Abstract
Hydroxyl radicals induce hinge cleavage in a human IgG1 molecule via initial radical formation at the first hinge Cys231 followed by electron transfer to the upper hinge residues. To enable engineering of a stable monoclonal antibody hinge, we investigated the role of the hinge His229 residue using structure modeling and site-directed mutagenesis. Direct involvement of His229 in the reaction mechanism is suggested by a 75–85% reduction of the hinge cleavage for variants in which His229 was substituted with either Gln, Ser, or Ala. In contrast, mutation of Lys227 to Gln, Ser, or Ala increased hinge cleavage. However, the H229S/K227S double mutant shows hinge cleavage levels similar to that of the single H229S variant, further revealing the importance of His229. Examination of the hinge structure shows that His229 is capable of forming hydrogen bonds with surrounding residues. These observations led us to hypothesize that the imidazole ring of His229 may function to facilitate the cleavage by forming a transient radical center that is capable of extracting a proton from neighboring residues. The work presented here suggests the feasibility of engineering a new generation of monoclonal antibodies capable of resisting hinge cleavage to improve product stability and efficacy.
Keywords: Antibodies, Electron Transfer, Histidine, Mass Spectrometry (MS), Radicals, Hinge, Hydrogen Bond
Introduction
Availability of the full-length human IgG1 crystal structure has enriched the structural information around the region that connects the Fab and Fc domains (1, 2). The hinge consists of three parts: an upper hinge (Asp-Lys-Thr-His-Thr), a core hinge (Cys-Pro-Pro-Cys), and a lower hinge (Pro-Ala-Glu-Leu-Leu-Gly-Gly). The hinge region contains interchain disulfide bonds and provides structural flexibility that facilitates Fab movement. The hinge is required for the function of an IgG molecule, in particular, binding to immune effector molecules (e.g. ADCC, CDC) (3–5). The importance of the core hinge residues was demonstrated by studies that showed a negative impact to C1q binding and complement activation if Cys or Pro was mutated (3, 5, 6). In contrast, the upper hinge has no significant impact on the effector functions of an IgG1 molecule as demonstrated recently in a systematic study (7). The exposed and flexible upper hinge has been found vulnerable to various degradation mechanisms such as papain cleavage and β-elimination reactions (8). Our previous study revealed the hydroxyl radical-mediated hinge cleavage of a human IgG1 molecule (9). In this radical reaction, the first hinge disulfide bond between the two Cys231 residues was broken, followed by the formation of a thiyl radical at one of the two Cys231 residues that initiated an electron transfer (ET)2 to an upper hinge residue where cleavage was observed.
Although our previous study sheds light on the hydroxyl radical attack, some critical questions remained pertaining to the mechanism by which the hinge is cleaved. Radical formation at amino acid residues involves the loss of both a proton and an electron that can be transferred among the residues in the protein via either a single-step superexchange or a multistep hopping process (10–15). The ET process can be influenced by the nature of the amino acid side chain, particularly those residues with oxidizable side chains such as Tyr, His, and Trp (10, 15). His residues have been found to be involved in the radical-mediated degradation of proteins, despite the precise pathways being poorly defined (16–18). In the radical-induced hinge cleavage, the upper hinge His229 residue is degraded into the pyruvyl derivative (CH3-CO-CO-) or converted to Asp, whereas other residues in the vicinity remain unmodified (9). Combining the observation that the major cleavage sites were in the upper hinge, 226DKTHT, rather than in the core hinge, 231CPPC, the results suggested that the side chains of these upper residues, particularly the imidazole ring, may play an important role in the reaction mechanism.
In this study, we evaluated the effect of the side chains in the radical reaction mechanisms using site-directed mutagenesis and structure analysis. Substitution of the hinge His229 residue with a polar residue improved the local conformational integrity and resistance to hydroxyl radical attack. The results suggest that the His229 imidazole ring participates in the hydrogen bond interaction that maintains the hinge local structure integrity. Furthermore, His229 may function to form a transient radical anion as the second radical center that is capable of transferring an electron by extracting a proton from its neighboring residues, leading to the hinge cleavage. Our study indicates the feasibility of engineering a new generation of monoclonal antibody that is capable of resisting oxidative degradations using rational design.
EXPERIMENTAL PROCEDURES
Materials
The antibody used in this study is a recombinant human antibody of the IgG1 subclass. The molecule was expressed in Chinese hamster ovary cells and chromatographically purified according to Shulka et al. (19).
Site-directed Mutagenesis
A total of seven mutants were generated in this study: K227S, K227Q, K227A, H229S, H229Q, H229A, and K227S/H229S. These mutations were introduced using a QuikChange site-directed mutagenesis kit (Stratagene). The primers used for each mutation were as follows: K227S, 5′-d(GAGCCCAAATCTTGT-GACAGCACTC-ACACATGCCCA)-3′; K227Q, 5′-d(GAGCCCA-AATCTTGTGACCAGA-CTCACACAT-GCCCA)-3′; K227A, 5′-d(GAGCCCAAATCTTGTGACG-CCACTCAC-ACATGCCCA)-3′; H229S, 5′-d(CTTGTGACAAAACTAGCACATGCCCACCGTGCCCA)-3′; H229Q, 5′-d(C-TTGTGACAA-AACTCAGACATGCCCACCGTGCCCA)-3′; H229A, 5′-d(CTTGTG-ACAAAACTGC-CACATGCCCACCGTGCCCA)-3′; and K227S/H229S, 5′-d(GAGCCCA-AATCTTGTGA-CTCTACTAGCACATGCCCA-CCGTGCCCA)-3′. These mutations were confirmed by DNA sequencing. The IgG1 heavy chain and light chain were cloned into the expression vector pTT5 transient mammalian expression constructs.
Antibody Expression and Purification
The mutant antibodies were expressed using a mammalian transient expression system and purified using protein A chromatography (5-ml column; Pierce) following the manufacturer's instructions. The purity of the human IgG1 mutants was greater than 95%, as characterized by size exclusion chromatography.
Size Exclusion Chromatography (SEC)
SEC was carried out using two 7.8 mm × 30-cm TSK G3000SW columns connected in series. The protein was eluted with a constant flow rate of 0.5 ml/min and was monitored at 220 nm. The SEC running buffer contained 50 mm sodium phosphate, 300 mm sodium chloride at pH 6.8. The cleavage was measured by the relative percentage of integrated peak area of partial molecules with respect to the total area.
Oxidation of Protein
The native IgG1 and its variants were incubated with 20 mm H2O2 at 25 °C in 1× phosphate-buffered saline buffer, pH 7.4. Catalase (250 milliunits; Sigma) was added into the reaction system to quench H2O2. In some cases, EDTA (50 mm) or 5,5-dimethyl-1-pyrroline N-oxide (DMPO (100 mm) was added to the sample prior to H2O2 treatment.
Reversed Phase Chromatography and Time-of-Flight Mass Analysis (RP-HPLC-TOF/MS)
RP-HPLC-TOF/MS was performed as described previously (9). Briefly, antibody was diluted to 1 mg/ml in 50 mm Tris-HCl at pH 8.0 with a final concentration of 4 m guanidine hydrochloride (Sigma). RP-HPLC was performed on an Agilent 1200 HPLC system. The mobile phase included water with 0.11% trifluoroacetic acid as solvent A and acetonitrile (Burdick Jackson) with 0.09% trifluoroacetic acid as solvent B. An Agilent Zorbax SB300 C8 column, 2.1 mm × 150 mm, was used for the RP-HPLC-TOF/MS analysis. The column eluent was analyzed by UV detection at 214 nm and then directed in-line to a TOF mass spectrometer. The initial mobile phase was 25% B for 5 min, and then a two-stage gradient was applied using an increase of 1.4% B/min from 25–32% B, followed by a second gradient of 0.24% B/min from 32–38% B. The separation was performed at 75 °C at a flow rate of 0.3 ml/min. The peak areas were calculated using ChemStation software (Agilent Technologies) by applying a tangent skimming technique for the integration of incompletely resolved peaks. Electrospray ionization TOF/MS was performed on an Agilent 6210 LC-TOF/MS spectrometer equipped with an Agilent 1200 HPLC system. The electrospray ionization mass spectra were analyzed using Agilent BioConfirm protein deconvolution software.
Protease Digestion and Peptide Maps
The antibodies were reduced by 5 mm dithiothreitol (Sigma) for 30 min at 55 °C. Alkylation was performed with 15 mm iodoacetamide (Sigma) at room temperature for 40 min in the dark. Recombinant sequencing grade Lys-C (Roche Applied Science) was added to samples at an enzyme to protein ratio of 1:10 (w/w), and the samples were digested for 4 h at 37 °C. Analytical peptide maps consisted of loading 50 μg of the digest onto a Phenomenex Jupiter Proteo C5 column, 2.0 mm × 250 mm, heated at 60 °C. The separation was performed by gradient elution on an Agilent HP 1200 HPLC system. The column was held at the initial condition of 0.5% solvent A (0.11% trifluoroacetic acid in water) at a flow rate of 0.2 ml/min for 5 min, and then the digest was eluted with a linear gradient to 60% solvent B (0.09% trifluoroacetic acid in acetonitrile) over 160 min. The peptides were identified by data-dependent tandem MS fragmentation using a Thermo LTQ mass spectrometer.
Gel Sieving Capillary Electrophoresis in SDS (CE-SDS) Analysis of Variants
Gel sieving CE-SDS was used to separate the IgG1 fragments based on differences in hydrodynamic size under denaturing, nonreducing conditions. The method was performed on a Beckman PA 800 CE system. The IgG1, at a final concentration of 1 mg/ml, was mixed with Beckman SDS sample buffer (50% v/v), 14 mm iodoacetamide (Sigman), and a 10-kDa internal standard (Beckman). The mixture was heated at 70 °C for 5 min, and the resulting negatively charged SDS-protein complex was electrokinetically injected into a 30-cm bare-fused silica capillary (Beckman) containing SDS sieving gel buffer (Beckman). Separation was achieved over 40 min by applying a −15-kV electrical voltage across the capillary. The migration of the sample peaks was monitored by 220-nm UV absorbance as they passed through the capillary window located 20 cm from the injection end of the capillary. Manual integration of the electropherogram peaks was performed using Beckman 32 Karat software, which calculates the percentage of corrected area of each integrated species as a function of its migration time and the capillary length to the detector.
Effector FcγR Binding Assay
The binding of the hinge variant antibody to FcγRIIIa protein was measured by a competition amplified luminescent proximity homogeneous assay (20). In this assay, the binding interaction between antibody and FcγRIIIa brings the donor and acceptor beads close in proximity, and under laser excitation at 680 nm the oxygen singlet generated by the donor beads reacts with the acceptor beads to generate chemiluminenscence at 520–620 nm. This light in turn transfers the energy to the chromophore inside the donor beads to generate fluorescence at ∼615 nm. Nonbiotinylated hinge variant antibodies were added to the reaction competing with the human IgG1/FcγRIIIa binding that resulted in a reduction of the fluorescence signal. A control human IgG1 was biotinylated using an EZ-Link NHS-POE4-biotinylation kit (Pierce). After FcγRIIIa (either Phe158 or Val158)-glutathione S-transferase-His6 in amplified luminescent proximity homogeneous universal assay buffer (phosphate-buffered saline, pH 7.5, 0.1% bovine serum albumin, and 0.01% Proclin-300) was added to 1 nm final concentration, the native and hinge variant antibodies or buffer control were prepared as 1:4 series of dilutions from 1,000 to 0.244 nm. After shaking at room temperature for 2 h and a brief centrifugation, 25 nm biotinylated human IgG1 and 100 μg/ml glutathione amplified luminescent proximity homogeneous acceptor beads (PerkinElmer Life Sciences) were added to each well to a final concentration of 5 nm and 20 μμg/ml, respectively. The plates were incubated overnight at room temperature with shaking, and then 100 μg/ml streptavidin donor beads (PerkinElmer Life Sciences) were added to a final concentration of 20 μg/ml. The plate was incubated by shaking at room temperature for 2 h. Fluorescence signals at 615 nm were read in an Envision Alpha-FP HT microplate reader (PerkinElmer Life Sciences). All of the tests were carried out in triplicate. The data were normalized to the highest signal (no competition) for each set and fit to a one-site competition model using nonlinear regression with the software GraphPad Prism 3.0.
Caspase-3 Assay
The in vitro biological activity of the native antibody and its variants were compared by measuring their ability to induce apoptosis in a human ascites colorectal adenocarcinoma cell line, Colo205. Caspase-3 activation was used as a positive marker for apoptosis. Briefly, in a 96-well plate, the samples were prepared at a starting concentration of 0.4 μg/ml with eight serial dilutions at 1:1.8 in the assay medium. These mixtures were incubated at 37 °C for 30 min and then applied to 2 × 10 4 cells/well of Colo205 cells. Luminescent caspase-3/7 substrates were added to each well after an additional incubation at 37 °C for 2 h, and then the plate was incubated for 1–2 h in a shaking incubator at room temperature. Luminescence, which is proportional to the amount of caspase activity present, was measured in a plate-reading luminometer. The biological activity of the test samples was determined by comparing test sample response to reference standard response.
IgG1 Antibody Hinge Region Structure Analysis
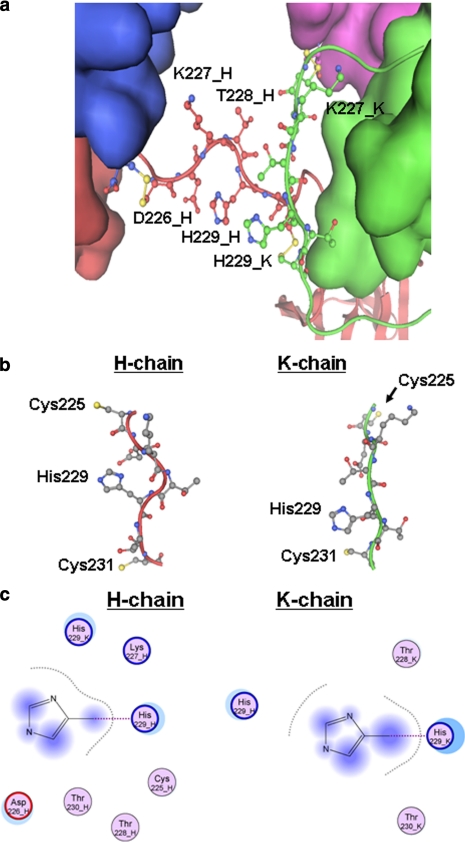
To examine the hinge structure of an IgG1 antibody, we used the human IgG1 b12 antibody crystal structure (Ref. 1; Protein Data Bank code 1HZH), which was determined at 2.7 Å resolution and had clear electron density for the hinge region except for the Thr228-His229-Thr230 peptide in one of the heavy chains (chain K in Fig. 1). The peptide (THT) that was missing the coordinates in the deposited coordinate file was modeled into the b12 structure using molecular operating environment software based on a Homology Modeling technique (21). With the lack of a template for the THT peptide, a fragment library generated from high resolution structures was used to model the fragment according to the molecular operating environment instructions. In all, 10 possible models were constructed, and the final model was selected based on the Generalized Born/Volume Integral score (22). The modeled structure was further minimized using the CHARMM22 forcefield (23), and the final structure was used as a template for examining the hinge region. For modeling the H229S substitution, only the Asp226–Cys231 segment in both heavy chains were allowed to move; all other atoms were constrained. The amino acid sequence used in the structure analysis was based on the deposited coordinate file in the Protein Data Bank (code 1HZH). It must also be noted that in this crystal structure, Cys234-H in the heavy chain does not form a disulfide bond with its partner (Cys234-K) in the adjacent heavy chain. However, this is unlikely to affect our analysis because we are interested in the upper hinge region only.
FIGURE 1.
IgG1 antibody hinge region structure analysis. a, a close-up view of the hinge region of IgG1b12 (Protein Data Bank code 1HZH). Thr228-His-Thr in the K chain was modeled into the structure, and the imidazole ring in the K-HC was only shown in a position close to the H-HC. An opposite position is possible but is not shown here. b, the local structure of each upper hinge including Cys231, only Cys (in yellow) and His are labeled. c, structural environments of the imidazole ring. The residues within a distance of 5 Å from the ring are shown here for the H chain and the K chain. The atoms in shadow are those exposed to the solvent.
RESULTS
Design and Assessment of the Hinge Variants
To explore the role of His229 in the radical mediated mechanism, we first examined its structural environment to determine whether any structural basis exists that may suggest whether the hinge cleavage is specific to one hinge over the other. We examined the local structure around the upper hinge region of the only available full-length human IgG1 structure (1, 2). This b12 IgG1 structure, however, lacks three upper hinge residues (228THT) in one chain because of the lack of traceable electron density. This disallowed a direct structural comparison between the two chains. Therefore, the three missing residues were modeled into the hinge region of the IgG structure using the homology modeling technique as described under “Experimental Procedures” (Fig. 1a). The region spanning the upper hinge along with Cys231 is shown in Fig. 1b. The residues surrounding the His229 side chain within a distance of 5 Å are illustrated in Fig. 1c. These examinations reveal that the local structure around 226DKTHT differs significantly between the two chains of the hinge.
The upper hinge residues (226DKTHT) in one of the heavy chains (H chain in Protein Data Bank entry 1HZH) are spatially proximal to its His229 imidazole ring, whereas in the other heavy chain (K chain) only the 228THT residues are within 5 Å of its His229 imidazole ring. It should be noted that the modeled imidazole ring in the K chain can be oriented differently, thus the local environments for each ring shown in Fig. 1c represent the extreme situation in which the two His229 side chains are in proximity. Interestingly, Cys225 in the H heavy chain (HC) that is in the interchain disulfide bond connecting the light chain (LC) and HC (HC-LC disulfide bond) is also proximal to the imidazole ring. In addition, Lys227 in the H heavy chain appears to be in a position to potentially interact with the imidazole ring. Based on these analyses, we decided to employ site-directed mutagenesis to substitute the two positively charged residues (His229 and Lys227) each with Gln, Ser, and Ala (Fig. 2). These modifications were predicted to maintain the flexibility and length of the hinge while reducing the size of the side chain with the Ser substitution, changing the charge state with the neutral charged Gln substitution, and abolishing the H-bonding capacity of the side chain with the nonpolar Ala substitution. In addition, a variant with the double mutations K227S and H229S was generated to further evaluate the combined effect of the His229 and Lys227 side chains.
FIGURE 2.

His and Lys variants generated for this study. Seven mutations were introduced into the upper hinge of the native sequence by site-directed mutagenesis using PCR by overlap extension followed by cloning into the heavy chain-encoding mammalian expression vector. The variants were transiently transfected and purified by a protein A column to a purity of greater than 95%. DNA sequencing and mass spectrometry analyses confirmed each mutation.
The mutations were confirmed by DNA sequencing and mass spectrometry analysis. A cell-based caspase bioassay indicated that all of the variants exhibited comparable potency levels at 94–102% of the native molecule (data not shown). The AlphaScreen assay was performed to measure the effect of the substitutions on Fcγ receptor binding, and the results showed a very similar function among these molecules at 91–105% of the native molecule when using Phe158 or Val158 Fcγ receptor variants (data not shown), suggesting that the substitutions did not affect Fcγ receptor function.
His229 Variants Inhibits the Hinge Cleavage
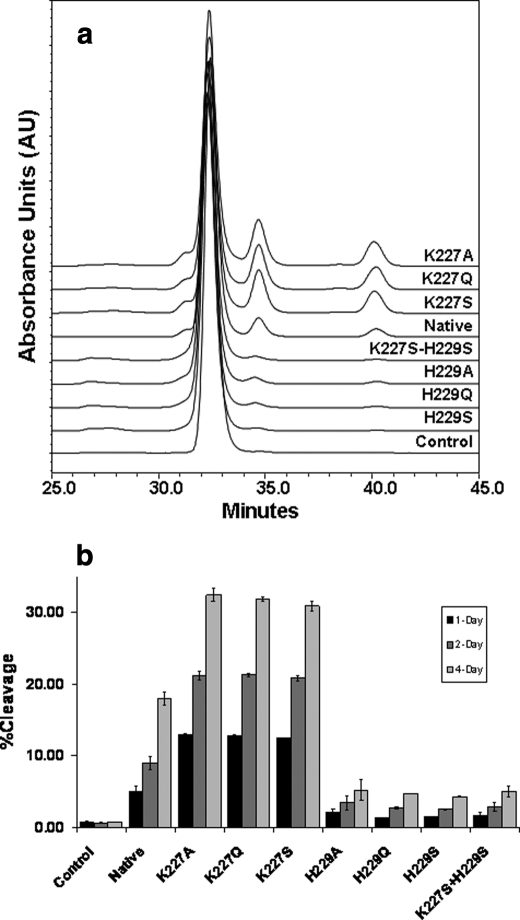
Hydroxyl radical-induced hinge cleavage generates only two products: the partial IgG1 missing a Fab and the Fab domain, both of which are detectable by SEC (9). The effect of the substitutions on the hinge cleavage was evaluated using the same approach. The cleavage products were monitored over time (1, 2, and 4 days) and reached a level of ∼18% for the native molecule after incubation with 20 mm H2O2 for 4 days at 25 °C. The His229 and Lys227 variants behaved very differently from each other (Fig. 3). The three His229 variants showed an ∼75–85% reduction in the hinge cleavage as compared with the native molecule (variant H229A at ∼5.5% cleavage, H229S at 3.5% cleavage, and H229Q at 3.7% cleavage). The three Lys227 variants resulted in an increased level of hinge cleavage to ∼71–85% (variant K227A at 32.6% cleavage, K227S at 30.8% cleavage, and K227Q at 31.6% cleavage). The critical role of His229 was demonstrated by examining the double mutant variant (H229S/K227S), which showed a 3.6% hinge cleavage, similar to that of the H229S single mutant. These results suggest the involvement of the two residues in the radical reaction and a key role for the His229 residue in the radical reaction mechanism.
FIGURE 3.
SEC measurements of the hinge cleavage of the native IgG1 and seven variants upon H2O2 incubation. a, SEC profiles of the variants after 4 days of H2O2 treatment. b, time-dependent response to the radical attack. The extent of the hinge cleavage was monitored at 220 nm and estimated by the percentage of the cleavage products to the intact molecule as a whole. The experimental conditions were as follows: IgG1 (1 mg/ml) in phosphate-buffered saline buffer, pH 7.4. The reaction was initiated by the addition of H2O2 to a final concentration of 20 mm, and the samples were incubated at 25 °C for 4 days. The reaction was stopped by catalase. Control is the native molecule incubated without H2O2.
RP-HPLC-TOF/MS Analysis of the Hinge Variants under Nonreducing Conditions
Redox chemistry is often involved in the ET pathway and could result in the formation of new redox products in the ET process (10, 15). In the hinge region of an IgG1, one of the two HC-LC disulfide bonds is proximal to the imidazole ring (Fig. 1c), highlighting the possibility of one of the His229 and Lys227 variants affecting the redox chemistry in the hinge region. To this end, we first examined the variants after 1 day of H2O2 treatment by RP-HPLC-TOF/MS under nonreducing conditions for any non-disulfide-bonded species.
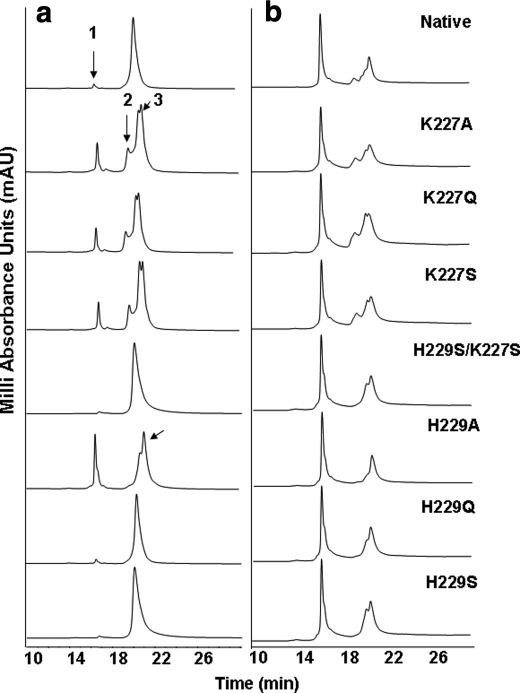
One species with a retention time of 16.5 min was observed in the products of the His229 and Lys227 variants and the native molecule (Fig. 4a). The total peak area for this species is <2% for the H229S and H229Q variants and 5% for the native molecule. Intriguingly, the peak area for the H229A variant was significantly higher at a level of ∼25%, 12 times more than the H229Q and H229S variants. The molecular mass of this species was determined to be 23,437.5 Da using in-line TOF/MS analysis, 48.5 Da heavier than the theoretical mass of 23,389.0 Da of the LC. This species from the H229A variant was separated using RP-HPLC and further characterized by the Lys-C peptide map. The result showed that only the C-terminal Cys215 residue in the LC was fully oxidized into sulfonic acid (+48 Da), and no other major oxidation site was observed (data not shown). These results indicate that the HC-LC disulfide bond was reduced, and Cys215 in the LC was further oxidized into Cys215-SO3H. Given a minimal difference in unpaired LC levels between the native molecule (∼3%) and the seven variants (1.5–2.0%) in untreated samples and that the only difference is an OH group in the side chain between Ala and Ser residue, the observation of 12 times more LC released from H229A than H229S suggests that the hydrogen bond interaction involving the imidazole ring is capable of maintaining the structural integrity in the upper hinge.
FIGURE 4.
RP-HPLC-TOF/MS analysis of native and the variants after 1-day (a) and 4-day (b) incubation with H2O2. Following incubation with H2O2, the samples were analyzed by RP-HPLC-TOF/MS under nonreducing conditions at 215 nm on a Zorbax SB-300 C8 column. Note that the His/Ala variant released a significant amount of LC (peak 1), and as a result, a partial IgG1 molecule (HHL) missing one LC was observed at a much greater level in the His/Ala variant compared with other His variants as indicated by an arrow. In the Lys variants, a partial form of the IgG1 missing the Fab domain (Fab-Fc, peak 2) and the Fab domain (peak 3) were observed.
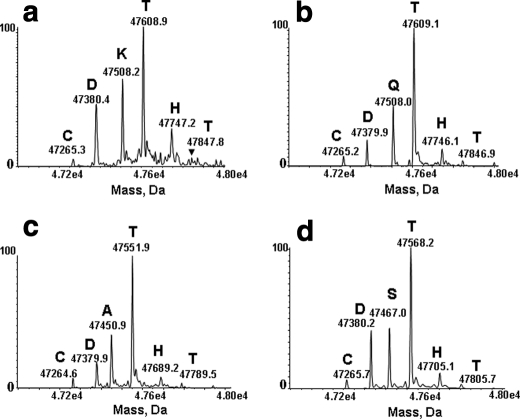
In contrast to the His229 variants, the Lys227 variants showed an increase in hinge cleavage (Fig. 4a). The products for each Lys227 variant include the oxidized LC (Peak 1) at ∼12% and a co-eluting partial IgG1 missing a Fab (Fab-Fc, Peak 2) and the Fab domain (Peak 3). TOF/MS analysis revealed heterogeneity in the C terminus of the Fab that featured a ladder of cleavage produced in the upper hinge residues of the native IgG1, whereas variants of the sequence of CDXTHT (where X = Q, S, or A for each variant, respectively) showed a similar cleavage pattern and ladder of cleavage as the native IgG1 (Fig. 5). The similarity of the C-terminal ladder between the native IgG1 and the Lys variants suggested the same mechanism drives the hinge cleavage. In addition, the H229S/K227S variant behaved in a very similar manner as compared with the H229S variant, suggesting that the imidazole ring of His229 plays a dominant role in the radical reaction mechanism, promoting cleavage. Similar cleavage profiles observed from the Lys variants suggested that the side chain of Lys227 may play an inhibitory role in the presence of the His229 residue.
FIGURE 5.
RP-HPLC-TOF/MS analysis of the Fab fragment released from the native IgG1 and the variants. Shown are the deconvoluted spectra of the Fab domain from the native molecule (a), and K227Q (b), K227A (c), and K227S (d) variants. For clarity, each major peak was labeled with the mass and the corresponding single amino acid code of the ending residue.
Additional information was obtained by examining the 4-day H2O2-treated samples (Fig. 4b). The oxidized LC was detected from all samples with a similar extent of 42–45%, differing from the 1-day results, suggesting that most of the HC-LC disulfide bonds were reduced after the exposure to the radical attack. With a typical mass ratio of LC and HC at ∼1:2, a non-disulfide-bonded LC level of >40% in all of the samples indicated a recovery issue with the HC-related species in the RP-HPLC. Davies and Delsignore (24) have demonstrated that radical attack could result in dramatic charge changes in a protein, which could alter hydrophobicity of some HC species, resulting in a recovery issue from RP-HPLC. This hypothesis was confirmed by CE-SDS analysis (see below).
The partial IgG1 (Fab-Fc) generated by 4 days of H2O2 treatment was resolved by RP-HPLC and was found to be heavily oxidized, attributed to the oxidation of five Met residues in the HC according to LC-MS/MS analysis (data not shown). However, TOF/MS analysis of each peak in the RP-HPLC profiles of the 4- and 1-day H2O2-treated samples showed that the species were identical in mass. The absence of any detectable level of the oxidized single chain of the HC observed suggests the interheavy chain disulfide bond between Cys234 residues in the core hinge (231CPPC234) remained intact.
CE-SDS Analysis
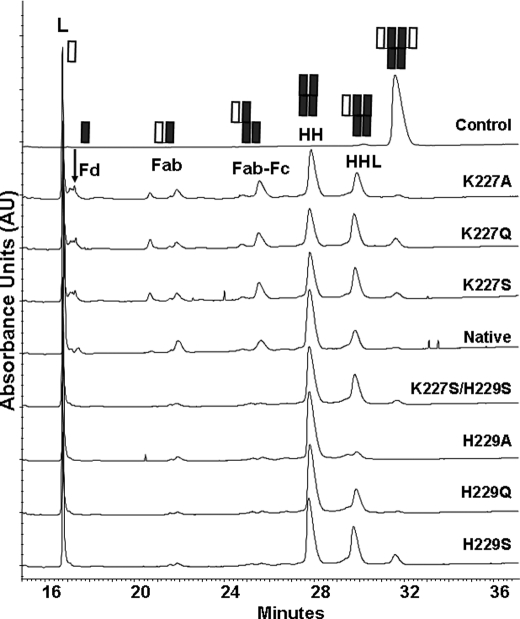
The presence of the non-disulfide-bonded LC and the absence of a single HC in the cleavage products after 4 days of H2O2 treatment demonstrated the unique nature of the radical-induced hinge cleavage, which was characterized by selective reduction of the interchain disulfide bonds. However, some unknowns remain because some HC was not recovered from the RP-HPLC, which might contain the single HC. To confirm the absence of the single HC chain in the products and assess the HC related species of the 4-day H2O2-treated samples, CE-SDS was employed to analyze these variants under nonreducing conditions (Fig. 6 and Table 1). The LC was detected from the native molecule and the seven variants that had a similar peak area of 30–32%; thus, intact IgG1 in each sample remained at a level of <5%, particularly for H229A, in which no intact IgG1 was detected. The partial molecules that were either missing one LC or both LCs were the main products and accounted for ∼46% of the products for the native molecule, 55–58% for the His229 variants, and 39–41% for the Lys227 variants. The fact that the LC and HC are at a normal level and that there was no detectable level of degraded HC in the CE-SDS confirmed our hypothesis of the HC recovery issue in RP-HPLC caused by the change in hydrophobicity not by the degradation of the HC.
FIGURE 6.
CE-SDS analysis of the native IgG1 and the variants after 4 days of H2O2 incubation. The control is the native molecule incubated without H2O2. The labels for the partial molecules are as follows: L, light chain; Fd, Fab portion of the heavy chain; Fab-Fc, partial molecule missing one Fab domain; HH, heavy chain dimer; HHL, partial molecule missing one light chain.
TABLE 1.
Summary of nonreducing CE-SDS results from the native and seven His229 and Lys227 variants, following 4-day H2O2 incubation
The component labels are as described for Fig. 6. The sum of the species is slightly less than 100% for most samples because of minimal levels of unclassified peaks with migration times of ∼18, 23, and 27 min in these samples. These species were not accounted for in the table because of their low amounts and are likely attributed to degradation products generated during sample preparation, as described by Michels et al. (38).
| Sample | L | Fd | Fab | Fab-Fc | HH | HHL | Whole |
|---|---|---|---|---|---|---|---|
| % | |||||||
| Control | 0.3 | 0.7 | 99.0 | ||||
| K227A | 26.9 | 8.2 | 7.6 | 12.2 | 23.3 | 15.5 | 2.0 |
| K227Q | 24.1 | 7.0 | 8.1 | 11.6 | 21.0 | 19.5 | 5.0 |
| K227S | 26.2 | 7.7 | 7.6 | 11.1 | 23.1 | 17.7 | 3.3 |
| Native | 29.7 | 4.8 | 7.6 | 8.7 | 33.6 | 12.1 | 1.2 |
| K227S/H229S | 28.5 | 1.9 | 2.7 | 4.3 | 35.0 | 21.4 | 3.0 |
| H229A | 32.5 | 2.2 | 3.3 | 4.0 | 46.1 | 9.1 | 0 |
| H229Q | 28.4 | 1.9 | 2.2 | 3.9 | 40.6 | 18.6 | 1.5 |
| H229S | 26.0 | 1.7 | 2.3 | 3.8 | 35.3 | 23.1 | 5.3 |
Other products from the hinge cleavage include the HC portion of Fab (Fd) (retention time at ∼17 min), two Fab species (∼20 and ∼21 min), and a partial IgG1 missing the Fab (Fab-Fc) (∼25 min). In contrast to the Lys227 variants that produced more cleavage products than the native molecule, the His229 variants generated very few cleavage products (Table 1). To confirm the two Fab species assignment, as opposed to a single HC and a Fab, the K227S variant was further characterized. Among the products, two major SDS-PAGE bands with a molecular mass of ∼50 kDa were separately subjected to N-terminal sequencing. The results revealed the N-terminal residues of the LC for both fragments, suggesting that the ∼50-kDa species contains more than one component and is not just LC or HC only species. These results confirm that the interchain disulfide bond between the HC-HC Cys234 residues remained intact. The lack of cleavage products from the His229 variants supports the key role of His229 in the radical reaction mechanism.
EDTA and DMPO Inhibit the Hinge Cleavage
We found that EDTA and DMPO are capable of blocking the hinge cleavage for all the variants. Only minimal degradation (<2%) was observed on SEC or RP-HPLC after these samples were treated with 20 mm H2O2 for 4 days at 25 °C in the presence of 50 mm EDTA or 10 mm DMPO (data not shown), similar to our previous observations (9).
DISCUSSION
The combined approaches of structure modeling and site-directed mutagenesis were employed in this study to explore the role of the hinge His229 in the radical-induced hinge cleavage of an IgG1. Our results indicated that the imidazole ring not only participates in the hydrogen bond that maintains the stability of the upper hinge region but also facilitates the radical reaction after localizing the electron transferred from the Cys231 radical. A typical ET, an energy-driven process (10, 25), usually does not result in the cleavage of a peptide bond. The rate of ET reduces exponentially with distance between the donor and acceptor (25). When encountering other nearby redox centers, the electron can tunnel from one center to another (12, 25) with bulk water playing a minor role in the ET reaction (26). The ET in the hinge of the IgG1 is localized because the upper hinge is framed by disulfide bonds at its N and C termini and lacks structurally proximal residues capable of forming a hydrogen bond network. The fact that EDTA and DMPO were capable of blocking cleavage suggests that the radical reaction mechanism is responsible for the hinge cleavage. The disulfide bonds that frame the upper hinge block the release of the elevated energy at a new radical site. Thus, we propose that the hinge cleavage and/or reduction of the interchain disulfide bond could be the outcome of an energy-driven process in the hinge region upon the radical attack.
The Role of the Hydrogen Bond
Reduction of the HC-LC disulfide bond suggested the involvement of redox chemistry in the ET process. The theoretical mass ratio of the LC and HC at ∼1:2 implies that a single LC accounts for 16–17% of the antibody mass (two LCs and two HCs). Because the RP-HPLC data shows a separate LC peak that is ∼25% of the whole antibody peak area after a 1-day H2O2 treatment of the H229A variant, both of the HC-LC disulfide bonds must have been attacked and reduced, suggesting a negative impact on the stability of the IgG1 from breaking the hydrogen bond involving His229.
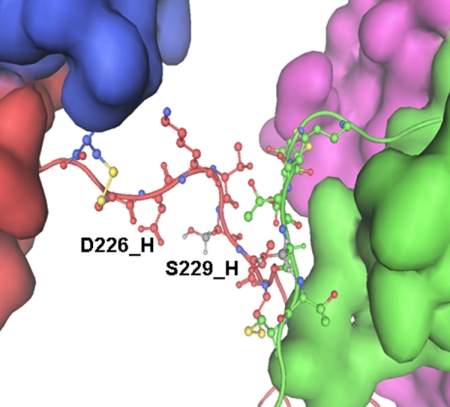
The distance of less than 3 Å between the side chains of His229 and Asp226 in IgG1 b12 allows the formation of a hydrogen bond. Structure modeling of H229S illustrated that the hydrogen bond could be maintained between Ser229 and Asp226, as shown in Fig. 7. Thus, the tunneling pathway for electron transport from the Cys231 radical remained largely intact with the imidazole ring being replaced by the Ser side chain. The distance between Cys231 and the HC-LC disulfide bond is ∼16 Å for one set and ∼17 Å for the other. Neither falls within the typical range of the tunneling limit for electron transfer in proteins, because Dutton and co-workers (12, 25) estimated an electron tunneling distance of 14 Å or less. Giese et al. (15) demonstrated that if the side chain of the central amino acid between the electron donor and acceptor contains an aliphatic group, single-step ET can occur, but with an oxidizable aromatic side chain as the central residue, a two-step hopping processes can take place. Because the rate of electron transfer decreases with distance (10, 12, 13), the electron transported from the Cys231 radical may not maintain enough energy to reduce the HC-LC disulfide bond in the H229S variant. Consequently, 12 times more LC released from the H229A variant suggests a new local conformation or conformational dynamic in the upper hinge region that brought the HC-LC disulfide bond more proximal to Cys231, allowing the reduction of the disulfide bond. Therefore, the initial resistance to the reduction of the HC-LC disulfide bond in the H229S and H229Q variants during 1-day of radical attack demonstrates that the hydrogen bond between the side chains of His229 and Asp226 is necessary to maintain the local hinge conformational integrity and the ability to resist radical induced degradation.
FIGURE 7.
The modeled structure of the His/Ser variant hinge for the IgG1b12. The distance between Ser229 and Asp226 was less than 3 Å in this structure, permitting formation of a hydrogen bond.
The Role of the His Residue in the Radical Reaction Mechanism
The electron chemistry is dominated by addition reactions with oxidizable rings, and the additional species formed are usually unstable and can rapidly undergo further reaction (27). In the case of the His residue, an electron could add to the C-2, C-4, or C-5 positions of the imidazole ring (28–30). In fact, the formation of the His radical at the C-5 position of the imidazole ring through electron transfer has been directly observed (29–31). Thus, we propose that the imidazole ring forms a new transient radical center that is capable of transferring the electron by extracting a proton from its neighboring residues, leading to hinge cleavage.
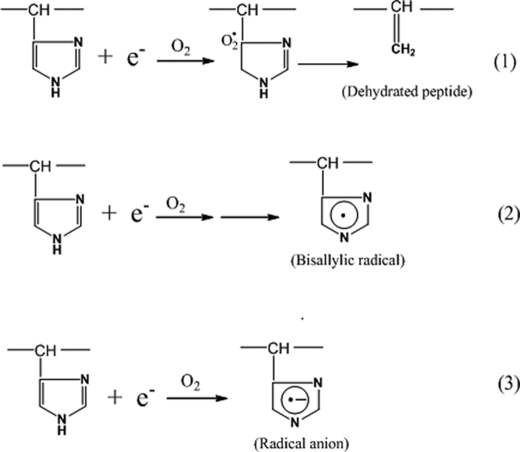
We consider three possible mechanisms for the role of the imidazole ring in the radical reaction, all of which could lead to hinge cleavage (Fig. 8): mechanism 1, electron attachment at the C-5 position of the imidazole ring in the presence of oxygen leads to the elimination of the ring, yielding a dehydropeptide that can be further degraded into the pyruvyl derivative (CH3-CO-CO-) (9); mechanism 2, the initial radical formed at the imidazole ring undergoes base-catalyzed loss of water to give rise to a stabilized bisallylic radical (32); and mechanism 3, the addition of the electron to the ring results in the formation of a transient radical anion, which can subsequently pick up a proton from its neighboring residues. The formation of the pyruvyl derivative (CH3-CO-CO-) supports mechanism 1 but contradicts the observed ladder of cleavage sites because this mechanism would yield only one cleavage site. Mechanism 2 explains the observations of the oxidative products of His into Asp, and the formation of the pyruvyl derivative (CH3-CO-CO-). However, the resultant stable radical of this mechanism is less capable of subtracting hydrogen from the neighboring residues, making it prone to degradation into a variety of products, including Asp, Asn, 2-Oxo-His, and pyruvyl derivatives (32–34).
FIGURE 8.
The three potential mechanisms of the imidazole ring functioning as the second radical center. Each of the three mechanisms was represented by a reaction between the imidazole ring and an electron. For clarity, only the imidazole ring and related intermediate radical/product are illustrated. More details for each mechanism are described in the text.
We propose that His229 forms a transient radical anion that is capable of picking up a proton from its neighboring residues (32), as shown in mechanism 3 in Fig. 8. The high degree of flexibility of the upper hinge allows greater conformational dynamics than in the rest of the IgG1 molecule (2) and facilitates the formation of the hydrogen bond involving the imidazole ring. The positive redox potential difference of 0.24 V between His (E° = 1.17V) and Cys (E° = 0.93V) (35) provides the driving force for the initial electron transfer from Cys231 to His229. The new radical center at His229 can initiate the redox chemistry to obtain a proton from its neighboring residue, leading to the radical induced cleavage of the hinge. It is possible that mechanism 1 or 2 represents intermediates in the later stages of the radical reaction.
We contend that the ET process in the hinge follows the hopping mechanism that was described previously (15). It is difficult to determine which of the two hinge Cys231 residues forms the radical, whereas once the Cys231 radical is formed the electron transfers, localizes at His229, and forms a new radical center. The fact that only one of the two chains of the hinge was cleaved suggests the coupling of one thiyl radical with one imidazole ring, and the chain being cleaved would be determined by the thiyl radical on the same chain. Although Thr230 precedes His229 in the sequence, His229 forms the radical center because it is spatially proximal to Cys231 as indicated in Fig. 1b and has a reaction rate constant of 6.4 × 107 m−1 s−1, faster than Ser, Thr, Gln, Ala, and Lys residues that have rate constants in a range of 1.0–2.0 × 107 m−1 s−1 at neutral pH (32). With the new radical center formed, the spatial proximity of the upper hinge residues to the imidazole ring determined the site and yield of the cleavage. In other words, the closer other upper hinge residues are to the imidazole ring, the more cleavage sites that are possible. In the case of the two His229 residues in proximity to each other, the nonradical His229 could be a proton donor. As illustrated in Fig. 1c, more cleavage occurring at Thr228 and Thr230 than Asp226 and Lys227 would be expected as the outcome, and indeed such is the observation in our previous study (9).
Possible Role of Lys Residue
Lys has a long side chain with a unique C-5 position (δ carbon) as compared with Ser, Gln, Ala, and His residues. It is known that the C-4 and C-5 positions in Lys are the major sites for electron attachment (36). Therefore, we explored the possible role of the C-5 position in mediating the hinge cleavage in the presence of His229. The three Lys227 variants showed a similar increase in the hinge cleavage, suggesting that the C-4 position did not play a role in the reaction because a Gln C-4 position is also capable of localizing an electron (32). As shown in Fig. 1c, Lys227 is proximal to the imidazole ring, so it is possible that the C-5 carbon of the Lys side chain plays a role in the reaction mechanism by influencing the protonation state of the imidazole ring or the electron transfer rate. Thus, the Lys227 variants removed these local constrains and facilitated the radical reaction.
In summary, our results indicate that the substitution of the hinge His229 residue with a polar residue (Ser or Gln) blocked the radical-mediated hinge cleavage. The variants were more stable than the native molecule and maintain the in vitro biological functions of the IgG1 (e.g. FcγR binding and caspase-3 assay). Although for some proteins, the presence of an oxidizable side chain in the tunneling pathway is the requirement for an increasing ET rate that enhances functional efficiency (37), in the case of IgG1, the presence of His229 facilitates the radical-mediated hinge cleavage. This suggests that the IgG1 upper hinge may not be optimized to prevent hinge cleavage. Our results indicate that it is feasible to design a new IgG1 with a hinge that is more resistant to radical induced degradation while maintaining its biological activity. Further characterization of a modified upper hinge and the potential impact to therapeutic applications would provide greater insight.
Acknowledgments
We are grateful to Drs. Michael Treuheit and Gerd Kleemann for critical comments on this manuscript and to Drs. Wei Yan and Michael Wittekind for support.
Footnotes
- ET
- electron transfer
- SEC
- size exclusion chromatography
- DMPO
- 5,5-dimethyl-1-pyrroline N-oxide
- RP-HPLC-TOF/MS
- reversed phase chromatography and time-of-flight mass spectrometry
- CE-SDS
- capillary electrophoresis in SDS
- HC
- heavy chain
- LC
- light chain.
REFERENCES
- 1.Stanfield R. L., Fieser T. M., Lerner R. A., Wilson I. A. (1990) Science 248, 712–719 [DOI] [PubMed] [Google Scholar]
- 2.Saphire E. O., Stanfield R. L., Crispin M. D., Parren P. W., Rudd P. M., Dwek R. A., Burton D. R., Wilson I. A. (2002) J. Mol. Biol. 319, 9–18 [DOI] [PubMed] [Google Scholar]
- 3.Coloma M. J., Trinh K. R., Wims L. A., Morrison S. L. (1997) J. Immunol. 158, 733–740 [PubMed] [Google Scholar]
- 4.Oi V. T., Vuong T. M., Hardy R., Reidler J., Dangle J., Herzenberg L. A., Stryer L. (1984) Nature 307, 136–140 [DOI] [PubMed] [Google Scholar]
- 5.Tan L. K., Shopes R. J., Oi V. T., Morrison S. L. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 162–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brekke O. H., Michaelsen T. E., Sandlie I. (1995) Immunol. Today 16, 85–90 [DOI] [PubMed] [Google Scholar]
- 7.Dall'Acqua W. F., Cook K. E., Damschroder M. M., Woods R. M., Wu H. (2006) J. Immunol. 177, 1129–1138 [DOI] [PubMed] [Google Scholar]
- 8.Cohen S. L., Price C., Vlasak J. (2007) J. Am. Chem. Soc. 129, 6976–6977 [DOI] [PubMed] [Google Scholar]
- 9.Yan B., Yates Z., Balland A., Kleemann G. R. (2009) J. Biol. Chem. 284, 35390–35402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reece S. Y., Nocera D. G. (2009) Annu. Rev. Biochem. 78, 673–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reece S. Y., Hodgkiss J. M., Stubbe J., Nocera D. G. (2006) Philos. Trans. R. Soc. Lond. B Biol. Sci. 361, 1351–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Page C. C., Moser C. C., Dutton P. L. (2003) Curr. Opin Chem. Biol. 7, 551–556 [DOI] [PubMed] [Google Scholar]
- 13.Gray H. B., Winkler J. R. (2003) Q. Rev. Biophys. 36, 341–372 [DOI] [PubMed] [Google Scholar]
- 14.Gray H. B., Winkler J. R. (1996) Annu. Rev. Biochem. 65, 537–561 [DOI] [PubMed] [Google Scholar]
- 15.Giese B., Graber M., Cordes M. (2008) Curr. Opin. Chem. Biol. 12, 755–759 [DOI] [PubMed] [Google Scholar]
- 16.Dean R. T., Wolff S. P., McElligott M. A. (1989) Free Radic. Res. Commun. 7, 97–103 [DOI] [PubMed] [Google Scholar]
- 17.Rodgers M. A., Sokol H. A., Garrison W. M. (1970) Biochem. Biophys. Res. Commun 40, 622–627 [DOI] [PubMed] [Google Scholar]
- 18.Uchida K., Stadtman E. R. (1993) J. Biol. Chem. 268, 6388–6393 [PubMed] [Google Scholar]
- 19.Shukla A. A., Hubbard B., Tressel T., Guhan S., Low D. (2007) J. Chromatogr. B 848, 28–39 [DOI] [PubMed] [Google Scholar]
- 20.Lazar G. A., Dang W., Karki S., Vafa O., Peng J. S., Hyun L., Chan C., Chung H. S., Eivazi A., Yoder S. C., Vielmetter J., Carmichael D. F., Hayes R. J., Dahiyat B. I. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 4005–4010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sali A., Blundell T. L. (1993) J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 22.Labute P. (2008) J. Comput. Chem. 29, 1693–1698 [DOI] [PubMed] [Google Scholar]
- 23.Mackerell A. D., Jr., Feig M., Brooks C. L., 3rd (2004) J. Comput. Chem. 25, 1400–1415 [DOI] [PubMed] [Google Scholar]
- 24.Davies K. J., Delsignore M. E. (1987) J. Biol. Chem. 262, 9908–9913 [PubMed] [Google Scholar]
- 25.Page C. C., Moser C. C., Chen X., Dutton P. L. (1999) Nature 402, 47–52 [DOI] [PubMed] [Google Scholar]
- 26.Crane B. R., Di Bilio A. J., Winkler J. R., Gray H. B. (2001) J. Am. Chem. Soc. 123, 11623–11631 [DOI] [PubMed] [Google Scholar]
- 27.Hart E. J. (1964) Science 146, 19–25 [DOI] [PubMed] [Google Scholar]
- 28.Lassmann G., Erikson L. A., Himo F., Lendzian F., Lubitz W. (1999) J. Phys. Chem. A 103, 1283–1290 [Google Scholar]
- 29.Lassmann G., Eriksson L. A., Lendzian F., Lubitz W. (2000) J. Phys. Chem. A 104, 9144–9152 [Google Scholar]
- 30.Rao P. S., Simic M., Hayon E. (1975) J. Phys. Chem. 79, 1260–1263 [Google Scholar]
- 31.Reece S. Y., Seyedsayamdost M. R., Stubbe J., Nocera D. G. (2007) J. Am. Chem. Soc. 129, 8500–8509 [DOI] [PubMed] [Google Scholar]
- 32.Davies M. J., Dean R. T. (1997) Radical Mediated Protein Oxidation, pp. 50–120, Oxford University Press [Google Scholar]
- 33.Garrison W. M. (1987) Chem. Rev. 87, 381–398 [Google Scholar]
- 34.Stadtman E. R., Levine R. L. (2003) Amino Acids 25, 207–218 [DOI] [PubMed] [Google Scholar]
- 35.Navaratnam S. P. (1998) J. Chem. Soc. Faraday Trans. 94, 2577–2581 [Google Scholar]
- 36.Hawkins C. L., Davies M. J. (1997) Biochim. Biophys. Acta 1360, 84–96 [DOI] [PubMed] [Google Scholar]
- 37.Shih C., Museth A. K., Abrahamsson M., Blanco-Rodriguez A. M., Di Bilio A. J., Sudhamsu J., Crane B. R., Ronayne K. L., Towrie M., Vlcek A., Jr., Richards J. H., Winkler J. R., Gray H. B. (2008) Science 320, 1760–1762 [DOI] [PubMed] [Google Scholar]
- 38.Michels D. A., Brady L. J., Guo A., Balland A. (2007) Anal. Chem. 79, 5963–5971 [DOI] [PubMed] [Google Scholar]