Abstract
Entamoeba histolytica cysteine proteinases (EhCPs) play a key role in disrupting the colonic epithelial barrier and the innate host immune response during invasion of E. histolytica, the protozoan cause of human amebiasis. EhCPs are encoded by 50 genes, of which ehcp4 (ehcp-a4) is the most up-regulated during invasion and colonization in a mouse cecal model of amebiasis. Up-regulation of ehcp4 in vivo correlated with our finding that co-culture of E. histolytica trophozoites with mucin-producing T84 cells increased ehcp4 expression up to 6-fold. We have expressed recombinant EhCP4, which was autocatalytically activated at acidic pH but had highest proteolytic activity at neutral pH. In contrast to the other amebic cysteine proteinases characterized so far, which have a preference for arginine in the P2 position, EhCP4 displayed a unique preference for valine and isoleucine at P2. This preference was confirmed by homology modeling, which revealed a shallow, hydrophobic S2 pocket. Endogenous EhCP4 localized to cytoplasmic vesicles, the nuclear region, and perinuclear endoplasmic reticulum (ER). Following co-culture with colonic cells, EhCP4 appeared in acidic vesicles and was released extracellularly. A specific vinyl sulfone inhibitor, WRR605, synthesized based on the substrate specificity of EhCP4, inhibited the recombinant enzyme in vitro and significantly reduced parasite burden and inflammation in the mouse cecal model. The unique expression pattern, localization, and biochemical properties of EhCP4 could be exploited as a potential target for drug design.
Keywords: Cysteine Protease, Drug Design, Enzyme Structure, Protease Inhibitor, Protozoan
Introduction
Entamoeba histolytica is the causative protozoan parasite of amebic colitis and liver abscesses and the second leading cause of death from parasitic diseases (1). Invasive E. histolytica trophozoites secrete 10–1,000-fold more cysteine proteinases than non-invasive Entamoeba dispar (2), whereas EhCP2-deficient trophozoites demonstrate reduced virulence in animal models (3). During amebic invasion, EhCPs degrade the mucus barrier and destroy the extracellular matrix (4) underlying the epithelium, thereby causing epithelial detachment and ulceration (4, 5). Amebic penetration through ulcers facilitates entry into the mucosa and subsequently spread to the liver (3–5). Furthermore, EhCPs interrupt the function of the host immune system by cleaving immune molecules, including IgA and IgG (6, 7), processing complement C3 (2) and inactivating complement C3a and C5a (8). They also generate mature IL-1β from pro-IL-1β (9) but inactivate pro-IL-18 (10). These alternations of the immune response contribute to ineffective host defenses and thereby excessive inflammation and tissue damage.
Genome-wide homology searches have identified 50 E. histolytica genes encoding cysteine proteases (11, 12), most of which belong to the C1 papain superfamily. These enzymes are expressed as zymogens with a hydrophobic pre-domain of 12–20 residues, a pro-domain of 55–148 residues, a catalytic domain of 190–488 residues, and six cysteine residues that form the disulfide bridges to stabilize the tertiary structure (13, 14). The EhCPs that have been characterized to date include EhCP1 (EhCP-A1) (15, 16), EhCP2 (EhCP-A2) (14), EhCP3 (EhCP-A3) (10, 14), EhCP5 (EhCP-A5) (15, 17), and EhCP112 (EhCP-B9) (18). These enzymes all undergo autocatalytic activation in which the inhibitory pro-domain is cleaved from the zymogen. They also have a cathepsin L-like structure, but like cathepsin B, these enzymes can also accommodate Arg at P2 (14). Among the remaining EhCPs, some are predicted to have putative transmembrane or glycosylphosphatidylinositol anchor attachment domains, suggesting that they are membrane-associated proteases (13). Additionally, a few of the predicted EhCPs are similar to calpain-like cysteine proteinases, ubiquitinyl hydrolase, Ulp1 peptidase, autophagin, and otubain (12). The biological functions of these non-papain family enzymes are not known.
In axenically cultured E. histolytica trophozoites, the papain family members EhCP1, EhCP2, and EhCP5 account for ∼90% of the total cysteine protease activity (19). However, the expression of EhCP genes in cultured trophozoites differs from those in vivo. Although some ehcp genes (e.g. ehcp4 (ehcp-a4), ehcp6 (ehcp-a6), and ehcp9 (ehcp-a8)) are expressed at low or undetectable levels in vitro (13), they are transcribed and significantly up-regulated during cecal infection in mice, especially ehcp4 (20). Moreover, in E. histolytica cysts from clinical isolates, ehcp4 and ehcp9 are also moderately up-regulated, whereas ehcp7 (ehcp-b1), ehcp18 (ehcp-b8), and a CP11-like ehcp (ehcp-b2) are highly up-regulated (21). These findings suggest that not only does the expression of EhCPs depend upon physiological context, but also the functions of these enzymes may be specific and/or synergistic. To this end, characterizing the biochemical properties and functions of these developmentally regulated EhCPs should shed light on how adaptation to the environmental changes promotes invasion by E. histolytica.
We chose to characterize EhCP4 because it is continuously expressed throughout all stages of invasion and colonization and is the most up-regulated during cecal invasion by E. histolytica. Upon expression of the recombinant zymogen of EhCP4 and refolding it to an active enzyme, we found that EhCP4 has unique substrate specificity based on synthetic peptidyl substrates, specific optimal pH values for autoactivation and proteolytic activities, and localization patterns that are distinct from the previously well studied EhCPs. Moreover, computer modeling and structure-based inhibitor design led to synthesis of an EhCP4-specific inhibitor, WRR605, which was both active in vitro and protective against invasive infection in a murine cecal model of amebiasis.
EXPERIMENTAL PROCEDURES
Entamoeba Cultures and Purification of Genomic DNA
E. histolytica strain HM1:IMSS was grown axenically (16). E. histolytica genomic DNA was purified from trophozoite nuclei with the DNeasy kit (Qiagen) as described previously (16).
Expression of Recombinant EhCP4
The sequence encoding the pro-domain and catalytic domain of EhCP4 (accession numbers XP656602 and EHI_050570) was cloned by PCR using PFU Ultra Hi Fi DNA polymerase (Stratagene) with Entamoeba genomic DNA as a template. The PCR product from the primer pair EhCP4 pQE80L5′ (GGA TCC GCT AAG AAC AAT AAA CAC TTC; BamHI site at the 5′-end) and EhCP4 pQE80L3′ (CTG CAG TTA ATT AGC ATC ATG AGC ACC; PstI site at the 5′-end) was cloned into pQE80L (Qiagen) for maximal N-terminal His-tagged protein expression for use in generating the EhCP4 antigen for antibody production. The PCR product from primer pair rEhCP-A4–5P (GGA TCC GGC TAA GAA CAA TAA ACA CTT CAC TG; BamHI site at the 5′-end) and rEhCP-A4–3P (CTC GAG TTA ATT AGC ATC ATG AGC ACC AGT; XhoI site at the 5′-end) was cloned into pET32b (Novagen) to generate a fusion protein with the Escherichia coli thioredoxin A and a His6 tag at the amino terminus of EhCP4 for use in refolding experiments. In-frame insertions of the constructs were confirmed by sequencing. pQE80LEhCP4 and pET32bEhCP4 were transfected into BL21 Condon Plus (DE3)-RIPL cells (Stratagene), and the fusion proteins were purified from inclusion bodies by nickel affinity chromatography as detailed previously (16). The total protein concentration was determined by Coomassie Plus (Pierce) and the purity by 12% SDS-PAGE following methanol/chloroform precipitation. The minimal purity of the His6-tagged rEhCP4 was 80% of the total protein.
Refolding and Purification of rEhCP4
Optimal refolding conditions were identified by following a modified screening protocol (22). Maximal active enzyme was obtained by reducing the denatured thioredoxin A-EhCP4 fusion protein with 25 mm DTT at 37 °C for 1 h followed by rapid dilution of the protein into ice-cold refolding buffer (100 mm Tris-Cl, pH 8.5, 100 mm NaCl, 20% glycerol, 250 mm arginine-HCl, 2 mm EDTA, pH 8, 10 mm GSH, 1 mm l-GSSG disodium salt) to reach a final concentration of the fusion protein of 0.6–0.8 μm. The refolding solution was incubated at 4 °C for 72 h and then in ambient temperature for 2–4 h. The insoluble protein was removed by centrifugation followed by filtration through a 0.45-μm surfactant-free cellulose acetate filter (Corning Glass). The soluble protein was concentrated with Amicon Ultra-15 (Ultracel-10K). The refolding buffer was desalted with 25 mm Tris-Cl, pH 8.0, using a PD-10 desalting column (GE Healthcare). The soluble protein was further purified by ion exchange fast protein liquid chromatography on a Hi-TrapTM HP-Q column (GE Healthcare). The fractions with cysteine proteinase activity were combined, concentrated, and stored at 4 °C.
Proteinase Activity Assay
Aliquots of the refolded recombinant EhCP4 were activated in the activation buffer (50 mm citric acid-Na2HPO4 buffer at pH 4 with 5 mm DTT) at 37 °C for 10–45 min. Proteinase activity was measured by the release of the fluorescent leaving group, 4-amino-7-methylcoumarin (AMC) from synthetic peptide substrates as described (16), using a Fluoroskan-Ascent fluorometer (Thermo Labsystems). Enzyme activity, initial velocity, and relative fluorescence units (RFU)/min (the amount of proteinase activity needed for the release of 1 pmol of AMC/min), and the Michaelis constant (Km) of rEhCP4 for synthetic peptide substrates were experimentally determined using standard Michaelis-Menten kinetics as described previously (16). Briefly, activated rEhCP4 (10–13 nm) was reacted with substrates Z-VVR-AMC or Z-LLVY-AMC in 50 mm citric acid-Na2HPO4 buffer, with 5 mm DTT, 0.005% Triton X-100, and 2 mm EDTA at pH 4, 7, and 8. The final concentrations of the substrates were 5.8–100 μm in serial dilutions. The measurement of the fluorescent signal (RFU) was performed on a Flex Station (Molecular Devices). For active site titration of enzyme, activated rEhCP4 was incubated with a series of concentrations of a pancysteine proteinase inhibitor, E-64, or the EhCP4-specific inhibitor, WRR605, in serial dilutions from 0–1.0 μm. The optimized inhibition time (100% inhibition) was predetermined. The linear portion of the plot (residual activity versus inhibitor concentration) was used to calculate the active enzyme concentration.
Mass Spectrum Analysis
The protein band of interest was excised from a 12% SDS-polyacrylamide gel. The gel slice was washed with 25 mm NH4HCO3, 50% acetonitrile (CH3CN), dehydrated with 100% CH3CN, dried, and digested overnight at 37 °C using Trypsin SinglesTM proteomics grade (Sigma). The reaction was quenched with an equal amount of 10% formic acid. Nanocapillary columns were packed at 600 p.s.i. to a length of 10 cm with C18 reverse-phase resin suspended in methanol. The column was equilibrated with 90% of solvent A (water, 0.1% acetic acid) and loaded with 10 μl of trypsin-digested rEhCP4 with 90% of solvent A and 10% of solvent B (CH3CN, 0.1% acetic acid). A gradient for eluting trypsin-digested peptides was established with a time-varying solvent mixture and directly electrosprayed into the LTQ-Fourier transform mass spectrometer (Thermo Fisher Scientific Inc.). Top intensity ions were selected for fragmentation by collision-induced dissociation. The LTQ was calibrated using the recommended Thermo Fisher calibration mix. rEhCP4 mass spectrometry data sets were searched against the NCBI non-redundant data base with the predicted rEhCP4 sequence. This search was facilitated by a DOS common-line version of the public software InSpecT (23). In addition to the NCBI non-redundant data base, a “phony” or reverse data base and a common contaminant data base was added to the search.
Amino-terminal Sequencing of the Recombinant Thioredoxin A Fusion EhCP4
Activated rEhCP4 was separated with a 12% SDS-acrylamide gel with 2 mm mercaptoacetic acid in the upper electrode buffer. The gel was blotted on an Immobilon-PSQ membrane with a semidry device (1 mA/cm2 for 100 min). The membrane was stained by 0.1% Coomassie Blue R, 50% methanol, 1% acetic acid for 5 min and destained with 50% methanol. The active rEhCP4 band (∼26 kDa) was cut from the membrane and sent to the PAN Facility (Stanford University Medical Center) for amino-terminal sequencing.
Substrate Specificity of rEhCP4
Activated rEhCP4 (100 nm) was used in screening a P1–P4 substrate library following the protocol as described previously (16) to determine the substrate specificities of the S1–S4 subsites.
pH Profiling
The pH activity profiles were established using activated rEhCP4 (10 nm) and substrate Z-VVR-AMC in the citric acid-Na2HPO4 buffers (from pH 2 to pH 8) with 5 mm DTT. Values were normalized with the highest activity (in RFU/min) set to 100% and represent three experiments.
EhCP4 Homology Modeling
The crystal structure of procathepsin L1 from Fasciola hepatica (FheCL1; Protein Data Bank code 2O6X) was used as the template for homology modeling of the mature domain of EhCP4 (24). Sequence alignment of the mature form of EhCP4 with the FheCL1 zymogen and homology modeling were performed in MODELLER (25).
Inhibitors and Inhibition Kinetics of rEhCP4
The cysteine proteinase inhibitors, WRR483, WRR605, and BODIPY-WRR-605, were synthesized using well established procedures for related compounds (26).3 Kinetic analyses of the irreversible cysteine protease inhibitors were performed by adding activated rEhCP1 (10 nm) to inhibitor dilutions with 2 μm Z-RR-AMC (Km = 2 μm) in 50 mm citric acid-Na2HPO4 buffer, pH 6.5, with 5 mm DTT, 0.005% Triton X-100, and 2 mm EDTA. Similarly, activated rEhCP4 (13 nm) was added to inhibitor dilutions with 30 μm Z-VVR-AMC (Km = 30 μm) in 50 mm citric acid-Na2HPO4 buffer, pH 7.0, with 5 mm DTT, 0.005% Triton X-100, and 2 mm EDTA. The measurement of the fluorescent signal (RFU) was performed on a Flex Station (Molecular Devices). Progress curves were obtained for 10 min at room temperature (less than 5% of substrate consumed) with 3-fold dilutions of inhibitors, starting at 33 μm and ranging down to 140 nm. Inhibitor dilutions, which gave simple exponential progress curves over a wide range of kobs, were used to determine kinetic parameters. The value of kobs was determined under pseudo-first order conditions using the progress curves method (27, 28) and calculated with Prism 4 (GraphPad) as reported previously (29).
DCG04 and BODIPY-WRR605 Labeling of Active Cysteine Proteinase
Aliquots of the refolded recombinant EhCP4 were activated in the activation buffer (50 mm citric acid-Na2HPO4 buffer at pH 4 with 5 mm DTT) at 37 °C for 10 min. The buffer was neutralized to pH 7 by 50 mm citric acid-Na2HPO4 buffer at pH 8. Active proteinase was detected with DCG04, a biotinylated epoxide inhibitor that binds most clan CA proteinases (kind gift from Dr. Doron Greenbaum, University of Pennsylvania) (30). DCG-04 was added to the activated EhCP4 to a final concentration of 2 μm at room temperature for 60 min. The labeled protein was detected by immunoblotting with alkaline phosphatase-conjugated streptavidin (Sigma). For BODIPY-WRR605 labeling, a 200 μm concentration of the recombinant EhCP4 was activated at 37 °C for 30 min. The activated enzyme was treated with 200 μm BODIPY-labeled WRR605 without or with increasing concentrations of unlabeled WRR605 (50 and 100 μm). The protein was separated on a 15% SDS-polyacrylamide gel, blotted onto a 0.22-μm polyvinylidene difluoride membrane, and scanned using a STORM860 PhosphorImager (GE Healthcare).
Identification and Cleavage of Host Proteins by Recombinant EhCP4
Potential host protein targets were identified by in silico data mining using the result of the P1–P4 library scan with a computational tool for modeling and predicting protease specificity (POPS, available from the Monash University web site). Except for human villin-1, candidates are commercially available. To purify endogenous human villin-1, isolation of soluble human villin-1 from HT-29 cells was performed as described (31). For immunoprecipitation, cell lysate (2 mg) was precleared with GammaBind Plus-Sepharose (15 μl; GE Healthcare). Monoclonal anti-villin antibody (12 μl, ID2C3 (AB739 lot: 581147); Abcam, Cambridge, UK) was incubated at 4 °C overnight with cell lysate and GammaBind Plus-Sepharose (22 μl). The Sepharose beads were washed twice with HNTP buffer (20 mm Hepes, pH 7.5, 150 mm NaCl, 0.1% Triton X-100, 10% glycerol), followed by three washes with PBS with 0.02% Triton X-100. The immunoblot of villin was detected with ID2C3 (1:500) and Mouse TrueBlot ULTRA (eBioscience). The cleavage of physiological proteins was performed as described (16). In brief, the rEhCPs were activated, and the DTT was removed by ultrafiltration with Amicon Ultra-15 (Ultracel-10K, Millipore Corp.) and phosphate buffers (16). Laminin-1 (4 μg; Roche Applied Science), villin, and C3 (2 μg; Quidel) were incubated for 1 h (for C3) and 4 h (for laminin and villin) at 37 °C, respectively, with rEhCP1 (as a control) and rEhCP4 in PBS alone or following preincubation of the proteinase for 15 min at room temperature with E-64. IgG (2 μg) and IgA (2 μg) (Sigma) were incubated with rEhCP4 without or with E-64 for 14 h at 37 °C. Pro-IL-18 was expressed and purified from E. coli and incubated with rEhCP4 for 1 h. The cleavage products were boiled in Laemmli buffer and separated with 15% or 4–20% SDS gradient polyacrylamide gels. The laminin-1 antibody (L9393, Sigma) was used to show laminin degradation.
Antibody Production
Anti-EhCP4 polyclonal antibodies were prepared by immunizing Rhode Island Red chickens with gel-purified pQE80LEhCP4 His6-tagged protein in Freund's complete adjuvant, followed by monthly boosting in Freund's incomplete adjuvant for 5 months (Robert Sargeant Antibody Laboratory, Ramona, CA). IgY was purified from egg yolks by a simple two-step procedure (32) and was further purified by affinity purification with its antigen to produce monospecific antibody.
Immunofluorescence Staining
Trophozoites were washed twice in cold Dulbecco's phosphate-buffered saline (DPBS), pH 7.4, and fixed in 4% paraformaldehyde for 60 min on ice, permeabilized in 0.1% Triton X-100, DPBS, 3% bovine serum albumin for 10 min, and blocked with 3% bovine serum albumin, 0.05% Triton X-100, in DPBS for 60 min at room temperature or 4 °C overnight. For LysoTracker labeling, LysoTrackerTM Red DND-99 (Invitrogen) was applied to the axenic co-culture of trophozoites and T84 cells at a final concentration of 2 μm for 4 h. BODIPY-WRR605 and free BODIPY-TMR (Invitrogen) were added to axenically cultured trophozoites at a final concentration of 50 μm for 3 h.
Infected mouse ceca were fixed with 4% paraformaldehyde in DPBS, pH 7.4, overnight at 4 °C, followed by three washes in cold DPBS. The fixed tissues were cryoprotected with 30% sucrose in 1× DPBS, followed by optimum cutting temperature compound (Sakura Finetek USA, Inc., Torrance, CA) embedding. 8-μm sections were made by Microm HM505E (Termo Scientific). The sections were air-dried, washed in DPBS, permeabilized in 0.1% Triton X-100, 3% bovine serum albumin, 2% normal goat serum, 0.5% fish skin gelatin in DPBS, for 10 min and blocked with 0.05% Triton X-100, 0.02% Tween 20, 3% bovine serum albumin, 2% normal goat serum (for tissue sections, 0.5% fish skin gelatin was added) in DPBS. Blocked tissues or cells were incubated with a monospecific polyclonal IgY to EhCP4 (1:500, 320 ng/ml) and a goat polyclonal IgG (sc-6467) to the ER marker, calreticulin (CRT) (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) (1:200), followed by staining with ALEXA 594-conjugated or ALEXA 488-conjugated secondary antibodies (1:800 dilution; Molecular Probes, Inc., Eugene, OR). The slides were mounted with Vectashield Hard Set (Vector Laboratories) and imaged with a Nikon Eclipse E800 epifluorescence microscope equipped with a DXM1200F digital camera or with a Leica TCS SP5 laser scanning microscope.
Co-culture of Trophozoites and Human Colon Cancer Cell Line T84
The human colon adenocarcinoma cell line, T84, was cultured in Dulbecco's modified Eagle's medium/F-12 medium with 5% newborn calf serum (Hyclone) to a confluent monolayer in T25 flasks (Corning Glass). Before the start of co-culture, T84 cells were treated with 300 ng/ml cholera toxin (Sigma), a weak secretagogue of mucin, for 8 h to induce mucus production. The T84 monolayer was then washed twice with culture medium to remove cholera toxin. Axenic HM-1 trophozoites (2.5 × 106) were laid over the confluent monolayer of T84 cells and incubated in culture medium in an anaerobic chamber box at 37 °C for 18 h when the monolayer of T84 was destroyed by E. histolytica trophozoites.
Real Time-PCR
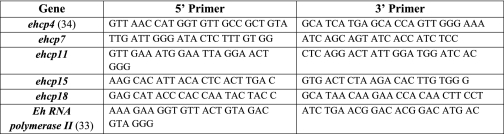
Total RNA of E. histolytica trophozoites was isolated with the PureLinkTM Micro-to-MidiTM Total RNA Purification System (Invitrogen). The reverse transcription reaction was done with Moloney murine leukemia virus reverse transcriptase and the hexadeoxynucleotides random primers (Promega). The PCR was performed using 2× SYBR Green Master Mix (Applied Biosystems) with the 7300 Real-Time PCR system (Applied Biosystems). The data were analyzed using the comparative Ct method. The primers used to amplify the different EhCP genes and the reference gene (EhRNA polymerase II) primers of E. histolytica are listed in Table 1 (20, 33, 34). For quantifying trophozoite numbers, the quantitative PCR primers of E. histolytica peroxiredoxin were forward primer (AAA TCA ATT GTG AAG TTA TTG GAG TGA) and the reverse primer (TCC TAC TCC TCC TTT ACT TTT ATC TGC T), as reported previously (16). The efficiencies of primers were 100% for SYBR Green-based quantitative PCR.
TABLE 1.
Primers for real-time PCR of ehcp genes
Nuclear and Cytoplasmic Protein Extraction
Axenic or cecum-passaged trophozoites were harvested at logarithmic phase for nuclear and cytoplasmic protein extraction. Crude nuclear extracts were prepared following the method described previously (35) with additional proteinase inhibitors (1 mm phenylmethylsulfonyl fluoride, 100 μm E64, Complete One protease inhibitor mixture (Roche Applied Science)).
E. histolytica Conditioned Media (CM)
Released proteinases were prepared as conditioned media as described previously (16). After incubation of trophozoites (1 × 107 cells/ml) at 37 °C for 1.5 h, viability with 0.2% trypan blue was >95%. Protein from the conditioned media was precipitated with trichloroacetic acid/acetone.
E. histolytica Infection of Mice
C3H/HeJ or CAB/J male mice (The Jackson Laboratory, Bar Harbor, ME) were maintained under specific pathogen-free conditions. To increase the infection rate, mice were pretreated with dexamethasone (0.2 mg intraperitoneally daily) for 4 days prior to surgery. Under general anesthesia, the cecum was externalized as described previously (36) and injected with 2 × 106 cecum-passed E. histolytica strain HM-1 trophozoites, which were preincubated with WRR605 (50 μm) or an equivalent volume of stock buffer alone for 30 min prior to infection. Following the inoculation, the mice were treated intraperitoneally with 50 mg/kg compound twice daily or PBS with 25% Cremophor EL for 7 days. The stock solution of the compound was first dissolved in 100% ethanol, diluted to 50% in Cremophor EL (Sigma), and injected with 25% of Cremophor EL in PBS. The extent of amebic infection and intensity of the host response were determined by histopathology, quantification of trophozoites in the cecum by real-time PCR (16), and myeloperoxidase (MPO) activity. The entire cecum was frozen, weighed, homogenized, and extracted with the QIAamp DNA Stool Mini kit (Qiagen). The number of amebic trophozoites in cecal tissues was determined by comparison with a standard curve generated with the DNA extracted from trophozoites added to an uninfected control cecum. Trophozoite-specific DNA was detected with primers to the E. histolytica peroxiredoxin gene using SYBR Green Quantitative PCR in a Step One Plus real-time PCR machine (Applied Biosystems) (16). MPO activity has been shown proportional to the number of neutrophils in the inflamed intestine (37). The activity was measured in the extracted cecal pellet and compared with a standard curve using pure MPO (Sigma) as described previously (9), with a range from 0.01 to 0.31 units. All animal studies were reviewed and approved by the University of California, San Diego, Institutional Animal Care and Use Committee.
RESULTS
Expression and Refolding of Recombinant EhCP4
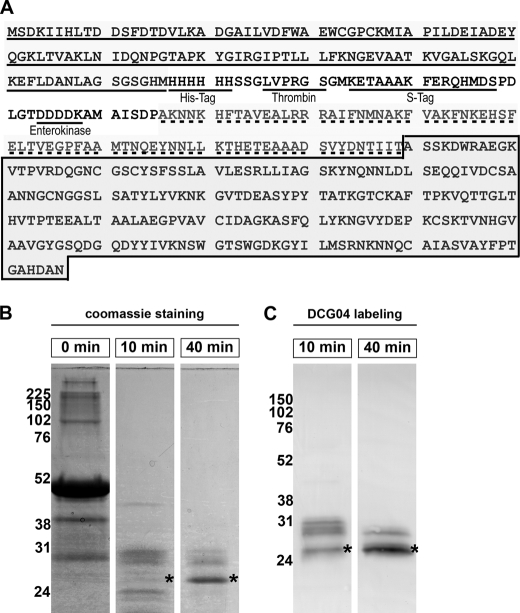
To define the biochemical properties of EhCP4, we cloned the zymogen, including the pro-domain and catalytic domain, into bacterial expression vectors with an N-terminal His6 affinity purification tag (Fig. 1A). Although the protein was not expressed in a soluble form, a relatively high level of expression (average 47 mg/liter of culture) was achieved in BL21 Codon Plus (DE3) RIPL (Stratagene) cells as inclusion bodies. Although existing refolding methods of EhCPs (16, 17, 38) failed to solubilize and recover active EhCP4, a refolding system that was modified from conditions used to refold Falcipain-2 (22) successfully produced active enzyme. Like other papain family cysteine proteinases (39), EhCP4 underwent autocatalytic activation in buffers containing 5 mm reducing reagent (e.g. DTT) (Fig. 1, B and C). Formation of mature enzyme during activation was demonstrated by Coomassie staining of protein fractionated on a 12% polyacrylamide gel (Fig. 1B) and the Western blot using an activity-based cysteine proteinase probe, DCG04 (Fig. 1C). The refolding process was relatively inefficient because <1% of the denatured zymogen refolded to active, mature proteinase. Multiple intermediate fragments were generated during the activation process (Fig. 1, B and C, and supplemental Fig. S1A) Mature rEhCP4 was unstable at acid pH and underwent further degradation (supplemental Fig. S1B). Protein identities of the intermediate fragments, mature enzyme, and degradation products were confirmed by mass spectrometry (supplemental Fig. S1). Amino acid sequencing identified the N-terminal peptides of mature rEhCP4 starting from ASSKD (Fig. 1A). The calculated molecular mass of the mature enzyme was 23 kDa, and the apparent molecular mass was 26 kDa shown on 12% SDS-polyacrylamide gels.
FIGURE 1.
Cloning and refolding of recombinant EhCP4. A, EhCP4 was cloned as a fusion protein with bacterial thioredoxin A (underline) with the His tag (underline) at the amino terminus of the prodomain of EhCP4 (dotted line). The mature enzyme is highlighted with gray in the box. B, time course of autocatalytic processing of rEhCP4 (Coomassie Blue stain). C, a Western blot shows formation of active enzyme with its active site labeled by the activity-based cysteine proteinase probe, DCG04. *, location of the mature enzyme on the gels.
Characterization of Recombinant EhCP4 Enzymatic Activity
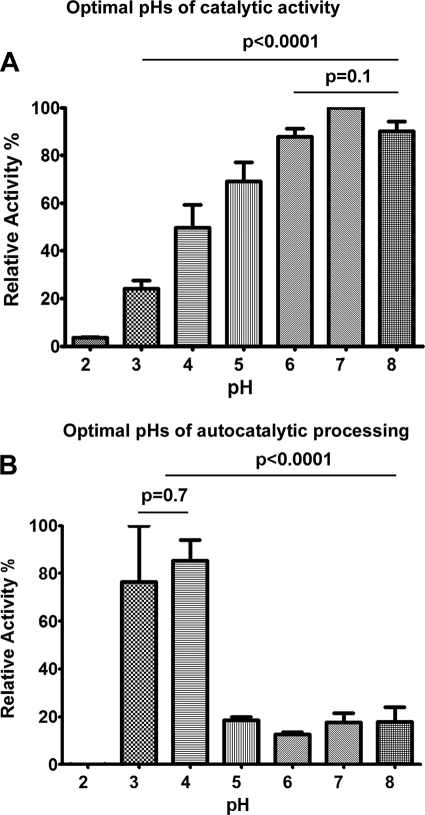
rEhCP4 did not cleave canonical, synthetic cathepsin B or L substrates (e.g. Z-RR-AMC or Z-FR-AMC peptides, the optimal substrates of EhCP-1, -2, -3, and -5) (14). Instead, it reacted with Suc-LLVY-AMC and Z-VVR-AMC. The optimum pH for substrate cleavage was 7 (Fig. 2A), whereas rEhCP4 was efficiently autoactivated at pH 3–4 with 5 mm DTT (Fig. 2B). At pH 7, the average Km with Suc-LLVY-AMC was 31 μm, and the average Vmax was 1.84 nm/s. With Z-VVR-AMC, the average Km was 30 μm, and the average Vmax was 6.39 nm/s. Using Z-VVR-AMC, the Km values of rEhCP4 at pH 4 (78 μm) and pH 8 (46 μm) were higher than that at pH 7.
FIGURE 2.
Optimal pH values of proteolytic activity and autocatalytic processing of rEhCP4. A, relative activities of rEhCP4 at different pH values are shown (the highest activity, set as 100%). The optimal pH of catalytic activity of rEhCP4 was pH ∼7. B, effect of pH on the autocatalytic processing of pro-rEhCP4 to produce mature enzyme (relative to the highest activity as 100% when assayed at pH 7). Data were collected from three experiments. p values, comparing multiple pH values, were calculated by two-way analysis of variance, and p values between two pH values were calculated using Student's t test. Error bars, S.E.
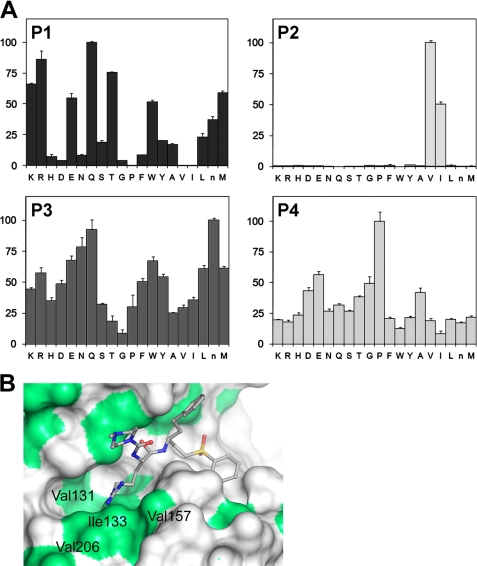
To confirm the preference of rEhCP4 for hydrophobic residues at the P2 position, we screened a P1–P4 substrate library (40) and identified Val and Ile as the preferred substrate of rEhCP4 at the P2 position (Val > Ile), with proline at P4 but a lack of specificity of residues at P1 and P3 (Fig. 3A).
FIGURE 3.
Substrate specificity of rEhCP4. A, P1–P4 substrate library screening. EhCP4 has a preference for hydrophobic amino acids, valine and isoleucine at the P2 position and broader specificities at the P1 and P3 positions. At P4, proline is relatively selective. B, homology modeling of EhCP4 shows the surface representation of the modeled EhCP4 active site. Residues lining the S2 pocket are labeled, and hydrophobic regions are colored green. The S2 pocket is located by superimposition of the model with the crystal structure of Cruzain complexed to a P2-Arg-containing inhibitor. This figure was generated in PyMOL (55). Error bars, S.E.
Homology Modeling of EhCP4
To understand the mechanism of the substrate preference of EhCP4, we performed homology modeling using a cathepsin L-like enzyme of known structure as the template. A BLAST search of the Protein Data Bank identified F. hepatica cathepsin L1, FheCL1, as the cathepsin L1-like template with the highest identity to EhCP4 (46%, E value = 2 × 10−48). Additionally, the FheCL1 structure was the most accurate of the top five templates with a resolution of 1.4 Å and an Rfree/Rfactor of 16.5%/12.9%. The homology model illustrates that EhCP4 is a member of the papain family of cysteine proteases. Although this was expected, given the high degree of primary sequence similarity found in sequence alignments of members of this family, modeling experiments allow for analytical consideration of biochemical observations based on a predicted three-dimensional model of the active site region of EhCP4. The active site lies at the interface between the two domains, and the highly conserved catalytic triad (Cys24-His159-Asn179) is in the expected position, similar to EhCP2 (14). The presence of an ERFNIN-like motif in their pro-domain sequences identifies papain family cysteine proteases of E. histolytica as cathepsin L-like. However, a common theme in a subset of EhCPs is a substrate specificity with a strong preference for basic residues (e.g. Arg at the P2 position) (14), a feature that is reminiscent of cathepsin B substrate specificity. In EhCP1, EhCP2, and EhCP3, the affinity for a positively charged residue can be correlated with the presence of a negatively charged Asp at the bottom of the predicted S2 pocket (41), a position that is often a key determinant of specificity in papain family cysteine proteinases (42). Superimposition of the EhCP4 model with Cruzain bound to a P2-Arg-containing inhibitor (2EFM, 191 matching a-carbons, root mean square deviation 1.0 Å) allows important residues in the S2 pocket to be identified (Fig. 3B). The hydrophobic nature of the S2 pocket, with Val206 in the base, explains the enzyme's tolerance for the hydrophobic valine and isoleucine at the P2 position, whereas the lack of any polar/charged residues around the bottom of the S2 pocket explains why residues of this type are poorly accepted at the P2 position of substrates.
Expression and Localization of Endogenous EhCP4 in E. histolytica Trophozoites
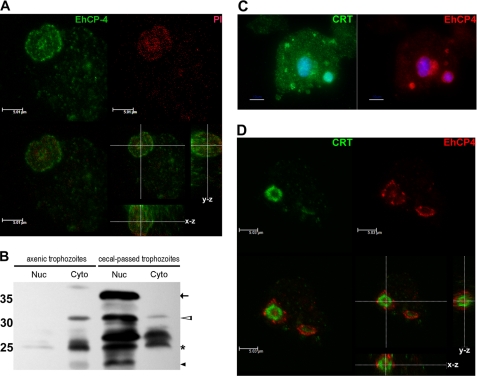
To identify the endogenous expression of EhCP4 in trophozoites, immunofluorescence imaging performed with anti-EhCP4 antibody revealed that EhCP4 was localized in the peri- and intranuclear regions as well as in variably sized vesicles in the cytoplasm (Fig. 4A). Immunoblots further confirmed this finding and identified bands consistent with mature EhCP4 in nuclear extracts and both zymogen and mature EhCP4 in the cytoplasmic fraction of all trophozoites. In axenic trophozoites, pro-EhCP4 and mature EhCP4 are primarily cytoplasmic (Fig. 4B). In contrast, in trophozoites isolated from infected mouse ceca, both nuclear and cytoplasmic extracts contained the EhCP4 zymogen (the sizes of pre- and pro-EhCP4), the mature enzyme, and degraded fragments of EhCP4, suggesting that processing of EhCP4 occurred in the nucleus and cytoplasm (Fig. 4B). The identities of the ∼25 kDa bands were confirmed by mass spectrum analysis of the excised gel bands. In samples from both the nuclear and cytoplasmic protein lanes, we detected two peptides, TVNHGVAAVGYGSQDGQDYYIVK and GVTDEASYPYTATK, from mature EhCP4 (coverage >5% in two independent runs). Moreover, compared with the same number of axenically cultured E. histolytica trophozoites, cecal trophozoites apparently had more EhCP4, which was located mostly in the nuclear fraction. This observation coincided with the fact that transcription of ehcp4 is up-regulated in cecal colitis (20), suggesting that the trophozoites require EhCP4, especially the nuclear form, to survive in the milieu of host environment.
FIGURE 4.
Nuclear and cytoplasmic localization of EhCP4 in E. histolytica trophozoites. A, an optical section (laser scan microscopy) of an E. histolytica trophozoite stained with anti-EhCP4 antibody (green). The nucleus was stained with propidium iodide (PI; red). Z-Stack thickness was 0.1 μm, scan depth was 13.1 μm, and scale bar is 5.01 μm. B, immunoblot with EhCP4 antibody to nuclear protein (Nuc) and cytoplasmic protein (Cyto) of E. histolytica trophozoites from axenic culture and trophozoites isolated from infected mouse ceca and separated on a 15% SDS-polyacrylamide gel (1 × 105 trophozoites/lane). Arrow, prepro-EhCP4 (∼34 kDa); hollow arrowheads, pro-EhCP4 (∼32 kDa); star, mature EhCP4 (∼26 kDa); black arrowhead, degraded EhCP4. C, localization of EhCP4 and calreticulin. Epifluorescence microscopy identified CRT (green) surrounding the nuclear area (4′,6-diamidino-2-phenylindole (DAPI), blue) and punctate staining in the cytoplasmic vesicles. EhCP4 (red) was associated with CRT+ structures. Scale bar, 10 μm. D, an optical section of the nuclear area showed that EhCP4 (red) was distributed in the nucleus and on the cytoplasmic side of the CRT+ structure (green). Z-Stack thickness was 0.3 μm, scan depth was 7.6 μm, and scale bar is 5.03 μm.
An ER-like structure that continuously distributes from nucleus to cytoplasm has been recently identified in Entamoeba (43). We hypothesized that the perinuclear localization of EhCP4 might be associated with the ER structure. CRT, an ER marker, was stained in axenic trophozoites and demonstrated a vesicular pattern in the cytoplasm and around the nucleus of trophozoites (Fig. 4C). We found that EhCP4 was distributed on either side of the CRT-stained structure that surrounded the nucleus (Fig. 4D). Similar staining was obtained in trophozoites isolated from infected ceca (data not shown). Co-localization of EhCP4 with CRT-stained ER structure suggests that synthesis and transportation of EhCP4 could be associated with the perinuclear ER.
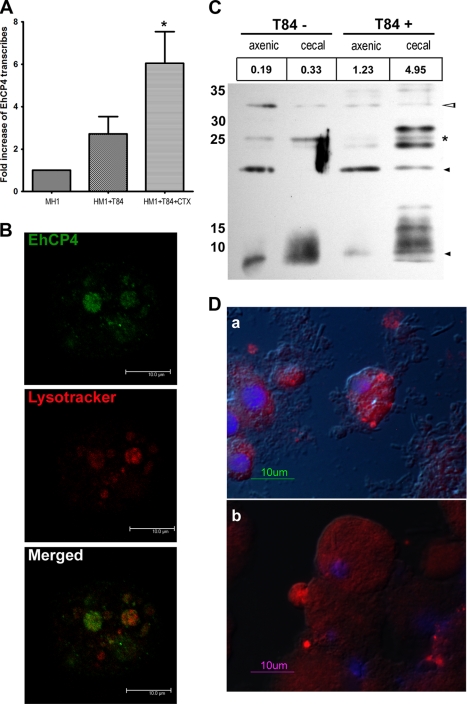
To examine the distribution and localization of EhCP4 in a milieu mimicking the early events of host-parasite interaction, we established a simpler in vitro model using an enteric cell line, T84. Following overnight co-culture of trophozoites with confluent T84 monolayers, the trophozoites were collected. Quantitative PCR analysis demonstrated that the expression level of ehcp4 mRNA was elevated by 3-fold (Fig. 5A). We hypothesized that mucin expressed in T84 cells may further induce EhCP4 expression. Indeed, when trophozoites were co-incubated with the T84 cells that had been pretreated with cholera toxin to stimulate mucin secretion, the mRNA level of ehcp4 was further increased to 6-fold compared with axenically cultured trophozoites alone (Fig. 5A). Noticeably, following co-culture with T84 cells, EhCP4 appeared in large vesicles that were stained by LysoTracker, indicating that these vesicles were acidic lysosome-like structures (Fig. 5B).
FIGURE 5.
Endogenous EhCP4 expression and release. A, up-regulation of ehcp4 mRNA following co-culture with T84 cells. Quantitative reverse transcription-PCR was performed on E. histolytica in culture medium alone, co-cultured with T-84 cells alone, and co-cultured with T-84 cells pretreated by cholera toxin to stimulate mucin secretion (*, p = 0.03, Student's t test). B, EhCP4 localizes to acidic vesicles. An optical section of an E. histolytica trophozoite stained with anti-EhCP4 antibody (green) and LysoTrackerTM Red DND-99 (red) showed EhCP4 localized to acidic vesicles, following co-culture with T84 monolayers for 2 h. Z-Stack thickness was 0.5 μm, scan depth was 10.1 μm, and scale bar is 10 μm. C, Western blot of EhCP4 released. CM proteins from 8.8 × 105 trophozoites were trichloroacetic acid-precipitated and resolved with a 15% SDS-polyacrylamide gel. EhCP4 protein release was detected in CM from E. histolytica trophozoites in medium alone (T84−) and from trophozoites following an overnight co-culture with a T84 monolayer (T84+). CM from axenically cultured E. histolytica trophozoites and the CM from trophozoites that infected mouse ceca are indicated as axenic and cecal, respectively. The numbers (RFU/min) above the lanes indicate the proteinase activity represented by Z-VVR-AMC. Hollow arrowhead, pro-EhCP4 (∼32 kDa); star, mature EhCP4 (∼26 kDa); black arrowhead, degraded EhCP4. D, EhCP4 release in vivo. EhCP4 (red) is shown in vesicles on the trophozoite surface by immunofluorescence staining with anti-EhCP4 antibody. Sections (8 μm) are from mouse ceca 1 week postinoculation (a) and 2 weeks postinoculation (b), respectively. Images were obtained by epifluorescence microscopy overlaid with differential interference contrast image.
Like EhCP1, -2, and -5, EhCP4 was also released into media. In the trophozoite CM, EhCP4 was found as both zymogen and mature enzyme. The trophozoites obtained from the infected mouse ceca released more EhCP4 than the same number of axenic trophozoites (Fig. 5C). In addition, when the trophozoites from the ceca were co-cultured with T84 overnight, the same number of trophozoites apparently released more EhCP4 into the CM than trophozoites in media alone, as indicated by the immunoblot and EhCP4-specific activities by Z-VVR-AMC (Fig. 5C). However, more degraded EhCP4 was also observed in this condition, indicating that EhCP4 might have a short half-life (Fig. 5C). As a control, bacterial flora alone did not degrade the EhCP4 substrate in this condition. Release of EhCP4 was further confirmed in vivo, on sections from infected mouse ceca. The cecal trophozoites exhibited cell surface patches and vesicles that were heavily stained by EhCP4 antibody (Fig. 5D).
Host Proteins Digested by EhCPs
To identify potential host proteins degraded by EhCP4, we first examined the substrates that are known to be degraded by other released EhCPs. Because rEhCP4 showed a strong preference for hydrophobic residues at the P2 position of the substrates, we predicted that the fragments digested by rEhCP4 would be different from those reported previously. EhCP1 cleaves the third component of complement, one amino acid residue proximal to the C3 convertases, forming an active C3b molecule (2, 16). When C3 was incubated with EhCP4, the primary product was a fragment of 70 kDa, a size similar to that of C3b (supplemental Fig. S2A). The C3b-like fragment was further proteolytically degraded by EhCP4 (supplemental Fig. S2A), which was unlike EhCP1-mediated C3 processing. Like EhCP1 (16), EhCP4 also degraded IgA (supplemental Fig. S2B) and IgG (supplemental Fig. S2C) as well as pro-IL-18 (supplemental Fig. S2D). In silico analysis of potential physiologic proteins based on the peptide substrate specificity of rEhCP4, identified two additional potential targets, laminin and villin, as amebic cysteine proteinases degrade extracellular matrix (e.g. laminin) and disrupt the brush border of intestinal epithelial cells (e.g. villin) (4, 44). Both villin-1 (supplemental Fig. S3A) and laminin-1 (supplemental Fig. S3B) were degraded, but different fragments were detected following cleavage by EhCP4 compared with EhCP1.
Specific Inhibitors of EhCP4
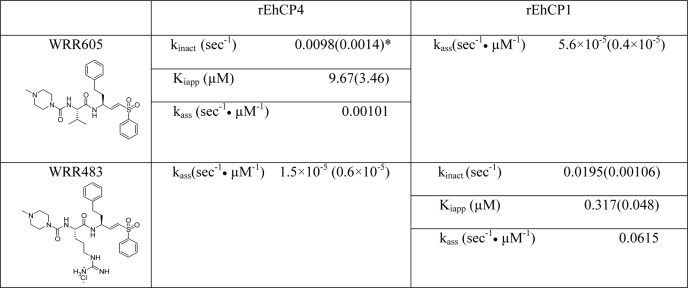
In order to study the biological function of EhCP4, we synthesized an inhibitor, WRR605, based on the substrate preference and homology modeling, to target the specific S2 binding pocket of EhCP4. The compound was derived from a vinyl sulfone cysteine proteinase inhibitor, K11777, with a dipeptidyl moiety occupying the S2 subsite of the targeted enzyme (16). As expected, WRR605 selectively inhibited rEhCP4, whereas WRR483, which targeted the S2 binding pocket of EhCP1, selectively inhibited rEhCP1. The irreversible inhibitor kinetics of these inhibitors to rEhCP4 and rEhCP1 were summarized in Table 2. For the panpapain-like cysteine proteinase inhibitor, E-64, the ka value (s−1 μm−1) for rEhCP4 was 0.017, and the value for rEhCP1 was 0.00026. To demonstrate that WRR605 bound intracellular EhCP4, we used BODIPY-labeled WRR605 in localization experiments. BODIPY-WRR605 co-localized with nuclear EhCP4 (supplemental Fig. S4A). The same nuclear localization was not seen with free BODIPY-TMR alone, which was pinocytosed by the amebic trophozoites and located in the cytoplasm (supplemental Fig. S4B). In addition, BODIPY-WRR605 and WRR605 competitively bound to rEhCP4 in vitro (supplemental Fig. S4C), indicating that modification of WRR605 by BODIPY did not alter the specificity toward EhCP4.
TABLE 2.
Inhibitor kinetics of rEhCP4 and rEhCP1
kass = kinact/Kiapp, the apparent rate of association or inhibition; second-order rate constants defining the efficiency of enzyme inactivation by the inhibitor.
*, Values in parentheses are S.E.
Based on computer modeling, we would predict that four other cysteine proteinases might have a substrate specificity similar to that of EhCP4 and be inhibited by WRR605: EhCP11, with a Val at the base of the S2 pocket, and EhCP7, EhCP15, and EhCP18, which have an Ile at the base. mRNA levels of these proteinases were measured by quantitative PCR and found to be significantly lower than ehcp4; ehcp11 had 14% of ehcp4 levels, ehcp15 had 25%, and ehcp18 had 4%. Levels of ehcp7 mRNA could not be accurately determined because of the high homology of ehcp7 and ehcp11 sequences. These results were supported by the previous microarray data (21), which showed that transcripts of these enzymes, including ehcp7 in trophozoites, were only 0.2–0.8% of EhCP4.
Efficacy of the Specific EhCP4 Inhibitor in a Mouse Cecal Model of Amebiasis
Because only humans and higher primates are naturally susceptible to E. histolytica infection, we had to adapt a murine amebic colitis model to investigate the role of EhCP4 during invasion and colonization. Houpt's group (36) had previously established the model with C3H/HeJ or CAB/J mice by inoculating cecum-passed E. histolytica trophozoites cultured with enteric flora into a surgically exposed cecum. By pretreating the C3H/HeJ mice with 0.2 mg of dexamethasone intraperitoneally daily for 4 days prior to surgery and using trophozoites isolated from infected ceca within a month, we obtained an infection rate of 97.5% (40 of 41 mice) by the end of week one.
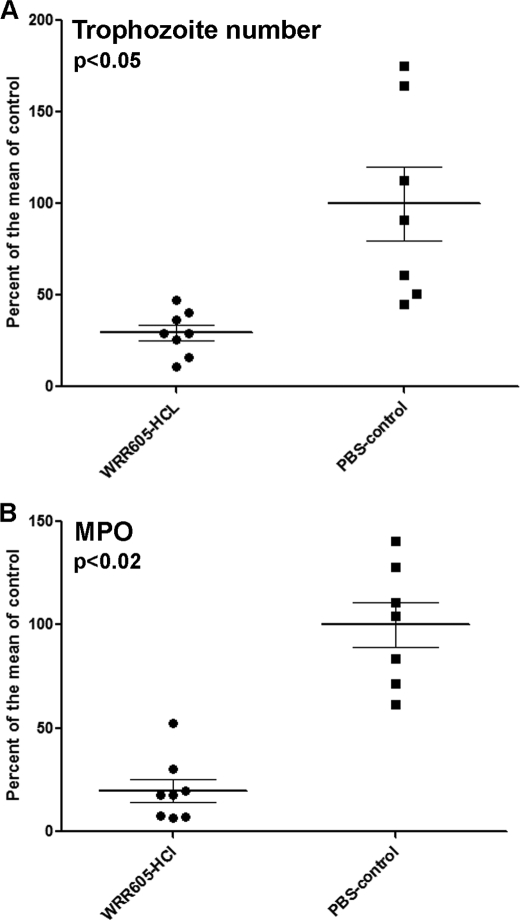
In order to target both the initial invasion and the subsequent established infection, trophozoites were pretreated with WRR605 before inoculation. The inoculated mice were then treated twice daily by intraperitoneal injection of WRR605 for 7 days. Treatment with WRR605 significantly decreased the trophozoite burden as well as the intensity of cecal inflammation as measured by MPO activity (Fig. 6). These results indicate that EhCP4 plays an important role in amebic colitis, which can be blocked with specific peptide inhibitors.
FIGURE 6.
WRR605 decreases the parasite burden and inflammation in the infected cecum. The effect of treatment of mice with WRR605 is presented as the percentage of trophozoites/g of tissue or MPO units/g of tissue compared with the means of infected controls (as 100%). A, treatment with WRR605 significantly reduced the number of cecal trophozoites at 1 week (p < 0.05). B, MPO levels were also significantly decreased (p < 0.02) in the WRR605-treated group. p values were generated by Student's t test.
DISCUSSION
Cysteine proteinases are well described virulence factors of the human enteric pathogen, E. histolytica (5, 11, 14, 17). Recent reports identified differential expression of the cysteine proteinase genes in cysts and trophozoites obtained from in vitro culture versus in vivo infection (20, 21). The functions of most of these EhCPs have not been investigated. Here, we focused on EhCP4, the most up-regulated gene when trophozoites invade and colonize murine cecal tissue (20).
EhCP4 has a number of unique features compared with the well characterized amebic cysteine proteinases. Sequence alignment and computer modeling (Fig. 3B) confirmed that EhCP4 is a member of the Clan CA, C1A subfamily with a cathepsin L-like structure (13, 14, 45), which is similar to previously characterized EhCPs (14, 16, 45). However, EhCP4 does not cleave any canonical, synthetic cathepsin B or L substrates. Both peptide mapping and computer modeling confirmed that EhCP4 has a substrate preference for small hydrophobic residues at the P2 position (Fig. 3). Indeed, the dipeptidyl vinyl sulfone inhibitor, WRR605, which has Val (P2) and Phe (P1) as the probe moiety binding to the enzyme, has a specific inhibitory effect against EhCP4 but not EhCP1, whereas WRR483, which has Arg (P2) and Phe (P1) as the probe, inhibited EhCP1 (16) but not EhCP4 (Table 2). Based on computer modeling of the S2 pocket, four other papain-like EhCPs could have a substrate preference similar to that of EhCP4. However, EhCP7, EhCP11, EhCP15, and EhCP18 are expressed at <25% of the level of mRNA for EhCP4 and are highly expressed in Entamoeba cysts (21) rather than in trophozoites Thus, EhCP4 should be the primary, endogenous target of WRR605 in trophozoites.
Another distinguishing property of EhCP4 is an absolute requirement of an acidic pH to undergo autocatalytic conversion from a zymogen to a mature enzyme (Fig. 2B), a biochemical behavior similar to papain and cathepsins (39). In contrast, activation of recombinant EhCP1, -2, -5, and -112 is conducted either at a slightly alkaline condition (15, 17, 18) or neutral pH (16). Consistent with this feature of EhCP4, the endogenous enzyme appeared in LysoTracker-labeled acidic vesicles (Fig. 5B) so that the acidic environment could promote not only autocatalytic activation but also subsequent autodegradation of the mature proteinase. These data may explain the observation that polyclonal anti-EhCP4 antibody was able to detect multiple truncated EhCP4 bands in cellular and secreted protein samples (Figs. 4B and 5C). These results suggest that the endogenous zymogen of EhCP4 could be activated in lysosomes where the acidic pH leads to autocatalytic activation, but it may have a short half-life. Indeed, the amount of EhCP4 in amebic phagosomes changes over time (46). During phagosome acidification and maturation, the pH drops rapidly by the first 15 min (47). The peptide coverage of EhCP4 in phagosomes identified by mass spectrometry falls from >5% to 1–5% by 30 min (46). In late stage phagosomes, the amount of EhCP4 increases again, probably due to new enzymes transported into the phagosome by lysosome-phagosome fusion. Thus, this apparently rapid turnover of EhCP4 may allow dynamic post-translational control of enzyme levels.
EhCP4 is also a multifunctional proteolytic enzyme. First, its localization was not limited to acidic proteolytic vacuoles inside trophozoites because EhCP4 was released into culture medium, where its optimal proteolytic activity was obtained at physiological pH. Indeed, in the infected murine cecum, immunofluorescence staining of EhCP4 demonstrated strong signals in vesicle-like structures located in discrete surface areas. Thus, EhCP4 was apparently secreted into the microenvironment in vivo. During the cross-talk between enterocytes and trophozoites, villin proteolysis is one of the early events causing disruption of microvilli, due to EhCP activity (44). Although we found that either EhCP1 or EhCP4 could digest human villin-1 and laminin (supplemental Fig. S3), the digestion fragments were different, as would be expected from their substrate specificities. At focal areas where trophozoites and epithelial cells made contact, the secreted EhCP4 could be at relatively high concentrations, so that it could play an important part in destroying the integrity of host tissue synergistically with other EhCPs. In addition to the intestinal structural proteins, IgA and IgG, pro-IL-18, and complement C3 were also targets of EhCP4 (supplemental Fig. S2). Thus, like EhCP1, EhCP4 may be involved in evading the host immune system and contribute to the inflammatory response in amebic lesions (2).
Immunoblotting and confocal optical sectioning both confirmed the presence of EhCP4 in the nuclear region of trophozoites. The nuclear localization pattern of EhCP4 appeared identical to that of a nuclear cathepsin L at the G1/S transition phase of mammalian cells (48). However, the exact physiological function of the nuclear EhCP4 is not clear. At least five cysteine proteinases in higher eukaryotic cells have been shown to have nuclear localization: a cathepsin L isoform missing its regular signal peptide (48, 49), a cathepsin B-like cysteine proteinase (50), cathepsin F (51), mouse cathepsin 7 (52), and a plant papain-like cysteine proteinase RD19 (53). These enzymes regulate events such as DNA replication, chromatin assembly, activation or deactivation of transcription factors, and cell cycle-related proteins. Their nuclear localization is usually associated with cell cycle or differentiation. It is possible that nuclear proteins of trophozoites can be substrates of EhCP4 and that it functions in chromatin organization and transcription regulation during invasion and colonization in the cecal tissue. Interestingly, EhCP4 has no typical nuclear localization signal; nor do most other nuclear cysteine proteinases. The mechanisms of nuclear transport are still unknown. Chaperones and cofactors that mediate a co-transportation into the nucleus may be involved.
Unlike the most abundant cysteine proteinases, EhCP1, -2, and -5, expression of ehcp4 responds to the environmental changes dramatically, as shown by Gilchrist et al. (20) in mice and by our in vitro studies with a mucin-producing colonic cell line (54), suggesting that EhCP4 is regulated following exposure to intestinal cells. Therefore, the functions of EhCP4 cannot be demonstrated simply by in vitro culture. Indeed, incubation of the EhCP4-specific inhibitor, WRR605, with axenically cultured trophozoites did not inhibit cellular proliferation or endocytosis (data not shown). To study the function of EhCP4 in vivo, we adapted a murine cecal model of infection (36) using C3H/HeJ male mice. Systematic delivery of WRR605 by intraperitoneal injection for 7 days resulted in a significant attenuation of trophozoite burden and decrease of inflammation in the infected cecum (Fig. 6). Pretreatment of trophozoites with WRR605 before inoculation did not completely block invasion as was seen with the cysteine proteinase inhibitors, K11777 and WRR483, that target EhCP1, -2, -3, and -5 and block acute invasion in human colonic xenografts (16). We hypothesize that E. histolytica may utilize different cysteine proteinases during initial invasion and subsequent maintenance of infection.
In summary, we have cloned and characterized an amebic cysteine proteinase, EhCP4, with a unique substrate specificity and nuclear location. It is up-regulated following exposure to colonic cells, active in phagosomes, and released extracellularly. Substrate mapping led to the design of a specific inhibitor, which attenuated infection in a mouse model of cecal amebiasis. These observations prove that cysteine proteinases can be targeted with specific inhibitors and strongly support their potential as effective drug targets for the treatment of amebiasis.
Supplementary Material
Acknowledgments
We are grateful for the technical support and the initial cecum-passed E. histolytica trophozoites from S. Stroup and Dr. X. T. Guo (University of Virginia). We thank the mouse surgery supporting team, G. Hwang, Y. Anderson, J. Scholtz, and H. Yoo (University of California, San Diego). We thank Dr. J. Engel (University of California, San Francisco) and J. Steinauer and Dr. L. Ellies (University of California, San Diego) for technical advice and support of microscopy; Dr. M. Diccianni (University of California, San Diego) for fluorescence scan; Dr. M. Ramos and Dr. S. Meléndez-López (Universidad Autónoma de Baja California, México) for general advice; and Dr. S. J. Bark (University of California, San Diego) for identification of the purified recombinant EhCP4 antigen for antibody production. In addition, we thank Dr. C. Davis for critical review of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants 1-UO1-AI077822 (to S. L. R., J. M., and W. R. R.) and R01 CA128765 (to E. L. S. and C. S. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
C. D. Emal, E. Hansell, A. Alvarez-Hernandez, P. Y. Chong, B. Knapp-Reed, J. H. McKerrow, and W. R. Roush, submitted for publication.
- EhCP
- E. histolytica cysteine proteinase
- ER
- endoplasmic reticulum
- IL
- interleukin
- CRT
- calreticulin
- MPO
- myeloperoxidase
- CM
- conditioned medium
- AMC
- 4-amino-7-methylcoumarin
- Z-
- N-benzyloxycarbonyl-
- Suc
- N-succinyl
- DTT
- dithiothreitol
- RFU
- relative fluorescence units
- rEhCP
- recombinant EhCP
- PBS
- phosphate-buffered saline
- DPBS
- Dulbecco's PBS.
REFERENCES
- 1.Stanley S. L., Jr. (2003) Lancet 361, 1025–1034 [DOI] [PubMed] [Google Scholar]
- 2.Reed S. L., Keene W. E., McKerrow J. H., Gigli I. (1989) J. Immunol. 143, 189–195 [PubMed] [Google Scholar]
- 3.Ankri S., Stolarsky T., Bracha R., Padilla-Vaca F., Mirelman D. (1999) Infect. Immun. 67, 421–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keene W. E., Petitt M. G., Allen S., McKerrow J. H. (1986) J. Exp. Med. 163, 536–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moncada D., Keller K., Chadee K. (2003) Infect. Immun. 71, 838–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kelsall B. L., Ravdin J. I. (1993) J. Infect. Dis. 168, 1319–1322 [DOI] [PubMed] [Google Scholar]
- 7.Tran V. Q., Herdman D. S., Torian B. E., Reed S. L. (1998) J. Infect. Dis. 177, 508–511 [DOI] [PubMed] [Google Scholar]
- 8.Reed S. L., Ember J. A., Herdman D. S., DiScipio R. G., Hugli T. E., Gigli I. (1995) J. Immunol. 155, 266–274 [PubMed] [Google Scholar]
- 9.Zhang Z., Wang L., Seydel K. B., Li E., Ankri S., Mirelman D., Stanley S. L., Jr. (2000) Mol. Microbiol. 37, 542–548 [DOI] [PubMed] [Google Scholar]
- 10.Que X., Kim S. H., Sajid M., Eckmann L., Dinarello C. A., McKerrow J. H., Reed S. L. (2003) Infect. Immun. 71, 1274–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clark C. G., Alsmark U. C., Tazreiter M., Saito-Nakano Y., Ali V., Marion S., Weber C., Mukherjee C., Bruchhaus I., Tannich E., Leippe M., Sicheritz-Ponten T., Foster P. G., Samuelson J., Noël C. J., Hirt R. P., Embley T. M., Gilchrist C. A., Mann B. J., Singh U., Ackers J. P., Bhattacharya S., Bhattacharya A., Lohia A., Guillén N., Duchêne M., Nozaki T., Hall N. (2007) Adv. Parasitol. 65, 51–190 [DOI] [PubMed] [Google Scholar]
- 12.Tillack M., Biller L., Irmer H., Freitas M., Gomes M. A., Tannich E., Bruchhaus I. (2007) BMC Genomics 8, 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bruchhaus I., Loftus B. J., Hall N., Tannich E. (2003) Eukaryot. Cell 2, 501–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Que X., Brinen L. S., Perkins P., Herdman S., Hirata K., Torian B. E., Rubin H., McKerrow J. H., Reed S. L. (2002) Mol. Biochem. Parasitol. 119, 23–32 [DOI] [PubMed] [Google Scholar]
- 15.Sato D., Nakada-Tsukui K., Okada M., Nozaki T. (2006) FEBS Lett. 580, 5306–5312 [DOI] [PubMed] [Google Scholar]
- 16.Meléndez-López S. G., Herdman S., Hirata K., Choi M. H., Choe Y., Craik C., Caffrey C. R., Hansell E., Chávez-Munguía B., Chen Y. T., Roush W. R., McKerrow J., Eckmann L., Guo J., Stanley S. L., Jr., Reed S. L. (2007) Eukaryot. Cell 6, 1130–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hellberg A., Nowak N., Leippe M., Tannich E., Bruchhaus I. (2002) Protein Expr. Purif. 24, 131–137 [DOI] [PubMed] [Google Scholar]
- 18.Quintas-Granados L. I., Orozco E., Brieba L. G., Arroyo R., Ortega-López J. (2009) Protein Expr. Purif. 63, 26–32 [DOI] [PubMed] [Google Scholar]
- 19.Bruchhaus I., Jacobs T., Leippe M., Tannich E. (1996) Mol. Microbiol. 22, 255–263 [DOI] [PubMed] [Google Scholar]
- 20.Gilchrist C. A., Houpt E., Trapaidze N., Fei Z., Crasta O., Asgharpour A., Evans C., Martino-Catt S., Baba D. J., Stroup S., Hamano S., Ehrenkaufer G., Okada M., Singh U., Nozaki T., Mann B. J., Petri W. A., Jr. (2006) Mol. Biochem. Parasitol. 147, 163–176 [DOI] [PubMed] [Google Scholar]
- 21.Ehrenkaufer G. M., Haque R., Hackney J. A., Eichinger D. J., Singh U. (2007) Cell Microbiol. 9, 1426–1444 [DOI] [PubMed] [Google Scholar]
- 22.Sijwali P. S., Brinen L. S., Rosenthal P. J. (2001) Protein Expr. Purif. 22, 128–134 [DOI] [PubMed] [Google Scholar]
- 23.Tanner S., Shu H., Frank A., Wang L. C., Zandi E., Mumby M., Pevzner P. A., Bafna V. (2005) Anal. Chem. 77, 4626–4639 [DOI] [PubMed] [Google Scholar]
- 24.Stack C. M., Caffrey C. R., Donnelly S. M., Seshaadri A., Lowther J., Tort J. F., Collins P. R., Robinson M. W., Xu W., McKerrow J. H., Craik C. S., Geiger S. R., Marion R., Brinen L. S., Dalton J. P. (2008) J. Biol. Chem. 283, 9896–9908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sali A., Blundell T. L. (1993) J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 26.Roush W. R., Cheng J., Knapp-Reed B., Alvarez-Hernandez A., McKerrow J. H., Hansell E., Engel J. C. (2001) Bioorg. Med. Chem. Lett. 11, 2759–2762 [DOI] [PubMed] [Google Scholar]
- 27.Bieth J. G. (1995) Methods Enzymol. 248, 59–84 [DOI] [PubMed] [Google Scholar]
- 28.Beynon R., Bond J. S. (eds) (2001) Proteolytic Enzymes: A Practical Approach, pp. 144– 147,Oxford University Press, New York [Google Scholar]
- 29.Kerr I. D., Lee J. H., Farady C. J., Marion R., Rickert M., Sajid M., Pandey K. C., Caffrey C. R., Legac J., Hansell E., McKerrow J. H., Craik C. S., Rosenthal P. J., Brinen L. S. (2009) J. Biol. Chem. 284, 25697–25703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greenbaum D., Medzihradszky K. F., Burlingame A., Bogyo M. (2000) Chem. Biol. 7, 569–581 [DOI] [PubMed] [Google Scholar]
- 31.Athman R., Louvard D., Robine S. (2003) Mol. Biol. Cell 14, 4641–4653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Camenisch G., Tini M., Chilov D., Kvietikova I., Srinivas V., Caro J., Spielmann P., Wenger R. H., Gassmann M. (1999) FASEB J. 13, 81–88 [DOI] [PubMed] [Google Scholar]
- 33.Beck D. L., Boettner D. R., Dragulev B., Ready K., Nozaki T., Petri W. A., Jr. (2005) Eukaryot. Cell 4, 722–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davis P. H., Schulze J., Stanley S. L., Jr. (2007) Mol. Biochem. Parasitol. 151, 118–128 [DOI] [PubMed] [Google Scholar]
- 35.Gilchrist C. A., Mann B. J., Petri W. A., Jr. (1998) Infect. Immun. 66, 2383–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Houpt E. R., Glembocki D. J., Obrig T. G., Moskaluk C. A., Lockhart L. A., Wright R. L., Seaner R. M., Keepers T. R., Wilkins T. D., Petri W. A., Jr. (2002) J. Immunol. 169, 4496–4503 [DOI] [PubMed] [Google Scholar]
- 37.Lih-Brody L., Powell S. R., Collier K. P., Reddy G. M., Cerchia R., Kahn E., Weissman G. S., Katz S., Floyd R. A., McKinley M. J., Fisher S. E., Mullin G. E. (1996) Dig. Dis. Sci. 41, 2078–2086 [DOI] [PubMed] [Google Scholar]
- 38.Mitra B. N., Saito-Nakano Y., Nakada-Tsukui K., Sato D., Nozaki T. (2007) Cell Microbiol. 9, 2112–2125 [DOI] [PubMed] [Google Scholar]
- 39.Brömme D., Nallaseth F. S., Turk B. (2004) Methods 32, 199–206 [DOI] [PubMed] [Google Scholar]
- 40.Choe Y., Leonetti F., Greenbaum D. C., Lecaille F., Bogyo M., Brömme D., Ellman J. A., Craik C. S. (2006) J. Biol. Chem. 281, 12824–12832 [DOI] [PubMed] [Google Scholar]
- 41.Brinen L. S., Que X., McKerrow J. H., Reed S. L. (2000) Arch. Med. Res. 31, S63–64 [DOI] [PubMed] [Google Scholar]
- 42.McGrath M. E. (1999) Annu. Rev. Biophys. Biomol. Struct. 28, 181–204 [DOI] [PubMed] [Google Scholar]
- 43.Teixeira J. E., Huston C. D. (2008) Eukaryot. Cell 7, 1222–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lauwaet T., Oliveira M. J., Callewaert B., De Bruyne G., Saelens X., Ankri S., Vandenabeele P., Mirelman D., Mareel M., Leroy A. (2003) J. Biol. Chem. 278, 22650–22656 [DOI] [PubMed] [Google Scholar]
- 45.Jacobs T., Bruchhaus I., Dandekar T., Tannich E., Leippe M. (1998) Mol. Microbiol. 27, 269–276 [DOI] [PubMed] [Google Scholar]
- 46.Okada M., Huston C. D., Oue M., Mann B. J., Petri W. A., Jr., Kita K., Nozaki T. (2006) Mol. Biochem. Parasitol. 145, 171–183 [DOI] [PubMed] [Google Scholar]
- 47.Mitra B. N., Kobayashi S., Saito-Nakano Y., Nozaki T. (2006) Exp. Parasitol. 114, 57–61 [DOI] [PubMed] [Google Scholar]
- 48.Goulet B., Baruch A., Moon N. S., Poirier M., Sansregret L. L., Erickson A., Bogyo M., Nepveu A. (2004) Mol. Cell 14, 207–219 [DOI] [PubMed] [Google Scholar]
- 49.Duncan E. M., Muratore-Schroeder T. L., Cook R. G., Garcia B. A., Shabanowitz J., Hunt D. F., Allis C. D. (2008) Cell 135, 284–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pizzorno M. C. (2001) Biochim. Biophys. Acta 1536, 31–42 [DOI] [PubMed] [Google Scholar]
- 51.Maubach G., Lim M. C., Zhuo L. (2008) Mol. Biol. Cell 19, 4238–4248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Screen M., Dean W., Cross J. C., Hemberger M. (2008) Development 135, 3311–3320 [DOI] [PubMed] [Google Scholar]
- 53.Bernoux M., Timmers T., Jauneau A., Brière C., de Wit P. J., Marco Y., Deslandes L. (2008) Plant Cell 20, 2252–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Göttke M. U., Keller K., Belley A., Garcia R. M., Hollingsworth M. A., Mack D. R., Chadee K. (1998) J. Eukaryot. Microbiol. 45, 17S–23S [DOI] [PubMed] [Google Scholar]
- 55.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.