Abstract
MDM2 plays a major role in cancer development and progression via both p53-dependent and -independent functions. One of its p53-independent functions is the induction of the ubiquitin-independent proteasomal degradation of p21Waf1. The present study was designed to characterize the mechanism(s) by which MDM2 induces p21Waf1 degradation. We first determined the regions of MDM2 required for p21Waf1 degradation using pulldown assays and Western blotting and then examined the mechanisms using limited proteolysis and fluorescence resonance energy transfer assays. We found that the MDM2-p21Waf1 interaction depended on the central domain of MDM2 and that nuclear localization of both proteins was necessary for p21Waf1 degradation. Specifically, amino acids 226–250 of MDM2 were required for p21Waf1 binding and degradation, and amino acids 251–260 were necessary for p21Waf1 degradation. The latter region induced a conformation change in p21Waf1, increasing its interaction with the C8 subunit of the proteasome, leading to its degradation. When MDM2 lacked either segment (aa 226–250 or aa 251–260), its capacity to promote p21Waf1 degradation and cell cycle progression was significantly reduced. In summary, the present study elucidated a previously unknown mechanism by which MDM2 promotes the degradation of an intact protein (p21Waf1) through an ubiquitin-independent proteasomal degradation pathway. Because MDM2 also increases the degradation of other proteins in a ubiquitin-independent manner, this mechanism may underlie part of its tumorigenic properties.
Keywords: Cell/Checkpoint, Cell/Apoptosis, Cell/Cycle, Oncogene, Protein/Protein-Protein Interactions, Protein/Degradation, Protein/Domains, Protein/Stability, Cell Cycle
Introduction
The MDM2 (mouse double minute 2) oncogene is overexpressed in many human cancers, and overexpression of MDM2 correlates with a poor prognosis in cancer patients (reviewed in Refs. 1 and 2). The tumorigenicity of MDM2 has mainly been attributed to the autoregulatory loop it forms with tumor suppressor p53 (3–6). However, there also is increasing evidence supporting that the tumorigenicity of MDM2 is partially dependent upon its p53-independent activities. For instance, clinical studies have demonstrated overexpression of the MDM2 protein and amplification of the MDM2 gene in human cancers, regardless of p53 status (2, 7). Additionally, although p53 transactivates MDM2 transcription as part of the autoregulatory loop (8, 9), the expression of MDM2 can also be increased by the growth factor-coupled Ras-Raf-mitogen-activated protein kinase (MAPK) and phosphatidylinositol 3-kinase-mTOR signaling pathways, independent of p53 (10, 11). Further demonstrating the importance of the p53-independent activities of MDM2, >40 other proteins have been found to interact with MDM2 (reviewed in Ref. 12). Moreover, through these other proteins, MDM2 can exert p53-independent effects on numerous processes, including cell cycle progression, cell differentiation, apoptosis, signal transduction, migration, and angiogenesis (12). These activities can all be exploited by cancer cells and may underlie the aggressive phenotypes of human cancers with MDM2 overexpression.
Among the many MDM2 interactive proteins is p21Waf1 (13, 14). The interaction between MDM2 and p21Waf1 results in the ubiquitin-independent proteasomal degradation of p21Waf1 (13, 15), leading to unchecked cell cycle progression and proliferation. However, the underlying mechanism(s) by which MDM2 promotes p21Waf1 degradation has not been fully established. As an E3 ubiquitin ligase, MDM2 catalyzes the ubiquitination of several proteins, including p53 (16, 17). However, the increase in p21Waf1 degradation induced by MDM2 does not depend upon its ubiquitination (13, 14). Instead, it has been shown that the central part of MDM2 (spanning aa5 180–299) is essential for the ubiquitin-independent proteasomal degradation of p21Waf1 (14). Although 14-3-3-τ recently has been suggested as a cofactor for promoting MDM2-p21Waf1-C8 binding and MDM2-mediated p21Waf1 degradation (18), the mechanism by which MDM2 can induce the ubiquitin-independent degradation of the intact p21Waf1 protein remains unclear.
We herein report that a small region of the acidic domain (AD) of MDM2 induces a conformational change in the p21Waf1 protein upon binding. This effect is essential for the MDM2-mediated increase in p21Waf1 recognition by the proteasome and subsequent p21Waf1 degradation. Our study provides new insights into the degradation and regulation of p21Waf1 and also reveals a new mechanism that may be important in other p53-independent functions of MDM2.
EXPERIMENTAL PROCEDURES
Plasmids
The pcDNA3-FLAG-MDM2 vectors for expression of human MDM2 were kindly provided by Dr. Z. Ronai (Burnham Institute). Additional constructs expressing deletions of the MDM2 protein were generated by proofreading PCR. GST-C8 was a gift from Dr. M. J. Allday (Ludwig Institute for Cancer Research). GST-p21Wafl, GST-MDM2, and pcDNA3-p21Waf1-HA were described previously (14). The positive control for FRET, EYFP-ECFP, was a kind gift from Dr. B.K. Berdiev (University of Alabama at Birmingham) and was generated by fusing ECFP to the EYFP-C1 vector using XhoI and BamHI restriction sites through a seven-amino acid linker (SGLRSRA). The human p21Wafl cDNA insert was subcloned into the BspEI and XhoI sites of EYFP-ECFP after deletion of the seven-amino acid linker. The pEGFP vectors for expression of human MDM2 and additional constructs expressing deletions of the MDM2 protein were generated by proofreading PCR.
Cell Lines
PC3 cells were maintained as described previously (14). HCT116 (p53−/−) cells were kindly provided by Dr. B. Vogelstein (The Johns Hopkins University) and were maintained in McCoy's 5A medium. To establish p21Waf1 stable knockdown HCT116 (p53−/−) cell lines, HCT116 (p53−/−) cells were transfected with pBabe-U6-p21Waf1. Positive cell clones were selected, maintained with puromycin (1.2 μg/ml), and confirmed by immunoblotting. COS-7 cells were cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum and 1% penicillin/streptomycin.
Immunofluorescence
After fixation in 3% formaldehyde, cells were permeabilized with 0.5% Triton X-100 followed by blocking in 3% bovine serum albumin for 15 min. The cells were then sequentially incubated with mouse FLAG antibody (F-3165, Sigma), goat anti-mouse IgG Alexa Fluor 488 (Molecular Probes), rabbit p21Waf1 (H-164, sc-756, Santa Cruz Biotechnology), and goat anti-rabbit IgG Alexa Fluor 594 (Molecular Probes) antibodies. After each round of incubation with antibody, cells were washed in phosphate-buffered saline four times for 5 min each. Finally, the coverslips were washed, counterstained with Hoechst 33342 for 4 min and mounted in 0.2% n-propyl gallate. Immunofluorescence images were captured by confocal microscopy.
Generation of GST Fusion Proteins and Pulldown Assays
The pGEX-2T-C8, pGEX-2T-MDM2, and pGEX-2T-p21Waf1 plasmids were transformed into Escherichia coli BL-21 (DE3) (Promega) and induced with 0.1 mm isopropyl-1-thio-β-d-galactopyranoside (Sigma). Bacterial pellets were lysed in lysis buffer (20 mm Tris, pH 8.0, 100 mm NaCl, 1 mm EDTA, 0.5% Nonidet P-40, and 5% (v/v) protease inhibitor mixture (Sigma)), followed by sonication (12 × 10 s). The fusion and GST proteins were purified using glutathione-agarose beads. Direct physical binding in vitro was assessed using GST fusion proteins incubated with proteins transcribed and translated in vitro in binding buffer (50 mm Tris, pH 7.5, 137 mm NaCl, 0.5% Nonidet P-40, 10% glycerol, 5% (v/v) protease inhibitors) at 4 °C for 5 h. The bound proteins were captured by glutathione-agarose beads, which were washed three times with binding buffer, eluted with 5× SDS sample buffer at 100 °C for 10 min, resolved on SDS-PAGE, and detected with the appropriate antibody.
FRET Imaging
The acceptor photobleaching approach was performed as described previously (19, 20). Briefly, images were acquired using a Leica confocal scanning system (Exton, PA) TCS SP2 outfitted with a Leica DMRXE upright microscope, a blue argon laser, and a 63 × 1.4 numerical aperture plan apochromatic oil immersion objective. ECFP (donor) and EYFP (acceptor) fluorescence were excited with 458 and 514 nm laser light, respectively. Dual excitation was achieved by a dual dichroic (DD458 nm/514 nm) mirror in the excitation path. The emissions spectra for ECFP and EYFP were collected using 465–500 and 525–600 nm bandpass windows, respectively. Regions of interest were created on a cell image expressing both fluorophores, and the EYFP fluorescence channel was photobleached using 514 nm laser light to 35% of the original intensity. ECFP and EYFP images were taken both before and after acceptor photobleaching. FRET efficiency (E) was calculated using Leica software: FRETEff = (Dpost − Dpre)/Dpost, for all Dpost > Dpre, where Dpre and Dpost are the ECFP emission spectra before and after regional photobleaching (21).
Cell Cycle Distribution Assay
Cells were trypsinized, washed with phosphate-buffered saline, and fixed in 1.5 ml 95% ethanol at 4 °C overnight and then incubated with RNase and stained by propidium iodide. DNA contents were determined by flow cytometry.
Statistical Analysis
Student's t test was used to analyze the significance of the differences in cell cycle phase between the GFP control, wild-type MDM2, and the different MDM2 mutant HCT116 p53−/− and p53/p21Waf1 double knockdown cells.
RESULTS
The Central Part of MDM2 Is Essential for Its Effects on p21Waf1 Protein Degradation
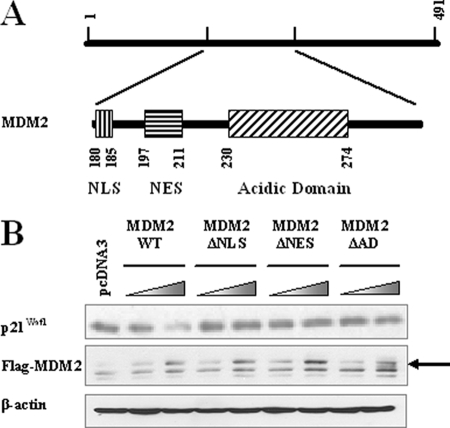
We have demonstrated previously that the central part of MDM2 (aa 180–299) is important for MDM2-mediated p21Waf1 degradation (14). There are several functional domains in the central region of MDM2, including the nuclear localization signal (NLS, spanning aa 180–185), nuclear exportation signal (NES, spanning aa 197–211) and the acidic domain (AD, spanning aa 230–274) (Fig. 1A). As the first step in elucidating the mechanism by which MDM2 induces the proteasomal degradation of p21Waf1, we investigated the importance of these three motifs. As shown in Fig. 1B, mutants of MDM2 lacking the NLS, NES, or AD were incapable of promoting p21Waf1 degradation, suggesting that all three of these motifs are involved in the MDM2-mediated degradation of p21Waf1.
FIGURE 1.
The NLS, NES, and AD of MDM2 are essential for promoting p21Waf1 degradation. A, the structure of the central domain of the MDM2 protein. B, MDM2 wild-type (WT) and MDM2 mutant proteins without the NLS, NES, or AD were overexpressed in PC3 cells. After 24 h, cell lysates were collected and examined for levels of p21Waf1 expression.
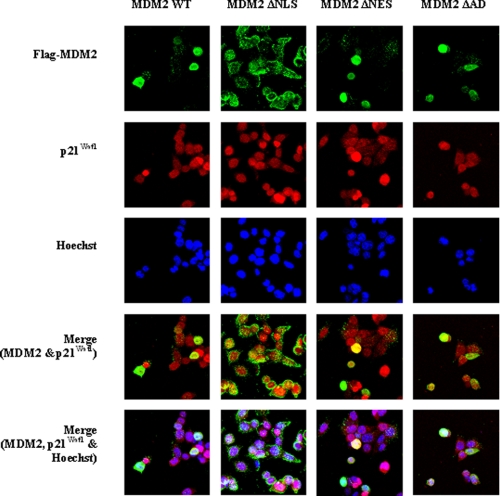
MDM2 and p21Waf1 Co-localize in the Nucleus
The involvement of the NLS and NES indicates that the subcellular localization of the proteins is important for p21Waf1 degradation. HCT 116 (p53−/−) human colon carcinoma cells were used to evaluate the localization of the MDM2 and p21Waf1 proteins. As shown in Fig. 2, the proteins co-localize within the nucleus. To further examine the effects of MDM2 on p21Waf1 localization, HCT116 (p53−/−) cells were transfected with mutants of MDM2 without the NLS (ΔNLS), NES (ΔNES), or AD (ΔAD). As expected, when the MDM2 mutant without the NLS was overexpressed, it localized in the cytoplasm of the HCT116 (p53−/−) cells. In contrast, MDM2 mutants with mutations to the NES or AD still co-localized with p21Waf1 in the nucleus (Fig. 2). The fact that deletion of the NLS abrogated p21Waf1 degradation suggests that the nuclear localization of MDM2 is required for degradation of p21Waf1. In addition, it also appears that disruption of the dynamics of the shuttling of MDM2 between the cytoplasm and nucleus leads to interference with the degradation of p21Waf1 (as indicated by the lack of degradation when the NES mutant of MDM2 was examined).
FIGURE 2.
MDM2 and p21Waf1 co-localize in the nucleus, and mutations to the NLS, NES, and AD of MDM2 prevent its degradation of p21Waf1. HCT116 (p53−/−) cells were grown on coverslips for 24 h and transfected with either wild-type MDM2 or mutants without the NLS, NES, or AD. The subcellular localization of the wild-type (WT) or mutant MDM2 and endogenous p21Waf1 proteins was determined by immunofluorescence.
While it is logical that the nuclear localization/transport of MDM2 (and thus the NLS and NES of the protein) is important for its degradation of p21Waf1, it is not clear why the AD is required for efficient degradation of p21Waf1. Even MDM2 with mutations to the AD co-localized with p21Waf1 within the nucleus, indicating that while this region is important for p21Waf1 degradation, it is not necessary for nuclear localization (Fig. 2).
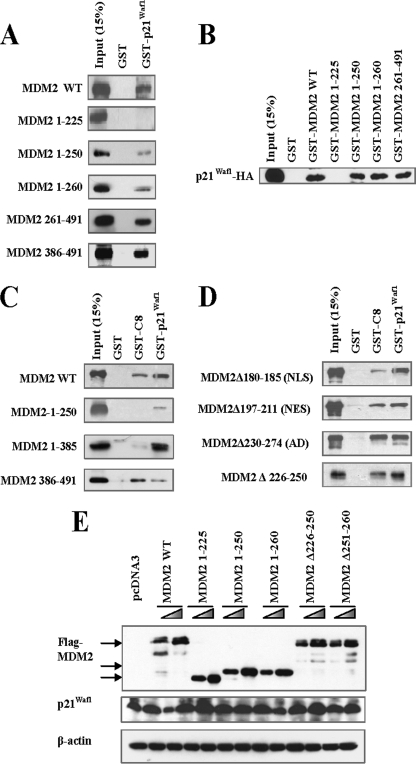
The AD Is Dispensable for MDM2 Binding to p21Waf1, but Amino Acids 226–250 Are Important for the MDM2-p21Waf1 Interaction and p21Waf1 Degradation
To address the role of the AD in MDM2-mediated p21Waf1 degradation, we examined whether it was required for MDM2- p21Waf1 binding. We constructed a series of MDM2 deletions and performed pulldown assays to determine their extent of p21Waf1 binding (Fig. 3, A–C). We observed that two segments spanning amino acids 226–250 and 386–491 were involved in the binding of MDM2 to p21Waf1, whereas MDM2 mutants without the NLS and NES were still capable of binding (Fig. 3D). Because amino acids 226–250 contain more than half of the AD, we hypothesized that MDM2 AD mutants are unable to facilitate the degradation of p21Waf1 because they are unable to bind to the protein. However, we observed that mutant MDM2 without the acidic domain was still capable of binding to p21Waf1, suggesting that the acidic domain is not required for either localization or binding of MDM2. Nevertheless, it is still necessary for p21Waf1 degradation. Further investigation into the AD indicated that a smaller segment spanning amino acids 251–260 is the region that is crucial for p21Waf1 degradation (Fig. 3E).
FIGURE 3.
The N-terminal of MDM2 (aa 1–260) is sufficient for promoting p21Waf1 degradation. A, GST-p21Waf1 or GST was incubated with MDM2 wild-type (WT) or deletion proteins in vitro. The binding between GST-p21Waf1 and the MDM2 proteins was determined by pulldown assay. B, GST and GST-tagged MDM2 or MDM2 deletions were incubated with HA-p21Waf1 in vitro. The binding between GST-MDM2 and p21Waf1 proteins was determined by pulldown assay. C and D, GST-p21Waf1, GST-C8, or GST were incubated with MDM2 or MDM2 deletions in vitro. The binding between GST-p21Waf1 or GST-C8 and the MDM2 deletions was determined by pulldown assay. E, MDM2 deletion proteins were overexpressed in HCT116 (p53−/−) cells. After 24 h, cell lysates were collected for examination of p21Waf1 expression.
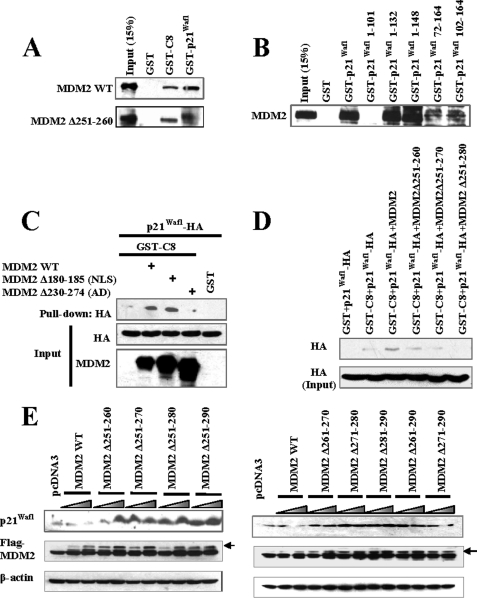
Amino Acids 251–260 in MDM2 Facilitate Binding of p21Waf1 to the C8 Subunit of the Proteasome
Proteasome binding via the C8 subunit is important for the degradation of p21Waf1 (22). Because MDM2 has been demonstrated to bind to the C8 subunit, we examined whether MDM2-C8 binding might be required for p21Waf1 degradation or for the MDM2-p21Waf1 interaction. Our results indicated that the C terminus of MDM2 is involved in its interaction with C8 (Fig. 3C). However, the acidic domain and the segment from amino acids 251–260, which we demonstrated is important for p21Waf1 degradation (but not binding or localization), is not involved in the MDM2-C8 interaction (Fig. 4A). The MDM2 binding site on p21Waf1 (amino acids 102–132) does not overlap with its C8 binding site (Fig. 4B), further indicating that the binding of MDM2-C8 is not involved in the MDM2-p21Waf1 interaction.
FIGURE 4.
Amino acids 251–260 of MDM2 promote the binding between p21Waf1 and C8. A, an MDM2 mutant without amino acids 251–260 was incubated with GST-p21Waf1 or GST-C8 in vitro, and the protein interaction was determined by pulldown assay. B, GST, GST-p21Waf1, or GST-p21Waf1 deletions were incubated with MDM2. The binding between GST-p21Waf1 and GST- p21Waf1 deletions and MDM2 was determined by pulldown assay. C and D, GST or GST-C8 protein was incubated with p21Waf1 in the presence of MDM2 or MDM2 mutants in vitro. The binding between GST-C8 and p21Waf1 was determined by pulldown assay. E, MDM2 or MDM2 deletions were overexpressed in PC3 cells. After 24 h, cell lysates were collected to examine the expression of p21Waf1. WT, wild-type.
Our previous study also suggested that the binding of MDM2 to C8 is not necessary for p21Waf1 degradation (14). Therefore, it is unlikely that MDM2 presents p21Waf1 directly to the proteasome by forming a ternary complex with p21Waf1 and C8. Interestingly, although the region of MDM2 spanning amino acids 251–260 is not involved in MDM2-C8 binding, it does appear to facilitate p21Waf1 recognition by C8 (Fig. 4, C and D). Therefore, MDM2 mutants without amino acids 251–260 (but not without other amino acids of the acidic domain of MDM2) lose the ability to promote the C8 binding and subsequent degradation of p21Waf1 (Fig. 4E).
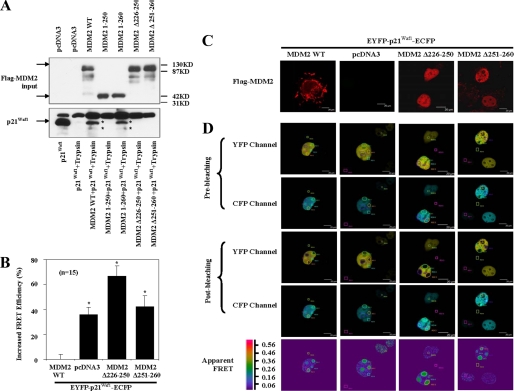
Amino Acids 251–260 in MDM2 Induce a Change in p21Waf1 Protein Conformation That Is Necessary for Its Degradation
p21Waf1 is a loosely folded protein that is directly degraded by the 20S proteasome, independent of ubiquitination (22). NMR studies have revealed that MDM2 is a flexible protein and that its conformation changes upon substrate binding (23, 24). It is therefore possible that the MDM2-p21Waf1 interaction results in a conformational change in p21Waf1, leading to the formation of a motif recognized by the 20S proteasome. A limited proteolysis assay provided evidence to support this hypothesis. As shown in Fig. 5A, the profile of p21Waf1 incubated alone with trypsin was notably different from that when it was incubated in the presence of MDM2. In contrast, MDM2 mutants either without aa 251–260, or aa 226–250, or retaining only aa 1–250 have no effect on the profile of p21Waf1 proteolytic breakdown (Fig. 5A). Additionally, MDM2 mutants with only aa 1–260 had effects similar to full-length MDM2 (Fig. 5A). These observations suggest that the segment in MDM2 spanning aa 251–260 changes the conformation of p21Waf1 when it is bound by MDM2. Further supporting evidence comes from FRET studies in which a plasmid encoding p21Waf1 flanked by YFP and CFP was employed. FRET can be used to indicate when there are changes in the conformation of a target protein. In this case, when there was a conformational change in the p21Waf1 protein, there was an accompanying change in the relative efficiency of the p21Waf1 FRET. Confirming that MDM2 alters p21Waf1 conformation, the presence of overexpressed wild type MDM2, but not mutants of MDM2 either without aa 226–250 or without aa 251–260, significantly decreased the FRET efficiency of EYFP- p21Waf1-ECFP (Fig. 5, B–D). All of these observations suggest that MDM2 induces a conformational change in p21Waf1 that is important for its degradation.
FIGURE 5.
MDM2 (aa 251–260) induces changes in p21Waf1 conformation. A, p21Waf1 and MDM2 or MDM2 deletions were incubated with trypsin in vitro for 35 min at 20 °C. The proteolytic profiles of p21Waf1 were determined by immunoblotting with p21Waf1 antibodies (Ab-1, Calbiochem; H-164, 187, and F-5, Santa Cruz Biotechnology). B, FRET efficiencies from COS7 cells transfected with EYFP-p21Waf1-ECFP or from cells transfected with combinations of MDM2, MDM2Δ226–250, or MDM2Δ251–260 with EYFP-p21Waf1-ECFP were measured in 15 cells from each group and are representative of five distinct experiments. The increases in relative FRET efficiency are shown in the figure (*, p < 0.05). Error bars represent S.D. C and D, COS7 cells were transfected to overexpress proteins as in the above panels. C, the subcellular localization of the transfected wild-type and mutant MDM2 proteins was determined by immunofluorescence. D, FRET was measured as the ratiometric increase in ECFP fluorescence upon photobleaching of EYFP. ECFP and EYFP images were taken both before and after acceptor photobleaching. The areas of photobleaching are labeled as regions of interest (ROI) 1, 2, and 3. The areas of the background are labeled as regions of interest 4, 5, and 6. FRET efficiency was displayed as a pseudocolor representation. Scale bar, 20 μm. WT, wild-type.
Wild-type MDM2, but Not Mutants Lacking Amino Acids 226–250 or 251–260, Promotes Cell Cycle Progression, Independent of p53
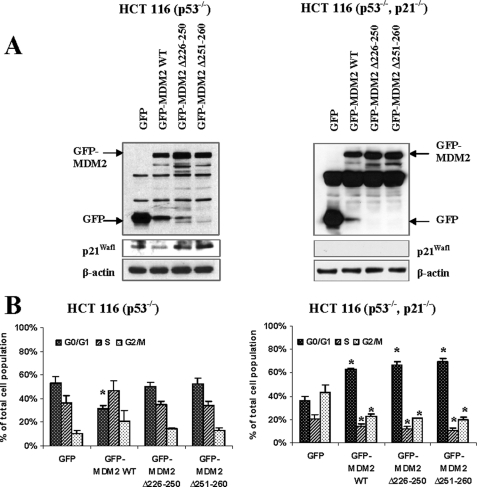
To explore the functional consequences of the MDM2-p21Waf1 interaction, we examined the growth of HCT116 cells with p53 knock-out that were transfected with wild-type MDM2 or mutants (without either amino acids 251–260 or 226–250). Cells were serum-deprived for 48 h to synchronize them, and then they were released for 6 h prior to assessing cell cycle distribution. As shown in Fig. 6A (left panel), p21Waf1 was decreased by overexpression of wild-type MDM2, but not by overexpression of either of the MDM2 mutants. In agreement with this decrease in p21Waf1 expression, overexpression of wild-type MDM2 promoted cell cycle progression, as indicated by the significant decrease in the number of cells in the G0/G1 phase (Fig. 6B, left panel). This significant increase in cell cycle progression was not observed for the mutants.
FIGURE 6.
The region spanning amino acids 251–260 of MDM2 is important for p21Waf1 degradation and cell cycle regulation. HCT116 (p53−/−) cells and HCT116 (p53−/−, p21Waf1−/−) cells overexpressing GFP, GFP-MDM2, or GFP-MDM2 mutants were synchronized by incubation in serum-free medium for 48 h, followed by a 6-h release. The protein expression in these cells was examined by immunoblotting (A), and the GFP-gated cell populations in each phase of the cell cycle were evaluated by cell cycle distribution assay (B). The percentages shown represent an average of three independent experiments. The statistical analyses of the results were performed using the Student's t test, with p values < 0.05 being considered significant. *, p < 0.05. WT, wild-type.
As illustrated in Fig. 6 (right panels), when p21Waf1 was knocked out in these cells (generating p53−/−, p21Waf1−/− double knock-out cells), there was a significant increase in the percentage of cells in the G2/M phase, especially compared with the HCT116 p53−/− cells. This is because when p21Waf1 is absent, the cells are no longer arrested in G1, resulting in more cells that are actively dividing (progressing to G2 after release). When MDM2 was overexpressed in these cells, there was a significant increase in the number of cells in the G0/G1 phase, and significant decreases in the number of cells in the S and G2 phases compared with those transfected with GFP alone (Fig. 6B, right panel). This indicates that MDM2 overexpression leads to further increases in cell cycle progression/cell division, increasing the speed with which the cells pass through the various phases of the cell cycle. Thus, while cells expressing normal levels of MDM2 are proliferating, those with overexpression of MDM2 have a further shortened doubling time.
In the double knock-out cells, overexpression of the MDM2 mutants without amino acids 251–260 or 226–250 had the same effect as the wild-type protein (a significant increase in the percentage of cells in the G1 phase and significant decreases in G2 and S compared with the GFP-transfected cells). It is possible that these mutants are still capable of interacting with the other molecules that MDM2 regulates (12), allowing for the increased division/proliferation that was observed. These results demonstrate that the segment between amino acids 251–260 is important in MDM2-mediated p21Waf1 degradation and confirm that the MDM2-p21Waf1 interaction is involved in regulating cell cycle progression during the G1 phase.
DISCUSSION
Although it is still best known for its regulation of p53, MDM2 is now recognized as having numerous p53-independent activities that may be equally (or more) important as its regulation of p53. MDM2 has been shown to bind to and regulate the stability or activity of >40 proteins (12). One of the major mechanisms by which MDM2 regulates the level of its binding partners is to promote their proteasomal degradation. As an ubiquitin E3 ligase, MDM2 catalyzes the ubiquitination of various molecules, including the insulin-like growth factor 1 receptor (25), G-protein-coupled receptor kinase 2 (26), the β2-adrenergic receptor, and β-arrestin (27). However, the proteasomal degradation of some substrates, including p21Waf1 and Rb, can be promoted by MDM2, independent of their ubiquitination (13, 14, 28). The molecular mechanisms underlying this process remain unclear. In the present study, we have uncovered part of this mechanism and demonstrated that MDM2 promotes the ubiquitin-independent degradation of p21Waf1 by inducing a change in its conformation. This activity may also be applicable to other MDM2 substrates.
Because p21Waf1 is a loosely folded protein (29) and the interaction between p21Waf1 and the proteasomal C8 subunit is sufficient for its degradation (22), our observation that the interaction of p21Waf1 with C8 increases in the presence of MDM2 could explain why MDM2 induces p21Waf1 proteasomal degradation and how p21Waf1 is recognized by the proteasome. We believe that the MDM2-p21Waf1 interaction may induce a disorder-to-order transition in the p21Waf1 protein upon binding. This conformational change in p21Waf1 then results in the formation of a characteristic motif that can bind to C8. Accumulating evidence has demonstrated the importance of such transitions (30). For example, >15,000 of the 91,000 proteins in the Swiss Protein Database were identified as having long regions of intrinsic disorder, including p53 and pRb (31). Of note, intrinsic disorder is more frequently observed in tumor suppressors and oncoproteins than in other proteins (32).
One of the characteristic secondary structures inducing disorder-to-order transitions is the presence of an α-helix (30). Our study has defined a region in MDM2-spanning amino acids 251–260 as promoting the conformational change in p21Waf1. This region is made up of predominantly hydrophobic amino acids that would be predicted to form part of an α-helix. Thus, MDM2 apparently functions as a chaperone-like protein to facilitate the degradation of intact p21Waf1. This activity may be involved in the regulation of other substrates of MDM2. Supporting this idea is the fact that the region of MDM2 identified in this study has also been reported to be important for the MDM2-mediated ubiquitin-independent proteasomal degradation of retinoblastoma protein (28).
Although C8 has functions in substrate recognition, tethering the substrate to the proteasome and facilitating assembly of the 20S proteasome, it has no proteolytic activity (22, 33). The crystal structure of the 20S proteasome shows that the entrance to the proteasome is not normally open (34). Thus, it is unclear how p21Waf1 enters the 20S proteasome after being recognized by C8. Providing clues about this process, recent studies have identified another proteasome activator, PA28 (11S regulator, REG), that facilitates ubiquitin-independent proteasomal degradation of substrates (35). Given the importance of subcellular localization to MDM2-mediated p21Waf1 degradation, the γ-isoform of PA28 may be important for this process, because PA28γ is localized within the nucleus (36). PA28γ may activate the 20S proteasome to degrade p21Waf1 in the nucleus after it is recognized by C8. Consistent with this concept, it was recently demonstrated that PA28γ promotes p21Waf1 degradation through the 20S proteasome (15, 37). In addition, an interaction between MDM2 and PA28γ has also been demonstrated (38). It is therefore possible that MDM2 has dual roles in the proteasomal degradation of p21Waf1, inducing its recognition by the proteasome and promoting proteasome activation via an interaction with the proteasome activator. The interaction between MDM2, p21Waf1, C8, and 14–3-3-τ may also impact proteasomal recognition or binding of p21Waf1 (18).
The induction of p21Waf1 degradation in MDM2-associated cell cycle progression was further demonstrated in our experiments with p53 knock-out cells (Fig. 6, left panels). MDM2 mutants lacking the segments associated with p21Waf1 binding and degradation (226–250 and 251–260) failed to prevent p21Waf1-induced cell cycle arrest in the G0/G1 phase. To further demonstrate the dependence of the cell cycle checkpoint on p21Waf1 and the inhibitory function of MDM2, we used p53 and p21Waf1 double knock-out cells to examine the cell cycle distribution. Of interest, when both p53 and p21waf1 were knocked out, the overexpression of wild-type MDM2 failed to increase the number of cells in the G2/M phase as seen in GFP-transfected control cells. As a result, an increase in the percentage of cells in the G1 phase was observed (Fig. 6B, right panel). This indicates that MDM2 has a role in promoting the passage of cells through the G2/M phase, independent of p53 or p21Waf1. Supporting evidence also included the fact that there were no differences in the cell cycle distribution patterns among the double knock-out cells transfected with wild-type MDM2 or either of the MDM2 mutants Although one early study indicated that MDM2 might have an inhibitory effect on the G1-S transition in normal cells (39), it is more likely that the overexpression of MDM2 accelerates cell growth/division in cancer cells (especially those lacking both p53 and p21Waf1). Thus, without normal checkpoint control, the MDM2 overexpressing cells divide more rapidly than cells with normal levels of MDM2. As a result of this increased division/progression, the double knock-out cells overexpressing MDM2 were found to be increased in the G1 phase at 6 h after release from serum starvation. Considering that the MDM2 overexpressing cells would have a shorter doubling time, the increase in the cells in G0/G1 may be associated with our sampling time. Future studies should examine the cell cycle distribution at a longer time after release from serum starvation in these cells.
Although the evidence presented here describes the molecular mechanism underlying the MDM2-mediated degradation of p21Waf1, it is unclear why the protein is ubiquitinated if it can be degraded via a ubiquitin-independent process. Despite the fact that the ubiquitination of proteins is most frequently associated with their degradation, it can serve other purposes. Thus, the ubiquitination of p21Waf1 may have alternative functions (40–42). For example, ubiquitination can regulate the subcellular localization of proteins and can alter the transcription of particular genes (41, 42). The ubiquitination of p21Waf1 may regulate its translocation between the cytoplasm and nucleus, an effect that has been observed for p53 (42). This may also help keep the p21Waf1-MDM2 interaction at optimal levels.
In addition to elucidating a previously uncharacterized mechanism responsible for p21Waf1 degradation, the present study also supports a growing body of work that demonstrates the importance of the MDM2 oncoprotein as a therapeutic target. Although MDM2 has been the target of numerous therapeutic strategies, nearly all of them have focused on the interaction between MDM2 and p53 (43). Because it is estimated that more than half of all human cancers, especially advanced cancers, have lost expression of functional p53 (44), it is no surprise that these strategies have been criticized and mostly unsuccessful. Strategies directly targeting MDM2 expression (e.g. siRNA or antisense approaches) or targeting other functions of MDM2 (beyond its E3 ligase activity) would ensure that the agent(s) have wider applications and would be effective for more patients.
As an example of a new potential target, the present study indicates that amino acids 251–260 in MDM2 induce a change in p21Waf1 conformation. This region of MDM2 is also important for its degradation of retinoblastoma protein, which also occurs independently of ubiquitination. It is possible that this region has general chaperone-like activity, not only toward p21Waf1 and pRb, but also other molecules. If this is the case, developing small molecule inhibitors or peptides targeting this small region of MDM2 would be promising for human cancer therapy or prevention.
In summary, our results demonstrate a novel mechanism by which MDM2 induces the ubiquitin-independent proteasomal degradation of an intact protein. This previously unknown chaperone-like function of MDM2 induces a conformational change in p21Waf1, leading to its increased binding to the C8 subunit of the proteasome and subsequent degradation. It is possible that this mechanism may also be active for other substrates. In addition, the present study provides further evidence supporting the importance of the p53-independent activities of MDM2 and suggests a potential novel target for therapy.
Acknowledgments
We thank Dr. E. R. Rayburn for help with preparing the manuscript and Dr. D. Chen and X. Zhang for helpful discussions. We also appreciate the technical assistance with FRET imaging and immunofluorescence from Shawn Williams at the University of Alabama at Birmingham High Resolution Imaging Facility within the Department of Vision Science at University of Alabama at Birmingham.
This work was supported in part by NCI, National Institutes of Health Grants R01CA112029 and R01CA121211 (to R. Z.).
- aa
- amino acids
- FRET
- fluorescence resonance energy transfer
- AD
- acidic domain
- GST
- glutathione S-transferase
- GFP
- green fluorescent protein
- NLS
- nuclear localization signal
- HA
- hemagglutinin
- NES
- nuclear exportation signal
- EYFP
- enhanced yellow fluorescent protein
- ECFP
- enhanced cyan fluorescent protein.
REFERENCES
- 1.Onel K., Cordon-Cardo C. (2004) Mol. Cancer Res. 2, 1–8 [PubMed] [Google Scholar]
- 2.Rayburn E., Zhang R., He J., Wang H. (2005) Curr. Cancer Drug Targets 5, 27–41 [DOI] [PubMed] [Google Scholar]
- 3.Clegg H. V., Itahana K., Zhang Y. (2008) Cell Cycle 7, 287–292 [DOI] [PubMed] [Google Scholar]
- 4.Haupt Y., Maya R., Kazaz A., Oren M. (1997) Nature 387, 296–299 [DOI] [PubMed] [Google Scholar]
- 5.Momand J., Zambetti G. P., Olson D. C., George D., Levine A. J. (1992) Cell 69, 1237–1245 [DOI] [PubMed] [Google Scholar]
- 6.Ohkubo S., Tanaka T., Taya Y., Kitazato K., Prives C. (2006) J. Biol. Chem. 281, 16943–16950 [DOI] [PubMed] [Google Scholar]
- 7.Momand J., Jung D., Wilczynski S., Niland J. (1998) Nucleic Acids Res. 26, 3453–3459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barak Y., Gottlieb E., Juven-Gershon T., Oren M. (1994) Genes Dev. 8, 1739–1749 [DOI] [PubMed] [Google Scholar]
- 9.Lahav G. (2008) Adv. Exp. Med. Biol. 641, 28–38 [DOI] [PubMed] [Google Scholar]
- 10.Ries S., Biederer C., Woods D., Shifman O., Shirasawa S., Sasazuki T., McMahon M., Oren M., McCormick F. (2000) Cell 103, 321–330 [DOI] [PubMed] [Google Scholar]
- 11.Li M., Zhang Z., Hill D. L., Wang H., Zhang R. (2007) Cancer Res. 67, 1988–1996 [DOI] [PubMed] [Google Scholar]
- 12.Zhang Z., Zhang R. (2005) Curr. Cancer Drug Targets. 5, 9–20 [DOI] [PubMed] [Google Scholar]
- 13.Jin Y., Lee H., Zeng S. X., Dai M. S., Lu H. (2003) EMBO J. 22, 6365–6377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Z., Wang H., Li M., Agrawal S., Chen X., Zhang R. (2004) J. Biol. Chem. 279, 16000–16006 [DOI] [PubMed] [Google Scholar]
- 15.Li X., Amazit L., Long W., Lonard D. M., Monaco J. J, O'Malley B. W. (2007) Mol. Cell 26, 831–842 [DOI] [PubMed] [Google Scholar]
- 16.Honda R., Tanaka H., Yasuda H. (1997) FEBS Lett. 420, 25–27 [DOI] [PubMed] [Google Scholar]
- 17.Poyurovsky M. V., Priest C., Kentsis A., Borden K. L., Pan Z. Q., Pavletich N., Prives C. (2007) EMBO J. 26, 90–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang B., Liu K., Lin H. Y., Bellam N., Ling S., Lin W. C. (2010) Mol. Cell Biol. 30, 1508–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karpova T. S., Baumann C. T., He L., Wu X., Grammer A., Lipsky P., Hager G. L., McNally J. G. (2003) J. Microsc. 209, 56–70 [DOI] [PubMed] [Google Scholar]
- 20.Meltzer R. H., Kapoor N., Qadri Y. J., Anderson S. J., Fuller C. M., Benos D. J. (2007) J. Biol. Chem. 282, 25548–25559 [DOI] [PubMed] [Google Scholar]
- 21.Berdiev B. K., Cormet-Boyaka E., Tousson A., Qadri Y. J., Oosterveld-Hut H. M., Hong J. S., Gonzales P. A., Fuller C. M., Sorscher E. J., Lukacs G. L., Benos D. J. (2007) J. Biol. Chem. 282, 36481–36488 [DOI] [PubMed] [Google Scholar]
- 22.Touitou R., Richardson J., Bose S., Nakanishi M., Rivett J., Allday M. J. (2001) EMBO J. 20, 2367–2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schon O., Friedler A., Freund S., Fersht A. R. (2004) J. Mol. Biol. 336, 197–202 [DOI] [PubMed] [Google Scholar]
- 24.Wallace M., Worrall E., Pettersson S., Hupp T. R., Ball K. L. (2006) Mol. Cell 23, 251–263 [DOI] [PubMed] [Google Scholar]
- 25.Girnita L., Girnita A., Larsson O. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 8247–8252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salcedo A., Mayor F., Jr., Penela P. (2006) EMBO J. 25, 4752–4762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shenoy S. K., McDonald P. H., Kohout T. A., Lefkowitz R. J. (2001) Science 294, 1307–1313 [DOI] [PubMed] [Google Scholar]
- 28.Sdek P., Ying H., Chang D. L., Qiu W., Zheng H., Touitou R., Allday M. J., Xiao Z. X. (2005) Mol. Cell 20, 699–708 [DOI] [PubMed] [Google Scholar]
- 29.Kriwacki R. W., Hengst L., Tennant L., Reed S. I., Wright P. E. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 11504–11509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oldfield C. J., Cheng Y., Cortese M. S., Romero P., Uversky V. N., Dunker A. K. (2005) Biochemistry 20, 12454–12470 [DOI] [PubMed] [Google Scholar]
- 31.Romero P., Obradovic Z., Kissinger C. R., Villafranca J. E., Garner E., Guilliot S., Dunker A. K. (1998) Pac. Symp. Biocomput. 437–448 [PubMed] [Google Scholar]
- 32.Iakoucheva L. M., Brown C. J., Lawson J. D., Obradović Z., Dunker A. K. (2002) J. Mol. Biol. 323, 573–584 [DOI] [PubMed] [Google Scholar]
- 33.Gerards W. L., de Jong W. W., Bloemendal H., Boelens W. (1998) J. Mol. Biol. 275, 113–121 [DOI] [PubMed] [Google Scholar]
- 34.Groll M., Ditzel L., Löwe J., Stock D., Bochtler M., Bartunik H. D., Huber R. (1997) Nature 386, 463–471 [DOI] [PubMed] [Google Scholar]
- 35.Dick T. P., Ruppert T., Groettrup M., Kloetzel P. M., Kuehn L., Koszinowski U. H., Stevanović S., Schild H., Rammensee H. G. (1996) Cell 86, 253–262 [DOI] [PubMed] [Google Scholar]
- 36.Rechsteiner M., Hill C. P. (2005) Trends Cell Biol. 15, 27–33 [DOI] [PubMed] [Google Scholar]
- 37.Chen X., Barton L. F., Chi Y., Clurman B. E., Roberts J. M. (2007) Mol. Cell 26, 843–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Z., Zhang R. (2008) EMBO J. 27, 852–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brown D. R., Thomas C. A., Deb S. P. (1998) EMBO J. 17, 2513–2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rousseau D., Cannella D., Boulaire J., Fitzgerald P., Fotedar A., Fotedar R. (1999) Oncogene 18, 4313–4325 [DOI] [PubMed] [Google Scholar]
- 41.Joo H. Y., Zhai L., Yang C., Nie S., Erdjument-Bromage H., Tempst P., Chang C., Wang H. (2007) Nature 449, 1068–1072 [DOI] [PubMed] [Google Scholar]
- 42.Li M., Brooks C. L., Wu-Baer F., Chen D., Baer R., Gu W. (2003) Science 302, 1972–1975 [DOI] [PubMed] [Google Scholar]
- 43.Vassilev L. T. (2007) Trends Mol. Med. 13, 23–31 [DOI] [PubMed] [Google Scholar]
- 44.Bártek J., Bártková J., Vojtĕsek B., Stasková Z., Lukás J., Rejthar A., Kovarík J., Midgley C. A., Gannon J. V., Lane D. P. (1991) Oncogene 6, 1699–1703 [PubMed] [Google Scholar]