Abstract
Essential in mitosis, the human Kinesin-5 protein is a target for >80 classes of allosteric compounds that bind to a surface-exposed site formed by the L5 loop. Not established is why there are differing efficacies in drug inhibition. Here we compare the ligand-bound states of two L5-directed inhibitors against 15 Kinesin-5 mutants by ATPase assays and IR spectroscopy. Biochemical kinetics uncovers functional differences between individual residues at the N or C termini of the L5 loop. Infrared evaluation of solution structures and multivariate analysis of the vibrational spectra reveal that mutation and/or ligand binding not only can remodel the allosteric binding surface but also can transmit long range effects. Changes in L5-localized 310 helix and disordered content, regardless of substitution or drug potency, are experimentally detected. Principal component analysis couples these local structural events to two types of rearrangements in β-sheet hydrogen bonding. These transformations in β-sheet contacts are correlated with inhibitory drug response and are corroborated by wild type Kinesin-5 crystal structures. Despite considerable evolutionary divergence, our data directly support a theorized conserved element for long distance mechanochemical coupling in kinesin, myosin, and F1-ATPase. These findings also suggest that these relatively rapid IR approaches can provide structural biomarkers for clinical determination of drug sensitivity and drug efficacy in nucleotide triphosphatases.
Keywords: Allosteric Regulation, ATPases, Drug Action, Drug Resistance, Fourier Transform IR (FTIR), Kinesin, Molecular Motors, Protein Structure, Multivariate Data Analysis
Introduction
Allostery is important in controlled catalysis, signal transduction, and apoptosis (1). The classic view of proteins demonstrating this property (2) asserts that binding of a ligand at one site provokes conformational changes at a remote, second site. Recent studies (3) evaluating underlying mechanisms of allostery alternatively suggest that ligand binding results in selection of preexisting conformational substates. Implicit in the latter model is the principle that interactions between the orthosteric and allosteric sites are tightly linked through structure and thermodynamics (4). Active challenges in structural biology, which are central to this work, are deciphering the chemical nature of the ligand-protein interactions as well as how energy is transduced through protein structures to transmit allosteric events.
Our experimental model, the human Kinesin-5 motor protein (Eg5 or KSP), plays key roles in bipolar mitotic spindle formation and is a protein target for allosteric compounds (5–7) that alter catalytic ATPase activity of the protein (8, 9). Biochemical studies demonstrate a wide concentration range of inhibition by these compounds (10–12); there may be differences in the kinetic mechanism of allostery (13–15), and even allosteric activation (16) is possible. The best characterized inhibitors, monastrol (10) and S-trityl-l-cysteine (STC)2 (11), were uncovered from independent chemical screens.
Interest in these allosteric compounds has been acute because they are potential anticancer agents. Additionally, these compounds serve as research tools to probe the fundamental mechanism by which Eg5 and perhaps all motor proteins convert and transduce energy to conformational changes in distal regions of the protein. Insights concerning allosteric conformational states of Eg5 result principally from diffraction- and microscopy-based techniques. The Eg5 motor domain is an arrowhead-shaped structure with a central β-sheet flanked by three helices on each side. In crystallographic studies of Eg5·ADP complexed with monastrol (17, 18) and other allosteric inhibitors (12, 19–23), the wild type Kinesin-5 motor domain adopts a similar conformer, irrespective of the chemical nature of the allosteric drug. This suggests a single mode of allosteric communication, a conclusion that conflicts with the body of kinetic data.
The most notable conformational change observed is the adoption of a “closed” conformation by the insertion loop (L5) within the α2 helix that cradles the allosteric compound. This is in contrast with the “open” conformation observed in the absence (24) of an allosteric ligand. However, conformational transitions of the L5 loop are found not only in response to drug binding but also in normal motor function. The Eg5 conformer trapped by allosteric agents is proposed to be an intermediate state of its normal ATP hydrolysis cycle (25). The L5 loop is observed in the closed conformation in cryo-electron microscopy experiments with a Drosophila melanogaster homologue of Eg5 bound to microtubules using non-hydrolyzable substrate analogues (26) and in Eg5·AMPPNP crystals in the absence of allosteric agents (27). These data suggest that surface loops in the motor domain are indispensable for controlling catalysis in native kinesins.
No consensus of L5 residues key for transmitting allosteric information has been established, primarily due to interdependence of the allosteric and active-site interactions and the lack of rapid methods to observe structures of mutant motor proteins. Simple sequence conservation analysis (Fig. 1A) does not reveal how the L5 loop specifically recognizes and binds an allosteric molecule. Studies (15, 17, 28, 29) using targeted mutagenesis to measure the contribution of specific contacts in Eg5 allostery and chemical-kinetic measurements of mutant motor ensembles in solution on the whole concluded that their data were consistent with the crystallographic observation of the wild type Eg5·inhibitor complexes. However, neither these reports nor x-ray structural analyses of allosterically inhibited Eg5 proteins arrive at a common set of contact residues required for the inhibition by different chemical partners.
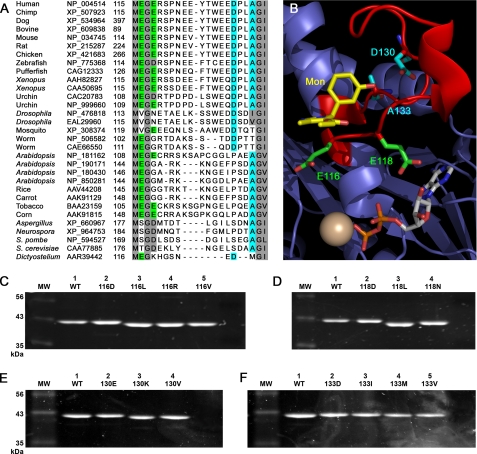
FIGURE 1.
Substitution of L5 loop residues in Eg5 kinesin, with known sequence and spatial organization, can result in gel shifting in denaturing SDS-PAGE analysis. A, sequence alignment of Eg5 and other members of the Kinesin-5 family was generated using the ClustalW method in MegAlign 6.1.2 (DNASTAR). Conserved residues of interest are colored. Scored in green and aqua are Glu-116/Glu-118 and Asp-130/Ala-133 residues in the insertion loop, respectively. B, PyMOL representation of the crystallographic data from Ref. 18. Highlighted are the N-terminal Glu-116 and Glu-118 (green) and C-terminal Asp-130 and Ala-133 (aqua) residues in the L5 loop (red) of Eg5 kinesin. Also shown are monastrol (yellow), Mg2+ (gold sphere), and ADP. Substitutions of Glu-116 (C), Glu-118 (D), Asp-130 (E), and Ala-133 (F) were SDS-PAGE-analyzed with a 10% acrylamide gel. The gel migration of the double mutant, E116D/E118D (DD), matched wild type (WT) Eg5 samples (data not shown). Protein samples were expressed, purified, and used in two different laboratories; equivalent results were observed in both settings.
This work focuses on the unaddressed question of why there are differing efficacies in drug inhibition of Eg5. Our hypothesis is that there is more than one pathway for allosteric communication that initiates from the L5 loop of Eg5; differences in the inhibitory power of monastrol and STC among the Kinesin-5 orthologues result from these deviations in allosteric communication. We demonstrate an integrated approach to build, test, and refine a model of how allostery is propagated from the surface L5 loop through the Eg5 kinesin motor domain in solution. We do so by measuring not only biochemical behavior of mutant proteins but also protein secondary structure in solution. Unique to this work is a systems approach to examine multivariate data representing protein structure in bulk solution; such discovery methods can organize and uncover patterns at the atomic level in the spectroscopic data. These global and integrated analyses can provide new hypotheses; the resulting model reveals that allosteric conformational changes at the protein surface can promote two different types of intraprotein contacts at the central core of the kinesin motor domain. These changes discriminate between the inhibitory power of allosteric drugs.
EXPERIMENTAL PROCEDURES
Generation of Single and Double Mutants of Eg5
Starting cDNA was the truncated form of wild type Eg5 (residues 1–370) (30). Appropriate primers were synthesized and used for site-directed mutagenesis with the QuikChange II XL kit (Stratagene, La Jolla, CA). Construction of the Eg5 motor domain with substitutions of Asp for Glu-116 and Glu-118 was performed using two rounds of mutagenesis. The expected mutations were confirmed by a single-read sequencing reaction. A subset of five mutations was completely sequenced on both strands to ensure that inadvertent coding errors were not introduced.
Motor Protein Expression and Purification
The wild type Eg5 kinesin motor domain as well as single and double site substitutions, were expressed in BL21-Codon Plus (DE3)-RIL cell lines (Stratagene) and purified by cation exchange chromatography as described (30). The purity of wild type and all Eg5 substitutions was determined by SDS-PAGE analysis using a 10% acrylamide, 0.27% bisacrylamide gel with a 0.38 m Tris-HCl, pH 8.8, resolving gel and a 0.125 m Tris-HCl, pH 6.8, stacking gel component. SDS sample buffer was added, adjusting the sample to a final concentration of 2% SDS, 0.29 m 2-mercaptoethanol, 0.05 m Tris-HCl, pH 6.8, 10% glycerol, 0.0013% (w/v) bromphenol blue. SDS-PAGE samples were then boiled at 100 °C to ensure denaturing conditions. Following boiling, samples were immediately run along an unstained molecular weight ladder (New England Biolabs) and were visualized using Sypro Tangerine (Molecular Probes, Inc., Eugene, OR). Protein purity is estimated to be >90%. All protein concentrations were determined by Bradford assay (Coomassie Plus Protein Assay Reagent, ThermoScientific), using bovine serum albumin as the standard; concentrations ranged from 0.5 to 6.0 mg/ml. Final storage buffer for the purified Eg5 proteins was 125 mm NaCl, 50 mm HEPES, pH 7.4, 0.1 mm MgATP, 1 mm dithiothreitol, and 10% glycerol. Purified motor protein was flash-frozen in liquid nitrogen and stored at −80 °C until use.
In Vitro ATPase Assays
Basal ATPase activities were measured with a coupled enzymatic assay (31) with 5 μm motor protein and 1 mm MgATP in a SpectraMax 2E spectrophotometer for high throughput kinetic measurements (32). Reactions were supplemented with monastrol (0–600 μm; Sigma) or STC (0–200 μm; Sigma) as appropriate, and control reactions contained equivalent DMSO. Reactions used for calculation of the IC50 values contained between 3 and 22.5 mm NaCl. Data were reproducible with protein samples purified from differing batches of cultured cells from two different laboratories.
Infrared Spectroscopy
All spectroscopic data were acquired at room temperature (30) on a Vertex 80v (Bruker Optics, Billerica, MA) with the use of a BioATR (Bruker Optics). The spectrometer was equipped with a liquid nitrogen-cooled MCT or photovoltaic detector and a KBr beam splitter. The mirror velocity was 20 kHz, the spectral resolution was 2 cm−1, and 1000 mirror scans were acquired for each double-sided interferogram. A 6-mm aperture, gain of 1 on the preamp A setting, Blackman-Harris 3-Term apodization function, and two levels of zero filling were used. A minimum of three spectra and two independent protein purifications per Eg5 sample were used to generate the averaged data.
Protein samples were 2H2O-exchanged using Zeba desalting spin columns (Pierce) or SpectraPor dialysis membranes (Spectrum Laboratories) and equilibrated with 2H2O buffer (10 mm HEPES, p2H 7.4, 0.5 mm MgCl2, and 0.1 mm MgATP). Protein samples utilized in IR experiments had final concentrations of 0.9–6 mg/ml, as determined by Bradford assays. Stock concentrations of monastrol and STC were 100 mm in straight DMSO (99.9% American Chemical Society spectrophotometric grade; Sigma). Positive displacement pipettes were used for all DMSO aliquotting to ensure accurate volumetric measurement of the viscous solvent. For Eg5·inhibitor samples, a 20-μl sample was achieved by mixture of 19 μl of Eg5 protein with 1 μl of inhibitor in equimolar concentrations, resulting in a final DMSO concentration of 5%. Inhibitor stocks were serially diluted in DMSO, using a maximum 1:5 ratio, until the desired concentration was achieved. Of this 20-μl mixture, a sample volume of 12–15 μl was dispensed onto the ATR crystal and allowed to equilibrate to room temperature for 10–15 min before an interferogram was collected.
Spectral Analyses
Each sample spectrum was subtracted from the spectrum of 2H2O buffer or 2H2O buffer plus DMSO when samples contained drug. After base-line correction, spectra were area-normalized. Savitsky-Golay second-derivative, Fourier self-deconvolution, and spectral fitting analyses were performed with Grams/AI (ThermoFisher Scientific). Self-deconvolution was performed with a Bessel apodization function, and peak fitting utilized the Levenberg-Marquardt method with Lorentzian components. Identical parameters were used for all spectra with an amide I′ fitting range of 1710 to 1594 cm−1. Bandwidth, frequency, and amplitude of each spectral component were iterated until a minimum in the fit to the experimental data was obtained. The frequencies of the resulting spectral components and their amplitudes generated good fits, as judged by residuals and reduced χ2 parameters.
The frequencies, obtained from the second derivative and Fourier self-deconvolution analyses, were used as the first set of inputs for the multivariate data analyses. Area-normalized data sets of infrared spectra, in comparison with wild type Eg5 samples, also were used as the second set of inputs for the multivariate data analyses. Principal component analysis (PCA) was performed with Matlab software (Mathworks). PCA plots presented were scaled on the x and y axes (−1,1). The point spread on the PCA indicates that our infrared measurements sample excellent variances in vector space, and there is high signal-to-noise in the infrared spectra and calculations. All figures were generated using IGOR Pro software (Wavefunction).
Crystallization and X-ray Structure Solution
The Eg5 motor domain (residues 1–369) was expressed in bacteria and purified as described previously (27). Prior to crystallization experiments, purified Eg5 (∼250 μm) was mixed with 1 mm STC (Sigma). Original crystals obtained in 250 mm NH4SO4, 25% polyethylene glycol 3350, 100 mm MES, pH 6.0, and 10 mm trimethylamine hydrochloride were used to streak seed crystals grown in 250 mm NH4SO4, 25% polyethylene glycol 3350, 100 mm MES, pH 6.0, 10 mm trimethylamine hydrochloride, and 10–15% glycerol. Crystals were grown at 4 °C in sitting drops consisting of 2 μl of protein/drug mixture and 1 μl of well solution. Cubic crystals appeared several days after streak seeding. The crystals were allowed to grow for several weeks before being flash-frozen for data collection. Diffraction data were collected at 100 K using a Bruker Microstar x-ray generator equipped with Helios optics and a Proteum 4K Platinum 135 CCD camera and were integrated and scaled with PROTEUM2 software (Bruker AXS). Crystals belonged to space group I2(1)3 with cell parameters of a = b = c = 157.93 Å. The Eg5·STC complex structure was determined via molecular replacement methods (AMoRe) using a search model based on molecule A of the Eg5·ADP·monastrol complex (Protein Data Bank (PDB) ID 1X88). Successive rounds of manual model building (33) and automated refinement were performed using O and CNS. The final model is refined to R and Rfree values of 24.07 and 27.46, respectively (all |F| > 0; 25.0–2.5 Å), and contains residues 18–271 and 288–366, along with the Mg-ADP complex, 74 waters, one STC molecule, and one trimethylamine molecule.
RESULTS
Substitutions of L5 Residues Result in Altered SDS-PAGE Mobility
The initial step of this study was to assess how perturbations of the L5 loop affect Eg5 steady-state kinetics and solution structure. The first type of perturbation involves alteration of side chain chemistry. We sampled 15 mutations at four sites in the L5 loop: two at the N terminus and two at the C terminus of the insertion loop. Prior studies scrutinizing the allosteric pocket typically have assumed that point mutations have little effect beyond the localized change of a particular functional group. However, this stance can underestimate the ability of the point mutation to remodel the binding surface or transmit long range effects that are dependent on the nature of the substitution. Therefore, our mutations ranged from conservative to non-conservative substitutions, allowing us to see a wide range of chemical and structural outcomes propagating from specific positions.
The N-terminal L5 residues chosen were Glu-116 and Glu-118. In the Eg5·monastrol structure (18), the Glu-116 side chain is nestled at the edge of the allosteric pocket (Fig. 1B), in contrast to the Glu-118 side chain orientation toward the orthosteric site. C-terminal L5 residues studied were Asp-130 and Ala-133, embedded within short stretches of negatively charged and hydrophobic side chains and interrupted by a proline (Fig. 1A). There was no apparent interaction between the side chains of any of these four residues and the allosteric inhibitors in the co-crystal structure of Eg5·ADP and monastrol (Fig. 1B). However, we note that the E116V and E118N substitutions are present in the Drosophila Kinesin-5 protein that is insensitive to allosteric inhibitors and that the D130V and A133D mutations were found in human cell lines resistant to Eg5 allosteric inhibitors (16).
Expression plasmids with these and other substitutions in Eg5 cDNA were successfully constructed, and the mutant motor proteins were readily expressed and purified, as evident from SDS-PAGE (Fig. 1, C–F). Wild type Eg5 has an apparent and expected molecular mass of 42 kDa. Migration of the mutant Eg5 proteins in SDS-PAGE, however, did not correlate with the calculated molecular weights. Both forms of anomalous gel mobility were observed for mutant proteins. Faster migration of purified proteins was detected for non-conservative substitutions of the L5 carboxylates (Fig. 1, C and D). C-terminal mutant proteins overall had slower gel mobility in SDS-PAGE (Fig. 1, E and F).
The observed gel shift behavior of the Eg5 mutant proteins most likely originated from altered detergent binding in the L5-modified proteins. A recent study (34) demonstrates that PAGE migration, SDS aggregation number, hydrodynamic radius, and protein conformation are directly linked. Observed variations in gel shifting in Fig. 1 suggest that more than one type of modification in intraprotein contacts can be found in these mutant Eg5 motor domains that affects the overall protein fold.
Preservation of N-terminal Local Structure and C-terminal Chemistry in the L5 Loop Is Required for Allostery and Catalysis
There are marked kinetic differences between the N-terminal and C-terminal substitutions of L5 loop residues. Compared with wild type samples, single site substitutions of Glu-116 decreased the steady-state, basal ATP hydrolysis rate of Eg5 kinesin monomers (Fig. 2A, open boxes). However, mutants of Glu-118 displayed increased activity (Fig. 2A, filled boxes). These results were consistent, whether or not the amino acid substitutions were synonymous with carboxylates. These data demonstrate that the native interactions of Glu-116 and/or Glu-118 in the motor domain influence ATP hydrolysis rates achievable by Eg5. Moreover, the faithful difference in mutant kinetic behavior suggests that the native side chains play a predominantly structural role, propagating effects to the nucleotide site rather than electrostatic interactions.
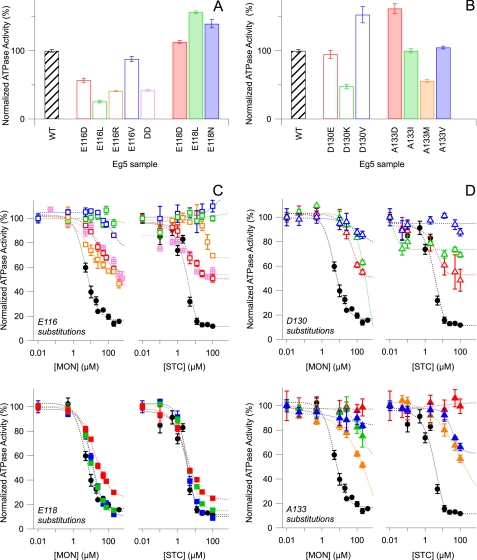
FIGURE 2.
Kinetic effects of N-terminal L5 residue substitutions are residue-dependent, but C-terminal L5 residue substitutions have no clear kinetic correlation. A and B, steady-state, basal ATP hydrolysis rates, for which mean Vmax for wild type (WT) (black) is 0.144 ± 0.003 s−1. For wild type, n = 101, whereas n for all substitutions ranges from 20 to 51. C and D, normalized Eg5 ATP hydrolysis rates were plotted against increasing concentrations of monastrol (left) or STC (right). WT kinetic data (black) are superimposed on Glu-116 (C, top), Glu-118 (C, bottom), Asp-130 (D, top), and Ala-133 mutants (D, bottom). The double mutant E116D/E118D (DD) data are shown in pink hatched boxes (C, top). All reactions contain 3–22.5 mm NaCl. Colors correspond to protein samples in A and B, respectively. ATP hydrolysis rates for each substitution are normalized with respect to the 0 mm monastrol or 0 mm STC measurements of the parent substitution. All data points in the figure reflect the averaged normalized rate from 2–4 separate protein preparations and the S.E. (error bars).
In contrast, the single site C-terminal L5 substitutions did not have such uniform kinetic outcomes; rates of ATP hydrolysis for these residues were dependent on side chain chemistry (Fig. 2B). For both Asp-130 and Ala-133, substitutions resulted in either increased or decreased ATPase activity. D130E samples showed basal rates similar to those of wild type Eg5 protein. Substitution of a positively charged side chain in lieu of aspartate at residue 130 decreased Eg5 activity, whereas replacement with a hydrophobic side chain increased basal rates by ∼50%. A133I and A133V Eg5 motor domains had ATP hydrolysis rates comparable with those of wild type samples, but A133M was not as kinetically active. Only the substitution of a polar, charged side chain for Ala-133 produced an increase in enzymatic activity. We note that mutations known to confer ispinesib resistance (16) had opposing changes in chemical character and resulted in the greatest change in basal rates. Therefore, the chemical nature of these C-terminal L5 residues is key in their catalytic role.
The second type of perturbation to the L5 loop was caused by allosteric small-molecule inhibitors. Specifically, we examined the effects of two commonly used inhibitors of Eg5, monastrol and S-trityl-l-cysteine. Wild type Eg5 had an IC50 of 8 and 4 μm for monastrol and STC, respectively (Fig. 2C, black filled circles). Altering the chemistry of the Glu-116, Glu-118, Asp-130, and Ala-133, despite the paucity of direct contacts observed in crystal structures (Fig. 1B), changed the effect of small molecule ligands on Eg5, encompassing the kinetic range from neutral to inhibited. For Glu-118 substitutions (Fig. 2C), all variants of Eg5 were allosterically inhibited in equivalent measure to wild type protein, suggesting that this carboxylate side chain is not involved in allosteric inhibition.
In general, loss of basal rate was correlated with loss of drug sensitivity in N-terminal mutations. However, C-terminal mutations did not have a clear relationship between basal activity and drug sensitivity. All Glu-116 (Fig. 2C), Asp-130 (Fig. 2D), and Ala-133 (Fig. 2D) mutants showed reduced sensitivity to monastrol and STC but to varying degrees. Only the E116R and D130K motor proteins displayed differing levels of inhibition between the two compounds used, the former being more sensitive to monastrol and the latter to STC. Of the C-terminal L5 mutants, D130V (open blue triangles) and A133D (filled red triangles) were the most insensitive to either inhibitor, as anticipated from studies on ispinesib (16). For the N-terminal L5 substitutions, loss of allosteric compound sensitivity was more marked in samples with aliphatic residue substitutions at position 116, compared with Eg5 proteins with charged residues at these positions. Thus, the capacity for hydrogen bond formation and the side chain length of Glu-116 are singled out to be essential for basal catalytic activity and for effective allosteric inhibition of Eg5.
Secondary Structural Changes of L5-perturbed Kinesins in Solution Can Be Rapidly and Quantitatively Measured by Infrared Spectroscopy
To understand the role protein structure plays in allosteric interactions, conformational transitions throughout a globular protein need to be monitored in response to localized perturbations. There are no published structural descriptions of Eg5 mutants. Crystallographic reports of how allosteric small compounds perturb wild type Eg5·ADP complexes (12, 19–23) note conformational changes in the L5 loop but no pronounced changes in the nucleotide pocket or in other regions of the motor domain. Although x-ray diffraction of protein crystals provides detailed atomic information, it can only provide a static single frame, or “snapshot,” of Eg5 inhibition.
Methods for determining the protein structure in solution can provide an ensemble conformational picture of polypeptides in a nearly native environment. However, to date, there is no published method, rapid and quantitative, for screening conformational alterations (35) resulting from peptide sequence variations that parallels high throughput measurement of kinetic activity or genetic microarray profiling. Thus, our experimental hurdle is to develop high throughput methods toward analyzing protein structure in solution. Infrared spectroscopy is widely used as a technique for the analysis of protein secondary structure (36). The spectral region most commonly examined is the amide I′ band, centered at 1650 cm−1, which arises from the delocalized C=O vibrations of the peptide linkage. Changes in the amide I′ region can be directly correlated with variation in the percentage of protein occupied in α helix, β sheet, or other structural motifs. We have obtained infrared data on Eg5 proteins in solution using reflectance spectral acquisition methods: spectra are collected using only 12–15-μl volumes and 20 minutes of time.
Fourier transform IR (FTIR) spectra were acquired on wild type and all mutant Eg5 proteins alone and in the equimolar presence of monastrol or STC. Shown in Fig. 3A is a subset of the averaged spectral data on 2H2O-exchanged Eg5 proteins in the amide I′ region (1720–1600 cm−1). The normalized data share broadly similar line shapes, arguing for similar overall secondary structure composition. Secondary structure contributions can be accurately measured through band narrowing and regression analysis (37, 38) on averaged FTIR spectra (supplemental Fig. S1). Differences in amplitude for each frequency component measure proportionate changes in net secondary structure motifs between wild type and mutant proteins. These quantitative changes are revealed after digital subtraction of the parent spectra and can be decisively assigned to the net gain or loss of structural motifs (Fig. 3B).
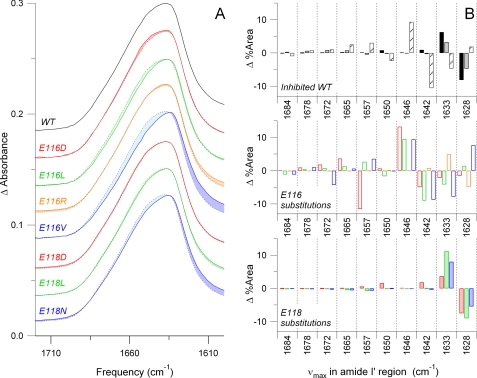
FIGURE 3.
FTIR spectra illustrate quantifiable changes in secondary structure resulting from challenges to L5, either by drug inhibition or residue substitution. A, averaged FTIR spectra of the amide I′ region of wild type (WT), Glu-116 substitutions, and Glu-118 substitutions are plotted. The wild type spectra are superimposed (dotted line) on each trace, and differences in line shape are highlighted, showing changes in secondary structure. FTIR spectra were acquired from two independent purifications and averaged. B, band narrowing of the amide I′ region of the N-terminal L5 mutant proteins resulted in 10 frequency bins. Positive amplitudes indicate a gain of secondary structure at a given frequency from wild type. Negative amplitudes indicate a loss of structure. Wild type was inhibited by both monastrol in 1:1 (black box, dark gray fill) and 1:20 (light gray fill) ratios and STC (striped fill). Other bar colors correspond to those for a given substitution shown in A.
FTIR spectra of wild type Eg5 proteins in the presence of allosteric inhibitors record structural changes that occur as a result of perturbing the L5 loop. Eg5·monastrol samples (Fig. 3B, gray bars) had a 6% increased content for the 1633 cm−1 spectral component and an 8% decreased content for the 1628 cm−1 constituent compared with uninhibited wild type. From correlations of amide I′ band frequency with secondary structural elements in proteins in 2H2O (39, 40), both 1628 and 1633 cm−1 were assigned to the low frequency component of β-sheets. These changes are well above the level of noise; variation from wild type to wild type sample was typically less than 1% at a given frequency (data not shown).
In contrast, the major changes between wild type Eg5·STC and ligand-free wild type samples (Fig. 3B, striped black boxes) were measured for the 1646 and 1642 cm−1 spectral components, attributable to a 9% increase in disordered structure and 10% decrease in 310 helical content, respectively. Thus, interaction of wild type Kinesin-5 protein with monastrol results in spectral amide I′ components that differ from those from STC interaction. Changes in the high frequency β-sheet components at 1684, 1678, and 1672 cm−1 were minimal in both wild type Eg5·STC- and Eg5·monastrol-bound samples. The 1665, 1657, and 1650 cm−1 modes, assignable to turns and α helices, showed alterations in these structural elements between Eg5 bound with STC and Eg5 bound with monastrol.
Alterations in peak height and shape of the broad amide I′ envelope were detected also in the mutant Eg5 proteins (Fig. 3A). Difference spectra comparing mutant and wild type data were obtained in a fashion similar to those described above and were digitally subtracted (Fig. 3B). In general, Glu-116 mutants had the largest absorbance changes at 1646, 1642, 1633, and 1628 cm−1 and they showed vibrational markers similar to those for STC-inhibited wild type samples. Glu-118 mutants had the largest changes in area at 1633 and 1628 cm−1, and they showed vibrational markers similar to those for monastrol-inhibited wild type samples. Because the overall pattern of secondary structural changes fell into either STC-like or monastrol-like spectrotypes (Fig. 3B), these data suggest two different populations of Eg5 conformers for these N-terminal L5 mutations. This interpretation is supported by the anomalous gel migration in Fig. 1. Thus, the N-terminal L5 substitutions resulted in modular changes in the secondary structure of Eg5 in solution, irrespective of mutation or drug binding.
Similar analyses were conducted for IR spectra collected from C-terminal L5 mutants in solution, in both their free and ligand-bound forms. Substitution of Ala-133 or Asp-130 resulted in a greater number of possible conformational outcomes, as observed in our vibrational data (supplemental Fig. S2). Overall, there is not a clear catalogue of conformational changes in the C-terminal L5 mutants, compared with the N-terminal L5 mutants. The C-terminal L5 mutants have the largest absorbance changes at 1646, 1642, 1633, and 1628 cm−1 and additional absorbance changes at 1665, 1657, and 1650 cm−1 (supplemental Fig. S2). Some of these C-terminal Ala-130 and Asp-133 substitutions exhibit changes in secondary structural content similar to those of the Glu-116 mutants (Fig. 3B). However, other mutants in the C terminus of the L5 loop had perturbations that are unique. Thus, because the C-terminal L5 mutants did not fall uniformly into the same spectrotype as STC- or monastrol-bound samples, these results suggest that the long range conformational effects are dependent on the chemical nature of the of the C-terminal residue side chain.
Exploratory PCA Identifies That N-terminal L5 Mutants Occupy a Bimodal Structural Distribution in Solution
To create a model of how Eg5 undergoes allosteric conformational changes propagated from the L5 loop, we used computationally driven multivariate data analysis to extract structural information from our IR data. Techniques for deconvoluting multivariate data are common analytic tools in microarray data analyses, systems biology studies, and chemometric detection (41–45), but their implementation for discovering atomic-level information is novel to this work. The vibrational spectrum is fundamentally multivariate and complex in nature. Within a series of discrete measurements at defined frequency intervals found in each spectrum, the vast number of normal modes in the Kinesin-5 motor domain (>15,000 vibrational degrees of freedom) can have frequency overlap.
Our initial foray into multivariate data analysis uses PCA. We apply this non-parametric method that uses eigenvectors to decompose vibrational spectra to a smaller set of computer-derived variables and to form models for predicting factors causing responses. Although PCA does not directly relate cause and effect, it serves to highlight the larger variations in the input data and find patterns in a set of spectra that on the surface can appear similar. Last, the use of statistical algorithms to categorize our FTIR data provides an unbiased method that is mathematically grounded.
Spectroscopic data from the N-terminal and C-terminal L5 mutants were subjected to exploratory PCA. For proper formation of an initial model and for proper evaluation of the model's predictions, we did not use cross-validation methods. Instead, we partitioned the data into two spectral data sets to be evaluated independently. By not using a sample before testing it, we eliminate bias in our prediction accuracy (46). Wild type Eg5 and the N-terminal L5 mutants serve as the training set for PCA. The query or test set is the C-terminal L5 mutant spectra, used to verify and refine the predictive model. We emphasize that this evaluation step, which included analysis of an independently purified second wild type sample, was physically and temporally distinct from development of the initial training set.
The spectroscopic inputs are the band-narrowed amide I′ frequencies from Eg5 protein in the absence of inhibitor, in the presence of monastrol, and in the presence of STC. In the outputs from these multivariate analyses (Fig. 4), the principal components (PC1 and PC2), or variables of the projection space, are linear combinations of the original frequencies.
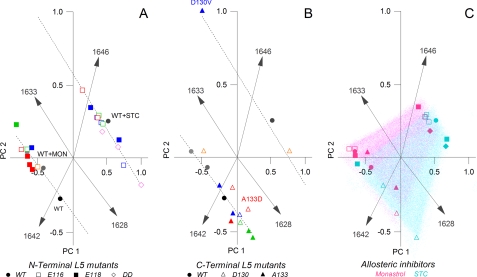
FIGURE 4.
PCA of secondary structural elements in the Eg5 motor domain reveals distinct conformer populations, resulting from L5-initiated effects. The score, or spatial positioning, of the data points is based on changes in percentage area of the secondary structure profiles determined by IR band narrowing. A, PCA clusters the majority of Glu-118 (■) and WT (●) data points together based on secondary structure. The Glu-116 (□) polymorphisms fall into a separate line of degeneracy with a larger disordered component. The 1642 and 1646 cm−1 vectors separate these two populations. B, PCA of C-terminal L5 substitutions, Ala-133 (▴) and Asp-130 (▵), falls predominantly into the same population occupied by the N-terminal Glu-118 mutations and can be differentiated by the 1633 and 1628 cm−1 vectors, respectively. C, a composite of both N- and C-terminal polymorphisms are plotted. Highlighted are the monastrol (blue) and STC (pink) data points.
Using wild type and N-terminal L5 mutant Kinesin-5 proteins, we demonstrate that distinct patterns were obtained from PCA. The 10-dimensional PCA output of IR frequencies was projected on a two-dimensional plane formed by PC1 and PC2 (Fig. 4), which accounted for 90% of the variance in our spectra. The dominant four vectors are defined by the 1642, 1646, 1633 and 1628 cm−1 IR frequencies (Fig. 4A, gray rays). These patterns define the amide I' frequencies associated with the Glu-118 and Glu-116 mutations and separate the motor proteins into two populations of conformers with high correlation. The first group contains Glu-118 mutants (filled boxes) and wild type Eg5 (filled circles) in a tightly clustered population. The second group is predominantly composed of Glu-116 mutants (open boxes) and the E116D/E118D double mutant (open diamonds).
The 1642 and 1646 cm−1 vectors (Fig. 4A) are the most influential in distinguishing the two mutant populations. Principal component analysis emphasizes the strong connection between residue substitution in the N terminus of the L5 loop and changes in 310 helical content and disordered structures, respectively. The 1633 and 1628 cm−1 vectors (Fig. 4A) also discriminate between the mutant populations, linking changes in Eg5 β-sheets with perturbations of the L5 loop.
The results from PCA were further supported by examination of additional protein samples that were not used in the initial model-building step. Independent PCA analysis of the C-terminal L5 mutants (Fig. 4B) accounted for ∼85% of the variance in these spectra. PCA of these test FTIR spectra (Fig. 4B) place the Asp-130 (open triangles) and Ala-133 mutants (filled triangles) within identical structural populations of the N-terminal L5 mutants and predominantly fall into line with the wild type/Glu-118 data points. The fidelity of the training and test sets for this PCA experiment argues that the initial model accurately predicts the conformer populations of Eg5 motor domains in which the L5 loop is challenged. Additionally, the high PCA correlation makes a case that our collection of Eg5 structures samples a sufficient number of conformational possibilities to inform reasonable allosteric models.
Monastrol and STC Produce Similar Local, but Dissimilar Distal, Structural Changes
We also show that statistical algorithms can be applied to infrared data to reveal features that are the basis for discriminating between the effects of monastrol and STC. The presence of monastrol (Fig. 4C, pink symbols) or STC (Fig. 4C, blue symbols) also resulted in coincident, steady-state structural changes in mutant kinesin proteins; protein-inhibitor samples are separated by the 1646 and 1642 cm−1 components and fall into the same two PCA populations as free protein. This highlights that analogous modifications in disordered and 310 helical content are reproducibly created in both Eg5 mutants and in allosteric inhibition.
The 1633 and 1628 cm−1 components differentiate the clustering of Eg5 samples incubated with monastrol or with STC (Fig. 4C). Regardless into which population the data points fall, STC samples of the parent mutant generally have a greater 1628 cm−1 component (Fig. 4C, pink), whereas monastrol samples have a greater 1633 cm−1 component (Fig. 4C, blue). The 1633 and 1628 cm−1 frequencies arise from changes in β-sheet structures, which are found primarily at the core of all kinesin motor domains. This PCA analysis underscores that, by IR spectroscopy, we have directly measured long distance changes to β-sheets within the kinesin protein. These distal alterations are linked with high correlation to perturbations to the surface-exposed L5 site, a requirement for allostery. Moreover, the 5 cm−1 difference between the β-sheet components indicates that each frequency arises from a unique perturbation of this secondary structure. Thus, there is more than one means by which L5-initiated perturbations are coupled to distant secondary elements.
These observations by vibrational methods are corroborated by comparison of crystal structures of wild type Eg5·ADP bound with monastrol (17) or STC. At the initiation of this work, there was no published report of an Eg5·ADP·STC structure, despite the widespread usage of the inhibitor. Our 2.5 Å x-ray crystallographic determination of Eg5·ADP bound with a single STC molecule (Table 1) shows the canonical arrowhead-shaped globular structure (supplemental Fig. S3A) seen in other Eg5 structures. We observe that STC and monastrol stabilize similar overall states of the motor domain with the neck linker in the “docked” conformation and with similar conformations adopted by the switch I and II regions.
TABLE 1.
X-ray refinement statistics for the wild type Eg5·ADP·STC structure
| Parameters | Values |
|---|---|
| Resolution (Å) | 2.5 |
| Rwork/Rfree | 0.241/0.275 |
| No. of atoms | |
| Total | 2749 |
| Protein | 2617 |
| Ligand/ion | 58 |
| Water | 74 |
| B-Factors | |
| Protein | 25.537 |
| Ligand/ion | 26.120 |
| Water | 18.767 |
| Root mean square deviations | |
| Bond lengths (Å) | 0.008 |
| Bond angles (degrees) | 1.426 |
As expected from other crystallographic studies of Eg5 inhibitors, there are no large differences in the local structure of the allosteric site in our Eg5·ADP·STC structure. The overall fold of the L5 loop is similar to that of the two published monastrol-bound Eg5 structures (supplemental Fig. S3B). However, we note that STC binding results in a slightly greater restriction of the allosteric site by the L5 loop; STC binding occludes ∼80 Å2 more of the Eg5 surface from solvent access than monastrol. Of the residues examined in this study, only the rotamer configuration of Glu-118 in Eg5·ADP·STC differs slightly from monastrol-bound Eg5 (supplemental Fig. S3B).
We also draw attention to long range structural effects in the β-sheet core of the motor domain due to binding of L5-directed inhibitors, which have been disregarded in prior crystallographic reports. The central β-sheet structure of all kinesin motor domains consists of a parallel β-sheet, with a topology (or β-strand order) of 2183, adjacent to an antiparallel β-sheet with a topology of 7645. There are clear distortions of the antiparallel β-sheet (Fig. 5) that probably impose novel strain on the motor domain core in the presence of these allosteric inhibitors.
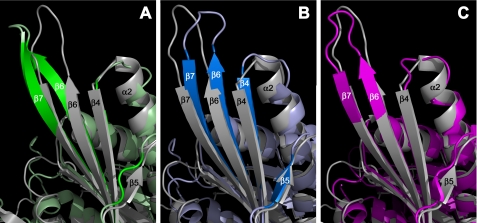
FIGURE 5.
Comparison of x-ray crystallographic structures of the Eg5 motor domain bound with L5-directed inhibitors shows distortions in β-sheets. Shown is a region of the antiparallel β-sheet of the Eg5 motor domain; other structural motifs that cover the central β-sheet were removed in PyMOL for visual clarity. The wild type Eg5·ADP structure (PDB ID 1II6) in gray is overlaid on the ternary structure in complex with STC (PDB ID 3KEN) in green (A), monastrol (PDB ID 1X88) in blue (B), and a tetrahydroisoquinoline carboxamide in pink (PDB ID 2FME) (C). Appropriate β-strands are labeled.
Strands that undergo the largest changes are β4, β5, β6, and β7. Although all strands typically exhibit a right-handed twist in higher order structures, the N terminus of the β4 strand is pulled closer to the α2 helix in both STC-bound (Fig. 5A, green ribbons) and monastrol-bound (Fig. 5B, blue ribbons) structures, compared with the native Eg5 structure (Fig. 5, gray ribbons). The β5 strand is not formed in the Eg5·STC structure. The β6 and β7 strands have large distortions that differ in magnitude and direction, dependent on whether STC or monastrol was bound to the L5 loop. Direct comparison of the STC- and monastrol-bound Eg5 structures (supplemental Movie S1) highlights the divergence in the twist of the β6 and β7 strands. Exaggeration in the twist of the β6 and β7 strands is also observed in other structures of Eg5 co-crystallized with L5-directed inhibitors (Fig. 5C).
DISCUSSION
Important questions in protein biochemistry are whether and how conformational flexibility is linked to catalytic function. Although the atomic level detail of the 20 PDB entries for wild type Eg5 is fairly complete, whether these static “snapshots” accurately reflect functional states that these amino acid assemblies can achieve is more difficult to determine. Thus, obtaining all needed conformational possibilities in kinesin mechanotransduction and its allosteric inhibition is a significant challenge.
Herein we provide novel approaches to query the conformational component in the term “structure-function relationship” and to apply computational analysis when intuitive reasoning becomes insufficient. We used wild type protein and 15 different mutants in three different protein conditions: drug-free, monastrol-bound, and STC-bound. The resulting 48 different structural analyses exceeds the current number of Eg5 structures in the RCSB from nearly a decade of work. This large data set of Eg5 solution structures permits fresh appreciation of structural linkages and also allows quantification of the relationships.
Our multivariate analysis of these structural measurements yields a model, in which there is more than one linked outcome between conformational changes from the surface L5 loop to the β-sheet core of the motor domain. Questions we have addressed are (i) the prediction accuracy between vibrational spectroscopy and protein crystallography, (ii) the unique detection of force coupled with chemistry (or mechanochemistry) by IR measurements, (iii) whether the solution structures sampled via inhibitors and mutation reflect preexisting conformational states in native protein, and (iv) how novel use of IR spectroscopy and PCA can be applied to clinical practice.
There Is High Prediction Accuracy between IR and X-ray Diffraction Determination of Eg5 Helical Structures
Foremost in this novel application of IR spectroscopy is the question of where these techniques intersect and where they diverge. Current IR data do not inherently contain information on the precise site(s) of origin within the Eg5 motor domain. Inspection of relevant crystal structures can facilitate the further assignment of an IR signature to a specific protein segment. Attempts at mutant protein crystallography, despite prior success in our hands (27), did not garner L5 mutant protein crystals that diffract well (data not shown).
However, examination of fractional secondary structure content of wild type Eg5 motor proteins determined from existing x-ray structures supports our multivariate findings of Kinesin-5 in solution. From DSSP analysis (47) of Eg5 structures in the PDB, increased 310 helix content is observed when comparing ligand-bound versus drug-free Eg5 proteins (Table 2). In the Eg5·ADP crystal structure (24), four 310 helices are found within the α3, α5, and α6 helices and in the switch I loop, each of which is 3 residues long. Thus, 3.3% of residues in the Eg5 motor domain are found in 310 helices in the inhibitor-free crystal structures. In inhibitor-complexed Eg5 proteins, there is generally a gain of two 310 helices in the L5 loop; as 6 residues undergo a random coil-310 helix transition, the fractional content of 310 helices increases to 4.9%.
TABLE 2.
Comparison of fractional secondary structure composition in inhibitor-free and inhibitor-bound crystal structures of Eg5
Secondary structure assignments in crystallographic data are obtained from DSSP analysis. Fractional compositions were calculated for each protein within the asymmetric unit cell and averaged, when appropriate. Subtraction values of inhibitor-bound from inhibitor-free Eg5 (1II6) structures are shown.
| Compared structures | Percentage change in secondary structure |
|||||
|---|---|---|---|---|---|---|
| β-Sheet | α helix | 310 helix | Turn | Bend | β bridge | |
| % | % | % | % | % | % | |
| (STC) 3KEN versus 1II6 | −0.7 | 1.5 | 0.0 | −1.5 | 0.8 | 0.0 |
| (monastrol) 1Q0B versus 1II6 | 3.0 | −1.4 | 1.6 | 0.3 | −0.7 | 0.0 |
| (monastrol) 1X88 versus 1II6 | 2.3 | −1.5 | 1.6 | 0.9 | −1.6 | 0.0 |
| 1YRS versus 1II6 | 0.0 | −2.4 | 1.2 | 1.2 | 0.9 | −0.3 |
| 2IEH versus 1II6 | 2.7 | −2.6 | 0.0 | 2.4 | 0.7 | −0.3 |
Although the IR absorbance spectrum is a reflection of the motor protein ensemble, we surmise that the above IR-detectable changes at 1642 and 1646 cm−1 are localized to the L5 loop because it is the only protein segment that shows a conformational isomerization between 310 helices and a random loop in the crystal structures. Moreover, this stringent comparison between our solution structural determinations and crystallographic data calls attention to the sensitivity of the vibrational methods employed here; even changes in 6 residues can be accurately detected within the amide I′ envelope.
IR Spectroscopy Is Uniquely Suited to Measure Mechanochemical Changes in Motor Proteins
Despite the initial similarities, the experimental distinction between the two biophysical methods becomes evident in their respective examination of Eg5 β-sheet structures. Protein crystallography defines the spatial organization of 90–100 Eg5 residues (or 24–27% of the motor domain) into β-strands or sheets, based on distance and dihedral angle guidelines. In contrast, infrared frequencies are a direct function of the force constants of the vibrating bonds and of the reduced mass of a chemical group. They are therefore correlated to a number of other physicochemical parameters, such as bond length and strength, and as such are reporters of not only group structure but also the environment surrounding the functional group.
From the arguments above, we highlight here that IR spectroscopy can provide a direct measure of mechanochemical coupling in motor proteins. Likewise, comparison of infrared and crystallographic analysis may reveal parallels between them but need not always show complete accord. Examination of Eg5 crystallographic data shows that L5-directed inhibitors can increase β-sheet content by 3% of the total residues, decrease β-sheet content by 0.7%, or yield no change (Table 2) in comparison with Eg5·ADP complexes (24). In contrast, we observe changes in two frequencies associated with β-sheet carbonyls in our IR analysis of Eg5 proteins that are biochemically or molecularly perturbed at the allosteric site. Measurement of the 1633 and 1628 cm−1 modes showed there is 5–10% change in β-sheet carbonyls in all Eg5 conditions sampled (wild-type and mutants in drug-free, monastrol-, or STC-bound states). Although there is reasonable qualitative agreement, discernment of two different types of β-sheet perturbations and the quantitative difference show that there is no simple one-to-one correspondence between the biophysical measurements.
There are several potential biochemical causes for observing two differing infrared frequencies for β-sheet carbonyls in these analyses. According to the simple two-atomic oscillator model, factors affecting the force constant between the atoms and/or the reduced mass of the chemical group will alter the C=O stretching frequency in the amide I′ region. Changes in strength or length of the hydrogen bonds (48) alter the C=O stretching frequency: distinctions in hydrogen bonding patterns between β-sheets, such as those between parallel and antiparallel β-strands, are detectable by IR spectroscopy. Differences in the exposure of β-sheet structures to the aqueous environment or to other structural elements within the motor domain can also be monitored via infrared frequencies.
It is also equally probable that, because bond energy and length are directly related, bond distortions can be monitored with accuracy (48). The force constant can report strain arising via distortions of secondary structure. From our data, we conclude that monastrol inhibition produces a distortion of the central β-sheet structure and/or hydrogen-bonding pattern between β-strands that differs from STC. When the more potent inhibitor (STC) is bound to Eg5, the β6 and β7 strands pull toward the face of the motor domain containing the MT-binding site, whereas binding of monastrol results in these β-strands pulling into the L5 face. Thus, crystallographic data on wild type Kinesin-5 corroborate our assertion that IR spectroscopy can directly measure mechanochemical coupling in motor proteins.
Reports that describe these structures typically conclude that observed alterations in the β-strands are artifacts due to crystal packing in the lattice (e.g. see Ref. 23). Alternatively, computational analysis of empirical experimental observations can uniquely draw mechanistic inferences of how biochemical systems function without bias, such as in the well developed area of enzyme kinetics. In this work, only from PCA analysis of the IR frequencies could we capture the dominant modes of motion within Eg5 during allostery. These results compel unique conclusions in the fundamental understanding of ATPase mechanisms and in novel biomedical applications.
Distortions in β-Sheets Are a Conserved Means of Mechanochemical Communication in Eg5 Kinesin, Myosin, and F1-ATPases
In our model of Eg5 allostery, distortions in secondary structures are a means of communication across the motor domain and may comprise a portion of the force component in kinesin mechanotransduction. Our data show that 310 helices in the L5 loop and β-strands in the central sheet undergo mechanochemical changes upon drug binding or sequence mutation. We do not anticipate that Eg5 undergoes unitary conformational transitions, in which the whole protein changes its structure simultaneously, but rather that there are probably stepwise or hierarchical changes across the L5 loop and the β-sheet.
At the local allosteric site, the 310 helices within the L5 loop and the α2 helix, encapsulating the loop, can impose torque on the L5 structure. The stored potential energy resulting from this stress and strain allows for the springlike motion of the L5 loop to enclose a small molecule inhibitor within the allosteric pocket. We speculate that observed reduction of 310 helix content results in diminished strain, decreased energy, and loss of L5 rigidity, thereby significantly reducing drug sensitivity in Eg5 mutants.
Intramolecular interactions between the hydrogen-bonding edges of β-strands can constitute a fundamental form of force propagation in Eg5. Twisting or distortion of the central β-sheet may propagate changes of the protein structure that result in lower steady-state, basal ATPase rates in general. The β6 and β7 strands immediately follow and precede switch I and switch II, respectively. This suggests a mechanism by which the allosteric compounds can impact ATP hydrolysis. These switch regions are critical in the binding of the nucleophilic water and catalytic base (27). Large conformational modification of the switch regions will directly affect catalytic rates. In addition, there is a deformation of the microtubule-binding site observable in Kinesin-5 crystal structures with a bound allosteric molecule (data not shown), suggesting a rationale for the observed interruption of cytoskeletal interactions (49).
This model offers an alternate hypothesis from commonly adopted views that spatial translation of loops, such as the L5 loop and the neck linker, influence the organization of residues involved in productive chemical transformations and movement. However, there is a growing recognition of the importance of intramolecular interactions among β-strands. Such twisting-shearing motions in the central β-sheet have been reported for the allosteric integrin I domains (50). Reorganization of the contacts within the central β-sheet has also been observed in cryo-electron microscopy studies of Kar3 (51) and in crystal structures of nucleotide-free myosins (52, 53).
Despite considerable evolutionary divergence, there is apparent conservation of structural requirements in mechanotransduction across motor proteins and F1-ATPase. For myosin, the Sweeney and Houdusse laboratories (54) also have proposed that the myosin β-sheet is a mechanochemical “transducer”; distortions of this structural element control communication between the cytoskeletal interface and the nucleotide-binding site. Second, they speculate that there must be some form of communication from a surface loop in myosin, analogous to the kinesin L5 loop, to the central β-sheet.
Our data demonstrate that β-sheet distortions in Kinesin-5 resemble states of myosin that have been crystallized, and they also show the full range of distortions observed in F1-ATPase (55). Thus, three classes of ATPase proteins have maintained similar structural elements and similar means of allosteric communication. In keeping with the original proposal in Ref. 55 and echoed in Ref. 54, we speculate that distortion of β-sheets is responsible for sequential product release from the orthosteric site during force generation. This idea is experimentally supported by real-time observation of conformational changes during Eg5 catalysis in solution (56). Moreover, we provide the first experimental proof that there is direct coupling of the conformational transitions in the L5 loop with the kinesin β-sheet transducer.
In summary, these above studies suggest that small changes in backbone conformation may be responsible for allosteric effects and may reflect events in the normal functional cycle of kinesins. Specifically, our results show that the impact of mutations on enzyme activity is due to changes in the equilibrium conformations sampled and hence the overall free energy barrier for the ensemble rather than non-equilibrium dynamic factors. In other words, we argue that the effects observed for L5-challenged Kinesin-5 proteins may be explained by increasing or decreasing the relative proportion of preexisting conformational substates found in native catalysis.
These IR Spectrotypes May Serve as Novel Structural Biomarkers to Predict Drug Sensitivity
This model can be used not only to better understand mechanisms but also to motivate new experiments. These experiments demonstrate that inhibitor efficacy can be measured structurally; we predict that an even larger 1628 cm−1 component would be observed in PCA of more potent inhibitors of Eg5. The uncovered interactions essential for drug sensitivity in Eg5 mutants in vitro can be extended to predict how human polymorphisms can affect clinical responsiveness.
We also demonstrate that particular structural signatures are correlated with inhibitor resistance in Eg5 variants. The first condition for inhibitor resistance in Eg5 is the conformational selection of the L5 loop in solution. For the N-terminal L5 substitutions, there is a clear difference in the pattern of secondary structure changes observed in the Glu-116 mutant proteins when compared with those of the Glu-118 substitutions (Figs. 3 and 4A). The kinetic sensitivity of Glu-118 mutants to drug inhibition (Fig. 2) is correlated with adoption of 310 helical structure in the L5 loop and a large 1642 cm−1 vector (Fig. 4A, filled boxes). In line with this conclusion, the clustering of the Glu-116 mutant samples in PCA argues that a large 1646 cm−1 component is strongly correlated with drug resistance. Thus, loss of the 310 helix in the L5 loop unilaterally prohibits binding of small molecules to the allosteric site, as shown by the D130V mutation that displays a large 1646 cm−1 component and confers resistance in Eg5 to the clinical trial drugs in purified form, in tumor cell culture, and in xenograft model systems (16).
The second proviso for inhibitor resistance in Kinesin-5 proteins is the mechanochemical state of the antiparallel β-sheet. For Eg5 variants that populate the 1646 cm−1 cluster in the PCA, the lack of 310 helix functionally overrides any conformational changes observed in the β-sheet. Therefore, there is a hierarchy of structural changes that dictate inhibitor resistance. For Eg5 variants that populate the 1642 cm−1 cluster in PCA, only genotypes that result in strong 1633 cm−1 vectors remain sensitive to drug inhibition. If there is a twist of the β6 and β7 strands that resembles the STC-bound Eg5 (Fig. 5A) and thus exhibits a strong 1628 cm−1 component in multivariate analysis (Fig. 4B), this torque on the antiparallel β-sheet negates drug sensitivity, even if 310 helices are adopted. This is the case for the established ispinesib-resistant mutant, A133D.
We speculate that isomerization of the L5 loop and distortions of the central β-sheet can also serve as novel structural biomarkers to predict drug sensitivity of Eg5. This biomarker provides correlation between protein structure and therapeutic outcome, which is advantageous because the final target for clinical drugs is typically polypeptides. As a case in point, more than half of the mutants examined had catalytic rates comparable or greater than those of wild type samples. We anticipate that seemingly normal Eg5 kinesin function could harbor a polymorphism that would confer resistance to clinical drugs, and our biomarker could discriminate against Eg5-directed inhibitors. To our knowledge, this is the first report of rapid and quantitative methods for assaying conformational alterations, resulting from peptide sequence variations, of protein ensembles in solution. This biophysical approach does not supplant genomic efforts but rather addresses complementary questions; it answers the “why” and “how much” an individual protein variant is resistant to a drug, in comparison with the binary answer to the sole question “if” supplied by correlations with genomic biomarkers. Future studies will focus on whether these structural biomarkers based on conformational changes due to the sequence polymorphism can be used in the clinic for appropriate treatment options for kinesin-related diseases.
Supplementary Material
Acknowledgments
Prof. Timothy Mitchison, Christine Fields, and the Department of Systems Biology at Harvard Medical School are gratefully recognized for encouragement and support of E. J. W. and S. K. Catherine D. Kim, Nathan Stevens, and Randy Saunders are thanked for assistance with the mutant activity assays.
This work was supported, in whole or in part, by National Institutes of Health Grant GM066328 (to E. J. W.). This work was also supported by grants from the Louisiana Board of Regents (to S. K. and to D. K. W.), the OSER/Carilion Biomedical Institute (to E. J. W. and S. K.), ASPIRES (to S. K., R. A. W., and E. J. W.), and the National Science Foundation (to R. A. W.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Movie S1 and Figs. S1–S3.
The atomic coordinates and structure factors (code 3KEN) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- STC
- S-trityl-l-cysteine
- AMPPNP
- 5′-adenylyl-β,γ-imidodiphosphate
- PCA
- principal component analysis
- PDB
- Protein Data Bank.
REFERENCES
- 1.Gunasekaran K., Ma B., Nussinov R. (2004) Proteins 57, 433–443 [DOI] [PubMed] [Google Scholar]
- 2.Monod J., Wyman J., Changeux J. P. (1965) J. Mol. Biol. 12, 88–118 [DOI] [PubMed] [Google Scholar]
- 3.Kern D., Zuiderweg E. R. (2003) Curr. Opin. Struct. Biol. 13, 748–757 [DOI] [PubMed] [Google Scholar]
- 4.Cooper A., Dryden D. T. (1984) Eur. Biophys. J. 11, 103–109 [DOI] [PubMed] [Google Scholar]
- 5.Hotha S., Yarrow J. C., Yang J. G., Garrett S., Renduchintala K. V., Mayer T. U., Kapoor T. M. (2003) Angew. Chem. Int. Ed. Engl. 42, 2379–2382 [DOI] [PubMed] [Google Scholar]
- 6.DeBonis S., Skoufias D. A., Lebeau L., Lopez R., Robin G., Margolis R. L., Wade R. H., Kozielski F. (2004) Mol. Cancer Ther. 3, 1079–1090 [PubMed] [Google Scholar]
- 7.Sunder-Plassmann N., Sarli V., Gartner M., Utz M., Seiler J., Huemmer S., Mayer T. U., Surrey T., Giannis A. (2005) Bioorg. Med. Chem. 13, 6094–6111 [DOI] [PubMed] [Google Scholar]
- 8.Maliga Z., Kapoor T. M., Mitchison T. J. (2002) Chem. Biol. 9, 989–996 [DOI] [PubMed] [Google Scholar]
- 9.DeBonis S., Simorre J. P., Crevel I., Lebeau L., Skoufias D. A., Blangy A., Ebel C., Gans P., Cross R., Hackney D. D., Wade R. H., Kozielski F. (2003) Biochemistry 42, 338–349 [DOI] [PubMed] [Google Scholar]
- 10.Mayer T. U., Kapoor T. M., Haggarty S. J., King R. W., Schreiber S. L., Mitchison T. J. (1999) Science 286, 971–974 [DOI] [PubMed] [Google Scholar]
- 11.Brier S., Lemaire D., Debonis S., Forest E., Kozielski F. (2004) Biochemistry 43, 13072–13082 [DOI] [PubMed] [Google Scholar]
- 12.Cox C. D., Breslin M. J., Mariano B. J., Coleman P. J., Buser C. A., Walsh E. S., Hamilton K., Huber H. E., Kohl N. E., Torrent M., Yan Y., Kuo L. C., Hartman G. D. (2005) Bioorg. Med. Chem. Lett. 15, 2041–2045 [DOI] [PubMed] [Google Scholar]
- 13.Cochran J. C., Gatial J. E., 3rd, Kapoor T. M., Gilbert S. P. (2005) J. Biol. Chem. 280, 12658–12667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skoufias D. A., DeBonis S., Saoudi Y., Lebeau L., Crevel I., Cross R., Wade R. H., Hackney D., Kozielski F. (2006) J. Biol. Chem. 281, 17559–17569 [DOI] [PubMed] [Google Scholar]
- 15.Lad L., Luo L., Carson J. D., Wood K. W., Hartman J. J., Copeland R. A., Sakowicz R. (2008) Biochemistry 47, 3576–3585 [DOI] [PubMed] [Google Scholar]
- 16.Luo L., Parrish C. A., Nevins N., McNulty D. E., Chaudhari A. M., Carson J. D., Sudakin V., Shaw A. N., Lehr R., Zhao H., Sweitzer S., Lad L., Wood K. W., Sakowicz R., Annan R. S., Huang P. S., Jackson J. R., Dhanak D., Copeland R. A., Auger K. R. (2007) Nat. Chem. Biol. 3, 722–726 [DOI] [PubMed] [Google Scholar]
- 17.Maliga Z., Mitchison T. J. (2006) BMC Chem. Biol. 6, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan Y., Sardana V., Xu B., Homnick C., Halczenko W., Buser C. A., Schaber M., Hartman G. D., Huber H. E., Kuo L. C. (2004) J. Mol. Biol. 335, 547–554 [DOI] [PubMed] [Google Scholar]
- 19.Fraley M. E., Garbaccio R. M., Arrington K. L., Hoffman W. F., Tasber E. S., Coleman P. J., Buser C. A., Walsh E. S., Hamilton K., Fernandes C., Schaber M. D., Lobell R. B., Tao W., South V. J., Yan Y., Kuo L. C., Prueksaritanont T., Shu C., Torrent M., Heimbrook D. C., Kohl N. E., Huber H. E., Hartman G. D. (2006) Bioorg. Med. Chem. Lett. 16, 1775–1779 [DOI] [PubMed] [Google Scholar]
- 20.Garbaccio R. M., Fraley M. E., Tasber E. S., Olson C. M., Hoffman W. F., Arrington K. L., Torrent M., Buser C. A., Walsh E. S., Hamilton K., Schaber M. D., Fernandes C., Lobell R. B., Tao W., South V. J., Yan Y., Kuo L. C., Prueksaritanont T., Slaughter D. E., Shu C., Heimbrook D. C., Kohl N. E., Huber H. E., Hartman G. D. (2006) Bioorg. Med. Chem. Lett. 16, 1780–1783 [DOI] [PubMed] [Google Scholar]
- 21.Garcia-Saez I., DeBonis S., Lopez R., Trucco F., Rousseau B., Thuéry P., Kozielski F. (2007) J. Biol. Chem. 282, 9740–9747 [DOI] [PubMed] [Google Scholar]
- 22.Kaan H. Y., Ulaganathan V., Hackney D. D., Kozielski F. (2010) Biochem. J. 425, 55–60 [DOI] [PubMed] [Google Scholar]
- 23.Zhang B., Liu J. F., Xu Y., Ng S. C. (2008) Biochem. Biophys. Res. Commun. 372, 565–570 [DOI] [PubMed] [Google Scholar]
- 24.Turner J., Anderson R., Guo J., Beraud C., Fletterick R., Sakowicz R. (2001) J. Biol. Chem. 276, 25496–25502 [DOI] [PubMed] [Google Scholar]
- 25.Cochran J. C., Gilbert S. P. (2005) Biochemistry 44, 16633–16648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bodey A. J., Kikkawa M., Moores C. A. (2009) J. Mol. Biol. 388, 218–224 [DOI] [PubMed] [Google Scholar]
- 27.Parke C. L., Wojcik E. J., Kim S., Worthylake D. K. (2010) J. Biol. Chem. 285, 5859–5867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brier S., Lemaire D., DeBonis S., Kozielski F., Forest E. (2006) Rapid Commun. Mass Spectrom. 20, 456–462 [DOI] [PubMed] [Google Scholar]
- 29.Marshall C. G., Torrent M., Williams O., Hamilton K. A., Buser C. A. (2009) Arch. Biochem. Biophys. 484, 1–7 [DOI] [PubMed] [Google Scholar]
- 30.Wojcik E. J., Dalrymple N. A., Alford S. R., Walker R. A., Kim S. (2004) Biochemistry 43, 9939–9949 [DOI] [PubMed] [Google Scholar]
- 31.Deavours B. E., Reddy A. S., Walker R. A. (1998) Cell. Motil. Cytoskeleton 40, 408–416 [DOI] [PubMed] [Google Scholar]
- 32.Learman S. S., Kim C. D., Stevens N. S., Kim S., Wojcik E. J., Walker R. A. (2009) Biochemistry 48, 1754–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Acta Crystallogr. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 34.Rath A., Glibowicka M., Nadeau V. G., Chen G., Deber C. M. (2009) Proc. Natl. Acad. Sci. U. S. A. 106, 1760–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Banci L., Bertini I., Cusack S., de Jong R. N., Heinemann U., Jones E. Y., Kozielski F., Maskos K., Messerschmidt A., Owens R., Perrakis A., Poterszman A., Schneider G., Siebold C., Silman I., Sixma T., Stewart-Jones G., Sussman J. L., Thierry J. C., Moras D. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 1208–1217 [DOI] [PubMed] [Google Scholar]
- 36.Barth A., Zscherp C. (2002) Q. Rev. Biophys. 35, 369–430 [DOI] [PubMed] [Google Scholar]
- 37.Goormaghtigh E., De Meutter J., Vanloo B., Brasseur R., Rosseneu M., Ruysschaert J. M. (1989) Biochim. Biophys. Acta 1006, 147–150 [DOI] [PubMed] [Google Scholar]
- 38.Hutchison R. S., Betts S. D., Yocum C. F., Barry B. A. (1998) Biochemistry 37, 5643–5653 [DOI] [PubMed] [Google Scholar]
- 39.Susi H., Byler D. M. (1986) Methods Enzymol. 130, 290–311 [DOI] [PubMed] [Google Scholar]
- 40.Surewicz W. K., Mantsch H. H., Chapman D. (1993) Biochemistry 32, 389–394 [DOI] [PubMed] [Google Scholar]
- 41.Misra J., Schmitt W., Hwang D., Hsiao L. L., Gullans S., Stephanopoulos G. (2002) Genome Res. 12, 1112–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paschou P., Ziv E., Burchard E. G., Choudhry S., Rodriguez-Cintron W., Mahoney M. W., Drineas P. (2007) PLoS Genet. 3, 1672–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bylesjö M., Eriksson D., Kusano M., Moritz T., Trygg J. (2007) Plant J. 52, 1181–1191 [DOI] [PubMed] [Google Scholar]
- 44.Lin Z., Altman R. B. (2004) Am. J. Hum. Genet. 75, 850–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beckwith-Hall B. M., Nicholson J. K., Nicholls A. W., Foxall P. J., Lindon J. C., Connor S. C., Abdi M., Connelly J., Holmes E. (1998) Chem. Res. Toxicol. 11, 260–272 [DOI] [PubMed] [Google Scholar]
- 46.Dupuy A., Simon R. M. (2007) J. Natl. Cancer Inst. 99, 147–157 [DOI] [PubMed] [Google Scholar]
- 47.Kabsch W., Sander C. (1983) Biopolymers 22, 2577–2637 [DOI] [PubMed] [Google Scholar]
- 48.Krimm S., Bandekar J. (1986) in Advances in Protein Chemistry (Anfinsen C. B., Edsall J. T., Richards F. M. eds) pp. 181–364, Academic Press, Inc., New York [Google Scholar]
- 49.Crevel I. M., Alonso M. C., Cross R. A. (2004) Curr. Biol. 14, R411–412 [DOI] [PubMed] [Google Scholar]
- 50.Jin M., Song G., Carman C. V., Kim Y. S., Astrof N. S., Shimaoka M., Wittrup D. K., Springer T. A. (2006) Proc. Natl. Acad. Sci. U. S. A. 103, 5758–5763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hirose K., Akimaru E., Akiba T., Endow S. A., Amos L. A. (2006) Mol. Cell 23, 913–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Coureux P. D., Wells A. L., Ménétrey J., Yengo C. M., Morris C. A., Sweeney H. L., Houdusse A. (2003) Nature 425, 419–423 [DOI] [PubMed] [Google Scholar]
- 53.Reubold T. F., Eschenburg S., Becker A., Kull F. J., Manstein D. J. (2003) Nat. Struct. Biol. 10, 826–830 [DOI] [PubMed] [Google Scholar]
- 54.Coureux P. D., Sweeney H. L., Houdusse A. (2004) EMBO J. 23, 4527–4537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Menz R. I., Walker J. E., Leslie A. G. (2001) Cell 106, 331–341 [DOI] [PubMed] [Google Scholar]
- 56.Jun B., Kim S. (2010) J. Biol. Chem. 285, 11073–11077 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.