Abstract
We here identify protein kinase D (PKD) as an upstream regulator of the F-actin-binding protein cortactin and the Arp actin polymerization machinery. PKD phosphorylates cortactin in vitro and in vivo at serine 298 thereby generating a 14-3-3 binding motif. In vitro, a phosphorylation-deficient cortactin-S298A protein accelerated VCA-Arp-cortactin-mediated synergistic actin polymerization and showed reduced F-actin binding, indicative of enhanced turnover of nucleation complexes. In vivo, cortactin co-localized with the nucleation promoting factor WAVE2, essential for lamellipodia extension, in the actin polymerization zone in Heregulin-treated MCF-7 cells. Using a 3-dye FRET-based approach we further demonstrate that WAVE2-Arp and cortactin prominently interact at these structures. Accordingly, cortactin-S298A significantly enhanced lamellipodia extension and directed cell migration. Our data thus unravel a previously unrecognized mechanism by which PKD controls cancer cell motility.
Keywords: Actin, Cell Migration, Cell Motility, Protein Kinases, Protein Phosphorylation, Arp, WAVE, Cortactin
Introduction
The mechanistic elucidation of signaling pathways regulating dynamic actin remodeling processes in migrating cells is pivotal to a comprehensive understanding of cancer cell metastasis. Protein kinase D (PKD)2 has recently been identified as a vital upstream regulator of polarized cell motility and F-actin organization (1–4). PKD localizes to sites of dynamic actin remodeling (1). The kinase activity is essential for the control of directed cell motility (1, 2), whereby active PKD1 inhibited, whereas kinase-inactive PKD1KD strongly enhanced motility and invasiveness (1–4). Mechanistically, a key role for PKD1 in controlling the activity of the ubiquitous F-actin depolymerizing- and severing factor cofilin via slingshot1L (SSH1L) cofilin phosphatase has been demonstrated. The activity of SSH1L is mainly regulated by its binding to filamentous actin (F-actin), which has been shown to strongly enhance its activity (5, 6). Phosphorylation of SSH1L at Ser978 by active PKD1, e.g. downstream of RhoA or oxidative stress, generates a 14-3-3 binding motif within an important F-actin binding region, thus resulting in the sequestration of SSH1L away from dynamic actin structures, reducing SSH1 activity and active non-S3-phosphorylated cofilin levels (2). By severing actin filaments, cofilin increases both the availability of G-actin monomers as well as the number of “barbed ends” for polymerization (7). Furthermore, severed filaments are the preferred substrate for dendritic nucleation by the Arp complex (8, 9). Localized induction of actin polymerization and the formation of branched actin networks constitute the basis for membrane protrusion and cell motility (2, 10, 11). In line with an upstream regulatory role in the control of these processes, PKD1 and -2 are also capable of binding to F-actin in vitro (1). In the case of PKD1, in vivo F-actin binding has been demonstrated as well, which most likely facilitates an interaction with actin regulatory proteins such as SSH1L (2). We now have identified a second key regulatory signaling pathway of PKD controlling the WAVE2-Arp2/3-complex-driven de novo actin polymerization and lamellipodia extension via the actin-regulatory protein cortactin. Cortactin is an actin-binding protein enriched in lamellipodia of motile cells and at dynamic actin structures such as membrane ruffles (12), circular dorsal ruffles (13), as well as invadopodia of invasive cancer cells (14). Cortactin was further shown to co-localize with proteins of the Arp2/3 complex at sites of actin polymerization within lamellipodia (12). It synergistically accelerates Arp complex-mediated actin polymerization (15) and has been shown to promote cell migration by enhancing lamellipodia persistence (16). We have thus investigated a role of PKD as an upstream regulator of cortactin and its function in actin organization as well as cell migration.
EXPERIMENTAL PROCEDURES
Cell Culture
Panc89 (PDAC) cells, MCF-7 cells, and HEK293T cells were maintained in RPMI supplemented with 10% fetal calf serum and penicillin/streptomycin (1:100). For microscopy, in the case of Panc89 cell lines, cells were seeded on coverslips coated with CollagenIV (Sigma), whereas MCF-7 cells were seeded on uncoated slips in sterile dishes (Barloworld Scientific) at 80,000 cells/coverslip. Time-lapse live imaging of MCF-7 cells was performed in 4-chamber Cellview glass-bottom dishes (Greiner Bio-one). Panc89 and MCF-7 cells were transfected with Lipofectamine 2000 (Invitrogen), and HEK293T cells using TransIT293T reagent (Mirus). For Heregulin stimulation cells were serum-starved overnight and then stimulated with 100 ng/ml of human Heregulin1 (PeproTech) for the indicated time points.
Plasmids, Antibodies, and Dye Reagents
GFP-tagged expression constructs for PKD1 and PKD1KD (K612W) have been described previously (1, 17). pEGFP-N1-cortactin and pCR3.V62Met-FLAG-cortactin constructs have been generated by inserting human cortactin transcript variant 1 (NM_005231) amplified from Panc89 cDNA via EcoRI/ApaI in respective vectors. Ser298 was mutated to an alanine residue by site-directed mutagenesis (Stratagene). The human pEGFP-C3-WAVE2-GFP construct was a gift from K. Rottner (Helmholtz Centre for Infection research, Braunschweig, Germany). pSuper PKD1 and PKD2 constructs have been described previously (2). LacZ (control) and PKD1/2 siRNAs have been described in Ref. 4. Cortactin was detected using cortactin-specific antibodies (H-191, Santa Cruz Biotechnology, BD Bioscience). Arp3 was probed with anti-Arp3 (BD Bioscience). PKD was probed with anti-pPKDS910 specific antibody (as described in Ref. 1) and PKD C20 (Santa Cruz Biotechnology). Anti-GFP specific antibody was purchased from Roche Applied Science and anti-FLAG-M2 and anti-actin antibodies were from Sigma. Anti-tubulin was acquired from NeoMarkers (CA). Rhodamine-phalloidin, Alexa 546-phalloidin, and Alexa 633-phalloidin dyes were acquired from Invitrogen.
Total Cell Lysates, GST Pulldown Assays, Co-immunoprecipitation, and PKD in Vitro Kinase Assays
GST 14-3-3β pulldown experiments, co-immunoprecipitation and in vitro kinase assays were performed as described previously (1, 18). Lysates were clarified by centrifugation at 13,000 × g for 10 min. Pulldown assays were performed by incubation of protein lysates with 10 μg of GST 14-3-3β-coupled to glutathione-Sepharose beads for 2 h at 4 °C. Beads were washed three times with lysis buffer. For immunoprecipitation equal amounts of proteins were incubated with specific antibodies for 1.5 h at 4 °C. Immune complexes were collected with protein G-Sepharose (GE Healthcare) and washed three times with lysis buffer. Precipitated proteins were released by boiling in sample buffer and subjected to SDS-PAGE. The proteins were blotted onto nitrocellulose membranes (Pall, Germany). Proteins were visualized with alkaline phosphatase-coupled secondary antibodies using nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate as substrate or horseradish peroxidase-coupled secondary antibodies using ECL. In vitro kinase assays were performed by incubating equal amounts of FLAG-M2 (Sigma) affinity purified cortactin and cortactin-S298A as substrate with ∼250 ng of purified PKD1 from Baculo-infected insect cells or purified His-tagged PKD1KD (kindly provided by A. Toker, Harvard). Kinase reactions were performed for 15 min at 37 °C in 30 μl of kinase buffer (50 mm Tris, pH 7.4, 10 mm MgCl2, and 2 mm dithiothreitol) containing 2 μCi of [α-32P]ATP. To terminate the reaction, 10 μl of 5× SDS sample buffer were added. Samples were resolved by SDS-PAGE, blotted onto nitrocellulose, and analyzed on a PhosphorImager (GE Healthcare). Quantification was done with ImageQuant software (GE Healthcare).
FLAG-M2 Affinity Purification of Cortactin and High-speed F-actin Binding Assays
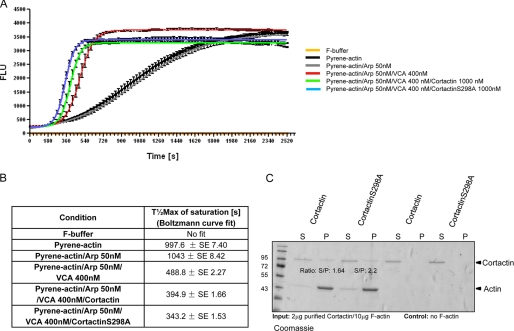
High-speed F-actin binding assays were essentially performed as described (1). FLAG-M2 affinity purification of cortactin and cortactin-S298A was performed from five 10-cm culture dishes. M2 affinity purification was performed with 100 μl of M2-agarose (Sigma) according to the manufacturer's description. Bound FLAG-cortactin was eluded after extensive washing steps with 3× 200 μl of 100 mm glycine (pH 3.5) in reaction tubes containing 1.5 m Tris (pH 8.8) to buffer the eluate. FLAG-cortactin/FLAG-cortactin-S298A concentrations were subsequently assessed by Bradford assay and purity was tested by SDS-PAGE. High-speed binding assays were done by incubating 2 μg/0.5 μg of cortactin and cortactin-S298A with 10 μg of in vitro polymerized F-actin (AKL99, Cytoskeleton Inc.) for 30 min at room temperature in F-buffer (10 mm imidazole, pH 7.2, 75 mm KCl, 5 mm MgCl2, 0.5 mm dithiothreitol, and 1 mm EGTA supplemented with Complete protease inhibitor (EDTA free) and PhosStop (Roche Applied Science)). Assays were subjected to centrifugation at 100,000 × g for 1 h at room temperature. The supernatant was removed and the pellet was incubated for 1 h in G-buffer (5 mm Tris-HCl, pH 8, 0.5 mm dithiothreitol, 0.2 mm CaCl2, and 0.2 mm ATP) to depolymerize F-actin. Equal amounts of supernatant and pellet fraction were analyzed by SDS-PAGE followed by Coomassie staining or Western blot. Integrated density of bands was quantified using NIH ImageJ.
In Vitro Pyrene-Actin Polymerization Assays
Pyrene-actin polymerization assays with purified proteins were performed as follows. Pyrene-labeled rabbit muscle G-actin (Cytoskeleton Inc.) was diluted to 3 μm in G-buffer (5 mm Tris-HCl, pH 8, 0.2 mm CaCl2, and 0.2 mm fresh ATP). In vitro polymerization assays were performed in black 96-well plates (Greiner Bio-one) in a volume of 100 μl. Triplicate wells per reaction condition were assayed: G-buffer; pyrene-actin; pyrene-actin, 50 nm Arp complex (Cytoskeleton Inc.); and pyrene-actin, 50 nm Arp, 400 nm GST-VCA (Cytoskeleton Inc.), as well as pyrene-actin, 50 nm Arp, 400 nm GST-VCA, and 1000 nm FLAG-purified cortactin or cortactin-S298A. Polymerization reactions were started immediately prior to measurement by the addition of 10 μl of 10× F-buffer (Cytoskeleton Inc.). The increase in pyrene fluorescence following polymerization was measured with a Tecan M200 plate reader: excitation, 360 nm, emission, 407 nm every 30 s. To quantify changes in polymerization rate, curves were fitted with Boltzmann sigmoidal equations (GraphPad, Prism). Half-maximal saturated polymerization values (T½max (s)) and % saturation of individual polymerization reactions were calculated from RAW data.
Migration Assays
Migration assays were performed using 8-μm pore diameter 24-well Transwell filter inserts (Corning). Panc89 cells were seeded at a density of 60,000 cells/insert. Migration was induced by a serum gradient of 10% fetal calf serum for 40 h. The inside of the Transwell insert was wiped with a cotton swab to remove non-migrated cells. Migrated cells were fixed using 4% paraformaldehyde stained with 4′,6-diamidino-2-phenylindole and visualized using a wide field fluorescence microscope (Leica) equipped with a CCD camera, and a monochromator light source at ×10 magnification. Results were calculated as the average number of migrated cells/visual field with at least 6 images per filter.
In Vitro Branching Assays
3 μm seed-free G-actin was polymerized in the presence of an equimolar amount Alexa 488-phalloidin in G-buffer to stabilize filaments. Reaction conditions were: 50 nm Arp complex (Cytoskeleton Inc.), 400 nm GST-VCA (Cytoskeleton Inc.), and 1000 nm FLAG-purified cortactin or cortactin-S298A. Polymerization reactions were started by the addition of ×10 F-buffer (Cytoskeleton Inc.). Reactions were terminated after 10 min by the addition of formaldehyde to a final concentration of 4% for 15 min. Samples were quenched with glycine and mounted as 1:10 dilution in Fluoromount G. Filaments were visualized at ×100 magnification by a Leica wide field fluorescence microscope using 510/30 filters. To enhance filament detection, 5 images each (cortactin/cortactin-S298A) were brightness and contrast enhanced, SPE-processed, segmented, and subjected to automatic detection by a 20-infinity filter (NIH ImageJ) after highly branched filaments with >2 branches were singled out manually and excluded from automatic detection. Quantified was the ratio of the total number of filaments (5 images)/filaments with branches >2 and % of branched filaments.
Immunohistochemistry and Confocal Microscopy
Panc89 and MCF-7 cells were fixed with 4% paraformaldehyde at room temperature for 20 min, washed, quenched with 0.1 m glycine, and then permeabilized with 0.1% Triton X-100. Samples were blocked and stained in phosphate-buffered saline supplemented with 5% fetal calf serum, 0.05% Tween 20. Primary as well as secondary Alexa dye antibodies (Invitrogen) were incubated for 2 h. Samples were mounted after extensive washing in Fluoromount G (Southern Biotechnology, AL) and analyzed by a Confocal Laser Scanning Microscope (TCS SP2, Leica, Germany) or a LSM 710 (Zeiss, Jena, Germany) equipped with the respective ×63 Plan Apo oil immersion objectives. GFP or Alexa 488 were excited with the 488-nm line and fluorescence was detected at 500–535 nm. Alexa 546 and Cy5 or Alexa 633 were excited with the 543 (Leica), 561 (Zeiss), and 633 nm line for Cy5/Alexa 633, respectively. Dyes were detected at 550–620 and 640–720 nm. Images were acquired in sequential scan mode and processed using NIH ImageJ.
Acceptor-photobleach FRET Experiments
Acceptor-photobleach FRET experiments were performed with MCF-7 cells stimulated with Heregulin. In brief, MCF-7 cells seeded on coverslips were transfected, serum-starved overnight, and stimulated with 100 ng/ml of Heregulin for 10 min. Samples were fixed with 4% paraformaldehyde and subjected to immunohistochemistry. FRET measurements were performed by acquiring a time-bleach image series using a LSM 710 confocal laser scanning microscope (Zeiss) equipped with a ×63 Plan Apo oil immersion objective. In experiments with Arp3-Alexa 546 as FRET acceptor, fluorescence of the acceptor was bleached using an intensive 561-nm laser line. For 3-dye “daisy chain” acceptor-photobleach-FRET experiments with WAVE2-GFP as the primary FRET donor, FLAG-cortactin stained with α-FLAG M2-Alexa 633 secondary antibody as the final FRET-acceptor, and F-actin (phalloidin-Alexa 546) as the FRET signal “transducer,” the final acceptor cortactin-FLAG-Alexa 633 was bleached with the 633 laser line. %FRET images were generated with ZEN Software (LSM 710 confocal microscope, Zeiss) using images immediately before and after the bleach. The Bezier bleach ROI and rectangle threshold ROI used to eliminate “off target-bleached pixels” are shown in figures. For the 3-dye daisy chain FRET experiments, RAW data and RAW %FRET values were also calculated by the mean of ROI analysis using ZEN Software.
Kymograph Analysis of Extending Lamellipodia Visualized by YFP-actin in MCF-7 Cells
Low level GFP-G-actin expression can be used to measure actin polymerization in vivo (19). We therefore used YFP-G-actin co-transfection to visualize actin polymerization and extension of lamellipodia in MCF-7 cells. For these experiments cells were seeded at a density of 80,000 cells/well in 4-chamber Cellview glass-bottom dishes. Cells were transfected at a ratio of 3:1 with FLAG-vector/FLAG-cortactin/FLAG-cortactin-S298A:YFP-G-actin to visualize polymerization, serum-starved overnight, and stimulated at T120 s after initiation of image acquisition with 100 ng/ml of Heregulin. Time-lapse videos of stimulated cells were generated by acquiring images of YFP fluorescence (excitation YFP, 500/20; emission, YFP, 535/30) every 10 s for 20 min at ×63 magnification with a Leica wide field fluorescence video microscope setup for live cell imaging with a 37 °C heating chamber as well as CO2 supply. Subsequently kymographs were generated with NIH ImageJ plug-ins by plotting the edge of the protruding lamellipodia visualized by YFP-G-actin against time (pixel/10 s) along a 1-pixel line perpendicular to the protrusion front for initially individual protrusions of 6 cells per construct (already showing statistical significance). The protrusion velocity (pixel/min) was calculated from kymographs during the linear phase of extension indicated by white lines in the kymogrames with a respective time difference of 1 min. Using the spatial calibration of images velocity was also calculated as [μm/min]. In line with literature (16), vector controls displayed reduced protrusion persistence when compared with cortactin and cortactin-S298A cells that did not allow for the same quantification and statistical analysis.
Statistic Analyses
Statistical analyses were performed using GraphPad Prism Software algorithms.
RESULTS
PKD Co-localizes and Interacts with Cortactin at Arp3-positive Structures
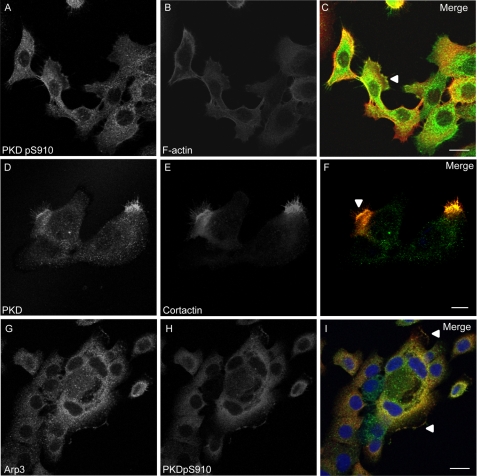
Using Panc89 cells derived from pancreatic ductal adenocarcinoma we have investigated the role of cortactin and the potential upstream regulatory function of PKD in the control of pancreatic cancer cell motility. Autophosphorylated endogenous PKD colocalized with F-actin at the edge of lamellipodia (Fig. 1, A–C) as well as with cortactin at lamellipodia, in membrane ruffles and at microspikes within the protrusion (Fig. 1, D–F). Autophosphorylated PKD also co-localized with Arp3 at the edge of lamellipodia and in ruffle zones (Fig. 1, G–I), pointing to a putative role of PKD together with cortactin in the Arp complex-mediated actin remodeling.
FIGURE 1.
Panels A–C, active autophosphorylated PKD co-localizes with F-actin at the edge of lamellipodia. Panels D and E, PKD co-localizes with cortactin in protruding lamellipodia-like structures. The co-localization of both proteins is enhanced at the edge of lamellipodia, in membrane ruffles and at microspikes within the protrusion. G and H, Arp3 and active autophosphorylated PKD (pS916) co-localize at the edge of protruding lamellipodia and in membrane ruffles. Panels A–I, depicted are confocal image sections of Panc89 cells. Relevant structures are marked with arrowheads. The scale bar represents 10 μm.
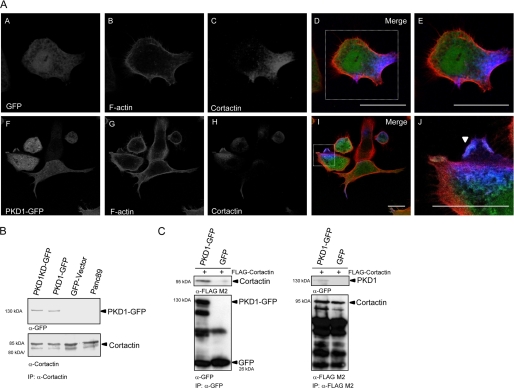
We next used Panc89 cells stably expressing GFP-tagged wild type PKD1 and kinase-inactive PKD1KD (1) to identify a potential function of PKD and cortactin in the Arp complex-mediated de novo actin polymerization as well as directed cell migration. As demonstrated for endogenous PKD, ectopically expressed PKD1-GFP showed enhanced co-localization with endogenous cortactin and F-actin at membrane ruffles of lamellipodia-like protrusions (Fig. 2A, panels A–J) in these cells. Moreover, both PKD1- and PKD1KD-GFP could be co-precipitated with endogenous cortactin (Fig. 2B). Reciprocal co-immunoprecipitation of ectopically expressed PKD1-GFP and FLAG-cortactin in HEK293T cells confirmed the specificity of this interaction (Fig. 2C).
FIGURE 2.
A, panels A–J, localization and interaction of PKD and cortactin. PKD1-GFP, F-actin, and cortactin co-localization is enhanced in membrane ruffles of lamellipodia-like protrusions. Confocal image sections of stable Panc89 cells expressing PKD1-GFP were stained for F-actin and cortactin. Relevant structures are marked with arrowheads. The scale bar represents 10 μm. B, co-immunoprecipitation (IP) of PKD1- and PKD1KD-GFP with endogenous cortactin from stable Panc89 cell lines. The characteristic p80/p85 double band of endogenous cortactin is shown in the lower blot panel. C, reciprocal co-immunoprecipitation PKD1-GFP and FLAG-cortactin from HEK293T cell lysates. GFP was used as specificity control. Ectopically expressed cortactin does not show explicit double bands.
PKD-mediated Phosphorylation of Cortactin at Ser298 Creates a 14-3-3 Binding Motif
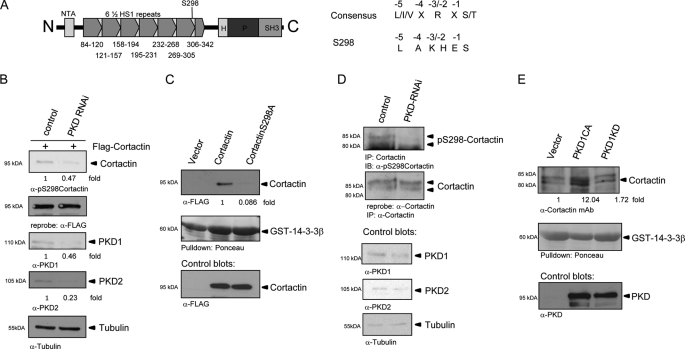
Because PKD and cortactin show overlapping subcellular localization and interaction in Panc89 as well as HEK293T cells, we were prompted to investigate whether cortactin is a direct substrate of PKD and whether respective phosphorylation sites are situated in important structural domains of the protein. Fig. 3A depicts a structural overview of cortactin. The N-terminal region of cortactin harbors an acidic motif (NTA) with DDW consensus sequences, which have been shown to mediate binding of cortactin to Arp3, a member of the 7-protein Arp complex (20). Cortactin also possesses an extended region of 6½ so-called HS-1 repeats containing dedicated sequence motifs for F-actin interaction. The C-terminal part of the protein harbors a helical domain (H), a proline-rich stretch of amino acids, and an SH3-binding motif (21) for recruitment of co-regulatory proteins such as WIP or WISH (21, 22). Following sequence analysis for PKD consensus phosphorylation motifs, we decided to focus our investigation on Ser298 as a potentially important PKD phosphorylation site with the consensus LAKHES* located in the 6th F-actin binding repeat. As shown in Fig. 3A, the cortactin consensus does not fully match the published PKD consensus sequence because there is no arginine at the −2/3 position (23). However, positively charged histidine and lysine residues at the −2/3 position might be able to compensate for the arginine. Recently, in vivo phosphorylation of this site by PKD has been published, however, neither the biological significance nor mechanistic consequences of this phosphorylation have been addressed in this study (24). We thus opted to investigate the biological relevance of this site because it has been demonstrated for a number of F-actin-binding proteins that phosphorylation alters the F-actin binding capacity impacting its specific function (2, 5, 25). The phosphorylation of respective consensus sites in some instances also generates 14-3-3 protein binding motifs impacting protein function and biological readouts (2, 18, 23, 26). Using purified PKD1 and FLAG affinity purified cortactin/cortactin-S298A proteins isolated from HEK293T cells, we show here that PKD1 phosphorylates Ser298 in cortactin in vitro (supplemental Fig. S1A). We, furthermore, demonstrate a PKD-dependent in vivo phosphorylation at this site using a site-specific pS298 antibody (24) following PKD knockdown, both for ectopically expressed FLAG-cortactin (Fig. 3B) as well as endogenous cortactin (Fig. 3D). Additional co-expression experiments with FLAG-cortactin, FLAG-cortactin-S298A, and active (CA)/kinase-inactive (KD) PKD1 constructs also support in vivo phosphorylation of this site (supplemental Fig. 1B). To assess, whether phosphorylation of Ser298 in cortactin generates a 14-3-3 binding motif, we performed pulldown assays with GST-14-3-3β-coated beads from HEK293T total cell lysates expressing FLAG-tagged cortactin as well as the cortactin-S298A mutant (Fig. 3C). The S298A mutation reduced pulldown of the respective mutant protein 11.6-fold indicating the generation of a functional 14-3-3 binding motif in cortactin upon phosphorylation. GST-14-3-3β-pulldown assays of endogenous cortactin following expression of active PKD1CA or kinase-inactive PKD1KD constructs (Fig. 3E) corroborate PKD-mediated phosphorylation and 14-3-3 binding to endogenous cortactin in vivo.
FIGURE 3.
A, structural overview of cortactin. NTA, N-terminal acidic region; 6½ HS-1-type F-actin binding repeats; H, helical domain; P, prolin-rich region. Amino acid positions are depicted below the structure plot. S298 marks the PKD phosphorylation site in HS repeat 6. PKD consensus sequence and consensus motif of Ser298 in cortactin are shown on the right-hand panel. B, characterization of cortactin pS298 phosphorylation using a site-specific pS298-cortactin antibody. Phosphorylation of FLAG-cortactin was assessed in total cell lysates following PKD1/2 knockdown. Control blots show knockdown efficiency and numbers depict quantification as fold-integrated bands density. C, phosphorylation of Ser298 generates a 14-3-3 binding motif. GST-14-3-3β pulldown assays were made from HEK293T lysates expressing FLAG-cortactin and FLAG-cortactin-S298A. Cortactin-S298A strongly reduces binding to GST-14-3-3β. Control blots show transgene expression. Numbers depict fold-integrated density normalized to control (1). D, characterization of endogenous cortactin pS298 phosphorylation. Immunoprecipitation (IP) of endogenous cortactin from HEK293T cells were probed for Ser298 phosphorylation following PKD1/2-knockdown and control blots for knockdown efficiency. E, GST-14-3-3β pulldown assays of endogenous cortactin from HEK293T cell lysates co-expressing active PKD1CA and kinase-inactive PKD1KD. Numbers depict fold-integrated density normalized to control. Control blots show transgene expression. IB, immunoblot.
Cortactin-S298A Enhances Polarized Cell Migration
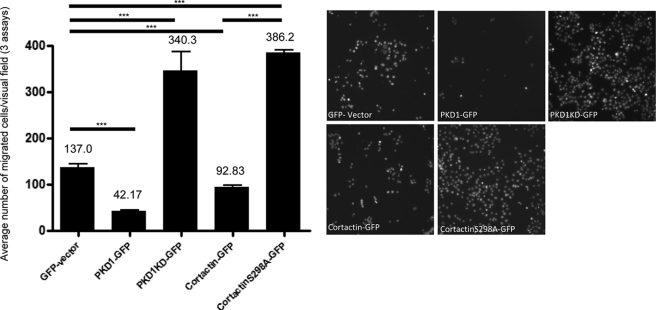
Because we have already shown that the activity status of PKD affects Panc89 as well as breast cancer cell motility (1–3) and cortactin has been published to affect cell migration (16) we were prompted to test whether cortactin or its respective S298A mutation would alter migration of Panc89 cells toward a chemotactic serum gradient in Transwell migration assays. In line with Ref. 1, the results summarized in Fig. 4 revealed that expression of PKD1-GFP inhibited migration relative to the GFP-transfected control cells (42.17 versus 137.0 cells/field, n = 3), whereas expression of kinase-inactive PKD1KD-GFP strongly enhanced motility (340.3 versus 137 cells/field, n = 3). Cortactin-GFP expressing cells displayed moderately reduced motility (92.83 cells/field n = 3) relative to GFP control, whereas cortactin-S298A-GFP (386.2 cell/field n = 3) strongly enhanced migration compared with cells expressing wild type cortactin or GFP, indicating that phosphorylation of Ser298 has a strong negative modulatory role in cancer cell motility. The equal expression of GFP fusion proteins for these experiments was evaluated by Western blots (supplemental Fig. 2) and fluorescence-activated cell sorter analysis, respectively (data not shown).
FIGURE 4.
Statistical analysis of 3 independent Transwell migration assays of stable Panc89 cells expressing GFP, PKD1-GFP, PKD1KD-GFP, cortactin-GFP, and cortactin-S298A-GFP. Cells were induced to migrate toward a serum gradient for 40 h. Cells migrated through the porous membrane were stained with 4′,6-diamidino-2-phenylindole and visualized by wide field fluorescence microscopy at ×10 magnification (right-hand panel). The diagram depicts the combined average number of cells per visual field and respective S.E.M. of three independent experiments. Three asterisks indicate p < 0.0001 of Student's t tests.
Cortactin and Cortactin-S298A Synergistically Enhance Arp Complex-mediated Actin Nucleation
To elucidate the molecular mechanism by which cortactin phosphorylation affects cell motility we studied cortactin in actin remodeling processes, especially in Arp complex-driven actin polymerization. To that end we performed in vitro actin polymerization assays with pyrene-labeled G-actin, purified Arp complex, purified GST-VCA minimal fragment (NPF), as well as FLAG affinity purified cortactin and cortactin-S298A, respectively, and the kinetic of polymerization was measured by increase in pyrene fluorescence (Fig. 5A). Half-maximal saturated polymerization values (T½max (s)) calculated from Boltzmann sigmoidal curve fits are summarized in Fig. 5B. T½max values for actin and actin/Arp reactions are 997.6 ± 7.4 and 1043 ± 8.42 s, respectively. This slow spontaneous polymerization as well as no nucleation in the absence of nucleation promoting factors, even if Arp complex is present, is in full accordance with literature (12, 15, 21). As expected, the GST-VCA minimal fragment strongly enhanced the polymerization rate with T½max of 488.8 ± 2.27 s. Weaver and colleagues (15) further proposed that cortactin can synergistically accelerate VCA/Arp-driven actin polymerization in vitro. According to their model, cortactin is able to alter the nucleation rate by competing for Arp3 binding via its N-terminal region, whereas VCA, or full-length NPFs, would bind to Arp2 and ARPC1 (p40) instead of Arp2/3 and p40 when no cortactin was present. Thus they suggested that under saturated conditions cortactin may be able to bridge the Arp2/3 complex to actin filaments via Arp3 and that VCA activates the Arp2/3 complex by binding Arp2 and/or ARPC1/p40. In support of this hypothesis, we observed enhanced polymerization rates in reactions where 1000 nm cortactin was present (VCA:cortactin, >1:2), assuring saturating concentrations for cortactin during polymerization (T½max of 394.9 ± 1.66 s). Cortactin-S298A was able to accelerate this polymerization rate even further with T½max values of 343.2 ± 1.53 s. A significant (p < 0.05) difference in synergistic polymerization rates between cortactin and cortactin-S298A became evident, too, when the amount of polymerized F-actin plotted as % of saturation of individual reactions was analyzed at two time points during the linear phase (T420/T450 s) (supplemental Fig. S3A). Cortactin reached 65.3 and 76.63% saturation, whereas cortactin-S298A showed 84.63 and 90.67% saturation for time points T420 and T450 s, respectively, amounting to a relative increase in polymerized F-actin of 19.6 (T420 s) or 14.04% (T450 s). Because Weaver et al. (15) proposed a mechanism by which cortactin would be able to bridge the Arp complex to actin filaments during synergistic nucleation, we investigated further to what extent in vitro F-actin binding capacities of purified FLAG-cortactin and cortactin-S298A proteins would be altered. We performed 100,000 × g high-speed F-actin sedimentation assays to study binding of purified cortactin/cortactin-S298A to in vitro polymerized F-actin. The supernatant and F-actin pellet fractions obtained following ultracentrifugation were resolved on 10% SDS-PAGE gels. Fig. 5C shows that the deletion of the Ser298 phosphorylation site reduced the in vitro F-actin binding capacity of cortactin. Wild type cortactin and cortactin-S298A displayed a ratio of supernatant/pellet fraction integrated band density (S/P) of 1.64 and 2.2, respectively, indicative of weaker binding of the mutated cortactin to F-actin (pellet fraction; S/P ratio difference, 25.5%). The experiment was also performed with lower amounts of cortactin (0.5/10 μg of actin) to rule out possible saturation effects. Under these conditions, an almost complete loss of the cortactin signal in the pellet fraction was observed (supplemental Fig. 3B), confirming the above data of reduced actin binding capacity of cortactin-S298A compared with wild type cortactin. By interfering with the F-actin binding capacity of cortactin, the ablation of Ser298 phosphorylation and concomitant loss of 14-3-3 binding might be able to alter the capability of cortactin to bridge the Arp complex to actin filaments during synergistic nucleation and thus enhance turnover of the nucleation complex at relevant sites. In vitro branching assays performed with cortactin and cortactin-S298A further suggest a reduced ability of cortactin-S298A to stabilize highly branched filament networks formed during in vitro nucleation, as the % of filaments with >2 branches is reduced by 29.2% with respect to wild type cortactin (supplemental Fig. S3C).
FIGURE 5.
A, pyrene-actin polymerization assay with purified proteins. Cortactin-S298A/GST-VCA/Arp synergistically accelerate Arp complex-driven actin polymerization in respect to cortactin/GST-VCA/Arp or GST-VCA/Arp solely induced polymerization. Assays were performed in triplicate with 3 μm pyrene-labeled G-actin, 50 nm purified Arp complex, 400 nm GST-VCA, as well as 1000 nm purified FLAG-cortactin and FLAG-cortactin-S298A from HEK293T cells. To quantify changes in polymerization, curves were fitted with Boltzmann sigmoidal equations (GraphPad Prism). Half-maximal saturated polymerization times (T½max (s)) are plotted in B. C, cortactin-S298A demonstrates reduced F-actin binding capacity. High-speed F-actin binding assays with purified FLAG-cortactin and FLAG-cortactin-S298A. Purified cortactin and cortactin-S298A were incubated with in vitro polymerized, purified F-actin. As specificity control for binding F-actin was omitted in control samples. Supernatant and 100,000 × g pellet fractions were analyzed by 10% SDS-PAGE and visualized by Coomassie staining. S/P values indicate ratios of supernatant (S) and pellet (P) fractions quantified by integrated densities of the respective bands.
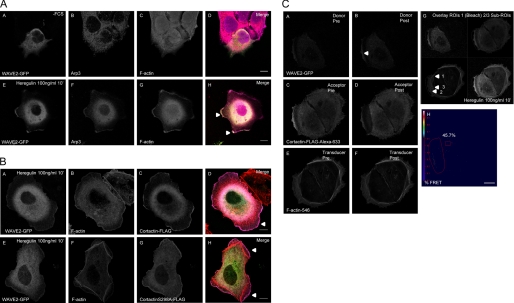
Cortactin and WAVE2 Co-localize and Interact in the Active Polymerization Zone at the Edge of Lamellipodia in Heregulin-stimulated MCF-7 Cells
To study biological implications of the enhanced actin polymerization rate and an enhanced turnover of Arp2/3 nucleation complexes by cortactin-S298A in vivo, we used MCF-7 breast cancer cells as a model system. In MCF-7 cells strong actin polymerization and extension of lamellipodia can be induced following stimulation with the growth factor Heregulin (27). During lamellipodia formation WAVE class nucleation promoting factors (NPFs), such as WAVE2, are major activators of the Arp complex in the polymerization zone at the edge of protruding lamellipodia (22). WAVE2, but not WAVE1, has also been shown to be essential for directed cell migration and lamellipodia formation in WAVE1/2-deficient mouse embryonic fibroblasts (28). Unlike N-WASP, an NPF existing in a closed autoinhibitory conformation and an open, active conformational state accessible for co-activators like CDC42 or phosphatidylinositol bisphosphate (21, 29), WAVE2 is not regulated by such conformation changes. Instead, WAVE2 is activated downstream of Rac, which is in turn activated by Heregulin stimulation in MCF-7 cells (27). WAVE2, however, does not interact with Rac directly. Its activity is rather modulated within multiprotein complexes by co-factors such as IRSp53, an essential intermediate signaling component of Rac to WAVE2 (22, 30). Because WAVE2 activation cannot be measured by applying intramolecular FRET biosensors similar to N-WASP (29), we have defined WAVE2 as being “active” once it is present in the nucleating Arp2/3 complex at the edge of protruding lamellipodia. We have investigated the localization of WAVE2-GFP in relation to markers Arp3 and F-actin in MCF-7 cells stimulated with 100 ng/ml of Heregulin for 10 min (panels A–D) and in serum-starved control cells (panels E–H) (Fig. 6A). WAVE2-GFP co-localized with Arp3 and F-actin at the edge of lamellipodia in membrane ruffles of MCF-7 breast cancer cells. The proteins also overlapped considerably within the cytoplasm in the perinuclear region following stimulation with Heregulin for 10 min. In serum-starved control cells, WAVE2-GFP, Arp3, and F-actin only demonstrated minimal, or no co-localization at all. Supplemental Fig. S4E depicts an exemplary acceptor-photobleach-FRET experiment with WAVE2-GFP as FRET donor and endogenous Arp3 detected by a monoclonal primary and Alexa 546 secondary antibody as FRET acceptor. Increase in donor fluorescence following bleaching of the FRET acceptor indicates FRET prior to bleaching and thus molecular interaction. Of particular relevance is the presence of WAVE2 within FRET distance (<10 nm) to Arp3 in the polymerization zone at the edge of extending lamellipodia, which meets prominently the above defined criteria as being present and active in nucleating Arp2/3 complexes during lamellipodia extension. In addition, FRET signals were also observed in a perinuclear region where both proteins are located as well. Respective pre- and post-bleach images of donor (A and B) and acceptor (C and D) are depicted in supplemental Fig. S4E. FRET was quantified by acquiring a time-bleach image series of cells. The threshold %FRET image depicting localization and FRET efficiency (14.6% mean for the entire bleach ROI) coded in Rainbow LUT as well as respective bleach and background ROIs (excluding pixels with “off target” acceptor-bleaching) used to generate the image are shown in E.
FIGURE 6.
A, panels A–H, WAVE2-GFP co-localizes with Arp3 and F-actin at the edge of lamellipodia in membrane ruffles of Heregulin-stimulated MCF-7 cells. The proteins also overlap considerably in the perinuclear, cytoplasmic region following stimulation with Heregulin for 10 min. In serum-starved control cells WAVE2-GFP, Arp3, and F-actin only show minimal or no co-localization. Single confocal image sections are shown. Relevant structures are marked with arrowheads. The scale bar represents 10 μm. B, WAVE2-GFP, F-actin, and FLAG-cortactin (A–D) or FLAG-cortactin-S298A (E–H) co-localize at the outer edge of MCF-7 lamellipodia stimulated with Heregulin, in membrane ruffles and in perinuclear structures (see also supplemental Fig. S6B). No difference in subcellular localization of cortactin and cortactin-S298A was evident. Single confocal sections are depicted. Respective relevant structures are highlighted with arrowheads. The scale bar represents 10 μm. WAVE2-GFP, F-actin, and FLAG-cortactin interact in a multimeric complex. C, exemplary 3-dye daisy chain acceptor-photobleach-FRET experiment with WAVE2-GFP as the primary FRET-donor, FLAG-cortactin stained with α-FLAG M2-Alexa 633 secondary antibody as the final FRET acceptor, and F-actin (phalloidin-Alexa 546) as FRET signal transducer. Pre- and post-bleach images of donor (A and B), acceptor (C and D), and transducer (E and F) are depicted. FRET was quantified by acquiring a time-bleach image series of cells. %FRET images (H) were generated using ZEN Software of the LSM 710 confocal microscope (Zeiss). The Bezier bleach ROI and rectangle threshold ROI are marked by red lines. The scale bar represents 10 μm. G, overlay images of bleach ROI and sub-ROIs for quantification of RAW data and calculation of RAW %FRET signals by mean of ROI analysis (supplemental Fig. S6C).
We further investigated the localization of FLAG-cortactin and FLAG-cortactin-S298A with respect to WAVE2-GFP and F-actin (phalloidin-Alexa 546). There was no difference in subcellular localization of cortactin and cortactin-S298A with respect to these markers. As shown in Fig. 6B WAVE2 and cortactin (panels A–D) as well as cortactin-S298A (panels E–H) co-localized with dynamic actin in the polymerization zone at the edge of lamellipodia in MCF-7 cells following Heregulin stimulation (panels A–H). The three proteins also co-localized in peripheral ruffles of lamellipodia as well as in the cytoplasmic perinuclear region (supplemental Fig. S4B, panels I–P), whereas co-localization in serum-starved control cells was limited to small ruffle zones and peripheral dot-like structures (supplemental Fig. S4A, panels A-H). In conclusion we were able to show a prominent co-localization of WAVE2 with FLAG-cortactin at highly dynamic actin structures at the edge of lamellipodia within the active polymerization zone or peripheral membrane ruffles, which are formed as a direct consequence of strong actin polymerization at these structures.
Cortactin Bridges Active WAVE2/Arp2/3 Complexes to Actin Filaments at Extending Lamellipodia in Heregulin-stimulated MCF-7 Cells
We next asked whether cortactin might be involved in bridging the Arp complex to actin filaments during synergistic polymerization (15), thereby effectively altering the branching status of the filament network. We performed a 3-dye daisy chain acceptor-photobleach-FRET experiment (Fig. 6C) in Heregulin-stimulated MCF-7 cells. WAVE2-GFP was used as the primary FRET donor, FLAG-cortactin was stained with α-FLAG-M2 and Alexa 633 secondary antibodies as the final FRET acceptor, whereas F-actin (phalloidin-Alexa 546) served as the intermediate FRET signal transducer. In this constellation, Foerster energy transfer to the final acceptor is only possible when all 3 dyes are within FRET distance, inferring simultaneous interaction of the three proteins. In Fig. 6C pre- and post-bleach images of donor (panels A and B), acceptor (panels C and D), and transducer (panels E and F) are depicted. Again FRET was quantified by acquiring a time-bleach image series of cells for the respective channels. The final daisy chain FRET-acceptor Alexa 633-labeled FLAG-cortactin was bleached with an intensive laser beam at 633 nm. Forester energy transfer of the primary donor (WAVE2-GFP) transduced via Alexa 546-labeled F-actin was visualized by an increase in primary donor fluorescence after bleaching. %FRET images (panel H) were thresholded using the ROI rectangle in panel H. The thresholded FRET pixels in panel H displayed a mean efficiency of 45.7% for the entire bleach region. To show that cortactin will only bridge active nucleation complexes as identified by WAVE2-GFP, we went on to evaluate the non-thresholded RAW data of the daisy chain FRET experiment by the mean of ROI analysis with sub-ROIs 2 and 3 located within the bleach region (ROI1) (supplemental Fig. S4C). Sub-ROI2 was placed at the peripheral dynamic ruffling structures at the edge of lamellipodia, whereas sub-ROI3 marked non-dynamic actin arc filaments with FLAG-cortactin and WAVE2-GFP still co-localizing within the ROI (panel G, see arrowheads). The transducer F-actin (phalloidin-Alexa 546) only mediated FRET signals to FLAG-cortactin in this setup at sub-ROI2 but not at sub-ROI3 as indicated by the mean of ROI intensity RAW data for ROIs 2 and 3 and the RAW non-thresholded mean %FRET values of 18.57% for the entire bleach ROI1, 15.2% for sub-ROI2 (dynamic actin), and no FRET transduced for sub-ROI3 (actin arc filaments). Analysis of a second cell is depicted in supplemental Fig. S4D. Uncoupling of the weak 3-dye dipole interaction by bleaching the final acceptor FLAG-cortactin-Alexa 633 also results in the increase in F-actin-546 transducer fluorescence, acting as donor for the Alexa 633 dye (supplemental Fig. S4, C and D). This further demonstrates the specificity of the used approach.
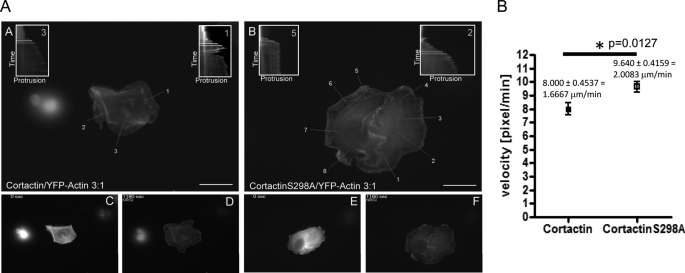
Cortactin-S298A Accelerates Actin Polymerization Kinetics and Extension of Lamellipodia in Heregulin-stimulated MCF-7 Cells
Based on these results, we studied next the dynamic changes in synergistic actin polymerization mediated by cortactin and cortactin-S298A during lamellipodia extension after Heregulin stimulation in MCF-7 cells. Low level co-expression of GFP-tagged G-actin can be used to visualize actin polymerization in vivo (19). Fig. 7A (panels C–F) depicts snapshot images of wide field fluorescence microscopy videos visualizing actin polymerization and lamellipodia extension of MCF-7 cells ectopically expressing cortactin/YFP-G-actin (panel C, T0; panel D, 20 min) and cortactin-S298A/YFP-G-actin (panel E, T0; panel F, 20 min) as well as FLAG-vector/YFP-G-actin (supplemental Fig. S5, panel B, T0; panel C, 20 min) at a ratio of 3:1. Time-lapse videos of YFP fluorescence were acquired at ×63 magnification every 10 s for 20 min. Cells were stimulated at T120 s after starting the acquisition. Then a kymography analysis of the protruding edge of the lamellipodia as visualized by polymerized YFP-actin was performed. In Fig. 7A (panels A and B) overlay images of cells with 1 pixel lines used to generate the kymograms are displayed. Protrusion velocities from kymograms were calculated as [pixel/min] along the highlighted lines oriented perpendicular to the protrusion front of initially individual protrusions. Sample kymograms used to calculate protrusion velocity at the indicated white lines are also shown in Fig. 7A. Fig. 7B displays the results of a statistical analysis (n = 6) for cortactin (n = 17 lamellipodia) and cortactin-S298A (n = 25 lamellipodia) already reaching statistical significance (p = 0.0127). Shown are protrusion velocities [pixel/min] with standard errors (S.E.) for cortactin amounting to 8.0000 ± 0.4537 [pixel/min] = 1.6667 μm/min and for cortactin-S298A 9.640 ± 0.4159 = 2.0083 μm/min, indicating that the S298A mutation enhanced the velocity of protrusion by 20.5%. Interestingly, actin polymerization in pyrene-actin assays at T420 s during the linear polymerization phase also demonstrated an increase in polymerized F-actin by 19.6% for cortactin-S298A compared with wild type cortactin (Fig. 5B). The latter can be explained by increased nucleation kinetics, which in turn could be responsible for the observed equivalent increase in lamellipodia extension velocity. In accordance with literature (16), vector-transfected cells displayed reduced persistence of protrusions (supplemental Fig. 5, A-C) when compared with cortactin or cortactin-S298A cells, which did not allow for a similar quantification and statistical analysis.
FIGURE 7.
A, cortactin-S298A accelerates lamellipodia extension of Heregulin-stimulated MCF-7 cells by ∼20.5%. MCF-7 cell were transfected at a ratio of 3:1 with FLAG-cortactin/FLAG-cortactin-S298A:YFP-G-actin to visualize actin polymerization. For kymograph analysis time-lapse images of the Heregulin-stimulated cell were acquired using a Leica wide field fluorescence microscope every 10 s at ×63 magnification. Kymographs were generated with NIH ImageJ by plotting the edge of the protruding lamellipodia visualized by YFP-G-actin against time (pixel/10 s) along a 1-pixel line perpendicular to the protrusion front for initially individual protrusions. The protrusion velocity was calculated from kymographs during the linear phase of extension. Vector controls (supplemental Fig. S7, A–C) show in line with Ref. 16 reduced the protrusion persistence when compared with cortactin cells. Plotted in the figure are overlay images with kymography lines and sample kymograms for cortactin (A, C, and D) and cortactin-S298A (B, E, and F)-transfected cells. White lines with a relative time difference of Δ60 s indicate the velocities measured. B, quantification and statistical analysis of kymograms. Cortactin (n = 6 cells, n = 17 lamellipodia kymographs) and cortactin-S298A (n = 6 cells, n = 25 lamellipodia kymographs) were analyzed. Shown in the graph are the protrusion velocities (pixel/min) with S.E. Velocities were also calculated as micrometer/min using the spatial calibration of images. The scale bar represents 10 μm. Statistical significance evaluated by Student's t test is marked by an asterisk (p = 0.0127).
DISCUSSION
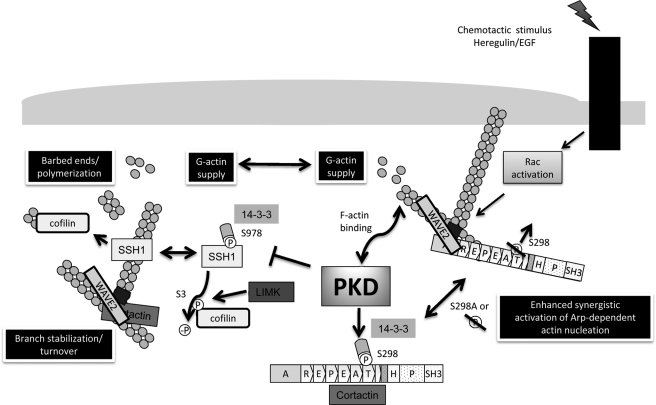
PKD has been described as a key regulator of the cofilin signaling pathway thereby controlling actin remodeling and polarized cell motility in cervix carcinoma, breast cancer, and pancreatic cancer cells (1–4, 31). We here describe a novel PKD-controlled pathway targeting Arp complex-mediated actin polymerization via the F-actin-binding protein cortactin. Our data fully support and expand the model proposed by Weaver et al. (15) for a potential role of cortactin in bridging the Arp2/3 complex to actin filaments at peripheral dynamic actin structures during synergistic polymerization, using WAVE2 as nucleation promoting factor in Heregulin-stimulated MCF-7 cells. In addition, we show that cortactin-S298A was able to enhance synergistic actin polymerization in vitro and, due to its reduced F-actin binding capacity, might also enhance turnover of nucleation complexes at the respective dynamic actin structures. By visualizing actin polymerization and lamellipodia protrusion velocities in the MCF-7 model system, we were further able to demonstrate the significance of cortactin and specifically of the PKD phosphorylation site at Ser298 in affecting actin polymerization and velocity of lamellipodia protrusion in vivo. Taken together, the above described molecular mechanism suffices to explain the enhanced motility of Panc89 pancreatic cancer cells expressing cortactin-S298A as opposed to wild type cortactin.
Thus, a new regulatory role for PKD in the modulation of Arp complex-mediated actin polymerization through cortactin-Ser298 phosphorylation becomes evident, which affects cancer cell motility and therefore adds to the previously described mechanism of PKD-mediated regulation of cell migration through control of SSH1 activity (2). Accordingly, localized PKD activity is able to modulate actin remodeling events at several, independent levels by phosphorylation of actin regulatory proteins, which control the polymerization and depolymerization processes of the actin cytoskeleton (Fig. 8). The modulation of both pathways by PKD fully accounts for the observed inhibition and enhancement, respectively, of cancer cell motility by active and kinase-inactive PKD (1, 2). Dynamic regulation of localized PKD activity, e.g. downstream of RhoA, during different stages of cell migration is necessary to fine-tune actin reorganization and polymerization events, providing the basis for altering cell motility. As proposed previously (8, 9), cofilin has important functions in controlling the initial directional response of cells after sensing a chemotactic gradient. Cofilin severs old filaments replenishing the G-actin pool of cells and it generates new “free barbed ends” after being recruited to the active polymerization zone at what will become the leading edge of cells oriented toward a growth factor gradient (7). By controlling cofilin activity via regulation of availability of the cofilin-phosphatase SSH1, PKD can vitally alter this early response of cells (2). Once polymerization has been initiated, Arp complex-mediated polymerization and the formation of branched actin networks is a second key factor supporting lamellipodia extension and eventually the chemotactic movement of cells (7). We have shown here that PKD can regulate this process by acting on WAVE2/Arp complex-mediated synergistic actin polymerization via phosphorylation of Ser298 of cortactin. Cortactin acts in conjunction with NPFs, like WAVE2 (22) and the Arp2/3 complex (20) to synergistically enhance actin polymerization kinetics and to bridge the nucleating Arp complex to actin filaments (15, 32). We show that bridging as demonstrated by the 3-dye FRET approach is restricted to dynamic actin structures and not to arc filaments at the cell periphery (polymerization zone). Our in vitro data further suggest that the stabilization of highly branched filament networks is reduced by the S298A mutation, concomitant with its reduced ability to bind to F-actin filaments, allowing for a more “flexible,” higher turnover rate of the branched network in lamellipodia.
FIGURE 8.
PKD-mediated effects of actin remodeling. PKD is a key novel regulator of polarized cell motility and F-actin organization. PKD is capable of binding to F-actin in vitro and in vivo, a feature of many actin regulatory proteins. PKD controls the activity of the ubiquitous F-actin depolymerizing and severing factor cofilin via slingshot family cofilin phosphatases. It modulates the supply of G-actin monomers and the generation of barbed ends at the leading edge of motile cells, which ultimately translates in membrane protrusion and cell migration. Cofilin is inactivated downstream of Rho-GTPases. Active RhoA, Rac, and CDC42 mediate the activation of Lim domain kinase via Rho-associated protein kinase or p21-activated kinases, respectively. Lim domain kinase then inactivates cofilin by phosphorylation at S3. Slingshot family phosphatases (SSH) regenerate cofilin activity by dephosphorylation. SSH1 is negatively regulated by direct upstream phosphorylation via protein kinase D downstream of RhoA, or oxidative stress. Phosphorylation of Ser978 mediates 14-3-3 binding and sequestration of SSH away from F-actin, which drastically reduces SSH cofilin-phosphatase activity. In line with this data we have identified the actin-binding protein cortactin as a second key F-actin regulatory protein controlled by PKD. Cortactin acts in conjunction with NPFs, like WAVE2, as well as the Arp2/3 complex to synergistically enhance actin polymerization rates and bridge the nucleating Arp complexes to actin filaments. PKD phosphorylates cortactin in vitro and in vivo at Ser298 generating a 14-3-3 binding motif. Ablation of Ser298 phosphorylation reduces F-actin binding of cortactin inferring increased turnover of nucleation complexes at relevant sites. Loss or ablation (SA mutation) of Ser298 phosphorylation synergistically accelerates WAVE2-Arp-driven actin polymerization, lamellipodia extension, and directed cell migration.
Recently, a comprehensive mass spectroscopy analysis identified 17 novel phosphorylation sites in cortactin, among them serine 113 (33). Interestingly, the serine/threonine kinase PAK1 was identified to directly phosphorylate cortactin on serine 113 thereby down-regulating F-actin binding to cortactin (34). We here describe a similar mechanism for PKD-mediated phosphorylation of cortactin on Ser298. In vivo phosphorylation of Ser298, which is located within the 6th F-actin binding repeat, was confirmed by mass spectrometry (24). On top of that, we provide evidence that phosphorylation of serine 298 generates a 14-3-3 binding motif. 14-3-3 proteins are scaffold proteins whose binding can alter localization, stability, phosphorylation state, activity, and/or molecular interactions of a target protein. Phosphorylation by PKD is not required for translocation of cortactin to lamellipodia or membrane ruffles. Instead, we demonstrate that ablation of Ser298 phosphorylation by the serine to alanine mutation reduces F-actin binding of cortactin, inferring increased turnover of nucleation complexes at relevant sites. Loss of 14-3-3 binding could thus induce changes in the three-dimensional structure of cortactin thereby masking the respective F-actin binding site.
In conclusion our previous findings, and the data presented herein provide a mechanistic understanding of the role of PKD as a negative regulator of cell migration affecting actin polymerization at different levels, as summarized in Fig. 8. In addition to controlling cancer cell motility, PKD is known as a key regulator of vesicular transport (35–37). Furthermore, in breast cancer cells PKD was shown to down-regulate the expression of a number of matrix metalloproteases, which are key factors for cancer cell invasiveness (3). Thus, a potentially important role of PKD in controlling cancer progression and metastasis can be postulated.
Supplementary Material
Acknowledgment
We thank Dr. Olaf Selchow (Carl Zeiss) for use of the DEMO-Microscopy facility (Zeiss).
This work was supported by grants from the Baden-Württemberg State Ministry for Science and Arts for the Center Systems Biology Stuttgart (project A1) (to K. P. and A. H.), the BMBF Systec-Program (to K. P.), and the Heidelberger Akademie der Wissenschaften (WIN-Kolleg) (to A. H.). This work was also supported by Grant G.0612.07 from FWO-Vlaanderen (Research Foundation—Flanders) (to J. V. L.), an Aspirtant Fellowship of the FWO-Vlaanderen (to L. D. K.), and Grant IAP 6/18 from the Interuniversity Attraction Poles Programme, initiated by the Belgian State Science Policy Office.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- PKD
- protein kinase D
- GFP
- green fluorescent protein
- YFP
- yellow fluorescent protein
- SSH1L
- slingshot1L
- FRET
- fluorescence resonance energy transfer
- ROS
- reactive oxygen species
- NPF
- nucleation promoting factor
- ROI
- region of interest
- WAVE
- SCAR (suppressor of cAMP receptor), Arp (actin-related protein), VCA (verprolin, constant, acidic)
- SH3
- Src homology domain 3
- EGFP
- enhanced green fluorescent protein
- GST
- glutathione S-transferase.
REFERENCES
- 1.Eiseler T., Schmid M. A., Topbas F., Pfizenmaier K., Hausser A. (2007) FEBS Lett. 581, 4279–4287 [DOI] [PubMed] [Google Scholar]
- 2.Eiseler T., Döppler H., Yan I. K., Kitatani K., Mizuno K., Storz P. (2009) Nat. Cell Biol. 11, 545–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eiseler T., Döppler H., Yan I. K., Goodison S., Storz P. (2009) Breast Cancer Res. 11, R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peterburs P., Heering J., Link G., Pfizenmaier K., Olayioye M. A., Hausser A. (2009) Cancer Res. 69, 5634–5638 [DOI] [PubMed] [Google Scholar]
- 5.Yamamoto M., Nagata-Ohashi K., Ohta Y., Ohashi K., Mizuno K. (2006) FEBS Lett. 580, 1789–1794 [DOI] [PubMed] [Google Scholar]
- 6.Kurita S., Watanabe Y., Gunji E., Ohashi K., Mizuno K. (2008) J. Biol. Chem. 283, 32542–32552 [DOI] [PubMed] [Google Scholar]
- 7.DesMarais V., Macaluso F., Condeelis J., Bailly M. (2004) J. Cell Sci. 117, 3499–3510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DesMarais V., Ichetovkin I., Condeelis J., Hitchcock-DeGregori S. E. (2002) J. Cell Sci. 115, 4649–4660 [DOI] [PubMed] [Google Scholar]
- 9.DesMarais V., Ghosh M., Eddy R., Condeelis J. (2005) J. Cell Sci. 118, 19–26 [DOI] [PubMed] [Google Scholar]
- 10.Sidani M., Wessels D., Mouneimne G., Ghosh M., Goswami S., Sarmiento C., Wang W., Kuhl S., El-Sibai M., Backer J. M., Eddy R., Soll D., Condeelis J. (2007) J. Cell Biol. 179, 777–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Rheenen J., Song X., van Roosmalen W., Cammer M., Chen X., Desmarais V., Yip S. C., Backer J. M., Eddy R. J., Condeelis J. S. (2007) J. Cell Biol. 179, 1247–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weed S. A., Karginov A. V., Schafer D. A., Weaver A. M., Kinley A. W., Cooper J. A., Parsons J. T. (2000) J. Cell Biol. 151, 29–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyle S. N., Michaud G. A., Schweitzer B., Predki P. F., Koleske A. J. (2007) Curr. Biol. 17, 445–451 [DOI] [PubMed] [Google Scholar]
- 14.Weaver A. M. (2008) Cancer Lett. 265, 157–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weaver A. M., Heuser J. E., Karginov A. V., Lee W. L., Parsons J. T., Cooper J. A. (2002) Curr. Biol. 12, 1270–1278 [DOI] [PubMed] [Google Scholar]
- 16.Bryce N. S., Clark E. S., Leysath J. L., Currie J. D., Webb D. J., Weaver A. M. (2005) Curr. Biol. 15, 1276–1285 [DOI] [PubMed] [Google Scholar]
- 17.Hausser A., Link G., Bamberg L., Burzlaff A., Lutz S., Pfizenmaier K., Johannes F. J. (2002) J. Cell Biol. 156, 65–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hausser A., Link G., Hoene M., Russo C., Selchow O., Pfizenmaier K. (2006) J. Cell Sci. 119, 3613–3621 [DOI] [PubMed] [Google Scholar]
- 19.Lorenz M., DesMarais V., Macaluso F., Singer R. H., Condeelis J. (2004) Cell Motil. Cytoskeleton 57, 207–217 [DOI] [PubMed] [Google Scholar]
- 20.Amann K. J., Pollard T. D. (2001) Nat. Cell Biol. 3, 306–310 [DOI] [PubMed] [Google Scholar]
- 21.Daly R. J. (2004) Biochem. J. 382, 13–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takenawa T., Miki H. (2001) J. Cell Sci. 114, 1801–1809 [DOI] [PubMed] [Google Scholar]
- 23.Hausser A., Storz P., Link G., Stoll H., Liu Y. C., Altman A., Pfizenmaier K., Johannes F. J. (1999) J. Biol. Chem. 274, 9258–9264 [DOI] [PubMed] [Google Scholar]
- 24.De Kimpe L., Janssens K., Derua R., Armacki M., Goicoechea S., Otey C., Waelkens E., Vandoninck S., Vandenheede J. R., Seufferlein T., Van Lint J. (2009) Cell Signal. 21, 253–263 [DOI] [PubMed] [Google Scholar]
- 25.Nishita M., Tomizawa C., Yamamoto M., Horita Y., Ohashi K., Mizuno K. (2005) J. Cell Biol. 171, 349–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vega R. B., Harrison B. C., Meadows E., Roberts C. R., Papst P. J., Olson E. N., McKinsey T. A. (2004) Mol. Cell. Biol. 24, 8374–8385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagata-Ohashi K., Ohta Y., Goto K., Chiba S., Mori R., Nishita M., Ohashi K., Kousaka K., Iwamatsu A., Niwa R., Uemura T., Mizuno K. (2004) J. Cell Biol. 165, 465–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takenawa T. (2005) Novartis Found. Symp. 269, 3–10; discussion 10–15, 30–14 [PubMed] [Google Scholar]
- 29.Lorenz M., Yamaguchi H., Wang Y., Singer R. H., Condeelis J. (2004) Curr. Biol. 14, 697–703 [DOI] [PubMed] [Google Scholar]
- 30.Miki H., Yamaguchi H., Suetsugu S., Takenawa T. (2000) Nature 408, 732–735 [DOI] [PubMed] [Google Scholar]
- 31.Storz P. (2009) Cell Cycle 8, 1975–1976 [PubMed] [Google Scholar]
- 32.Weaver A. M., Young M. E., Lee W. L., Cooper J. A. (2003) Curr. Opin. Cell Biol. 15, 23–30 [DOI] [PubMed] [Google Scholar]
- 33.Martin K. H., Jeffery E. D., Grigera P. R., Shabanowitz J., Hunt D. F., Parsons J. T. (2006) J. Cell Sci. 119, 2851–2853 [DOI] [PubMed] [Google Scholar]
- 34.Webb B. A., Zhou S., Eves R., Shen L., Jia L., Mak A. S. (2006) Arch. Biochem. Biophys. 456, 183–193 [DOI] [PubMed] [Google Scholar]
- 35.Díaz Añel A. M., Malhotra V. (2005) J. Cell Biol. 169, 83–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yeaman C., Ayala M. I., Wright J. R., Bard F., Bossard C., Ang A., Maeda Y., Seufferlein T., Mellman I., Nelson W. J., Malhotra V. (2004) Nat. Cell Biol. 6, 106–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hausser A., Storz P., Märtens S., Link G., Toker A., Pfizenmaier K. (2005) Nat. Cell Biol. 7, 880–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.