Abstract
Mammalian ALDH7A1 is homologous to plant ALDH7B1, an enzyme that protects against various forms of stress, such as salinity, dehydration, and osmotic stress. It is known that mutations in the human ALDH7A1 gene cause pyridoxine-dependent and folic acid-responsive seizures. Herein, we show for the first time that human ALDH7A1 protects against hyperosmotic stress by generating osmolytes and metabolizing toxic aldehydes. Human ALDH7A1 expression in Chinese hamster ovary cells attenuated osmotic stress-induced apoptosis caused by increased extracellular concentrations of sucrose or sodium chloride. Purified recombinant ALDH7A1 efficiently metabolized a number of aldehyde substrates, including the osmolyte precursor, betaine aldehyde, lipid peroxidation-derived aldehydes, and the intermediate lysine degradation product, α-aminoadipic semialdehyde. The crystal structure for ALDH7A1 supports the enzyme's substrate specificities. Tissue distribution studies in mice showed the highest expression of ALDH7A1 protein in liver, kidney, and brain, followed by pancreas and testes. ALDH7A1 protein was found in the cytosol, nucleus, and mitochondria, making it unique among the aldehyde dehydrogenase enzymes. Analysis of human and mouse cDNA sequences revealed mitochondrial and cytosolic transcripts that are differentially expressed in a tissue-specific manner in mice. In conclusion, ALDH7A1 is a novel aldehyde dehydrogenase expressed in multiple subcellular compartments that protects against hyperosmotic stress by generating osmolytes and metabolizing toxic aldehydes.
Keywords: Enzymes/Dehydrogenase, Enzymes/Structure, Gene/Structure, Protein/Structure, Enzyme Kinetics, ALDH7A1, Crystal Structure, Kinetics, Osmotic Stress
Introduction
The human aldehyde dehydrogenase (ALDH)3 superfamily contains 19 enzymes involved in the NAD(P)+-dependent oxidation of aldehydes to their corresponding carboxylic acids. These enzymes play crucial roles in a number of physiological processes by efficiently metabolizing a wide array of endogenous and exogenous aldehydes (1). Aldehydes are highly reactive molecules that can form adducts resulting in DNA damage and enzyme inactivation. As such, their removal is of the utmost importance. ALDH enzymes also couple the removal of these potentially toxic aldehydes to NAD(P)H production, which, in turn, helps maintain cellular redox balance. Finally, ALDH activity generates a number of important cellular molecules, including retinoic acid, the neurotransmitter γ-aminobutyric acid, the major dietary folate tetrahydrofolate, and the osmolyte betaine (1).
Human ALDH7A1 was originally identified as sharing 60% homology with the osmotic stress-induced 26g pea turgor protein (according to the official nomenclature now referred to as ALDH7B1), found in the common garden pea (Pisum sativum) (2). Initially named “antiquitin,” the ALDH7A1 protein has been highly conserved throughout evolution. Indeed, the degree of homology noted between ALDH7A1 and ALDH7B1 is comparable with that observed between the human and pea histone H2A proteins, which are among the most evolutionarily conserved of all eukaryotic proteins (2, 3). Such a high degree of sequence similarity between species often indicates an essential and functionally conserved role within the cell. Expression of ALDH7B1 increases in response to drought conditions and is thought to be involved in the regulation of osmotic pressure and protection against oxidative stress (4). A similar increase in mRNA has also been observed in canola, soybean, and tobacco plants, consistent with enhanced expression representing a general adaptive response to environmental stressors (5, 6). Ectopic expression in thale cress (Arabidopsis thaliana) and tobacco (Nicotiana tabacum) plants greatly enhanced their tolerance to either drought or high salinity, supporting an osmoprotective role. In addition, transgenic thale cress and tobacco plants showed decreased sensitivity to both hydrogen peroxide (H2O2) and paraquat, two reactive oxygen species-generating compounds (7). In rice, a T-DNA insertional mutant lacking functional ALDH7B1 enzyme showed increased sensitivity to cold, heat, salinity, dehydration, and paraquat. This protein was also required for proper seed maturation and viability (8).
An increase in extracellular solute concentration, or osmolarity, hinders cell metabolism and function through the passive loss of water by osmosis. Water loss leads to cell shrinkage and an increase in the intracellular concentrations of all inorganic ions and organic solutes as well as an increase in macromolecular crowding (9). Together, the changes associated with such osmotic stress disturb biochemical reaction rates and overall cell homoeostasis. Osmotic stress can also elevate reactive oxygen species levels, strain cytoskeletal networks, depolarize mitochondria, and damage both DNA and proteins (10). Through the process of evolution, cells have developed protective mechanisms to deal with the deleterious effects associated with hyperosmotic challenges. These osmoprotective mechanisms include increased uptake and synthesis of osmolytes and compensatory changes in gene expression, such as the up-regulation of enzymes that provide protection against oxidative stress and of molecular chaperones (e.g. heat shock proteins) (11).
In mammals, ALDH7A1 is known to play a primary role during lysine catabolism through the NAD+-dependent oxidative conversion of aminoadipate semialdehyde (AASA) to its corresponding carboxylic acid, α-aminoadipic acid (12). Lysine is an essential amino acid, and its catabolism helps maintain the cellular nitrogen pool as well as ketone body formation. Thus, ALDH7A1 serves as a critical step in this process. Recent studies have revealed mutations in ALDH7A1 as the underlying cause for both pyridoxine-dependent epilepsy (PDE) and folic acid-responsive seizures (13, 14). If left untreated, both disorders commonly result in status epilepticus and death. In PDE, ALDH7A1 mutations lead to AASA accumulation in plasma, urine, and cerebrospinal fluids (13). AASA forms a condensation product with pyridoxal-5′-phosphate to cause cofactor inactivation and acute vitamin B6 depletion. Additionally, elevated AASA levels in the brains of PDE patients may contribute to cerebral atrophy through the inherent toxicity of this aldehyde.
The osmoprotective properties of ALDH7A1, if any, have never been investigated, and, to date, the mechanism by which ALDH7 proteins protect against osmotic stress is unknown. The aims of this study were to test the hypothesis that human ALDH7A1 protects against osmotic stress and to determine the mechanism of such protection. The present study represents the most complete characterization of a mammalian ALDH7A1 protein, and results described herein indicate that ALDH7A1 is a major player in a diverse set of important physiological functions within the cell.
EXPERIMENTAL PROCEDURES
Bioinformatic Tools
Mouse and human transcript sequences were obtained from NCBI AceView (available at the NCBI web site). Alignments were performed using ClustalW (available at the European Bioinformatics Institute web site). Mitochondrial signal sequence predictions were performed using pTarget (available at the University at Albany web site), SignalP (available at the Center for Biological Sequence Analysis web site), Protein Prowler (available on the World Wide Web), and SherLoc (available at the Universität Tübingen web site). Both the mouse and human proteins were screened for putative nuclear localization signals and nuclear export signals using NLStradamus (available at the University of Toronto web site) and NetNES 1.1 (available at the Center for Biological Sequence Analysis web site), respectively.
Animals
C57BL/6J mice were maintained in a temperature-controlled room (21–22 °C) on a 12-h light/dark cycle and supplied with food and water ad libitum. Tissues were harvested from 60–90-day-old animals sacrificed by CO2 inhalation followed by cervical dislocation. All animal use was conducted in compliance with the Institutional Animal Care and Use Committee of the University of Colorado and performed according to published National Institutes of Health guidelines.
Cell Culture
Chinese hamster ovary (CHO) cells, human renal proximal tubular epithelial (HK-2) cells, and human umbilical vein endothelial cells (HUVEC) were cultivated as described previously (15, 16).
Western Immunoblotting
Tissue and cell lysates were prepared as previously described (17). Lysates (10–30 μg/well) were separated by SDS-PAGE (10%). Western blot analysis was performed as described previously using polyclonal anti-human ALDH7A1 antibody (1:2000 dilution) and horseradish peroxidase-conjugated secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) at 1:5000 (18).
Construction and Expression of Recombinant Plasmids
Human ALDH7A1_v1 cDNA was excised from the hALDH7A1/SK plasmid (19) with XhoI and BamHI and subsequently ligated at the same sites in the ΔpCEP4Δ mammalian expression vector to generate ΔpCEP4Δ-hALDH7A1_v1 (20). The insert was also subcloned into the baculovirus expression vector pBlueBac4.5 (Invitrogen) using the same strategy. Recombinant viruses were plaque-purified and amplified in Sf9 (Spodoptera frugiperda) insect cells as described previously (21, 22).
Generation of ALDH7A1-transfected Cell Lines
The SV-40-transformed CHO cell lines were stably transfected with either the mammalian expression vector ΔpCEP4Δ alone (CHO-Vector) or ΔpCEP4Δ-hALDH7A1_v1 cDNA (CHO-hALDH7A1_v1) using Lipofectamine Plus reagent (Invitrogen) as described previously (18). Stable cell populations were selected by incubation in medium containing hygromycin (0.4 mg/ml). ALDH7A1 expression was screened by Western blot (see above) and enzyme activity assays (see below). Two stable lines, CHO-hALDH7A1_v1 clones 14 and 35, were used in subsequent experiments.
Colony Formation Assay
CHO-Vector and CHO-hALDH7A1_v1 clone 35 cells (1 × 102) were plated in 6-well plates and allowed to attach for 24 h. The cultures were subsequently treated with a single dose of NaCl (25–500 mm) or sucrose (150–500 mm) for 4 h at 37 °C and thereafter monitored on a daily basis for colony formation. In general, colonies were visible 4–6 days after treatment with the solutions, and cells were fixed in buffered formalin and stained with 0.1% crystal violet. Colonies containing ≥50 cells were scored and counted.
DNA Fragmentation Assay
Culture plates of 1 × 106 CHO-Vector and CHO-hALDH7A1_v1 clone 35 cells were treated with either 200 or 400 mm NaCl for 4 h (as above). Following incubation, cells were harvested and processed as described previously (23).
ALDH7A1 Knockdown
HK-2 cells were seeded into 6-well plates. The following day when at ∼40% confluence, 20 nm Silencer Select siRNA (Ambion, Austin, TX) targeting human ALDH7A1 or Silencer Select Negative Control 1 (Ambion) siRNA were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Seventy h after transfection, the cells were either harvested for Western blotting or treated with NaCl for 3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide (MTT) assays.
MTT Assay
Seventy h after siRNA transfection, HK-2 cells were treated with 0–500 mm NaCl for 12 h. MTT assays were then performed as previously described (18).
Protein Purification
ALDH7A1 was purified by fast protein liquid chromatography using a 5′-AMP-Sepharose 4B affinity column as described previously by us for other ALDH proteins, including ALDH3A1 (22) and ALDH1A1 (21). Active fractions were subjected to SDS-PAGE and visualized by silver staining (Wako Chemicals, Richmond, VA). MALDI-TOF mass spectrometry was used to confirm the identity of the purified ALDH7A1 protein as described previously (24).
Antibody Production
Purified recombinant human ALDH7A1 was run on an SDS-polyacrylamide gel, stained using Coomassie stain (following the manufacturer's instructions), and destained in distilled water overnight at room temperature under gentle agitation. ALDH7A1 bands were cut out of the gel and shipped for antibody production (Cocalico Biologicals Inc., Reamstown, PA).
ALDH Enzymatic Activity and Kinetic Studies
The enzymatic activity and kinetic studies of recombinant ALDH7A1 were determined as previously described (25, 26). Aminoadipic semialdehyde was synthesized from allysine ethylene acetal (Chiralix B.V., Nijmegen, The Netherlands), as described previously (13). All other aldehydes used were purchased from Sigma.
Crystallography
For structural studies, a construct encoding residues 1–499 of hALDH7A1_v2 was cloned into the bacterial expression vector pNIC-Bsa4. The resultant plasmid encoded human ALDH7A1 lacking the mitochondrial leader sequence with an N-terminal hexahistidine tag and a tobacco etch virus protease recognition site. Recombinant human ALDH7A1 was expressed overnight at 18 °C in Escherichia coli BL21(DE3) in TB medium containing 50 μg/ml kanamycin and 0.5 mm isopropyl 1-thio-β-d-galactopyranoside. The protein was then purified by chromatography on Ni2+-Sepharose and Superdex S200 16/60 columns, and the affinity tag was removed by incubation with His-tagged tobacco etch virus protease. The digest was passed through a Ni2+-Sepharose column, and the unbound fraction was collected. The molecular mass of hALDH7A1 was determined by electrospray ionization-mass spectrometry, allowing confirmation that the tag had been removed. Prior to crystallization, the protein was concentrated to 30 mg/ml in 10 mm Hepes, 500 mm NaCl, 5% glycerol (pH 7.5); NADH was added to a final concentration of 11 mm. Crystals were grown in sitting drops at 20 °C by mixing 150 nl of protein solution with 150 nl of a well solution comprising 20% polyethylene glycol 3350, 10% ethylene glycol, 0.2 m NaBr, 0.1 m Bistris propane (pH 6.6), and equilibrating against a 50-μl reservoir of the well solution. A single crystal was transferred to a cryoprotectant (well solution supplemented with 20% ethylene glycol) and plunged into liquid nitrogen. A data set was collected at Swiss Light Source beamline X10SA and processed to 1.3 Å using the program XDS (27). All data to 1.3 Å were used in refinement; the data were >94% complete to 1.5 Å but decreased to 45% in the higher resolution shells. Initial phases were calculated by molecular replacement with the program PHASER (28) and using Protein Data Bank entry 1NZX as a search model. Two tetramers were located in the asymmetric unit. Refinement was performed in REFMAC (29), and manual rebuilding was done with the program COOT (30). Residue numbering in the deposited model is offset by +1 compared with the gene-derived sequence due to a vector-encoded serine at the N terminus that remains after tobacco etch virus cleavage.
Docking Methods
The program GOLD (31) was used to dock the substrates betaine aldehyde, nonanal, and AASA into the active site of human ALDH7A1. To reproduce a pose that would be conducive to theoretical catalysis, a distance restraint of 1.5–3.5 Å was applied between the carbonyl carbon of each substrate and the sulfur atom of Cys302. The top three solutions for each substrate were inspected, and, because they were all essentially equivalent, the top scoring pose was chosen for analysis.
Subcellular Fractionation
Liver or kidney tissue was isolated and placed in tissue homogenization buffer (1 ml/g of tissue). The tissues were then homogenized on ice using a Tissue Miser homogenizer gun (Fisher) and processed to isolate subcellular fractions as described previously (23). Ten μg of protein from each fraction was separated by SDS-PAGE, and ALDH7A1 was identified by Western blotting. Purity was verified by probing the blots with lamin B1 (ab16048, Abcam), protein-disulfide isomerase (ab2792, Abcam), VDAC1 (voltage-dependent anion channel 1) (ab15895, Abcam), and GCLM (glutamate cysteine ligase modifier subunit) antibodies for nuclear, microsomal, mitochondrial, and cytosolic fractions, respectively.
Immunohistochemistry
Liver, kidney, and heart were removed from C57BL/6J adult mice. Tissues were paraffin-embedded, sectioned, and processed as described elsewhere (32). Slides containing sectioned tissues were incubated with anti-ALDH7A1 antibody (1:20 dilution) followed by horseradish peroxidase-conjugated secondary antibody (goat anti-rabbit IgG) at a 1:100 dilution. After immunohistochemical processing, the sections were subjected to microscopic examination (Axioscop-2 microscope (Carl Zeiss Inc.) equipped with a DC 290 digital camera (Eastman Kodak Co.). HUVEC were seeded on 0.2% gelatin-coated glass coverslips and grown to confluence. Cells were fixed in 4% paraformaldehyde and then permeabilized in BD Perm/WashTM buffer (BD Biosciences) containing 0.1% saponin and 3% fetal bovine serum. Cells were then blocked in BD Perm/WashTM containing 5% goat serum and 1% bovine serum albumin and subsequently incubated with rabbit anti-ALDH7A1 serum or control preimmune serum (each at 1:200) in BD Perm/WashTM containing 1% bovine serum albumin. After washings, cells were incubated with a fluorescein isothiocyanate-conjugated goat anti-rabbit antibody (1:250 dilution) (Jackson ImmunoResearch Laboratories). DNA was counterstained with 4′,6-diamidino-2-phenylindole (DAPI) at 300 ng/ml. Slides were mounted in Fluoromount-G (SouthernBiotech, Hatfield, PA), and confocal fluorescence images were captured using a Zeiss laser-scanning microscope. Stable CHO-Vector and CHO-hALDH7A1_v1 clone 35 cells were grown on glass coverslips. MitoTracker® mitochondrial selective probe (Invitrogen) was used according to the manufacturer's protocol. The coverslips were processed as described previously (33).
ALDH7A1 Splice Variant RT-PCR
Tissues were isolated from euthanized C57BL/6J mice and immediately placed into RNAlater solution (Ambion) according to the manufacturer's protocol. Total RNA was isolated from the tissues using the RNeasy® minikit (Qiagen, Valencia, CA) according to the manufacturer's protocol. The RNA was then treated with TURBO DNA-freeTM DNase (Ambion). cDNA was synthesized using SuperScriptTM III First-Strand RT-PCR kit (Invitrogen). RT-PCR primers were designed to distinguish between the two mouse ALDH7A1 splice variants, mALDH7A1_v1 and mALDH7A1_v2. The primer sets utilize the same downstream primer (5′-CCGATATCTGCTCCACCACAAACCA-3′), which primes in exon 8 of mALDH7A1_v1 and mALDH7A1_v2. The mALDH7A1_v1 upstream primer (5′-CAGATCCAAGGAACGAGCATGCC-3′) anneals in exon 1b, whereas the mALDH7A1_v2 upstream primer (5′-CTCAGTATGTGGCGTGTGCCC-3′) anneals in the 5′-end of exon 1 upstream of the mALDH7A1_v1 splice site. This design generates a 937-bp and 779-bp product for mALDH7A1_v1 and mALDH7A1_v2, respectively, allowing for the differentiation between the two splice variants. Parallel reactions were run using β-actin primers as a control.
Data Analysis
Data are expressed as mean ± S.E. Comparison of results between different groups was performed by Student's unpaired t test using SigmaPlot (Version 7.0, 2001). Differences were considered significant at p < 0.05.
RESULTS
ALDH7A1 Expression Protects Chinese Hamster Ovary Cells and Human Renal Proximal Tubular Epithelial (HK-2) Cells from Hyperosmotic Stress
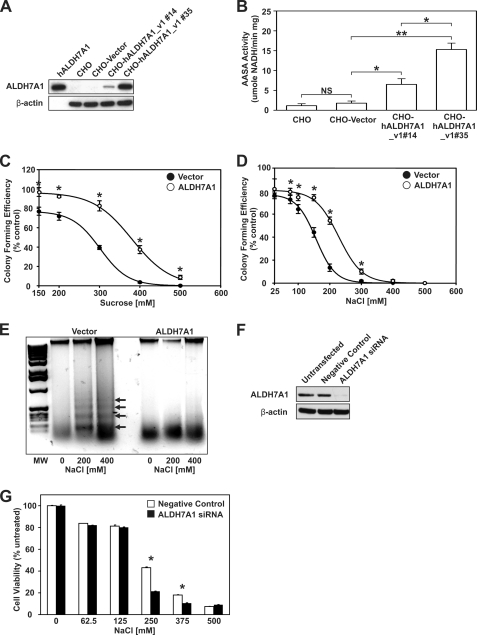
ALDH7B1 expression in plants provides significant protection from various types of hyperosmotic stress. To determine whether ALDH7A1 served a similar protective function in mammalian cells, CHO cells were stably transfected with either human ALDH7A1 (CHO-hALDH7A1_v1) or the empty ΔpCEP4Δ vector (CHO-Vector). Western blot analysis confirmed ALDH7A1 expression in both CHO-hALDH7A1_v1 clone 14 and 35 cell lines (Fig. 1A). Untransfected (CHO) and CHO-Vector cell lines lacked any detectable levels of ALDH7A1 protein (viz. negative controls). The CHO-hALDH7A1_v1 clone 14 cell line was identified as a low expressing line, as indicated by ALDH7A1 band intensity. By contrast, the CHO-hALDH7A1_v1 clone 35 cell line was considered a high expressing line. AASA activity in these cell lines confirmed the presence of enzymatically functional ALDH7A1 protein (Fig. 1B) and showed that the AASA activity roughly corresponds to the ALDH7A1 protein levels. The ability of ALDH7A1 to protect cells from hyperosmotic stress was tested using colony forming and DNA fragmentation assays. CHO-Vector and CHO-hALDH7A1_v1 clone 35 cells were grown under increasing concentrations of sucrose (Fig. 1C) and NaCl (Fig. 1D). The estimated EC50 values for inhibition of colony forming activity in vector and ALDH7A1 cell lines were 301 ± 3 and 380 ± 5 mm, respectively, for sucrose treatment and 156 ± 3 and 228 ± 5 mm, respectively, for NaCl treatment. These results are consistent with ALDH7A1 protein expression imbuing significant protection. DNA fragmentation assays were performed on CHO-Vector and CHO-hALDH7A1_v1 clone 35 cells treated with either 200 or 400 mm NaCl (Fig. 1E). Under these conditions, only the control cell line (CHO-Vector) showed substantial 200-bp DNA laddering commonly associated with apoptotic cell death. DNA laddering was almost completely absent from treated CHO-hALDH7A1_v1 clone 35 cells, further supporting the notion that ALDH7A1 expression protects cells from hyperosmotic stress. Finally, ALDH7A1 knockdown sensitizes human renal proximal tubular epithelial (HK-2) cells to hyperosmotic stress. HK-2 cells were transfected with siRNA targeting ALDH7A1. Endogenous protein expression was reduced to less than 10% (Fig. 1F). MTT assays revealed significantly reduced cell viability in ALDH7A1 siRNA-transfected HK-2 cells exposed to increasing concentrations of NaCl (Fig. 1G).
FIGURE 1.
Expression of ALDH7A1 in CHO cells protects against hyperosmotic stress. A, cell lysates (30 μg) from untransfected CHO cells and CHO cell lines stably transfected with ΔpCEP4Δ vector (CHO-Vector) or ΔpCEP4Δ-hALDH7A1_v1 (CHO-hALDH7A1_v1#13 and CHO-hALDH7A1_v1#35) were assayed for ALDH7A1 protein expression by Western blot analysis, using specific antibodies (as described under “Experimental Procedures”; top). The blot was reprobed for β-actin as loading correction. Purified recombinant human ALDH7A1 protein (50 ng) was used as positive control. B, cell lysates were assayed for AASA activity (as described under “Experimental Procedures”). C and D, CHO-Vector (filled circles) and CHO-hALDH7A1_v1 clone 35 (open circles) cells were treated with sucrose (C) or NaCl (D) for 4 h at 37 °C, and colonies were stained with crystal violet after 1 week. Mean colony numbers obtained with CHO-Vector and CHO-hALDH7A1_v1 clone 35 cells were normalized as a percentage of untreated control. E, CHO-Vector (Vector) and CHO-hALDH7A1_v1 clone 35 (ALDH7A1) cells were treated with 200 or 400 mm NaCl for 4 h at 37 °C. Agarose gel electrophoresis was performed on DNA isolated from untreated and treated cells. Only in CHO-Vector cells did NaCl promote the characteristic 200-bp DNA fragment laddering associated with apoptotic cell death (arrows). F, Western analysis of ALDH7A1 knockdown in HK-2 cell lysates (20 μg). Cells were transfected with 20 nm negative control or ALDH7A1-specific siRNA. Untransfected cells were used as an additional negative control. Blots were reprobed for β-actin. G, decreased cell viability in HK-2 cells lacking ALDH7A1. siRNA-transfected cells were treated with increasing concentrations of NaCl, and cell viability was analyzed via an MTT assay. Values represent mean ± S.E. (error bars) for triplicates in three separate experiments (NS, not significant (p > 0.05); *, p < 0.05; **, p < 0.01).
Biochemical Characterization of Human ALDH7A1
Purified human ALDH7A1 protein was used in kinetic studies to delineate substrate specificity. Silver staining revealed homogeneous purification as indicated by a single protein band at 55 kDa (data not shown). Subsequent MALDI-TOF mass spectrometry confirmed the protein's identity. Human ALDH7A1 was found to metabolize a wide range of aliphatic aldehydes. Table 1 shows calculated Km (μm) and Vmax (nmol of NADH/min/mg) values for a number of substrates when using NAD+ as a cofactor. As predicted, the enzyme efficiently metabolizes AASA, as reflected in its catalytic efficiency constant Vmax/Km. The enzyme also metabolized betaine aldehyde, presumably to betaine, a major cellular osmolyte and methyl donor (34). Human ALDH7A1 showed high specificity for the medium-chain saturated aldehydes hexanal, octanal, and nonanal, all of which are known as potentially toxic lipid peroxidation products (35). ALDH7A1 exhibited low affinity for benzaldehyde, an aromatic aldehyde; propanal, a short-chain aliphatic aldehyde; and glyceraldehyde, a short-chain aldose. The unsaturated aldehyde, trans-2-nonenal, another lipid peroxidation product, was also efficiently metabolized by ALDH7A1.
TABLE 1.
Kinetic properties of recombinant human ALDH7A1
| Substrate | Kma | Vmaxa | Vmax/Km |
|---|---|---|---|
| μm | nmol NADH/min/mg | ||
| Nonanal | 28.5 ± 4.8 | 364.9 ± 20.1 | 12.80 |
| trans-2-Nonenal | 5.3 ± 0.7 | 34.9 ± 0.9 | 6.58 |
| Hexanal | 39.1 ± 3.9 | 243.3 ± 5.9 | 6.22 |
| Octanal | 17.5 ± 2.1 | 72.3 ± 3.2 | 4.13 |
| Betaine aldehyde | 41.1 ± 9.3 | 101.4 ± 20.6 | 2.47 |
| AASA | 169.0 ± 9.5 | 276.2 ± 32.5 | 1.63 |
| Benzaldehyde | 530.2 ± 86.3 | 125.2 ± 6.6 | 0.24 |
| Propanal | 647.4 ± 38.5 | 69.9 ± 0.9 | 0.11 |
| Glyceraldehyde | 7374.3 ± 117.2 | 174.0 ± 15.5 | 0.02 |
a Apparent Km and Vmax values were determined by fitting the data to the Michaelis-Menten equation. Vmax/Km represents aldehyde-oxidizing capacity (expressed as nmol of NADH produced/min/mg of protein/nmol of aldehyde/ml). Data represent mean ± S.E. from three separate experiments.
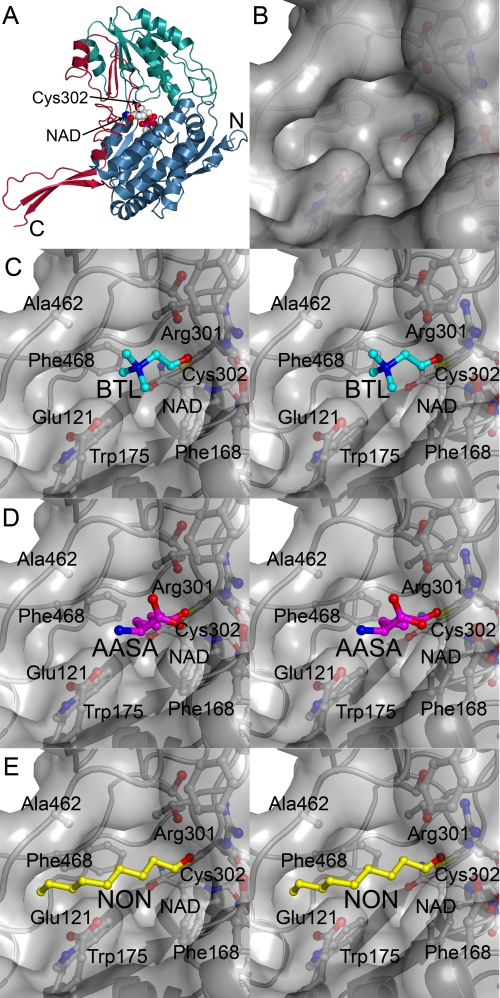
Crystal Structure of Human ALDH7A1
In order to elucidate how a wide range of aldehydes could be metabolized by human ALDH7A1, the protein was crystallized, and the structure was solved in the presence of NAD. Statistics for the final model appear in Table 2. The overall structure of human ALDH7A1 is similar to other ALDHs that have been structurally characterized. It consists of an N-terminal cofactor binding domain, a C-terminal catalytic domain, and an oligomerization domain composed from elements inserted into each of the larger domains (Fig. 2A). A well defined NAD occupies each active site, which resides at the interface of the two larger domains. The subunits were structurally very similar with root mean square deviations for α-carbons less than 0.4 Å in pairwise comparisons between individual chains and chain A. Although the active sites were mostly identical, Glu121 and Arg301 were more dynamic and were observed in alternate conformations in several subunits. A comparison with the ligand-bound structure of human ALDH2 (36) indicates that all NAD molecules in the current structure are in the extended hydride transfer conformation. The active site cysteine, Cys302, is positioned with the sulfur atom 2.7 Å from the NAD NC4 atom, and a crescent-shaped solvent-accessible pocket that is ∼10 Å deep and 12 Å wide extends from Cys302 to the surface (Fig. 2B). A ∼15-Å-long hydrophobic channel forms one end of the crescent, whereas the other side is polar and surrounded by Glu115, Glu121, and Arg301. The floor of this pocket is formed by Phe168, Cys302, and NAD. Structures of betaine aldehyde dehydrogenase complexed with betaine aldehyde (37) and hALDH2 complexed with the inhibitor daidzin (38) identify this region as the aldehyde binding site.
TABLE 2.
X-ray data collection and refinement statistics for ALDH7A1 (Protein Data Bank code 2J6L)
| Parameters | Values |
|---|---|
| Data collection | |
| Space group | C2 |
| Cell dimensions | |
| a, b, c (Å) | 155.29, 162.33, 159.02 |
| α, β, γ (degrees) | 90, 94.17, 90 |
| Resolutiona (Å) | 49.2–1.3 (1.4–1.3) |
| Rmergea | 0.090 (0.41) |
| I/σa (I) | 8.9 (1.6) |
| Completenessa (%) | 85.1 (44.0) |
| Redundancya | 3.3 (0.8) |
| Refinement | |
| Resolution (Å) | 50–1.3 |
| No. of reflections | 775,066 |
| Rwork/Rfree | 0.137/0.188 |
| No. of atoms | 36,106 |
| B-average (Å2) | 11.97 |
| Root mean square deviations | |
| Bond length (Å) | 0.015 |
| Bond angle (degrees) | 1.47 |
a Highest resolution shell shown in parentheses.
FIGURE 2.
Human ALDH7A1 crystal structure and docked substrates. A, α-carbon trace of human ALDH7A1 subunit A with the N-terminal cofactor binding domain (blue), the C-terminal catalytic domain (green), and the oligomerization domain (red). The NAD molecule is shown in spheres and the catalytic Cys302 is shown in a ball-and stick representation at the interface of the two larger domains. B, surface representation of active-site pocket. C–E are colored according to atom type with nitrogen blue, oxygen red, sulfur yellow, and protein carbon gray; stereoview close-up of betaine aldehyde (BTL; cyan carbons) (C), AASA (magenta carbons) (D), and nonanal (NON; yellow carbons) (E) docked into the active site. The orientation is the same as in A.
Docking Results
To gain an understanding of how various substrates may bind to human ALDH7A1, betaine aldehyde, AASA, and nonanal were docked into the active site (Fig. 2, C–E). The small and polar substrate betaine aldehyde can be accommodated in the central portion of the pocket where the positively charged amine has electrostatic interactions with Glu121 (3.94 Å). AASA is also predicted to bind in the polar core of the active site, where the amine can interact with Glu121 (3.0 Å) and the carboxylate forms electrostatic interactions with Arg301 (3.23 Å). In contrast, nonanal was placed with the aliphatic chain extending into the hydrophobic channel, where it forms van der Waals interactions with Trp175, Phe468, and Ala462. The terminal carbon of nonanal packs against the distal end of this hydrophobic binding site, which is delimited by the side chain of Ser480 and the main-chain carbonyl of Gly466. Trans-2-nonenal would probably be accommodated in this channel in a similar manner as nonanal, but longer aliphatic chains would not fit without introducing conformational strain due to the enclosed end of the pocket.
Tissue Distribution and Subcellular Localization of ALDH7A1
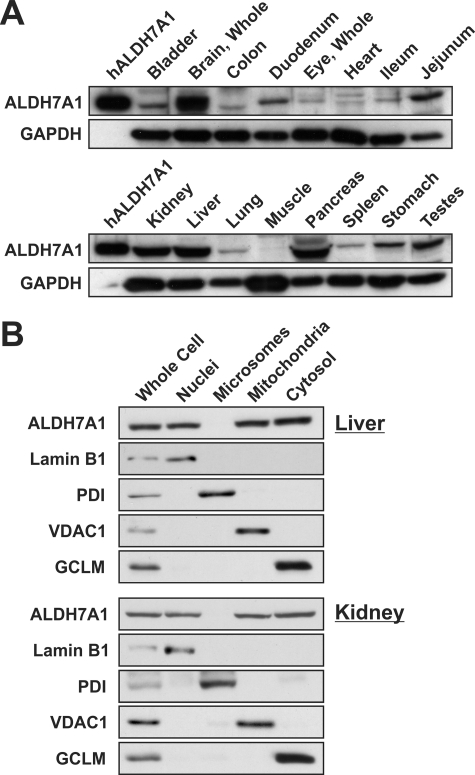
ALDH7A1 antibody specificity was assessed via Western blot analysis against recombinant ALDH7A1 as well as other closely related ALDH enzymes (data not shown). The antibody did not show cross-reactivity with recombinant human ALDH1A1, ALDH1B1, ALDH2, or ALDH3A1 protein. Tissue lysates from C57BL/6J/129/SvJ mice revealed highest ALDH7A1 expression in the liver, kidney, brain, and pancreas, followed by jejunum, duodenum, stomach, and testes (Fig. 3A). Low levels were observed in most other tissues evaluated, including eye, heart, bladder, colon, ileum, and lung. Subcellular fractionation of liver and kidney revealed ALDH7A1 localization in nuclei, mitochondria, and cytosol (Fig. 3B). Protein was not observed in microsomal fractions. Purity was verified by probing the blots with lamin B1, protein-disulfide isomerase, VDAC1, and GCLM antibodies for nuclear, microsomal, mitochondrial, and cytosolic fractions, respectively.
FIGURE 3.
Tissue and subcellular distribution of mouse ALDH7A1. Individual organs were isolated from adult C57BL/6J/129/SvJ mice and prepared for Western blot analysis of ALDH7A1 expression (as described under “Experimental Procedures”). A, tissue distribution of ALDH7A1. One hundred ng of purified human ALDH7A1 protein was loaded into lane 1 (left lane, hALDH7A1), and 30 μg of protein-soluble extract obtained from a variety of tissues was loaded into each additional lane. The same membranes were reprobed for glyceraldehyde-3-phosphate dehydrogenase (bottom panels) for loading correction. B, ALDH7A1 expression in subcellular fractions from liver and kidney was determined via Western blot analysis (as described under “Experimental Procedures”). Ten μg of whole cell or subcellular fraction lysate was loaded into each lane. Blots were probed with lamin B1 (Nuclei), protein-disulfide isomerase (Microsomes), VDAC1 (Mitochondria), and GCLM (Cytosol) to assess fraction purity.
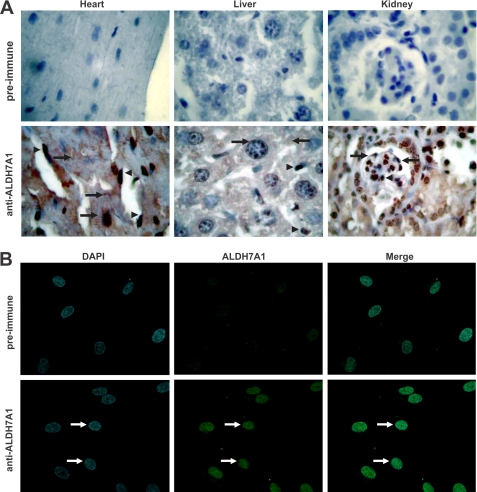
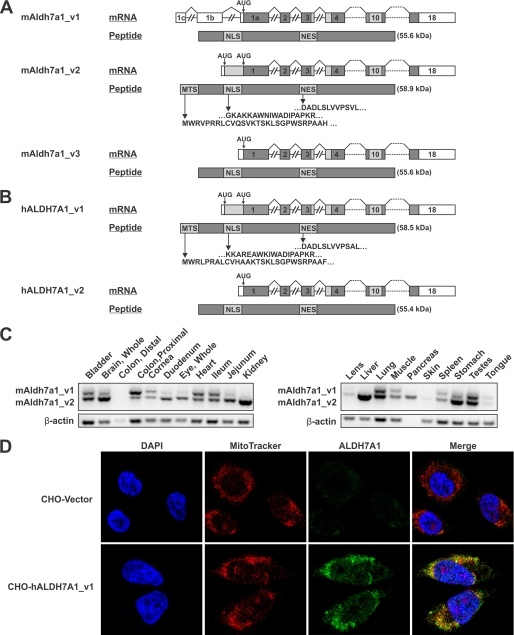
To further characterize ALDH7A1 distribution, immunohistochemical staining was performed on tissue sections from mouse heart, liver, and kidney. In the heart, ALDH7A1 staining was observed in the cytosol and nuclei of cardiomyocytes as well as the nuclei of cardiac endothelial cells (Fig. 4A). Liver sections also show pronounced nuclear localization in endothelial cells as well as intense cytosolic and nuclear staining in hepatocytes. ALDH7A1 in the kidney localizes to endothelial and podocyte cell nuclei. Cytoplasmic localization was also observed throughout the section. Little, if any, background staining was observed in tissue sections probed with preimmune serum. The intense nuclear staining in endothelial cells was reassessed using HUVEC in culture. Immunocytochemical staining with DAPI and ALDH7A1 antibody showed strong co-localization (when the images were merged), supporting a nuclear form of the protein (Fig. 4B). Using bioinformatics, we have identified putative nuclear localization signal and nuclear export signal in both mouse and human proteins (Fig. 5, A and B). These signals share high homology and map to accessible external α-helices on the ALDH7A1 crystal, structure supporting their potential role as targeting signals.
FIGURE 4.
ALDH7A1 immunochemical staining in mouse tissues and primary HUVEC. A, heart, liver, and kidney were isolated from C57BL/6J/129/SvJ adult mice. Five-μm sections (obtained and processed as described under “Experimental Procedures”) were incubated with preimmune serum (top) or human ALDH7A1 antibody (bottom), followed by streptavidin-conjugated goat anti-rabbit IgG secondary antibody. Representative staining patterns are shown at ×400 magnification. Arrowheads, intense nuclear staining in endothelial cells of all tissues. Additionally, strong ALDH7A1 staining was detected in the nuclei and cytosol of cardiomyocytes and hepatocytes as well as podocyte nuclei, in heart, liver, and kidney tissues sections, respectively (arrows). B, HUVEC grown in culture and probed with human ALDH7A1 antibody followed by fluorescein isothiocyanate-conjugated secondary antibody (green). Nuclei were stained with DAPI (blue). ALDH7A1 staining is predominantly nuclear (arrows). Co-localization was visualized by overlaying DAPI and ALDH7A1 images (Merge).
FIGURE 5.
Identification of mitochondrial ALDH7A1 transcript. A, diagram illustrating alternative splicing event at the 5′-end of the mouse Aldh7a1 gene, which gives rise to two splice variants, mAldh7a1_v1 and mAldh7a1_v2. Alternative splicing results in the removal of upstream AUG, which subsequently prevents translation of a mitochondrial localization signal. Another transcript, mAldh7a1_v3, lacks upstream exons present in mAldh7a1_v1 but utilizes the same start codon. B, the human ALDH7A1 gene contains two potential translation initiation sites. hALDH7A1_v1 and hALDH7A1_v2 have been sequenced and utilize different AUG start codons, resulting in either retention or removal of a mitochondrial targeting signal. Upstream AUG in both mAldh7a1_v2 and hALDH7A1_v1 are predicted to be non-optimal translation start sites, indicating potential use of a “leaky scanning” mechanism for translation initiation. To date, no homolog for mAldh7a1_v1 has been identified in humans. Both A and B also include predicted nuclear localization and nuclear export signals. Introns are not drawn to scale (MTS, mitochondrial targeting signal; NLS, nuclear localization signal; NES, nuclear export signal). C, RT-PCR of C57BL6 mouse tissues using splice variant-specific ALDH7A1 primers. Non-mitochondrial transcript is indicated by the 937 bp upper band. Mitochondrial transcript is identified by the 779 bp lower band. β-Actin primers were used as positive and loading controls. D, immunocytochemical staining of stable CHO cells transfected with mammalian expression vector comprising human ALDH7A1 containing the mitochondrial signal sequence (CHO-hALDH7A1_v1) or empty vector (CHO-Vector). Fixed cells were probed with polyclonal human ALDH7A1 antibody, followed by fluorescein isothiocyanate-conjugated secondary antibody (green). Mitochondria labeled with Invitrogen's MitoTracker® Orange CM-H2 TMRos probe (red). Nuclei were stained with DAPI (blue). The merged image denotes strong ALDH7A1 mitochondrial localization.
ALDH7A1 Gene Structure
The alignment of mouse transcripts indicated the presence of two splice variants, mALDH7A1_v1 and mALDH7A1_v2 (Fig. 5A). mALDH7A1_v1 included two additional non-coding upstream exons that increase the length of the 5′-untranslated region. Furthermore, the splicing event between exon 1b and exon 1a of mALDH7A1_v1 removed the upstream start codon utilized by mALDH7A1_v2. Translation initiation at this start codon results in the addition of a 28-amino acid mitochondrial signal sequence to mALDH7A1_v2. The alignments also indicated the presence of a third transcript, mALDH7A1_v3. This transcript appears similar to mALDH7A1_v2; however, the 5′ sequence is shorter, resulting in the use of the same downstream start codon as mALDH7A1_v1. Both variants, mALDH7A1_v1 and mALDH7A1_v3, produced identical proteins, and the transcripts differed only in their 5′-untranslated region sequences. mALDH7A1_v2 utilizes an alternate upstream start codon, resulting in the addition of a mitochondrial signal sequence to the amino terminus of the protein. To date, no human homologue has been identified for the mALDH7A1_v1 splice variant. The two human transcripts, hALDH7A1_v1 and hALDH7A1_v2, encode the mitochondrial and non-mitochondrial transcripts, respectively, and are homologous to mALDH7A1_v2 and mALDH7A1_v3 (Fig. 5B). Intron and exon lengths for both mouse and human transcripts can be found in Tables 3 and 4, respectively.
TABLE 3.
Comparison of mouse ALDH7A1 transcripts
Mouse exon and intron lengths were obtained from NCBI mouse build 37.1 locus NC_000084 (August 2008; strain C57BL/6J). An alternative splicing event occurs in mALDH7A1_v1, resulting in the use of a downstream start codon and subsequent loss of 28 amino acids form the N terminus of the protein. mALDH7A1_v2 contains an alternative upstream start codon in exon 1 leading to the addition of 28 amino acids to the amino terminus of the protein. mALDH7A1_v3 lacks the upstream exons found in mALDH7A1_v1 as well as the alternate upstream start codon found in mALDH7A1_v2.
| Number | mALDH7A1_v1 |
Number | mALDH7A1_v2 |
mALDH7A1_v3 |
|||
|---|---|---|---|---|---|---|---|
| Exon | Intron | Exon | Intron | Exon | Intron | ||
| bp | bp | bp | |||||
| 1c | 28 | 10,169 | |||||
| 1b | 193 | 278 | |||||
| 1a | 172 | 1203 | 1 | 228 | 1203 | 189 | 1203 |
| 2 | 54 | 1488 | 2 | ||||
| 3 | 66 | 7981 | 3 | ||||
| 4 | 81 | 740 | 4 | ||||
| 5 | 124 | 1716 | 5 | ||||
| 6 | 133 | 1498 | 6 | ||||
| 7 | 45 | 1861 | 7 | ||||
| 8 | 78 | 2642 | 8 | ||||
| 9 | 98 | 3482 | 9 | ||||
| 10 | 42 | 1402 | 10 | ||||
| 11 | 95 | 2929 | 11 | ||||
| 12 | 85 | 1595 | 12 | ||||
| 13 | 107 | 390 | 13 | ||||
| 14 | 117 | 172 | 14 | ||||
| 15 | 98 | 147 | 15 | ||||
| 16 | 74 | 3137 | 16 | ||||
| 17 | 76 | 1269 | 17 | ||||
| 18 | 260 | 18 | |||||
| Total | 2026 | 44,099 | 1861 | 33,652 | 1822 | 33,652 | |
TABLE 4.
Comparison of human ALDH7A1 transcripts
Human exon and intron lengths were obtained from NCBI human genome build 36 version 3 locus NC_000005 (October 2008). hALDH7A1_v1 transcript contains an alternative upstream start codon in exon 1 leading to the addition of 28 amino acids to the amino terminus of the protein. To date, no homolog corresponding to mALDH7A1_v1 has been identified in humans.
| Number | hALDH7A1_v1 |
hALDH7A1_v2 |
||
|---|---|---|---|---|
| Exon | Intron | Exon | Intron | |
| bp | bp | |||
| 1 | 242 | 1602 | 179 | 1602 |
| 2 | 54 | 621 | ||
| 3 | 66 | 8651 | ||
| 4 | 81 | 957 | ||
| 5 | 124 | 5639 | ||
| 6 | 133 | 1614 | ||
| 7 | 45 | 4560 | ||
| 8 | 78 | 2425 | ||
| 9 | 98 | 7134 | ||
| 10 | 42 | 1748 | ||
| 11 | 95 | 3224 | ||
| 12 | 85 | 1537 | ||
| 13 | 107 | 2149 | ||
| 14 | 117 | 1727 | ||
| 15 | 98 | 193 | ||
| 16 | 74 | 3529 | ||
| 17 | 76 | 1304 | ||
| 18 | 267 | |||
| Total | 1882 | 48,614 | 1819 | 48,614 |
Identification of Mitochondrial and Non-mitochondrial Variants of ALDH7A1
Mouse ALDH7A1 sequence alignments revealed a possible alternate start codon in the mALDH7A1_v2 transcript. Use of the alternate upstream start codon would result in the addition of 28 amino acids to the amino terminus of the translated protein. Bioinformatic screening identified with high probability that the additional sequence is a mitochondrial targeting signal. Corresponding mitochondrial and non-mitochondrial transcripts were also identified from sequenced human cDNA. Mouse transcript-specific RT-PCR results indicated that both transcripts were indeed expressed in mouse tissues and that the splice variants were differentially expressed in a tissue-specific manner (Fig. 5C). ALDH7A1 mRNA containing the mitochondrial signal was expressed in most tissues and was the predominant transcript in the liver, kidney, brain, duodenum, lens, jejunum, and pancreas. β-Actin primers were used as a positive control and to verify reverse transcription reaction efficiency. Furthermore, immunocytochemistry in CHO cells transfected with ΔpCEP4Δ-hALDH7A1_v1 (CHO-hALDH7A1_v1) confirmed mitochondrial localization of the ALDH7A1 protein. ALDH7A1 co-localized with the MitoTracker® stain, as indicated in the merged images, verifying the presence of a functional mitochondrial targeting signal on the 5′ sequence of the hALDH7A1_v1 transcript (Fig. 5D).
DISCUSSION
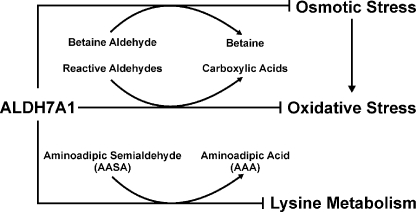
Herein, we report the novel finding that human ALDH7A1 plays an important role in protecting cells and tissues from hyperosmotic stress and suggest mechanisms by which such osmoprotection is likely to occur. CHO cells expressing ALDH7A1 were afforded significant protection from sucrose and sodium chloride-induced osmotic stress. The CHO-Vector cell line exhibited EC50 values of 301.4 and 156.2 mm for sucrose and NaCl, respectively. These EC50 values correspond to an overall increase in osmolarity of ∼300 mosm, suggesting that the two types of hyperosmotic stress affect the control cell line equally. Interestingly, the EC50 values obtained for sucrose- and NaCl-treated ALDH7A1-expressing cell lines were 380.2 and 228.6 mm, respectively, which correspond to increases in osmolarity of 380 and 457 mosm. In addition, ALDH7A1 knockdown experiments showed similar protective effects. The reduced expression of ALDH7A1 was associated with decreased cell viability after treatment with NaCl. These results indicate that ALDH7A1 significantly protects both cell lines but that the degree of protection is greater against NaCl. Inorganic ions, especially NaCl, at high concentrations are known to cause protein unfolding and perturb protein function and can result in enzyme inactivation (39, 40). Betaine belongs to the chemical class of osmolytes known as methylamines. Methylamines act as protein stabilizers and chemical chaperones by preventing protein misfolding and enhancing enzyme activity (41, 42). It is reasonable to postulate that the difference in protection may be due to betaine synthesis by ALDH7A1 directly counteracting NaCl-induced protein destabilization. It is also important to recognize that osmotic stress causes an increase in reactive oxygen and nitrogen species, leading to increased oxidative stress within the cell (43). Cellular protection against osmotic stress can be achieved through the generation organic osmolytes, including sorbital, inositol, taurine, betaine, and many others (34, 41), or by preventing the consequences of oxidative stress (11). Interestingly, we have also observed that both the mitochondrial and non-mitochondrial proteins significantly protect against oxidative stress in cells exposed to hydrogen peroxide (H2O2) or 4-hydroxynonenal (data not shown). Based on our biochemical and structural data provided herein, the role of human ALDH7A1 appears to be 3-fold (Fig. 6). First, ALDH7A1 metabolizes betaine aldehyde to betaine, an important cellular osmolyte and methyl donor. Second, the enzyme protects cells from oxidative stress by metabolizing a number of highly reactive LPO-derived aldehydes produced during oxidative stress. Finally, ALDH7A1 mediates lysine metabolism by converting AASA to the corresponding carboxylic acid, α-aminoadipic acid. As such, ALDH7A1 is a multifunctional enzyme that appears to exert important protective effects in mammalian cells.
FIGURE 6.
Proposed role of ALDH7A1 in the cell. ALDH7A1 protects cells and tissues from osmotic stress through the synthesis of the protective osmolyte betaine from betaine aldehyde. Furthermore, ALDH7A1 can directly metabolize a wide range of reactive aldehydes produced during lipid peroxidation, which potentiate oxidative stress within the cell. ALDH7A1 also plays a key role in lysine metabolism through the conversion of AASA to aminoadipic acid (AAA).
Betaine accumulation within cells counteracts extracellular hypertonicity without perturbing cell function even when at very high concentrations. Recent studies have shown that betaine serves as a chemical chaperone that prevents protein misfolding (42, 45). Moreover, the compound acts as a methyl group donor in the synthesis of methionine, which is, in turn, used to methylate DNA, protein, and lipids (46). Betaine aldehyde is primarily formed through the mitochondrial oxidation of choline in the liver and kidney (47, 48); hence, mitochondrial ALDH7A1 localization in liver and kidney is consistent with its role in betaine synthesis. To date, ALDH9A1 is the only other ALDH enzyme known to metabolize this substrate (1). ALDH9A1 purified from human liver exhibited a Km for betaine aldehyde of 260 μm with a corresponding catalytic efficiency value (Vmax/Km) equal to 0.03 (49). Human ALDH7A1 displayed a Km of 41 μm and Vmax/Km of 2.5, suggesting that ALDH7A1 may be the predominant betaine aldehyde metabolizing enzyme in mammals. Betaine concentrations in human serum fall between 20 and 70 μm (34), further supporting ALDH7A1, not ALDH9A1, as the major betaine aldehyde-metabolizing enzyme. Human and seabream ALDH7A1 enzymes have been described previously as possessing similar kinetic properties. However, the seabream ALDH7A1 protein reportedly does not exhibit any detectable activity toward betaine aldehyde (51), indicating species differences in the substrate selectivity of this enzyme. The metabolism of betaine aldehyde strongly supports a role for ALDH7A1 in protecting human cells against osmotic stress.
The metabolism of propanal and the aromatic aldehyde, benzaldehyde, by recombinant hALDH7A1 proved comparable with recombinant sbALDH7A1 (51). The similar kinetic characteristics of the two enzymes were no surprise, given their 83.3% homology. The efficient metabolism of hexanal, octanal, nonanal, and trans-2-nonenal suggest a significant role for hALDH7A1 in the metabolism of lipid peroxidation products created as a result of oxidative stress. These aldehydes are highly reactive and readily damage proteins, DNA, and other cellular constituents, making their removal a priority (52). ALDH7A1 joins a number of other ALDH enzymes known to effectively remove LPO-derived aldehydes, including ALDH1A1, ALDH1B1, ALDH2, ALDH3A1, and ALDH3B1 (1). This metabolic redundancy emphasizes the importance of the expeditious removal and detoxification of aldehydes in the cell.
Another important function of ALDH7A1 is its role in lysine catabolism through the oxidation of AASA. The recombinant human protein had a calculated Km of 169 μm for AASA, which is comparable with the value obtained from human liver homogenates (181 μm) and 2.5-fold higher than that of recombinant seabream ALDH7A1 (67 μm) (53, 54). The Km was also substantially higher than the value determined for ALDH7A1 partially purified from rat liver cytoplasm (20 μm) (12). Lysine catabolism is required for maintaining cellular nitrogen pools and also serves as a source for ketone bodies, an important energy resource in the heart and brain. Catabolism within mammalian cells can proceed through two pathways, the mitochondrial saccharopine pathway or the peroxisomal pipecolic acid pathway, both of which converge on ALDH7A1 through the production of AASA (55–57). Mutations in ALDH7A1 shunt both catabolic pathways and result in the buildup of AASA and pipecolic acid observed in PDE patients (13). Considering the inherent reactivity of aldehydes, elevated AASA levels in the brains of PDE patients may contribute to the observed cerebral atrophy (58, 59). ASAA is known to cross-link proteins (60), is produced during oxidative stress, and is a major carbonyl found in oxidized proteins (61). Recent evidence suggests that AASA, along with γ-glutamic semialdehyde, makes up 55–100% of total carbonyls formed during metal-catalyzed oxidation (61). Under these conditions, the levels of AASA and γ-glutamic semialdehyde are much higher than other carbonyls commonly associated with oxidative damage, such as 4-hydroxynonenal, MDA, and acrolein. Increased levels of AASA are observed in a number of pathologies, including diabetes, renal failure, sepsis, and aging (62, 63). The saccharopine pathway is the predominant pathway utilized in higher eukaryotes and is important for lysine degradation in the liver and kidney. The pipecolic acid pathway is the major route for lysine degradation in the brain (44, 64, 65). The ALDH7A1 tissue distribution described herein closely correlates with expected expression patterns based on regions of high lysine degradation, most notably high expression in the kidney, liver, and brain. The pipecolic acid and saccharopine pathways occur in different subcellular locations. Our subcellular fractionation and immunocytochemical results indicated specific subcellular localization of ALDH7A1 that paralleled those associated with the pathways. Taken together, these data suggest that cytosolic and mitochondrial ALDH7A1 may participate in the pipecolic acid and saccharopine pathways, respectively. Predominant expression of mitochondrial mALDH7A1_v2 mRNA in the liver and kidney lends further support for its role in the saccharopine pathway that is extensively utilized in these tissues. Both mitochondrial and cytosol transcripts were expressed in the brain where the pipecolic acid pathway is predominant.
For the first time, we have also shown nuclear localization of ALDH7A1 using subcellular fractionation, immunohistochemistry, and immunocytochemistry. Interestingly, ALDH7A1 expression is predominantly nuclear in endothelial cells. To date, nothing is known about the function of ALDH7A1 in the nucleus. Bioinformatic screening identified a single nuclear localization signal and a nuclear export signal (Fig. 5, A and B), indicating that the protein may shuttle between subcellular compartments. The predicted human nuclear localization signal and nuclear export signal mapped to accessible external α-helices on the ALDH7A1 crystal structure, supporting their potential role as targeting signals. Based on the broad substrate specificity of ALDH7A1 and lack of lysine degradation or betaine synthesis in the nucleus, it is possible that the enzyme's primary function is to metabolize substrates other than AASA or betaine aldehyde. Furthermore, the possibility cannot be excluded that the nuclear protein may have other non-catalytic functions.
In conclusion, we have shown that human ALDH7A1 attenuates hyperosmotic stress-induced apoptotic cell death, possibly through the synthesis of the osmolyte betaine. Results also support the involvement of ALDH7A1 in other vital physiological processes, including detoxification of reactive aldehydes generated during oxidative stress and lysine catabolism. This research greatly expands upon our understanding of mammalian ALDH7A1 and underscores the enzymes importance in maintaining cell homeostasis.
Acknowledgments
We thank Drs. David Thompson and Ying Chen for critical reading and discussion of the manuscript. Help in purification, crystallization, and refinement by Kunde Guo and Gabor Bunkoczi is gratefully acknowledged.
This work was supported, in whole or in part, by National Institutes of Health Grants EY11490 and EY17963. The Structural Genomics Consortium is a registered charity (number 1097737) that receives funds from the Canadian Institutes for Health Research, the Canadian Foundation for Innovation, Genome Canada through the Ontario Genomics Institute, GlaxoSmithKline, Karolinska Institutet, the Knut and Alice Wallenberg Foundation, the Ontario Innovation Trust, the Ontario Ministry for Research and Innovation, Merck & Co., Inc., the Novartis Research Foundation, the Swedish Agency for Innovation Systems, the Swedish Foundation for Strategic Research, and the Wellcome Trust. This work was also supported by the Oxford BIHR Biomedical Research Unit. Portions of this work were presented at the 48th Annual Meeting of the Society of Toxicology, Baltimore, MD, March 2009 (50).
The atomic coordinates and structure factors (code 2J6L) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- ALDH
- aldehyde dehydrogenase
- MALDI-TOF
- matrix-assisted laser desorption ionization time-of-flight
- Bistris propane
- 1,3-bis[tris(hydroxymethyl)methylamino]propane
- PDE
- pyridoxine-dependent epilepsy
- CHO
- Chinese hamster ovary
- HUVEC
- human umbilical vein endothelial cells
- DAPI
- 4′,6-diamidino-2-phenylindole
- RT
- reverse transcription
- siRNA
- small interfering RNA
- AASA
- aminoadipate semialdehyde
- MTT
- 3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide.
REFERENCES
- 1.Marchitti S. A., Brocker C., Stagos D., Vasiliou V. (2008) Expert Opin. Drug Metab. Toxicol. 4, 697–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee P., Kuhl W., Gelbart T., Kamimura T., West C., Beutler E. (1994) Genomics 21, 371–378 [DOI] [PubMed] [Google Scholar]
- 3.Malik H. S., Henikoff S. (2003) Nat. Struct. Biol. 10, 882–891 [DOI] [PubMed] [Google Scholar]
- 4.Guerrero F. D., Jones J. T., Mullet J. E. (1990) Plant Mol. Biol. 15, 11–26 [DOI] [PubMed] [Google Scholar]
- 5.Stroeher V. L., Boothe J. G., Good A. G. (1995) Plant Mol. Biol. 27, 541–551 [DOI] [PubMed] [Google Scholar]
- 6.Valente M. A., Faria J. A., Soares-Ramos J. R., Reis P. A., Pinheiro G. L., Piovesan N. D., Morais A. T., Menezes C. C., Cano M. A., Fietto L. G., Loureiro M. E., Aragão F. J., Fontes E. P. (2009) J. Exp. Bot. 60, 533–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodrigues S. M., Andrade M. O., Gomes A. P., Damatta F. M., Baracat-Pereira M. C., Fontes E. P. (2006) J. Exp. Bot. 57, 1909–1918 [DOI] [PubMed] [Google Scholar]
- 8.Shin J. H., Kim S. R., An G. (2009) Plant Physiol. 149, 905–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kültz D., Chakravarty D. (2001) Comp. Biochem. Physiol. A Mol. Integr. Physiol. 130, 421–428 [DOI] [PubMed] [Google Scholar]
- 10.Alfieri R. R., Petronini P. G. (2007) Pflugers Arch. 454, 173–185 [DOI] [PubMed] [Google Scholar]
- 11.Burg M. B., Ferraris J. D., Dmitrieva N. I. (2007) Physiol. Rev. 87, 1441–1474 [DOI] [PubMed] [Google Scholar]
- 12.Tsai C. H., Henderson L. M. (1974) J. Biol. Chem. 249, 5790–5792 [PubMed] [Google Scholar]
- 13.Mills P. B., Struys E., Jakobs C., Plecko B., Baxter P., Baumgartner M., Willemsen M. A., Omran H., Tacke U., Uhlenberg B., Weschke B., Clayton P. T. (2006) Nat. Med. 12, 307–309 [DOI] [PubMed] [Google Scholar]
- 14.Gallagher R. C., Van Hove J. L., Scharer G., Hyland K., Plecko B., Waters P. J., Mercimek-Mahmutoglu S., Stockler-Ipsiroglu S., Salomons G. S., Rosenberg E. H., Struys E. A., Jakobs C. (2009) Ann. Neurol. 65, 550–556 [DOI] [PubMed] [Google Scholar]
- 15.Chavakis T., Kanse S. M., Yutzy B., Lijnen H. R., Preissner K. T. (1998) Blood 91, 2305–2312 [PubMed] [Google Scholar]
- 16.Andreucci M., Fuiano G., Presta P., Lucisano G., Leone F., Fuiano L., Bisesti V., Esposito P., Russo D., Memoli B., Faga T., Michael A. (2009) Cell Prolif. 42, 554–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pappa A., Chen C., Koutalos Y., Townsend A. J., Vasiliou V. (2003) Free Radic. Biol. Med. 34, 1178–1189 [DOI] [PubMed] [Google Scholar]
- 18.Lassen N., Pappa A., Black W. J., Jester J. V., Day B. J., Min E., Vasiliou V. (2006) Free Radic. Biol. Med. 41, 1459–1469 [DOI] [PubMed] [Google Scholar]
- 19.Skvorak A. B., Robertson N. G., Yin Y., Weremowicz S., Her H., Bieber F. R., Beisel K. W., Lynch E. D., Beier D. R., Morton C. C. (1997) Genomics 46, 191–199 [DOI] [PubMed] [Google Scholar]
- 20.Bunting K. D., Townsend A. J. (1996) J. Biol. Chem. 271, 11884–11890 [DOI] [PubMed] [Google Scholar]
- 21.Manzer R., Qamar L., Estey T., Pappa A., Petersen D. R., Vasiliou V. (2003) DNA Cell Biol. 22, 329–338 [DOI] [PubMed] [Google Scholar]
- 22.Pappa A., Estey T., Manzer R., Brown D., Vasiliou V. (2003) Biochem. J. 376, 615–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pappa A., Brown D., Koutalos Y., DeGregori J., White C., Vasiliou V. (2005) J. Biol. Chem. 280, 27998–28006 [DOI] [PubMed] [Google Scholar]
- 24.Jia S., Omelchenko M., Garland D., Vasiliou V., Kanungo J., Spencer M., Wolf Y., Koonin E., Piatigorsky J. (2007) FASEB J. 21, 3318–3328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marchitti S. A., Orlicky D. J., Vasiliou V. (2007) Biochem. Biophys. Res. Commun. 356, 792–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lassen N., Estey T., Tanguay R. L., Pappa A., Reimers M. J., Vasiliou V. (2005) Drug Metab. Dispos. 33, 649–656 [DOI] [PubMed] [Google Scholar]
- 27.Schönbrunn E., Eschenburg S., Shuttleworth W. A., Schloss J. V., Amrhein N., Evans J. N., Kabsch W. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 1376–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Storoni L. C., McCoy A. J., Read R. J. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 432–438 [DOI] [PubMed] [Google Scholar]
- 29.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 30.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 31.Jones G., Willett P., Glen R. C., Leach A. R., Taylor R. (1997) J. Mol. Biol. 267, 727–748 [DOI] [PubMed] [Google Scholar]
- 32.Singh R. P., Sharma G., Dhanalakshmi S., Agarwal C., Agarwal R. (2003) Cancer Epidemiol. Biomarkers Prev. 12, 933–939 [PubMed] [Google Scholar]
- 33.Stewart B. J., Roede J. R., Doorn J. A., Petersen D. R. (2009) Biochim. Biophys. Acta 1791, 772–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Craig S. A. (2004) Am. J. Clin. Nutr. 80, 539–549 [DOI] [PubMed] [Google Scholar]
- 35.Esterbauer H., Zollner H. (1989) Free Radic. Biol. Med. 7, 197–203 [DOI] [PubMed] [Google Scholar]
- 36.Perez-Miller S. J., Hurley T. D. (2003) Biochemistry 42, 7100–7109 [DOI] [PubMed] [Google Scholar]
- 37.Gruez A., Roig-Zamboni V., Grisel S., Salomoni A., Valencia C., Campanacci V., Tegoni M., Cambillau C. (2004) J. Mol. Biol. 343, 29–41 [DOI] [PubMed] [Google Scholar]
- 38.Lowe E. D., Gao G. Y., Johnson L. N., Keung W. M. (2008) J. Med. Chem. 51, 4482–4487 [DOI] [PubMed] [Google Scholar]
- 39.Yancey P. H., Clark M. E., Hand S. C., Bowlus R. D., Somero G. N. (1982) Science 217, 1214–1222 [DOI] [PubMed] [Google Scholar]
- 40.Nosek T. M., Andrews M. A. (1998) Pflugers Arch. 435, 394–401 [DOI] [PubMed] [Google Scholar]
- 41.Yancey P. H. (2005) J. Exp. Biol. 208, 2819–2830 [DOI] [PubMed] [Google Scholar]
- 42.de Marco A., Vigh L., Diamant S., Goloubinoff P. (2005) Cell Stress Chaperones 10, 329–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schliess F., Görg B., Häussinger D. (2006) Biol. Chem. 387, 1363–1370 [DOI] [PubMed] [Google Scholar]
- 44.Chang Y. F. (1982) Neurochem. Res. 7, 577–588 [DOI] [PubMed] [Google Scholar]
- 45.Kharbanda K. K., Mailliard M. E., Baldwin C. R., Sorrell M. F., Tuma D. J. (2007) J. Hepatol. 46, 1119–1125 [DOI] [PubMed] [Google Scholar]
- 46.Ueland P. M., Holm P. I., Hustad S. (2005) Clin. Chem. Lab. Med. 43, 1069–1075 [DOI] [PubMed] [Google Scholar]
- 47.Li Z., Vance D. E. (2008) J. Lipid Res. 49, 1187–1194 [DOI] [PubMed] [Google Scholar]
- 48.Izaguirre G., Kikonyogo A., Pietruszko R. (1997) Comp. Biochem. Physiol. B Biochem. Mol. Biol. 118, 59–64 [DOI] [PubMed] [Google Scholar]
- 49.Chern M. K., Gage D. A., Pietruszko R. (2000) Biochem. Pharmacol. 60, 1629–1637 [DOI] [PubMed] [Google Scholar]
- 50.Brocker C., Cantore M., Lassen N., Pappa A., Vasiliou V. (2009) Toxicologist 108, 1556 [Google Scholar]
- 51.Tang W. K., Chan C. B., Cheng C. H., Fong W. P. (2005) FEBS Lett. 579, 3759–3764 [DOI] [PubMed] [Google Scholar]
- 52.Comporti M. (1998) Free Radic. Res. 28, 623–635 [DOI] [PubMed] [Google Scholar]
- 53.Chang Y. F., Ghosh P., Rao V. V. (1990) Biochim. Biophys. Acta 1038, 300–305 [DOI] [PubMed] [Google Scholar]
- 54.Tang W. K., Wong K. B., Lam Y. M., Cha S. S., Cheng C. H., Fong W. P. (2008) FEBS Lett. 582, 3090–3096 [DOI] [PubMed] [Google Scholar]
- 55.IJlst L., de Kromme I., Oostheim W., Wanders R. J. (2000) Biochem. Biophys. Res. Commun. 270, 1101–1105 [DOI] [PubMed] [Google Scholar]
- 56.Noda C., Ichihara A. (1978) Biochim. Biophys. Acta 525, 307–313 [DOI] [PubMed] [Google Scholar]
- 57.Rao V. V., Chang Y. F. (1990) Biochim. Biophys. Acta 1038, 295–299 [DOI] [PubMed] [Google Scholar]
- 58.Plecko B., Paul K., Paschke E., Stoeckler-Ipsiroglu S., Struys E., Jakobs C., Hartmann H., Luecke T., di Capua M., Korenke C., Hikel C., Reutershahn E., Freilinger M., Baumeister F., Bosch F., Erwa W. (2007) Hum. Mutat. 28, 19–26 [DOI] [PubMed] [Google Scholar]
- 59.O'Brien P. J., Siraki A. G., Shangari N. (2005) Crit. Rev. Toxicol. 35, 609–662 [DOI] [PubMed] [Google Scholar]
- 60.Siegel R. C. (1979) Int. Rev. Connect. Tissue Res. 8, 73–118 [DOI] [PubMed] [Google Scholar]
- 61.Akagawa M., Sasaki D., Ishii Y., Kurota Y., Yotsu-Yamashita M., Uchida K., Suyama K. (2006) Chem. Res. Toxicol. 19, 1059–1065 [DOI] [PubMed] [Google Scholar]
- 62.Akagawa M., Sasaki T., Suyama K. (2002) Eur. J. Biochem. 269, 5451–5458 [DOI] [PubMed] [Google Scholar]
- 63.Sell D. R., Strauch C. M., Shen W., Monnier V. M. (2007) Biochem. J. 404, 269–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hutzler J., Dancis J. (1968) Biochim. Biophys. Acta 158, 62–69 [DOI] [PubMed] [Google Scholar]
- 65.Chang Y. F. (1976) Biochem. Biophys. Res. Commun. 69, 174–180 [DOI] [PubMed] [Google Scholar]