed by using ChIP assay kits (QuickChIPTM, IMGENEX Corp., San Diego, CA), according to the manufacturer's instructions. Briefly, 5 × 106 cells were used for cross-linking by 1% formaldehyde, and then cells were lysed in 1 ml of SDS lysis buffer and sonicated by Bioruptor sonicator (Diagenode, UCD-200TM-EX) to shear the chromatin into 200–1000-bp fragments. The sonicated cell lysates (200 μl of the 1-ml supernatant from each cell line) and 4 μg of antibodies were used for each immunoprecipitation. The precipitated DNA was used as template for quantitative real-time PCR using the SYBR Green PCR kit (Qiagen, Germantown, MD). Antibodies used for ChIP were anti-menin (BL342; Bethyl Laboratories), anti-acetylated histone H3 (17–615, Millipore), anti-trimethylhistone H3K4 (17–614, Millipore), anti-histone H3 (ab1791, Abcam), anti-HDAC3 (ab7030, Abcam), anti E2F2 (sc-633, Santa Cruz Biotechnology), anti-E2F3 (sc-879, Santa Cruz), anti-polymerase II (MMS-126R, Covance), anti-NF-YB (sc-13045, Santa Cruz), anti-CBP (sc-369, Santa Cruz), and anti-IgG (ab46540-1, Abcam). Results were presented as the percent of input by quantifying the amount of chromatin obtained from immunoprecipitation relative to the amount in the input samples. For quantitative real-time PCR, results were representative of two independent ChIP experiments, each with triplicate reactions.
Fluorescence-activated Cell Sorting Analysis
Cells were pulsed with 1 mm 5′-bromo-2′-deoxyuridine-5′-triphosphate (Sigma), and 1 × 106 cells were harvested for incubation with anti-bromodeoxyuridine antibody (Alexa Fluor 488 conjugate, A21303, Invitrogen), stained with 5 μg/ml propidium iodide (Sigma), and analyzed on a FACSCalibur flow cytometer (342975, BD Biosciences) with Cell Quest Pro software. Results were representative of two independent experiments, each with triplicate samples.
Western Blotting
The whole cell lysate radioimmune precipitation assay buffer (Sigma) was separated on an SDS-polyacrylamide gel and transferred to polyvinylidene difluoride membranes. Antibodies used for Western blotting were anti-β-actin (Sigma), anti-menin (BL342; Bethyl Laboratories), and anti-cyclin B2 (sc-22776, Santa Cruz). The other antibodies used in Western blotting were the same as used in the ChIP assay. Results were representative of two independent experiments.
Transfection and Luciferase Assays
Cells were transfected using the Lipofectamine 2000 (Invitrogen). Dual-luciferase reporter assay (Promega, Madison, WI) kits were used according to the manufacturer's instructions. Both firefly luciferase and renilla luciferase activities were measured by Luminoskan Ascent (Thermo Fisher). Results were representative of two independent experiments.
RNA Extraction and RT-PCR
RNA was extracted by TRIzol (Invitrogen) and isolated with a Qiagen RNeasy mini kit according to manufacturer's instructions. The synthesized first-strand cDNA was used for qRT-PCR with a SYBR Green PCR kit (Qiagen) and the 7500 Fast Real-Time PCR system (Applied Biosystems). Gene expression levels were normalized to the level of glyceraldehyde-3-phosphate dehydrogenase mRNA expression and were derived from two independent experiments, each with triplicate reactions.
Lentivirus Packaging and Transducing Cells for Cyclin B2 Knockdown
shRNA targeting cyclin B2 in pLK0.1 plasmid or control pLK0.1 plasmid was co-transfected with PAX2 and PMD2 packaging plasmids by FuGENE 6 transfection reagent (Roche Applied Science) into 293T cells. Viruses were collected and used in infecting Men1−/− MEFs with 8 μg/ml Polybrene. Cells were selected with 2 μg/ml puromycin for 4 days.
Statistical Analysis and Quantification
For comparison between two groups, Student's t test was used. For comparison among multiple groups, one-way analysis of variance was used. p values less than 0.05 were considered statistically significant.
RESULTS
Menin Delays G2/M Phase Transition
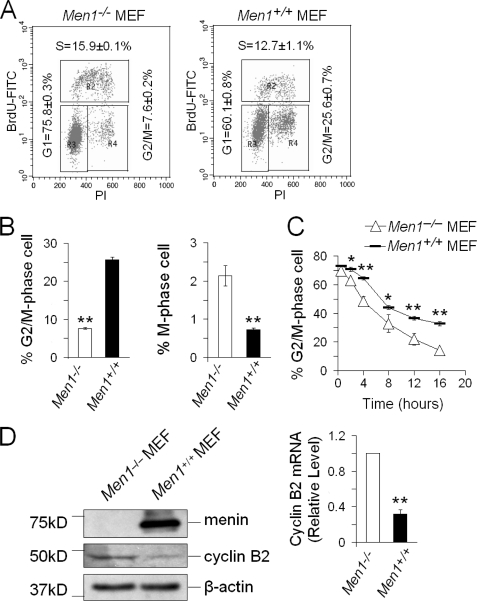
Our previous work has shown that expression of menin in MEN1-null MEFs increases the number of cells in G2/M phase (10, 12), raising the possibility that menin may control G2/M transition. To test this possibility, we analyzed bromodeoxyuridine incorporation and propidium iodide (PI) staining of MEN1-expressing and MEN1-null cells by flow cytometry and showed that MEN1 expression substantially increased the percentage of cells at G2/M phase (Fig. 1, A and B, left). To further examine the potential role of menin in controlling G2/M transition, we determined the impact of menin on the status of histone H3 serine 10 phosphorylation, which is tightly correlated with chromosomal condensation during mitosis and serves as a marker for M phase (29–31). We performed immunofluorescent staining for exponentially growing menin-expressing and menin-null cells using a phosphohistone H3 (Ser10)-specific antibody and found that the number of phosphorylated H3S10-positive cells was increased in menin-null cells (Fig. 1B, right, and supplemental Fig. S1). These results suggest that menin inhibits G2/M transition.
FIGURE 1.
Menin arrests G2/M and represses cyclin B2 expression. A, shown is the cell cycle profile of Men1−/− (left) and Men1+/+ MEFs (right) by fluorescence-activated cell sorting analysis. BrdU, bromodeoxyuridine; FITC, fluorescein isothiocyanate; PI, propidium iodide. B, shown is the percentage of G2/M phase cells in Men1−/− and Men1+/+ MEFs based on the fluorescence-activated cell sorter analysis (left); n = 3. The percentage of M-phase cells in Men1−/− and Men1+/+ MEFs is based on immunofluorescent staining for histone H3 S10 phosphorylation from supplemental Fig. S1 (right); n = 5. C, shown are the kinetics of the G2/M phase transition in Men1−/− and Men1+/+ MEFs. Cells were arrested at G2/M by 0.2 μg/ml nocodazole for 22 h and then released and harvested at the indicated times for flow cytometry analysis; n = 3. D, Western blotting analysis in Men1−/− and Men1+/+ MEFs (left) is shown. Shown is qRT-PCR analysis in Men1−/− and Men1+/+ MEFs (right); n = 3. *, p < 0.05; **, p < 0.01.
To further confirm the impact of menin on the kinetics of G2/M phase transition, we first blocked menin-expressing or menin-null cells at G2/M phase using nocodazole, an agent that interferes with the polymerization of microtubules and causes cell cycle arrest in the G2/M phase (32). Most of the nocodazole-treated cells were arrested at G2/M phase (∼70%) and then released from the block by removing nocodazole. The resulting cells were monitored for departure from G2/M using flow cytometry analysis. We found that menin-null cells transitioned through mitosis and left M phase more rapidly than menin-expressing cells (Fig. 1C). These results indicate a key role for menin in restricting G2/M transition of the cells.
Menin Down-regulates Expression of Cyclin B2
As it is well known that cyclin B/CDK1 is crucial for G2/M transition in the cell cycle (17, 18), we wondered whether menin delayed the G2/M phase transition via affecting certain genes that control G2/M transition. Gene Set Enrichment Analysis (GSEA) of gene expression in MEN1-expressing and MEN1-null primary pancreatic islets showed that cyclin B2 was up-regulated in Men1-excised pancreatic islets, which largely comprise beta cells from Men1-excised mice.3 Moreover, it has also been reported that insulinomas in MEN1-mutated mice expressed a higher level of cyclin B2 (33). However, it has been unclear whether menin regulates cyclin B2 in untransformed cells. Therefore, we performed qRT-PCR and Western blotting to examine the mRNA and protein levels of cyclin B2 in menin-expressing and menin-null cells and found that menin expression reduced the mRNA and protein levels of cyclin B2 (Fig. 1D).
MEN1 Disease-related Menin Point-mutants Lose the Function in Arresting G2/M Transition and Repressing Cyclin B2 Expression
Menin consists of 610 amino acid residues, and the N-terminal part of menin has been reported to interact with transcriptional factors, such as JunD and Smad3 (34). A large number of mutations in the MEN1 gene have been reported (1133 germ line and 203 somatic mutations) (4). The 1133 germ line mutations are scattered throughout the entire 1830-bp coding region and splice sites of the MEN1 gene and comprise 41% frameshift deletions or insertions, 23% nonsense mutations, and 20% missense mutations (4). The majority (>70%) of these mutations are predicted to lead to truncated forms of menin (4). As testing missense mutations could yield meaningful information regarding the minimal structural requirement for the function of menin and the potential importance of the N-terminal of menin, which is capable of interacting with DNA-binding proteins, we chose to test two N-terminal mutants, L22R and A242V, in regulating cyclin B2 expression. Menin mutants A242V and L22R are two naturally occurring germ line MEN1 missense mutations identified in affected inherited MEN1 disease (35). The A242V is the 242nd amino acid of menin mutated from alanine to valine, and the L22R is the 22nd amino acid of menin mutated from leucine to arginine.
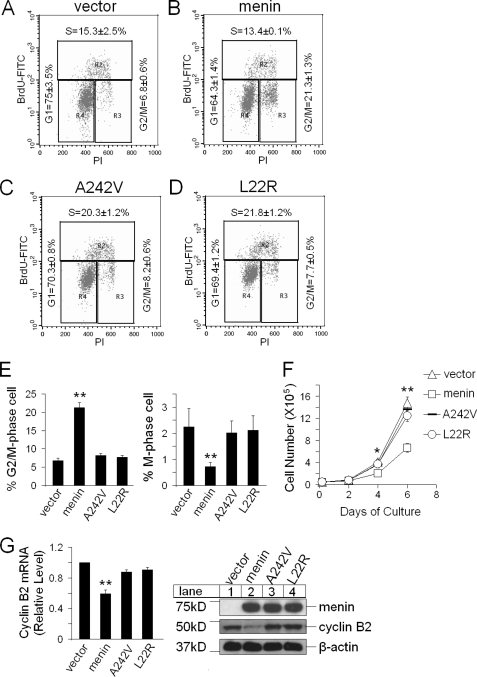
To assess the relevance of menin-mediated repression of cyclin B2 and the potential impact of menin point mutations on repressing cyclin B2 expression, we complemented menin-null cells with either wild-type menin or each of two menin mutants with a point mutation, L22R and A242V. We found that expression of wild-type menin, but not mutants L22R and A242V, in a Men1-excised cell line reduced the number of cells in G2/M phase, as shown by flow cytometry analysis (Fig. 2, A–D and E, left). Further analysis using fluorescent staining for S10-phosphorylated histone H3, a marker for M-phase, showed that expression of wild-type menin, but not its mutants, reduced the number of mitotic cells (Fig. 2E, right; supplemental Fig. S2). Therefore, MEN1 disease-related point mutants failed to effectively arrest cell cycle at the G2/M phase. In agreement with these results, we found that these two MEN1 disease-related point mutants, A242V and L22R, also lost the ability to inhibit cell proliferation (Fig. 2F).
FIGURE 2.
Menin mutants A242V and L22R fail to arrest G2/M and repress cyclin B2 expression. A–D, menin-null MEFs expressing control vector, menin, A242V, and L22R evaluated with flow cytometry analysis for cell distribution in various stages of cell cycle are shown. BrdU, bromodeoxyuridine; FITC, fluorescein isothiocyanate; PI, propidium iodide. E, shown are the percentage of G2/M phase cells in Men1−/− MEFs expressing control vector, menin, A242V, or L22R (left); n = 3. Shown is the percentage of M phase cells in the indicated cells (right), based on immunofluorescent staining for histone H3 S10 phosphorylation from supplemental Fig. S2, right; n = 5. F, shown is a cell growth curve of the indicated cells; n = 3. G, shown is qRT-PCR for cyclin B2 in the indicated cells (left); n = 3. Shown is a Western blotting analysis of cyclin B2 in the indicated cells (right). *, p < 0.05; **, p < 0.01.
Furthermore, we performed qRT-PCR and Western blotting to analyze the mRNA and protein levels of cyclin B2 in these four distinct cell lines and found that compared with the menin-null cells, expression of wild-type menin substantially reduced cyclin B2 expression (Fig. 2G). On the contrary, A242V and L22R expression failed to repress cyclin B2 expression (Fig. 2G). These results also confirm that the functions of menin, as observed in Fig. 1, were not because of the effect of variability of different cell lines used but, rather, because of the molecular expression of menin.
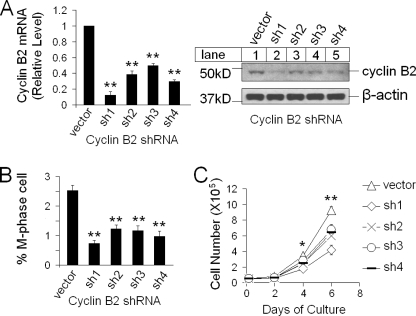
Cyclin B2 Knockdown Decreases the Number of Cells in M Phase and Reduces Cell Proliferation
Cyclin B is important for G2/M transition, and inhibition of cyclin B expression induces a block in cell division (36). To further determine the potential role of cyclin B2 in MEN1-mediated repression of G2/M transition, we knocked down expression of cyclin B2 using four distinct shRNAs targeting cyclin B2 in menin-null cells. Quantitative RT-PCR and Western blotting showed that transduction of the cells with each of the four distinct cyclin B2 shRNAs reduced the mRNA and protein levels of cyclin B2, as compared with the cells transduced with scrambled control vector (Fig. 3A). Immunofluorescent staining by phospho-H3S10 antibody detected fewer M-phase cells in the cyclin B2 knockdown cells as compared with the control cells (Fig. 3B and supplemental Fig. S3). Consistently, proliferation of cyclin B2 knockdown cells was also reduced (Fig. 3C). As a control, ectopic expression of cyclin B2 in cyclin B2 shRNA1-knocked-down cells failed to repress cell proliferation, suggesting that the impact of cyclin B2 knockdown on cell proliferation was not due to the off-target effect of the cyclin B2 shRNA (data not shown). Together, these results suggest that menin-induced repression of cyclin B2 plays an important role in menin-dependent inhibition of G2/M transition and cell proliferation.
FIGURE 3.
Knockdown of cyclin B2 reduces cell proliferation. A, shown is qRT-PCR analysis of cyclin B2 mRNA in cyclin B2 knockdown Men1−/− cell*s (left); n = 3. Shown is Western blotting for cyclin B2 (sh1-sh4) (right). B, the percentage of M-phase cells in Men1−/− MEFs and cyclin B2 knockdown Men1−/− MEFs, based on immunofluorescent staining for histone H3 S10 phosphorylation from supplemental Fig. S2 is shown; n = 5. C, cell growth curve of control Men1−/− cells and cyclin B2 knockdown cells (sh1–sh4) is shown; n = 3. *, p < 0.05; **, p < 0.01.
Menin Represses the Ccnb2 Promoter Activity but Not the Activity of the Promoter with Either a CDE or CHR Mutation
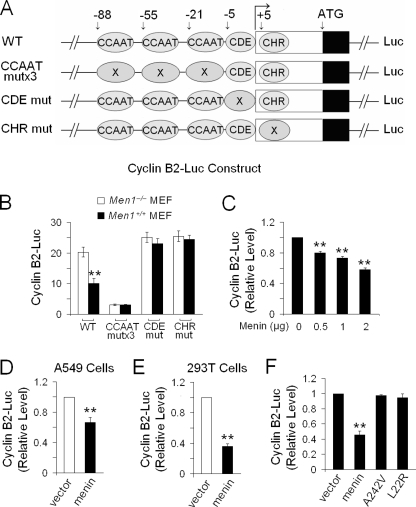
Three conserved CCAAT boxes are found in the Ccnb2 promoter, and the expression of cyclin B2 (encoded by Ccnb2) is largely driven by activation through binding of NF-Y to the CCAAT boxes (19). Cyclin B2 transcription is also controlled in a cell cycle-dependent manner through a repressive tandem transcriptional element composed of CDE/CHR (20). To explore the potential role of CCAAT boxes and CDE/CHR sites in menin-regulated expression of cyclin B2, we carried out a dual luciferase reporter assay to measure the transcriptional activity of wild-type or mutant Ccnb2 promoter (Fig. 4A). The reporter assay showed that menin expression repressed the transcriptional activity of wild-type Ccnb2 promoter but did not reduce the activity of the promoter with mutations in either CDE or CHR (Fig. 4B). As expected, the CCAAT-mutated promoter lost much of its activity (Fig. 4B), likely due to the inability of NF-Y to bind the mutated CCAAT box (37). As CDE and CHR were repressive elements of the Ccnb2 promoter (20), it is likely that the mutations of CDE or CHR might hamper the recruitment of menin or menin-interacting partners to repress cyclin B2. We performed a luciferase reporter assay with an increasing amount of menin cDNA in Men1-null MEFs and found that an increasing amount of menin cDNA progressively reduced the Ccnb2 promoter activity based on luciferase reporter assay (Fig. 4C). In addition, we also co-transfected the luciferase reporter driven by the wild-type Ccnb2 promoter with menin cDNA into A549 cells or 293T cells and showed that menin repressed the activity of the Ccnb2 promoter-driven reporter in these cells (Fig. 4, D and E). Furthermore, we showed that wild-type menin repressed the Ccnb2 promoter activity, but the A242V and L22R lost their ability to repress the transcriptional activity of the Ccnb2 promoter (Fig. 4F). As it has been previously reported that menin binds double-stranded DNA in a sequence-independent manner via its C-terminal positively charged residues (38), it is conceivable that menin contacts chromatin DNA, in concert with other factors.
FIGURE 4.
Menin represses the activity of Ccnb2 promoter. A, shown is a diagram of luciferase reporter constructs with wild-type (WT) Ccnb2 promoter or CCAAT-mut, CDE-mut, or CHR-mut Ccnb2 promoter with the gene. B, a dual luciferase assay was performed in Men1−/− and Men1+/+ MEFs transiently transfected with the Ccnb2 promoter-driven constructs; n = 3. C, a dual luciferase assay was performed in Men1−/− MEFs transiently co-transfected with the increasing amounts of menin and Ccnb2 promoter-driven constructs; n = 3. D, A549 cells were transiently co-transfected with pMX-menin, a wild-type Ccnb2 promoter for luciferase assay; n = 3. E, 293T cells were co-transfected with pMX-menin, a wild-type Ccnb2 promoter for luciferase assay; n = 3. F, shown is a dual luciferase assay in the menin-null MEFs expressing control vector, menin, A242V, and L22R; n = 3. *, p < 0.05; **, p < 0.01.
Menin Associates with the Ccnb2 Promoter
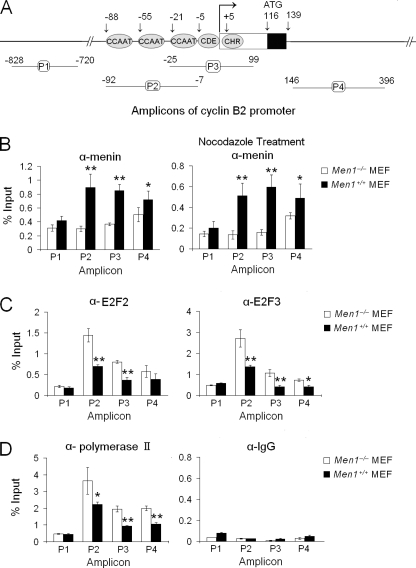
CCAAT boxes and CDE/CHR elements, which are important for binding of transcriptional regulators, are also important for menin-mediated repression of the Ccnb2 promoter (Fig. 4B). We reasoned that menin might associate with the regions or affect recruitment of other transcriptional regulators to repress cyclin B2 transcription. We initially designed three amplicons (P1, P2, and P3) to detect whether menin binds the promoter region of Ccnb2 and found that menin bound the regions detectable by P2 and P3 but not by P1 (Fig. 5, A and B). However, as menin also bound to the first intron, which is detectable by P4, these results suggest that a ChIP assay regarding menin is not sufficient to distinguish whether menin can bind to a particular element of the three CCAAT boxes or the adjacent CDE and CHR boxes. Nevertheless, a ChIP assay showed that E2F2/3 and polymerase II preferentially bound the regions that are detected by amplicons P2 and P3 but not P1 (Fig. 5, C and D). Therefore, menin may regulate cyclin B2 transcription by associating with the promoter region containing CCAAT boxes and CDE/CHR elements and the region near the first exon. As an important control, Men1 ablation reduced the detection of menin at the Ccnb2 promoter, indicating the specific detection of menin associating with the promoter in our assay (Fig. 5B, left). Cells treated with nocodazole and arrested at the G2/M phase were also used for ChIP assay, and again, menin was shown to be specifically associated with the Ccnb2 promoter (Fig. 5B, right).
FIGURE 5.
Cyclin B2 is a transcriptional target of menin. A, four amplicons were used to detect the indicated regions of the Ccnb2 promoter for ChIP assay; n = 3. B, shown is a ChIP assay using menin antibody for Men1−/− and Men1+/+ MEFs (left) or the cells treated with 0.2 μg/ml nocodazole for 22 h (right); n = 3. C, shown is a ChIP assay using the E2F2 antibody (left) or the E2F3 antibody (right); n = 3. D, shown is a ChIP assay using anti-Pol II antibody (left) or control IgG (right); n = 3.
Menin Prevents E2Fs and RNA Polymerase II from Binding to the Ccnb2 Promoter
The E2F transcription factors are important regulators that control expression of genes crucial for G1/S transition (39), G2/M transition, and mitosis (40, 41). Transcription of a vast majority of genes is catalyzed by RNA polymerase II (Pol II). In our CHIP assay we found that association of E2F2, E2F3, and Pol II with the Ccnb2 promoter was diminished in menin-expressing cells (Fig. 5, C and D). As a control, the total amounts of E2F2, E2F3, and Pol II were not altered in menin-expressing and menin-null cells (supplemental Fig. S5A). These findings suggest that menin may inhibit binding of the E2Fs and Pol II to the promoter, resulting in repression of cyclin B2 expression.
Menin Interferes with NF-Y Binding and Reduces Histone H3 Acetylation at the Ccnb2 Promoter
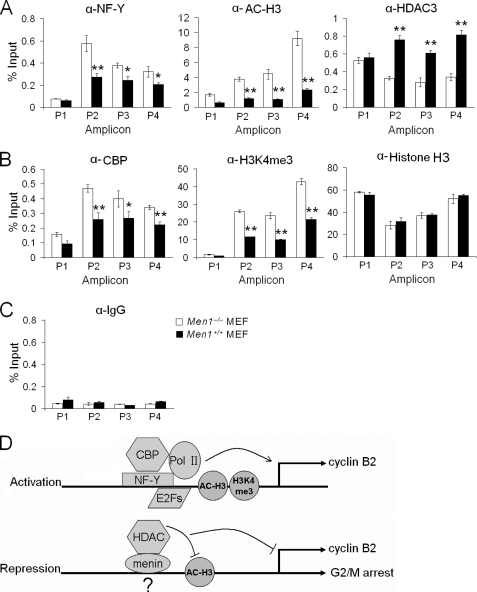
NF-Y has been well documented to bind to the CCAAT box in the Ccnb2 promoter in vitro by electrophoretic mobility shift assay and in vivo by ChIP assay (19,37). As menin associates with CCAAT boxes (Fig. 5B), we performed a ChIP assay in menin-expressing and menin-null cells to explore whether menin affects NF-Y binding to the Ccnb2 locus. We found that less NF-Y was associated with the Ccnb2 promoter in menin-expressing cells (Fig. 6A, left), suggesting that menin interferes with NF-Y binding to the Ccnb2 promoter. It has been reported that NF-Y is necessary for binding of p300/CBP, histone acetyltransferases, to the Ccnb2 promoter. Histone acetylation is crucial for cyclin B2 activation, whereas HDACs repress the Ccnb2 promoter (42–44). Thus, we performed the ChIP assay using menin-expressing and menin-null cells to explore whether menin affects histone acetylation at the Ccnb2 locus. Our results revealed that the level of acetylated histone H3 was reduced in menin-expressing cells (Fig. 6A, middle). This finding raised the possibility that menin recruits HDACs to deacetylate histone H3 or prevents cAMP-response element-binding proteins from acetylating histone H3 at the Ccnb2 promoter. Thus, we further determined if menin affects the binding of HDAC3 or CBP to the Ccnb2 promoter, as we found that menin co-immunoprecipitated with HDAC3.3 The ChIP assay showed that menin recruited HDAC3 to the Ccnb2 locus (Fig. 6A, right) and reduced the binding of CBP (Fig. 6B, left). These results suggest that menin represses cyclin B2 transcription at least partly through reducing histone acetylation at the locus.
FIGURE 6.
Menin reduces positive histone modifications at the Ccnb2 locus. A, shown is a ChIP assay for detection of NF-Y (left), acetylated-histone H3 (α-AC-H3, middle), and HDAC3 (right) at the Ccnb2 locus in Men1−/− and Men1+/+ MEFs; n = 3. B, shown is a ChIP assay using antibodies of CBP (left), trimethylated H3K4 (H3K4me3, middle), and histone H3 (right) in Men1−/− and Men1+/+ MEFs; n = 3. C, shown is a ChIP assay using IgG antibody as a control; n = 3. D, shown is a hypothetical model for menin-mediated repression of cyclin B2 and G2/M transition. In activated Ccnb2 promoter, NFY recruits CBP and Pol II to the locus and cooperates with E2Fs. The level of acetylated histone H3 and H3K4me3 is increased (top). In menin-repressed Ccnb2 promoter, menin may or may not bind to the promoter and reduces the binding of NFY, E2Fs, and CBP but increases recruitment of HDAC3 to deacetylate histone H3 (bottom).
As menin can recruit mixed lineage leukemia, a histone H3 lysine 4 (H3K4) methyltransferase, to the Hoxa9 promoter and increases the level of H3K4me3 at the locus to activate its transcription (45), we also examined the level of H3K4me3 at the Ccnb2 and Hoxa9 promoters in menin-expressing and menin-null cells. As expected, the H3K4me3 level was up-regulated on the Hoxa9 promoter in menin-expressing cells (supplemental Fig. S4) but down-regulated at the Ccnb2 promoter in menin-expressing cells (Fig. 6B, middle). We also performed a ChIP assay using anti-histone H3 antibody to determine whether menin affects the occupancy of nucleosome or modification of histones at the Ccnb2 locus. To this end we used antibody that recognizes total histone H3 (both modified and unmodified) for ChIP assay and found that Men1 excision did not affect the nucleosome occupancy at the Ccnb2 locus (Fig. 6B, right). Together, these results suggest that the impact of menin on the level of H3K4 trimethylation differs at distinct target genes, increasing H3K4m3 at menin-induced genes but decreasing H3K4m3 at menin-repressed genes. Perhaps the local histone epigenetic markers and associating factors at the chromatin play a crucial role in determining the impact of menin on H3K4m3 at a particular locus of the gene and, accordingly, the status of gene transcription. It is possible that menin affects recruitment of mixed lineage leukemia, which affects H3K4 methylation at the locus. On the other hand, we cannot rule out that menin reduces the binding of RNA polymerase II to the Ccnb2 locus and affects the status of H3K4 methylation at the locus. The detailed underlying mechanisms remain to be determined.
MEN1 Disease-related Menin Point Mutants Lose the Function of Reducing the Level of Acetylated Histone H3 at the Ccnb2 Locus
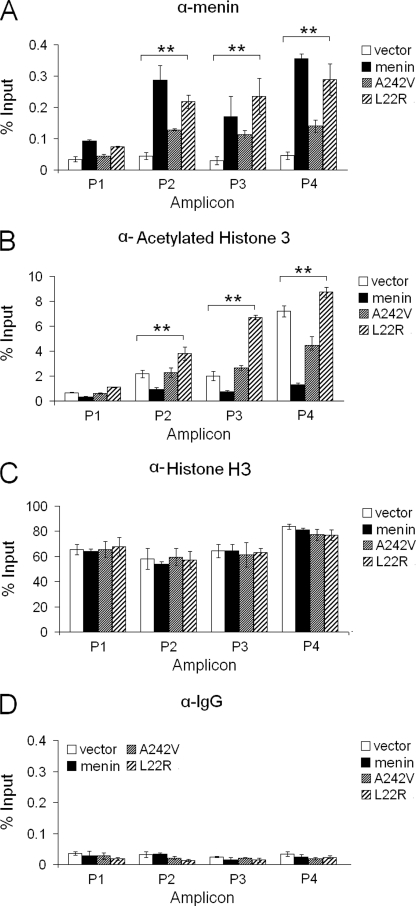
We then performed a ChIP assay to observe the impact of A242V and L22R mutations on menin binding to the cyclin B2 promoter. ChIP assay showed that A242V lost much ability to bind to the Ccnb2 promoter (Fig. 7A), whereas L22R bound to the Ccnb2 promoter almost as well as wild-type menin did. However, neither L22R nor A242V reduced the levels of acetylated histone H3 at the Ccnb2 locus (Fig. 7B), with little change of the occupancy of histone H3 (Fig. 7C). As a control, the expression of wild-type menin or menin mutants was similar (Fig. 2G, right, lanes 2–4). Together, these results strongly suggest that menin-mediated repression of cyclin B2 at least partly through repressing histone H3 acetylation at the Ccnb2 locus, and menin-induced repression of cyclin B2 expression is crucial for suppressing cell proliferation in MEN1 tumorigenesis.
FIGURE 7.
Menin mutants A242V and L22R fail to reduce the level of acetylated histone H3 at the Ccnb2 locus. A, shown is a ChIP assay using menin antibody in Men1−/− MEFs expressing control vector, menin, A242V, or L22R, n = 3. B, shown is a ChIP assay using acetylated-histone H3 antibody in the indicated cells, n = 3. C, shown is a ChIP assay using histone H3 antibody to see the level of occupancy of the histone H3 at the Ccnb2 locus in the indicated cells, n = 3. D, shown is a ChIP assay using IgG antibody as a control in the indicated cells, n = 3. *, p < 0.05; **, p < 0.01.
DISCUSSION
Although it has been shown that menin is crucial for repressing G1/S transition (12), it has been unclear whether menin also controls G2/M transition. We previously showed that Men1 excision in MEFs increased G1/0-S phase transition in serum-starved cells when the serum-starved cells were allowed to enter cell cycle after the addition of serum (12). It is noteworthy that these experimental conditions favored detection of a change of cell cycle transition from G0/1 to S phase, but not from G2 to M phase, because a majority of the serum-starved cells were arrested in G0/1 phase (12). Also using MEFs in our current studies, we used nocodazole to induce G2/M arrest, and this experimental approach gave us a better window to observe the impact of menin on G2/M transition. Therefore, our results suggest that menin can regulate both G1/0 to S and G2/M transition in the same type of cells. However, it cannot be ruled out that menin could also preferentially suppress either G0/1 to S or G2-M transition in certain types of cells. Our findings suggest that menin delays G2/M phase transition at least in part by repressing expression of cyclin B2. Menin represses transcription of cyclin B2 through antagonizing recruitment of the positive transcriptional regulator of cyclin B2 and promoting the recruitment of negative regulators. Thus, menin reduces the overall histone acetylation at the Ccnb2 locus, resulting in repression of gene transcription. Consistent with the notion that menin-repressed expression of cyclin B2 is crucial for blocking G2/M transition and cell proliferation, knockdown of cyclin B2 in MEN1-excised cells diminished the number of cells in M phase and reduced cell proliferation.
Menin is well known for its role in positive transcriptional regulation, but far less is known about how menin represses gene transcription. It has been shown that menin interacts with mixed lineage leukemia protein and up-regulates CDK inhibitors p18Ink4c and p27Kip1 via up-regulating H3K4 methylation (46, 47). In contrast, menin represses JunD via HDACs in vitro (8). However, it was previously unclear whether menin recruits HDACs to endogenous genes to repress their transcription. In our studies we showed that menin recruited HDAC3 to the Ccnb2 locus and repressed expression from the endogenous Ccnb2 gene. Menin-mediated repression of cyclin B2 may occur in part through recruiting HDAC3 and deacetylating histones at the Ccnb2 locus, as MEN1 ablation reduced HDAC3 binding to and histone H3 acetylation at the locus (Fig. 6). However, we cannot rule out that menin also affects recruitment of other HDACs. In contrast with the role of menin in recruiting HDAC3, menin repressed binding of CBP, a histone acetyltransferase, to the Ccnb2 locus. The differential role for menin in regulating HDAC3 and CBP may account partly for menin-dependent repression of cyclin B2 expression.
The CCAAT-mutated Ccnb2 promoter lost most of its activity in luciferase assays. This may be due to the failure of NF-Y to bind to the mutated CCAAT box, which accounts for >93% of the total activity of cyclin B2 (37). On the other hand, CDE or CHR-mutated Ccnb2 promoter remained active but was no longer effectively repressed by menin, suggesting that CDE/CHR is an important element for binding of menin or menin-interacting repressive complexes to repress cyclin B2 transcription. This is consistent with the role of CDE/CHR as a repressive tandem element of the Ccnb2 promoter (20). However, the identities of proteins potentially binding to the CDE/CHR elements have remained largely unknown (48). Although it has been reported that the CDE of cdc2 and Ccna2 promoters can bind cellular E2F ternary complexes in vitro with a low efficiency, other reports disagree with this result (22).
We observed that menin associated with the Ccnb2 promoter in the region containing three CCAAT boxes and CDE/CHR elements, which suggested that CCAAT boxes and CDE/CHR elements were important for menin-mediated repression of cyclin B2. It is possible that menin binds to this region through other factors that specifically recognize these conserved DNA elements, because menin only binds double-stranded DNA in a sequence-nonspecific manner (10). In our ChIP assay the resolution is not high enough to discern events that occur on the three CCAAT boxes or on the adjacent CDE and CHR boxes. Events in the region in the first intron detected by amplicon P4 followed the same trend as that in amplicons P2 and P3 (Figs. 5 and 6); this could be caused by a low resolution of the method as amplicons P1, P2, and P3 are close. However, at least the region containing three CCAAT boxes and CDE/CHR elements is important for menin in regulating the Ccnb2 promoter. Menin expression in cells reduced the association of E2Fs, NF-Y, CBP, and Pol II with these regions. It remains unclear how menin blocks binding of these proteins to the Ccnb2 locus. In addition, menin expression also reduced the levels of histone H3 acetylation and H3K4 methylation at the locus. One possibility is that menin-mediated repression of histone acetylation or H3K4 methylation results in a compact packaging of local nucleosomes, preventing recruitment of multiple positive transcriptional regulators. Together, these findings suggest that menin antagonizes binding of NF-Y and E2Fs and other crucial regulators to the Ccnb2 promoter and reduces positive histone acetylation and H3K4 methylation, leading to repression of gene transcription.
In contrast with the role of menin in maintaining an increased level of H3K4 trimethylation at the Hoxa9 gene (supplemental Fig. S4), menin expression reduced the level of H3K4 trimethylation at the Ccnb2 promoter, suggesting that the opposite role of menin in regulating expression of these two genes is gene-specific. This difference may in part reflect the different chromosomal environments surrounding the distinct genes.
MEN1 disease-related mutant A242V reduced its binding to the Ccnb2 promoter, and the L22R mutant remained capable of binding to Ccnb2, but both of the mutants failed to effectively repress expression of cyclin B2. Consistent with this observation, the level of acetylated histone H3 at the Ccnb2 locus was reduced in cells expressing either of the two mutants. Although L22R associates relatively well with the Ccnb2 promoter, it cannot repress histone H3 acetylation as does wild-type menin. It is possible that the L22R mutant loses the ability to recruit HDACs or to prevent CBP binding to the locus. Collectively, these results suggest that these mutants lost their ability to promote expression of cyclin B2 partly due to the failure to repress the histone H3 acetylation at the Ccnb2 promoter. We do not yet know why a conservative mutation, A242V, with only a difference of two methyl groups at side chain, has a stronger impact than a more drastic mutation, L22R, has on menin binding to the Ccnb2 locus even though both mutations have an obvious impact on reduction of histone acetylation at the Ccnb2 locus. Given that the crystal structure for menin remains lacking, it has been challenging to understand how these mutations have such unpredictable consequences. Although highly speculative, one possible scenario could be that the A242V mutation, with two additional hydrophobic methyl groups from the valine residue, may attenuate its interaction with a DNA-binding partner protein and, thus, reduce its own recruitment to the Ccnb2 locus. On the other hand, on a distinct surface, L22R mutation may not affect its own recruitment to the Ccnb2 locus; rather, with a gain of positive charges from the arginine residue, this mutant protein may affect its interaction with and recruitment of HDACs to reduce histone acetylation at the locus. We could not rule out that this mutant may also affect the enzyme activity of HDACs at the locus.
From our studies we propose a model (Fig. 6D) in which menin reduces transcription of cyclin B2 by repressing the transcriptional activity of the Ccnb2 promoter. Menin may associate with the promoter by direct or indirect binding to DNA and recruits HDACs to deacetylate histone H3. In addition, when menin associates with the promoter, it may interfere with the association of transcriptional factors such as E2Fs, NF-Y, and CBP to the Ccnb2 promoter. MEN1 disease-related menin mutants lose their ability to repress histone acetylation and cyclin B2 transcription.
The model proposed in Fig. 6D may represent only one of multiple scenarios where menin and other co-factors regulate the cyclin B2 promoter; our current assay is insufficient to distinguish whether menin binds to a particular element among the CCAAT, CDE, and CHR boxes. It remains speculative as to the mechanisms whereby menin affects association of other transcriptional factors, such as NF-Y and E2Fs with the Ccnb2 promoter. As both NF-Y and E2Fs have been reported to bind to CCAAT boxes and CDE, respectively (19, 37, 49, 50), and the C-terminal part of menin binds double-stranded DNA in a sequence-independent manner (27), it is possible that reduction of NF-Y and E2F binding to the Ccnb2 promoter in the presence of menin is due to the indirect effect from menin. This includes the impact of menin on acetylation of histones at the locus, and this effect could indirectly interfere with the binding of NF-Y and E2Fs. Nevertheless, together, our findings reveal a previously unappreciated mechanism to explain how menin controls G2/M transition and cell proliferation and how MEN1 mutations in MEN1 disease cripple menin function in repressing G2/M transition and cell proliferation.
Supplementary Material
Acknowledgments
We thank Kurt Engeland for providing the plasmids. We thank Hongxia Zhang, Buddha Gurung, and Peter Blessington for excellent technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants R01-CA-100912 and R01-CA-113962 (to Xianxin H.). This work was also supported by American Diabetes Association Grant 7-07-RA-60 (to Xianxin H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures and Figs. S1–S5.
T. Wu and Y. Yang, unpublished data.
- MEN1
- multiple endocrine neoplasia type 1
- H3K4
- histone H3 lysine 4
- NF-Y
- nuclear factor Y
- E2Fs
- E2 factors
- CBP
- histone acetyltransferase CREB (cAMP-response element-binding protein)-binding protein
- HDAC
- histone deacetylase
- MEF
- mouse embryonic fibroblast
- CDK
- cyclin-dependent kinase
- CDE
- cell cycle-dependent element
- CHR
- cell cycle gene homology region
- Pol II
- RNA polymerase II
- shRNA
- short hairpin RNA
- ChIP
- chromatin immunoprecipitation
- RT
- reverse transcription
- qRT
- quantitative real-time.
REFERENCES
- 1.Gardner D. G. (1997) Adv. Intern. Med. 42, 597–627 [PubMed] [Google Scholar]
- 2.Brandi M. L., Gagel R. F., Angeli A., Bilezikian J. P., Beck-Peccoz P., Bordi C., Conte-Devolx B., Falchetti A., Gheri R. G., Libroia A., Lips C. J., Lombardi G., Mannelli M., Pacini F., Ponder B. A., Raue F., Skogseid B., Tamburrano G., Thakker R. V., Thompson N. W., Tomassetti P., Tonelli F., Wells S. A., Jr., Marx S. J. (2001) J. Clin. Endocrinol. Metab. 86, 5658–5671 [DOI] [PubMed] [Google Scholar]
- 3.Chandrasekharappa S. C., Guru S. C., Manickam P., Olufemi S. E., Collins F. S., Emmert-Buck M. R., Debelenko L. V., Zhuang Z., Lubensky I. A., Liotta L. A., Crabtree J. S., Wang Y., Roe B. A., Weisemann J., Boguski M. S., Agarwal S. K., Kester M. B., Kim Y. S., Heppner C., Dong Q., Spiegel A. M., Burns A. L., Marx S. J. (1997) Science 276, 404–407 [DOI] [PubMed] [Google Scholar]
- 4.Lemos M. C., Thakker R. V. (2008) Hum. Mutat. 29, 22–32 [DOI] [PubMed] [Google Scholar]
- 5.Yang Y., Hua X. (2007) Mol. Cell. Endocrinol. 265–266, 34–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agarwal S. K., Guru S. C., Heppner C., Erdos M. R., Collins R. M., Park S. Y., Saggar S., Chandrasekharappa S. C., Collins F. S., Spiegel A. M., Marx S. J., Burns A. L. (1999) Cell 96, 143–152 [DOI] [PubMed] [Google Scholar]
- 7.Gobl A. E., Berg M., Lopez-Egido J. R., Oberg K., Skogseid B., Westin G. (1999) Biochim. Biophys. Acta 1447, 51–56 [DOI] [PubMed] [Google Scholar]
- 8.Kim H., Lee J. E., Cho E. J., Liu J. O., Youn H. D. (2003) Cancer Res. 63, 6135–6139 [PubMed] [Google Scholar]
- 9.Heppner C., Bilimoria K. Y., Agarwal S. K., Kester M., Whitty L. J., Guru S. C., Chandrasekharappa S. C., Collins F. S., Spiegel A. M., Marx S. J., Burns A. L. (2001) Oncogene 20, 4917–4925 [DOI] [PubMed] [Google Scholar]
- 10.La P., Silva A. C., Hou Z., Wang H., Schnepp R. W., Yan N., Shi Y., Hua X. (2004) J. Biol. Chem. 279, 49045–49054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao S. B., Feng Z. J., Xu B., Wu Y., Yin P., Yang Y., Hua X., Jin G. H. (2009) Oncogene 28, 4095–4104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schnepp R. W., Chen Y. X., Wang H., Cash T., Silva A., Diehl J. A., Brown E., Hua X. (2006) Cancer Res. 66, 5707–5715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Draetta G., Luca F., Westendorf J., Brizuela L., Ruderman J., Beach D. (1989) Cell 56, 829–838 [DOI] [PubMed] [Google Scholar]
- 14.Gautier J., Matsukawa T., Nurse P., Maller J. (1989) Nature 339, 626–629 [DOI] [PubMed] [Google Scholar]
- 15.Labbé J. C., Capony J. P., Caput D., Cavadore J. C., Derancourt J., Kaghad M., Lelias J. M., Picard A., Dorée M. (1989) EMBO J. 8, 3053–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gautier J., Minshull J., Lohka M., Glotzer M., Hunt T., Maller J. L. (1990) Cell 60, 487–494 [DOI] [PubMed] [Google Scholar]
- 17.Fisher D., Nurse P. (1995) Semin. Cell Biol. 6, 73–78 [DOI] [PubMed] [Google Scholar]
- 18.Nasmyth K. (1996) Science 274, 1643–1645 [DOI] [PubMed] [Google Scholar]
- 19.Bolognese F., Wasner M., Dohna C. L., Gurtner A., Ronchi A., Muller H., Manni I., Mossner J., Piaggio G., Mantovani R., Engeland K. (1999) Oncogene 18, 1845–1853 [DOI] [PubMed] [Google Scholar]
- 20.Lange-zu Dohna C., Brandeis M., Berr F., Mössner J., Engeland K. (2000) FEBS Lett. 484, 77–81 [DOI] [PubMed] [Google Scholar]
- 21.Zwicker J., Lucibello F. C., Wolfraim L. A., Gross C., Truss M., Engeland K., Müller R. (1995) EMBO J. 14, 4514–4522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fajas L., Le Cam L., Polanowska J., Fabbrizio E., Servant N., Philips A., Carnac G., Sardet C. (2000) FEBS Lett. 471, 29–33 [DOI] [PubMed] [Google Scholar]
- 23.Myslinski E., Gérard M. A., Krol A., Carbon P. (2007) Nucleic Acids Res. 35, 3453–3464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song N., Zhu X., Shi L., An J., Wu Y., Sang J. (2009) Sci. China C Life Sci. 52, 551–559 [DOI] [PubMed] [Google Scholar]
- 25.La P., Schnepp R. W., Petersen C. C., Silva A., Hua X. (2004) Endocrinology 145, 3443–3450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schnepp R. W., Mao H., Sykes S. M., Zong W. X., Silva A., La P., Hua X. (2004) J. Biol. Chem. 279, 10685–10691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.La P., Yang Y., Karnik S. K., Silva A. C., Schnepp R. W., Kim S. K., Hua X. (2007) J. Biol. Chem. 282, 31332–31340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin S., Mao H., Schnepp R. W., Sykes S. M., Silva A. C., D'Andrea A. D., Hua X. (2003) Cancer Res. 63, 4204–4210 [PubMed] [Google Scholar]
- 29.Goto H., Tomono Y., Ajiro K., Kosako H., Fujita M., Sakurai M., Okawa K., Iwamatsu A., Okigaki T., Takahashi T., Inagaki M. (1999) J. Biol. Chem. 274, 25543–25549 [DOI] [PubMed] [Google Scholar]
- 30.Hendzel M. J., Wei Y., Mancini M. A., Van Hooser A., Ranalli T., Brinkley B. R., Bazett-Jones D. P., Allis C. D. (1997) Chromosoma 106, 348–360 [DOI] [PubMed] [Google Scholar]
- 31.Preuss U., Landsberg G., Scheidtmann K. H. (2003) Nucleic Acids Res. 31, 878–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harper J. V. (2005) Methods Mol. Biol. 296, 157–166 [DOI] [PubMed] [Google Scholar]
- 33.Fontanière S., Tost J., Wierinckx A., Lachuer J., Lu J., Hussein N., Busato F., Gut I., Wang Z. Q., Zhang C. X. (2006) Endocr. Relat. Cancer 13, 1223–1236 [DOI] [PubMed] [Google Scholar]
- 34.Balogh K., Rácz K., Patócs A., Hunyady L. (2006) Trends Endocrinol. Metab. 17, 357–364 [DOI] [PubMed] [Google Scholar]
- 35.Agarwal S. K., Kester M. B., Debelenko L. V., Heppner C., Emmert-Buck M. R., Skarulis M. C., Doppman J. L., Kim Y. S., Lubensky I. A., Zhuang Z., Green J. S., Guru S. C., Manickam P., Olufemi S. E., Liotta L. A., Chandrasekharappa S. C., Collins F. S., Spiegel A. M., Burns A. L., Marx S. J. (1997) Hum. Mol. Genet 6, 1169–1175 [DOI] [PubMed] [Google Scholar]
- 36.Dagle J. M., Walder J. A., Weeks D. L. (1990) Nucleic Acids Res. 18, 4751–4757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wasner M., Haugwitz U., Reinhard W., Tschöp K., Spiesbach K., Lorenz J., Mössner J., Engeland K. (2003) Gene 312, 225–237 [DOI] [PubMed] [Google Scholar]
- 38.La P., Desmond A., Hou Z., Silva A. C., Schnepp R. W., Hua X. (2006) Oncogene 25, 3537–3546 [DOI] [PubMed] [Google Scholar]
- 39.Nevins J. R. (1992) Science 258, 424–429 [DOI] [PubMed] [Google Scholar]
- 40.Ishida S., Huang E., Zuzan H., Spang R., Leone G., West M., Nevins J. R. (2001) Mol. Cell. Biol. 21, 4684–4699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Polager S., Kalma Y., Berkovich E., Ginsberg D. (2002) Oncogene 21, 437–446 [DOI] [PubMed] [Google Scholar]
- 42.Caretti G., Salsi V., Vecchi C., Imbriano C., Mantovani R. (2003) J. Biol. Chem. 278, 30435–30440 [DOI] [PubMed] [Google Scholar]
- 43.Imbriano C., Gurtner A., Cocchiarella F., Di Agostino S., Basile V., Gostissa M., Dobbelstein M., Del Sal G., Piaggio G., Mantovani R. (2005) Mol. Cell. Biol. 25, 3737–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salsi V., Caretti G., Wasner M., Reinhard W., Haugwitz U., Engeland K., Mantovani R. (2003) J. Biol. Chem. 278, 6642–6650 [DOI] [PubMed] [Google Scholar]
- 45.Chen Y. X., Yan J., Keeshan K., Tubbs A. T., Wang H., Silva A., Brown E. J., Hess J. L., Pear W. S., Hua X. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 1018–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Milne T. A., Hughes C. M., Lloyd R., Yang Z., Rozenblatt-Rosen O., Dou Y., Schnepp R. W., Krankel C., Livolsi V. A., Gibbs D., Hua X., Roeder R. G., Meyerson M., Hess J. L. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 749–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karnik S. K., Hughes C. M., Gu X., Rozenblatt-Rosen O., McLean G. W., Xiong Y., Meyerson M., Kim S. K. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 14659–14664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tschöp K., Engeland K. (2007) FEBS J. 274, 5235–5249 [DOI] [PubMed] [Google Scholar]
- 49.Zerfass K., Spitkovsky D., Schulze A., Joswig S., Henglein B., Jansen-Dürr P. (1996) J. Virol. 70, 2637–2642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tommasi S., Pfeifer G. P. (1995) Mol. Cell. Biol. 15, 6901–6913 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.