Abstract
We previously characterized nucleoredoxin (NRX) as a negative regulator of the Wnt signaling pathway through Dishevelled (Dvl). We perform a comprehensive search for other NRX-interacting proteins and identify Flightless-I (Fli-I) as a novel NRX-binding partner. Fli-I binds to NRX and other related proteins, such as Rod-derived cone viability factor (RdCVF), whereas Dvl binds only to NRX. Endogenous NRX and Fli-I in vivo interactions are confirmed. Both NRX and RdCVF link Fli-I with myeloid differentiation primary response gene (88) (MyD88), an important adaptor protein for innate immune response. NRX and RdCVF also potentiate the negative effect of Fli-I upon lipopolysaccharide-induced activation of NF-κB through the Toll-like receptor 4/MyD88 pathway. Embryonic fibroblasts derived from NRX gene-targeted mice show aberrant NF-κB activation upon lipopolysaccharide stimulation. These results suggest that the NRX subfamily of proteins forms a link between MyD88 and Fli-I to mediate negative regulation of the Toll-like receptor 4/MyD88 pathway.
Keywords: Adaptor Proteins, Cellular Regulation, MyD88, NF-κB, Toll-like Receptors
Introduction
Thioredoxin (TRX)4 is a highly conserved redox-regulating protein which catalyzes disulfide reduction in various target proteins via two conserved cysteine residues. The disulfide reduction brought about by TRX and other similar proteins maintains cellular redox status (1). Nucleoredoxin (NRX) was originally discovered as a novel member of the TRX protein family (2). We previously identified NRX as a Dishevelled (Dvl)-binding protein (3). Dvl is an essential adaptor molecule for Wnt signaling, which is involved in early development, stem cell maintenance, and tumorigenesis (4). NRX interacts with the basic-PDZ domain of Dvl in a redox-dependent manner and mediates the redox-dependent activation of the Wnt/β-catenin pathway. NRX also regulates the Wnt/planar cell polarity pathway, another major branch of the Wnt signaling pathway (5).
NRX has three conserved domains. There are two (N-terminal and central) tryparedoxin (TryX)-like domains (a subclass of the TRX domain) and one C-terminal protein disulfide isomerase-b′-like domain (Fig. 1A). TryX is a TRX-like redox-regulating protein in Trypanosoma, a parasitic protozoan that kills thousands of people each year (6). We previously found that among the TRX-related proteins, NRX, Rod-derived cone viability factor (RdCVF), and C9orf121 possess TryX-like domains (7). Whether these proteins share common functionality remains unknown.
FIGURE 1.
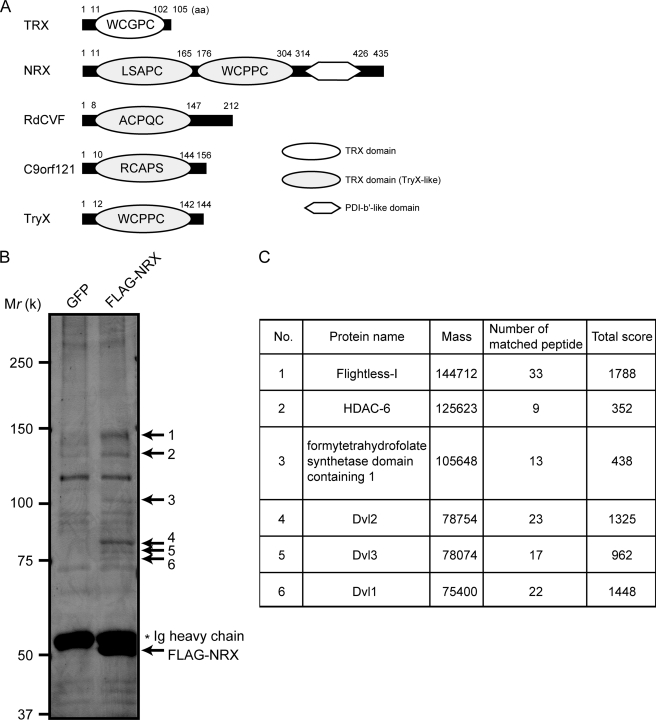
NRX co-precipitated proteins. A, schematics of TRX, NRX, RdCVF, C9orf121, and TryX. Amino acid residue numbers in each protein are also shown. B, lysate of NIH3T3 cells stably expressing FLAG-NRX or GFP immunoprecipitated with anti-FLAG antibody. The precipitated proteins were separated by SDS-PAGE and visualized by silver staining. The proteins corresponding to the bands (indicated by arrows) were subjected to MALDI-MS/MS analyses. C, results of the MALDI-MS/MS analyses. The names of the identified proteins, their calculated mass, the number of the peptides identified, and score are shown.
Flightless-I (Fli-I) was first identified in studies on Drosophila flightlessI mutants, which could not fly because of flight muscle defects (8, 9). It was subsequently discovered that Fli-I is conserved throughout species from nematodes to humans (10). Fli-I knock-out in mice leads to early embryonic lethality, indicating the vital importance of this molecule across species (11). Fli-I possesses a leucine-rich repeat in the N terminus and a gelsolin-like domain in the C terminus. Numerous cellular functions have been reported for Fli-I. In the nucleus, Fli-I acts as a cofactor in the nuclear receptor complex and facilitates the transcriptional activation of nuclear receptors such as the estrogen receptor and androgen receptor (12, 13). Cytosolic Fli-I binds to small GTPase Ras and localizes to the actin-based structures, probably through its gelsolin-like domain (14, 15). Fli-I also binds to proinflammatory caspases (caspase-1 and caspase-11) and suppresses caspase-1-mediated interleukin-1β maturation (16).
A comprehensive proteomic screen revealed that Fli-I is a major interacting partner with myeloid differentiation primary response gene (88) (MyD88) (17). MyD88 is known as an important adaptor protein for the Toll-like receptor (TLR) signaling pathway, essential to innate immunity and inflammation (18, 19). TLR4 recognizes lipopolysaccharide (LPS) and recruits IRAK1 and IRAK4 through MyD88, resulting in the activation of transcription factor NF-κB. Fli-I co-immunoprecipitates with MyD88 and inhibits the LPS-stimulated activation of the TLR4/MyD88 pathway (20), but its molecular detail is poorly characterized.
To search for a novel role for NRX, we performed a comprehensive screen for NRX-interacting proteins and identified Fli-I as a major binding partner of NRX as well as Dvl. We also found that recombinant Fli-I and MyD88 proteins do not bind directly in vitro, but do so only through NRX and that NRX potentiates the inhibitory effect of Fli-I on the TLR4/MyD88 pathway.
EXPERIMENTAL PROCEDURES
Mouse Embryonic Fibroblast (MEF) Isolation and Cell Culture
NRX-deficient (NRX−/−) mice were generated by homologous recombination to replace the latter half of exon 3 and whole exon 4 of mouse NRX gene with LacZ and neomycin resistance gene cassettes.5 We confirmed by immunoblotting that MEFs obtained from NRX−/− mice do not express NRX proteins (see Fig. 4E). MEFs were generated via a standard 3T3 method from C57BL/6J background wild type (WT) or NRX−/− mice. MEFs and COS7 cells were cultured in Dulbecco's modified Eagle's medium plus 10% fetal bovine serum and antibiotics.
FIGURE 4.
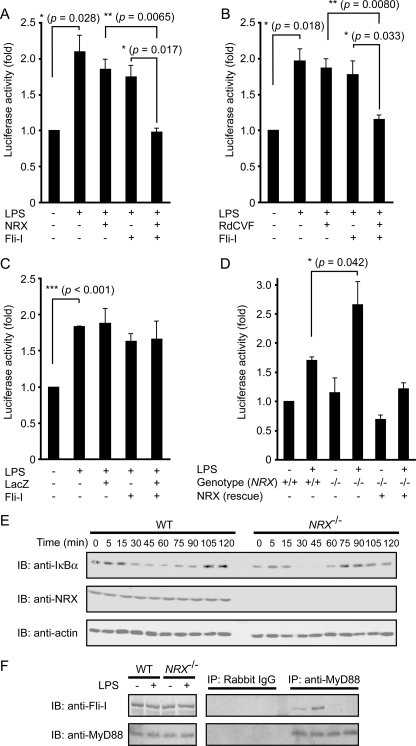
NRX synergistically suppresses the TLR4/MyD88 pathway with Fli-I. A–C, MEFs were transfected with the NF-κB reporter plasmid and the indicated expression constructs. The cells were stimulated with LPS (0.1 μg/ml, 6 h) before the reporter assays were performed. *, p < 0.05; **, p < 0.01; ***, p < 0.001 (Student's t test). Error bars, S.E. D, WT (+/+) or NRX-deficient (−/−) MEFs were transfected with the NF-κB reporter plasmid. *, p < 0.05 (Student's t test). E, WT or NRX−/− MEFs were treated with 0.1 μg/ml LPS for the indicated time periods, and their lysates were subjected to immunoblotting (IB) with anti-IκBα and anti-NRX antibodies. F, WT or NRX−/− MEFs were treated with LPS (1 μg/ml, 10 min), and their lysates were immunoprecipitated with anti-MyD88 antibody followed by immunoblotting with anti-Fli-I antibody. In this experiment, anti-MyD88 antibodies were covalently attached to the beads to avoid nonspecific signals.
Expression Constructs
Mouse NRX and TRX cDNAs were described previously (3). Murine MyD88 and human C9orf121 and RdCVF cDNAs were PCR-amplified from cDNA libraries derived from mouse liver (MyD88) and human embryonic kidney 293 cells (C9orf121 and RdCVF), respectively. The Fli-I construct was generated from a partial cDNA clone of human Fli-I, purchased from Invitrogen (IMAGE clone 4584634). The C-terminal region of Fli-I was filled up by PCR. All PCR products are sequence-verified.
NRX deletion mutants were generated by digestion with restriction enzymes. Amino acid residue numbers of the deletion constructs are as follows: N1 (1–176), N2 (1–302), M (177–327), and C (162–435). A C205S/C208S mutant of NRX (Mut) has been described previously (3). The LacZ expression vector (21) was a gift from Dr. K. Yoshikawa (Osaka University).
Recombinant Proteins
His-NRX proteins were purified as described (3). Recombinant GST-Fli-I proteins were expressed in Sf9 cells and purified with glutathione-Sepharose 4B (GE Healthcare). MBP-MyD88 was expressed in Escherichia coli strain BL21 DE3 and purified with amylose resin.
Antibodies
Anti-FLAG (mouse monoclonal) antibody was purchased from Sigma-Aldrich. Anti-Myc (rabbit polyclonal), anti-MyD88 (rabbit polyclonal, for immunoprecipitation), goat polyclonal (for immunoblotting), anti-IκBα (rabbit polyclonal), and anti-Fli-I (mouse monoclonal) antibodies were from Santa Cruz Biotechnology. Anti-Dvl and anti-NRX (rabbit polyclonal) antibodies were as described previously (3).
Modified Immunoprecipitation
To reduce IgG background levels, we covalently linked anti-NRX or anti-MyD88 antibody to protein A-agarose beads (Thermo Scientific) by incubating with 20 mm dimethylpimelimidate (Sigma) for 30 min at room temperature. Then, the beads were mixed with cell lysates. Immunoprecipitation experiments with other antibodies were performed using a standard method described elsewhere (22).
Reporter Assays
1 × 105 MEFs were seeded on 35-mm dishes and co-transfected with various expression constructs (Fli-I, NRX, RdCVF, or LacZ, or empty vector for controls) and with a pNF-κB reporter plasmid, using Lipofectamine 2000 (Invitrogen). pTK-Renilla was also transfected as a control to gauge the transfection efficiency in each experiment. 24 h later, the transfected cells were treated with 0.1 μg/ml LPS (Alexis Biochemical) for 6 h and harvested for luciferase assays using the Dual-Luciferase Reporter Assay System (Promega). The data are presented as mean ± S.E. (n = 3). Significance was determined by a two-tailed Student's t test.
Detection of Potential NRX-binding Proteins by Matrix-assisted Laser Desorption/Ionization Time-of-flight Tandem Mass Spectrometry (MALDI/TOF-MS/MS)
We generated an NIH3T3-derived cell line stably expressing FLAG-NRX or GFP. These cell lysates were used for immunoprecipitation with anti-FLAG M2-agarose (Sigma-Aldrich). Precipitated proteins were separated by SDS-PAGE and visualized with a SilverQuest™ silver staining kit (Invitrogen). The bands of interest were excised, trypsin-digested, and subjected to mass spectrometry as reported previously (3).
RESULTS
Identification of Novel NRX-binding Proteins
To discover potentially novel roles for NRX, we performed a comprehensive screen for NRX-binding proteins. We generated a cell line stably expressing FLAG-tagged NRX from NIH3T3 murine fibroblasts. Cell lysates were subjected to anti-FLAG immunoprecipitation followed by silver staining. We observed several bands of potential NRX-binding proteins (Fig. 1B). Six bands were excised and subjected to mass spectrometry. Based on the scores, which are the statistical parameters reflecting the probability that the experimentally obtained mass of peptides is in agreement with the calculated mass, we reliably identified all of the selected proteins (Fig. 1C). In addition to Dvl1–3, known binding partners of NRX, we also identified histone deacetylase-6, formyltetrahydrofolate synthetase domain containing 1, and Fli-I among the co-precipitates.
Association of Fli-I with NRX and Its Protein Subfamily
Among the identified proteins, we focused on Fli-I for further analyses based on its high scores and strong positive silver stain signal (Fig. 1, B and C; Fli-I was the only protein identified from this band 1). To confirm the interaction between the two proteins, we performed GST pulldown assays with purified Fli-I and NRX recombinant proteins. As expected, a positive Fli-I signal was observed when Fli-I was mixed with bead-immobilized GST-NRX, indicating their direct association (Fig. 2A).
FIGURE 2.
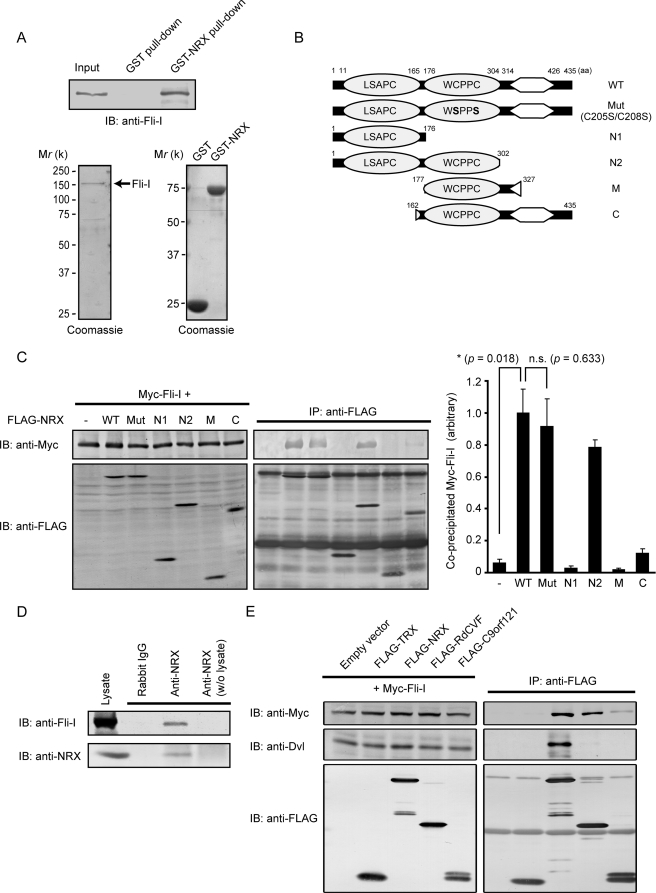
Interaction of Fli-I with NRX and its family member proteins. A, recombinant Fli-I proteins were incubated with GST- or GST-NRX-immobilized beads. After washing, the bound proteins were subjected to SDS-PAGE, followed by immunoblotting (IB) with anti-Fli-I antibody (upper panel). The purity of recombinant Fli-I, GST, and GST-NRX proteins, as determined by Coomassie staining, is also shown (lower panels). B, schematic representation of NRX mutants. C, Myc-Fli-I transfected into COS7 cells with FLAG-tagged WT or mutant forms of NRX. Cell lysates were immunoprecipitated (IP) with anti-FLAG antibody followed by immunoblotting. The relative amounts of co-precipitated Myc-Fli-I (right) were determined by densitometry (mean ± S.E. (error bars) of co-precipitated versus input from three independent experiments). *, p < 0.05 (Student's t test); n.s., not significant. D, MEF lysates subjected to immunoprecipitation with anti-NRX antibody. The precipitates were separated by SDS-PAGE followed by immunoblotting with anti-Fli-I antibody. In this experiment, anti-NRX antibodies were covalently attached to the beads to avoid nonspecific signals. E, lysates of COS7 cells transfected with Myc-Fli-I and FLAG-tagged TRX, NRX, RdCVF, and C9orf121 subjected to immunoprecipitation with anti-FLAG antibody, followed by immunoblotting with the indicated antibodies.
To characterize further the interaction between NRX and Fli-I, we prepared various FLAG-tagged deletion constructs of NRX (Fig. 2B) and expressed them with Myc-tagged Fli-I. We then performed immunoprecipitation with anti-FLAG antibody, which was followed by immunoblotting with anti-Myc antibody (Fig. 2C). Fli-I associated strongly with the WT and N2 constructs. In contrast, very little co-precipitation was observed with other deletion constructs, including N1, M, and C. We therefore concluded that both Try-X-like domains of NRX are required for the strong interaction with Fli-I. The redox-inactive mutant form of NRX (Mut C205S/C208S), in which two catalytically critical cysteine residues are mutated to serines, also formed protein complexes with Fli-I as did WT NRX.
We next examined complex formation between endogenous Fli-I and NRX. A positive Fli-I signal was clearly detected in anti-NRX precipitates, but not in the precipitates with control rabbit IgG (Fig. 2D). Hydrogen peroxide treatment did not significantly alter the amount of co-precipitated Fli-I (data not shown).
NRX belongs to the TRX family which, in mammals, is composed of more than 20 proteins. We previously found that among them, NRX, RdCVF, and C9orf121 can be classified in the same subfamily based on sequence homology (7). Thus, we performed immunoprecipitation analyses between Fli-I and TRX, NRX, RdCVF, or C9orf121 and found that both NRX and RdCVF bind firmly to Fli-I. A weak but significant interaction between Fli-I and C9orf121 was also observed, but TRX does not bind to Fli-I. Interestingly, Dvl specifically binds only to NRX (Fig. 2E). These results suggest that NRX and its subfamily members may have a common role through their interaction with Fli-I.
NRX Links Fli-I to MyD88
Recent reports have shown the involvement of Fli-I in the TLR4/MyD88 pathway (17, 20). As we identified Fli-I as a novel binding partner of NRX, we hypothesized that NRX may affect the TLR4/MyD88 pathway. First, to find any possible effect of NRX on the interaction between Fli-I and MyD88, we performed immunoprecipitation analyses. When FLAG-Fli-I and Myc-MyD88 were co-expressed in COS7 cells, we detected only a very weak signal of Myc-tagged MyD88 in the anti-FLAG precipitates. However, co-expression of Myc-NRX clearly enhanced the co-precipitation (Fig. 3A). Because we have shown that RdCVF also strongly binds to Fli-I (Fig. 2E), we tested the effect of RdCVF co-expression on the interaction between Fli-I and MyD88. As expected, the co-expression of RdCVF also significantly enhanced the co-precipitation (Fig. 3B).
FIGURE 3.
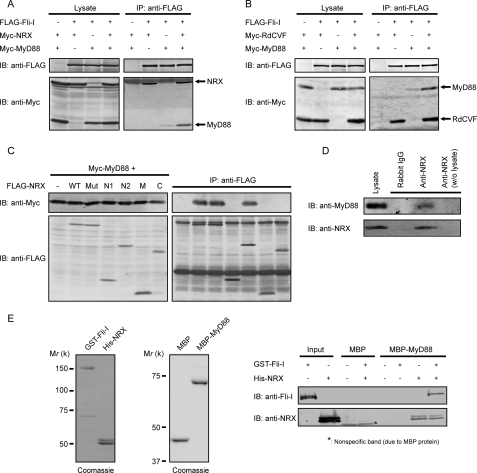
NRX links Fli-I to MyD88. A and B, COS7 cells were transfected with FLAG-Fli-I, Myc-MyD88, and Myc-NRX (A) or Myc-RdCVF (B). The cell lysates were subjected to anti-FLAG immunoprecipitation (IP). IB, immunoblotting. C, Myc-MyD88 and FLAG-tagged WT or mutant forms of NRX were transfected into COS7 cells, and their lysates were immunoprecipitated with anti-FLAG antibody followed by immunoblotting. D, lysates from MEFs were subjected to immunoprecipitation with anti-NRX antibody. The precipitates were separated by SDS-PAGE followed by anti-MyD88 immunoblotting. In this experiment, anti-NRX antibodies were covalently attached to the beads to avoid nonspecific signals. E, purified MBP-MyD88 proteins were immobilized onto beads and then incubated with purified GST-Fli-I and/or His-NRX proteins. The bound proteins were detected by immunoblotting with the indicated antibodies. The purity of recombinant GST-Fli-I, His-NRX, MBP, and MBP-MyD88, determined by Coomassie staining, is shown.
The above results suggest that NRX may also interact with MyD88 to enhance Fli-I·MyD88 complex formation. Therefore, we performed immunoprecipitation analyses by expressing FLAG-NRX and Myc-MyD88 in COS7 cells. The strong positive signal of Myc-MyD88 was observed in the precipitates of WT NRX (Fig. 3C). Further immunoprecipitation experiments with various deletion/point mutant proteins of NRX revealed that, similar to the NRX/Fli-I interaction, Mut and N2 constructs of NRX showed strong MyD88 binding comparable with WT NRX (Fig. 3C). We also confirmed the complex formation between endogenous NRX and MyD88 (Fig. 3D).
To explore the relationship among Fli-I, MyD88, and NRX in detail, we purified recombinant proteins of these three and performed pulldown assays. Surprisingly, when we immobilized MBP-MyD88 on beads and incubated them with GST-Fli-I alone, GST-Fli-I was not detected in the MBP-MyD88 precipitate. A positive signal of GST-Fli-I was detected only when His-NRX was added to the mixture (Fig. 3E), clearly indicating that NRX is essential for Fli-I·MyD88 complex formation. These results suggest that the interaction between Fli-I and MyD88 is indirect and requires NRX as an adaptor to link the two.
NRX Synergistically Inhibits the TLR4/MyD88 Signaling Pathway with Fli-I
Because NRX is required for Fli-I·MyD88 complex formation, we next performed NF-κB reporter assays to evaluate any functional role of NRX in the TLR4/MyD88 signaling pathway, which is reportedly suppressed by Fli-I (20). Ectopic expression of either Fli-I or NRX alone resulted in a marginal suppression of LPS-induced NF-κB activation. However, when Fli-I and NRX were co-expressed, NF-κB activity was clearly inhibited to basal levels (Fig. 4A). A similarly synergistic effect was observed between Fli-I and RdCVF (Fig. 4B). Co-expression of LacZ with Fli-I, as a control for artificial protein expression, showed no significant effect on LPS-induced NF-κB activation (Fig. 4C). These data clearly show that co-expression of NRX or RdCVF, which strengthens the complex formation between Fli-I and MyD88, is important to suppress the function of MyD88.
To confirm the importance of endogenous NRX as a suppressor of the TLR4/MyD88 pathway, we performed reporter assays with NRX−/− MEFs. MEFs were used instead of immune cells because NRX−/− mice die around birth,5 and it is difficult to obtain sufficient immune cells from embryos. We found that NRX−/− MEFs showed elevated NF-κB activity upon LPS stimulation, thus supporting the suppressive role of NRX. To confirm that this is due to the lack of NRX, we ectopically expressed NRX in NRX−/− MEFs and found that it significantly reduced reporter activity to levels similar to those of WT MEFs (Fig. 4D).
To substantiate further the effect of NRX on the TLR4/MyD88 pathway, we also examined the amount of IκBα, which is known to be degraded via activation of the TLR4/MyD88 pathway. Compared with WT MEFs, the amount of IκBα of LPS-treated NRX−/− MEFs was more drastically reduced (Fig. 4E), consistent with the results of the reporter assays. It was also noted that recovery of IκBα to basal levels appeared to occur more quickly; this phenomenon is similar to that of IRAK-M knock-outs, IRAK-M also being known as a negative regulator of MyD88-mediated signaling (23). The quick recovery may be due to faster activation of the negative feedback system, which is mediated by suppressors of cytokine signaling (SOCS) proteins (24).
It is reported that co-immunoprecipitation between Fli-I and MyD88 was enhanced upon LPS stimulation (20). Therefore, we performed immunoprecipitation analyses with LPS-treated WT or NRX−/− MEFs to see the complex formation between endogenous MyD88 and Fli-I in these cells. Predictably, the amount of Fli-I co-immunoprecipitated with MyD88 was significantly increased in WT MEFs upon LPS stimulation (Fig. 4F). In contrast, the complex formation between Fli-I and MyD88 in NRX−/− MEFs was very weak, and it was not strengthened by LPS stimulation (Fig. 4F). These results agree well with the results that NRX is required for the strong complex formation between Fli-I and MyD88 (Fig. 3) and that NRX−/− MEFs activate the TLR4/MyD88 pathway more strongly than WT MEFs (Fig. 4, D and E). Collectively, we concluded that NRX functions as a suppressor of the TLR4/MyD88 pathway by linking Fli-I to MyD88.
DISCUSSION
We have reported that NRX, RdCVF, and C9orf121 resemble each other (7). We found that they all interact with Fli-I, and both NRX and RdCVF augment complex formation between Fli-I and MyD88 and synergistically suppress LPS-induced NF-κB activity with Fli-I (Figs. 3 and 4). Collectively, NRX subfamily proteins may share a common function as suppressors of the TLR4/MyD88 pathway by linking Fli-I to MyD88. In contrast, we found that Dvl binds to NRX, but not to RdCVF or C9orf121 (Fig. 2E). Thus, it seems that NRX functions in both the Wnt signaling pathway and the TLR4/MyD88 pathway, whereas RdCVF specifically represses the TLR4/MyD88 pathway.
On the basis of the results of co-immunoprecipitation analyses, Wang et al. (20) showed that MyD88 and Fli-I form a complex; however, whether the interaction is direct or not was not determined. Using purified recombinant proteins, we clearly showed that the association between MyD88 and Fli-I is not direct but requires NRX as a link (Fig. 3E). We have thus defined a novel and unexpected function for NRX as an adaptor (Fig. 5), which might be helpful in understanding other TRX family proteins with unknown function.
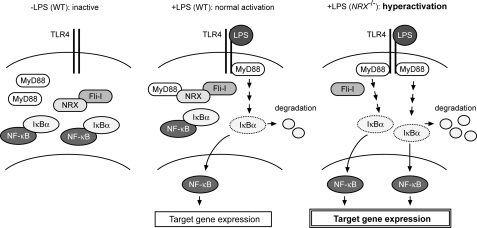
FIGURE 5.
Schematic illustration of the role of NRX, Fli-I, and MyD88 on TLR4 signaling. Left, in resting state (−LPS), IκBα associates with NF-κB and retains it in the cytosol. Center, LPS binding to TLR4 induces the recruitment of MyD88 to TLR4, which leads to the degradation of IκBα. NF-κB free from IκBα then moves into the nucleus and activates the transcription of various target genes. In WT cells, Fli-I forms a complex with MyD88 through NRX and avoids the unnecessary activation. Right, in NRX-deficient (NRX−/−) cells, Fli-I cannot sequester MyD88 from TLR4, and thus the hyperactivation of TLR4/MyD88 signaling occurs upon LPS stimulation.
It should be noted that, unlike RdCVF and C9orf121, NRX possesses two tandem TryX-like domains, neither one of which is able to interact with Fli-I by itself (Fig. 2C). This is similar to the case of Syndecan, which binds to both Syntenin (25) and CASK (26). Syntenin possesses two tandem PDZ domains, both of which are required for interaction with Syndecan, whereas CASK has only one PDZ domain that is sufficient for its binding. Structural analyses revealed that the first PDZ domain of Syntenin plays a supportive role for the second PDZ domain to interact with Syndecan (27). NRX may bind to Fli-I in a similar fashion, which will be answered in future structural analyses.
NRX is ubiquitously expressed with abundant expression in skin, testis, skeletal muscle, and moderately in thymus and spleen (2), whereas RdCVF expression is limited to the retina (28). NRX, therefore, appears to play a general role in regulation of the TLR4/MyD88 pathway, including innate immunity, but RdCVF may play a special role in the retina. A study utilizing TLR4 knock-out mice revealed that TLR4 and MyD88 suppress the proliferation of retinal progenitor cells (29). Moreover, a variant of TLR4 is reportedly associated with susceptibility to age-related macular degeneration, a degenerative retinal disease that causes defects in sharp central vision in elderly people (30). RdCVF was originally identified as a cone cell viability factor (28). Therefore, its function in the retinal cells may be explained by its suppression of the TLR4/MyD88 pathway. It would be interesting to evaluate whether the possible regulation of the TLR4/MyD88 pathway through RdCVF in the retina is important in the pathogenesis of age-related macular degeneration.
Acknowledgments
We thank Dr. S. Ohmi (University of Tokyo) and Dr. T. Takao (Osaka University) for the MALDI/TOF-MS/MS analyses and Dr. K. Yoshikawa (Osaka University) for the LacZ expression plasmid.
This study was supported in part by a grant-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science and Technology-Japan and the Japan Society for the Promotion of Science.
Y. Funato, M. Yoshida, and H. Miki, unpublished findings.
- TRX
- thioredoxin
- NRX
- nucleoredoxin
- Dvl
- Dishevelled
- TryX
- tryparedoxin
- RdCVF
- Rod-derived cone viability factor
- Fli-I
- Flightless-I
- MyD88
- myeloid differentiation primary response gene (88)
- TLR
- Toll-like receptor
- LPS
- lipopolysaccharide
- MEF
- mouse embryonic fibroblast
- WT
- wild type
- GST
- glutathione S-transferase
- MALDI/TOF-MS/MS
- matrix-assisted laser desorption/ionization time-of-flight tandem mass spectrometry
- MBP
- maltose-binding protein.
REFERENCES
- 1.Lillig C. H., Holmgren A. (2007) Antioxid. Redox Signal. 9, 25–47 [DOI] [PubMed] [Google Scholar]
- 2.Kurooka H., Kato K., Minoguchi S., Takahashi Y., Ikeda J., Habu S., Osawa N., Buchberg A. M., Moriwaki K., Shisa H., Honjo T. (1997) Genomics 39, 331–339 [DOI] [PubMed] [Google Scholar]
- 3.Funato Y., Michiue T., Asashima M., Miki H. (2006) Nat. Cell Biol. 8, 501–508 [DOI] [PubMed] [Google Scholar]
- 4.Clevers H. (2006) Cell 127, 469–480 [DOI] [PubMed] [Google Scholar]
- 5.Funato Y., Michiue T., Terabayashi T., Yukita A., Danno H., Asashima M., Miki H. (2008) Genes Cells 13, 965–975 [DOI] [PubMed] [Google Scholar]
- 6.Barrett M. P., Burchmore R. J., Stich A., Lazzari J. O., Frasch A. C., Cazzulo J. J., Krishna S. (2003) Lancet 362, 1469–1480 [DOI] [PubMed] [Google Scholar]
- 7.Funato Y., Miki H. (2007) Antioxid. Redox Signal. 9, 1035–1057 [DOI] [PubMed] [Google Scholar]
- 8.Perrimon N., Smouse D., Miklos G. L. (1989) Genetics 121, 313–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miklos G. L., De Couet H. G. (1990) J. Neurogenet. 6, 133–151 [DOI] [PubMed] [Google Scholar]
- 10.Campbell H. D., Schimansky T., Claudianos C., Ozsarac N., Kasprzak A. B., Cotsell J. N., Young I. G., de Couet H. G., Miklos G. L. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 11386–11390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campbell H. D., Fountain S., McLennan I. S., Berven L. A., Crouch M. F., Davy D. A., Hooper J. A., Waterford K., Chen K. S., Lupski J. R., Ledermann B., Young I. G., Matthaei K. I. (2002) Mol. Cell. Biol. 22, 3518–3526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeong K. W., Lee Y. H., Stallcup M. R. (2009) J. Biol. Chem. 284, 29298–29309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee Y. H., Campbell H. D., Stallcup M. R. (2004) Mol. Cell. Biol. 24, 2103–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goshima M., Kariya K., Yamawaki-Kataoka Y., Okada T., Shibatohge M., Shima F., Fujimoto E., Kataoka T. (1999) Biochem. Biophys. Res. Commun. 257, 111–116 [DOI] [PubMed] [Google Scholar]
- 15.Davy D. A., Campbell H. D., Fountain S., de Jong D., Crouch M. F. (2001) J. Cell Sci. 114, 549–562 [DOI] [PubMed] [Google Scholar]
- 16.Li J., Yin H. L., Yuan J. (2008) J. Cell Biol. 181, 321–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang T., Gu S., Ronni T., Du Y. C., Chen X. (2005) J. Proteome Res. 4, 941–949 [DOI] [PubMed] [Google Scholar]
- 18.Kawai T., Akira S. (2006) Cell Death Differ. 13, 816–825 [DOI] [PubMed] [Google Scholar]
- 19.O'Neill L. A., Bowie A. G. (2007) Nat. Rev. Immunol. 7, 353–364 [DOI] [PubMed] [Google Scholar]
- 20.Wang T., Chuang T. H., Ronni T., Gu S., Du Y. C., Cai H., Sun H. Q., Yin H. L., Chen X. (2006) J. Immunol. 176, 1355–1362 [DOI] [PubMed] [Google Scholar]
- 21.Kuwajima T., Taniura H., Nishimura I., Yoshikawa K. (2004) J. Biol. Chem. 279, 40484–40493 [DOI] [PubMed] [Google Scholar]
- 22.Terabayashi T., Itoh T. J., Yamaguchi H., Yoshimura Y., Funato Y., Ohno S., Miki H. (2007) J. Neurosci. 27, 13098–13107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobayashi K., Hernandez L. D., Galán J. E., Janeway C. A., Jr., Medzhitov R., Flavell R. A. (2002) Cell 110, 191–202 [DOI] [PubMed] [Google Scholar]
- 24.Dalpke A., Heeg K., Bartz H., Baetz A. (2008) Immunobiology 213, 225–235 [DOI] [PubMed] [Google Scholar]
- 25.Grootjans J. J., Zimmermann P., Reekmans G., Smets A., Degeest G., Dürr J., David G. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 13683–13688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsueh Y. P., Yang F. C., Kharazia V., Naisbitt S., Cohen A. R., Weinberg R. J., Sheng M. (1998) J. Cell Biol. 142, 139–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang B. S., Cooper D. R., Jelen F., Devedjiev Y., Derewenda U., Dauter Z., Otlewski J., Derewenda Z. S. (2003) Structure 11, 459–468 [DOI] [PubMed] [Google Scholar]
- 28.Léveillard T., Mohand-Saïd S., Lorentz O., Hicks D., Fintz A. C., Clérin E., Simonutti M., Forster V., Cavusoglu N., Chalmel F., Dollé P., Poch O., Lambrou G., Sahel J. A. (2004) Nat. Genet. 36, 755–759 [DOI] [PubMed] [Google Scholar]
- 29.Shechter R., Ronen A., Rolls A., London A., Bakalash S., Young M. J., Schwartz M. (2008) J. Cell Biol. 183, 393–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zareparsi S., Buraczynska M., Branham K. E., Shah S., Eng D., Li M., Pawar H., Yashar B. M., Moroi S. E., Lichter P. R., Petty H. R., Richards J. E., Abecasis G. R., Elner V. M., Swaroop A. (2005) Hum. Mol. Genet. 14, 1449–1455 [DOI] [PubMed] [Google Scholar]