Abstract
The clade B human immunodeficiency virus, type 1 (HIV-1) Tat (trans-acting regulatory protein) induces interleukin-10 (IL-10) production in monocytes. IL-10, an anti-inflammatory cytokine, down-regulates proinflammatory cytokines and suppresses the immune response, leading to a rapid progression from HIV-1 infection to AIDS. Nine clades of HIV-1 are responsible for the majority of infections worldwide. Recent studies demonstrate that different HIV-1 clades have biological differences in relation to transmission, replication, and disease progression. In this study, we show that the cysteine to serine mutation at position 31, found in >90% of HIV-1 clade C Tat proteins, results in a marked decrease in IL-10 production in monocytes compared with clade B Tat. Additionally, the C31S mutation found in C Tat is responsible for the inability of these Tat proteins to produce high IL-10 levels in monocytes due to its inability to induce intracellular calcium flux through L-type calcium channels. Moreover, we show that p38α/p38β and phosphoinositide 3-kinase are crucial to Tat-induced IL-10 production. These findings provide further evidence that HIV-1 clades differ in their biological properties that may impact HIV-1 pathogenesis and disease progression.
Keywords: Calcium, HIV, p38 MAPK, Phosphatidylinositol 3-kinase, ERK, Clade C, IL-10, L-type Calcium Channel, Monocyte, Tat
Introduction
The human immunodeficiency virus type 1 (HIV-1)3 trans-acting regulatory protein (Tat) is an 86–101-residue protein involved in initiating viral transcription and RNA chain elongation. In addition to its primary role as a transcriptional activator of viral gene expression, Tat is actively released from unruptured, HIV-1-infected cells and is detectable in ex vivo culture supernatants and in the serum of HIV-1 infected individuals (1, 2). Most exogenous Tat studies use a truncated 86-residue HIV-1 clade B Tat with few studies examining the functions of other clades or isotypes (3). However, these studies have shown that exogenous B Tat induces the production of cytokines, such as tumor necrosis factor, chemokine (C-C motif) ligand 2 (CCL2), interleukin-6 (IL-6), and interleukin-10 (IL-10) from monocytes and macrophages (4–9).
The expression of IL-10-specific mRNA and the production of IL-10 are both increased in HIV-1-infected individuals (10–12). IL-10 is an anti-inflammatory cytokine produced by a wide variety of cells including monocytes, macrophages, T cells, natural killer cells, and B cells (13) that down-regulates major histocompatibility complex class II (13) and inhibits T cell proliferation while reducing the production of proinflammatory cytokines. Elevated IL-10 levels are found in individuals with rapid progression to AIDS (14–16), and individuals with higher plasma levels of IL-10 have more severely compromised T helper cell function (14–16) combined with lower T helper cell counts (17). Interestingly, in samples from patients chronically infected with HIV, blocking the IL-10/IL-10 receptor pathway in vitro using specific antibodies enhanced CD4+ T cell responses (14–16). Therefore, maintaining low levels of IL-10 may slow HIV-1 disease progression.
Recent studies have shown that HIV-1 clades possess biological differences in relation to transmission, replication, and disease progression (18–20). HIV-1 clade C accounts for 50% of infections worldwide and has a full-length 101-residue Tat protein. 90% of sequenced HIV-1 clade C Tat possess a C31S mutation not found in clade B Tat that disrupts the 30C-C motif (21). This motif is essential for the induction of a Ca2+ flux in, and chemotaxis of, monocytes and the induction of inflammatory cytokines and chemokines. Conversely, disruption of this motif actually increases its trans-activational activity (6, 7, 22, 23). To date, no study has assessed the capacity of different full length HIV-1 Tat clades to induce IL-10 production in monocytes. In this study, we compared the abilities of HIV-1 clade C Tat and the long form of clade B Tat (22, 24) to induce IL-10 production in monocytes. In this study, we show that clade C Tat has a reduced ability to induce IL-10 production in monocytes.
EXPERIMENTAL PROCEDURES
Cells and Reagents
Human peripheral blood mononuclear cells (PBMC) were isolated from healthy HIV-1-negative donors using density centrifugation over Ficoll-PaqueTM premium (GE Healthcare). Monocytes were isolated from PBMC by adherence as described previously (25, 26).
Full-length synthetic Tat proteins were kindly provided by Dr. Erwann Loret of the Université de la Méditerranée (Marseille, France) and were synthesized and purified as described previously (27). Peptides corresponding to residues 1–45 of clade B TatHXB2 (Tat1–45) and residues 1–45 of clade B TatHXB2 with a C31S mutation (Tat1–45C31S) were custom synthesized and purified by Bionexus (Oakland, CA).
The p38α/p38β inhibitor, SB 203580, the phosphoinositide 3-kinase (PI3K) inhibitor wortmannin, MAPK kinase-1 (MKK1) inhibitors U0126 and PD 98059, and FS2 from Dendroaspis polylepis polylepis venom, an angusticeps type III L-type Ca2+ channel (CaL) inhibitor, were all purchased from Sigma. The p38α/p38β inhibitor doramapimod and the CCR5 antagonist maraviroc were purchased from LC Laboratories (Woburn, MA) and Toronto Research Chemicals (North York, ON, Canada), respectively. The MKK1 inhibitor PD 0325901, the PI3K inhibitor PI-103, the CaL inhibitor nimodipine, the ryanodine receptor inhibitor 3,4,5-trimethyloxybenzoic acid 8-(diethylamino)octyl ester (TMB-8), and the inositol-1,4,5-triphosphate receptor inhibitor xestospongin C were purchased from EMD Chemicals. The cytotoxic effect of the different inhibitors was assessed using the cytotoxicity detection kit (Roche Applied Science) and/or the trypan blue exclusion assay, and none were found to be cytotoxic.
Determination of Cytokine Production
IL-10 present in monocyte supernatants was quantified using a sensitive enzyme-linked immunosorbent assay (ELISA) kit (Invitrogen) according to the manufacturer's protocol.
IL-10 mRNA expression in monocytes was measured by real-time PCR. Monocytes were stimulated with Tat in a 2:1 ratio of AIM-V/Iscove's modified Dulbecco's media (both from Invitrogen) then harvested. Total cellular RNA was prepared with the RNeasy mini kit using the optional DNase step in accordance with the manufacturer's directions (Qiagen). IL-10 mRNA expression was determined using the LightCycler system and the FastStart RNA Master SYBR Green I kit (both from Roche Applied Science) with RNA polymerase II expression measured as an internal standard according to the manufacturer's instructions. Primers were synthesized by Integrated DNA Technologies and were as follows; IL-10, 5′-ATGCTTCGAGATCTCCGAGA-3′ (sense) and 5′-AAATCGATGACAGCGCCGTA-3′ (anti-sense); and RNA polymerase II, 5′-GCACCACGTCCAATGACAT-3′ (sense) and 5′-GTGCGGCTGCTTCCATAA-3′ (anti-sense). Amplification was performed for 40 cycles with the following cycle parameters: 5 s denaturation at 95 °C, 20 s primer annealing at 53 °C, and 20 s fragment elongation at 72 °C. mRNA levels were normalized using the RelQuant software (Roche Applied Science). All results are expressed as the ratio between the normalized expression of the target gene in treated cells and the normalized expression of the target gene in untreated or control cells, so that IL-10 mRNA expression in unconditioned cells equals 1.00.
Calcium Mobilization
The increase in intracellular cytoplasmic Ca2+ concentration in monocytes in response to 50 nm Tat was evaluated by flow cytometry using PBMC loaded with Fluo-4 acetoxymethylester and Fura Red acetoxymethylester (both from Molecular Probes) as described previously (27). Calcium mobilization is reported as the ratio of Fluo-4 to Fura Red fluorescence intensity over time as calculated using FCSPress version 1.4 (Ray Hicks, Department of Medicine, University of Cambridge).
Statistical Analysis
All p values were obtained using a paired two-tailed Student's t test. A p value of ≤ 0.05 was considered statistically significant.
RESULTS
Tat Induces IL-10 in a Time- and Dose-dependent Manner in Monocytes
In this study, we initially used the HIV-1 clade B TatHXB2, which was described by K. T. Jeang et al. (24), and a clade C Tat93In derived from HIV-193In905 isolated in India (Fig. 1). Tat93In is typical of clade C isolated, in that it possesses a C31S substitution in the cysteine-rich region (28). Of the seven highly conserved cysteines in Tat, only substitutions of Cys31 do not affect its trans-activational ability (21, 23, 24, 29).
FIGURE 1.
Sequences of the Tat variants and Tat peptides. TatHXB2 is used as the primary reference genome for HIV-1 at the Los Alamos HIV Databases. HIV-1HXB2 is a specific clone from the French isolate HIV-1LAI (formerly HIV-1BRU), which is also referred to as HIV-1IIIB or LAV. It was one of the first published nucleotide sequences of HIV-1. The clade C Tat93In was derived from HIV-193In905 isolated in India. Tat93In is typical of clade C isolated in that it possesses a C31S substitution in the cysteine-rich region (28). Tat96Bw, also a clade C protein, was derived from the Botswanian isolate HIV-196Bw0504 and possesses an intact 30C-C motif (29, 31). Tat1–45 and Tat1–45C31S are truncated peptides of TatHXB2 that differ only in a C31S mutation.
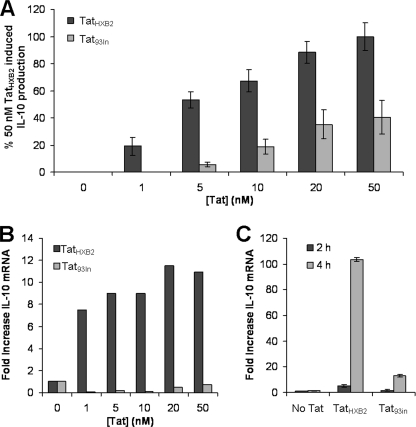
Studies have shown that HIV-1 clade B Tat induces IL-10 production from human monocytes (4, 30). Therefore, we first examined whether the different clades of Tat have a differential effect on IL-10 secreted from freshly isolated primary human monocytes. For these experiments, monocytes were treated with increasing concentrations of TatHXB2 or Tat93In. Supernatants and cells were then harvested and analyzed for secreted IL-10 by ELISA and IL-10 mRNA by real-time PCR. The secreted IL-10 response to both clades of Tat was both time- and dose-dependent and was maximal at 24 h post-treatment (Fig. 2A and data not shown). Although donor-specific variations did occur with regards to absolute concentrations of IL-10 produced, at all concentrations tested, clade B TatHXB2 consistently induced significantly greater quantities of IL-10 than the clade C Tat93In (p < 0.05, Fig. 2A; data not shown). 1 nm of TatHXB2, the lowest concentration tested, was sufficient to induce significantly more IL-10 than untreated cells (p = 0.02, Fig. 2A), whereas the minimum amount of Tat93In required to elicit a significant response was 5 nm (p = 0.02, Fig. 2A). We next assessed the IL-10 mRNA response of these cells at 24 h post-Tat treatment. Donor specific absolute fold-increase variations were observed with regards to IL-10 mRNA production; but, at all concentrations tested, we observed significantly greater quantities of IL-10 mRNA being produced post-TatHXB2 treatment than post-Tat93In treatment (p < 0.05; Fig. 2B), consistent with the observed IL-10 protein concentrations present in the supernatants (Fig. 2A).
FIGURE 2.
Clade B TatHXB2 induces more IL-10 from monocytes than the clade C Tat93In. Monocytes from HIV-1-negative subjects were incubated with increasing concentrations of Tat. After 24 h, the supernatants were collected and analyzed for IL-10 by ELISA (A), the cells were harvested and processed for total RNA and analyzed by real-time PCR for IL-10 mRNA content as described under “Experimental Procedures” (B). At all concentrations tested, the clade B TatHXB2 (black bars) induced both significantly more secreted IL-10 (p < 0.02) and more IL-10 mRNA (p < 0.001) from monocytes compared with the clade C Tat93In (gray bars). C, RNA was also extracted from donor cells at 2 (black bars) and 4 h (gray bars) post-Tat (10 nm) treatment and analyzed for IL-10 mRNA as described above. TatHXB2 induced significantly more IL-10 mRNA at these time points than Tat93In. The bars represent means ± S.E. from three independent experiments carried out in triplicate.
The mRNA response precedes the secreted protein response; therefore, we also examined the IL-10 mRNA response to 10 nm Tat at 2 and 4 h post-treatment. We found that TatHXB2 produced a 4-fold increase at 2 h, and a 100-fold increase at 4 h post-treatment compared with untreated cells (Fig. 2C). Conversely, although Tat93In induced more IL-10 mRNA than untreated cells at both time points, those levels were significantly below those of B Tat (p < 0.0003; Fig. 2C).
The C31S Mutation Found in Clade C Tat Is Responsible for the Decreased IL-10 Response
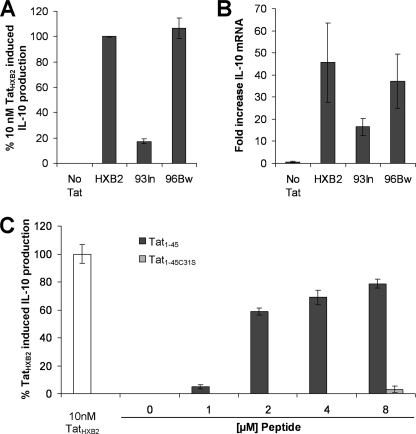
To determine the role of the C31S mutation in Tat93In in the induction of IL-10, we first assessed the ability of another clade C Tat, Tat96Bw derived from the Botswanian isolate HIV-196Bw0504 (29, 31), to stimulate an IL-10 response from monocytes. Tat96Bw is an atypical clade C Tat protein as it differs from Tat93In in that it does not possess the C31S mutation representative of clade C Tat (Fig. 1). We found that monocytes stimulated with 10 nm clade B TatHXB2 and clade C Tat96Bw produced 6-fold more IL-10 than cells incubated with clade C Tat93In (Fig. 3A; p < 0.01) with no significant difference observed between TatHXB2 and Tat96Bw induced IL-10 production (p = 0.99).
FIGURE 3.
The C31S mutation found in clade C Tat proteins is responsible for their reduced ability to induce IL-10 from monocytes. A, monocytes were incubated with 10 nm Tat for 24 h after which the supernatants were collected and analyzed for IL-10. Alternatively, the cells were harvested after 4 h and analyzed for IL-10 mRNA content, B. The clade B TatHXB2 and the clade C Tat96Bw induced comparable levels of IL-10 and IL-10 mRNA. Conversely, the clade C Tat93In induced IL-10 and IL-10 mRNA was significantly inferior (p < 0.05). C, monocytes were incubated with increasing concentrations of Tat peptides. After 24 h, supernatants were collected and analyzed for IL-10 by ELISA. Tat1–45 but not Tat1–45C31S induced significant levels of IL-10. The bars represent means ± S.E. from three independent experiments carried out in triplicate.
We then investigated the IL-10 mRNA response to 10 nm Tat at 4 h post-treatment. In the representative donor shown, clade B TatHXB2 up-regulated IL-10 mRNA expression 45-fold (Fig. 3B). Similarly, cells treated with Tat96Bw increased IL-10 mRNA expression 37-fold (Fig. 3B). In contrast, monocytes incubated with Tat93In failed to induce comparable IL-10 mRNA levels, suggesting a possible role for the C31S mutation in IL-10 induction.
Although Tat96Bw and Tat93In are both clade C variants, the differences between the two proteins extends to more than the C31S mutation. However, a previous study showed that the IL-10-inducing effect of Tat is exerted at the membrane and that the active domain is located within the N-terminal residues 1–45 (4). Therefore, to show that the difference in IL-10 induction is due to the C31S mutation, we synthesized two peptides using the first 45 residues of TatHXB2: Tat1–45 and Tat1–45C31S, that are identical except for the C31S mutation in Tat1–45C31S (Fig. 1). At all concentrations tested, Tat1–45C31S was unable to elicit a significant IL-10 response from primary monocytes. Conversely, Tat1–45 induced a significant IL-10 response (Fig. 3C).
PI3K and p38α/p38β MAPK Are Required for HIV-1 Tat-induced IL-10 Production in Monocytes
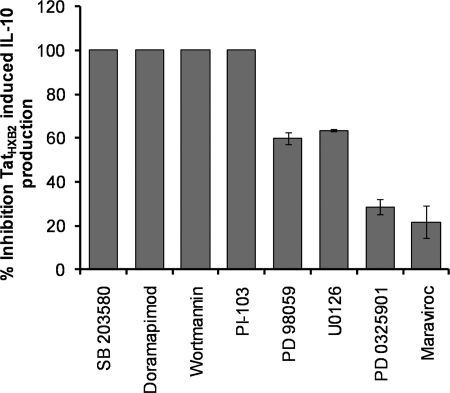
Tat-induced IL-10 production in monocytes has been shown to be dependent upon the MAPK pathway (4, 30, 32, 33). To determine which MAPKs are involved in Tat-induced IL-10 production, we initially pretreated monocytes with cell-permeable chemical inhibitors for p38α/p38β, PI3K, and MKK1 (34, 35) for 1 h and then subsequently treated monocytes with clade B TatHXB2 in the continuing presence of the inhibitors for an additional 24 h. Supernatants were then collected and analyzed by ELISA for the presence of IL-10. As a control, the CCR5 antagonist maraviroc was also used. The specificity and guidelines for the use of the MAPK inhibitors used has been reported by Bain et al. (35). Both p38α/p38β inhibitors, SB 203580 and doramapimod, and both class I PI3K inhibitors PI-103 and wortmannin, abolished Tat-induced IL-10 production (p < 0.001). The MKK1 inhibitors PD 98059, U0126, and PD 0325901 reduced IL-10 levels by 60, 63, and 29%, respectively, indicating a lesser role for this kinase (Fig. 4).
FIGURE 4.
MAPK inhibitors suppress TatHXB2-induced IL-10 production from monocytes. Monocytes were pretreated with 30 μm SB 203580, 100 nm doramapimod (both p38α/p38β inhibitors), 500 nm PI-103, 10 nm wortmannin (both class I PI3K inhibitors), 20 μm PD 98059, 100 nm PD 0325901, 10 μm U0126 (all MKK1 inhibitors), 10 nm maraviroc (CCR5 inhibitor), or vehicle control for 1 h before incubation with TatHXB2 for 24 h. Supernatants were then collected and analyzed for IL-10 by ELISA. The p38α/p38β and class I PI3K inhibitors completely abrogated the IL-10 response. The bars represent means ± S.E. from three independent experiments carried out in triplicate.
Tat93In Does Not Induce [Ca2+]i Flux in Human Monocytes
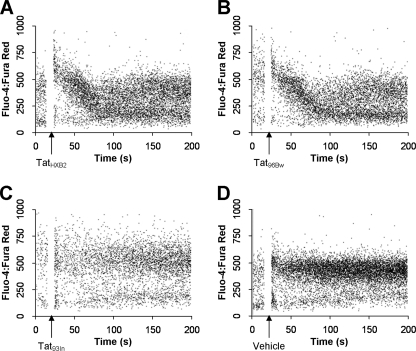
We have shown previously that clade B Tat, but not a clade C Tat possessing the C31S mutation, induces an increase in monocyte [Ca2+]i (27). Therefore, to determine whether the mutations present in Tat96Bw played a role in inducing a transient [Ca2+]i flux in monocytes, PBMC from healthy blood donors were loaded with the cell-permeant calcium fluorescent probes Fluo 4 acetoxymethylester and Fura Red acetoxymethylester, stimulated with Tat and the variations in [Ca2+]i measured by flow cytometry. Both TatHXB2 (Fig. 5A) and Tat96Bw (Fig. 5B) elicited a transient increase in [Ca2+]i in monocytes that returned to base-line 1 min after the initial flux. Tat93In failed to induce a measurable increase in [Ca2+]i (Fig. 5C) and was comparable to the vehicle control (Fig. 5D).
FIGURE 5.
Analysis of intracellular calcium mobilization by flow cytometry. PBMC were loaded with Fluo 4 acetoxymethyl ester and Fura Red acetoxymethyl ester. Cells were treated with 10 nm Tat at the time points indicated (A–D) and monitored for calcium mobilization. Clade B TatHXB2 and clade C Tat96Bw induced a Ca2+ flux in monocytes, whereas the clade C Tat93In and vehicle control did not.
B Tat-induced IL-10 Production Is Reliant upon L-type Calcium Channels
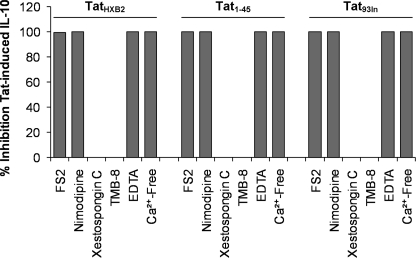
Previous studies have shown that an increase in cytoplasmic Ca2+ concentration ([Ca2+]i) is critical to Tat-induced IL-10 production in monocytes (5, 30). These data, combined with the complete abrogation of TatHXB2-induced IL-10 production by the class I PI3K inhibitors PI-103 and wortmannin, prompted us to examine the role of a [Ca2+]i flux in B Tat-induced IL-10 production. A previous study using 2-aminoethoxydiphenyl borate as a specific inhibitor of inositol 1,4,5-trisphosphate mediated Ca2+ release noted that the source was specifically intracellular from inositol 1,4,5-trisphosphate mediated Ca2+ release (30). However, 2-aminoethoxydiphenyl borate is not specific for this action, and, in our own work (27) and that of Contreras et al. (6) using the specific xestospongin C (36), we observed that inhibition of this store has no effect on Tat-induced monocyte [Ca2+]i flux. We have shown previously that the increase in [Ca2+]i is instead reliant upon L-type calcium channels (CaL) using the highly specific inhibitor for CaL, nimodipine (27). Therefore, we investigated the role of Ca2+ and CaL in Tat-induced IL-10 production from monocytes. Monocytes were pretreated for 1 h with 100 nm FS2, a specific CaL inhibitor (37), and subsequently incubated with B Tat and FS2 for 4 h. Monocytes were then washed and harvested for IL-10 mRNA production or were further incubated with media alone for an additional 20 h, after which supernatants were harvested and analyzed for IL-10 by ELISA. FS2-treated cells exposed to B Tat released significantly lower amounts of IL-10 compared with non-FS2-treated cells (mean 3 pg/ml versus 367 pg/ml; p = 0.005, Fig. 6), suggesting an important role for CaL in Tat-induced IL-10 production in monocytes. Furthermore, FS2 rendered the IL-10 mRNA response to B Tat treatment to levels comparable to untreated cells (data not shown), suggesting a crucial role of CaL in Tat-induced IL-10 production. To confirm these findings, we repeated the experiment using nimodipine and observed the complete abrogation of the IL-10 response (p < 0.001, Fig. 6). We then treated primary monocytes with TatHXB2 in the presence of 2 mm ethylenediaminetetraacetic acid or in calcium-free Dulbecco's modified Eagle medium for 4 h followed by a 2:1 ratio of AIM-V/Iscove's modified Dulbecco's medium for a further 20 h. The complete abrogation of the IL-10 response (Fig. 6) was observed in each case. These findings suggest that an extracellularly sourced [Ca2+]i flux through CaL is essential for Tat-induced IL-10 production. Finally, we investigated the involvement of intracellular stores of calcium using xestospongin C and TMB-8, inhibitors of intracellular calcium stores stimulated by inositol-1,4,5-triphosphate (36) and ryanodine, respectively. Neither had an inhibitory effect on TatHXB2 induced IL-10 production (Fig. 6). We also performed these experiments using both Tat1–45 and Tat93In and observed similar results (Fig. 6).
FIGURE 6.
Extracellular Ca2+ is required for TatHXB2-induced IL-10 production from monocytes. Monocytes were pretreated with 10 nm FS2, 1 μm nimodipine (both CaL inhibitors), 1 μm xestospongin C, 1 μm TMB-8 (inhibitors of intracellular calcium stores stimulated by inositol 1,4,5-triphosphate and ryanodine, respectively), 2 mm ethylenediaminetetraacetic acid (EDTA) or vehicle control or placed in Ca2+-free media for 1 h prior to incubation with 10 nm TatHXB2, 2 μm Tat1–45, or 10 nm Tat93In for 4 h, after which cells were washed and incubated in fresh complete media for a further 20 h, and supernatants were collected and analyzed for IL-10. The CaL inhibitors, the Ca2+ chelator EDTA, and incubation in Ca2+-free media all abrogated the IL-10 response elicited by TatHXB2, Tat93In, and Tat1–45, whereas the inhibitors of Ca2+ from inositol 1,4,5-triphosphate-regulated stores or caffeine-sensitive ryanodine receptor-regulated intracellular stores had no effect. The bars represent means ± S.E. from three independent experiments carried out in triplicate.
DISCUSSION
IL-10 plays an important role in HIV-1 infection and replication (38–42), with elevated IL-10 levels associated with immunosuppression and progression to AIDS (14–16). Tat has been shown to induce IL-10 production in monocytes through a calcium dependent mechanism (9). Most research on HIV-1 Tat use a peptide corresponding to the first exon (72 residues) or a truncated 86-residue form of the clade B TatHXB2 (24, 43). However, the majority of Tat proteins sequenced from clinical isolates possess the longer 101-residue form of Tat, which has an extra 14 residues located at its C terminus (24). In a previous study, we have shown that these 14 residues contribute to a greater trans-activational ability combined with a reduced ability to induce apoptosis of CD4+ T cells (22). HIV-1 clade C accounts for >50% of infections worldwide (44) and is unique among group M viruses, in that they have lower replicative fitness in PBMC, which may be associated with slower disease progression (45); thus, understanding this clade is important in understanding HIV-1 pathogenesis. 90% of clade C viruses possess a C31S substitution in the cysteine-rich region of Tat (21), disrupting the 30C-C-motif. We have shown previously that the C31S mutation found in clade C Tat renders it unable to bind chemokine (C-C motif) receptor 2b (CCR2b) and fails to induce a transient calcium flux and activate signaling modules that regulate monocyte chemotaxis (27). Moreover, clade C Tat is unable to up-regulate CXCR4 on the surface of CD4+ T cells through a CCR2b-dependent mechanism (46), while having no impact upon its ability to trans-activate at low concentrations (27, 29). However, in this trans-activation model, Tat is added as an exogenous protein that must traverse the cell membrane before it can bind the trans-activation responsive element and trans-activate HIV-1 long terminal repeat gene expression. A study using endogenously produced Tat showed that clade C Tat displayed higher affinities for both the trans-activation responsive element and for the positive transcription elongation factor b complex than clade B Tat and displayed a greater trans-activation potential (23). A recent study showed that the serine residues in Tat are phosphorylated by a cyclin-dependent kinase-2 mechanism and that this phosphorylation is important for HIV-1 transcription and the activation of integrated HIV-1 provirus (47). It is possible that the C31S mutation found in clade C Tat gives it a potential new phosphorylation site, augmenting its trans-activational activity. Moreover, the C31S mutation may have other functions that have not yet been identified.
In this study, we explored the ability of clade C and clade B Tat to up-regulate IL-10 from monocytes. We confirmed and expanded previously published findings that B Tat up-regulates IL-10 in monocytes in a dose- and time-dependent manner (4, 5, 30). Moreover, we also show that a clade C Tat protein lacking Cys31 induces significantly less IL-10 secretion than one possessing Cys31. The aforementioned studies on IL-10 and B Tat also demonstrated that, without traversing the cell membrane, B Tat acts through a surface receptor/channel on human monocytes to induce a [Ca2+]i flux that regulates IL-10 transcription through calmodulin and calmodulin-dependent protein kinase-II-activated p38 MAPK that activates the downstream cAMP-responsive element binding protein 1 and Sp-1 transcription factors (5, 30). In this study, we demonstrated that the Tat-induced [Ca2+]i flux essential for IL-10 production is extracellular in origin and is dependent upon CaL channels (27). Different regions of Tat have been implicated in the interactions with surface proteins: the first nine residues in the N terminus of Tat with CD26, the 78RGD motif with adhesion molecule receptors, mainly integrins such as α5β1 and αvβ3, the basic region with membrane lipids and vascular endothelial factor receptors and the 30C-C motif with CCR2b (48). Interestingly, the N terminus and basic regions of Tat93In and Tat96Bw are identical and both possess a R78Q mutation that disrupts the 78RGD motif. However, these two Tat proteins do differ in that Tat96Bw possesses an intact 30C-C motif, whereas Tat93In has a C31S mutation (Fig. 1).
A previous study reported no calcium involvement in Tat-induced IL-10 transcription (4). This study monitored the intracellular calcium flux for 30 min using microspectrofluorimetry with images captured every 5 s, while our study observed the intracellular calcium flux over 3 min using real time flow cytometry. Using this method, we detected a rapid and transient calcium flux following B Tat treatment that is consistent with other studies using similar methods (5, 9, 30, 48) that was possibly missed using a time-lapse method. Our results also show that CaL and PI3K inhibitors reduce the ability of clade B Tat to induce IL-10 production in monocytes. Together, these suggest that extracellularly sourced calcium is required for TatHXB2 to induce IL-10 production from monocytes. However, Tat93In, although inducing significantly less IL-10 transcription than B TatHXB2, was still able to significantly up-regulate IL-10 despite its inability to induce a measurable [Ca2+]i flux. This may be explained by the ability of exogenous, soluble Tat to penetrate cells and activate the transcription factor nuclear factor κ-light chain enhancer of activated B cells either directly or through upstream effector kinases leading to the production of many cytokines, including IL-10 (5, 49–52), suggesting an alternative pathway by which Tat may up-regulate IL-10 production. Alternatively, it is possible that Tat93In induces a [Ca2+]i flux independently of the 30C-C motif, but this flux is too small, too rapid, or occurs much later than 200 s to be detected by our methods, as demonstrated by the inhibition of Tat93In-induced IL-10 production from monocytes by the highly specific nimodipine and also by stimulation in Ca2+-free media.
We examined the role of p38α/p38β MAPK and class I PI3K in Tat-induced IL-10 production from monocytes using small, cell-permeable chemical inhibitors according to current recommendations (35). Our results suggest that both p38α/p38β and the calcium-dependent class I PI3K are crucial to Tat-induction of IL-10 in monocytes. Small, cell-permeable chemical inhibitors have the serious limitation in that many possess an inherent lack of specificity. However, following the guidelines specified in Bain et al. (35), we addressed the role of specific kinases in Tat-induced IL-10 production by using inhibitors that are selective for specific kinases and in parallel with another inhibitor that interacts with the same target kinase in a different manner with little overlap in nontarget inhibition. Despite their lack of specificity, if used correctly, these small cell-permeable chemical inhibitors do have advantages over small interfering RNA. The first is that the inhibitory effect on the cell occurs within minutes. In contrast, small interfering RNA has the disadvantage that the target has been “knocked down” for 18 h or more before Tat treatment. Therefore, the effects that we may observe are oftentimes indirect and result from long term alterations in gene expression. Secondly, small interfering RNA are recognized by the Toll-like receptor 8 and retinoic acid-inducible protein 1 that are present in monocytes (53). This recognition triggers downstream signaling via Toll-interleukin-1 receptor domains activating the interferon regulatory factor, nuclear factor κ-light chain enhancer of activated B cells and MAPK pathways leading to the expression of interferon, proinflammatory cytokines, as well as IL-10 (54).
The concentrations of Tat used in our experiments are comparable to those found in the plasma of HIV-1-infected patients (1, 2, 55, 56), suggesting that effective Tat concentrations might be reached in vivo. Moreover, higher concentrations than those present in the plasma may be found in lymphoid tissue, where productively infected cells are most frequent and where Tat may act locally after secretion from HIV-1-infected cells (2).
In summary, we observed a differential effect of HIV-1 clade B and clade C Tat on the expression of the anti-inflammatory cytokine IL-10 in monocytes. B Tat induces IL-10 production from monocytes through a calcium-dependent mechanism that involves the 30C-C motif. The C31S mutation found in the majority of clade C variants abrogates its ability to induce a measurable [Ca2+]i flux and is responsible for the marked decrease in IL-10 production. Furthermore, we also show that p38α/p38β and class I PI3K are crucially involved in clade B Tat-induced IL-10 production. These data demonstrate the importance of studying the regulation of cytokines and chemokines by different HIV-1 clades and the impact that these differences may have on HIV-1 pathogenesis and disease progression.
Acknowledgments
We thank Erwann P. Loret, Jennifer D. Watkins, and Daniel Lafitte (INSERM U911, Université de la Méditerranée, Marseille, France) for the synthesis and purification of the HIV-1 Tat proteins. We also thank Carol Mundy (Pediatrics, University of California, San Diego (UCSD)) for assistance in obtaining peripheral blood mononuclear cells and Dennis Young (Flow Cytometry Core Facility, UCSD) for assistance in fluorescence-activated cell sorter analysis.
This work was supported by NIAID/National Institutes of Health Grant AI068632 to the IMPAACT Network.
- HIV-1
- human immunodeficiency virus, type 1
- CaL
- L-type calcium channel
- CCR2b
- chemokine (C-C motif) receptor 2b
- CCL2
- chemokine (C-C motif) ligand 2
- IL-10
- interleukin-10
- MAPK
- mitogen-activated protein kinase
- MKK1
- MAPK kinase-1
- TMB-8
- 3,4,5-trimethyloxybenzoic acid 8-(diethylamino)octyl ester
- ELISA
- enzyme-linked immunosorbent assay
- PBMC
- peripheral blood mononuclear cells
- PI3K
- phosphoinositide 3-kinase.
REFERENCES
- 1.Westendorp M. O., Frank R., Ochsenbauer C., Stricker K., Dhein J., Walczak H., Debatin K. M., Krammer P. H. (1995) Nature 375, 497–500 [DOI] [PubMed] [Google Scholar]
- 2.Xiao H., Neuveut C., Tiffany H. L., Benkirane M., Rich E. A., Murphy P. M., Jeang K. T. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 11466–11471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell G. R., Loret E. P. (2009) Retrovirology 6, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Badou A., Bennasser Y., Moreau M., Leclerc C., Benkirane M., Bahraoui E. (2000) J. Virol. 74, 10551–10562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bennasser Y., Bahraoui E. (2002) FASEB J. 16, 546–554 [DOI] [PubMed] [Google Scholar]
- 6.Contreras X., Bennasser Y., Chazal N., Moreau M., Leclerc C., Tkaczuk J., Bahraoui E. (2005) Virology 332, 316–328 [DOI] [PubMed] [Google Scholar]
- 7.Park I. W., Wang J. F., Groopman J. E. (2001) Blood 97, 352–358 [DOI] [PubMed] [Google Scholar]
- 8.Rautonen J., Rautonen N., Martin N. L., Wara D. W. (1994) AIDS Res. Hum. Retroviruses 10, 781–785 [DOI] [PubMed] [Google Scholar]
- 9.Bennasser Y., Yamina B., Contreras X., Xavier C., Moreau M., Marc M., Le, Clerc C., Catherine L., Badou A., Abdallah B., Bahraoui E. (2001) J. Soc. Biol. 195, 319–326 [PubMed] [Google Scholar]
- 10.Akridge R. E., Oyafuso L. K., Reed S. G. (1994) J. Immunol. 153, 5782–5789 [PubMed] [Google Scholar]
- 11.Fauci A. S. (1996) Nature 384, 529–534 [DOI] [PubMed] [Google Scholar]
- 12.Kedzierska K., Crowe S. M. (2001) Antivir. Chem. Chemother. 12, 133–150 [DOI] [PubMed] [Google Scholar]
- 13.Moore K. W., de Waal Malefyt R., Coffman R. L., O'Garra A. (2001) Annu. Rev. Immunol. 19, 683–765 [DOI] [PubMed] [Google Scholar]
- 14.Stylianou E., Aukrust P., Kvale D., Müller F., Frøland S. S. (1999) Clin. Exp. Immunol. 116, 115–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ameglio F., Cordiali Fei P., Solmone M., Bonifati C., Prignano G., Giglio A., Caprilli F., Gentili G., Capobianchi M. R. (1994) J. Biol. Regul. Homeost. Agents 8, 48–52 [PubMed] [Google Scholar]
- 16.Clerici M., Wynn T. A., Berzofsky J. A., Blatt S. P., Hendrix C. W., Sher A., Coffman R. L., Shearer G. M. (1994) J. Clin. Invest. 93, 768–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orsilles M. A., Pieri E., Cooke P., Caula C. (2006) APMIS 114, 55–60 [DOI] [PubMed] [Google Scholar]
- 18.Bhoopat L., Rithaporn T. S., Khunamornpong S., Bhoopat T., Taylor C. R., Thorner P. S. (2006) Mod. Pathol. 19, 255–263 [DOI] [PubMed] [Google Scholar]
- 19.Kaleebu P., Ross A., Morgan D., Yirrell D., Oram J., Rutebemberwa A., Lyagoba F., Hamilton L., Biryahwaho B., Whitworth J. (2001) AIDS 15, 293–299 [DOI] [PubMed] [Google Scholar]
- 20.Vasan A., Renjifo B., Hertzmark E., Chaplin B., Msamanga G., Essex M., Fawzi W., Hunter D. (2006) Clin. Infect. Dis. 42, 843–852 [DOI] [PubMed] [Google Scholar]
- 21.Ranga U., Shankarappa R., Siddappa N. B., Ramakrishna L., Nagendran R., Mahalingam M., Mahadevan A., Jayasuryan N., Satishchandra P., Shankar S. K., Prasad V. R. (2004) J. Virol. 78, 2586–2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Campbell G. R., Watkins J. D., Esquieu D., Pasquier E., Loret E. P., Spector S. A. (2005) J. Biol. Chem. 280, 38376–38382 [DOI] [PubMed] [Google Scholar]
- 23.Desfosses Y., Solis M., Sun Q., Grandvaux N., Van Lint C., Burny A., Gatignol A., Wainberg M. A., Lin R., Hiscott J. (2005) J. Virol. 79, 9180–9191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeang K. T., Xiao H., Rich E. A. (1999) J. Biol. Chem. 274, 28837–28840 [DOI] [PubMed] [Google Scholar]
- 25.Bennett W. E., Cohn Z. A. (1966) J. Exp. Med. 123, 145–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson W. D., Jr., Mei B., Cohn Z. A. (1977) J. Exp. Med. 146, 1613–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Campbell G. R., Watkins J. D., Singh K. K., Loret E. P., Spector S. A. (2007) J. Virol. 81, 5919–5928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lole K. S., Bollinger R. C., Paranjape R. S., Gadkari D., Kulkarni S. S., Novak N. G., Ingersoll R., Sheppard H. W., Ray S. C. (1999) J. Virol. 73, 152–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Opi S., Péloponèse J. M., Jr., Esquieu D., Watkins J., Campbell G., De, Mareuil J., Jeang K. T., Yirrell D. L., Kaleebu P., Loret E. P. (2004) Vaccine 22, 3105–3111 [DOI] [PubMed] [Google Scholar]
- 30.Gee K., Angel J. B., Mishra S., Blahoianu M. A., Kumar A. (2007) J. Immunol. 178, 798–807 [DOI] [PubMed] [Google Scholar]
- 31.Novitsky V. A., Montano M. A., McLane M. F., Renjifo B., Vannberg F., Foley B. T., Ndung'u T. P., Rahman M., Makhema M. J., Marlink R., Essex M. (1999) J. Virol. 73, 4427–4432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J. C., Lee D. C., Cheung B. K., Lau A. S. (2005) FEBS Lett. 579, 3055–3062 [DOI] [PubMed] [Google Scholar]
- 33.Li J. C., Lau A. S. (2007) Immunology 121, 337–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davies S. P., Reddy H., Caivano M., Cohen P. (2000) Biochem. J. 351, 95–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bain J., Plater L., Elliott M., Shpiro N., Hastie C. J., McLauchlan H., Klevernic I., Arthur J. S., Alessi D. R., Cohen P. (2007) Biochem. J. 408, 297–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ta T. A., Feng W., Molinski T. F., Pessah I. N. (2006) Mol. Pharmacol. 69, 532–538 [DOI] [PubMed] [Google Scholar]
- 37.Albrand J. P., Blackledge M. J., Pascaud F., Hollecker M., Marion D. (1995) Biochemistry 34, 5923–5937 [DOI] [PubMed] [Google Scholar]
- 38.Ancuta P., Bakri Y., Chomont N., Hocini H., Gabuzda D., Haeffner-Cavaillon N. (2001) J. Immunol. 166, 4244–4253 [DOI] [PubMed] [Google Scholar]
- 39.Kootstra N. A., van, 't Wout A., Huisman H. G., Miedema F., Schuitemaker H. (1994) J. Virol. 68, 6967–6975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Masood R., Lunardi-Iskandar Y., Moudgil T., Zhang Y., Law R. E., Huang C. L., Puri R. K., Levine A. M., Gill P. S. (1994) Biochem. Biophys. Res. Commun. 202, 374–383 [DOI] [PubMed] [Google Scholar]
- 41.Naif H. M., Chang J., Ho-Shon M., Li S., Cunningham A. L. (1996) AIDS Res. Hum. Retroviruses 12, 1237–1245 [DOI] [PubMed] [Google Scholar]
- 42.Wang J., Roderiquez G., Oravecz T., Norcross M. A. (1998) J. Virol. 72, 7642–7647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ratner L., Haseltine W., Patarca R., Livak K. J., Starcich B., Josephs S. F., Doran E. R., Rafalski J. A., Whitehorn E. A., Baumeister K., Ivanoff L., Petteway S. R., Pearson M. L., Lautenberger J. A., Papas T. S., Ghrayeb J., Chang N. T., Gallo R. C., Wong-Staal F. (1985) Nature 313, 277–284 [DOI] [PubMed] [Google Scholar]
- 44.Hemelaar J., Gouws E., Ghys P. D., Osmanov S. (2006) AIDS 20, W13–23 [DOI] [PubMed] [Google Scholar]
- 45.Tebit D. M., Nankya I., Arts E. J., Gao Y. (2007) AIDS Rev. 9, 75–87 [PubMed] [Google Scholar]
- 46.Campbell G. R., Loret E. P., Spector S. A. (2010) J. Biol. Chem. 285, 1681–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ammosova T., Berro R., Jerebtsova M., Jackson A., Charles S., Klase Z., Southerland W., Gordeuk V. R., Kashanchi F., Nekhai S. (2006) Retrovirology 3, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Albini A., Benelli R., Giunciuglio D., Cai T., Mariani G., Ferrini S., Noonan D. M. (1998) J. Biol. Chem. 273, 15895–15900 [DOI] [PubMed] [Google Scholar]
- 49.Conant K., Ma M., Nath A., Major E. O. (1996) J. Virol. 70, 1384–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cota-Gomez A., Flores N. C., Cruz C., Casullo A., Aw T. Y., Ichikawa H., Schaack J., Scheinman R., Flores S. C. (2002) J. Biol. Chem. 277, 14390–14399 [DOI] [PubMed] [Google Scholar]
- 51.Leghmari K., Bennasser Y., Bahraoui E. (2008) Eur. J. Cell Biol. 87, 947–962 [DOI] [PubMed] [Google Scholar]
- 52.Ott M., Lovett J. L., Mueller L., Verdin E. (1998) J. Immunol. 160, 2872–2880 [PubMed] [Google Scholar]
- 53.Goodchild A., Nopper N., King A., Doan T., Tanudji M., Arndt G. M., Poidinger M., Rivory L. P., Passioura T. (2009) BMC Immunol. 10, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saruta M., Michelsen K. S., Thomas L. S., Yu Q. T., Landers C. J., Targan S. R. (2009) Eur. J. Immunol. 39, 2195–2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chang H. C., Samaniego F., Nair B. C., Buonaguro L., Ensoli B. (1997) AIDS 11, 1421–1431 [DOI] [PubMed] [Google Scholar]
- 56.Ensoli B., Barillari G., Salahuddin S. Z., Gallo R. C., Wong-Staal F. (1990) Nature 345, 84–86 [DOI] [PubMed] [Google Scholar]