Abstract
The ETS protein TEL, a transcriptional repressor, contains a PNT domain that, as an isolated fragment in vitro, self-associates to form a head-to-tail polymer. How such polymerization might affect the DNA-binding properties of full-length TEL is unclear. Here we report that monomeric TEL binds to a consensus ETS site with unusually low affinity (Kd = 2.8 × 10−8 m). A deletion analysis demonstrated that the low affinity was caused by a C-terminal inhibitory domain (CID) that attenuates DNA binding by ∼10-fold. An NMR spectroscopically derived structure of a TEL fragment, deposited in the Protein Data Bank, revealed that the CID consists of two α-helices, one of which appears to block the DNA binding surface of the TEL ETS domain. Based on this structure, we substituted two conserved glutamic acids (Glu-431 and Glu-434) with alanines and found that this activated DNA binding and enhanced trypsin sensitivity in the CID. We propose that TEL displays a conformational equilibrium between inhibited and activated states and that electrostatic interactions involving these negatively charged residues play a role in stabilizing the inhibited conformation. Using a TEL dimer as a model polymer, we show that self-association facilitates cooperative binding to DNA. Cooperativity was observed on DNA duplexes containing tandem consensus ETS sites at variable spacing and orientations, suggesting flexibility in the region of TEL linking its self-associating PNT domain and DNA-binding ETS domain. We speculate that TEL compensates for the low affinity, which is caused by autoinhibition, by binding to DNA as a cooperative polymer.
Keywords: DNA-binding Protein, Ets Family Transcription Factor, Protein-DNA Interaction, Transcription Factors, Transcription Repressor, Autoinhibition, Polymerization
Introduction
The ETS transcription factors play roles in normal cellular processes as well as a variety of human malignancies. For example, chromosome rearrangements at specific ets loci are associated with Ewing sarcoma (1), prostate cancer (2), and certain leukemias. Specifically, the human chromosome translocation t(12;21), linked with B-cell acute lymphoblastic leukemia, encodes a TEL/AML1 fusion protein (3, 4). This oncogenic fusion retains the PNT (or SAM) domain of the ETS protein TEL (or ETV6) and the DNA binding domain of RUNX1 (or AML1). The PNT domain of TEL is implicated in transcriptional repression (5–9). Thus, it is postulated that in the aberrant, oncogenic context, the TEL PNT domain causes repression of target genes selected by the RUNX1 DNA binding domain (10, 11). Despite these significant biological phenomena, the DNA-binding properties of TEL and the TEL/AML1 fusion protein are not understood, nor are there any well characterized transcriptional targets.
TEL, one of the 27 ETS transcription factors encoded by the human genome, binds DNA through its highly conserved ETS domain. All ETS proteins recognize a consensus 5′-GGAA/T-3′ motif within the context of a 9- to 10-bp DNA sequence (12, 13). The conservation of DNA-binding properties among ETS proteins necessitates additional mechanisms that can regulate selection of specific transcriptional targets within biological contexts. Heterotypic protein partnerships with non-ETS proteins that stabilize ternary complexes are found frequently and help provide such additional DNA-binding specificity. For example, SRF associates with the ETS proteins ELK-1, ELK-4 (SAP1), or ELK-3 (NET) in ternary complexes (14), ETS protein PU.1 binds DNA with IRFx (Pip) (15), and Ets1 partners with RUNX1 on DNA (16–20).
A possible uniqueness of the DNA-binding properties of TEL comes from the biophysical studies indicating that its PNT domain can form a very stable head-to-tail polymer (21–23), and, thus most likely, the full-length protein also polymerizes (see Fig. 1). We speculated that the DNA binding mode of full-length TEL may be affected by self-association, enabling the interaction with multiple ETS binding sites. The PNT domain retained in the TEL/AML1 fusion protein may similarly affect its DNA-binding properties. Although several cell-based assays suggest that wild-type TEL self-associates (24, 25), whether or not the polymeric state exists in full-length, wild-type TEL has not been shown definitively in vitro by biochemical or biophysical approaches. In direct relevance to this report, no DNA binding studies have determined how self-association of TEL might impact its recognition of promoter or enhancer elements.
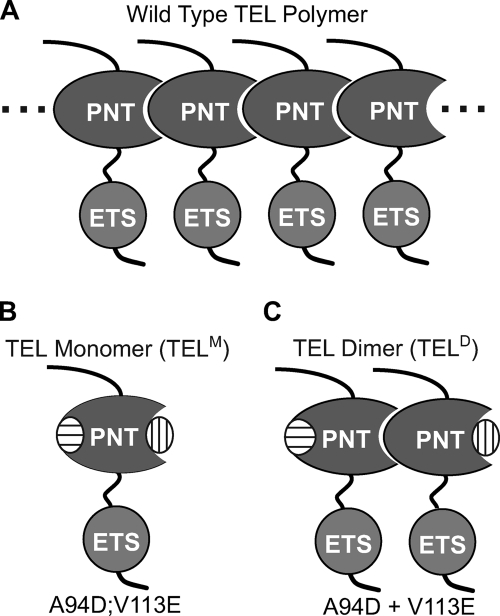
FIGURE 1.
Engineering of TEL monomeric and dimeric configurations. A, proposed model of wild-type TEL forming open-ended polymers via head-to-tail PNT domain interactions. The ETS domain, which mediates DNA binding, is also shown attached by a proposed flexible linker (this report). B, two point mutations (A94D, oval with horizontal lines, or V113E, oval with vertical lines) in the PNT domain block polymerization by disrupting one of the two surfaces necessary for subunit interaction (21). TEL species bearing both point mutations are monomeric, denoted here TELM. C, a combination of the two mutant proteins each bearing a single point mutation yields a stable dimer, denoted here TELD.
In this study, we demonstrated that TEL DNA binding is strongly repressed by an autoinhibitory mechanism that appears to involve steric blockage of the monomeric ETS domain DNA-binding interface by a flanking C-terminal helix. A conformational change of this inhibitory region is likely necessary for DNA binding. Next, using a dimeric TEL variant generated with PNT domain point mutations, we demonstrated that dimerization of TEL facilitates cooperative DNA binding. We also defined the spacing and orientation requirements of tandem ETS sites for this binding. These results demonstrated that self-association compensates for the low affinity of the autoinhibited TEL monomer. Our findings can predict promoter and enhancer architecture of downstream targets of TEL, several highly related ETS proteins, and the TEL/AML1 fusion protein, which all potentially can self-associate through their respective PNT domains.
EXPERIMENTAL PROCEDURES
Expression Plasmids
Murine TEL cDNA was cloned into the bacterial expression vector pET28B (Novagen) and used for generation of mutant versions (A94D, V113E, and A94D/V113E) by site-directed mutagenesis. The pET-based pAED vector, which carries a high copy ori (26), was used for cloning and expression of the deletion species ΔC436, ΔC426, ΔN127, ΔN331, ΔN334:ΔC436 (wild-type, E431A, E434A, and E431A/E434A), and ΔN331:ΔC426. Cloning by PCR-based strategies used the primer pairs listed in supplemental Table 1. The PCR-generated fragments were inserted into the NdeI and KpnI sites of the pAED4 vector to enable expression of fragments of TEL with the only non-TEL residue being an N-terminal Met.
Expression and Purification of TEL Species
pET-based vectors were used for inducible expression in Escherichia coli BL21(λDE3). Cells were grown at 37 °C to an A600 of 0.8, and induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside for 2 h. The cell pellet was resuspended in 50 mm Tris, pH 7.9, 1 m NaCl, 1 mm EDTA, 1 mm DTT,4 and 1 mm phenylmethylsulfonyl fluoride, then lysed by sonication. The cleared lysate was subjected to an overnight dialysis at 4 °C into 20 mm sodium citrate, pH 5.3, 150 mm KCl, 1 mm EDTA, 1 mm DTT, 0.2 mm phenylmethylsulfonyl fluoride, and 10% glycerol. The recombinant protein was then purified to near homogeneity using conventional chromatography. Briefly, the TEL species passed through a DEAE column, bound to an S-Sepharose column in the 150 mm KCl buffer (above), and then eluted at ∼500 mm KCl. After pooling appropriate fractions, the protein was concentrated and further purified on a Superdex 75 column (20 mm sodium citrate, pH 5.3, 500 mm KCl, 1 mm EDTA, 1 mm DTT, and 10% glycerol). Purified proteins were stored at −80 °C in 50-μl aliquots for one-time use. TELD preparations were made by mixing equal molar amounts of two mutant forms.
DNA Duplexes for EMSAs
To generate the DNA duplex used to investigate autoinhibition, complementary oligonucleotides 27 bases in length were designed such that a 9-bp, high affinity consensus ETS binding site (underlined) was positioned in the middle of a 23-bp double-stranded duplex: 5′-TCGACGGCCAAGCCGGAAGTGAGTGCC-3′ (top strand), 5′-TCGAGGCACTCACTTCCGGCTTGGCCG-3′ (bottom strand). To generate DNA duplexes used to test TEL binding to direct and inverted binding site repeats the following oligonucleotides were used for the direct repeat and inverted repeats, respectively: 5′-TCGACGTGTGCGCCTTAAGTGTACTTCCGGCCGTGTTACTTCCGGCATGCCGG-3′ (top strand) and 5′-TCGACGTGTGCGCCGGAAGTGTACTTCCGGCCGTGTTACTTAAGGCATGCCGG-3′ (top strand). End labeling was performed using an equimolar mixture of the two oligonucleotides, T4 polynucleotide kinase (Invitrogen) and [γ-32P]ATP. The oligonucleotides were then annealed by boiling for 5 min, followed by slow cooling to room temperature.
DNA duplexes used to test TEL binding to a direct ETS binding site repeat spaced by 54, 65, 76, and 108 bp (or 5, 6, 7, and 10 helical turns, respectively) were generated using a two-step cloning process. First, complimentary oligonucleotides were designed to contain a single ETS binding site (underlined) and no additional GGA sequences: 5′-CACGCCGCATGTATGTAGCCTGTTGCTAGCTGCCGGAAGTAATACGTAACGCAGCTATTGCTACTAACTATTGTGCTGCA-3′ (top strand). Once annealed, the DNA duplex was digested with SacI and PstI and inserted into the pKS Bluescript vector to generate pKS-EBS1. To create the second binding site in the DNA duplex, a subsequent round of cloning was conducted. As before, complimentary oligonucleotides were designed to contain a single ETS binding site and no additional GGA sequences (supplemental Table 2). Once annealed, the DNA duplexes were digested with PstI (5′) and KpnI (3′) and inserted into the pKS-EBS1 to generate a series of pKS-EBS2 vectors with appropriately spaced binding sites. After the second round of cloning, DNA duplexes were radiolabeled by PCR using the following primers: 5′-GAGCTCACGCCGCATGTATGT-3′ (forward) and 5′-GGTACCGTTAGCATTAAGCTA-3′ (reverse). Radiolabeled PCR products were gel-purified.
EMSAs
EMSAs were performed as previously described (27). In brief, equilibrium binding conditions were set up with a series of protein dilutions (10−13 to 10−7 m) mixed with radiolabeled DNA duplexes (2.5 × 10−12 m) and incubated at room temperature for 1 h to reach equilibrium. The aliquots were then loaded onto a running 6% native gel, which was subsequently dried and analyzed by phosphorimaging. To determine equilibrium dissociation constants (Kd), a nonlinear least squares fitting of the free protein concentration [P] versus the fraction of DNA bound ([PD]/[Dt]) to the equation, [PD]/[Dt] = 1/(1 + (Kd/[P])), was conducted, using KaleidaGraph (v. 3.51, Synergy Software). Unbound [P] is assumed to be the total [Pt] due to the excess of protein in all reactions. This approach was not suitable for determining the affinity of the cooperative TELD dimer, because the dimerization equilibrium constants for the full-length protein in its free and DNA-bound forms were not measured.
Dissociation Rate Experiments
Dissociation rate experiments were conducted as previously described (28). Binding reactions consisting of protein (10−8 m) and radiolabeled DNA duplex (10−10 m) were incubated at room temperature for ∼1 h. A control aliquot was removed and mixed with 1/10 volume of EMSA buffer (25 mm Tris, pH 7.9, 1 mm EDTA, 1 mm DTT, 10% glycerol, 6 mm MgCl2, 50 mm KCl, 0.1 μg/μl bovine serum albumin, and 50 ng/μl poly(dI-dC)). To the remaining reaction mixture, a 1000-fold excess unlabeled 27-bp DNA duplex containing a single ETS binding site in 1/10 volume of EMSA buffer was added. After brief vortexing, aliquots were removed at timed intervals and immediately loaded onto a running 6% native gel. The ratio of the shifted signal, quantified by phosphorimaging, at each time point to the signal in the control lane was presented as the fraction of original complex remaining at time t, ([PDt]/[PD0]). The dissociation rate constant, k−1 (and corresponding half-life t½ = ln(2)/k−1), was determined by least-squares analyses (KaleidaGraph) of the equation, ln([PDt]/[PD0]) = −k−1t.
Limited Proteolysis
TEL species (30 μm) ΔN334:ΔC436 and ΔN334:ΔC436 (E431A/E434A) were digested for 2 min at room temperature with increasing amounts of trypsin (2–500 ng) in 25 mm Tris, pH 8.8, 175 mm NaCl, 10 mm CaCl2, and 1 mm DTT. The reactions were stopped by adding SDS sample buffer, followed by boiling for at least 5 min. Reaction mixtures were electrophoresed on 15% SDS gels and visualized by Coomassie staining. For analysis by electrospray ionization/mass spectrometry (ESI/MS), TEL species ΔN334:ΔC436, and ΔN334:ΔC436 (E431A and E434A) were digested with 50 ng of trypsin under conditions described above, and the reactions were stopped by the addition of 1% trifluoroacetic acid.
RESULTS
Monomeric TEL Binds DNA with Low Affinity
To obtain a soluble form of full-length TEL we exploited two point mutations, A94D and V113E, which are well known to disrupt polymerization of the PNT domain (21) (Fig. 1). Recombinant TEL with either single or double substitutions displayed a dramatic increase in solubility relative to native TEL, which otherwise was highly insoluble when expressed in E. coli. As expected, the double mutant (A94D/V113E), termed TELM, appeared to be monomeric based on its chromatographic properties (supplemental Fig. S1). These findings suggested that wild-type, full-length TEL has self-association properties similar to the well characterized PNT domain-only fragment and that the monomeric TELM species was suitable for further analysis.
The equilibrium dissociation constant (Kd) of TELM for DNA was quantified by EMSAs. We used a consensus binding site with the core sequence 5′-GCCGGAAGT-3′, which has been well characterized as a high affinity binding site for several ETS proteins (Kd values in the 10−9 to 10−10 m range) (19, 28). The core sequence is consistent, except for the first G position, with the reported DNA sequence preferences for TEL (29). Unexpectedly, TELM displayed an unusually low affinity for this binding site (Kd = 28 × 10−9 m) (Fig. 2 and Table 1).
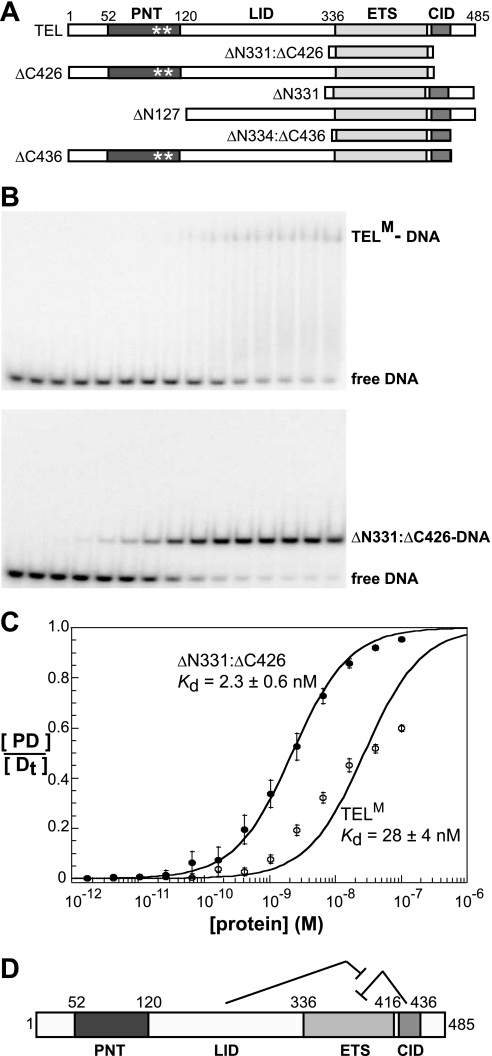
FIGURE 2.
TEL DNA binding is autoinhibited by sequences C-terminal to the ETS domain. A, schematic representation of TEL deletion mutants. PNT domain with polymer-blocking point mutations A94D and V113E (*), linker inhibitory damper (LID), ETS domain, and C-terminal inhibitory domain (CID) indicated. B, equilibrium DNA binding studies of TELM (upper) and ΔN331:ΔC426 (lower). Proteins were titrated over a concentration range of 10−13 to 10−7 m with each lane representing a 2.5-fold increase. The leftmost lane is a DNA-only control. C, DNA-binding isotherms were generated, as described under “Experimental Procedures,” for TELM (open circles) and ΔN331:ΔC426 (filled circles). Data points with error bars and Kd values represent the mean ± S.D. from four independent experiments. D, schematic of TEL autoinhibition by the CID with dampening by the LID.
TABLE 1.
DNA binding affinity of monomeric TEL species
Fig. 2 provides the schematic of all mutant species. Kd values are determined by binding isotherms, as described in Fig. 2. Reported Kd values are mean ± S.D. determined from four independent experiments. The extremely low affinity of ΔN334:ΔC436 and ΔN331 provides less accurate measurements and, thus, -fold inhibition is an estimate (footnote a). The -fold inhibition is calculated as the ratio of Kd (TELx)/Kd (ΔN331:ΔC426). Species bearing PNT domains, full-length TEL, ΔC426, and ΔC436 were monomeric due to polymer-blocking mutations (see Figs. 1 and 2).
| Protein | Kd | -fold inhibition |
|---|---|---|
| 10−9m | ||
| TEL (full-length, monomeric) | 28 ± 4 | 10 |
| ΔN331:ΔC426 (ETS domain-only) | 2.8 ± 0.4 | 1 |
| ΔC426 (monomeric) | 2.3 ± 0.6 | 1 |
| ΔN331 | 110 ± 37 | ∼50a |
| ΔN127 | 21 ± 3 | 7.5 |
| ΔC436 (monomeric) | 26 ± 2 | 9.2 |
| ΔN334:ΔC436 (structurally described) | 270 ± 73 | ∼110a |
| ΔN334:ΔC436 (E431A) | 1.6 ± 0.2 | 1 |
| ΔN334:ΔC436 (E434A) | 1.6 ± 0.8 | 1 |
| ΔN334:ΔC436 (E431A/E434A) | 1.3 ± 0.1 | 1 |
a Estimated.
TEL DNA Binding Is Autoinhibited
Several ETS proteins, including Ets-1 (26, 30–34), Elk-1 (35, 36), Elf-3 (37), and ETV4 (38), display autoinhibition of DNA binding that is mediated by elements outside of the ETS domain. Therefore, we hypothesized that TEL may be regulated in this manner. Indeed, the minimal ETS domain (ΔN331:ΔC426) exhibited ∼10-fold higher affinity (Kd = 2.3 × 10−9 m) than full-length TELM for the consensus binding site (Fig. 2 and Table 1). These findings demonstrated that TEL DNA binding is indeed repressed by an autoinhibition mechanism.
Through further deletion mapping, inhibitory sequences were better defined (Fig. 2 and Table 1). Deletion of the region C-terminal to the ETS domain (ΔC426) activated DNA binding by TELM to the level observed with the minimal ETS domain. This result identified a C-terminal inhibitory domain, which we termed the CID. Surprisingly, deletion of the N-terminal residues yielded a fragment (ΔN331) with even lower affinity than that of the full-length protein (Kd = 110 × 10−9 m). This finding suggested that the CID can actually repress DNA binding by the ETS domain ∼50-fold, but that the sequences N-terminal of the ETS domain interfere with its full inhibitory potential. ΔN127, which lacks the PNT domain, retained this dampening activity. Furthermore, because ΔC426 and the minimal ETS domain had the same affinity for the consensus DNA duplex, the de-repression was dependent on the presence of the CID. We have termed the region (within residues 127–331) that displays this activity the linker inhibitory damper (LID). In conclusion, sequences flanking the ETS domain of TEL regulate its DNA-binding properties (Fig. 2D).
The CID Includes an α-Helix Positioned to Block DNA Binding
An undocumented, NMR-based tertiary structure of a TEL fragment corresponding to residues 334–436, deposited in the Protein Data Bank (2DAO.pdb), provided insights into the possible mechanism of autoinhibition. In addition to the expected ETS domain “winged helix-turn-helix” fold between residues 334 and 426, two C-terminal helices (H4 and H5) are also present in this fragment (Fig. 3A). NMR spectroscopic analyses of ΔN331:ΔC458 verified these reported secondary structural elements (supplemental Figs. S2 and S3). Conformation of the tertiary structure of this TEL fragment awaits additional analyses. The DNA-binding activities of ΔN334:ΔC436 and ΔC436 were shown to be repressed ∼100- and ∼10-fold, respectively, and thus are similar to ΔN331 and wild-type TEL (Fig. 2A and Table 1). This finer mapping confirmed that the CID function was intact in the structurally characterized fragment. In comparison to high resolution models of ETS domain-DNA complexes, illustrated with Ets-1 (Fig. 3B), helix H5 of TEL would block the DNA binding surface. These structural data suggest that TEL DNA binding requires displacement, possibly via local unfolding of helix H5 of the CID. Thus, in our model of TEL autoinhibition the protein exists in two conformations: a closed, inhibited form and an open, active form that is competent to bind to DNA.
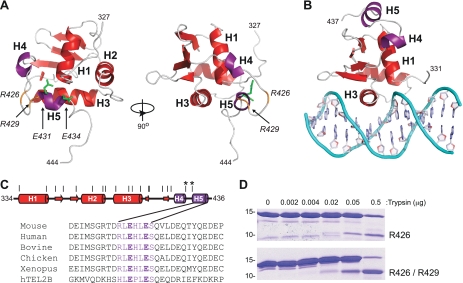
FIGURE 3.
Inhibitory helix H5 blocks the TEL DNA binding interface. A, unpublished NMR-derived tertiary structure of ΔN334:ΔC436 in Protein Data Base (2DAO.pdb). The three α-helices (H1–H3) and four β-strands (S1–S4) that comprise the ETS domain (red), as well as two additional C-terminal helices (H4 and H5) (purple) of the CID are indicated. Conserved acidic residues Glu-431 and Glu-434 are shown as sticks (green). Arg-426 and Arg-429, which are sites of trypsin cleavage, are highlighted (orange). B, the DNA binding surface of an ETS domain is illustrated by a crystal structure of an Ets-1-DNA complex (selected residues 331–437 from 1MDM.pbd (42)): helix H3 lying in the major groove and the N terminus of helix H1 contacting the phosphodiester backbone, ETS domain (red); helix H4 and H5 of Ets-1 inhibitory module (purple), additional inhibitory helices N-terminal to the ETS domain are not illustrated for simplicity. C, schematic representation of ΔN334:ΔC436, with helices (cylinders) and β-strands (arrows) identified. Potential trypsin cleavage sites at all arginine and lysine residues are depicted as vertical lines, and Arg-426 and Arg-429 are indicated (*). A sequence alignment of C-terminal TEL residues (426–436) shows the position of helix H5 and the negatively charged residues Glu-431 and Glu-434. D, partial trypsin proteolysis of ΔN334:ΔC436 (upper) and ΔN334:ΔC436(E431A/E434A) (lower) with increasing amounts of trypsin. The positions of 10- and 15-kDa molecular mass markers are shown. Fragments with cleavage at Arg-426 and Arg-429 were identified via ESI/MS.
Testing the Conformational Change Mechanism
To test the hypothesized mechanism of autoinhibition, we sought to identify residues in the CID that stabilize the inhibited conformation of TEL. By aligning TEL sequences from five vertebrate species and the closely related human TEL2b (Fig. 3C), we found high conservation among the residues in helix H5. Of particular interest were the negatively charged Glu-431 and Glu-434, which are positioned in the TEL fragment structure to interact with the positively charged N terminus of helix H1 of the ETS domain (Fig. 3A). Mutant proteins with alanine substitutions at Glu-431 and Glu-434 were analyzed for structural changes by protease sensitivity. Partial trypsin proteolysis, followed by ESI/MS, was used to identify sites most susceptible to cleavage. When assayed over the same time period, fragment(s) appeared at lower trypsin concentrations in the mutant proteins than in wild-type species (Fig. 3D). For example, a trypsin cleavage site at Arg-426, which lies in an unstructured loop between helices H4 and H5, was observed in both proteins (Fig. 3A). In contrast, cleavage at Arg-429 at the N terminus of helix H5 was observed only in the mutant, potentially accounting for its overall enhanced trypsin sensitivity. These findings indicate that the CID adopts different conformations in the wild-type, parental versus mutant species.
We next measured the DNA binding affinity of the CID mutants. Each single and the double mutant bound DNA with significantly higher affinity than the parental fragment (ΔN334:ΔC436). Thus, the binding affinity of the CID mutants was approximately equivalent to the high affinity, minimal ETS-domain fragment (ΔN334:ΔC426) (Table 1). In support of a conformational change mechanism of autoinhibition, Glu-431 and/or Glu-434 could function in intramolecular interactions that stabilize a closed, inhibited conformation of the TEL ETS domain. Furthermore, we hypothesize that the altered conformation of the mutant proteins mimics the active, bound conformation of TEL.
TEL Dimers Display Higher DNA Binding Affinity
In considering the relatively low affinity of monomeric TEL for the consensus binding site, we speculated that polymerization would enhance the overall DNA binding of wild-type, full-length TEL. Because full-length, wild-type TEL is insoluble and not tractable for in vitro analysis, we took advantage of known PNT domain mutants with disrupted polymer interfaces (Fig. 1, B and C). Each of the single substitution mutant proteins (A94D and V113E) retains one of the native surfaces required for self-association, and thus a mixture of the two in the context of the isolated PNT domain results in a very stable TEL PNT dimer (21). Based on chromatographic properties, a mixture of full-length TEL versions of the two PNT domain mutants also yielded a stable dimer, which we have termed TELD (supplemental Fig. S1).
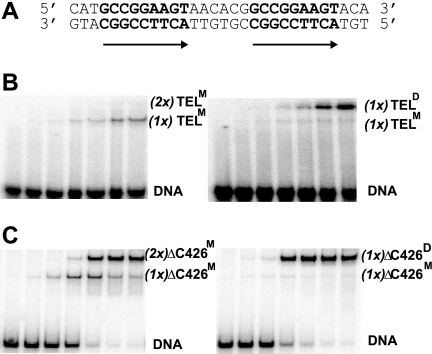
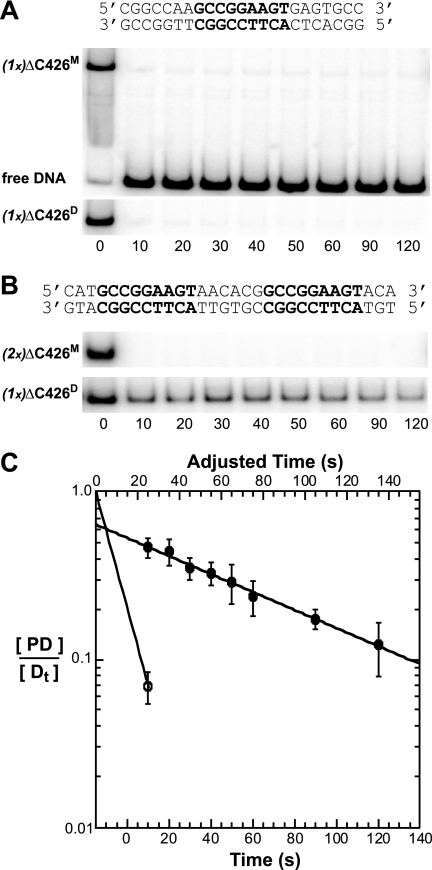
The DNA-binding properties of TELM and TELD were assayed on DNA duplexes bearing two ETS binding sites in tandem (Fig. 4A). With increasing concentrations of each TEL species, DNA-protein complexes of two different mobilities were observed (Fig. 4B). Notably, for TELM, a single protein-DNA species was detected, which we propose corresponded to one low affinity protein monomer, (1x)TELM, bound to one DNA duplex, (Fig. 4B, left). In contrast, two shifted bands appeared in the TELD assay, with the slower mobility one being dominant. We propose this slower migrating band has the two subunits of the TELD dimer, (1x)TELD, bound to the DNA duplex (Fig. 4B, right). The faint, faster migrating species would be due to the monomeric species, presumably present due to incomplete dimer formation. To enhance the sensitivity of the EMSA approach, we used the high affinity species ΔC426, in which the CID is deleted. In this case, ΔC426M displayed two complexes, which we propose corresponded to single and double occupancy by independent TEL ΔC426 monomers, (1x)- or (2x)ΔC426M (Fig. 4C, left). In contrast, ΔC426D, similar to TELD, showed predominantly one complex, which we propose represents cooperative binding by dimeric ΔC426, (1x)ΔC426D (Fig. 4C, right). In analyses of both wild-type TEL and ΔC426, the relative dominance of the slower TELD-DNA complexes suggested that the dimer binds a tandem DNA site with higher affinity than the monomeric species, highly suggestive of a cooperative dimer. Lack of information regarding the Kd for the dimer dissociation on or off of DNA precluded additional quantitative analysis of these results.
FIGURE 4.
TEL dimers bind cooperatively on DNA duplexes with a double ETS site. A, the ETS binding sites (bold) within the 57-bp DNA duplex used for EMSAs. Arrows indicate the orientation of 5′-GGA-3′ core sequences in DNA. B, DNA binding patterns of full-length, autoinhibited TELM (left) and TELD (right) as described (Fig. 2), except here each lane represents a reaction with a 10-fold higher protein concentration. Labels, (1x)TELM, (2x)TELM, and (1x)TELD, refer to the proposed identity (M, monomer or D, dimer) and number (1x or 2x) of independent binding species. The darker appearance of the right panel represents a longer exposure of the radioactive gel and is inconsequential as all assays had equivalent amounts of DNA. C, analysis of ΔC426 species, as in B. This fragment lacks autoinhibition and, thus, exhibits higher affinity DNA binding.
Cooperative DNA Binding Stabilizes Complexes
To quantify the proposed difference in DNA-binding affinity between monomeric and dimeric TEL species and to test whether the tandem ETS binding sites are necessary, we measured dissociation rates. We used the CID-deleted fragment ΔC426, because its inherent higher affinity enabled more robust measurements. The dissociation rate constants (k−1) and half-lives (t½) of the protein-DNA complexes were determined by following the kinetics of dissociation from the labeled DNA in the presence of a large excess of unlabeled DNA. Both ΔC426M and ΔC426D displayed rapid dissociation from a DNA duplex containing a single ETS binding site (Fig. 5, A and C, and Table 2). Similar to results with a direct repeat in equilibrium binding studies (Fig. 4A), ΔC426M achieved double occupancy. However, doubly bound monomers were unstable similar to the single occupancy complex (Fig. 5B, upper, Fig. 5C, and Table 2). In contrast, ΔC426D formed a more stable ternary complex with DNA, as evidenced by a longer half-life of 75 s (Fig. 5B, lower, Fig. 5C, and Table 2). These results demonstrated that TEL dimerization stabilizes the binding to a direct repeat of the ETS binding site, but not to a single ETS binding site. In conclusion, dimerization, mediated by the PNT domain, provides DNA binding cooperativity between bound TEL species.
FIGURE 5.
TEL dimers form more stable complexes than TEL monomers on tandem ETS binding sites. Dissociation of protein-DNA complexes were measured by EMSAs. Labels, (1x)ΔC426M, (2x)ΔC426M, and (1x)ΔC426D, defined as in Fig. 4. Free DNA is only shown (A, upper). A, stability of ΔC426M (upper) and ΔC426D (lower) bound to a single ETS consensus site (shown). B, stability of ΔC426M (upper) and ΔC426D (lower) bound to a direct repeat of an ETS consensus site (shown). C, exponential decay plots of data from A and B. The upper x axis is adjusted by 15 s to account for the time required for the mixing of the sample and the sample to enter the gel. ΔC426D bound to a direct repeat (filled circles) and ΔC426D bound to a single ETS binding site (open circle) are shown. Similar to ΔC426D on a single site, dissociation plots for ΔC426M indicate a half-life of <10 s on either a direct repeat or a single site. These data are not shown for clarity of the graph. See Table 2 for t½ values of ΔC426D on a single ETS site or a variety of double ETS site duplexes.
TABLE 2.
DNA determinants for TEL dimeric cooperativity
DNA duplexes contained ETS binding sites with the core 5′-CCGGAAGT-3′, as described under “Experimental Procedures.” Values in parentheses indicate either the number of base pairs between the first G of the GGA of each of the two binding sites or the number of helical turns with one turn equal to 10.5 bp. The dissociation rate, k−1, was determined from exponential decay plots of fraction of starting signal remaining ([PDt]/[PD0]) plotted against time (seconds), as illustrated in Fig. 5. The half-life of each complex was calculated as t½ = 0.693/k−1. All experiments were performed with the dimeric ΔC426D bearing polymer-blocking mutations (see Figs. 1 and 2).
| Binding site | k−1 | t½ |
|---|---|---|
| s−1 | s | |
| Single GGA | >0.07 | <10 |
| Direct GGA (+15 bp) | 0.009 | 75 |
| Inverted GGA (+13 bp) | 0.013 | 52 |
| Direct GGA (+3 turns) | 0.008 | 86 |
| Direct GGA (+5 turns) | 0.020 | 35 |
| Direct GGA (+6 turns)) | >0.07 | <10 |
| Direct GGA (+7 turns) | >0.07 | <10 |
| Direct GGA (+10 turns) | >0.07 | <10 |
Determinants for TEL Cooperative DNA Binding
To characterize the DNA determinants for cooperativity, we measured the kinetics of dissociation of ΔC426D from DNA duplexes bearing two ETS binding sites with variable spacing and orientations (Table 2). ΔC426D displayed similar dissociation rates from DNA duplexes bearing either a direct or an inverted repeat, corresponding to half-lives of 75 and 52 s, respectively. Furthermore, ΔC426D formed these stable complexes on DNA containing a direct repeat spaced up to five helical turns apart (t½ = 35 s). Cooperative, stable complexes were not detected on binding sites with more extended spacing. In conclusion, ΔC426D binds cooperatively to double ETS consensus sites of variable spacing and orientation, within a limit of five or less DNA helical turns.
DISCUSSION
We have demonstrated that TEL DNA binding is repressed by autoinhibition. Furthermore, TEL self-association facilitates cooperative binding to DNA. Notably, cooperativity was observed on DNA duplexes displaying variable spacing and orientation of two ETS binding sites. These findings demonstrate that the dimerization enhances the DNA-binding activity of TEL and provides a route to compensate for the relatively low affinity of the autoinhibited TEL ETS domain.
A Model for TEL Autoinhibition
Combining our biochemical data with the reported structure of the TEL ETS domain, we propose that TEL displays a conformational equilibrium between two different structural states. One state is an inhibited conformation in which the CID interferes with DNA binding. The other state is an uninhibited conformation where the CID is displaced or disrupted and, thus, is no longer an impediment for DNA binding. Based on an undocumented structure of a TEL fragment in the Protein Data Base and the established mode of ETS domain DNA binding (39–43), we propose a helix within the CID could directly block the binding surface of the ETS domain in two complementary ways. First, due to its proximity to the DNA recognition helix (H3) in the inhibited conformation, helix H5 prevents helix H3 from entering the DNA major groove. Second, helix H5 also prevents the N terminus of helix H1 from contacting the negatively charged DNA backbone, an interaction known to be important in ETS domain DNA binding (27). Thus, the possible conformational change is a change in the position of helix H5. Unfolding of helix H5 is a simple form of displacement that is consistent with the enhanced sensitivity of the CID to partial proteolysis upon mutation of Glu-431 and Glu-434. Attempts to detect a conformational change of this helix in the activated mutant (ΔN331:ΔC458) versus wild-type forms of TEL by CD spectroscopy were not successful (data not shown). However, helix H5 is relatively small and may not contribute substantially to the overall CD spectrum of the protein. One possible mechanism for de-repression of autoinhibition could be an interaction of helix H5 with the LID. A CID-LID interaction could compete with the CID-ETS domain interaction noted in the inhibited state, thus explaining the dampening of autoinhibition by the LID. Testing this proposed model will require additional structural and dynamic measurements of a series of TEL deletion fragments.
The steric mechanism proposed for TEL autoinhibition differs from the predominantly allosteric mechanism described for Ets-1 autoinhibition (26, 28, 33, 34). Four inhibitory helices flank the ETS domain of Ets-1 and function cooperatively as an inhibitory module. This module is positioned distal to the DNA recognition helix and allosterically regulates DNA binding by dampening the dynamics of an internal core of hydrophobic residues, thus stabilizing a rigid, inhibited conformation. However, similar to the mechanism proposed for TEL, a conformational change accompanies DNA binding. In the case of Ets-1, unfolding of an inhibitory helix is observed, yielding a more flexible, active conformation of the ETS domain. Ets-1 autoinhibition is re-enforced by stabilizing the inhibitory module through multisite phosphorylation of an adjacent serine-rich region (33). In contrast, autoinhibition is counteracted via interactions with the partner protein RUNX1 (16–18), which occurs in vivo at many composite promoter and enhancer sites (19, 20). This route to counteracting the autoinhibition of Ets-1 led us to explore what could overcome the relatively low affinity of monomeric TEL for a single consensus DNA sequence.
TELD Counteracts Autoinhibition by Binding Tandem DNA Sites as a Cooperative Dimer
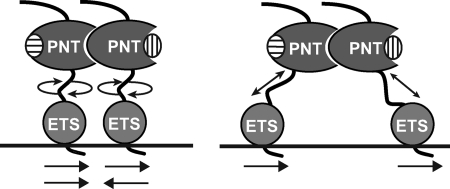
We discovered that TEL self-association via the PNT domain facilitates cooperative DNA binding, thus providing one route to counteract autoinhibition. Cooperativity of DNA binding by dimeric TELD was observed on DNA duplexes containing two ETS binding sites. Furthermore, this cooperativity was observed with variable spacing and orientations of the two sites, suggesting a substantial degree of flexibility in the middle region of TEL linking its PNT and ETS domains (Fig. 6). However, cooperativity was lost when binding sites were spaced farther than five DNA helical turns apart, indicating that this is a limit to this flexibility. Based on these findings, we speculate that a wild-type TEL polymer may not require a well ordered array of target sites for cooperative binding in vivo. Nevertheless, there would be a requirement for relatively close proximity of two or more of these sites.
FIGURE 6.
Model of dimeric TEL binding to DNA with a double ETS site. Determinants of cooperative dimeric binding suggest flexibility of TELD between the self-associating PNT domain and the DNA-binding ETS domain. Horizontal arrows indicate binding site positions and orientation of the core 5′-GGA-3′ recognition sequence. Domains and point mutations are illustrated as in Fig. 1.
We speculate that TEL may function only at promoters with multiple binding sites that can accommodate a cooperative polymer. TEL has been shown to repress luciferase reporters driven by promoter elements of the stromelysin-1 (9, 44, 45) and Bcl-XL (46) genes. Notably, both of these promoters contain multiple consensus ETS binding sites, implying that a cooperative TEL polymer may be required for repression. Indeed, TEL-mediated repression of stromelysin-1 is reported to be dependent upon dimerization properties, because mutants unable to self-associate failed to repress (45). Based on these data, we hypothesize that autoinhibition and polymerization may define TEL specificity by targeting the ETS proteins to promoters bearing multiple binding sites.
PNT Domain Provides Specificity in the ETS Family
The polymerization properties of TEL distinguish it from most other ETS family members, in part because only one-third of the ETS proteins have PNT domains. However, even those with PNT domains display surprising differences (47). For example, in Ets-1 and Ets-2, the monomeric PNT domain has a docking site for the ERK2 kinase that phosphorylates nearby phosphoacceptor sites (48). The subsequent phosphorylation acts in concert with the PNT domain to enhance binding of the co-activator CBP (49). FLI1 and TEL2 have been suggested to associate with TEL, and thus some type of polymerization may be a common property for a limited number of PNT domain-containing ETS proteins (5, 50). Also, there is an interesting network of homotypic and heterotypic interactions among ETS proteins in Drosophila that involves their PNT domains (51, 52). The Drosophila protein Yan functions as a repressor to counter the activity of the activating Pnt-P2, possibly by alternative binding to common downstream transcriptional targets (53, 54). Yan is the apparent ortholog of TEL, and, self-association properties have been implicated in its biological activity (55). Similar to TEL, wild-type and engineered dimeric versions of full-length Yan show no difference in DNA binding affinity on a single ETS binding site (51). In addition, there is a PNT domain-only protein in Drosophila, Mae, which has polymerization-blocking properties, because it has only one active association surface. Mae binds to both Pnt-P2 and Yan to regulate their biological activities (51, 56). Thus, the PNT domain and its diverse competencies represent a route to specificity and regulation with the ETS transcription factor family.
TEL Self-association and Cancer
The mechanism of TEL/AML1-mediated oncogenesis is unclear, although it is likely related to transcriptional effects on downstream targets. Because the PNT domain of TEL is retained in this fusion, we speculate that the self-association properties of the PNT domain may facilitate cooperative DNA-binding activities in TEL/AML1 similar to TEL. Such properties may enable the fusion protein to bind cooperatively to promoters not normally bound by monomeric AML1, thus contributing to the gain of function activity of the oncogenic fusion protein.
Supplementary Material
Acknowledgment
We thank Hong Wang, who made several of the TEL constructs used in this report during her tenure in the Graves laboratory.
This work was supported, in whole or in part, by National Institutes of Health Grants R01GM38663 (to B. J. G.) and P50CA42014 (to the Huntsman Cancer Institute for support of shared resources). This work was also supported by funds from the Canadian Cancer Society Research Institute (to L. P. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Tables 1 and 2.
- DTT
- dithiothreitol
- CID
- C-terminal inhibitory domain
- EMSA
- electrophoretic mobility shift assay
- LID
- linker inhibitory damper
- ESI/M
- electrospray ionization mass spectrometry.
REFERENCES
- 1.May W. A., Gishizky M. L., Lessnick S. L., Lunsford L. B., Lewis B. C., Delattre O., Zucman J., Thomas G., Denny C. T. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 5752–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tomlins S. A., Rhodes D. R., Perner S., Dhanasekaran S. M., Mehra R., Sun X. W., Varambally S., Cao X., Tchinda J., Kuefer R., Lee C., Montie J. E., Shah R. B., Pienta K. J., Rubin M. A., Chinnaiyan A. M. (2005) Science 310, 644–648 [DOI] [PubMed] [Google Scholar]
- 3.Golub T. R., Barker G. F., Bohlander S. K., Hiebert S. W., Ward D. C., Bray-Ward P., Morgan E., Raimondi S. C., Rowley J. D., Gilliland D. G. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 4917–4921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Golub T. R., Barker G. F., Stegmaier K., Gilliland D. G. (1997) Curr. Top. Microbiol. Immunol. 220, 67–79 [DOI] [PubMed] [Google Scholar]
- 5.Kwiatkowski B. A., Bastian L. S., Bauer T. R., Jr., Tsai S., Zielinska-Kwiatkowska A. G., Hickstein D. D. (1998) J. Biol. Chem. 273, 17525–17530 [DOI] [PubMed] [Google Scholar]
- 6.Chakrabarti S. R., Nucifora G. (1999) Biochem. Biophys. Res. Commun. 264, 871–877 [DOI] [PubMed] [Google Scholar]
- 7.Fenrick R., Amann J. M., Lutterbach B., Wang L., Westendorf J. J., Downing J. R., Hiebert S. W. (1999) Mol. Cell. Biol. 19, 6566–6574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lopez R. G., Carron C., Oury C., Gardellin P., Bernard O., Ghysdael J. (1999) J. Biol. Chem. 274, 30132–30138 [DOI] [PubMed] [Google Scholar]
- 9.Fenrick R., Wang L., Nip J., Amann J. M., Rooney R. J., Walker-Daniels J., Crawford H. C., Hulboy D. L., Kinch M. S., Matrisian L. M., Hiebert S. W. (2000) Mol. Cell. Biol. 20, 5828–5839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hiebert S. W., Sun W., Davis J. N., Golub T., Shurtleff S., Buijs A., Downing J. R., Grosveld G., Roussell M. F., Gilliland D. G., Lenny N., Meyers S. (1996) Mol. Cell. Biol. 16, 1349–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uchida H., Downing J. R., Miyazaki Y., Frank R., Zhang J., Nimer S. D. (1999) Oncogene 18, 1015–1022 [DOI] [PubMed] [Google Scholar]
- 12.Graves B. J., Petersen J. M. (1998) in Advances in Cancer Research ( Woude G. V., Klein G. eds) pp.1– 55, Academic Press, San Diego [Google Scholar]
- 13.Sharrocks A. D. (2001) Nat. Rev. Mol. Cell Biol. 2, 827–837 [DOI] [PubMed] [Google Scholar]
- 14.Ling Y., Lakey J. H., Roberts C. E., Sharrocks A. D. (1997) EMBO J. 16, 2431–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brass A. L., Kehrli E., Eisenbeis C. F., Storb U., Singh H. (1996) Genes Dev. 10, 2335–2347 [DOI] [PubMed] [Google Scholar]
- 16.Kim W. Y., Sieweke M., Ogawa D., Wee H. J., Englemeier U., Graf T., Ito Y. (1999) EMBO J. 18, 1609–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goetz T. L., Gu T. L., Speck N. A., Graves B. J. (2000) Mol. Cell. Biol. 20, 81–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gu T. L., Goetz T. L., Graves B. J., Speck N. A. (2000) Mol. Cell. Biol. 20, 91–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hollenhorst P. C., Shah A. A., Hopkins C., Graves B. J. (2007) Genes Dev. 21, 1882–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hollenhorst P. C., Chandler K. J., Poulsen R. L., Johnson W. E., Speck N. A., Graves B. J. (2009) PLoS Genet. 5, e1000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim C. A., Phillips M. L., Kim W., Gingery M., Tran H. H., Robinson M. A., Faham S., Bowie J. U. (2001) EMBO J. 20, 4173–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tran H. H., Kim C. A., Faham S., Siddall M. C., Bowie J. U. (2002) BMC Struct. Biol. 2, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qiao F., Bowie J. U. (2005) Science STKE 2005, RE7. [DOI] [PubMed] [Google Scholar]
- 24.Roukens M. G., Alloul-Ramdhani M., Vertegaal A. C., Anvarian Z., Balog C. I., Deelder A. M., Hensbergen P. J., Baker D. A. (2008) Mol. Cell. Biol. 28, 2342–2357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roukens M. G., Alloul-Ramdhani M., Moghadasi S., Op den Brouw M., Baker D. A. (2008) Mol. Cell. Biol. 28, 4394–4406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petersen J. M., Skalicky J. J., Donaldson L. W., McIntosh L. P., Alber T., Graves B. J. (1995) Science 269, 1866–1869 [DOI] [PubMed] [Google Scholar]
- 27.Wang H., McIntosh L. P., Graves B. J. (2002) J. Biol. Chem. 277, 2225–2233 [DOI] [PubMed] [Google Scholar]
- 28.Jonsen M. D., Petersen J. M., Xu Q. P., Graves B. J. (1996) Mol. Cell. Biol. 16, 2065–2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szymczyna B. R., Arrowsmith C. H. (2000) J. Biol. Chem. 275, 28363–28370 [DOI] [PubMed] [Google Scholar]
- 30.Hagman J., Grosschedl R. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 8889–8893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lim F., Kraut N., Framptom J., Graf T. (1992) EMBO J. 11, 643–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wasylyk B., Hahn S. L., Giovane A. (1993) Eur. J. Biochem. 211, 7–18 [DOI] [PubMed] [Google Scholar]
- 33.Pufall M. A., Lee G. M., Nelson M. L., Kang H. S., Velyvis A., Kay L. E., McIntosh L. P., Graves B. J. (2005) Science 309, 142–145 [DOI] [PubMed] [Google Scholar]
- 34.Lee G. M., Pufall M. A., Meeker C. A., Kang H. S., Graves B. J., McIntosh L. P. (2008) J. Mol. Biol. 382, 1014–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang S. H., Shore P., Willingham N., Lakey J. H., Sharrocks A. D. (1999) EMBO J. 18, 5666–5674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang S. H., Bumpass D. C., Perkins N. D., Sharrocks A. D. (2002) Mol. Cell. Biol. 22, 5036–5046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kopp J. L., Wilder P. J., Desler M., Kinarsky L., Rizzino A. (2007) J. Biol. Chem. 282, 3027–3041 [DOI] [PubMed] [Google Scholar]
- 38.Bojović B. B., Hassell J. A. (2001) J. Biol. Chem. 276, 4509–4521 [DOI] [PubMed] [Google Scholar]
- 39.Batchelor A. H., Piper D. E., de la Brousse F. C., McKnight S. L., Wolberger C. (1998) Science 279, 1037–1041 [DOI] [PubMed] [Google Scholar]
- 40.Mo Y., Vaessen B., Johnston K., Marmorstein R. (2000) Nat. Struct. Biol. 7, 292–297 [DOI] [PubMed] [Google Scholar]
- 41.Garvie C. W., Hagman J., Wolberger C. (2001) Mol Cell 8, 1267–1276 [DOI] [PubMed] [Google Scholar]
- 42.Garvie C. W., Pufall M. A., Graves B. J., Wolberger C. (2002) J. Biol. Chem. 277, 45529–45536 [DOI] [PubMed] [Google Scholar]
- 43.Agarkar V. B., Babayeva N. D., Wilder P. J., Rizzino A., Tahirov T. H. (2010) J. Mol. Biol. 397, 278–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang L., Hiebert S. W. (2001) Oncogene 20, 3716–3725 [DOI] [PubMed] [Google Scholar]
- 45.Wood L. D., Irvin B. J., Nucifora G., Luce K. S., Hiebert S. W. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 3257–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Irvin B. J., Wood L. D., Wang L., Fenrick R., Sansam C. G., Packham G., Kinch M., Yang E., Hiebert S. W. (2003) J. Biol. Chem. 278, 46378–46386 [DOI] [PubMed] [Google Scholar]
- 47.Mackereth C. D., Schärpf M., Gentile L. N., MacIntosh S. E., Slupsky C. M., McIntosh L. P. (2004) J. Mol. Biol. 342, 1249–1264 [DOI] [PubMed] [Google Scholar]
- 48.Seidel J. J., Graves B. J. (2002) Genes Dev. 16, 127–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Foulds C. E., Nelson M. L., Blaszczak A. G., Graves B. J. (2004) Mol. Cell. Biol. 24, 10954–10964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Poirel H., Lopez R. G., Lacronique V., Della Valle V., Mauchauffé M., Berger R., Ghysdael J., Bernard O. A. (2000) Oncogene 19, 4802–4806 [DOI] [PubMed] [Google Scholar]
- 51.Qiao F., Song H., Kim C. A., Sawaya M. R., Hunter J. B., Gingery M., Rebay I., Courey A. J., Bowie J. U. (2004) Cell 118, 163–173 [DOI] [PubMed] [Google Scholar]
- 52.Qiao F., Harada B., Song H., Whitelegge J., Courey A. J., Bowie J. U. (2006) EMBO J. 25, 70–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O'Neill E. M., Rebay I., Tjian R., Rubin G. M. (1994) Cell 78, 137–147 [DOI] [PubMed] [Google Scholar]
- 54.Rebay I., Rubin G. M. (1995) Cell 81, 857–866 [DOI] [PubMed] [Google Scholar]
- 55.Zhang J., Graham T. G., Vivekanand P., Cote L., Cetera M., Rebay I. (2010) Mol. Cell. Biol. 30, 1158–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Song H., Nie M., Qiao F., Bowie J. U., Courey A. J. (2005) Genes Dev. 19, 1767–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.