Abstract
Suppressor of cytokine signaling 3 (SOCS3) regulates the proinflammatory cytokine signaling mediated by the JAK/STAT signaling pathway. SOCS3 is rapidly induced and then targeted to the ubiquitin-proteasome pathway via a mechanism that requires the C-terminal SOCS box. Due to its rapid turnover, the intracellular stores of SOCS3 seem insufficient to control acute or protracted inflammatory diseases. Previously, we developed an intracellular protein therapy that uses a recombinant cell-penetrating form of SOCS3 (CP-SOCS3) to inhibit the JAK/STAT pathway and prevent cytokine-mediated lethal inflammation and apoptosis of the liver (Jo, D., Liu, D., Yao, S., Collins, R. D., and Hawiger, J. (2005) Nat. Med. 11, 892–898). The potent anti-inflammatory and cytoprotective activity of CP-SOCS3 prompted us to analyze its intracellular turnover, as compared with that of endogenous SOCS3 protein induced in macrophages by the proinflammatory agonists, interferon-γ and lipopolysaccharide. We found that the half-life (t½) of endogenous SOCS3 is 0.7 h in activated macrophages, compared with a t½ of 6.2 h for recombinant CP-SOCS3. Deletion of the SOCS box in CP-SOCS3 renders it more resistant to proteasomal degradation, extending its t½ to 29 h. Consequently, this SOCS box-deleted form of CP-SOCS3 displays persistent inhibitory activity for 24 h toward interferon-γ- and lipopolysaccharide-induced cytokine and chemokine production. Compared with the wild-type suppressor, this gain-of-function CP-SOCS3 mutant provides a longer acting inhibitor of cytokine signaling, a feature that offers a clear advantage for the intracellular delivery of proteins to treat acute or protracted inflammatory diseases.
Keywords: Cytokine Action, Inflammation, Intracellular Trafficking, Protein Motifs, Protein Turnover, Signal Transduction, Ubiquitination, Intracellular Delivery, Intracellular Protein Therapy
Introduction
Inflammation constitutes a fundamental mechanism of diseases caused by microbial, autoimmune, and metabolic factors. These inducers evoke production of cytokines, chemokines, and other mediators of the host immune and inflammatory response. The inflammatory response depends on tightly regulated intracellular signal transduction by stress-responsive transcription factors as positive effectors of proinflammatory signaling in the nucleus (1). The genome can respond physiologically to proinflammatory cues by expressing a set of suppressors that extinguish inflammation when the homeostatic balance is not excessively tipped in favor of proinflammatory agonists (e.g. IL-1, IL-6, TNF-α, and IFN-γ).4 Overproduction of these agonists contributes to runaway systemic inflammation dubbed “cytokine storm” that underlies life-threatening sepsis. Moreover, they mediate chronic tissue injury in inflammatory bowel disease, rheumatoid arthritis, multiple sclerosis, and other autoimmune diseases (2, 3). To counteract the deleterious action of proinflammatory cytokines and chemokines, a set of extracellular anti-inflammatory molecules including transforming growth factor-β, IL-10, and IL-1 receptor antagonist are produced. In addition, an intracellular negative feedback system has evolved to limit the duration and strength of proinflammatory signaling. This system is comprised of intracellular inhibitory proteins such as, interleukin 1-receptor-associated kinase (IRAK)-M, an inhibitory member of the IRAK family, inhibitors of transcription factor NF-κB (IκB), proteins that inhibit activated STAT (PIAS), suppressors of cytokine signaling (SOCS), and ubiquitin-modifying enzyme A20 (4–7). Although SH2-containing inositol 5-phosphatases (SHIP and SHIP 1) counteract signaling events based on tyrosine phosphorylation, SOCS proteins prevent cytokine receptor signaling by binding to the cytoplasmic tail of cytokine receptors and/or catalytic sites on JAK kinases.
SOCS proteins are encoded by immediate early genes and they influence the extent and outcome of proinflammatory cytokine signaling (4). The SOCS family is composed of eight cytoplasmic SH2 domain-containing proteins: SOCS1 to SOCS7 and cytokine-inducible SH2. These physiologic suppressors uniquely disrupt proinflammatory signaling by either inhibiting the activity of JAK kinases or interacting with the cytoplasmic segment of ligand-occupied cytokine receptors (8). In addition, SOCS proteins contain a C-terminal SOCS box that associates with elongins B/C and cullin to form a ubiquitin E3 ligase that targets SOCS proteins and their signaling complexes for proteasomal degradation (9). Several members of the SOCS family, including SOCS1 and SOCS3, contain a proline, glutamine, serine, threonine (PEST) motif, which targets proteins for rapid intracellular proteolysis by calpain proteases (10). Among the SOCS family members, SOCS1 and SOCS3 are the best characterized in terms of their abilities to regulate proinflammatory cytokine signaling. Although structurally similar to SOCS1, SOCS3 does not inhibit cytokine signaling by binding directly to JAK, rather it inhibits JAK only in the presence of gp130 (11).
Using a new technology platform, we recently harnessed these two physiologic inhibitors of inflammation and apoptosis for intracellular delivery (12, 13). Functional studies demonstrate that cell-penetrating (CP) forms of SOCS1 and SOCS3 potently inhibit the JAK/STAT signaling pathway in cultured cells and CP-SOCS3 suppresses inflammation and protects vital organs from failure in mice challenged with the superantigenic staphylococcal enterotoxin B or endotoxic lipopolysaccharide (12, 13). Unexpectedly, when fluorescently tagged CP-SOCS3 was administered in vivo we noted its striking persistence in blood leukocytes, lymphocytes, and spleen cells (12). These findings led us to investigate in this present work, the intracellular turnover of CP-SOCS3 as compared with endogenous SOCS3 induced by proinflammatory agonists. We also investigated the role of the PEST motif and SOCS box in the turnover of CP-SOCS3 and its endogenous counterpart. Our results indicate a remarkable half-life prolongation for CP-SOCS3 as compared with endogenous wild-type SOCS3 and provide compelling evidence that protein degradation motifs play an important role in the turnover of full-length SOCS3. Moreover, deletion of the SOCS box, which controls proteasomal degradation, led to a much longer acting (t½ = 29 h) suppressor of proinflammatory agonist-induced cytokine and chemokine production.
EXPERIMENTAL PROCEDURES
Cell Culture
RAW 264.7 macrophages (a murine peritoneal macrophage cell line) and AMJ2.C8 macrophages (a murine alveolar macrophage cell line) were cultured in DMEM (Mediatech) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Atlanta Biologicals), and 100 units/ml of penicillin, 100 μg/ml of streptomycin (Mediatech) under standard conditions. Primary bone marrow-derived macrophages (BMDM) were prepared as follows: female 8-week-old C3H/HeJ mice were sacrificed and their femurs removed with the hip and knee joints intact. Femurs were sterilized by rinsing in 70% ethanol and the hip and knee joints were removed. Bone marrow cells were collected by inserting a 27-gauge needle into the open end of bone and flushing the marrow with 10 ml of DMEM. The cell suspension was filtered through a 70-μm nylon membrane and pelleted by centrifugation. Cells were resuspended in DMEM supplemented with 10% FBS, 10 mm HEPES (Mediatech), 50 units/ml of penicillin, 50 μg/ml of streptomycin, and 20% L-cell-conditioned medium to direct differentiation of naive bone marrow cells to macrophages. On day 3 of culture, fresh medium was replaced. On day 7, the purity of the macrophage culture was determined to be ≥95% as measured by FACS analysis gating on macrophage-specific cell surface markers. BMDM were then plated and used in the experiments as indicated.
SOCS3 Plasmid Constructs
Full-length murine SOCS3 was provided by M. Shong, Chungnam National University, Korea. The hydrophobic membrane translocating motif (MTM) was derived from a hydrophobic region of the signal sequence of fibroblast growth factor 4 (14). The MTM and/or His6 tag were added to SOCS3 using standard PCR conditions. The following primers were used to engineer full-length and SOCS3 deletion mutant constructs: non-CP-SOCS3, forward, 5′-CG GGA TCC GCC ATG GCC CAT CAT CAC CAT CAC CAT AAT GCC CAT ACC GGT ATG GTC ACC CAC AGC AAG TTT CCC G-3′ and reverse, 5′-GA GAA TTC TTA AAG TGG AGC ATC ATA CTG ATC CAG G-3′; CP-SOCS3, forward, 5′-CG GGA TCC GCC ATG GCC CAT CAT CAC CAT CAC CAT AAT GCC CAT ACC GGT GCA GCT GTG CTT CTC CCT GTG C-3′ and reverse, 5′-GA GAA TTC TTA AAG TGG AGC ATC ATA CTG ATC CAG G-3′; CP-SOCS3ΔSB, forward, 5′-CG GGA TCC GCC ATG GCC CAT CAT CAC CAT CAC CAT AAT GCC CAT ACC GGT GCA GCT GTG CTT CTC CCT GTG C-3′ and reverse, 5′-GA GGA TTC TTA GGA GGA GAG AGG TCG GCT CAG TAC CAG C-3′.
Production of Recombinant Proteins
Non-CP-SOCS3, CP-SOCS3, and CP-SOCS3ΔSB constructs were cloned into the pET21a(+) vector using standard engineering techniques. Plasmid constructs were then transformed into BL21(DE3) recombinant protein Escherichia coli cells and positive clones were identified and verified by DNA sequencing. Positive clones were grown up in liquid Luria broth cultures, containing ampicillin. Expression of proteins was induced by incubating bacterial cells for 3 h with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside. After 3 h, cells were collected by centrifugation, cell pellets were weighed and resuspended in 5 ml/g weight of resuspension buffer (100 mm sodium phosphate, monobasic, 10 mm Tris base, 8 m urea, pH 8.0). Bacterial cells were lysed by sonication and recombinant SOCS3 proteins were purified with histidine affinity column by fast protein liquid chromatography (AKTA Purifier, GE Healthcare, Piscataway, NJ) using nickel-nitrilotriacetic acid resin (Qiagen). The protein was then refolded through a two-step dialysis to remove denaturant (13). The final diluent was DMEM supplemented with penicillin/streptomycin (concentrations listed above). Identification of purified proteins was done by Western blot analysis (Fig. 1B). Protein concentration of CP-SOCS3 and CP-SOCS3ΔSB was determined by the Bradford assay. Proteins were stored at −40 (long term) or 4 °C (short term) until used in assays.
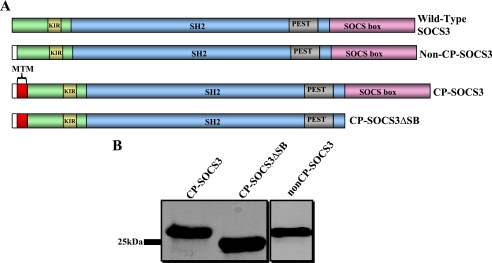
FIGURE 1.
Design of recombinant CP-SOCS3 proteins for bacterial expression and affinity purification. A, schematic representation of full-length wild-type SOCS3, showing the different functional domains of the protein: KIR (kinase inhibitory region), SH2 domain, PEST motif, and SOCS box; non-CP-SOCS3, the non-cell penetrating SOCS3 that lacks the MTM, but contains an N-terminal His6 tag (white); CP-SOCS3, cell-penetrating full-length SOCS3 with a 12-amino acid MTM (red) at the NH2-terminal, and His6 tag (white); CP-SOCS3ΔSB, cell-penetrating SOCS3 deletion mutant lacking the C-terminal SOCS box, but possessing the MTM (red) and His6 tag (white) at the N terminus. B, immunoblot displaying expressed and purified non-CP-SOCS3 (26.6 kDa), CP-SOCS3 (27.9 kDa), and CP-SOCS3ΔSB (23.4 kDa).
Half-life Determination
The half-life of endogenous SOCS3 was determined as follows: RAW cells were stimulated with 100 units/ml of IFN-γ (EMD Biosciences) and 250 ng/ml of LPS (Sigma) for 4 h to induce SOCS3 expression. This 4-h post-stimulation time point represents t = 0 h, from which cells were harvested following the indicated treatments. In the designated experiments, cells were treated with 15 μg/ml of cycloheximide (Sigma) 4 h after LPS/IFN-γ stimulation, 1 μm epoxomicin (Sigma) 2 h after LPS/IFN-γ stimulation, and 40 μg/ml of calpeptin (VWR) was added 3.5 h post-stimulation. Following treatment, cells were harvested at the indicated time points, and lysed with 1× CBA lysis buffer (BD Biosciences). Samples were then heated at 100 °C for 20 min, and centrifuged to clear lysates of cellular debris. Supernatants were then snap frozen and stored at −40 °C until immunoblotting was performed. The half-life of recombinant full-length CP-SOCS3 or CP-SOCS3ΔSB proteins was determined as follows: RAW cells were stimulated for 1 h with 100 units/ml of IFN-γ and 250 ng/ml of LPS, and during the same time interval, cells were treated with 1 μm CP-SOCS3 or CP-SOCS3ΔSB while incubating at 37 °C. Cells were rinsed 3 times with PBS (Mediatech) warmed to 37 °C, and treated with 10 μg/ml of proteinase K (Sigma) for 15 min to degrade any non-internalized protein attached to the outside of the cell. Cells were rinsed again 3 times with warm PBS, and incubated with DMEM supplemented with FBS and penicillin/streptomycin (see above). At the indicated time intervals, cells were harvested, lysed, and prepared for immunoblotting, as described above. The medium was also collected, snap frozen at −40 °C, and later analyzed by immunoblot for SOCS3 protein.
Immunoblotting
Lysates that were collected from half-life assays, cytokine/chemokine assays, and STAT1 phosphorylation assays were mixed with 6× SDS loading buffer and boiled at 100 °C for 5 min. Samples were resolved on 12% SDS-PAGE and transferred to nitrocellulose membrane. Membranes were probed with rabbit anti-SOCS3 (Abcam), which recognizes a C-terminal epitope of SOCS3, or rabbit anti-His6 tag antibody (Rockland), or mouse anti-pSTAT1 (Y701) antibody (BD Biosciences) and mouse anti-β-actin (Abcam) according to the manufacturer's protocol. Bands were developed using the following secondary antibodies: donkey anti-rabbit IR Dye 800 (LI-COR Biosciences) and donkey anti-mouse IR Dye 680 (LI-COR Biosciences). Probing was performed according to the individual manufacturer's protocol. Bands were visualized using Licor's Odyssey Infrared Imaging System. SOCS3 and His6 tag protein bands were normalized against the levels of expressed β-actin. Quantification and analysis of bands were performed using Odyssey software (version 3.0).
STAT1 Phosphorylation Assay
BMDM or AMJ2.C8 macrophages (plated the previous day with 5 × 106 cells/well in a 12-well plate) were pre-treated for 1 h with 10 μm CP-SOCS3 or 10 μm CP-SOCS3ΔSB. Medium containing the added protein was removed and replaced with DMEM supplemented with FBS, HEPES, penicillin/streptomycin, and L-cell-conditioned medium (for BMDM) or DMEM supplemented with FBS and penicillin/streptomycin (for AMJ2.C8 macrophages). Cells were then stimulated with 100 units/ml of IFN-γ and 0.2 μg/ml of LPS for 15 min. Cells were lysed with 1× CBA lysis buffer. Lysates were assayed to determine the levels of phosphorylated STAT1 using the phospho-STAT1 (Tyr-701) Flex Set Cytometric Bead Array (BD Biosciences) according to the manufacturer's protocol. Flow cytometry was performed using BD FACS Calibur, and data acquisition and analysis was performed using BD Pro CellQuest Software and BD 4-Bead analysis software. AMJ2.C8 and BMDM lysates were also subjected to Western blot analysis to verify phosphorylated STAT1 levels. Lysates were prepared for immunoblotting as described above. At the end of the stimulation period, cell viability was ≥95% after staining cells with fluorescein and ethidium bromide to detect live and dead cells (15).
Cytokine and Chemokine Analysis
Cultured primary cells (BMDM) or an established cell line (AMJ2.C8 macrophages), plated the previous day with 1 × 106 cells/well in a 96-well plate, were pre-treated for 1 h with 10 μm of the indicated cell-penetrating protein (CP-SOCS3 or CP-SOCS3ΔSB). Medium containing added protein was then removed and replaced with fresh DMEM supplemented with FBS, HEPES, penicillin/streptomycin, and L-cell-conditioned medium (for BMDM) or FBS and penicillin/streptomycin (for AMJ2.C8 macrophages). At 6 or 24 h after CP-SOCS3 or CP-SOCS3ΔSB treatments, cells were stimulated with 100 units/ml of IFN-γ and 500 ng/ml of LPS for 6 h. Supernatants (50 μl) were collected before CP-proteins or diluent were added (0 h) and at the end of the 6-h activation with proinflammatory agonists. This means that test samples were analyzed at 12 and 30 h following CP-protein treatment. Samples were assayed for the presence of inflammatory cytokine/chemokine using the Mouse Inflammatory Cytokine Bead Array (CBA) kit (BD Biosciences). Cytokine analysis was performed according to the manufacturer's protocol and flow cytometry was performed using BD FACS Calibur. Data acquisition and analysis were done using BD Pro CellQuest Software and BD 6-bead analysis software. Cells were also harvested at the end of the stimulation period (12 and 30 h), and lysates were immunoblotted to determine the level of cell-penetrating proteins remaining in the cell. Lysates were prepared for immunoblotting as described above. Additionally, at the end of the stimulation period, cell viability was ≥95% after staining cells with fluorescein and ethidium bromide to detect live and dead cells (15).
Fluorescein Isothiocyanate (FITC) Labeling of Proteins
Recombinant SOCS3 proteins were labeled with FITC (Pierce) as previously reported (12), and briefly described here. Approximately 1 mg of CP-SOCS3 or CP-SOCS3ΔSB was added to 0.5 ml of conjugation buffer (0.4 m carbonate, 0.1 m bicarbonate final concentration, pH 9.0). FITC was dissolved into dimethylformamide to a final concentration of 30 mg/ml. A 2-fold excess of FITC solution was added to the cell-penetrating protein/conjugation buffer, and the mixture was gently stirred for 1 h at room temperature in the dark. FITC-CP-protein solution was additionally incubated at 37 °C in the dark to ensure labeling. After labeling, proteins were dialyzed in the dark against DMEM (no FBS or penicillin/streptomycin supplement) for 2–4 h to remove excess dye. The relative fluorescence of the FITC-labeled proteins was determined using a Fusion Universal Microplate Analyzer (PerkinElmer Life Sciences) at 485 nm excitation, 535 nm emission, and 20 nm band pass. Protein solutions with equivalent fluorescence units were used in all experiments. A solution of FITC only was used as a control for labeling. FITC-labeled proteins were stored at 4 °C until added to RAW cells for intracellular delivery and subcellular localization experiments.
Protease Accessibility Assay and Confocal Microscopy
RAW cells were plated at 1 × 105 cells in a MAT-TEK 35-mm plate with a number 1.5 coverglass, and incubated overnight at 37 °C. The following day, medium was replaced with DMEM (no FBS supplement), and ∼1 μm FITC-labeled recombinant proteins or FITC only (based on equivalent fluorescence) was added to cells, and incubated at 37 °C for 1 h. Cells were gently rinsed three times with warm PBS, and treated for 15 min at 37 °C with 10 μg/ml of proteinase K to degrade any protein not internalized into cells. Cells were again rinsed three times with PBS, and cold fresh medium supplemented with FBS was placed on the cells. RAW cells were kept cold until microscopy could be completed. In experiments to determine intracellular localization of recombinant FITC-labeled SOCS3 proteins, cells were additionally treated with 5 μm FM595 (Invitrogen), a fluorescent marker for endosomal/plasma membrane. Cells were labeled for 5 min at 25 °C following proteinase K treatment to remove non-internalized FITC-labeled CP-SOCS3, CP-SOCS3ΔSB, and non-CP-SOCS3 (control). Cells were then rinsed three times with cold PBS, and incubated with DMEM (supplemented with 10% FBS), and kept on ice. Microscopy examination was completed immediately after labeling. Confocal microscopy was performed using a Zeiss LSM 310 META inverted confocal microscope, and results were analyzed with Zeiss LSM Image Browser (version 4.2.0.121).
Statistical Analysis
Experimental data were graphed using GraphPad Prism 4 (Version 4.03) software. Two-way analysis of variance was used to determine the significance of difference between groups of data. Data are expressed as mean ± S.E.
RESULTS AND DISCUSSION
The outcome of inflammation depends on the genome-orchestrated balance between proinflammatory mediators and anti-inflammatory suppressors. Prior studies have firmly established that SOCS3 inhibits pro-inflammatory signaling at the level of the JAK/STAT pathway (4, 16). However, excessive pro-inflammatory signaling can overwhelm this protective mechanism, leading to SOCS3 degradation via the Ub-proteasome pathway, depletion of intracellular SOCS3 stores, and attendant pathological consequences. In this regard, we have engineered CP forms of wild-type SOCS3 that are persistently expressed in primary, immunocompetent cells and that function as potent anti-inflammatory suppressors in vivo (12). We investigated the possibility of further reinforcing the intracellular pool of SOCS3 and extending its anti-inflammatory potential by engineering a degradation-resistant form of the protein. The development and characterization of this mutant led to experimental proof of its significantly prolonged inhibition of proinflammatory signaling in an inflammation-relevant cell type.
Endogenous SOCS3 Is Rapidly Degraded in Stimulated RAW Macrophages
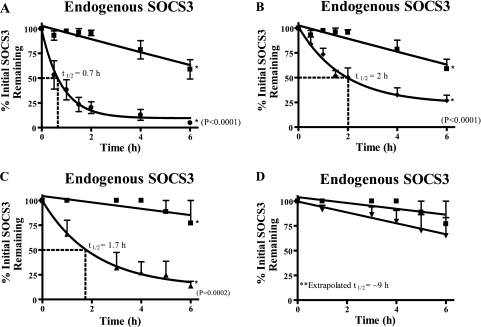
Previous protein turnover studies of transfected SOCS3 indicated a t½ of 1.6 h when the protein is overexpressed in monkey COS cells (17). To determine the t½ of endogenously expressed SOCS3 in the murine peritoneal macrophage cell line, RAW 264.7, we established conditions for quantitative measurement of its expression upon stimulation with proinflammatory agonists. RAW macrophages were stimulated for 4 h with LPS and IFN-γ to induce SOCS3 protein that was readily measured by quantitative immunoblotting using the infrared Odyssey system. At this time point, cycloheximide (15 μg/ml) was added to inhibit de novo protein synthesis and the cells were sampled at specified time intervals to determine SOCS3 protein levels. Endogenous SOCS3 was rapidly degraded as documented by its t½ of 39 min or 0.7 h (Fig. 2A). This rate of SOCS3 turnover is similar to that observed in the murine pro-B cell line Ba/F3 (18) but faster than that of ectopically expressed SOCS3 in COS cells (17). Rapid turnover of SOCS3 depends on its two protein degradation motifs, a C-terminal SOCS box and a PEST motif (10) (Fig. 1). The proteasome-independent degradation mechanism is based on recognition and cleavage of PEST sequences by calpain proteases (19–22). In turn, the SOCS box functions as a platform for E3 ligase formed by elongin B/C and cullin 5 to target the SOCS3 protein for ubiquitin-mediated proteasome degradation (23, 24). SOCS box-dependent proteasomal degradation can be attenuated by the irreversible inhibitor epoxomicin. Therefore, we analyzed endogenous SOCS3 turnover in RAW macrophages treated with inhibitors of proteasome- and calpain-based proteolysis to determine the role of these degradative pathways in the stability of endogenous SOCS3. Treatment with epoxomicin extended the t½ of SOCS3 from 0.7 to 2 h (Fig. 2B). When cells were treated with calpeptin to inhibit the activity of calpain proteases that recognize the PEST motif, the rate of SOCS3 turnover was also increased from 0.7 to 1.7 h (Fig. 2C). Combined treatment of RAW macrophages with both inhibitors extended the t½ over 10-fold for endogenous SOCS3 to ∼9 h (Fig. 2D). Taken together, these results indicate that inhibitors of proteasomes and calpain act synergistically to inhibit degradation of endogenous SOCS3 mediated by its SOCS box and PEST motif.
FIGURE 2.
Endogenous SOCS3 turnover in RAW macrophages stimulated with proinflammatory agonists. RAW 264.7 cells were stimulated with 100 units/ml of IFN-γ and 250 ng/ml of LPS for 4 h to induce SOCS3 expression (t = 0). After the treatments, samples were collected at 0, 0.5, 1, 1.5, 2, 4, and 6 h. SOCS3 protein levels were quantified by immunoblotting (IB) using the infrared Odyssey Li-Cor system software. A, RAW cells were incubated without (squares) or with (circles) 15 μg/ml of cycloheximide. B, RAW cells were incubated without (squares) or with (diamonds) 15 μg/ml of cycloheximide and 1 μm epoxomicin. C, RAW cells incubated without (squares) or with (triangles) 15 μg/ml of cycloheximide plus 40 μg/ml of calpeptin. D, RAW cells were incubated without (squares) or with (inverted triangle) all three inhibitors: 15 μg/ml of cycloheximide, 40 μg/ml of calpeptin, and 1 μm epoxomicin. The bars represent the mean ± S.E. of n = 3 (A and B) or n = 4 (C and D) independent experiments.
Intracellular Delivery and Turnover of Recombinant Cell-penetrating SOCS3
The rapid turnover of endogenous SOCS3 in macrophages exceeds that of the previously reported value for ectopically expressed SOCS3 in a COS cell line (17). Although the forced expression of genes that encode intracellular signal transducers and their regulators has provided valuable information about the mechanism of intracellular action of these molecules, this method is subject to variable efficiency of transfection and an inability to control the abundance of the expressed proteins. In contrast, intracellular delivery of physiologic proteins based on the attachment of a MTM to a recombinant intracellular anti-inflammatory protein allows its controlled delivery in terms of time and concentration to analyze and inhibit signal transduction. We demonstrated previously that recombinant CP-SOCS3 inhibits the JAK/STAT pathway and prevents cytokine-mediated lethal inflammation and apoptosis of the liver (12). We hypothesized that CP-SOCS3 may have an extended t½ relative to endogenous SOCS3, as FITC-labeled CP-SOCS3 persists for 8 h in blood leukocytes, lymphocytes, and spleen cells in vivo (12). To investigate this possibility, we engineered not only full-length CP-SOCS3 but also a deletion mutant in which the SOCS box (amino acids 185–225) had been deleted (Fig. 1A). This mutant, CP-SOCS3ΔSB, was comprised of amino acids 1–184 of murine SOCS3, including the recently discovered PEST motif (10). We added a 12-amino acid MTM at the NH2-terminal end of the recombinant protein, which enabled recombinant SOCS3 to cross the plasma membrane in cultured cells and in vivo (12) (Fig. 1A).
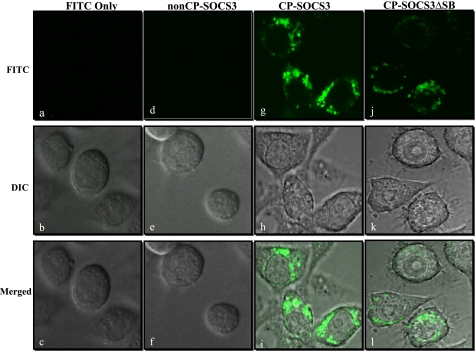
Cellular uptake of full-length CP-SOCS3 and CP-SOCS3ΔSB proteins labeled with FITC was analyzed using the protease accessibility assay (12). In this assay, FITC-labeled purified recombinant proteins are added to RAW macrophages for 1 h to allow entry of proteins into the cells. After washing the cells to remove extracellular pools of FITC-labeled proteins, proteinase K was added to the medium to degrade any membrane-bound proteins not translocated into cells. Proteinase K-treated cells were analyzed directly (without fixation) by confocal microscopy to visualize fluorescent signals indicative of internalized proteins (Fig. 3). As a control, SOCS3 lacking the MTM (non-CP-SOCS3) (Fig. 1A) was not detected in the cells following the protease accessibility assay (Fig. 3, d–f). In striking contrast, full-length CP-SOCS and CP-SOCS3ΔSB deletion mutants produced strong fluorescent signals in RAW macrophages (Fig. 3, g–l). Significantly, the fluorescent signal was detected in the cytoplasm and not in the nuclei of RAW macrophages indicating that deleting the SOCS box did not alter intracellular distribution of CP-SOCS3ΔSB.
FIGURE 3.
Intracellular delivery of recombinant SOCS3 proteins. Fluorescence confocal laser scanning microscopy of proteinase K-treated and non-fixed RAW macrophages shows intracellular localization of FITC-labeled CP-SOCS3 proteins (green). a–l, images presented represent a 13-μm midcell cross-section. a–c, confocal images of RAW cells incubated with FITC alone. a, FITC image: no fluorescent signal observed. b, differential interference contrast (DIC) image of the RAW cells depicted. c, merged view of DIC and FITC images. d–f, confocal images of RAW cells incubated with FITC-labeled non-CP-SOCS3. d, FITC image, no fluorescent signal detected. e, DIC image of the RAW cells depicted. f, merged view of DIC and FITC images. g–i, confocal images of RAW cells incubated with FITC-labeled CP-SOCS3. g, FITC image, strong fluorescence throughout the cytoplasm. h, DIC image of RAW cells depicted. i, merged view of DIC and FITC images showing localization of FITC-labeled CP-SOCS3 throughout the cytoplasm of the RAW cells. j–l, confocal images of RAW cells incubated with FITC-labeled CP-SOCS3ΔSB. j, FITC image, strong fluorescence throughout the cytoplasm. k, DIC image of RAW cells depicted. l, merged view of DIC and FITC images showing localization of FITC-labeled CPSOCS3ΔSB throughout the cytoplasm. All images are representative of multiple unfixed cells from three independent experiments.
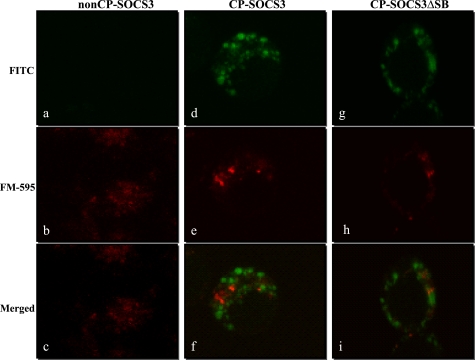
Our previous study of the mechanism of intracellular delivery of short peptides (Mr = 2,800) documented an endocytosis-independent process of crossing the plasma membrane mediated by hydrophobic MTM (15). In particular, the helical hairpin design of the MTM allows for its insertion directly into the plasma membrane, and plausibly the “looping-unlooping” of the hairpin allows for the movement of attached peptides through the phospholipid bilayer to the interior of the cell (15). However, it is possible that a larger “cargo,” such as that of SOCS3 proteins (Mr = 27,000), may induce uptake through the endosomal pathway. Therefore, to address the potential role of endocytosis in the intracellular delivery of CP-SOCS3, we analyzed its subcellular distribution as compared with CP-SOCS3ΔSB in RAW macrophages that had also been labeled with FM-595, an endosomal/plasma membrane marker. Confocal microscope analysis revealed that the FITC-labeled recombinant cell-penetrating SOCS3 proteins (CP-SOCS3 and CP-SOCS3ΔSB), and the endosomal marker FM-595 exhibited distinct distribution throughout the cytoplasm, and did not appear to co-localize with one another (Fig. 4). Interestingly, both CP-SOCS3 and CP-SOCS3ΔSB displayed a punctuate pattern of dispersal throughout the cytoplasm of RAW macrophages, possibly suggesting that these proteins self-aggregate or interact with large signaling complexes in the cytosol. This is of potential significance because aggregates of CP-SOCS3 or CP-SOCS3ΔSB could much more efficiently sequester target proteins in the cytosol thereby interfering with pro-inflammatory signaling pathways (i.e. JAK/STAT pathway). Consistent with the initial results shown in Fig. 3, FITC-labeled non-CP-SOCS3 did not enter the cell, and only the endosomal marker signal was detected in these samples (Fig. 4, a–c). These results suggest that CP-SOCS3 and CP-SOCS3ΔSB are delivered to the cytoplasm of RAW macrophages by crossing the plasma membrane independently of the endosomal pathway, thereby avoiding its influence on intracellular turnover of recombinant SOCS3 proteins.
FIGURE 4.
Intracellular delivery of CP-SOCS3 and CP-SOCS3ΔSB is mostly independent of endosomal membrane compartment. Fluorescence confocal laser scanning microscopy of RAW macrophages incubated with FITC-labeled recombinant proteins (green), and the endosomal marker FM-595 (red). a–i, images presented represent a 10-μm midcell cross-section. a–c, RAW cells incubated with FITC-labeled non-CP-SOCS3 (green) and FM-595 (red). a, FITC image, no fluorescent signal detected. b, FM-595 only, endosomes detected throughout the cell. c, merged view of FITC and FM-595 images. d–f, confocal images of RAW cells incubated with FITC-labeled CP-SOCS3 and FM-595. d, FITC image, fluorescent signal throughout the cytoplasm. e, FM-595 only, endosomes detected throughout the cell. f, merged view of FITC and FM-595 images, no overlapping green and red fluorescent signals. g–i, confocal images of RAW cells incubated with FITC-labeled CP-SOCS3ΔSB and FM-595. g, FITC images, fluorescent signal throughout the cytoplasm. h, FM-595 only, endosomes detected throughout the cell. i, merged view of FITC and FM-595 images, no overlapping green and red fluorescent signals. All images are representative of multiple unfixed cells from three independent experiments.
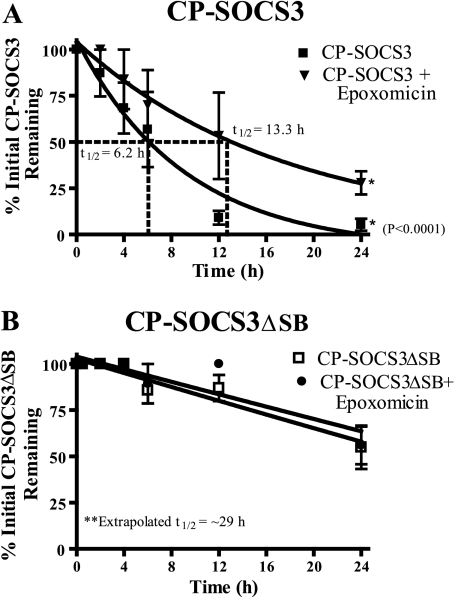
Having established intracellular delivery of recombinant CP-SOCS3 and CP-SOCS3ΔSB that appears to bypass the endocytic pathway, we determined their t½ in RAW cells under the same conditions as employed in the t½ measurements of the endogenous SOCS3 (see above). Proinflammatory agonist-stimulated RAW macrophages were pulsed with CP-SOCS3 or CP-SOCS3ΔSB (1 μm final concentration), and samples collected at regular intervals were analyzed by quantitative immunoblotting. Because these experiments were performed in a stimulated cell line known to express SOCS3 under the same conditions (Fig. 2), blots were probed with an anti-His6 tag antibody to distinguish recombinant SOCS3 proteins from endogenous SOCS3. As a control, samples from RAW macrophages that did not receive recombinant cell-penetrating protein treatment, but were stimulated with IFN-γ and LPS were probed with the anti-His6 tag antibody. No band corresponding to the molecular weight of SOCS3 was detected (data not shown). We found that CP-SOCS3 had a t½ of 6.2 h (Fig. 5A) as compared with the much shorter t½ of 0.7 h for endogenous SOCS3. Hence, in the absence of protease inhibitors, recombinant CP-SOCS3 displays a significantly extended half-life. These results help explain the previous in vivo observations in which FITC-labeled CP-SOCS3 was detectable in the blood leukocytes and lymphocytes and spleen cells of mice 8 h after intraperitoneal administration (12). The t½ of CP-SOCS3 is ∼9 times longer than that of endogenous SOCS3 (0.7 h), but is similar to that of endogenous SOCS3 in RAW macrophages treated with inhibitors of proteolysis mediated by calpain and proteasomes (Fig. 2D). Under the experimental conditions employed in these experiments, CP-SOCS3 appears to be more resistant to these two intracellular protein degradation mechanisms than endogenous SOCS3. Moreover, the t½ of CP-SOCS3 is extended to 13.3 h, when the proteasomal pathway of proteolysis is inhibited with epoxomicin (Fig. 5A). This result indicates that the CP-SOCS3 turnover is in part regulated by the proteasomal pathway of degradation.
FIGURE 5.
Proteasomal inhibitor extends the half-life of CP-SOCS3 and deletion of the SOCS box dramatically improves the intracellular stability of CP-SOCS3. A, stimulated RAW macrophages were treated for 1 h with CP-SOCS3 in the presence (inverted triangle) or absence (squares) of 1 μm epoxomicin. Half-life was determined by immunoblot analysis of samples collected at 0, 0.5, 2, 4, 6, 12, and 24 h. B, stimulated RAW macrophages were treated for 1 h with CP-SOCS3ΔSB in the absence (open squares) or presence (solid circles) of 1 μm epoxomicin. Half-life was determined by immunoblot analysis of samples collected at 0, 0.5, 2, 4, 6, 12, and 24 h. The bars represent the mean ± S.E. of three independent experiments (n = 3).
The role of the proteasomal pathway in CP-SOCS3 degradation was further explored by analysis of the CP-SOCS3 mutant, CP-SOCS3ΔSB, in which the SOCS box has been deleted (Fig. 5B). The t½ of CP-SOCS3ΔSB was extended to ∼29 h, a remarkable 41-fold increase in stability relative to endogenous SOCS3. In comparison, NH2-truncated SOCS3 that lacked Lys-6 displayed only a 4-fold gain in stability following retroviral transduction of the pro-B lymphocyte Ba/F3 cell line (18). Importantly, the t½ of CP-SOCS3ΔSB remained virtually unchanged in the presence of epoxomicin (Fig. 5B), thereby providing additional proof that CP-SOCS3ΔSB is resistant to proteasomal degradation.
CP-SOCS3ΔSB Inhibits STAT1 Phosphorylation
It has been firmly established that SOCS3 regulates the JAK/STAT pathway by binding to both activated JAK kinase, and/or the cytoplasmic domain of the phosphorylated gp130 receptor, which inhibits docking and subsequent activation of STAT proteins (25–30). SOCS3, induced by Toll-like receptor 4 stimulation, indirectly influences the outcome of this signaling pathway by modulating the secondary effects of LPS-induced cytokines that act through MyD88/TRAF 6 or the JAK/STAT pathway (16). Moreover, SOCS3 deficiency in cells causes a significant increase in STAT1 phosphorylation and an IFN-γ-like gene response (31–33). We previously reported that full-length CP-SOCS3 can inhibit STAT1 phosphorylation in AMJ2.C8 macrophages (12). Because we use IFN-γ and LPS to stimulate proinflammatory signaling in macrophages, we ascertained whether CP-SOCS3ΔSB retains inhibitory activity toward STAT1 phosphorylation. Therefore, we treated AMJ2.C8 macrophages with either CP-SOCS3 or CP-SOCS3ΔSB for 1 h. After replacing the medium, the macrophages were stimulated with IFN-γ and LPS for 15 min to induce STAT1 phosphorylation. The cells were harvested and lysates were assayed for phosphorylated STAT1 using a flow cytometric bead-based assay. We determined that CP-SOCS3 or CP-SOCS3ΔSB reduced STAT1 phosphorylation in AMJ2.C8 macrophages (Fig. 6A). Western blot analysis of the lysates verified lower levels of phosphorylated STAT1 in CP-SOCS3- and CP-SOCS3ΔSB-treated samples as compared with untreated controls (Fig. 6B). Thus, the lack of SOCS box in CP-SOCS3ΔSB did not impede its ability to inhibit STAT1 phosphorylation in IFN-γ- and LPS-stimulated cells.
FIGURE 6.
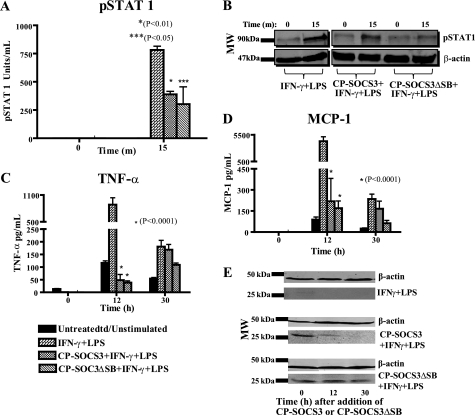
CP-SOCS3ΔSB inhibits STAT1 phosphorylation and displays prolonged anti-inflammatory activity associated with intracellular persistence as compared with full-length CP-SOCS3 in AMJ2.C8 macrophage cell line. A and B, after a 1-h pre-treatment of macrophages with cell-penetrating proteins, cells were stimulated with 100 units/ml of IFN-γ and 0.2 μg/ml of LPS for 15 min. Cells were harvested with 1× CBA lysis buffer and analyzed for phosphorylated STAT1 levels by CBA. A, pSTAT1 (units/ml). B, immunoblotting results of pSTAT1 in AMJ2.C8 macrophages treated with CP-SOCS3 or CP-SOCS3ΔSB for 1 h and stimulated for 15 min. C and D, the cells were treated for 1 h with CP-SOCS3 or CP-SOCS3ΔSB. Six or 24 h following protein treatment, cells were stimulated with 100 units/ml of IFN-γ and 0.5 μg/ml of LPS for 6 h. Supernatants were collected before treatment (t = 0 h) and after the 6-h stimulation, at 12 and 30 h, respectively. Samples analyzed for inflammatory cytokine/chemokine production by CBA. C, TNF-α (pg/ml). D, MCP-1 (pg/ml). E, immunoblotting results of CP-SOCS3 or CP-SOCS3ΔSB protein levels in cells after 6 h stimulation, at 12 and 30 h. The bars represent the mean ± S.E. of n = 3 (A and B) or n = 4 (C–E) independent experiments.
This analysis was extended to primary macrophages. BMDM were treated with CP-SOCS3 or CP-SOCS3ΔSB and stimulated with IFN-γ and LPS according to the same protocol as outlined above. These primary cells displayed a heightened response to IFN-γ and LPS as attested by the higher level of STAT1 phosphorylation. Nevertheless, STAT1 phosphorylation was reduced in CP-SOCS3- or CP-SOCS3ΔSB-treated BMDM (Fig. 7A). These results were verified by immunoblot analysis of phosphorylated STAT1 (Fig. 7B). Cumulatively, these functional data demonstrate that the SOCS box deletion mutant, CP-SOCS3ΔSB, functions through a similar mechanism as a full-length CP-SOCS3. Both inhibit STAT1 phosphorylation through the interaction with the cytokine receptor and/or JAK kinase. It is thus apparent that the SOCS box in CP-SOCS3 is dispensable for its cytokine signaling suppressor function, whereas its intracellular turnover is greatly reduced.
FIGURE 7.
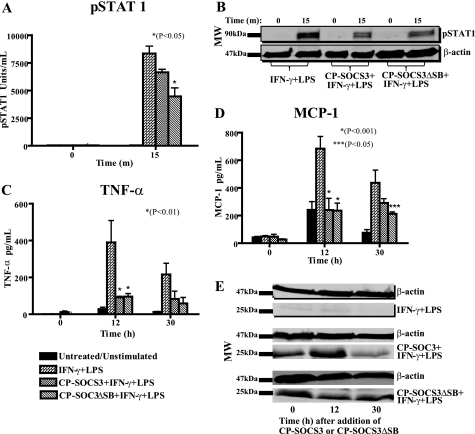
CP-SOCS3ΔSB inhibits STAT1 phosphorylation and displays prolonged anti-inflammatory activity associated with intracellular persistence in primary macrophages. BMDM obtained from C3H/HeJ mice were treated for 1 h with 10 μm CP-SOCS3 or 10 μm CP-SOCS3ΔSB. A and B, after a 1-h pre-treatment of macrophages with cell-penetrating proteins, cells were stimulated with 100 units/ml of IFN-γ and 0.2 μg/ml of LPS for 15 min. Cells were harvested with 1× CBA lysis buffer and analyzed for phosphorylated STAT1 by CBA. A, pSTAT1 (units/ml). B, immunoblots of pSTAT1 levels in BMDM treated with CP-SOCS3 or CP-SOCS3ΔSB for 1 h and stimulated for 15 min. C and D, 6 or 24 h following protein treatment, cells were stimulated with 100 units/ml of IFN-γ and 0.5 μg/ml of LPS for 6 h. Supernatants were collected before treatment (t = 0 h) and after the 6-h stimulation, at 12 and 30 h, respectively. Samples were analyzed for inflammatory cytokine/chemokine production by CBA. C, TNF-α (pg/ml). D, MCP-1 (pg/ml). E, immunoblots of CP-SOCS3 or CP-SOCS3ΔSB protein levels in cells after a 6-h stimulation, at 12 and 30 h. The bars represent the mean ± S.E. of n = 3 (A and B) or n = 4 (C–E) independent experiments.
Deletion of the SOCS Box Extends Cytokine/Chemokine Suppression Mediated by CP-SOCS3
We next assessed whether deletion of the SOCS box would change the inhibitory activity of the CP-SOCS3ΔSB mutant toward production of proinflammatory cytokines and chemokines upon intracellular delivery. To that end, AMJ2.C8 macrophages were treated with either CP-SOCS3 or CP-SOCS3ΔSB for 1 h; cells were then rinsed and replaced with fresh medium. At 6 or 24 h following cell-penetrating protein treatment, cells were stimulated with IFN-γ and LPS for 6 h (i.e. 12 and 30 h after CP-protein treatment, respectively), and samples were analyzed for proinflammatory cytokine and chemokine production. As shown in Fig. 6, both CP-SOCS3 and CP-SOCS3ΔSB inhibit proinflammatory agonist-induced production of the cytokine, TNF-α, and the chemokine, MCP-1, at 12 h post cell-penetrating protein treatment (Fig. 6, C and D). In contrast, at 30 h, only CP-SOCS3ΔSB maintained its inhibitory activity although the CP-SOCS3 anti-inflammatory effect was negligible. These functional results are consistent with the persistence of CP-SOCS3ΔSB in AMJ2.C8 macrophages at 30 h, as detected by immunoblotting (Fig. 6E). We extended these experiments to BMDM obtained from C3H/HeJ mice. These freshly obtained primary cells were treated with CP-SOCS3 or CP-SOCS3ΔSB for 1 h, followed by a 6-h stimulation with IFN-γ and LPS at 6 and 24 h post cell-penetrating protein treatment (see above). We found that both CP-SOCS3 and CP-SOCS3ΔSB significantly reduced the production of TNF-α and MCP-1 (Fig. 7, C and D) at 12 h post cell-penetrating protein treatment. At 30 h post-protein treatment, both proteins, CP-SOCS3 and CP-SOCS3ΔSB, suppressed the production of these inflammatory mediators as compared with untreated (non-cell-penetrating protein) controls, although inhibition by CP-SOCS3ΔSB was more significant than that of CP-SOCS3 (Fig. 7, C and D). Immunoblot analysis confirmed that CP-SOCS3ΔSB also persists for 30 h in BMDM, a fact that supports the functional data (Fig. 7E). Taken together, these results demonstrate that CP-SOCS3ΔSB, which has a significantly increased t½, also retains its inhibitory function following intracellular delivery, as evidenced by the reduced production of TNF-α and MCP-1.
Overall, our results identify key mechanisms that play a role in intracellular turnover of endogenous SOCS3 and recombinant CP-SOCS3. In addition, we developed and characterized a SOCS box-deleted form of CP-SOCS3 that has a greatly extended t½, which prolongs its ability to suppress proinflammatory cytokine signaling. The extended anti-inflammatory activity of CP-SOCS3ΔSB supports a recently published report that identified a SOCS box-independent mechanism of cytokine signaling suppression (24). Furthermore, the unexpectedly extended t½ of CP-SOCS3 suggests that addition of the MTM to recombinant CP-SOCS3ΔSB may provide a protective “shield,” against intracellular protein degradation mediated by the PEST domain and possibly other putative protein degradation sites in SOCS3. The truncated form of CP-SOCS3 (CP-SOCS3ΔSB) offers a particularly attractive form for in vivo testing of its prolonged anti-inflammatory action in preclinical studies. The feasibility of such studies was demonstrated previously in acute inflammation and apoptosis of the liver in which rapid delivery of CP-SOCS3 to blood cells and organs, such as liver, spleen, kidneys, lung, and heart, was documented (12).
In summary, we have developed a SOCS box-deleted form of CP-SOCS3 that suppresses proinflammatory cytokine signaling much more effectively than its wild-type counterpart. Deletion of the SOCS box from CP-SOCS3 greatly extends the half-life of CP-SOCS3, whereas endogenous wild-type SOCS3 is rapidly degraded following its induction with proinflammatory agonists in macrophages. This increased stability, coupled with the capacity for rapid, intracellular delivery, renders the SOCS3 mutant an attractive candidate for protein therapeutic approaches to suppress pathologic inflammation. Further in vivo studies of long-acting CP-SOC3 forms in relevant models of acute and chronic inflammation will expand our understanding of the global role of SOCS3 in modulating signals generated by a variety of proinflammatory agonists in multiple organ systems. In principle, the results presented here may also be applicable to the conversion of other conditionally labile suppressors into more stable, persistently acting forms for use in intracellular therapy.
Acknowledgments
We thank Maria Lima, Shirley Russell, Ruth Ann Veach, Ifeanyi Arinze, Dean Ballard, Gautaum Chaudhuri, Daniel Moore, and Jozef Zienkiewicz for critically reading the manuscript. We thank Danya Liu, Lukasz Wylezinski, Ruth Ann Veach, Jozef Zienkiewicz, Qing (Andrew) Lin, Jennifer Easterling, and Chad Sethman for experimental advice. We also thank Susanna Richards for editorial assistance in preparing this manuscript. The use of core facilities in this study was supported by National Institutes of Health Grants CA68485, DK20593, DK58404, HD15052, DK59637, and EY08126 to the Vanderbilt University Medical Center Cell Imaging Shared Resource. We also thank the Vanderbilt DNA Sequencing Facility of the Vanderbilt Institute for Integrative Genomics.
This work was supported, in whole or in part, by National Institutes of Health Grants HL069452 and AA015752 from the United States Public Health Service, and Training Grants T32 AI007281 and T32 HL069765.
- IL
- interleukin
- JAK
- Janus kinase
- STAT
- signal transducers and activators of transcription
- TNF-α
- tumor necrosis factor-α
- NF-κB
- nuclear factor κB
- SH2
- Src homology domain 2
- gp130
- glycoprotein 130
- DMEM
- Dulbecco's modified Eagles's medium
- SOCS
- suppressor of cytokine signaling
- CP
- cell-penetrating form
- FBS
- fetal bovine serum
- BMDM
- bone marrow-derived macrophages
- FACS
- fluorescence-activated cell sorter
- MTM
- membrane translocating motif
- PBS
- phosphate-buffered saline
- FITC
- fluorescein isothiocyanate.
REFERENCES
- 1.Hawiger J. (2001) Immunol. Res. 23, 99–109 [DOI] [PubMed] [Google Scholar]
- 2.Dinarello C. A. (2000) Chest 118, 503–508 [DOI] [PubMed] [Google Scholar]
- 3.Opal S. M., DePalo V. A. (2000) Chest 117, 1162–1172 [DOI] [PubMed] [Google Scholar]
- 4.Alexander W. S., Hilton D. J. (2004) Annu. Rev. Immunol. 22, 503–529 [DOI] [PubMed] [Google Scholar]
- 5.Liew F. Y., Xu D., Brint E. K., O'Neill L. A. (2005) Nat. Rev. Immunol. 5, 446–458 [DOI] [PubMed] [Google Scholar]
- 6.Rakesh K., Agrawal D. K. (2005) Biochem. Pharmacol. 70, 649–657 [DOI] [PubMed] [Google Scholar]
- 7.Coornaert B., Carpentier I., Beyaert R. (2009) J. Biol. Chem. 284, 8217–8221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nicholson S. E., Willson T. A., Farley A., Starr R., Zhang J. G., Baca M., Alexander W. S., Metcalf D., Hilton D. J., Nicola N. A. (1999) EMBO J. 18, 375–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kamura T., Sato S., Haque D., Liu L., Kaelin W. G., Jr., Conaway R. C., Conaway J. W. (1998) Genes Dev. 12, 3872–3881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Babon J. J., McManus E. J., Yao S., DeSouza D. P., Mielke L. A., Sprigg N. S., Willson T. A., Hilton D. J., Nicola N. A., Baca M., Nicholson S. E., Norton R. S. (2006) Mol. Cell. 22, 205–216 [DOI] [PubMed] [Google Scholar]
- 11.Kubo M., Hanada T., Yoshimura A. (2003) Nat. Immunol. 4, 1169–1176 [DOI] [PubMed] [Google Scholar]
- 12.Jo D., Liu D., Yao S., Collins R. D., Hawiger J. (2005) Nat. Med. 11, 892–898 [DOI] [PubMed] [Google Scholar]
- 13.DiGiandomenico A., Wylezinski L. S., Hawiger J. (2009) Sci. Signal. 2, ra37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hawiger J. (1999) Curr. Opin. Chem. Biol. 3, 89–94 [DOI] [PubMed] [Google Scholar]
- 15.Veach R. A., Liu D., Yao S., Chen Y., Liu X. Y., Downs S., Hawiger J. (2004) J. Biol. Chem. 279, 11425–11431 [DOI] [PubMed] [Google Scholar]
- 16.Dimitriou I. D., Clemenza L., Scotter A. J., Chen G., Guerra F. M., Rottapel R. (2008) Immunol. Rev. 224, 265–283 [DOI] [PubMed] [Google Scholar]
- 17.Siewert E., Müller-Esterl W., Starr R., Heinrich P. C., Schaper F. (1999) Eur. J. Biochem. 265, 251–257 [DOI] [PubMed] [Google Scholar]
- 18.Sasaki A., Inagaki-Ohara K., Yoshida T., Yamanaka A., Sasaki M., Yasukawa H., Koromilas A. E., Yoshimura A. (2003) J. Biol. Chem. 278, 2432–2436 [DOI] [PubMed] [Google Scholar]
- 19.Barnes J. A., Gomes A. V. (1995) Mol. Cell. Biochem. 149–150, 17–27 [DOI] [PubMed] [Google Scholar]
- 20.Molinari M., Anagli J., Carafoli E. (1995) J. Biol. Chem. 270, 2032–3035 [DOI] [PubMed] [Google Scholar]
- 21.Chen B. B., Mallampalli R. K. (2007) J. Biol. Chem. 282, 33494–33506 [DOI] [PubMed] [Google Scholar]
- 22.Badi I., Cinquetti R., Frascoli M., Parolini C., Chiesa G., Taramelli R., Acquati F. (2009) FEBS Lett. 583, 2486–2492 [DOI] [PubMed] [Google Scholar]
- 23.Babon J. J., Sabo J. K., Soetopo A., Yao S., Bailey M. F., Zhang J. G., Nicola N. A., Norton R. S. (2008) J. Mol. Biol. 381, 928–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Babon J. J., Sabo J. K., Zhang J. G., Nicola N. A., Norton R. S. (2009) J. Mol. Biol. 387, 162–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sasaki A., Yasukawa H., Suzuki A., Kamizono S., Syoda T., Kinjyo I., Sasaki M., Johnston J. A., Yoshimura A. (1999) Genes Cells 4, 339–351 [DOI] [PubMed] [Google Scholar]
- 26.Nicholson S. E., De Souza D., Fabri L. J., Corbin J., Willson T. A., Zhang J. G., Silva A., Asimakis M., Farley A., Nash A. D., Metcalf D., Hilton D. J., Nicola N. A., Baca M. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 6493–6498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hörtner M., Nielsch U., Mayr L. M., Heinrich P. C., Haan S. (2002) Eur. J. Biochem. 269, 2516–2526 [DOI] [PubMed] [Google Scholar]
- 28.Hörtner M., Nielsch U., Mayr L. M., Johnston J. A., Heinrich P. C., Haan S. (2002) J. Immunol. 169, 1219–1227 [DOI] [PubMed] [Google Scholar]
- 29.Dunn S. L., Björnholm M., Bates S. H., Chen Z., Seifert M., Myers M. G., Jr. (2005) Mol. Endocrinol. 19, 925–938 [DOI] [PubMed] [Google Scholar]
- 30.Kiu H., Greenhalgh C. J., Thaus A., Hilton D. J., Nicola N. A., Alexander W. S., Roberts A. W. (2009) Growth Factors 27, 384–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Croker B. A., Krebs D. L., Zhang J. G., Wormald S., Willson T. A., Stanley E. G., Robb L., Greenhalgh C. J., Förster I., Clausen B. E., Nicola N. A., Metcalf D., Hilton D. J., Roberts A. W., Alexander W. S. (2003) Nat. Immunol. 4, 540–545 [DOI] [PubMed] [Google Scholar]
- 32.Lang R., Pauleau A. L., Parganas E., Takahashi Y., Mages J., Ihle J. N., Rutschman R., Murray P. J. (2003) Nat. Immunol. 4, 546–550 [DOI] [PubMed] [Google Scholar]
- 33.Costa-Pereira A. P., Tininini S., Strobl B., Alonzi T., Schlaak J. F., Is'harc H., Gesualdo I., Newman S. J., Kerr I. M., Poli V. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 8043–8047 [DOI] [PMC free article] [PubMed] [Google Scholar]