Abstract
It has been assumed that R5 and X4 HIV utilize similar strategies to support viral cDNA synthesis post viral entry. In this study, we provide evidence to show that R5 and X4 HIV have distinct requirements for host cell uracil DNA glycosylase (UNG2) during the early stage of infection. UNG2 has been previously implicated in HIV infection, but its precise role remains controversial. In this study we show that, although UNG2 is highly expressed in different cell lines, UNG2 levels are low in the natural host cells of HIV. Short interfering RNA knockdown of endogenous UNG2 in primary cells showed that UNG2 is required for R5 but not X4 HIV infection and that this requirement is bypassed when HIV enters the target cell via vesicular stomatitis virus envelope-glycoprotein-mediated endocytosis. We also show that short interfering RNA knockdown of UNG2 in virus-producing primary cells leads to defective R5 HIV virions that are unable to complete viral cDNA synthesis. Quantitative PCR analysis revealed that endogenous UNG2 levels are transiently up-regulated post HIV infection, and this increase in UNG2 mRNA is ∼10–20 times higher in R5 versus X4 HIV-infected cells. Our data show that both virion-associated UNG2 and HIV infection-induced UNG2 expression are critical for reverse transcription during R5 but not X4 HIV infection. More importantly, we have made the novel observation that R5 and X4 HIV have distinct host cell factor requirements and differential capacities to induce gene expression during the early stages of infection. These differences may result from activation of distinct signaling cascades and/or infection of divergent T-lymphocyte subpopulations.
Keywords: HIV, Reverse Transcription, RNA Viruses, siRNA, Viral Replication, Virus Entry, DNA Repairing Enzyme, HIV Uncoating, Viral Tropism
Introduction
Human immunodeficiency virus type 1 (HIV-1)2 uses one of two cell surface chemokine receptors, CCR5 and CXCR4 (for reviews see Refs. 1–6) as a co-receptor for infection, and these viruses are referred to as R5 and X4 HIV-1, respectively. Isolates that are able to use both co-receptors with comparable efficiency are referred to as R5X4 (7, 8). R5 HIV-1 is characteristic of the asymptomatic stage of infection and is thought to be the predominant strain transmitted between individuals (9–15). X4 HIV-1 is generally detected during the late stages of disease when the immune system is waning. Tropism switch from R5 to R5X4 or X4 HIV-1 occurs in ∼40–50% of infected individuals and has been associated with CD4+ T-cell decline and disease progression (15–17).
Once receptor-mediated fusion has occurred between the viral and target cell membranes, it is thought that the viral capsid core is released into the cytoplasm, where it undergoes a process known as uncoating. HIV-1 uncoating is poorly understood, and it is generally assumed that both X4 and R5 HIV-1 share the same uncoating mechanism that enables the synthesis of viral cDNA. Efficient uncoating is critical for viral replication, and recent work has shown that the uncoating of retroviruses is a major battleground in the struggle between the virus and its host (18, 19). Mammalian cells have evolved a number of strategies to damage invading viruses, several of which rely on the introduction of uracil into the viral genome (20, 21).
Uracil is both a natural pyrimidine base in RNA and a common base lesion in genomic DNA. Uracil can be introduced into DNA either by misincorporation of deoxyuridine triphosphate (dUTP) during DNA polymerization or repair, or by deamination of cytosine residues in DNA. Unless removed, these uracil residues can have numerous detrimental effects (22, 23). Uracil DNA glycosylase (UNG) enzymes are DNA repair enzymes involved in base excision repair pathway and are found in most eukaryotic and prokaryotic cells. Members of the UNG protein family function to specifically remove uracil from both single and double-stranded DNA. The removal of uracil is achieved by cleaving the N-glycosyl bond that connects the base to the deoxyribose sugar leaving an abasic site in the DNA. The subsequent repair of these abasic sites is mediated by an apurinic/apyrimidinic endonuclease enzyme followed by the action of nucleic acid polymerases and DNA ligases (for review see Ref. 24).
The human UNG protein family consists of a number of cellular enzymes with uracil-excising activity, including UNG1, UNG2, and SMUG1 (single strand selective monofunctional uracil-DNA glycosylase) (25). The human UNG gene encodes both the nuclear (UNG2) and mitochondrial (UNG1) isoforms of the enzyme (26). Both UNG and SMUG1 prefer single-stranded DNA as a substrate, but are also capable of removing uracil from double-stranded DNA. In humans, the patterns of UNG1, UNG2, and total UNG activity differ among cell and tissue types (27), with UNG expression and activity being generally higher in proliferating as compared with non-proliferating cells (27–29). UNG2 expression is cell cycle-regulated with the highest levels found in early S phase (30) followed by subsequent degradation in a proteasome-dependent manner (31). UNG2 is the major species in most tissues, representing ∼70% of the total UNG activity (26).
The capacity to prevent uracil accumulation in genomic DNA is important to pathogens as well as to their host cells, and it is now appreciated that mammalian cells actively make use of uracilation to suppress retroviral infection (20). Many viral pathogens have developed an array of mechanisms to lower the uracil content in their DNA genomes. Similar to humans, viral pathogens (such as the poxviruses (32) and herpesviruses (33)) have evolved to encode their own UNG. In addition, several families of viruses have developed additional strategies to reduce the uracil content within their genomes. It has long been appreciated that many members of the retrovirus and herpesvirus families encode dUTP pyrophosphatase to lower the local dUTP concentration during viral DNA synthesis, which limits uracil misincorporation into the viral genome (21, 34, 35). The introduction of uracil into viral genomes is a potent and effective means to suppress viral propagation. Recent work has shown that mammalian cells express a family of nucleic acids editing enzymes (cytosine deaminases, APOBECs), which function to introduce uracil (and ultimately mutations) into retroviral genomes. HIV-1 counteracts this antiviral activity by encoding a viral protein, Vif, which functions to induce the degradation of these APOBEC proteins (20, 36, 37). The conserved requirement for methods to lower uracil in genomic DNA among various viral families could reflect a general principle employed by the host to control viral invasion. Because UNG2 has been shown to bind to HIV-1 proteins in vitro (38, 39) and to be packaged into HIV-1 virions (40), many have questioned whether UNG2 acts in concert with APOBEC and Vif to regulate the levels of uracilation within the viral genome.
UNG2 was first recognized as an HIV-1-interacting protein via a yeast two-hybrid screen (38), and subsequent work has shown that UNG2 binds specifically to both HIV-1 and simian immunodeficiency virus Vpr proteins in vitro (41). Using UNG2 mutants that are unable to bind Vpr, UNG2 has been shown to suppress the occurrence of mutations in the HIV-1 viral genome (42). More specifically, it is thought that UNG2 is able to regulate the HIV-1 mutation rate during infection of macrophages (43). Reports by the Quérat and Sire laboratories are also consistent with the theory that UNG2 is important for HIV-1 replication (44). Priet et al. further suggest that the HIV-1 reverse transcriptase protein displays a previously unknown apurinic endonuclease activity, which functions in the completion of UNG2-initiated nucleic acid repair (44). In contrast, Emerman and colleagues have used a UNG2-defective B-lymphocytic cell line and a UNG inhibitor to suppress the enzymatic activity of UNG2 in immortalized T-cell lines to show that UNG2 is dispensable for HIV-1 replication (45). By overexpressing both UNG2 and HIV-1 viral proteins in an embryonic kidney carcinoma cell, two laboratories have reached a different conclusion, that UNG2 is harmful for HIV-1 replication (46, 47). It has also been suggested that HIV-1 counteracts the detrimental effects of UNG2 function by actively degrading cellular UNG2 during viral assembly (46, 47).
Here we have used primary cells to investigate the role of UNG2 in HIV-1 replication. Immunoblotting for endogenous UNG2 and quantitative PCR for UNG2 mRNA transcripts show that, although UNG2 is highly expressed in several commonly used laboratory cell lines, it is poorly expressed in human peripheral blood mononuclear cells (PBMCs) and monocyte-derived macrophages (MDMs). siRNA knockdown of UNG2 in PBMCs (both as virus-producing and target cells) showed that expression of UNG2 is important for the production of infectious R5 HIV-1 and the synthesis of viral cDNA during R5 HIV-1 infection. Expression of host cell UNG2 mRNA is transiently up-regulated post HIV-1 infection, and R5 HIV is able to induce UNG2 expression 10–20 times more efficiently than X4 HIV. UNG2 expression was also found to be up-regulated in primary T-lymphocytes following treatment with the natural CCR5 ligand RANTES. Our data provide direct evidence to reconcile the conflicting data on the role of UNG2 in HIV-1 replication in the current literature and show a specific requirement for UNG2 during R5 HIV replication. Furthermore, we have demonstrated for the first time that X4 and R5 HIV-1 have distinct post entry infection mechanisms and have identified UNG2 as a potential tool to reveal these individual processes during HIV-1 infection.
EXPERIMENTAL PROCEDURES
Plasmid DNA
The wild-type HIV-1 proviral DNA NL4.3 was obtained through the National Institutes of Health AIDS Reagents Program from Dr. Malcolm Martin (48). The pNLAD8 HIV-1 AD8 Macrophage-Tropic R5 clone was obtained through the AIDS Research and Reference Reagent Program from Dr. Eric O. Freed (49). The vesicular stomatitis virus (VSV) envelope glycoprotein expression plasmid pHCMV-G was generously provided by Dr. Jane Burns (University of California, San Diego). The UGi expression plasmid pUGi-FBG has been described previously (50).
Cell Culture
PBMCs were isolated from buffy coats of HIV-1 seronegative blood donors (supplied by the Red Cross Blood Bank Service, Melbourne) by density gradient centrifugation over Ficoll-Paque Plus (Amersham Biosciences) as previously described (34). PBMCs were then either used for isolation of monocytes and peripheral blood lymphocytes (PBLs) or stimulated in media (2 × 106 cells/ml) supplemented with 10 μg/ml phytohemagglutinin (PHA, Murex Diagnostics, Dartford, Kent, UK) for 3 days in Teflon-coated jars. Monocytes and PBLs were isolated by plastic adherence as previously described (51). Monocytes were then cultured in media containing 10% human serum adherent to plastic and allowed to differentiate into MDMs for 5–7 days before infection. Cell medium was changed weekly.
Virus Production
Virus stocks were produced either by transfection of 293T cells using poly(ethylenimine) (Polysciences Inc., Warrington, PA) or infection of PHA-stimulated PBLs. VSV-G-pseudotyped HIV-1 was produced by co-transfection of the NL4.3- and NLAD8-based plasmids with pHCMV-G at a 2:1 molar ratio. Viruses produced in the presence of the UNG inhibitor UGi were produced by co-transfection of the proviral DNA with pUGi-FBG. Cells were washed twice in PBS 12 h post transfection, and fresh medium was added. Supernatants were collected 36 h post transfection.
PHA-stimulated PBLs were infected with VSV-G-pseudotyped HIV-1 (NL4.3 or NLAD8) 48 h post nucleofection. 12 h later cells were washed twice with PBS and maintained in media supplemented with 10 units/ml human interleukin-2 (Roche Applied Science) at a concentration of 2 × 106 cells/ml. Supernatants were collected 4 days post infection and centrifuged at 1462 × g for 30 min at 4 °C to remove cellular debris.
Viral particles from either transfection or infection cell culture supernatants were purified and concentrated by filtration through a 0.45-μm sterile syringe filter (Sartorius, Goettingen, Germany) and ultracentrifugation through a 20% sucrose cushion using an L-90 ultracentrifuge (SW-41 rotor, Beckman Coulter, Fullerton, CA) at 100,000 × g for 1 h at 4 °C. Pellets were then resuspended in Benzonase buffer (20 mm Tris-HCl, pH 8.0, 2 mm MgCl2, and 20 mm NaCl) and treated with 90 units/ml Benzonase (Sigma-Aldrich) for 30 min at 37 °C to remove any contaminating plasmid DNA. The concentrated viral stocks were quantified using the Vironostika HIV-1 antigen (p24 CA) MicroELISA assay (bioMérieux, Boxtel, the Netherlands) according to the manufacturer's instructions and frozen in single-use aliquots at −80 °C.
UNG Enzymatic Activity Assay
Virus stocks containing 10 ng of p24 protein, as determined by an HIV-1 antigen (p24 CA) MicroELISA assay (bioMérieux) were lysed in virion lysis buffer (10 mm Tris, pH 7.6, 150 mm NaCl, 2 mm EDTA, 0.5% Triton X-100). Virus lysates were then mixed with a 32P-labeled DNA oligonucleotide (5′-32P-TTTTTTTTTTTTUTTTTTTTTTTTT-3′) substrate in UNG reaction buffer (20 mm Tris-HCl, 1 mm EDTA, 1 mm dithiothreitol, pH 8.0) and incubated at 37 °C for 1 h. Apurinic sites were cleaved by adding 0.5 volume of 0.5 m NaOH and 0.5 volume of 30 mm EDTA and then heating to 99 °C for 10 min. Samples were then resolved on a non-denaturing 20% polyacrylamide gel by electrophoresis at 150 V for 1 h. The cleaved substrate was then analyzed by phosphorimaging on a BioImaging Analyzer (Fuji Photo Film Co.).
Immunoblot Analysis
Cells were lysed in TBS lysis buffer at a concentration of ∼10 × 106 cells per ml. Cell debris was subsequently removed by centrifugation at 20,200 × g for 20 min at 4 °C. Total protein content of the clarified lysates was quantified using an EZQ protein assay (Molecular Probes/Invitrogen) according to the manufacturer's instructions. Lysates containing equivalent amounts of total protein were mixed with 5× loading buffer (100 mm Tris-HCl, pH 6.8, 1.6% β-mercaptoethanol, 3% SDS, 33% glycerol, and 0.3% bromphenol blue), incubated at 95 °C for 5 min, and resolved by 10% SDS-PAGE. Resolved proteins were transferred to nitrocellulose membranes and blocked overnight in 5% skim milk dissolved in 1× TBS containing 0.3% Tween 20 (TBST) at 4 °C followed by a 4-h incubation with the anti-UNG2 antibody Ab225 (31, 52) or anti-α-tubulin antibody (Sigma) diluted 1:2000 in 5% skim milk dissolved in 1× TBST. Membranes were washed in TBST and incubated for 1 h with a 1:10,000 dilution of IR800 goat anti-rabbit IgG (Rockland Immunochemicals) or AlexaFluor 680 rabbit anti-mouse IgG, respectively (Molecular Probes), in 5% skim milk dissolved in 1× TBST. Membranes were washed again in TBST, and then proteins were visualized using the Odyssey Infrared Imager (LI-COR, Lincoln, NE). Band intensities were quantified using the Odyssey Application Software Version 1.2 (LI-COR).
Detection and Quantification of HIV and UNG2 mRNA
Poly(A)+ RNA was purified from cells using GenoPrep magnetic oligo(dT) beads (GenoVision, Oslo, Norway) as per the manufacturer's instructions. The mRNA was converted into cDNA in a 20-μl reaction mixture containing the bead/mRNA complex, 1× First-Strand Buffer (Invitrogen), 5 mm dithiothreitol, 0.5 mm dNTPs, 1 unit/μl RNasin (Promega), and 10 units/μl SuperScript III reverse transcriptase (Invitrogen). Samples were incubated for 1 h at 55 °C, and then the reaction was inactivated by incubation at 70 °C for 15 min. The RT mix was removed, and the beads were resuspended in 50 μl of 10 mm Tris-HCl (pH 7.5). The cDNA was stored at 4 °C until use in subsequent PCR reactions.
cDNA quantification was performed using quantitative PCR (qPCR) with an iCycler Real Time PCR machine (Bio-Rad). Each PCR reaction contained 1× SYBR Green I Master mix (Qiagen), 400 nm of each primer, and 2 μl of cell lysate in a 25-μl reaction volume. The HIV-1-specific primer pair M661/M667 (53) was used to detect HIV cDNA. HIV-1 PCR conditions were an initial denaturation at 95 °C for 10 min followed by 40 rounds of cycling at 95 °C for 15 s, then 61 °C for 30 s. UNG2 cDNA was detected using the specific primers, 5′-GCC AGA AGA CGC TCT ACT CC-3′ (sense) and 5′-TCG CTT CCT GGC GGG-3′ (antisense). cDNA was standardized for the human GAPDH gene using the primers 5′-TGG TAT CGT GGA AGG ACT CAT GAC-3′ (sense) and 5′-ATG CCA GTG AGC TTC CCG TTC AGC-3′ (antisense). UNG2 and GAPDH PCR conditions were an initial denaturation at 95 °C for 3 min followed by 40 rounds of cycling at 95 °C for 15 s, 55 °C for 30 s, and then 72 °C for 30 s.
Detection and Quantification of HIV cDNA
Cells were infected with equivalent amounts of virus, as determined by a HIV-1 antigen (p24 CA) MicroELISA assay. A heat-inactivated virus control (2 h at 56 °C) was used to confirm efficient removal of plasmid DNA for each sample. Cells were lysed at a concentration of 1 × 106 cells per 100 μl in PCR lysis buffer containing 1× PCR buffer (Roche Applied Science) with 0.5% v/v Triton X-100, 0.5% v/v Nonidet P-40, and 75 μg/ml proteinase K (Roche Applied Science). Samples were incubated at 56 °C for 1 h to allow proteinase K digestions before inactivation was carried out at 95 °C for 10 min. Samples were then stored at −20 °C. Cell lysates were diluted 10× in PCR-grade H2O before PCR analysis.
Quantification of HIV-1 reverse transcription products and standardization of cell numbers was performed using qPCR. qPCR was carried out on an iCycler Real Time PCR detection system (Bio-Rad). Each PCR reaction contained 1× SYBR Green I Master mix (Bio-Rad), 400 nm of each primer, and 2.5 μl of diluted cell lysate in a 25-μl reaction volume. The HIV-1-specific primer pair M661/M667 (53) was used to detect HIV cDNA. HIV-1 PCR conditions were an initial denaturation at 95 °C for 10 min followed by 40 rounds of cycling at 95 °C for 15 s, then 61 °C for 30 s. Cell numbers were standardized for the human CCR5 gene using the primers LK46 (sense, 5′-GCT GTG TTT GCG TCT CTC CCA GGA-3′) and LK47 (antisense, 5′-CTC ACA GCC CTG TGC CTC TTC TTC-3′). CCR5 PCR conditions were an initial denaturation at 95 °C for 10 min followed by 45 rounds of cycling at 94 °C for 20 s, 58.3 °C for 30 s, and 72° for 30 s.
Immunostimulation Assays
Tumor necrosis factor α (TNFα) and interferon α production was measured by ELISA of cell culture supernatants as previously described (54). Supernatants were collected either 24 h post nucleofection of freshly isolated PHA-stimulated PBMCs, or 16 h following DOTAP transfection of RAW-ELAM cells. siRNAs were complexed to DOTAP in a 3.74 μg/μl of siRNA (80 μm) ratio and applied to RAW-ELAM cells to a final concentration of 750 nm.
RNA Interference
Freshly isolated PBMCs were stimulated at a concentration of 2 × 106 cells/ml in media supplemented with 10 μg/ml PHA in Teflon-coated jars. After 48 h cells were washed and cultured in fresh media supplemented with 10 units/ml interleukin-2 (Roche Applied Science) for a further 24 h before nucleofection. Monocytes were nucleofected immediately after isolation. Cells were either mock nucleofected or nucleofected with 100 pmol of either AllStars Negative Control siRNA (Qiagen) or a UNG2-specific siRNA (Qiagen) using a Nucleofector II machine (Amaxa, Cologne, Germany) and protocol T-23 (PBMCs) or Y-001 (monocytes). After nucleofection, PBMCs were maintained in media supplemented with 10 units/ml interleukin-2 in 6-well plates for 2 days before infection. Monocytes were plated onto 6-well plates in media supplemented with 10% human serum and allowed to differentiate into MDMs for 5 days before infection.
FACS Analysis
Cells were incubated with anti-CCR5-PE and anti CXCR4-APC or isotype-matched control antibodies (BD Biosciences) for 60 min at 37 °C. Cells were then washed in PBS and fixed with 4% formaldehyde. After fixation cells were washed in PBS and permeabilized in 0.5% Triton X-100. Cells were then washed again in PBS and incubated with anti-UNG2 antibody Ab225 (31, 52) for 30 min at room temperature followed by anti-rabbit Alexa Fluor 488 (Invitrogen).
Flow cytometry was performed (FACSCalibur; BD Biosciences, Franklin Lakes, NJ) and analyzed using Cell Quest or Cell Quest Pro analysis software (BD Biosciences). For analysis, the entire PBMC population was initially gated based on size (forward light scatter) and granularity (side angle light scatter) characteristics. Flow cytometric analysis was also used to determine delivery efficiency of AlexaFluor 480-labeled siRNAs 3 h post nucleofection.
Statistical Analysis
Data from infection studies and qPCR analysis of HIV-1 reverse transcription was analyzed by paired two-tailed Student's t test. A p value of <0.05 was considered statistically significant for all tests.
RESULTS
The Enzymatic Activity of UNG Is Not Required for HIV-1 Infection in a Reporter Cell Line
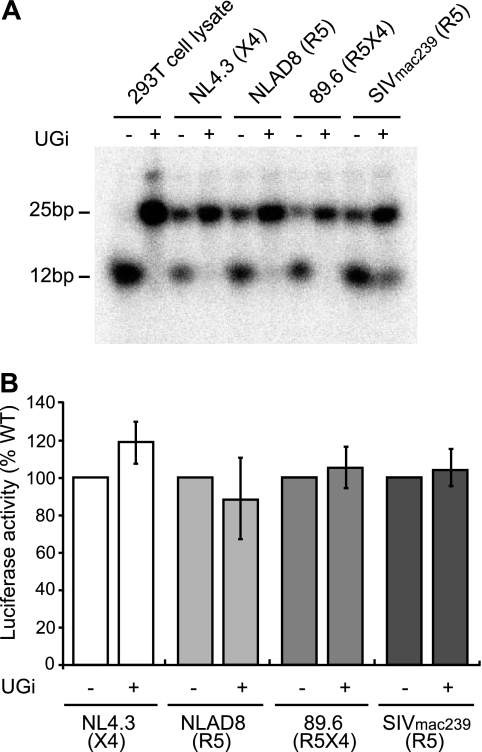
To investigate any potential role for UNG2 in HIV-1 replication, virus-associated UNG enzymatic activity levels were modified using the UNG inhibitor, UGi (55), which is a potent and specific inhibitor of both UNG1 and UNG2 enzymatic activity. HIV-1 (NL4.3, NLAD8, and 89.6) and SIVmac239 viruses were produced by transfection of the relevant proviral DNA into 293T cells, either in the presence or absence of UGi. The UNG enzymatic activity of the purified viruses was measured using an oligonucleotide-based UNG activity assay. All viruses produced in the absence of UGi were able to cleave the oligonucleotide substrate, demonstrating the presence of UNG enzymatic activity in the purified virus stocks, whereas viruses produced in the presence of UGi showed a significant reduction in virion-associated UNG activity (Fig. 1A).
FIGURE 1.
The enzymatic activity of UNG is not required for HIV-1 or SIV infection in a reporter cell line. HIV-1 (NL4.3, NLAD8, and 89.6) and SIVmac239 virions were produced from 293T cells in either the presence or absence of the UNG inhibitor, UGi. Virion-associated UNG activity was measured using an oligonucleotide-based UNG enzymatic activity assay (A). Equivalent amounts of virus were used to infect TZM-bl reporter cells, and the level of luciferase activity was used as an indicator of viral infectivity (B). Results are reported as a percentage of the WT control. Data shown are means (±S.E.) from either five (NL4.3 and 89.6) or three (NLAD8 and SIVmac239) independent experiments.
Equivalent amounts of virus, as measured by HIV-1 p24 CA protein, were used to infect the TZM-bl reporter cell line. TZM-bl cells contain integrated copies of the luciferase and β-galactosidase genes under control of the HIV-1 LTR promoter (56). Consequently, the production of HIV-1 Tat upon successful infection can be quantified using the level of luciferase activity. Two days after infection the cells were lysed, and the level of luciferase activity was measured as an indicator of viral infectivity. Luciferase activities for all viruses tested indicated no difference in infectivity between viruses produced in the presence or absence of the UNG inhibitor, UGi (Fig. 1B). These data demonstrate that virion-associated UNG enzymatic activity is not required for infection of the TZM-bl reporter cell line using either the X4 NL4.3 strain of HIV-1, the R5 NLAD8 strain of HIV-1, the dual tropic X4R5 primary HIV-1 isolate 89.6, or SIVmac239.
UNG2 Is Poorly Expressed in the Natural Target Cells of HIV-1
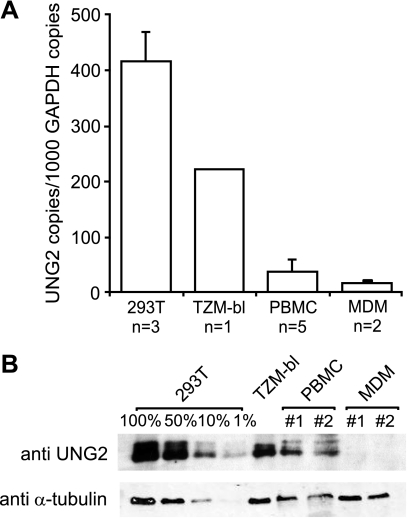
As model cell lines do not always reveal the true biology of viral pathogens (19, 57, 58), it is possible that abnormal UNG2 and other host cell protein expression levels in model cell systems could bias experimental outcomes. To understand how UNG2 levels in cell model systems compare with UNG2 levels in the natural target cells of HIV-1, UNG2 expression levels were examined in two primary cell subsets (PBMCs and MDMs) and two laboratory cell lines (293T and TZM-bl) using quantitative RT-PCR for UNG2 mRNA transcripts and immunoblot analysis for endogenous UNG2 protein. Analyses of UNG2 mRNA levels show that, although UNG2 is highly expressed in the human embryonic kidney cell line 293T and (to a lesser extent) in the HeLa derived TZM-bl cell line, UNG2 is very poorly expressed in PHA-stimulated PBMCs and MDMs (Fig. 2A). Protein expression levels closely mirrored the observed mRNA levels for all cell types except MDMs, where UNG2 was undetectable at the protein level despite low levels of UNG2 mRNA being detected (Fig. 2B). These data suggest that cell line model systems may not be an appropriate tool for examining the role of UNG2 in HIV-1 replication.
FIGURE 2.
UNG2 is poorly expressed in the natural target cells of HIV-1. UNG2 expression levels in two primary cell subsets (PBMCs and MDMs) and two laboratory cell lines (293T and TZM-bl) were measured by quantitative RT-PCR for UNG2 mRNA (A) and immunoblot analysis using the UNG2-specific antibody Ab225. Blots were also probed with an anti-α-tubulin antibody as a loading control. Independent blood donors are labeled #1 and #2 (B). Data shown are means (±S.E.) from independent experiments or independent blood donors for PBMCs and MDMs.
siRNA Knockdown of UNG2 in Primary Lymphocytes
Due to the fact that UNG2 expression levels in the model cell lines examined did not reflect UNG2 expression in the natural target cells of HIV-1 and the implications that this may have on data interpretation, we have investigated the role of UNG2 during HIV-1 infection of primary cells. For these experiments RNA interference was used to manipulate UNG2 levels in human primary lymphocytes, and then these cells were used either as virus-producing cells or target cells for infection. Two short interfering RNAs (siRNAs) were designed to target the unique 5′-region of the UNG2 mRNA (siUNG2a and siUNG2b) (Fig. 3A). A non-silencing siRNA (siRandom) was used as a negative control in all experiments. All siRNAs were AlexaFluor488-tagged to assess their capacities to enter the target cells via nucleofection. The nucleofection efficiency was measured by FACS analysis 3 h post nucleofection (Fig. 3B), where both the UNG2-specific siRNA and the control siRNA were shown to enter PBMCs with similar efficiency (73.2–83.6%) (Fig. 3B).
FIGURE 3.
siRNA knockdown of UNG2. Two siRNAs were designed to target the 5′ region that is unique to the human UNG2 mRNA (siUNG2a and siUNG2b) (A), in addition a non-silencing siRNA (siRandom) was used as a control in all experiments. All siRNAs were AlexaFluor488-labeled. Nucleofection efficiency was measured by FACS analysis 3 h post nucleofection (B). To monitor any recruitment of the innate immune system by the siRNAs, TNFα, and interferon α production was measured by ELISA 24 h after either nucleofection of PHA-stimulated PBMCs (C) or transfection of RAW-ELAM cells (D). Cells were also treated with the control compounds lipopolysaccharide (TLR4 agonist at 500 ng/ml), ODN2216 (CpG-containing DNA, TLR9 agonist at 3 μm), or CL075 (TLR8 agonist at 1 μg/ml). To confirm UNG2 knockdown in PBMCs, protein levels were measured by immunoblot using the UNG2-specific antibody Ab225 and normalized to α-tubulin levels (E). The data are representative of three independent blood donors (C), an aggregate of two independent experiments in triplicate (D) or six independent blood donors (E). Data shown are means (±S.E.).
A growing concern when using siRNAs for experimental knockdown of specific proteins is the potential for unanticipated nonspecific effects, in particular, initiation of innate immune responses via toll-like receptor (TLR) activation. Recent studies report that siRNAs can be potent inducers of interferons and inflammatory cytokines both in vivo and in vitro (59–63). To ascertain whether the siRNAs used in this study stimulated an innate immune response, the ability of these siRNAs to induce interferon α and TNFα production was tested in two different experimental systems.
Delivery of siRNAs by electroporation, which, like nucleofection, opens up pores in the cell allowing siRNA molecules to enter directly into the cytoplasm, has been shown to bypass activation by the endosomally localized TLRs (63). To confirm the absence of any immunostimulatory effects of the siRNAs used in this study when delivered via nucleofection, interferon α and TNFα production was measured by ELISA 24 h post nucleofection of freshly isolated PHA-stimulated PBMCs (Fig. 3C). Cells were also treated with the control compounds lipopolysaccharide (TLR4 agonist) and ODN2216 (CpG-containing DNA, TLR9 agonist). Results show that delivery of siRNA into cells via nucleofection did not result in an increase in the production of either interferonα or TNFα above the level observed for the untreated cell control or the mock nucleofection (Fig. 3C), demonstrating that neither the nucleofection procedure nor the siRNAs induced an innate immune response.
However, preliminary data indicated that siUNG2b strongly recruited endosomal TLR7/8 sequence-dependent pathways in human PBMCs when delivered using DOTAP liposomal transfection reagent, a delivery method that results in internalization via endocytosis and therefore endosomal localization and exposure to TLRs (data not shown). To further investigate the immunostimulatory potential of both siRNA sequences, we used a murine cell model allowing for the recruitment of TLR7 by short RNAs in a sequence-dependent fashion (54, 60). In accordance with our preliminary data in human blood, siUNG2b significantly induced TNFα production, a hallmark of TLR7 recruitment. However, siUNG2a did not trigger any cytokine production in PBMCs (data not shown) or murine macrophages (Fig. 3D), indicating that this siRNA sequence is not immunostimulatory. Due to the immunostimulatory effects of siUNG2b, this siRNA was not used for further knockdown experiments. The knockdown efficiency of siUNG2a was measured 48 h post nucleofection of freshly isolated PHA-stimulated PBMCs by immunoblot (Fig. 3E), with an average knockdown of 75% being observed.
UNG2 Is Required for R5 but Not X4 HIV-1 Infection in Primary Cells
Previous studies utilizing a number of different experimental systems have reported that UNG2 is either required for (43, 44), detrimental to (46), or dispensable for HIV-1 replication (45). In most of these studies overexpressed and epitope-tagged UNG2 proteins were used in experimental systems that were not the natural target cells of HIV-1. In this study the role of endogenous UNG2 during HIV-1 replication was examined in primary cell populations that are the authentic host cells of HIV-1. PBMCs were nucleofected with either a UNG2-specific siRNA (siUNG2a), a random control siRNA (siRandom), or mock nucleofected, and these cells were used either as virus-producing cells or target cells for infection.
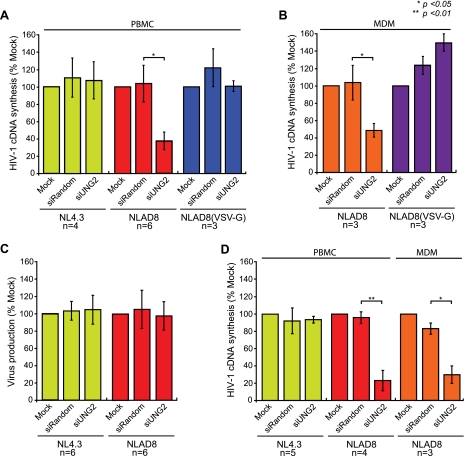
Infection of UNG2-depleted PBMCs with 293T cell-derived NL4.3 (X4) HIV virus showed no significant difference in the production of late reverse transcription cDNA products compared with mock nucleofection or siRandom-treated control cells (Fig. 4A). Unexpectedly, when UNG2-depleted PBMCs were infected with 293T cell-derived NLAD8 (R5) HIV in parallel experiments, a significant decrease in reverse transcription products in the UNG2-depleted cells was observed in comparison to the mock and siRandom-treated control cells (Fig. 4A). However, wild-type levels of reverse transcription were restored when infections were carried out using VSV-G-pseudotyped HIV-1 NLAD8 (Fig. 4A), which allows viral entry to take place though endocytosis bypassing CD4 and CCR5 receptor-mediated entry.
FIGURE 4.
UNG2 is required for HIV-1 NLAD8 (R5) but not HIV-1 NL4. 3 (X4) infection and viral cDNA synthesis. PBMCs from pooled donors (A) or freshly isolated monocytes (B) were either mock nucleofected, nucleofected with a non-silencing control siRNA (siRandom), or nucleofected with a UNG2-specific siRNA (siUNG2). Monocytes were then cultured on plastic and differentiated to macrophages for 5 days. siRNA-treated cells were then used as target cells for infection with either 293T cell-derived NL43, NLAD8, or NLAD8 virus pseudotyped with the VSV envelope glycoprotein. Cells were lysed either 24 h (A) or 48 h (B) post infection, and HIV-1 cDNA synthesis was quantified by qPCR and normalized to cell number. Virus production from siRNA-treated PBMCs from pooled donors infected with VSV-G-pseudotyped NL4.3 and NLAD8 was measured was measured 4 days post infection by quantification of HIV-1 p24 (CA) protein in cell culture supernatants (C). Infectivity of NL4.3 and NLAD8 virus produced from siRNA-treated PBMCs was examined in freshly isolated PHA-stimulated PBMCs (NL43 and NLAD8) or MDMs (NLAD8) (D). Equivalent amounts of virus, as determined by HIV-1 p24 CA, were used for all infections. Cells were lysed either 24 h (PBMC) or 48 h (MDM) post infection and HIV-1 cDNA was quantified by qPCR and normalized to cell number. Data shown are means (±S.E.) from independent blood donors.
A parallel experiment was carried out in which HIV NLAD8 (R5) and VSV-G-pseudotyped HIV NLAD8 were used to infect MDMs. Similar to infection with primary T-lymphocytes, siUNG2-treated MDMs were unable to support the synthesis of viral cDNA during R5 HIV infection, whereas VSV-G-pseudotyped R5 HIV cDNA synthesis was unaffected (Fig. 4B). These data further support a role for UNG2 during the early stages of R5 HIV but not X4 HIV infection.
It has been reported that UNG2 is packaged into HIV-1 virions during viral assembly (39, 40). Because our data show that UNG is required for the completion of the early stages of infection by R5 but not X4 HIV, we also assessed the capacity of UNG2 knockdown cells to produce infectious HIV-1 virions. Because VSV-G pseudotyping of NLAD8 HIV-1 was shown to overcome the observed defects in the early stages of viral replication (Fig. 4C), UNG2-depleted PBMCs were infected with VSV-G-pseudotyped HIV-1 (either NL4.3 or NLAD8) as a means to deliver the viral genomes into primary cells for virus production. The p24 (CA) levels in cell culture supernatants were measured 4 days post infection and serve as an indicator of virus particle production. Our data show that UNG2 is not required for the production of virion particles with either HIV-1 NL4.3 or NLAD8 (Fig. 4C).
The functional impact of UNG2 knockdown on X4 and R5 progeny HIV was also assessed. Viruses were harvested from cell culture supernatants, normalized by p24 CA, and subsequently used to infect freshly isolated PHA-stimulated PBMCs or MDMs (Fig. 4D) derived from the same donor. Infection with UNG2 knockdown cell-derived NL43 virus showed no significant difference in the production of late reverse transcription products compared with virus produced from mock or siRandom-nucleofected cells (Fig. 4D), whereas infection with UNG2 knockdown cell-derived NLAD8 virus showed a significant decrease in reverse transcription products in both PBMCs and MDMs (Fig. 4D), despite the same PBMCs and same donor-derived MDMs being used in the NL43 and NLAD8 viral infections.
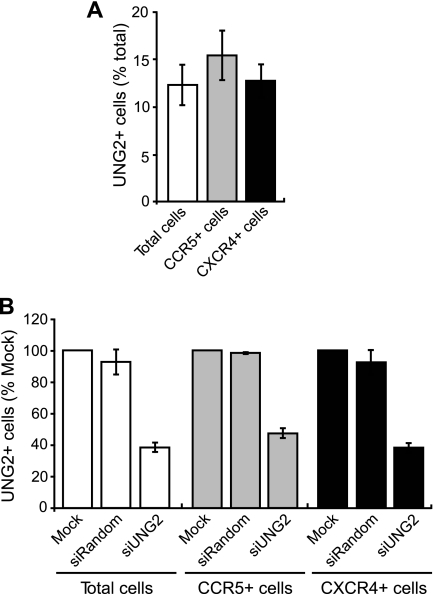
To rule out any differences in UNG2 expression levels between CCR5- and CXCR4-expressing cells causing our observed results, PHA-stimulated PBMCs were stained for cell surface CCR5 and CXCR4 in addition to intracellular UNG2. FACS analysis of protein expression showed that the percent UNG2+ cells did not significantly differ between the total, CCR5+, and CXCR4+ PBMC populations (Fig. 5A). In addition, UNG2 knockdown in the CCR5+ and CXCR4+ cell populations was also measured by FACS. Knockdown was also found to be equivalent across the PBMC populations (Fig. 5B).
FIGURE 5.
UNG2 in CCR5- and CXCR4-expressing primary T-lymphocytes. Freshly isolated PHA-stimulated PBLs were stained for CCR5, CXCR4, and UNG2. UNG2 expression levels in the total, CCR5-expressing, and CXCR4-expressing cell populations was analyzed by FACS (A). Freshly isolated PHA-stimulated PBLs were either mock nucleofected, nucleofected with a non-silencing control siRNA (siRandom), or nucleofected with a UNG2-specific siRNA (siUNG2). 48 h post infection cells were stained for CCR5, CXCR4, and UNG2. UNG2 expression levels in the total, CCR5-expressing, and CXCR4-expressing cell populations was analyzed by FACS (B). Data shown are means (±S.E.) from four independent blood donors.
Virus for infection was normalized by HIV p24 CA protein, because normalization by multiplicity of infection in primary cells is inherently difficult due to donor variability. To eliminate the addition of greater amounts of NL4.3 versus NLAD8 virus as the cause of our observed results, virus stocks were titrated in TZM-Bl-reported cells. NLAD8 virus stocks were found to contain higher infectious particles per ng/p24 than the NL43 virus stocks suggesting that virus normalization is unlikely to account for the observed results (data not shown). These data illustrate that both virion-associated UNG2 and target cell UNG2 are needed to support the synthesis of viral cDNA during R5 HIV infection.
UNG2 Expression Is Transiently Up-regulated during HIV Infection
Because both primary T-lymphocytes and MDMs have very low levels of UNG2 expression, it is unclear how the siRNA-mediated protein knockdown causes a defect in viral cDNA synthesis. It is possible that siUNG2 blocks R5 HIV infection either by knocking down the already low level of UNG2 mRNA or blocking the induction of UNG2 expression during infection. To examine this quantitative PCR analysis was carried out to determine the dynamics of UNG2 mRNA levels during HIV infection.
Our data show that UNG2 mRNA expression is transiently up-regulated at 6 h post synchronized infection and returns back to background levels at 12 h post infection (Fig. 6A). This transient induction of UNG2 mRNA is conserved for both X4 and R5 HIV infection (Fig. 6A). These data suggest that the observed impairment of siUNG2 on R5 HIV infection is likely a result of the suppression of the induction of host cell UNG2 that is critical for HIV reverse transcription. However, an equivalent increase in UNG2 mRNA was observed upon infection with NL4.3 (X4) HIV-1, yet this virus strain is not affected by treatment with the UNG-specific siRNA.
FIGURE 6.
HIV-1 infection induces a transient increase in UNG2 expression in primary T-lymphocytes. Freshly isolated PHA stimulated PBLs were infected with equivalent amounts, as determined by HIV-1 p24 (CA) protein, of either NL4.3 or NLA8 HIV-1 virus. Cells were harvested 6 and 12 h post infection (A) or 48 h post infection (B) and UNG2 mRNA expression levels were measured by quantitative RT-PCR and normalized to GAPDH copy numbers. HIV-1 mRNA expression levels 48 h post infection were measured by quantitative RT-PCR (C). UNG2 mRNA levels normalized to HIV-1 RNA copies 48 h post infection (D). Freshly isolated PHA-stimulated PBLs were incubated with either RANTES or SDF1α at either 10 or 20 ng/ml. 12 h post addition of chemokines, cells were fixed and stained for CCR5 and UNG2. UNG2 expression levels in the total and CCR5-expressing cell populations was analyzed by FACS (E). Data shown are means (±S.E.) from either 4 (A–D) or 2 (E) independent blood donors. F, model of host cell UNG2 expression during HIV infection by X4 and R5 HIV.
Because X4 and R5 HIV target different primary T-lymphocyte cell subsets during infection, which compose different proportions of the total CD4+ T-cell population (64, 65), it is expected that infection levels achieved with X4 and R5 HIV-1 also greatly differ. In an attempt to correct for this discrepancy in infection levels the capacity of X4 and R5 HIV to induce UNG2 expression in PBMCs was measured and normalized to HIV infection levels. Quantitative RT-PCR was used to determine the number of UNG2 (Fig. 6B) and HIV (Fig. 6C) mRNA copies in the cell population 48 h post infection. Infected cells were cultured for 48 h to allow secondary (or tertiary) infection. HIV mRNA levels were used to estimate the relative amount of X4- and R5 HIV-infected cells in the primary cell population. The level of UNG2 mRNA during X4 and R5 HIV infection are presented as UNG2 copies per 1000 HIV copies (Fig. 6D). When normalized for HIV infection levels our analysis shows that R5 HIV is 10–20 times more efficient than X4 HIV at inducing UNG2 expression upon infection.
Because the observed induction of UNG2 mRNA expression upon HIV infection differed depending on co-receptor usage we also investigated the ability of natural CCR5 and CXCR4 ligands to induce UNG2 expression in primary T-lymphocytes. PHA-stimulated PBMCs were cultured in the presence of either the CCR5 ligand RANTES or the CXCR4 ligand SDF1α. 12 h post addition of chemokines, cells were stained for cell surface CCR5 then fixed and stained for intracellular UNG2. Protein expression was analyzed by FACS. Analysis of UNG2 expression (% UNG2+ cells) in the total cell population showed no effect of either chemokine on the level of UNG2-expressing cells. However, when the CCR5-only-expressing population was analyzed data showed that the percentage of UNG2-expressing cells was significantly increased in RANTES-, but not SDF1α-, treated cells in a dose-dependent manner (Fig. 6E).
These data are consistent with a role for UNG2 in the synthesis of viral cDNA during R5 HIV infection in primary cells. Collectively, our data demonstrate that UNG2 is required for R5 but not X4 HIV to establish infection in primary lymphocytes and macrophages.
DISCUSSION
Previous studies examining UNG2 in HIV-1 biology have failed to reach a consensus on its role in, or even its requirement for, viral replication. It has been reported that UNG2 is either required for (43, 44), detrimental to (46), or dispensable for HIV-1 replication (45). Results presented here suggest that these discrepancies are most likely due to differences in the model systems used in previous studies, which may not accurately reflect the conditions found during natural HIV-1 infection. By manipulating the levels of endogenous UNG2 in primary cells, we have provided direct evidence to reconcile this dispute, showing that UNG2 is required to support the synthesis of viral cDNA during R5, but not X4, HIV infection. More importantly, we have made a novel observation that the requirement for UNG2 in HIV-1 replication is associated with virus co-receptor usage. Our work provides the first evidence that X4 and R5 HIV-1 have distinct host cell factor requirements during uncoating/reverse transcription post viral entry.
In this study experiments using the UNG-specific inhibitor UGi and siRNA depletion of UNG2 from either virus-producing or target cells have demonstrated that the requirement for UNG2 in HIV-1 replication is context-dependent. The enzymatic activity of UNG was not required for HIV or SIV infection in a reporter cell line (Fig. 1), which expresses very high levels of UNG2 (Fig. 2). However, UNG2 was required for efficient cDNA synthesis during R5 but not X4 HIV infection in primary cells. This cell type and virus strain-dependent requirement for UNG may help to explain the currently conflicting data in the literature. Interestingly, a previous study that found UNG2 to be dispensable for HIV replication (45) used an X4 strain of HIV-1 (LAI), whereas previous studies that found UNG2 to be required for HIV replication (43, 44) used R5 HIV-1 virus strains (AD8 and YU-2).
The fact that sequence differences between the X4 and R5 HIV-1 molecular clones used here occur predominantly in the envelope gene, plus the finding that the UNG2 requirement observed for R5 HIV-1 can be overcome by pseudotyping with the VSV envelope glycoprotein, suggest that our observed differences in the requirement for UNG2 are primarily entry-dependent. The major difference between X4 and R5 HIV-1 is their co-receptor usage. PBMCs isolated from whole blood consist of a number of immune cell subsets with differing cell surface receptor expressions, which results in distinct susceptibilities of different T-cell subsets to viral entry by X4 and R5 using viruses. Our observed difference in the requirement for UNG2 in HIV-1 replication may reflect a difference in the primary cell subsets infected by X4 compared with R5 HIV-1. Although no differences were found in the levels of endogenous UNG (Fig. 5A) other differences in the cellular environment of CCR5- and CXCR4-expressing subsets, such as cellular dNTP levels, may dictate whether UNG2 is required for viral replication.
Although it has been assumed that CCR5- and CXCR4-mediated HIV-1 infection only differ in terms of cell tropism, the limited data on the events that immediately follow HIV-1 entry mean it has been previously unclear if post entry events also differ between R5 and X4 viruses. Binding of HIV-1 to cell surface receptors is known to trigger signaling cascades (66–68), and it is becoming increasingly clear that activation of different signaling cascades by CXCR4 versus CCR5 binding may impact on downstream stages viral infection (for review see Ref. 69). The identification of a CD4+ T-cell subset known as TEMRA cells, which display a specific post entry block to R5 HIV-1 despite expressing high levels of CCR5 but are susceptible to X4 HIV-1 infection (70), suggests that the mechanisms controlling CCR5- and CXCR4-mediated HIV-1 infection may differ even within the same cell type. The notion that different receptor-mediated entry pathways can lead to distinct biological outcomes is also supported by a recent report (71) showing that HIV envelope and VSV glycoprotein can impact the ability of HIV to establish infection in resting CD4 T cells. Results from our study provide direct evidence, through their differential requirement of host cell factor UNG2 (Fig. 4), that X4 and R5 HIV undergo different post entry events. Furthermore, the differential levels UNG2 induction between X4 and R5 HIV infection and the natural CCR5 and CXCR4 ligands RANTES and SDF1α strongly suggest that distinct signaling cascades have been activated by these two strains of HIV upon viral entry. Interestingly, removal of the requirement for CD4 and either of the HIV-1 co-receptors also removed the requirement for UNG2 during R5 HIV-1 infection. Although the exact mechanism behind this is unclear, it may be that entry through VSV-G-mediated endocytosis allows indiscriminate infection of all cell populations regardless of their cell surface receptor expression or subset, resulting in a masking of the observed requirement for UNG2 in R5-expressing cells.
The transient induction of UNG2 mRNA post HIV infection further highlights its importance for efficient reverse transcription of HIV genomes (Fig. 6F). One of the most likely roles for the stronger capacity to induce UNG2 expression at the early stages of infection exhibited by R5 HIV compared with X4 HIV is to support the synthesis of viral cDNA in macrophages. It has been suggested that UNG2 may function in the removal of uracil bases from HIV-1 DNA during synthesis to regulate the levels of uracilation within the viral genome. It has also been proposed that the higher concentration of dUTP in macrophages could result in a higher rate of uracil misincorporation into the viral genome, and the induction of UNG2 expression during R5 HIV infection can help to correct these errors to facilitate viral replication (44). This strategy is reminiscent of how feline immunodeficiency virus and caprine arthritis encephalitis virus encode a viral dUTP pyrophosphatase protein to reduce viral mutation rates in macrophages (72, 73). Because R5-mediated HIV infection in macrophages is generally less robust than in primary T-lymphocytes, a highly sensitive detection and separation method will need to be developed to isolate HIV-infected macrophages to discern the dynamics of HIV infection and UNG2 function in these natural target cells.
Previous studies have suggested that UNG2 may be detrimental to the HIV-1 replication process by facilitating the degradation of APOBEC3G-edited viral DNA and thereby potentiating the anti-HIV-1 effect of APOBEC3G (46, 47). It has also been reported that the expression of HIV-1 accessory protein Vpr is able to induce the proteasomal degradation of cellular UNG and the closely related SMUG1 protein and that this degradation may function to protect the viral genome (47, 74). By overexpressing UNG2 and HIV proteins in laboratory adopted cell lines, we have also observed that the stability of UNG2 is impaired upon HIV protein expression (data not shown). Because the observed induction of UNG2 expression in this study was transient and not constant (Fig. 6A), our results are in line with the proposal that excess UNG2 could have a negative impact on the later stages of HIV infection (47). This interpretation is consistent with our pulse-chase analyses in 239T cells showing that the half-life of UNG2 is reduced in the presence of both full-length HIV-1 and HIV-1 Env and accessory proteins (data not shown). One of the major functions of the Vpr during HIV-1 replication is to induce cell cycle arrest, halting cells in the G2 phase of the cell cycle (75–78), which ultimately enhances the production of viral particles (79). It is important to note that UNG2 expression is cell cycle-regulated with UNG2 levels at their highest during early S phase followed by a pronounced reduction in late S and G2 phase (27, 30, 80, 81). Because HIV-1 Vpr causes arrest of cells in G2 and cellular UNG2 level are naturally low in G2 due to proteasomal degradation during late S phase (31), the observed UNG2 degradation in HIV-1-infected cells may be an indirect consequence of cell cycle arrest.
In summary, results presented here provide evidence to unify the apparently conflicting data in the current literature. Our results suggest that differences in both the HIV-1 strain and the cell type used in experimental investigations into the role of UNG2 in HIV-1 replication may result in diverse conclusions. We have also directly shown that UNG2 is required for the synthesis of viral cDNA during R5 HIV infection. Furthermore the fact that the requirement for UNG2 during HIV-1 replication is co-receptor usage-dependent illustrates, for the first time, that X4 and R5 HIV-1 have distinct post entry mechanisms. Host cell factors, such as UNG2, will be an invaluable tool to dissect the distinct uncoating processes of HIV-1 during viral infection.
Footnotes
- HIV-1
- human immunodeficiency virus, type 1
- UNG
- Uracil DNA glycosylase
- PBMC
- peripheral blood mononuclear cell
- MDM
- monocyte-derived macrophage
- siRandom
- non-silencing siRNA
- RANTES
- regulated on activation normal T cell expressed and secreted
- siRNA
- short interfering RNA
- VSV-G
- vesicular stomatitis virus envelope glycoprotein
- PBL
- peripheral blood lymphocyte
- PHA
- phytohemagglutinin
- ELISA
- enzyme-linked immunosorbent assay
- PBS
- phosphate-buffered saline
- TBS
- Tris-buffered saline
- RT
- reverse transcription
- qPCR
- quantitative PCR
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- TNF
- tumor necrosis factor
- FACS
- fluorescence-activated cell sorting
- TLR
- Toll-like receptor
- UGi
- UNG inhibitor
- CA
- capsid protein
- APOBEC
- apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like
- dUTP
- deoxyuridine triphosphate.
REFERENCES
- 1.Verani A., Lusso P. (2002) Curr. Mol. Med. 2, 691–702 [DOI] [PubMed] [Google Scholar]
- 2.Alkhatib G., Berger E. A. (2007) Eur. J. Med. Res. 12, 375–384 [PubMed] [Google Scholar]
- 3.Berger E. A., Murphy P. M., Farber J. M. (1999) Annu. Rev. Immunol. 17, 657–700 [DOI] [PubMed] [Google Scholar]
- 4.Moore J. P., Kitchen S. G., Pugach P., Zack J. A. (2004) AIDS Res. Hum. Retroviruses 20, 111–126 [DOI] [PubMed] [Google Scholar]
- 5.Dragic T. (2001) J. Gen. Virol. 82, 1807–1814 [DOI] [PubMed] [Google Scholar]
- 6.Zaitseva M., Peden K., Golding H. (2003) Biochim. Biophys. Acta 1614, 51–61 [DOI] [PubMed] [Google Scholar]
- 7.Berger E. A., Doms R. W., Fenyö E. M., Korber B. T., Littman D. R., Moore J. P., Sattentau Q. J., Schuitemaker H., Sodroski J., Weiss R. A. (1998) Nature 391, 240. [DOI] [PubMed] [Google Scholar]
- 8.Goodenow M. M., Collman R. G. (2006) J. Leukoc. Biol. 80, 965–972 [DOI] [PubMed] [Google Scholar]
- 9.Choe H., Farzan M., Sun Y., Sullivan N., Rollins B., Ponath P. D., Wu L., Mackay C. R., LaRosa G., Newman W., Gerard N., Gerard C., Sodroski J. (1996) Cell 85, 1135–1148 [DOI] [PubMed] [Google Scholar]
- 10.Deng H., Liu R., Ellmeier W., Choe S., Unutmaz D., Burkhart M., Di Marzio P., Marmon S., Sutton R. E., Hill C. M., Davis C. B., Peiper S. C., Schall T. J., Littman D. R., Landau N. R. (1996) Nature 381, 661–666 [DOI] [PubMed] [Google Scholar]
- 11.Liu R., Paxton W. A., Choe S., Ceradini D., Martin S. R., Horuk R., MacDonald M. E., Stuhlmann H., Koup R. A., Landau N. R. (1996) Cell 86, 367–377 [DOI] [PubMed] [Google Scholar]
- 12.Dragic T., Litwin V., Allaway G. P., Martin S. R., Huang Y., Nagashima K. A., Cayanan C., Maddon P. J., Koup R. A., Moore J. P., Paxton W. A. (1996) Nature 381, 667–673 [DOI] [PubMed] [Google Scholar]
- 13.Alkhatib G., Combadiere C., Broder C. C., Feng Y., Kennedy P. E., Murphy P. M., Berger E. A. (1996) Science 272, 1955–1958 [DOI] [PubMed] [Google Scholar]
- 14.Doranz B. J., Rucker J., Yi Y., Smyth R. J., Samson M., Peiper S. C., Parmentier M., Collman R. G., Doms R. W. (1996) Cell 85, 1149–1158 [DOI] [PubMed] [Google Scholar]
- 15.Connor R. I., Sheridan K. E., Ceradini D., Choe S., Landau N. R. (1997) J. Exp. Med. 185, 621–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Björndal A., Deng H., Jansson M., Fiore J. R., Colognesi C., Karlsson A., Albert J., Scarlatti G., Littman D. R., Fenyö E. M. (1997) J. Virol. 71, 7478–7487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koot M., Keet I. P., Vos A. H., de Goede R. E., Roos M. T., Coutinho R. A., Miedema F., Schellekens P. T., Tersmette M. (1993) Ann. Intern. Med. 118, 681–688 [DOI] [PubMed] [Google Scholar]
- 18.Stremlau M., Owens C. M., Perron M. J., Kiessling M., Autissier P., Sodroski J. (2004) Nature 427, 848–853 [DOI] [PubMed] [Google Scholar]
- 19.Sheehy A. M., Gaddis N. C., Choi J. D., Malim M. H. (2002) Nature 418, 646–650 [DOI] [PubMed] [Google Scholar]
- 20.Harris R. S., Liddament M. T. (2004) Nat. Rev. Immunol. 4, 868–877 [DOI] [PubMed] [Google Scholar]
- 21.Payne S. L., Elder J. H. (2001) Curr. Protein Pept. Sci. 2, 381–388 [DOI] [PubMed] [Google Scholar]
- 22.Pu W. T., Struhl K. (1992) Nucleic Acids Res. 20, 771–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boiteux S., Guillet M. (2004) DNA Repair 3, 1–12 [DOI] [PubMed] [Google Scholar]
- 24.Dianov G. L., Sleeth K. M., Dianova, Allinson S. L. (2003) Mutat. Res. 531, 157–163 [DOI] [PubMed] [Google Scholar]
- 25.Haushalter K. A., Todd Stukenberg M. W., Kirschner M. W., Verdine G. L. (1999) Curr. Biol. 9, 174–185 [DOI] [PubMed] [Google Scholar]
- 26.Slupphaug G., Markussen F. H., Olsen L. C., Aasland R., Aarsaether N., Bakke O., Krokan H. E., Helland D. E. (1993) Nucleic Acids Res. 21, 2579–2584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haug T., Skorpen F., Aas P. A., Malm V., Skjelbred C., Krokan H. E. (1998) Nucleic Acids Res. 26, 1449–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vilpo J. A. (1988) Mutat. Res. 193, 207–217 [DOI] [PubMed] [Google Scholar]
- 29.Aprelikova O. N., Tomilin N. V. (1982) FEBS Lett. 137, 193–195 [DOI] [PubMed] [Google Scholar]
- 30.Slupphaug G., Olsen L. C., Helland D., Aasland R., Krokan H. E. (1991) Nucleic Acids Res. 19, 5131–5137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fischer J. A., Muller-Weeks S., Caradonna S. (2004) DNA Repair 3, 505–513 [DOI] [PubMed] [Google Scholar]
- 32.Upton C., Stuart D. T., McFadden G. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 4518–4522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caradonna S., Worrad D., Lirette R. (1987) J. Virol. 61, 3040–3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGeehan J. E., Depledge N. W., McGeoch D. J. (2001) Curr. Protein Pept. Sci. 2, 325–333 [DOI] [PubMed] [Google Scholar]
- 35.Studebaker A. W., Balendiran G. K., Williams M. V. (2001) Curr. Protein Pept. Sci. 2, 371–379 [DOI] [PubMed] [Google Scholar]
- 36.Jones K. L., Mak J. (2005) Curr. Pharmacogen. 3, 97–117 [Google Scholar]
- 37.Holmes R. K., Malim M. H., Bishop K. N. (2007) Trends Biochem. Sci. 32, 118–128 [DOI] [PubMed] [Google Scholar]
- 38.Bouhamdan M., Benichou S., Rey F., Navarro J. M., Agostini I., Spire B., Camonis J., Slupphaug G., Vigne R., Benarous R., Sire J. (1996) J. Virol. 70, 697–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Priet S., Navarro J. M., Gros N., Querat G., Sire J. (2003) Virology 307, 283–289 [DOI] [PubMed] [Google Scholar]
- 40.Willetts K. E., Rey F., Agostini I., Navarro J. M., Baudat Y., Vigne R., Sire J. (1999) J. Virol. 73, 1682–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Selig L., Benichou S., Rogel M. E., Wu L. I., Vodicka M. A., Sire J., Benarous R., Emerman M. (1997) J. Virol. 71, 4842–4846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mansky L. M., Preveral S., Selig L., Benarous R., Benichou S. (2000) J. Virol. 74, 7039–7047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen R., Le Rouzic E., Kearney J. A., Mansky L. M., Benichou S. (2004) J. Biol. Chem. 279, 28419–28425 [DOI] [PubMed] [Google Scholar]
- 44.Priet S., Gros N., Navarro J. M., Boretto J., Canard B., Quérat G., Sire J. (2005) Mol. Cell 17, 479–490 [DOI] [PubMed] [Google Scholar]
- 45.Kaiser S. M., Emerman M. (2006) J. Virol. 80, 875–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang B., Chen K., Zhang C., Huang S., Zhang H. (2007) J. Biol. Chem. 282, 11667–11675 [DOI] [PubMed] [Google Scholar]
- 47.Schröfelbauer B., Yu Q., Zeitlin S. G., Landau N. R. (2005) J. Virol. 79, 10978–10987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adachi A., Gendelman H. E., Koenig S., Folks T., Willey R., Rabson A., Martin M. A. (1986) J. Virol. 59, 284–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Freed E. O., Englund G., Martin M. A. (1995) J. Virol. 69, 3949–3954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Begum N. A., Kinoshita K., Kakazu N., Muramatsu M., Nagaoka H., Shinkura R., Biniszkiewicz D., Boyer L. A., Jaenisch R., Honjo T. (2004) Science 305, 1160–1163 [DOI] [PubMed] [Google Scholar]
- 51.Sonza S., Maerz A., Deacon N., Meanger J., Mills J., Crowe S. (1996) J. Virol. 70, 3863–3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fischer J. A., Muller-Weeks S., Caradonna S. J. (2006) Cancer Res. 66, 8829–8837 [DOI] [PubMed] [Google Scholar]
- 53.Zack J. A., Arrigo S. J., Weitsman S. R., Go A. S., Haislip A., Chen I. S. (1990) Cell 61, 213–222 [DOI] [PubMed] [Google Scholar]
- 54.Gantier M. P., Tong S., Behlke M. A., Xu D., Phipps S., Foster P. S., Williams B. R. (2008) J. Immunol. 180, 2117–2124 [DOI] [PubMed] [Google Scholar]
- 55.Mol C. D., Arvai A. S., Sanderson R. J., Slupphaug G., Kavli B., Krokan H. E., Mosbaugh D. W., Tainer J. A. (1995) Cell 82, 701–708 [DOI] [PubMed] [Google Scholar]
- 56.Platt E. J., Wehrly K., Kuhmann S. E., Chesebro B., Kabat D. (1998) J. Virol. 72, 2855–2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hill M. K., Shehu-Xhilaga M., Campbell S. M., Poumbourios P., Crowe S. M., Mak J. (2003) J. Virol. 77, 8329–8335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jones K. L., Sonza S., Mak J. (2008) Nucleic Acids Res. 36, 1578–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hornung V., Guenthner-Biller M., Bourquin C., Ablasser A., Schlee M., Uematsu S., Noronha A., Manoharan M., Akira S., de Fougerolles A., Endres S., Hartmann G. (2005) Nat. Med. 11, 263–270 [DOI] [PubMed] [Google Scholar]
- 60.Judge A. D., Sood V., Shaw J. R., Fang D., McClintock K., MacLachlan I. (2005) Nat. Biotechnol. 23, 457–462 [DOI] [PubMed] [Google Scholar]
- 61.Karikü K., Bhuyan P., Capodici J., Ni H., Lubinski J., Friedman H., Weissman D. (2004) Cells Tissues Organs 177, 132–138 [DOI] [PubMed] [Google Scholar]
- 62.Marques J. T., Williams B. R. (2005) Nat. Biotechnol. 23, 1399–1405 [DOI] [PubMed] [Google Scholar]
- 63.Sioud M. (2005) J. Mol. Biol. 348, 1079–1090 [DOI] [PubMed] [Google Scholar]
- 64.Blaak H., van't Wout A. B., Brouwer M., Hooibrink B., Hovenkamp E., Schuitemaker H. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 1269–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van Rij R. P., Blaak H., Visser J. A., Brouwer M., Rientsma R., Broersen S., de Roda Husman A. M., Schuitemaker H. (2000) J. Clin. Invest. 106, 1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schmid-Antomarchi H., Benkirane M., Breittmayer V., Husson H., Ticchioni M., Devaux C., Rossi B. (1996) Eur. J. Immunol. 26, 717–720 [DOI] [PubMed] [Google Scholar]
- 67.Davis C. B., Dikic I., Unutmaz D., Hill C. M., Arthos J., Siani M. A., Thompson D. A., Schlessinger J., Littman D. R. (1997) J. Exp. Med. 186, 1793–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Briant L., Coudronnière N., Robert-Hebmann V., Benkirane M., Devaux C. (1996) J. Immunol. 156, 3994–4004 [PubMed] [Google Scholar]
- 69.Wu Y., Yoder A. (2009) PLoS Pathog. 5, e1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Oswald-Richter K., Grill S. M., Leelawong M., Tseng M., Kalams S. A., Hulgan T., Haas D. W., Unutmaz D. (2007) PLoS Pathog. 3, e58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu D., Wang W., Yoder A., Spear M., Wu Y. (2009) PLoS Pathog. 5, e1000633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lerner D. L., Wagaman P. C., Phillips T. R., Prospero-Garcia O., Henriksen S. J., Fox H. S., Bloom F. E., Elder J. H. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 7480–7484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Turelli P., Guiguen F., Mornex J. F., Vigne R., Quérat G. (1997) J. Virol. 71, 4522–4530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schröfelbauer B., Hakata Y., Landau N. R. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 4130–4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rogel M. E., Wu L. I., Emerman M. (1995) J. Virol. 69, 882–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Re F., Braaten D., Franke E. K., Luban J. (1995) J. Virol. 69, 6859–6864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jowett J. B., Planelles V., Poon B., Shah N. P., Chen M. L., Chen I. S. (1995) J. Virol. 69, 6304–6313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.He J., Choe S., Walker R., Di Marzio P., Morgan D. O., Landau N. R. (1995) J. Virol. 69, 6705–6711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Goh W. C., Rogel M. E., Kinsey C. M., Michael S. F., Fultz P. N., Nowak M. A., Hahn B. H., Emerman M. (1998) Nat. Med. 4, 65–71 [DOI] [PubMed] [Google Scholar]
- 80.Nagelhus T. A., Slupphaug G., Lindmo T., Krokan H. E. (1995) Exp. Cell Res. 220, 292–297 [DOI] [PubMed] [Google Scholar]
- 81.Hagen L., Kavli B., Sousa M. M., Torseth K., Liabakk N. B., Sundheim O., Pena-Diaz J., Otterlei M., Hørning O., Jensen O. N., Krokan H. E., Slupphaug G. (2008) EMBO J. 27, 51–61 [DOI] [PMC free article] [PubMed] [Google Scholar]