Abstract
Amyotrophic lateral sclerosis (ALS) is a debilitating and fatal late-onset neurodegenerative disease. Familial cases of ALS (FALS) constitute ∼10% of all ALS cases, and mutant superoxide dismutase 1 (SOD1) is found in 15–20% of FALS. SOD1 mutations confer a toxic gain of unknown function to the protein that specifically targets the motor neurons in the cortex and the spinal cord. We have previously shown that the autosomal dominant Legs at odd angles (Loa) mutation in cytoplasmic dynein heavy chain (Dync1h1) delays disease onset and extends the life span of transgenic mice harboring human mutant SOD1G93A. In this study we provide evidence that despite the lack of direct interactions between mutant SOD1 and either mutant or wild-type cytoplasmic dynein, the Loa mutation confers significant reductions in the amount of mutant SOD1 protein in the mitochondrial matrix. Moreover, we show that the Loa mutation ameliorates defects in mitochondrial respiration and membrane potential observed in SOD1G93A motor neuron mitochondria. These data suggest that the Loa mutation reduces the vulnerability of mitochondria to the toxic effects of mutant SOD1, leading to improved mitochondrial function in SOD1G93A motor neurons.
Keywords: Amyotropic Lateral Sclerosis (Lou Gehrig's Disease), Dynein, Mitochondria, Mitochondrial Transport, Neurodegeneration, ALS, Axonal Transport, Loa, Motor Neuron Disease, Superoxide Dismutase 1 (SOD1)
Introduction
Amyotrophic lateral sclerosis (ALS)2 is a debilitating late-onset, progressive form of motor neuron disease that primarily targets motor neurons in the cortex, brainstem, and the anterior horn of the spinal cord, leading to paralysis and eventually death, typically within 5 years of diagnosis (1–5). The majority of ALS cases are sporadic, and ∼10% are familial, although the etiology of ALS is largely unknown. Between 15 to 20% of familial ALS and ∼1% of sporadic ALS are caused by dominant gain-of-toxic function mutations in the gene coding for Cu,Zn superoxide dismutase 1 (SOD1) (6–8).
Transgenic mice overexpressing mutant forms of human SOD1 have been invaluable in providing insights into the pathogenesis of ALS and in highlighting the cellular functions that are targeted in the disease (for reviews, see Refs 9 and 10). Formation of intracellular protein aggregates in motor neurons is a common characteristic of all ALS cases and is also observed in SOD1 transgenic mouse models. Biochemical and immunohistochemical studies have shown that SOD1 and ubiquitin aggregates are present in SOD1-mediated familial ALS cases and their transgenic mouse models and that TDP-43 and ubiquitin aggregates are seen in most ALS inclusions, including sporadic ALS (3, 10–12).
Mutant SOD1-mediated motor neuron death is thought to be due to a gain of aberrant chemistry in copper and zinc active sites, making them highly reactive with subsequent damage to other proteins, but Wang et al. (13) showed that mutant SOD1 proteins with significantly reduced affinity to copper, but with propensity to aggregation, induced disease similar to those variants that stably bind copper. However, mutant SOD1 toxicity could be the result of toxicity of the intracellular aggregates through aberrant chemistry, sequestration of other proteins into the aggregates, proteasome overload, and damage to specific organelles such as mitochondria (3, 14).
In addition, impaired axonal transport has been highlighted in motor neuron death in ALS, and we and others have shown that axonal transport defects are one of the earliest pathological events observed in motor neurons of mutant SOD1 transgenic mice (15–20). Furthermore, in vivo experiments involving injection of a neurotracer have shown that transport from muscle to motor neurons is impaired in SOD1G93A mice and that there is an association of dynein with mutant SOD1 aggregates in the motor neurons of these mice (21). Two recent studies have reported that there is a direct “gain-of-interaction” between aggregate-prone variants of mutant, but not wild-type, SOD1 (including SOD1G93A), and cytoplasmic dynein in glutathione S-transferase (GST) pulldown and immunoprecipitation assays and that disruption of cytoplasmic dynein function abolishes this interaction (22, 23).
Cytoplasmic dynein is a motor protein involved in diverse cellular processes including nuclear movement, positioning of mitotic spindles, Golgi apparatus and endoplasmic reticulum, and retrograde axonal transport of mitochondria and endocytic membranes containing neurotrophic factors in neurons (24–28). This multisubunit protein complex consists of two homodimerized heavy chains, DYNC1H1, here referred to as DHC, and several intermediates (DYNC1I), here referred to as DIC, light intermediates (DYNC1LI), and light (DYNL) chains (25, 29). Dynein function is regulated by another multisubunit protein, dynactin, which through its p150 subunit also plays a significant role as a docking protein for some of the various cargos transported by dynein (30). We have previously shown that an autosomal dominant point mutation causing F580Y substitution in DHC gives rise to a progressive motor deficit in heterozygous Loa (Dync1h1Loa/+) mice (31–33) and that in cultured motor neurons isolated from E13.5 homozygous embryos this mutation impairs retrograde axonal transport leading to motor neuron degeneration and death of the pups within a day after birth (31). In addition, others have shown that the Loa mutation causes γ-motor neuron and proprioceptive sensory neuron degeneration in heterozygous Dync1h1Loa/+ which is thought to be the cause of the phenotype in these mice (34, 35). Interestingly, when we crossed Dync1h1Loa/+ with SOD1G93A mice to investigate a possible link between dynein and mutant SOD1, the SOD1G93A;Dync1h1Loa/+ progeny from this cross showed a wild-type phenotype with regard to muscle force, motor unit, and motor neuron survival at 120 days when their SOD1G93A littermates exhibited severe defects (17). Moreover, the average lifespan of SOD1G93A;Dync1h1Loa/+ mice increased by 28% compared with their SOD1G93A littermates (p ≤ 0.001). In addition, the Loa allelic mutation Y1055C in Dync1h1Cra1/+ mice has also been shown to delay disease onset and increase the life span of SOD1G93A mice by 14% (36). Subsequently Chen et al. (34) and Ilieva et al. (35) replicated these in mice bearing the Loa mutation and found increases in life span of 21% (p < 0.01) and 9% (p = 0.002), respectively.
Both (34) and Ilieva et al. (35) also reported significant loss of proprioceptive sensory neurons in Dync1h1Loa/+ mice. The loss of sensory neurons in Dync1h1Loa/+ mice prompted Ilieva et al. (35) to propose that the rescue of SOD1G93A is a result of reduced glutamate excitotoxicity brought about by the loss of the glutamatergic proprioceptive sensory neurons (35). However, the Sprawling (Dync1h1Swl1/+) mouse with a G1040-T1043delinsA mutation in the dynein heavy chain shows even greater proprioceptive sensory neuron loss than the Loa, but it is not able to rescue the SOD1G93A phenotype (34). Moreover, the W1206R mutation in Abnormal rear leg (Arl) mouse, which is allelic to Loa, confers loss of sensory neuron fibers in the Dync1h1Arl/+ mice, but it has no effect on the disease onset or life span in the double mutant SOD1G93A;Dync1h1Arl/+ mice.3 These findings, therefore, do not support the hypothesis that the protection of motor neurons in SOD1G93A/Dync1h1Loa/+ mice is solely the result of reduced glutamatergic sensory neuron input by the Loa mutation.
The above findings do, however, suggest a link between cytoplasmic dynein and SOD1G93A toxicity. In this report we present evidence to suggest that the Loa mutation in dynein affects the subcellular distribution of mutant SOD1 protein in SOD1G93A;Dync1h1Loa/+ mice after, but not before, the onset of the disease. Furthermore, the Loa mutation in dynein weakens the association of SOD1G93A protein with the mitochondria in the cortex of the brain and spinal cord. We also present data showing severe defects in respiration and membrane potential of SOD1G93A mitochondria, which are ameliorated in mitochondria isolated from SOD1G93A;Dync1h1Loa/+ mice. In addition, the results of this study show that there is no direct interaction between dynein and SOD1G93A and that SOD1G93A protein does not disrupt the dynein complex.
EXPERIMENTAL PROCEDURES
Animals and Tissue Collection
Congenic heterozygous Dync1h1Loa/+ female mice, maintained on a C57BL/6 (Harlan UK) genetic background, were crossed with male transgenic mice expressing human SOD1G93A, maintained on an F1 (SJL x C57BL/6) genetic background, to produce four genetically distinct groups of littermates: Dync1h1+/+, Dync1h1Loa/+ heterozygote, SOD1G93A hemizygote, and SOD1G93A;Dync1h1Loa/+ double-heterozygote (represented in the figures as +/+, Loa/+, SOD1G93A, and SOD1G93A/Loa, respectively) mice. The SJL mice were purchased from Harlan UK. All mice were identified by genotyping for the Loa mutation in the cytoplasmic dynein heavy chain gene Dync1h1 and the human SOD1 transgene from tail genomic DNA (31, 37). Tissues were harvested from mice at different ages and/or different stages of disease. The early stage was when the mice showed a body weight loss of less than 10% accompanied by shaky hind limbs; the late stage was characterized by a 10–15% reduction in body weight accompanied by an apparent muscle wasting and paralysis of hind limbs; the end stage was when the mice lost their righting reflex and showed 20% reduction of their body weight compared with that before becoming symptomatic. Mice were killed by a schedule 1 killing, and brains and spinal cords were dissected, washed with appropriate ice-cold buffers, and either used fresh or snap-frozen in liquid nitrogen and stored at −80 °C. All animal experiments were conducted in accord with the UK Animal (Scientific Procedures) Act (1986).
Chemicals, Reagents, and Antibodies
All chemicals and reagents were obtained from Sigma unless otherwise stated. Phosphate-buffered saline (PBS−Ca−Mg) was from Invitrogen; RIPA was from Upstate Biotechnology; protein A-Sepharose 4B beads were from Zymed Laboratories Inc.; protein A- and protein B-agarose beads were from Roche Applied Science; proteinase K was from New England Biolabs; BS3 and the BCA protein assay kit were from Pierce. The following antibodies were used in this study: mouse monoclonal anti-dynein intermediate chain 74.1 (generously provided by Dr. K. Pfister, University of Virginia or Santa Cruz, sc-13524), FL-154 rabbit polyclonal anti-SOD1, H-300 rabbit polyclonal anti-dynactin p150Glued, R-325 rabbit polyclonal anti-dynein heavy chain, 20-E8 mouse monoclonal anti-Cox4, C-20 goat polyclonal anti-calnexin, C-15 goat polyclonal anti-TGN38, (Santa Cruz Biotechnology, sc-11407, sc-11363, sc-9115, sc-58348, sc-6465, sc-27680, respectively), NCL-SOD1 mouse monoclonal anti-SOD1 (Novo Castra), mouse monoclonal anti-p150Glued (BD Transduction Laboratories, 610473), and mouse monoclonal anti-α-tubulin (Upstate Biotechnology, 05-829).
Immunoprecipitation and Cross-linking
Homogenization of the mouse brain or spinal cord tissues and immunoprecipitation experiments were carried out after either the procedure we developed in our laboratories (see “Results”) or as described elsewhere (22). Brains and spinal cords were homogenized in a range of homogenization buffers (HBs) (see “Results” for details). The homogenates were clarified by centrifugation at 16,000 × g for 15 min at 4 °C, and their protein contents were determined using the BCA kit. Approximately 2.0 mg of homogenate proteins were cleared by incubation with beads for 1 h with shaking at 4 °C, and the beads then removed by centrifugation. The cleared homogenates were then incubated with a range of different beads (protein G-agarose, protein A-agarose, or protein A-Sepharose 4B beads or TrueBlot™ anti-mouse Ig IP beads from eBioscience) linked with anti-DIC, anti-dynactin p150, or anti-SOD1 antibodies or with anti-hemagglutinin antibody or IgG as the negative control for time periods that varied between 3 h to overnight with shaking at 4 °C. The immunoprecipitated complexes on beads were retrieved by brief centrifugation and washed four times with HB and once with water. Protein complexes on beads were resuspended in 1× SDS loading buffer, heated for 5 min at 95 °C, and analyzed by SDS-PAGE and Western blotting detecting for DIC, DHC, p150, and SOD1. Chemical cross-linking with BS3, the water soluble analogue of disuccinimidyl suberate, was performed according to the manufacturer's recommendations and Zhang et al. (22).
Sucrose Density Gradient Analysis
A 4.8-ml 5–40% continuous sucrose gradient in PBS−Ca/Mg was established overnight at 4 °C in Beckman Ultra-Clear Centrifuge tubes (Beckman, Palo Alto, CA). Brains and spinal cords of Dync1h1+/+, Dync1h1Loa/+, SOD1G93A, and SOD1G93A;Dync1h1Loa/+ mice at different postnatal stages were homogenized in PBS−Ca/Mg containing 0.1% Triton X-100 and protease and phosphatase inhibitors. Homogenates were spun at 800 × g for 15 min. Protein contents of the supernatants were determined, and the equivalent to 1.5–2 mg of protein of tissue homogenate was layered onto the gradient. After centrifugation at 237,000 × g in a SW-55 Ti rotor (Beckman) for 4 h at 4 °C, 16 equal-volume fractions were collected, and equal volumes of each fraction were loaded onto 4–12% gradient SDS-PAGE gels and analyzed by immunoblotting.
Buoyant Density Analysis
Buoyant density analysis was carried out essentially as described by Vande Velde et al. (38). Briefly, cerebral cortices and spinal cords of late-stage SOD1G93A and SOD1G93A;Dync1h1Loa/+ mice were collected and washed with PBS−Ca/Mg and then placed in an ice-cold HB containing 250 mm sucrose, 10 mm Hepes-NaOH (pH 7.4), 1 mm EDTA plus protease, and phosphatase inhibitors. Tissues were homogenized on ice with seven strokes of a glass-pestle homogenizer. Homogenates were centrifuged at 1000 × g for 10 min. Supernatants were collected, and the pellets were washed with 0.5 volumes HB and centrifuged again. Supernatants were pooled and centrifuged at 16,000 × g for 15 min to yield a crude mitochondrial pellet and post-mitochondrial supernatant. Crude mitochondria were gently resuspended in HB and then adjusted to 1.204 g/ml Optiprep (iodixanol) and loaded in the bottom of Beckman Ultra-Clear centrifuge tubes. Crude mitochondria were overlaid with an equal volume of 1.175 g/ml and 1.078 g/ml Optiprep in HB and centrifuged at 50,000 × g for 4 h in a Beckman SW-55 Ti rotor. During centrifugation, mitochondria along with any proteins tightly associated with them float up to their corresponding buoyant density of 1.13–1.15 g/ml, which bands at the interface between the 1.078- and 1.175-g/ml layers of the gradient. Under these conditions, soluble or aggregated proteins (density 1.26 g/ml) would not be able to break through the density barrier and should sediment downward. After centrifugation the mitochondria-enriched bands were collected and washed once with HB to remove the Optiprep. The protein contents of these fractions were then determined. Samples of the SOD1G93A and SOD1G93A;Dync1h1Loa/+ cerebral cortex and spinal cord buoyant density gradient (BDG) fractions containing equal amounts of proteins were subjected to treatments with salt, alkali, and digestion with proteinase K in the presence and absence of detergents and then analyzed by SDS-PAGE and immunoblotting.
Preparation of the Microsomes
The post-crude mitochondrial supernatant obtained during sample preparation for the buoyant density analysis was used to obtain the microsomal fraction. The pellet (microsomal fraction) obtained after spinning the post-mitochondrial supernatant at 60,000 × g at 4 °C in the Beckman TL-100 ultracentrifuge was washed once with the HB, and the protein content was determined. Samples of the SOD1G93A and SOD1G93A;Dync1h1Loa/+ cerebral cortex and spinal cord microsomal fractions containing equal amounts of proteins were subjected to chemical insults with salt and alkali and digestion with proteinase K in the presence and absence of detergents and then analyzed by SDS-PAGE and immunoblotting.
Treatment with Alkali
Samples from the BDG fractions and the microsomal fraction were incubated with 0.1 m Na2CO3 (pH 11.5) for 30 min on ice and then centrifuged at 16,000 × g for 15 min. Supernatants were collected, and pellets were washed once and then resuspended in homogenization buffer. Supernatants and pellets were analyzed by SDS-PAGE and immunoblotting.
Extraction with High Salt
Samples from the BDG fractions and the microsomal fraction containing equal amounts of proteins were incubated sequentially in 0.2, 0.5, and 1.0 m KCl for 20 min on ice then centrifuged at 16,000 × g for 15 min. Pellets were washed once. Pellets and supernatants were analyzed by SDS-PAGE and immunoblotting.
Treatment with Proteinase K
Samples from the BDG fractions and the microsomal fraction containing equal amounts of proteins were treated with 100 μg/ml proteinase K for 15 min at room temperature in the absence or presence of differing combinations of 0.5–1% Triton X-100 and 1% SDS. Proteinase K was inactivated by adding 10 mm phenylmethylsulfonyl fluoride for 10 min on ice. Digestions were analyzed by SDS-PAGE and immunoblotting.
Preparation of Mitochondria-enriched Fractions for Polarographic Studies and Measurement of Oxygen Consumption
Brain cortex and spinal cord tissues were dissected and washed once in ice-cold tissue wash buffer (TWB; 0.3 m mannitol, 20 mm MOPS, 2 mm EDTA (pH 7.5)) then homogenized with a loose pestle in ∼1 ml of TWB supplemented with 1% bovine serum albumin and 7 mm l-cysteine hydrochloride monohydrate. Homogenates were centrifuged at 800 × g for 10 min at 4 °C. The supernatants were transferred into new tubes and centrifuged at 10,000 × g for 10 min at 4 °C. The pellets were gently resuspended in ∼1 ml of TWB supplemented with 1% bovine serum albumin but without l-cysteine and re-centrifuged as above; the pellets (mitochondrial fractions) were then gently resuspended in the minimum amount of TWB supplemented with 1% bovine serum albumin, and protein concentrations were determined.
Oxygen consumption was measured polarographically in a Rank oxygen electrode containing 0.4 ml of 0.3 m mannitol, 10 mm potassium phosphate, 10 mm KCl, 5 mm MgCl2, and 10 mm HEPES (pH 7.4). Cytochrome c oxidase activity was determined after the addition of 10 mm sodium ascorbate and 1 mm tetramethyl-p-phenylenediamine. Rates are expressed as nmol of O2·min−1.mg of protein−1.
Subfractionation of the Mitochondria
Three freshly isolated mouse cortices of each genotype were homogenized and subjected to buoyant density gradient centrifugation to isolate floated mitochondria that were free from protein aggregates, as described above. We then used Pallotti and Lenaz (39) method to fractionate the mitochondria. Briefly, the mitochondria were resuspended in hypotonic 15 mm KCl and allowed to swell for 10 min on ice before centrifugation at 105,000 × g for 15 min. The supernatants, intermembrane space (IMS) fractions, were kept, and the pellets (mitoplasts) were resuspended in 150 mm KCl followed by incubation on ice for 10 min. The mitoplast suspensions were then subjected to centrifugation at 105,000 × g for 15 min. The supernatants (S2) were kept, and the mitoplast pellets were then resuspended in 250 mm sucrose and 10 mm Hepes-NaOH (pH7.4) with 1 mm EDTA plus proteases and phosphatase inhibitors followed by drastic sonication at 150 Watt for a total of 10 min, with 30-s bursts and 30-s off cycles. The sonicated suspensions were then centrifuged at 26,000 × g for 15 min to remove large particles. The resulting supernatants were pooled and ultracentrifuged at 152,000 × g for 40 min. The supernatants (matrix fractions) were collected, and the pellets (membrane fractions) were resuspended in the above sucrose buffer and kept frozen at −80 °C for Western blot analysis.
Primary Motor Neuron Cultures
Mixed motor neuron cultures were prepared from SOD1G93A mice (40). Briefly, embryonic spinal cords (E13) were removed, and the ventral horns were isolated. Genotype was determined, and wild type, Loa/+, SOD1G93A, or SOD1G93A/Loa ventral horns were pooled and cultured separately. Motor neurons were seeded at 2.5 × 104 cells/cm2. Cells were maintained in complete neurobasal medium (supplemented with 2% B-27 supplement, 0.5 mm glutamine, 0.05% mercaptoethanol, and 2% horse serum (all Invitrogen), 0.1 ng/ml glial-derived neurotrophic factor, 0.5 ng/ml ciliary neurotrophic factor, and 0.1 ng/ml brain derived neurotrophic factor (all Caltag, Silverstone, UK), 50 units/ml penicillin, 50 μg/ml streptomycin, and 2.5 μg/ml amphotericin B (all Sigma)) in a 37 °C, 5% CO2 humidified incubator for 7 days. Motor neurons were distinguished from interneurons using the following morphological criteria: a cell body diameter ≥15 μm and the presence of a minimum of three neuritic processes (for details see Bilsland et al. (41).
Fluorescent Measurement of Mitochondrial Membrane Potential (ΔΨm)
Confocal images were obtained using a Zeiss 510 confocal laser scanning microscope (Oberkocken, Germany) equipped with 40× and 63× plan-apochromat objective lenses. Cells were loaded with 30 nm tetramethylrhodamine methyl ester (TMRM from Molecular Probes, Paisley, UK) for 30 min and incubated at 37 °C in a HEPES-buffered salt solution (recording medium) composed of 156 mm NaCl, 3 mm KCl, 2 mm MgSO4, 1.25 mm KH2PO4, 2 mm CaCl2, 10 mm glucose, and 10 mm HEPES (pH 7.35; all Sigma) for the measurement of ΔΨm. Fluorescence was excited using the 543 nm laser line, and emitted fluorescence was measured above 560 nm. Measurements were made from a compressed z stack to avoid bias from confocal sampling from a single image plane (41).
Image Analysis
ΔΨm was analyzed using LSM software (Carl Zeiss GmbH in association with EMBL, Heidelberg, Germany). In all experiments the analysis was made by measuring the mean TMRM fluorescence intensity in mitochondria excluding all background signals, so that the signal was not influenced by mitochondrial mass.
GST Tagging, Bacterial Expression, and GST Pulldown of SOD1
Wild type SOD1, SOD1A4V, SOD1G37R, and SOD1G93A in the pET28 expression vector (42) were subcloned in-frame into the GST pGEX-4T-1 (Amersham Biosciences) bacterial expression vector using BamHI/XhoI restriction sites. Escherichia coli BL21 (DE3) bacteria were transfected with the GST-tagged SOD1 constructs or pGEX-4T-1 vector, and the proteins were expressed by isopropyl 1-thio-β-d-galactopyranoside induction (43). After lysing the cells, the expressed GST and GST-tagged SOD1 proteins were purified using S-linked glutathione-agarose beads (Sigma). The captured GST and GST-SOD1 proteins on beads were then incubated with brain homogenate prepared from a non-transgenic C57BL/6 mouse. Beads were then washed 4 times with homogenization buffer then heated in 1× SDS sample loading buffer at 95 °C for 5 min. The pulldown assays were then analyzed by SDS-PAGE and Western blotting.
Statistical Analysis
We used GraphPad Prism and SigmaStat to analyze our data by two-way analysis of variance followed by Bonferroni post tests, for mitochondrial respiratory function studies, Wilcoxon matched-pairs test, for Western blots, the Mann-Whitney U test, Student's t test and analysis of variance, for motor neuron studies. Significance was set at p < 0.05.
RESULTS
The Integrity and Sucrose Sedimentation Pattern of the Dynein Complex Are Not Compromised in SOD1G93A and SOD1G93A;Dync1h1Loa/+
We have previously shown that dynein-mediated retrograde axonal transport is impaired in cultured motor neurons isolated from SOD1G93A embryos and that the Loa mutation rescues this defect (17). We, therefore, crossed Dync1h1Loa/+ mice with the SOD1G93A transgenic mice to generate SOD1G93A;Dync1h1Loa/+ double mutants, SOD1G93A, Dync1h1Loa/+, and Dync1h1+/+ progeny and examined whether mutant SOD1 disrupts the dynein complex and, if so, whether the Loa mutation confers any resistance to this disruption in SOD1G93A;Dync1h1Loa/+ mice, giving rise to delayed disease and extended life span. To investigate this possibility we analyzed the sedimentation of the dynein complex using 5–40% continuous sucrose gradient density sedimentation assays on brain and spinal cord homogenates of late-stage SOD1G93A (120 days old) and SOD1G93A;Dync1h1Loa/+ (150 days old) and Dync1h1Loa/+ and Dync1h1+/+ at 150 days of age (Fig. 1A).
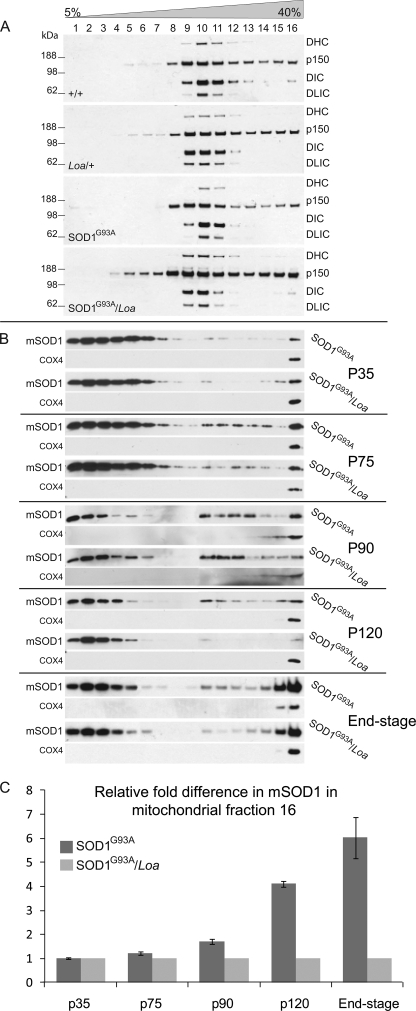
FIGURE 1.
The integrity and sucrose sedimentation pattern of the dynein complex is not compromised in SOD1G93A and SOD1G93A/Loa, but the Loa mutation in dynein alters the sedimentation of mutant SOD1 in SOD1G93A/Loa mice. A, brain homogenates of equal protein contents prepared from +/+, Loa/+; SOD1G93A, and SOD1G93A/Loa mice (at end stage of disease in the SOD1G93A and SOD1G93A/Loa mice) were sedimented in 5–40% continuous sucrose gradient by centrifugation at 237,000 × g for 4 h at 4 °C as described under “Experimental Procedures.” Then 16 equal-volume fractions. were collected from top to bottom and analyzed by SDS-PAGE and Western blotting detection for DHC, p150, DIC, and dynein light intermediate chain (DLIC) as representatives of the dynein-dynactin complex. B, shown are brain homogenates of equal protein contents prepared from SOD1G93A and SOD1G93A/Loa littermates at the postnatal stages; P35 (before disease onset), P75 (onset of disease), P90 (early stage of disease), P120 (late stage of disease), and end-stage, were sedimented in 5–40% continuous sucrose gradient by centrifugation at 237,000 × g for 4 h at 4 °C as described under “Experimental Procedures.” Then 16 equal-volume fractions were collected from top to bottom and analyzed by SDS-PAGE and Western blotting detection for SOD1 (to avoid overexposure caused by much higher levels of mutant SOD1 (mSOD1) protein in the lighter fractions of the gradient, we loaded 3 μl of fractions 1–3, 8 μl of fractions 5–7, 10 μl of fractions 8–9, and 20 μl of fractions 11–16; supplemental Fig. 3 shows the true representations of equal loading of the fractions on SDS-PAGE immunoblots). C, -fold difference (n = 3–6) in mutant SOD1 protein in fraction 16 of the SOD1G93A mice was quantified relative to that in SOD1G93A/Loa mice using COX4 as the internal control.
These data show that the bulk of the dynein complex, represented by DHC, DIC, and dynein light intermediate chain, sediments in fractions 9, 10, and 11 and that their sedimentation patterns in wild-type (+/+) brain matched those of SOD1G93A brain, and likewise, the sedimentation patterns of these polypeptides from Dync1h1Loa/+ brain matched those from SOD1G93A;Dync1h1Loa/+. However, we note that there is a slight Loa-dependent shift of the dynein complex toward lighter fraction 9 in Dync1h1Loa/+ and SOD1G93A;Dync1h1Loa/+, as evident from DIC and dynein light intermediate chain signals in fractions 11 and 12 versus those in fraction 9 of all the genotypes (Fig. 1A). We will discuss these data further under “Discussion.” As only a subpopulation of the p150 subunit of dynactin is associated with the dynein complex, there was a wider sucrose gradient density distribution of this polypeptide, but there were no significantly reproducible differences between the genotypes for p150. Together, these data indicate that the dynein complex in brain and spinal cord of transgenic mice is neither perturbed by the presence of mutant SOD1 nor is it involved in higher molecular weight complexes, such as SOD1G93A aggregates.
The Loa Mutation in Dynein Alters the Sedimentation of Mutant SOD1 in SOD1G93A;Dync1h1Loa/+ Mice
As the mutant SOD1 did not affect the sedimentation pattern of the dynein complex, we decided to look at the effect of mutant dynein on the sedimentation pattern of SOD1G93A in brains and spinal cords at P35 (postnatal day 35; before symptom onset), P75 (at symptom onset), P90 (early stage disease), P120 (late stage disease), and end-stage (when both SOD1G93A and SOD1G93A;Dync1h1Loa/+ mice show hind limb paralysis and delayed righting reflex). Fig. 1B shows representative data from brain homogenates and demonstrates that the mutant dynein does not affect the sedimentation of SOD1G93A at P35 (i.e. before the onset of the disease). However, at P75, the sedimentation of mutant SOD1 protein in SOD1G93A brain homogenate starts to show a different pattern than that from SOD1G93A;Dync1h1Loa/+ in which the amount of the mutant SOD1 protein sedimented in the denser fractions of the gradient was less than that in the equivalent fractions from SOD1G93A brain homogenate. Interestingly, this difference increased as the disease progressed from the early stage toward the end-stage, when the difference was most prominent (see P90, p120, and end-stage in Fig. 1B). It is noteworthy that to avoid overexposure of the films due to significantly more mutant SOD1 in the lighter fractions, we loaded smaller amounts of protein in lanes 1–10 for SDS-PAGE (Fig. 1B).
Quantitative analyses of these data using the mitochondrial marker COX4 as the internal control are shown in Fig. 1C. Similar results were obtained from BDG centrifugation assays (data not shown) and from spinal cords (see supplemental Fig. 1), but the effect was more profound in the brains.
To test that this difference in sedimentation was not due to differences in expression of SOD1G93A protein in SOD1G93A;Dync1h1Loa/+ and SOD1G93A or in the protein contents loaded onto the sedimentation columns, various amounts of proteins from the same brain homogenates used for sedimentation experiments were analyzed by SDS-PAGE followed by Western blotting using antibodies against SOD1 and α-tubulin. As shown in supplemental Fig. 2, the expression levels of SOD1G93A protein are almost identical in SOD1G93A;Dync1h1Loa/+ and SOD1G93A brains, as judged by the signal from the loading control protein, α-tubulin. Together these data suggest that mutant dynein alters the sucrose gradient density distribution of SOD1G93A, clearing the protein away from the densest fractions of the gradient. This effect, as shown in Fig. 1, B and C, becomes more evident with the progression of the disease.
The Loa Mutation Reduces the Amount of Mutant SOD1 in Subcellular Fractions Containing Mitochondria and Endoplasmic Reticulum (ER) Microsomes
Next, we used more organelle-specific markers to look at the sedimentation patterns of different cell organelles in the above continuous sucrose gradients. As supplemental Fig. 3 shows, the distribution of the trans-Golgi marker TGN38 (type I integral trans-Golgi membrane protein) was confined to the lightest fractions of the gradient (supplemental Fig. 3, lanes 1–4). ER microsomes were spread across most of the gradient as judged by the signal from calnexin (type I integral ER membrane protein), an ER-specific marker (supplemental Fig. 3, lanes 3–16). Detection for Cox4 (an integral inner mitochondrial membrane protein), on the other hand, revealed that the mitochondria were sedimented solely in the densest fractions of the gradient (Fig. 1B and supplemental Fig. 3). The pattern of SOD1G93A and the distribution of the mitochondria and ER in the sucrose gradients suggest that the mitochondria and/or ER could be involved in the effect exerted by Loa on SOD1G93A distribution.
Effect of the Loa Mutation on the Association of SOD1G93A Protein with Mitochondria and ER in the Brain Cortex
The co-enrichment of mutant SOD1 aggregates with organelles such as mitochondria during fractionation, in which the samples were layered on top of a continuous gradient, has been suggested to be a contamination rather than genuine association of the mutant protein with this organelle (38). Thus, we have used a discontinuous iodixanol BDG centrifugation to circumvent this potential contamination problem. After centrifugation, the enriched floated mitochondrial fractions were collected. The iodixanol was removed, and the protein contents of these fractions and the 60,000 × g “microsomal” fraction obtained from the post-mitochondrial supernatant (for details, see “Experimental Procedures”) were then determined. Subsequently, the association of SOD1G93A with the organelles contained in these fractions was tested by subjecting samples of equal protein content from all fractions to salt extraction and alkali treatment. These treatments dissociate mutant SOD1 and other peripheral membrane proteins that form electrostatic or hydrogen bonds with the cytoplasmic face of the mitochondrial outer membrane.
Fig. 2A shows representative results of the sequential salt (KCl) or alkali (Na2CO3) extractions from the mitochondria-enriched fractions to release the peripheral membrane proteins. These samples, containing equal amounts of protein, were subjected to increasing concentrations of 0.2, 0.5, and 1.0 m KCl or 0.1 m Na2CO3 (pH 11.5). After incubation with each salt or alkali concentration, the samples were centrifuged down to obtain soluble and insoluble fractions. All fractions were then analyzed by SDS-PAGE and Western blotting. The Western blotting analyses in Fig. 2A show positive signals for Cox4 and calnexin (Fig. 2A, lanes 4, 8, 10, and 12), thus indicating the presence of mitochondria as well as the microsomal ER in the mitochondria-enriched fraction of the buoyant density gradient. Moreover, treatment of these membranes with 0.2 m KCl followed by quantification analysis of the signals using COX4 as the internal control revealed that compared with SOD1G93A mice, the release of membrane-associated mutant SOD1 protein from SOD1G93A;Dync1h1Loa/+ brain cortex was significantly higher (p = 0.03, n = 6) (Fig. 2, A, lanes 1 and 5, and B, left panel). Increasing the salt concentration did not induce further release of SOD1G93A from brain cortex in any of the samples (Fig. 2A, lanes 2 and 3 and lanes 6 and 7).
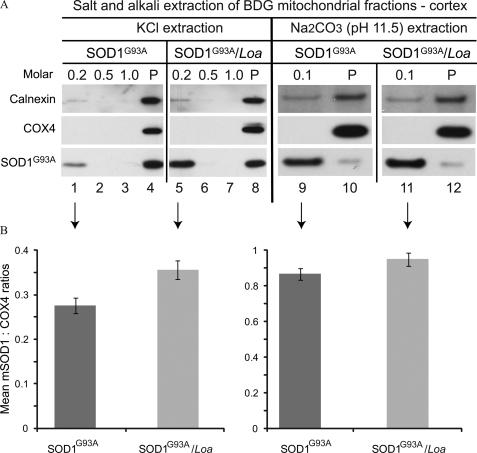
FIGURE 2.
SOD1G93A is less strongly associated with the mitochondria and ER in the brain cortex of SOD1G93A/Loa mice. A, samples containing equal protein amounts of the BDG mitochondrial fractions prepared from brain cortices of SOD1G93A and SOD1G93A/Loa littermates at the late-stage of disease were sequentially incubated with 0.2, 0.5, and 1.0 m KCl or 0.1 m Na2CO3 (pH 11.5) for 20 or 30 min, respectively, on ice then centrifuged at 16,000 × g for 15 min after each incubation to obtain soluble and insoluble (P) fractions that were then analyzed by SDS-PAGE and Western blotting detection for calnexin, Cox4, and SOD1. B, the amounts of mutant SOD1 released after treatment were quantified (n = 4) using COX4 as the internal control for salt extraction, 0.2 m KCl, and 0.1 m Na2CO3 as indicated by the arrows. Wilcoxon matched-pairs analysis showed that more mutant SOD1 is released from SOD1G93A/Loa compared with SOD1G93A, reaching statistical significance for salt (p = 0.03), but not for alkali (p = 0.13) extractions.
Representative data showing the alkali extraction of SOD1G93A are demonstrated in Fig. 2 (right panels). Quantification analysis of these data (Fig. 2B, right panel) showed increased levels of mutant SOD1 released from the membranes of SOD1G93A;Dync1h1Loa/+ (Fig. 2A, lanes 11 and 12) than from SOD1G93A (Fig. 2A, lanes 9 and 10). This increase, however, did not reach statistical significance (p = 0.1, n = 4).
Thus, the mutant SOD1 protein association with the organelle membranes in the mitochondrial-enriched fraction seems to be more sensitive to salt and alkali in the cerebral cortex of SOD1G93A;Dync1h1Loa/+ compared with that from SOD1G93A mice. Moreover, this fraction contains microsomal-ER as indicated by signals from calnexin (Fig. 2A). Some of calnexin, or at least its cytoplasmic domain, can be seen stripped off the membranes and is, therefore, detected in the soluble fractions (Fig. 2A, lanes 1 and 5 and lanes 9 and 11). It is worth noting that calnexin is a type I integral membrane protein with its COOH terminus exposed on the cytoplasmic side, and the antibody used in these experiments was raised against the cytoplasmic domain, hence, suggesting that the cytoplasmic domain of calnexin might have been cleaved off the membranes by the salt and alkali insults. Together these data suggest that the SOD1G93A protein associates with mitochondria and/or microsomes and that the Loa mutation weakens this association.
Organelle-associated SOD1G93A Is Largely Protease- and Detergent-resistant
The organelle-associated SOD1G93A protein in BDG fractions isolated from SOD1G93A and SOD1G93A;Dync1h1Loa/+ spinal cord and brain homogenates was also tested for its protease and detergent sensitivity. Supplemental Fig. 4A shows representative data obtained by incubating the mitochondria-enriched fractions of these samples with proteinase K (PK) in the absence (lanes 1 and 3) or presence (lanes 2 and 4) of 0.5% Triton X-100, which solubilizes the organelle membranes. Incubating the organelles present in this fraction with PK alone did not have any significant effect on SOD1G93A protein or organelles as indicated by the presence of signals from mutant SOD1 and the organelle-specific integral membrane proteins calnexin, TGN38, and Cox4 (supplemental Fig. 4A, lanes 1 and 3). However, in the presence of 0.5% Triton X-100, PK digested the organelle specific markers, but it was still unable to digest mutant SOD1 protein (supplemental Fig. 4A, lanes 2 and 4). The presence of Triton X-100 at this concentration was clearly not sufficient to completely solubilize the inner mitochondrial membrane, as some of the Cox4 protein could still be detected in the presence of the detergent (supplemental Fig. 4A, lanes 2 and 4). Moreover, the Loa mutation did not appear to have any effect on the PK sensitivity of mutant SOD1 in the presence or absence of 0.5% Triton X-100.
Thus, we analyzed the PK digestion of mitochondrial SOD1G93A in two BDG fractions, 1 and 3, of spinal cord by increasing the concentration of Triton X-100 to 1% and adding SDS (1%) to the reaction. Under these conditions the inner mitochondrial membrane was completely dissolved, thereby making the intramitochondrial SOD1G93A accessible to PK digestion (supplemental Fig. 4B). As a result, the intramitochondrial SOD1G93A protein was largely but not totally digested compared with the reactions without the detergents (supplemental Fig. 4B, lanes 2, 4, 6, and 8). These data show that some PK-sensitive SOD1G93A protein is in the matrix or associated with mitochondrial membranes; however, there is still a large proportion of the mutant SOD1 protein, most likely in an aggregated form, that is not degraded by PK. Moreover, we did not detect any difference between SOD1G93A and SOD1G93A;Dync1h1Loa/+ genotypes in these assays (supplemental Fig. 4B), suggesting that the Loa mutation does not alter the nature of mutant SOD1 protein in the mitochondria.
Loa Mutation Alters SOD1G93A Localization in the Mitochondria
To examine the effect of the Loa mutation on the redistribution of the mutant SOD1 protein, we focused on the mitochondria. Floated mitochondria were isolated from cerebral cortex of three SOD1G93A or SOD1G93A;Dync1h1Loa/+ mice and fractionated into IMS, matrix, or membrane fractions, as described under “Experimental Procedures.”
As shown in Fig. 3, in SOD1G93A mitochondria most of the mutant protein is in the matrix. In contrast, the mutant SOD1 is mainly in the IMS of the SOD1G93A;Dync1h1Loa/+ mitochondria (Fig. 3A). Quantitative analysis of these data using COX4 as internal control revealed that the IMS of SOD1G93A contained significantly less mutant protein compared with that in SOD1G93A;Dync1h1Loa/+ mitochondria (p = 0.03, n = 3). The matrix and membrane fractions of SOD1G93A mitochondria, however, contained higher levels of the SOD1G93A protein, reaching statistical significance for the matrix (p = 0.02 and 0.25, respectively, n = 3) (Fig. 3B).
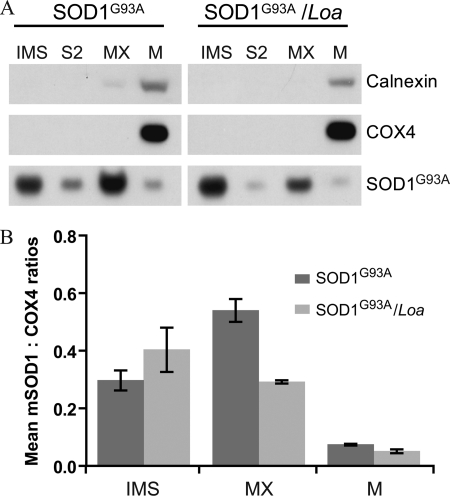
FIGURE 3.
Loa mutation alters the localization of mutant SOD1 in the mitochondria. A, suspensions of freshly isolated mitochondria from cerebral cortex of three mice of each genotype were suspended in 0.015 m KCl and allowed to swell for 10 min on ice before centrifugation at 105,000 × g for 15 min. The supernatants containing mutant SOD1 in the mitochondrial IMS were kept, and the pellets were resuspended in 0.15 m KCl for 10 min on ice followed by centrifugation at 105,000 × g for 15 min. The supernatants (S2) were kept, and the pellets (mitoplasts) were then resuspended in 0.25 m sucrose and 10 mm Hepes-NaOH (pH 7.4) with 1 mm EDTA plus proteases and phosphatase inhibitors. The mitoplasts were then harshly sonicated with 30-s bursts followed by 30-s off cycles at 150 Watt for a total of 10 min. The sonicated suspension was centrifuged at 26,000 × g for 15 min to remove large particles. The resulting supernatants containing mitochondrial matrix (MX) were ultracentrifuged at 152,000 × g for 40 min. The supernatants were collected as matrix (MX), and the pellets, membranes (M), were suspended in the above sucrose buffer followed by SDS-PAGE and Western blot analysis. B, quantitative analysis (n = 3) using COX4 as the internal control and the Wilcoxon matched-pairs test of the mitochondrial subfractions revealed the presence of significantly higher (p < 0.02) and lower (p = 0.016) levels of mutant SOD1 (mSOD1) in the IMS and matrix, respectively, of SOD1G93A/Loa mitochondria compared with those in SOD1G93A. The mutant SOD1 associated with the membrane subfractions tended to be higher in the SOD1G93A, but it did not reach statistical significance (p = 0.25).
Mitochondrial Respiration and Membrane Potential Are Severely Impaired in SOD1G93A Mice, but They Are Ameliorated in SOD1G93A;Dync1h1Loa/+ Mice
Collectively, the above data suggested differential association of mutant SOD1 with mitochondria from SOD1G93A;Dync1h1Loa/+ versus those from SOD1G93A mice. Moreover, there is growing evidence that the mitochondrial localization of mutant SOD1 may play an important role in the pathogenesis of ALS (14, 41). Therefore, given the limited amount of the protein in mitochondrial preps and as cytochrome c oxidase (complex IV) activity is a sensitive indicator of mitochondrial function, we assessed mitochondrial function at the cytochrome c oxidase level in mitochondrial fractions from brain cortex and spinal cord of the four possible genotypes (wild-type Dync1h1+/+, heterozygous Dync1h1Loa/+, hemizygous SOD1G93A, and double mutants SOD1G93A;Dync1h1Loa/+). In these experiments cytochrome c oxidase activity was assayed polarographically by measuring oxygen consumption rate in the presence of ascorbate and tetramethyl-p-phenylenediamine. Our data indicate that the respiratory rate of the mitochondria from 120-day (late stage) SOD1G93A brain is substantially reduced compared with that of their wild-type littermates (gray bars in Fig. 4A).
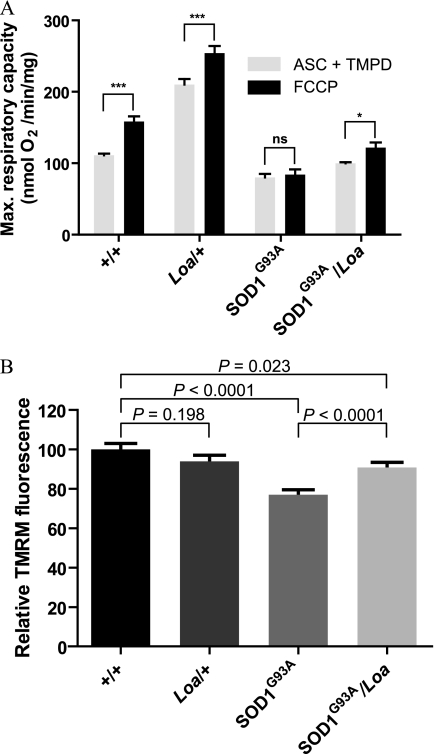
FIGURE 4.
Loa mutation ameliorates defects in oxygen consumption rate and membrane potential of SOD1G93A mitochondria. A, brain mitochondria were purified from homogenates in TWB buffer supplemented with 1% bovine serum albumin and 7 mm l-cysteine hydrochloride monohydrate as described under “Experimental Procedures.” Oxygen consumption rates of mitochondria were measured in an oxygen electrode in the presence of ascorbate (ASC) and tetramethyl-p-phenylenediamine (TMPD; gray bars). Respiration was stimulated by the addition of 1 μm FCCP (black bars). As expected, the addition of FCCP significantly stimulated oxygen consumption in +/+ (p < 0.001) and Dync1h1Loa/+ (p < 0.001). SOD1G93A mitochondria, on the other hand, failed to produce any significant response to FCCP. However, SOD1G93A/Loa mitochondria showed increased respiration after the addition of FCCP, which was statistically significant (p < 0.05). Two-way analysis of variance (degrees of freedom = 15) followed by Bonferroni post tests were used for statistical analysis of these data. ns, not significant. *, p < 0.05; ***, p < 0.001. B, mitochondria in motor neurons from SOD1G93A mice are depolarized, but this effect is abolished in motor neurons of SOD1G93A/Loa mice. TMRM is a potentiometric indicator with a single delocalized positive charge, and thus, it becomes sequestered in mitochondrial as a result of the electrochemical potential gradient that exists between the cytosol and mitochondria. Using TMRM and confocal microscopy, mitochondrial membrane potentials (ΔΨm) were measured in cultured motor neurons isolated from E13 embryos of each genotype. The ΔΨm of SOD1G93A motor neurons was 23% lower than the ΔΨm of motor neurons from wild-type littermates (p < 0.001), indicating that mitochondria from SOD1 motor neurons are depolarized. However, in motor neurons from SOD1G93A/Loa mice, ΔΨm was 10% lower (p = 0.023) than that in motor neurons of wild-type littermates, indicating that the Loa mutation is able to ameliorate the defect in ΔΨm present in SOD1G93A motor neurons.
Interestingly, mitochondrial oxygen consumption in Dync1h1Loa/+ exceeds that of the wild type and it is improved in SOD1G93A;Dync1h1Loa/+ relative to SOD1G93A (gray bars in Fig. 4A). The subsequent addition of FCCP to obtain the maximal respiratory activity resulted in significant respiratory stimulations in mitochondria from the wild type, Dync1h1Loa/+, and SOD1G93A;Dync1h1Loa/+ but not with the SOD1G93A mitochondria (black bars in Fig. 4A). Statistical analysis of these data using two-way analysis of variance followed by Bonferroni post tests revealed that the mitochondrial response to FCCP stimulation is not consistent for all genotypes (p = 0.012, degrees of freedom = 15) and that this stimulatory response is significant in wild type (p < 0.001), Dync1h1Loa/+ (p < 0.001), and SOD1G93A;Dync1h1Loa/+ (p < 0.05) but not in SOD1G93A (p > 0.05).
These data indicate that in SOD1G93A mitochondria cytochrome c oxidase activity is impaired and that the Loa mutation appears to partially restore this defect. This amelioration of complex IV function by the Loa mutation is consistent with our confocal microscopy imaging data obtained from examining the mitochondrial membrane potential in cultured primary motor neurons using TMRM. The data from these assays are shown in Fig. 4B and reveal that, compared with motor neurons from wild-type littermates (n = 55), the mitochondrial membrane potential (ΔΨm) in SOD1G93A motor neurons (n = 54) is significantly depolarized (p < 0.0001) and that this defect is ameliorated in motor neurons of SOD1G93A;Dync1h1Loa/+ mice (n = 71). In contrast, the ΔΨm of Dync1h1Loa/+ motor neurons (n = 48) was not significantly different from the ΔΨm of wild type motor neurons (n = 55).
Mutant SOD1G93A Does Not Show Interactions with Cytoplasmic Dynein in Immunoprecipitation (IP) Assays
The redistribution of mutant SOD1 protein, the amelioration of mitochondrial function, and the previously observed increased life span and delayed disease onset in SOD1G93A;Dync1h1Loa/+ double mutant mouse strains suggested a possible interaction between components of the dynein complex and SOD1G93A protein. We, therefore, analyzed the mutant SOD1 protein and components of the dynein complex in the spinal cord and brain homogenates from SOD1G93A;Dync1h1Loa/+ double mutants, SOD1G93A, Dync1h1Loa/+, and Dync1h1+/+ mice at varying stages of the disease. The interaction between dynein-dynactin and SOD1G93A was investigated by IP assays.
Co-IP analysis of the brain homogenate of an end-stage SOD1G93A mouse (125 days old) revealed that the polyclonal anti-SOD1 antibody could precipitate only a small amount of the SOD1G93A protein, but considerable amounts of p150 and DIC were present in the IP sample (supplemental Fig. 5A, lane 3). On the other hand, neither anti-dynactin p150 nor anti-DIC antibody was successful in co-immunoprecipitating SOD1G93A from the same brain homogenate (supplemental Fig. 5A, lanes 8 and 13). A weaker signal from DIC and p150 was also found in the negative control IP, in which the brain homogenate was incubated with beads only (supplemental Fig. 5A, lane 4). However, the difference in signal intensities obtained from sample and negative control IPs was big enough to suggest the pulldown may be specific, at least for the p150 (supplemental Fig. 5A, lane 3).
To test this further, the IP experiment was repeated on brain homogenate prepared from another end-stage mouse using mouse monoclonal or rabbit polyclonal anti-SOD1 and mouse monoclonal anti-p150 as IP antibodies. As shown in supplemental Fig. 5B, lanes 4 and 8, only the polyclonal and not the monoclonal anti-SOD1 antibody co-immunoprecipitated p150, suggesting that the interaction of SOD1G93A and p150 is not direct and may be in part due to temporal entrapment of p150 protein and possibly other soluble proteins within SOD1G93A aggregates. Once again, the anti-p150 antibody was not able to pull down SOD1G93A (supplemental Fig. 5B, lane 12).
To verify the specificity of SOD1-DIC interaction observed in supplemental Fig. 5A, we conducted more IP experiments on brain homogenates prepared from several end-stage SOD1G93A mice using antibodies against DIC and SOD1. The anti-DIC antibody was only able to specifically co-IP some of the SOD1G93A protein with dynein in one mouse brain homogenate (supplemental Fig. 5C, lane 3). On the other hand, when a reciprocal IP was conducted, a mouse monoclonal anti-SOD1 antibody failed to co-IP any dynein along with SOD1G93A from the same brain homogenate (supplemental Fig. 5C, lane 5).
Lack of reproducible data for interaction between SOD1G93A and dynein prompted us to try a series of different homogenization and binding buffers and conditions. Supplemental Fig. 5D shows that none of these conditions was able to co-IP SOD1 with dynein (lanes 3–5) even when all dynein present in the sample was immunoprecipitated (lane 9).
This lack of dynein and SOD1G93A interaction contradicts the report by Zhang et al. (22) in which IP assays show a progressive interaction between these two proteins, which correlated with the disease progression in SOD1G93A transgenic mice. Thus, we sought to investigate these interactions in brains and spinal cords of end-stage SOD1G93A mice when, according to the above report, dynein and SOD1G93A interact with high affinity (22).
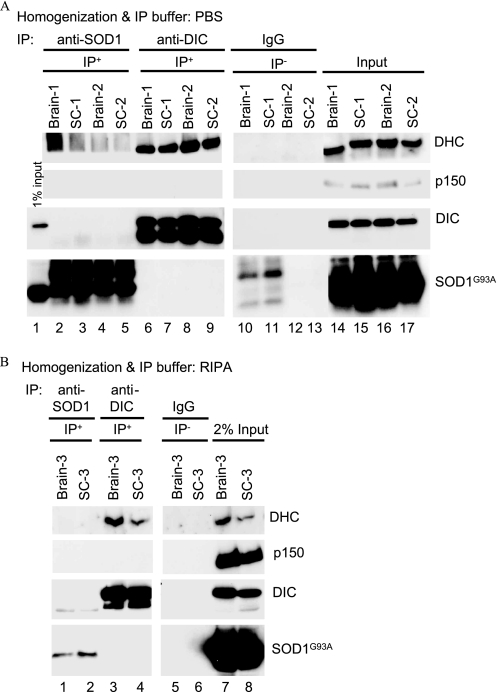
We used the Zhang et al. (22) RIPA buffer for homogenization and binding reactions as well as our IP protocol that utilizes PBS−Ca/Mg. Fig. 5 shows representative data from these assays on brain and spinal cord homogenates from end-stage SOD1G93A mice. As shown in Fig. 5A, we successfully pulled down DHC and DIC using the 74.1 antibody against DIC in PBS as homogenization and binding buffer, but no SOD1G93A protein could be detected in the pulldown assays (Fig. 5A, lanes 6–9). Likewise, the anti-SOD1 antibody did not IP DHC, DIC, or p150 (Fig. 5A, lanes 2–5). In addition, using the same RIPA buffer and IP antibody and conditions as reported by Zhang et al. (22), DHC and DIC were successfully immunoprecipitated, but we did not detect any co-IP of SOD1G93A protein with dynein (Fig. 5B, lanes 3 and 4). The anti-SOD1 antibody did not IP DHC, DIC, or p150 in this buffer either (Fig. 5B, lanes 1 and 2). It is noteworthy that the DIC74.1 antibody is currently the most efficient antibody available for IP of the dynein complex, but as its epitope is in the p150 binding domain of DIC, it appears to disrupt dynein-p150 interactions (44–46), and hence, as shown in Fig. 5, this antibody does not efficiently pull down p150.
FIGURE 5.
Immunoprecipitation of the dynein-dynactin complex and mutant SOD1 from brains and spinal cords of end-stage SOD1G93A mice using two different IP buffers. Homogenates of brains and spinal cords (SC) from three end-stage transgenic mice (SC-1–3) prepared in PBS without Ca2+ and Mg2+ (A) or in RIPA buffer containing 50 mm Tris-HCl (pH 7.4), 150 mm NaCl, 0.25% deoxycholic acid, 1% Nonidet P-40, 1 mm EDTA (B) were incubated with anti-DIC, anti-SOD1 antibody, or with IgG as a negative control, and the IP procedure was conducted as described by Zhang et al. (22). The IP products along with 2% of the input were analyzed by SDS-PAGE and Western blotting.
To ensure that the above lack of interaction between dynein and SOD1G93A in our IP experiments is not due to the dissociation of DIC from the dynein complex and to ascertain the oligomerization of mutant SOD1, we analyzed SOD1G93A and SOD1G93A;Dync1h1Loa/+ brain and spinal cord homogenates prepared in PBS or RIPA buffer in non-denaturing and non-reducing gradient PAGE (4–15%) (supplemental Fig. 6). As shown in the left panel of supplemental Fig. 6, DIC in these samples is part of large molecular weight complexes indicating that the DHC-DIC interactions are intact in these samples. Moreover, SOD1G93A shows stretched ladders/smears that are consistent with its tendency to form oligomers and aggregates (supplemental Fig. 6, right panel). These data clearly show that the methods used to prepare the samples for IP did not affect the complexity of dynein or the heterogeneity of the mutant SOD1 protein.
We also tested the likelihood of interactions between dynein and SOD1G93A in the spinal cord of symptomatic (hind limbs paralyzed) against asymptomatic (no apparent muscle wasting or paralysis in the hind limbs) SOD1G93A and non-transgenic mice aged 131, 118, and 100 days, respectively. Supplemental Fig. 7 shows representative examples of the data using anti-DIC antibody to pull down the dynein complex from the spinal cord homogenates in RIPA buffer. The anti-DIC antibody efficiently pulled down the dynein complex represented by DIC and DHC in all samples, but no SOD1G93A was pulled down, as shown in supplemental Fig. 7, lanes 2, 5, and 8.
Cross-linking Buffer Causes Disruption of Dynein Complex and Nonspecific Pulldown of Spinal Cord SOD1G93A Protein
Cross-linking and immunoprecipitation of proteins in cell lysates from SOD1A4V-expressing NSC34 motor neuron-like cells has been reported to co-IP DHC with the SOD1A4V-containing high molecular weight complexes (22), suggesting a possible transient/weak interaction between mutant SOD1 and dynein that could be stabilized by cross-linking. To test this possibility we cross-linked spinal cord proteins using BS3 followed by IP with the DIC74.1 antibody and immunoblotting to detect DHC, DIC, and SOD1G93A (supplemental Fig. 8A).
Immunoprecipitation was also carried out on spinal cord homogenates prepared for cross-linking but lacking the cross-linker BS3, i.e. the cross-linking conditions were met but without including the cross-linker itself. We found that cross-linking had two effects on the IP of the dynein complex. First, the IP of DIC was less efficient from the cross-linked versus the un-cross-linked samples (supplemental Fig. 8A, DIC bands in lanes 2 versus 3; and 5 versus 6). Second, in the cross-linked samples, DHC was not co-immunoprecipitated by anti-DIC antibody (supplemental Fig. 8A, lanes 3 and 6). Moreover, SOD1G93A was non-specifically precipitated whether the samples were cross-linked or not, suggesting that the SOD1G93A precipitation was solely due to the presence of the cross-linking buffer (supplemental Fig. 8A, lanes 5–7).
This nonspecific pulldown of SOD1G93A seems to be intrinsic to the mutant human SOD1 as mouse SOD1 protein was not found in the pulldown of the cross-linked homogenates from non-transgenic mice (supplemental Fig. 8A, lanes 2 and 3). These data indicate that BS3 cross-linking disrupts DHC-DIC interactions and that the cross-linking buffer used in this assay promotes nonspecific pulldown of SOD1G93A. Furthermore, the dissociation of the DHC-DIC complex by cross-linking was independent of the presence of the mutant SOD1 as it also occurred in the samples prepared from non-transgenic mice.
In the above experiments we followed the procedure recommended by Zhang et al. (22) in which after incubation of the homogenates with the antibodies for 3 h to overnight, the beads were washed 3 times with the IP buffer. To further test the specificity of the SOD1G93A that was co- immunoprecipitated with dynein in the cross-linking buffer (supplemental Fig. 8A), we repeated the above experiments in duplicate; spinal cord homogenates from 100 day old transgenic mice were prepared in cross-linking buffer lacking BS3. In one set of the IP samples the beads were washed three times, as recommended by Zhang et al. (22), and in the other set they were washed four times instead of three. Supplemental Fig. 8B shows that the non-specifically pulled down SOD1G93A in lanes 2 and 3 was completely removed simply by introducing a fourth wash as shown in lanes 5 and 6. These data confirm that IP assays do not detect specific interactions between SOD1G93A and dynein.
Human Wild-type and Different Mutant SOD1-GST Fusion Proteins Do Not Pull Down Dynein in Vitro
We expressed GST-tagged human wild-type SOD1, SOD1A4V, SOD1G37R, SOD1G85R, and SOD1G93A in bacteria for in vitro pulldown experiments. After purification, the GST or the GST-SOD1 proteins captured on glutathione-agarose beads were incubated with brain homogenate prepared from a non-transgenic mouse. After washing four times with homogenization buffer, the pulled-down proteins on beads were analyzed by SDS-PAGE and Western blotting, detecting for SOD1 and DIC. In these pulldown experiments neither the wild-type SOD1 (see supplemental Fig. 9, lane 5) nor any of the mutant SOD1 proteins (lanes 6–9) was able to pull down dynein.
The Loa Mutation Does Not Induce Novel Interaction between Dynein and SOD1G93A
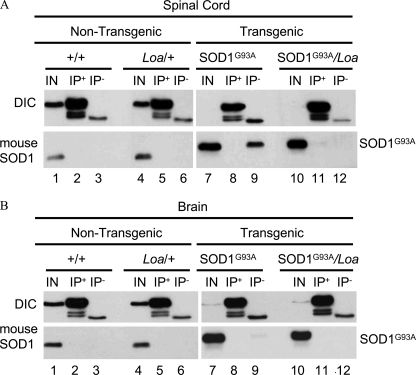
To investigate whether the Loa mutation promotes any interaction between dynein and SOD1, we tested brain and spinal cord homogenates prepared from SOD1G93A, SOD1G93A;Dync1h1Loa/+, Dync1h1Loa/+, and Dync1h1+/+ littermates at a late stage (120 days) of the disease using the IP protocol described by Zhang et al. (22). Fig. 6 shows a representative of our IP assays using DIC74.1 antibody for IP of the dynein complex and its associated proteins. As shown in Figs. 6, A and B, lanes 2, 5, 8, and 11), although DIC is successfully pulled down, neither the endogenous mouse SOD1 nor the transgenic mutant SOD1G93A co-IP with DIC. Interestingly, we also noted that in some cases SOD1G93A protein is pulled down non-specifically in the negative controls containing anti-hemagglutinin antibody only from spinal cord (see lane 9 in Fig. 6A) and to some extent from brain homogenates of the SOD1G93A but not SOD1G93A;Dync1h1Loa/+ mice.
FIGURE 6.
Dynein Loa mutation does not induce novel interaction between dynein and SOD1G93A. Spinal cords (A) and brains (B) of 100-day-old mice representing the four different genotypes produced by crossing SOD1G93A with Loa/+ mice were homogenized in RIPA buffer then subjected to IP as described (22) using anti-DIC antibody or anti-hemagglutinin antibody as the negative control. The IP products along with 1% or 0.2% of the input (IN) from non-transgenic and transgenic homogenates, respectively, were analyzed by SDS-PAGE and Western blotting.
As our earlier data had shown that the Loa mutation in SOD1G93A;Dync1h1Loa/+ mice rescued the retrograde axonal transport defect observed in embryonic SOD1G93A motor neurons in culture (17), we examined the possibility of a likely interaction between SOD1 and dynein during embryonic development. We tested this possibility at embryonic stage E16.5 in IP assays using brain tissues and an antibody against human SOD1 to pull down the mutant SOD1 and any associated components of the dynein-dynactin complex (supplemental Fig. 10). The anti-SOD1 antibody efficiently pulled down SOD1G93A protein from the samples, but DHC, DIC, and p150 were not co- immunoprecipitated with SOD1G93A (see supplemental Fig. 10, lanes 12 and 16). Although a faint signal of DHC was observed in IP from the non-transgenic Dync1h1Loa/+ and from SOD1G93A homogenates (supplemental Fig. 10, lanes 4 and 12), these were regarded as nonspecific as a similar signal for DHC was also found in the IP from the negative control IP in which the IP antibody was not included (data not shown). Collectively these data indicate that there is no IP-detectable interaction between SOD1G93A and dynein in brain homogenates prepared from E16.5 embryos of the Dync1h1Loa/+ x SOD1G93A mouse cross.
DISCUSSION
Data from studies on conditional transgenic mice with cell-specific expression of mutant SOD1 suggest that motor neuron death in ALS is non-cell autonomous. These studies have shown that the deleterious effects of mutant SOD1 within motor neurons determines disease onset, but it is the expression of mutant SOD1 in astrocytes and microglia that accelerates later disease progression independent of the disease onset (47–50). Therefore, because the Loa mutation delays disease onset without changing disease progression, it is possible that the amelioration of the disease induced by the Loa mutation occurs at the level of the motor neuron, suggesting that there may be a functional interaction between mutant SOD1G93A and the Loa mutation within motor neurons.
Moreover, Perlson et al. (51) have recently shown that the retrograde trafficking of trophic factors is defective in both Dync1h1Loa/+ and SOD1G93A mice, but a significant shift from survival retrograde signaling to stress/death signaling is possibly responsible for the rapid demise of the motor neurons in SOD1G93A mice.
The recent proposition (35) that the amelioration of the disease in SOD1G93A-;Dync1h1Loa/+ mice might be a result of reduced excitotoxicity brought about by the loss of sensory neurons in Dync1h1Loa/+ is unlikely to be the sole explanation, as Dync1h1Swl/+, Dync1h1Cra1/+, and Dync1h1Arl/+ mice also show similar or greater sensory neuronal loss but do not ameliorate the SOD1G93A disease phenotype.
The shift in the sedimentation of the dynein complex to the lighter fractions (Fig. 1A) in sucrose gradient density is only in the presence of the Loa mutant dynein. We have also observed this in newborn mice from the Loa colony and have examined and confirmed this difference in detail. A manuscript describing these data and the role of the Loa mutation in the assembly of the dynein complex is in preparation.
There is mounting evidence that mutant SOD1 protein associates with mitochondria, leading to mitochondrial dysfunction in familial ALS and its rodent transgenic models of familial ALS (Ref. 38; for review, see Manfredi and co-workers (52)). Moreover, it has been shown that mitochondrial respiration is generally impaired in SOD1G93A mice (53, 54), and increased levels of mutant SOD1 in mitochondria of transgenic mice appear to adversely affect disease progression (55). In addition, Mattiazzi et al. (56) found deficits in complex I, II, III, and IV activities in spinal cord mitochondria from symptomatic SOD1G93A mice, and Kirkinezos et al. (57) observed losses in complex IV activity within the brain and spinal cord of paralyzed SOD1G93A mice. However, the mechanism by which mitochondrial dysfunction plays a role in the death of motor neurons in SOD1 transgenic mice is not yet fully understood.
It is, therefore, intriguing that, as shown here, decreased levels of mutant SOD1 in the mitochondrial matrix and membranes (Fig. 3) plus a reduction in the association of total mutant SOD1 protein with SOD1G93A;Dync1h1Loa/+ mitochondria, compared with those of SOD1G93A mice (Fig. 1, B and C), correlates with amelioration of the defects in mitochondrial respiration and membrane potential in these mice. Therefore, the amelioration of disease in SOD1G93A mice by the Loa mutation could be due to some as yet unknown gain-of-function by mutant cytoplasmic dynein, which is manifested by better equipped mitochondria and possibly ER to resist the toxic effects of mutant SOD1 protein in motor neurons.
Mitochondria are one of many transported cargos whose distribution in the cell body and axons is regulated by retrograde and anterograde motors. Recently, it has been reported that the anterograde mitochondrial transport is defective in motor neurons of SOD1G93A mice (20). This defect has been shown to elicit an imbalance in the anterograde and retrograde transport of the mitochondria along motor axons, giving rise to more sparse mitochondrial populations in the axons. Thus, an explanation for the rescue of the SOD1G93A phenotype by Loa could be the partial restoration of the balance in retrograde and anterograde axonal transport by slowing the retrograde transport of the mitochondria.
The data in the present study together with those from other groups show that mutant SOD1 not only accumulates within the mitochondrial matrix and the intermembrane space but also associates with mitochondrial outer membrane (38). As a result, mutant SOD1 could change mitochondrial structure and membrane composition and render the interaction of the molecular motors with their docking site on the mitochondrial membrane less efficient. By reducing the amount of mutant SOD1 in the mitochondria, as we have shown here, the Loa mutation could improve the docking of the molecular motors to the mitochondria, and this could subsequently restore their trafficking.
However, in our earlier studies, using a fluorescently labeled fragment of the tetanus toxin, we have shown that in cultured embryonic motor neurons retrograde transport is also impaired in the SOD1G93A motor neurons (17). But in the presence of heterozygous Loa mutation the speed of retrograde axonal transport in SOD1G93A motor neurons improved dramatically (17). Analyses of the mitochondrial transport in motor neurons of SOD1G93A;Dync1h1Loa/+ and SOD1G93A are, therefore, required to shed further light on the possible restoration of the balance between the anterograde and retrograde transports in these mice.
Varadi et al. (60) show that the disruption of dynein function results in the dissociation of dynamin-related protein (Drp1) from the mitochondrial outer membrane, causing significant morphological changes to the mitochondria and the excessive accumulation of the mitochondria in the perinuclear region of the cells. Drp1 is involved in mitochondrial fission, and it has been shown to interact with dynein-dynactin, suggesting that the delivery of Drp1 to the mitochondrial membrane could be mediated by dynein (58, 59, 60). Our data and the evidence from the Varadi et al. study (60) could, therefore, provide a plausible explanation for the improvement in disease phenotype induced in SOD1G93A mice by the Loa mutation.
The data presented in this paper clearly indicate that, contrary to the conclusion of Zhang et al. (22), the interaction between mutant SOD1G93A and dynein is not direct or at least is not detectable by immunoprecipitation.
According to Zhang et al. (22), there are direct interactions between cytoplasmic dynein and mutant SOD1, but not wild-type SOD1, and that these interactions increase with disease progression. However, analyzing brain and spinal cord tissue homogenates isolated from animals at different stages of the disease and using a range of IP antibodies, buffers, and protocols, including those described by Zhang et al. (22), we did not detect any evidence that could conclusively indicate direct interactions between SOD1G93A protein and cytoplasmic dynein in SOD1G93A or SOD1G93A;Dync1h1Loa/+ mice. This is despite ensuring that the dynein complex is intact in our experiments, as tested in native gel and sucrose density gradient sedimentation assays. In fact when we used the cross-linking method used by Zhang et al. (22) to show the interaction between dynein and SOD1A4V, the cross-linking buffer dissociated dynein heavy chain from the intermediate chain, and more importantly, the cross-linking buffer in the absence of dynein or SOD1 antibody mediated nonspecific pulldown of SOD1G93A protein.
In conclusion, our data suggest that mutant dynein in SOD1G93A;Dync1h1Loa/+ mice protects motor neurons and delays disease onset at least in part by conferring subtle morphological and structural modifications to the mitochondria, rendering them less prone to association with toxic SOD1G93A and consequently resulting in a significant improvement in mitochondrial respiration and the overall function of this vital organelle. Future studies aimed at understanding the underlying molecular mechanisms of the mutant-dynein-mediated protection of the mitochondria from toxic SOD1G93A protein and restoration of mitochondrial function in SOD1G93A;Dync1h1Loa/+ will be invaluable in elucidating the molecular pathology of ALS, which in turn may highlight novel therapeutic pathways.
Supplementary Material
Acknowledgments
We thank Dr. Kevin Pfister (Dept. of Cell Biology, School of Medicine, University of Virginia) for providing the anti-DIC antibody and the detailed protocol for immunoprecipitating the dynein complex. We also thank Dr. Haining Zhu (Dept. of Molecular and Cellular Biochemistry, University of Kentucky) for providing the detailed protocol for co-immunoprecipitating dynein-SOD1.
This work was supported by the Medical Research Council (to M. H., A. M. E.-K., and E. S.), the Biotechnology and Biological Sciences Research Council (to M. H., W. D., and A. L. M.), the Amyotrophic Lateral Sclerosis Association and Robert Packard Center for ALS Research at Johns Hopkins (to M. H., L. G., E. M. C. F., and V. B.-F.), the Brain Research Trust (to L. G.), The Wellcome Trust (to G. B. and E. M. C. F.), and the University of Sussex (to A. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–10.
V. Bros-Facer, M. Golding, D. Boërio-Guéguen, N. Nirmalananthan, R. Chia, A. Philpott, A. M. Flenniken, I. Vukobrodovic, L. R. Osborne, S. L. Adamson, J. Rossant, E. Stoddart, E. M. C. Fisher, H. Bostock, G. Schiavo, L. Greensmith, and M. Hafezparast, manuscript in preparation.
- ALS
- amyotrophic lateral sclerosis
- SOD1
- Cu,Zn superoxide dismutase 1
- Dync1h1 and DHC
- cytoplasmic dynein heavy chain 1
- DIC
- dynein intermediate chain
- p150
- dynactin p150Glued
- Loa
- Legs at odd angles
- TGN38
- trans-Golgi network 38
- HB
- homogenization buffer
- IP
- immunoprecipitation
- BDG
- buoyant density gradient
- PK
- proteinase K
- IMS
- intermembrane space
- FCCP
- cyanide-p-trifluoromethoxyphenylhydrazone
- TMRM
- tetramethylrhodamine methyl ester perchlorate
- GST
- glutathione S-transferase
- ER
- endoplasmic reticulum
- TWB
- tissue wash buffer
- RIPA buffer
- radioimmune precipitation assay buffer
- MOPS
- 4-morpholinepropanesulfonic acid
- PBS
- phosphate-buffered saline.
REFERENCES
- 1.Rowland L. P., Shneider N. A. (2001) N. Engl. J. Med. 344, 1688–1700 [DOI] [PubMed] [Google Scholar]
- 2.Brown R. H., Jr. (1995) Curr. Opin. Neurobiol. 5, 841–846 [DOI] [PubMed] [Google Scholar]
- 3.Bruijn L. I., Miller T. M., Cleveland D. W. (2004) Annu. Rev. Neurosci. 27, 723–749 [DOI] [PubMed] [Google Scholar]
- 4.Cleveland D. W., Rothstein J. D. (2001) Nat. Rev. Neurosci. 2, 806–819 [DOI] [PubMed] [Google Scholar]
- 5.Shaw P. J. (1999) BMJ 318, 1118–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deng H. X., Hentati A., Tainer J. A., Iqbal Z., Cayabyab A., Hung W. Y., Getzoff E. D., Hu P., Herzfeldt B., Roos R. P. (1993) Science 261, 1047–1051 [DOI] [PubMed] [Google Scholar]
- 7.Rosen D. R., Siddique T., Patterson D., Figlewicz D. A., Sapp P., Hentati A., Donaldson D., Goto J., O'Regan J. P., Deng H. X. (1993) Nature 362, 59–62 [DOI] [PubMed] [Google Scholar]
- 8.Pasinelli P., Brown R. H. (2006) Nat. Rev. Neurosci. 7, 710–723 [DOI] [PubMed] [Google Scholar]
- 9.Shibata N. (2001) Neuropathology 21, 82–92 [DOI] [PubMed] [Google Scholar]
- 10.Turner B. J., Talbot K. (2008) Prog. Neurobiol. 85, 94–134 [DOI] [PubMed] [Google Scholar]
- 11.Shaw B. F., Valentine J. S. (2007) Trends Biochem. Sci. 32, 78–85 [DOI] [PubMed] [Google Scholar]
- 12.Banks G. T., Kuta A., Isaacs A. M., Fisher E. M. (2008) Mamm. Genome 19, 299–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J., Caruano-Yzermans A., Rodriguez A., Scheurmann J. P., Slunt H. H., Cao X., Gitlin J., Hart P. J., Borchelt D. R. (2007) J. Biol. Chem. 282, 345–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manfredi G., Xu Z. (2005) Mitochondrion 5, 77–87 [DOI] [PubMed] [Google Scholar]
- 15.Breuer A. C., Lynn M. P., Atkinson M. B., Chou S. M., Wilbourn A. J., Marks K. E., Culver J. E., Fleegler E. J. (1987) Neurology 37, 738–748 [DOI] [PubMed] [Google Scholar]
- 16.Jablonka S., Wiese S., Sendtner M. (2004) J. Neurobiol. 58, 272–286 [DOI] [PubMed] [Google Scholar]
- 17.Kieran D., Hafezparast M., Bohnert S., Dick J. R., Martin J., Schiavo G., Fisher E. M., Greensmith L. (2005) J. Cell Biol. 169, 561–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rao M. V., Nixon R. A. (2003) Neurochem. Res. 28, 1041–1047 [DOI] [PubMed] [Google Scholar]
- 19.Williamson T. L., Cleveland D. W. (1999) Nat. Neurosci. 2, 50–56 [DOI] [PubMed] [Google Scholar]
- 20.De Vos K. J., Chapman A. L., Tennant M. E., Manser C., Tudor E. L., Lau K. F., Brownlees J., Ackerley S., Shaw P. J., McLoughlin D. M., Shaw C. E., Leigh P. N., Miller C. C., Grierson A. J. (2007) Hum. Mol. Genet. 16, 2720–2728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ligon L. A., LaMonte B. H., Wallace K. E., Weber N., Kalb R. G., Holzbaur E. L. (2005) Neuroreport 16, 533–536 [DOI] [PubMed] [Google Scholar]
- 22.Zhang F., Ström A. L., Fukada K., Lee S., Hayward L. J., Zhu H. (2007) J. Biol. Chem. 282, 16691–16699 [DOI] [PubMed] [Google Scholar]
- 23.Ström A. L., Shi P., Zhang F., Gal J., Kilty R., Hayward L. J., Zhu H. (2008) J. Biol. Chem. 283, 22795–22805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.King S. M. (2000) Biochim. Biophys. Acta 1496, 60–75 [DOI] [PubMed] [Google Scholar]
- 25.Pfister K. K., Shah P. R., Hummerich H., Russ A., Cotton J., Annuar A. A., King S. M., Fisher E. M. (2006) PLoS Genet 2, e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vale R. D. (2003) Cell 112, 467–480 [DOI] [PubMed] [Google Scholar]
- 27.Vallee R. B., Williams J. C., Varma D., Barnhart L. E. (2004) J. Neurobiol. 58, 189–200 [DOI] [PubMed] [Google Scholar]
- 28.Yano H., Lee F. S., Kong H., Chuang J., Arevalo J., Perez P., Sung C., Chao M. V. (2001) J. Neurosci. 21, RC125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pfister K. K., Fisher E. M., Gibbons I. R., Hays T. S., Holzbaur E. L., McIntosh J. R., Porter M. E., Schroer T. A., Vaughan K. T., Witman G. B., King S. M., Vallee R. B. (2005) J. Cell Biol. 171, 411–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schroer T. A. (2004) Annu. Rev. Cell Dev. Biol. 20, 759–779 [DOI] [PubMed] [Google Scholar]
- 31.Hafezparast M., Klocke R., Ruhrberg C., Marquardt A., Ahmad-Annuar A., Bowen S., Lalli G., Witherden A. S., Hummerich H., Nicholson S., Morgan P. J., Oozageer R., Priestley J. V., Averill S., King V. R., Ball S., Peters J., Toda T., Yamamoto A., Hiraoka Y., Augustin M., Korthaus D., Wattler S., Wabnitz P., Dickneite C., Lampel S., Boehme F., Peraus G., Popp A., Rudelius M., Schlegel J., Fuchs H., Hrabe de Angelis M., Schiavo G., Shima D. T., Russ A. P., Stumm G., Martin J. E., Fisher E. M. (2003) Science 300, 808–812 [DOI] [PubMed] [Google Scholar]
- 32.Hafezparast M., Ahmad-Annuar A., Wood N. W., Tabrizi S. J., Fisher E. M. (2002) Lancet Neurol. 1, 215–224 [DOI] [PubMed] [Google Scholar]
- 33.Rogers D. C., Peters J., Martin J. E., Ball S., Nicholson S. J., Witherden A. S., Hafezparast M., Latcham J., Robinson T. L., Quilter C. A., Fisher E. M. (2001) Neurosci. Lett. 306, 89–92 [DOI] [PubMed] [Google Scholar]
- 34.Chen X. J., Levedakou E. N., Millen K. J., Wollmann R. L., Soliven B., Popko B. (2007) J. Neurosci. 27, 14515–14524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ilieva H. S., Yamanaka K., Malkmus S., Kakinohana O., Yaksh T., Marsala M., Cleveland D. W. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 12599–12604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Teuchert M., Fischer D., Schwalenstoecker B., Habisch H. J., Böckers T. M., Ludolph A. C. (2006) Exp. Neurol. 198, 271–274 [DOI] [PubMed] [Google Scholar]
- 37.Gurney M. E., Pu H., Chiu A. Y., Dal Canto M. C., Polchow C. Y., Alexander D. D., Caliendo J., Hentati A., Kwon Y. W., Deng H. X. (1994) Science 264, 1772–1775 [DOI] [PubMed] [Google Scholar]
- 38.Vande Velde C., Miller T. M., Cashman N. R., Cleveland D. W. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 4022–4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pallotti F., Lenaz G. (2007) Methods Cell Biol. 80, 3–44 [DOI] [PubMed] [Google Scholar]
- 40.Arce V., Garces A., de Bovis B., Filippi P., Henderson C., Pettmann B., deLapeyrière O. (1999) J. Neurosci. Res. 55, 119–126 [DOI] [PubMed] [Google Scholar]
- 41.Bilsland L. G., Nirmalananthan N., Yip J., Greensmith L., Duchen M. R. (2008) J. Neurochem. 107, 1271–1283 [DOI] [PubMed] [Google Scholar]
- 42.Stevens J. C., Chia R., Hendriks W. T., Bros-Facer V., van Minnen J., Martin J. E., Jackson G. S., Greensmith L., Schiavo G., Fisher E. M. C. (2010) PLoS ONE 5, e9541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guan K. L., Dixon J. E. (1991) Anal. Biochem. 192, 262–267 [DOI] [PubMed] [Google Scholar]
- 44.King S. M., Barbarese E., Dillman J. F., 3rd, Patel-King R. S., Carson J. H., Pfister K. K. (1996) J. Biol. Chem. 271, 19358–19366 [DOI] [PubMed] [Google Scholar]
- 45.Brill L. B., 2nd, Pfister K. K. (2000) Methods 22, 307–316 [DOI] [PubMed] [Google Scholar]
- 46.Pfister K. K., Benashski S. E., Dillman J. F., 3rd, Patel-King R. S., King S. M. (1998) Cell Motil. Cytoskeleton 41, 154–167 [DOI] [PubMed] [Google Scholar]
- 47.Boillée S., Yamanaka K., Lobsiger C. S., Copeland N. G., Jenkins N. A., Kassiotis G., Kollias G., Cleveland D. W. (2006) Science 312, 1389–1392 [DOI] [PubMed] [Google Scholar]
- 48.Clement A. M., Nguyen M. D., Roberts E. A., Garcia M. L., Boillée S., Rule M., McMahon A. P., Doucette W., Siwek D., Ferrante R. J., Brown R. H., Jr., Julien J. P., Goldstein L. S., Cleveland D. W. (2003) Science 302, 113–117 [DOI] [PubMed] [Google Scholar]
- 49.Yamanaka K., Boillee S., Roberts E. A., Garcia M. L., McAlonis-Downes M., Mikse O. R., Cleveland D. W., Goldstein L. S. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 7594–7599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamanaka K., Chun S. J., Boillee S., Fujimori-Tonou N., Yamashita H., Gutmann D. H., Takahashi R., Misawa H., Cleveland D. W. (2008) Nat. Neurosci. 11, 251–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Perlson E., Jeong G. B., Ross J. L., Dixit R., Wallace K. E., Kalb R. G., Holzbaur E. L. (2009) J. Neurosci. 29, 9903–9917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hervias I., Beal M. F., Manfredi G. (2006) Muscle Nerve 33, 598–608 [DOI] [PubMed] [Google Scholar]
- 53.Browne S. E., Bowling A. C., Baik M. J., Gurney M., Brown R. H., Jr., Beal M. F. (1998) J. Neurochem. 71, 281–287 [DOI] [PubMed] [Google Scholar]
- 54.Martin L. J., Liu Z., Chen K., Price A. C., Pan Y., Swaby J. A., Golden W. C. (2007) J. Comp. Neurol. 500, 20–46 [DOI] [PubMed] [Google Scholar]
- 55.Deng H. X., Shi Y., Furukawa Y., Zhai H., Fu R., Liu E., Gorrie G. H., Khan M. S., Hung W. Y., Bigio E. H., Lukas T., Dal Canto M. C., O'Halloran T. V., Siddique T. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 7142–7147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mattiazzi M., D'Aurelio M., Gajewski C. D., Martushova K., Kiaei M., Beal M. F., Manfredi G. (2002) J. Biol. Chem. 277, 29626–29633 [DOI] [PubMed] [Google Scholar]
- 57.Kirkinezos I. G., Bacman S. R., Hernandez D., Oca-Cossio J., Arias L. J., Perez-Pinzon M. A., Bradley W. G., Moraes C. T. (2005) J. Neurosci. 25, 164–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Karbowski M., Youle R. J. (2003) Cell Death Differ. 10, 870–880 [DOI] [PubMed] [Google Scholar]
- 59.Smirnova E., Griparic L., Shurland D. L., van der Bliek A. M. (2001) Mol. Biol. Cell 12, 2245–2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Varadi A., Johnson-Cadwell L. I., Cirulli V., Yoon Y., Allan V. J., Rutter G. A. (2004) J. Cell Sci. 117, 4389–4400 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.