Abstract
Autophagy is being increasingly implicated in both cell survival and death. However, the intricate relationships between drug-induced autophagy and apoptosis remain elusive. Here we demonstrate that a tubulin-binding noscapine analog, (R)-9-bromo-5-((S)-4,5-dimethoxy-1,3-dihydroisobenzofuran-1-yl)-4-methoxy-6-methyl-5,6,7,8-tetrahydro-[1,3]-di-oxolo[4,5-g]isoquinoline (Red-Br-nos), exerts a novel autophagic response followed by apoptotic cell death in human prostate cancer PC-3 cells. Red-Br-nos-induced autophagy was an early event detectable within 12 h that displayed a wide array of characteristic features including double membranous vacuoles with entrapped organelles, acidic vesicular organelles, and increased expression of LC3-II and beclin-1. Red-Br-nos-triggered release of reactive oxygen species (ROS) and attenuation of ROS by tiron, a ROS scavenger, reduced the sub-G1 population suggesting ROS-dependent apoptosis. Abrogation of ROS also reduced autophagy indicating that ROS triggers autophagy. Pharmacological and genetic approaches to inhibit autophagy uncovered the protective role of Red-Br-nos-induced autophagy in PC-3 cells. Direct effects of the drug on mitochondria viz. disruption of normal cristae architecture and dissipation of mitochondrial transmembrane potential revealed a functional link between ROS generation, autophagy, and apoptosis induction. This is the first report to demonstrate the protective role of ROS-mediated autophagy and induction of caspase-independent ROS-dependent apoptosis in PC-3 cells by Red-Br-nos, a member of the noscapinoid family of microtubule-modulating anticancer agents.
Keywords: Anticancer Drug, Apoptosis, Autophagy, Mitochondria, Reactive Oxygen Species (ROS)
Introduction
Chemotherapy remains the mainstay of treatment for both early stage as well as metastatic tumors. The ultimate goal of successful chemotherapy is to strategically induce robust apoptosis with simultaneous suppression of survival signaling circuitries in cancer cells with minimal toxicity to normal cells. Various chemotherapeutic agents induce cell death by halting cell cycle progression, enhancing expression of pro-apoptotic molecules whereas down-regulating survival molecules that impede apoptosis. However, beyond the provocation of death-inducing signals, chemotherapeutic drugs unfavorably up-regulate apoptosis-inhibitory molecules such as survivin and XIAP (1), and trigger various stress-response-activated pathways that promote cell survival and adaptation (2). One such stress-response mechanism is autophagy, a well conserved ancient mechanism of cellular self-destruction that mediates survival (3–5). Regulation of autophagy is highly complex with inputs from the cellular environment that promote cell survival through the phosphatidylinositol 3-OH kinase (PI3K)2/Akt/mammalian target of rapamycin pathway, members of the Bcl2 family, and p53 (6–9). Although autophagy has recently gained much attention for its paradoxical roles in cell survival and cell death, the true identity of autophagy lies in its adaptive cellular homeostatic and housekeeping mechanisms (10). Induction of autophagy thus constitutes a protective mechanism that renders tumor cells resistant to death (11).
Drug-induced autophagy is being increasingly implicated in modulating cell death responses. When considering treatment options, it is conceivable that autophagy could limit the effects of cytotoxic anticancer drugs through its ability to clear damaged organelles and proteins. It may also help cancer cells to survive chemotherapy-induced stress. In these scenarios, inhibiting autophagy would be beneficial to treatment outcome. Equally plausible, however, is the notion that drug-induced autophagy might contribute to tumor cell demise and in this case inhibition of autophagy would lead to enhanced tumor growth. Several anticancer agents that induce autophagy include tamoxifen, arsenic trioxide, temozolomide, HDAC inhibitors, etoposide, vitamin D analogs, and tubulin-binding drugs (paclitaxel and 2-methoxyestradiol) (12–17). In these examples, autophagy has been shown to be pro-survival or death inducing in a context-dependent manner. Thus, manipulation of autophagy in favor of death induction in cancer cells would be contingent on context, such as cell-type, nature, and duration of stress etc.
Ongoing efforts in our laboratory are actively focused on expansion of a novel class of anticancer agents, noscapinoids, based upon the parent noscapine (18–21). Unlike conventional tubulin-binding agents, these novel molecules are weak affinity tubulin binders, and subtly attenuate microtubule dynamics without appreciably perturbing the steady state monomer/polymer ratio of tubulin (22, 23). This unique property perhaps underlies the “kinder and gentler” nature of noscapine and its analogs and represents a unique edge over conventional anti-tubulin agents (24). A novel noscapinoid, (R)-9-bromo-5-((S)-4,5-dimethoxy-1,3-dihydro-isobenzofuran-1-yl)-4-methoxy-6-methyl-5,6,7,8-tetrahydro-[1,3]di-oxolo[4,5-g]isoquinoline (hereon referred to as Red-Br-nos) has been shown to be significantly more potent than the founding molecule, noscapine (22). Previous reports demonstrate that Red-Br-nos arrests cell cycle at the G2/M phase and induces apoptosis in ovarian cancer cells in vitro and in vivo without detectable toxicity (25, 26). The identity of key determinants that arbitrate life and death decisions upon Red-Br-nos exposure and contribute to the final outcome of cell death are still unknown.
The present study reports a novel protective autophagic response in PC-3 human prostate cancer cells upon Red-Br-nos treatment. The induction of autophagy preceded the onset of apoptosis. Both opposing responses, autophagy and apoptosis, were ROS triggered. Attenuation of ROS reduced autophagic protection and enhanced apoptosis induction, suggesting that ROS production was upstream of these two events. Red-Br-nos directly affected the mitochondria that generates ROS to trigger a protective autophagic response that perhaps finally sets the stage for apoptosis in the presence of overwhelming damage. This is the first report to identify the induction of autophagy by a novel noscapine family member, as a protective defense mechanism against apoptotic cell death.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
Acridine orange (AO), 2′,7′-dichlorofluorescein diacetate (DCFDA), chloroquine, 3-methyl adenine (3-MA), and anti-β-actin antibody were purchased from Sigma. Dihydroethidium (DHE), 4,5-dihydroxy-1,3-benzenedisulfonic acid disodium salt monohydrate (tiron), 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolyl carbocyanine iodide (JC-1), and cyclosporin A were from Fisher Scientific. The concentrations of the above reagents used in the study were: 25 μm Red-Br-nos, 25 μg/ml of AO, 5 μm DHE, 25 μm DCFDA, 2.5 μg/ml of JC-1, 0.5 mm 3-MA; 1 mm tiron, and 5 μm cyclosporin A. Primary antibodies for beclin-1, light chain 3 (LC3), caspase-2, caspase-3, caspase-7, caspase-8, caspase-9, cytochrome c, and apoptosis inducing factor (AIF) were from Cell Signaling (Beverly, MA). Endo-G was from Abcam (Cambridge, MA). Horseradish peroxidase-conjugated secondary antibodies were from Santa Cruz Biotechnology, Inc. Alexa 488- or 555-conjugated secondary antibodies were from Molecular Probes (Carlsbad, CA). Control and beclin-1 siRNA were from Invitrogen.
Detection and Quantification of Acidic Vesicular Organelles (AVOs)
PC-3 cells (1 × 105) were grown on coverslips followed by treatment with DMSO (control) or 25 μm Red-Br-nos for the specified time periods. Cells were stained with 25 μg/ml of AO for 15 min, washed with PBS, fixed using 100% methanol followed by examining under a Zeiss (Axioplan-2) fluorescence microscope using a ×63 objective. AVO were also quantified by flow cytometry after the cell pellet was stained with AO for 15–20 min followed by washing the cells with PBS and resuspension in 500 μl of PBS. Green (510–530 nm) and red (650 nm) fluorescence emission from 2500 cells illuminated with blue (488 nm) excitation light was measured using a FACS Calibur flow cytometer. The red:green fluorescence ratio for individual cells was calculated using FlowJo software (27).
Detection of GFP-LC3
PC-3 cells grown on coverslips were transfected with 0.5 μg of GFP-LC3 plasmid using Lipofectamine in a 24-well format. After 6 h, cells were treated with DMSO or Red-Br-nos for 24 h followed by fixation in 100% methanol. The fluorescence of GFP-LC3 was viewed under the microscope and the number of GFP-LC3 dots were counted.
Measurement of ROS
Following treatment with Red-Br-nos or DMSO, PC-3 cells were stained with either DHE (which is oxidized by ROS into ethidium bromide and fluoresces red) or DCFDA (which is oxidized by ROS to DCF). Fluorescently labeled cells were analyzed flow cytometrically for quantification. Cells that were labeled on the coverslips were examined using fluorescence microscopy. ROS was also measured in labeled cells using fluorimetry.
JC-1 Staining
PC-3 cells were treated with 25 μm Red-Br-nos for 24 h. The cells were then labeled with the JC-1 reagent for 15 min at 37 °C. After washing, cell fluorescence was measured on a flow cytometer using orange-red emission filters. Stained cells were also observed using a Zeiss (Axioplan-2) fluorescence microscope with a ×63 objective. In addition, JC-1 aggregates were detected with a fluorimeter at excitation/emission = 540/570 nm.
Apoptosis Measurement
PC-3 cells were seeded in culture dishes and grown until ∼70% confluence. After various treatments, cells were centrifuged, washed twice with ice-cold PBS, and fixed in 70% ethanol. Tubes containing the cell pellets were stored at 4 °C for at least 24 h. Cells were then centrifuged for 10 min, and the supernatant was discarded. Pellets were washed twice with PBS and stained with propidium iodide in the presence of RNase A for 45 min in the dark. Samples were analyzed by flow cytometry. Apoptosis was quantitated by measuring the sub-G1 population on the cell cycle profile data acquired by flow cytometry.
Western Blotting
The protein lysates collected from control or 25 μm Red-Br-nos-treated cells were resolved by SDS-PAGE and transferred onto polyvinylidene difluoride membrane. The membrane was incubated with the desired primary antibody overnight at 4 °C and the appropriate secondary antibody for 1 h at room temperature. The immune-reactive bands were visualized by the chemiluminescence detection kit (Pierce). β-Actin was used as loading control.
Electron Microscopy
Cells were collected and fixed in 2% paraformaldehyde, 0.1% glutaraldehyde in 0.1 m sodium cacodylate for 2 h, post-fixed with 1% osmium tetroxide for 1.5 h, washed, and stained en bloc for 1 h in 1% aqueous uranyl acetate (pH 3.3). The samples were then washed again, dehydrated with a graded ethanol series (through 3 × 100%) and embedded in Spurr epoxy resin (Electron Microscopy Sciences). Ultrathin sections were cut on a RMC-MYX ultramicrotome, counterstained with lead citrate, and examined on a LEO 906e transmission electron microscope.
Immunofluorescence Microscopy
PC-3 cells were grown on glass coverslips for immunofluorescence microscopy. After treatment with 25 μm Red-Br-nos, cells were fixed with cold (−20 °C) methanol for 10 min and blocked by incubating with 2% bovine serum albumin/PBS at 37 °C for 1 h. Endo-G/AIF/beclin-1/cyctochrome c antibodies (1:100 dilution) were incubated with coverslips for 2 h at 37 °C. The cells were washed with 2% bovine serum albumin/PBS for 10 min at room temperature before incubating with a 1:500 dilution of Alexa 488- or 555-conjugated secondary antibodies. Cells were mounted with Prolong Gold antifade reagent that contains 4′,6-diamidino-2-phenylindole (Invitrogen).
Caspase Activity Assay
Cells were treated with or without 25 μm Red-Br-nos for 24 h. Cell lysates were examined for caspase-3-like activity using a specific substrate, Ac-DEVD-7-amino-4-trifluoromethyl-coumarin, which detects the activities of caspase-3 and caspase-7 according to a standard protocol (Calbiochem). The results were evaluated using a fluorescence microplate reader.
Statistical Analysis
All experiments were repeated three times. The data were expressed as mean ± S.D. Statistical analysis was performed using Student's t test. The criterion for statistical significance was p < 0.05. For immunoblotting data, band intensities were measured using ImageJ and normalized to β-actin.
RESULTS
Red-Br-nos Induces Robust Autophagy in Prostate Cancer Cells
Formation of Double Membranous Autophagosomes in Red-Br-nos-treated PC-3 Cells
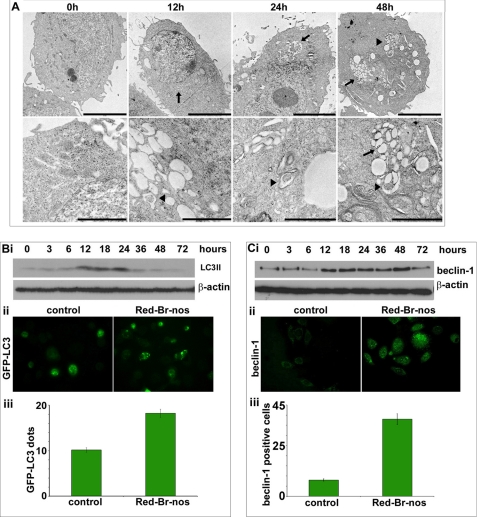
Several members of the noscapinoid family (EM011, EM015) activate a mitochondrially mediated intrinsic apoptotic pathway to induce cell death in lymphoma and breast cancer cells (18–21). Because mitochondrial damage has been widely implicated in the induction of autophagy, we asked if Red-Br-nos can also induce autophagy. Classically, electron microscopy has been considered as the gold standard to demonstrate autophagosomes in cells (28). Thus, the ultrastructure of control and 25 μm Red-Br-nos-treated PC-3 cells was first examined using transmission electron microscopy. The choice of drug concentration (25 μm) was based upon dose response (supplemental Fig. S1) and time course (supplemental Fig. S2) flow cytometric experiments that determined the sub-G1 population that is indicative of apoptosis. As can be seen in Fig. 1A, PC-3 cells treated with Red-Br-nos revealed the appearance of large double-membranous cytoplasmic vacuoles (black arrowheads) as early as 12 h that progressively accumulated upon increasing the drug exposure time. These vacuoles resembled autophagosomes and several of them showed entrapped intracellular organelles such as mitochondria or endoplasmic reticulum, and digested residual material (Fig. 1A, black arrows). On the contrary, control PC-3 cells displayed large nuclei with finely dispersed chromatin material surrounded by cytoplasm with a normal complement of healthy looking mitochondria at all time points (12, 24, and 48 h) studied. After 48 h of drug exposure, cells with fragmented nuclei were observable indicating induction of apoptosis (data not shown). These results indicated that Red-Br-nos treatment caused formation of autophagosome-like structures and this cellular response preceded the onset of apoptosis in PC-3 cells.
FIGURE 1.
Red-Br-nos induces robust autophagy in prostate cancer PC-3 cells. Panel A, representative transmission electron micrographs showing the ultrastructure of PC-3 cells treated with DMSO (controls) or 25 μm Red-Br-nos for the indicated time points. Note the abundance of double membrane vacuoles (black arrowheads) in Red-Br-nos-treated PC-3 cells, which were infrequently seen in controls. Remnants of organelles, including mitochondria (black arrows) were evident in some of these double membrane autophagic vacuoles. Upper panels, scale bar = 5 μm; lower panels, scale bar = 2 μm. Panel B, i, immunoblot analysis of LC3-II expression levels in lysates from PC-3 cells treated with 25 μm Red-Br-nos for the indicated time points. β-Actin was used to ensure equal protein loading. Similar results were observed in at least two independent experiments. Panel ii, immunofluorescence micrographs showing GFP-LC3 plasmid-transfected cells treated in the absence or presence of Red-Br-nos. Panel iii, GFP-LC3 dots were counted to quantify autophagic cells (24-h drug treatment) from 6 to 8 random image fields totaling 200 cells and reported as mean ± S.D. (p < 0.05, compared with controls). Panel C, i, immunoblot analysis of beclin-1 expression levels in drug-treated PC-3 cells for the indicated time points. β-Actin was a loading control. Panel ii, immunofluorescence micrographs showing beclin-1-stained PC-3 cells treated in the presence (right) or absence of Red-Br-nos (left). Panel iii, quantitation of beclin-1 positive cells in control and 24-h drug-treated PC-3 cells from random image fields totaling 200 cells and reported as mean ± S.D. (p < 0.05, compared with controls).
Recruitment of LC3-II into Autophagolysosomes and Up-regulation of Beclin-1 Expression
More recently, the microtubule-associated protein-1 LC3 has been used as a marker of autophagy. LC3 exists in two forms, an 18-kDa cytosolic protein (LC3-I) and a processed 16-kDa form (LC3-II) that is membrane-bound and increased during autophagy by conversion from LC3-I (28). To gain insights into the mechanism of Red-Br-nos-induced autophagy, we examined drug effects on LC3, the mammalian homolog of the yeast autophagy protein Apg8/Aut7p (29, 30). Red-Br-nos treatment caused processing of full-length LC3-I (18 kDa) to LC3-II (16 kDa) as evident by immunoblotting data using lysates from PC-3 cells treated with either DMSO (control) or 25 μm Red-Br-nos for the indicated time points (Fig. 1B, i). An immunoreactive band corresponding to processed LC3-II (16 kDa) was present with a weak intensity (3 and 6 h) in drug-treated lysates. However, at 12 h, LC3-II levels were increased by ∼6-fold compared with controls, suggesting induction of LC3-II, the form that is recruited to autophagosomes (Fig 1B, i). The autophagic response was not restricted to the lower drug concentration of 25 μm. Higher dose levels of Red-Br-nos (50 and 100 μm) also showed induction of autophagy as seen by increased LC3-II expression at 24 h post-treatment (supplemental Fig. S3).
Next, we confirmed autophagy by quantifying GFP-LC3-tagged compartments by fluorescence microscopy. To this end, transient transfectants of PC-3 cells expressing GFP-LC3 were generated. Although there was a diffused localization of GFP-LC3 in control cells (Fig. 1B, ii), treatment of cells with Red-Br-nos for 24 h produced a punctate pattern for GFP-LC3 fluorescence, indicating recruitment of LC3 to autophagosomes during Red-Br-nos-induced autophagy (Fig. 1B, ii). GFP-LC3 compartments at 24 h post-treatment were quantified by counting GFP-LC3 dots in control and Red-Br-nos-treated cells (Fig. 1B, iii). Our results show that there was a significant increase in the number of GFP-LC3 dots by ∼80% in 24-h drug-treated cells compared with controls (Fig. 1B, ii). This clearly suggested increased induction of autophagy over time following Red-Br-nos treatment.
An essential autophagy effector, beclin-1, is a central player in autophagosome formation (31). Immunoblotting data showed a time-dependent increase of beclin-1 expression upon drug exposure suggesting autophagic induction (Fig. 1C, i). Quantitation of Western data using ImageJ revealed a ∼3-fold increase in beclin-1 expression at 12 h compared with controls. There was an increase of beclin-1 positive cells by ∼3.7-fold at 24 h post-treatment as revealed by quantitation of at least 200 cells from random fields (Fig. 1C, iii).
Red-Br-nos Causes Formation of AVOs
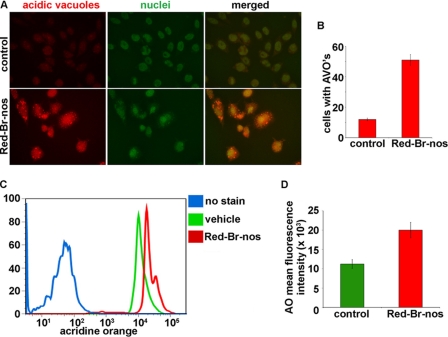
Yet another characteristic feature of cells engaged in autophagy is the formation of AVOs following treatment with different stimuli (32, 33). Thus, we visualized the effect of Red-Br-nos treatment on formation of AVOs in PC-3 cells using fluorescence microscopy upon staining with the lysosomotropic agent AO (Fig. 2A). Essentially, AO is a weak base that traverses freely across biological membranes in an uncharged state characterized by green fluorescence. Its protonated form accumulates as aggregates in acidic compartments characterized by red fluorescence. As is visually evident in Fig. 2A, control cells primarily displayed green fluorescence with minimal red fluorescence, indicating a lack of AVOs. On the other hand, drug-treated PC-3 cells showed a ∼3.2-fold increase in red fluorescent AVOs at 24 h post-treatment compared with controls (Fig. 2B). We also quantitated AO fluorescence using flow cytometry (Fig. 2C). Histogram profiles show the mean fluorescence intensity of unstained cells (no AO, blue profile), control cells (AO, green profile), and drug-treated cells (profile), and drug-treated cells (AO, red profile) (Fig. 2C). There was an increase in red fluorescence intensity upon drug treatment indicating an enhancement of AVOs. Fig. 2D is a bar graph quantitation showing a ∼78% increase in red fluorescent cells upon drug treatment for 24 h compared with controls. These results provided further evidence to conclude that Red-Br-nos treatment induced autophagy in PC-3 cells.
FIGURE 2.
Panel A, immunofluorescence microscopy of acridine orange-stained PC-3 cells treated for 24 h with DMSO (control) or 25 μm Red-Br-nos. Increase in number of cells with AO accumulating acidic vesicular organelles (orange-red fluorescence) in Red-Br-nos-treated cells was evident. Panel B, quantitation of cells with AVOs from 6 to 8 random image fields totaling ∼200 cells and reported as mean ± S.D. (p < 0.05). Panel C, histogram profiles of control and Red-Br-nos-treated cells that were read flow cytometrically. Blue profile shows unstained cells without AO as negative controls. Green profile depicts control cells that were stained with AO and the red profile shows drug-treated cells with increased AO fluorescence, indicative of numerous red AVOs. Panel D, bar graph representation of the quantitation of the mean fluorescence intensity in control and drug-treated PC-3 cells. Columns, mean ± S.D. (p < 0.05, compared with controls).
Red-Br-nos Triggers ROS Generation
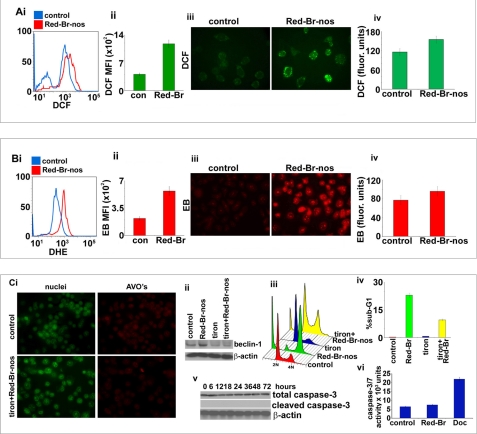
Several reports provide strong evidence for the involvement of ROS in the induction of autophagy as well as apoptosis (2, 34). To test whether Red-Br-nos induced ROS production, we stained cells using DCFDA and read them flow cytometrically (Fig. 3A, i). DCFDA is cell permeable, cleaved by nonspecific cellular esterases, and reacts mainly with H2O2 and other peroxides to yield fluorescent DCF (35, 36). Red-Br-nos-treated PC-3 cells (red profile) exhibited a statistically significant increase in DCF mean fluorescence intensity by ∼1.8-fold compared with controls (blue profile) (Fig. 3A, i and ii). Microscopic inspection of DCFDA-stained drug-treated cells showed a significant increase in intensity of DCF staining compared with controls (Fig. 3A, iii). The increase in DCF fluorescence was also measured fluorimetrically using excitation and emission wavelengths at 485 and 535 nm, respectively. As shown in Fig. 3A, iv, drug treatment resulted in a ∼28% increase of fluorescence signal compared with controls, at 24 h post-treatment.
FIGURE 3.
Red-Br-nos-triggered ROS generation in PC-3 cells. Panel A, i, histogram profiles of control and drug-treated cells that were read flow cytometrically upon DCFDA staining (an indicator of ROS generation). Panel ii, bar graph representation of the quantitation of the increase in the mean fluorescence intensity (DCF-positive cells) in PC-3 cultures treated with DMSO (control) or Red-Br-nos for 24 h. Columns, mean ± S.D. (*, p < 0.05, compared with controls). Panel iii, microscopic visualization of DCF fluorescence in control and drug-treated PC-3 cells. Panel iv, fluorimetric data showing an increase in DCF fluorescence upon drug treatment compared with controls. Panel B, i, flow cytometric histogram profiles of control (blue profile) and drug-treated (red profile) cells upon DHE staining (another indicator of ROS generation). Panel ii, bar graph representation of the quantitation of the increase in the mean fluorescence intensity (EtBr-positive cells) in PC-3 cultures treated with DMSO (control) or Red-Br-nos for 24 h. Panel iii, microscopic visualization of EtBr (EB) fluorescence in control and drug-treated PC-3 cells. Panel iv, fluorimetric data showing an increase in EtBr fluorescence upon drug treatment compared with controls. Panel C, i, attenuation of ROS by tiron reduced the autophagic response as seen by a decrease in the number of red acidic compartments (AVOs), a characteristic feature of autophagic vacuoles. Panel ii, abrogation of ROS by tiron also decreased beclin-1 expression levels upon Red-Br-nos treatment for 24 h, suggesting a clear involvement of ROS in induction of autophagy. Panel iii, ROS plays a role in inducing Red-Br-nos-induced apoptosis. Attenuating ROS levels by tiron reduced the sub-G1 population as shown in the three-dimensional disposition of cell-cycle profiles. Panel iv, bar graph representation of the quantitation of the sub-G1 population upon drug treatment in the presence or absence of tiron. Panel v shows an immunoblot analysis for total and cleaved caspase-3 at the noted time points. β-Actin was used as a loading control. Panel vi is a bar graph representation for caspase-3/7 activity measured at 24 h upon treatment with vehicle control, Red-Br-nos and docetaxel. Columns, mean ± S.D. (*, p < 0.05, compared with controls).
The results observed using DCF fluorescence were next confirmed with another ROS probe, DHE that yields ethidium bromide (EtBr) upon DHE oxidation and accumulates as red fluorescence in the nucleus (36). Flow cytometry data showed a ∼1.6-fold increase of mean fluorescence intensity of EtBr upon drug treatment (red profile) compared with controls (blue profile) (Fig. 3B, i and ii). Indeed, upon drug treatment, nuclear staining with ethidium bromide was evident microscopically, indicating that PC-3 cells accumulated peroxides (Fig. 3A, iii). Fluorimetric data also showed an enhanced fluorescence emission at 580 nm correlating with a ∼27% increase of EtBr-stained cells at 24 h post-treatment compared with controls (Fig. 3B, iv).
Red-Br-nos-induced ROS Triggers Autophagy
ROS serve as signaling molecules in many cellular processes including growth, differentiation, and apoptosis (37–39). ROS specifically, H2O2, and superoxide anion O2˙̄ have been reported to induce autophagy (37, 39, 40). Having identified that Red-Br-nos induced generation of ROS, we next asked if ROS triggered autophagy in PC-3 cells. To investigate a functional link between ROS production and autophagy induction, we used tiron, a ROS scavenger, and examined its effect on AVO formation and beclin-1 expression over the Red-Br-nos treatment time. Attenuation of ROS levels by tiron significantly decreased the number of AVOs upon Red-Br-nos treatment (Fig. 3C, i). This was in contrast to Red-Br-nos treatment alone that caused a significant induction of cells with AVOs (Fig. 2A, i). In addition, treatment with tiron in the absence or presence of Red-Br-nos did not induce ROS production as evident by DCF staining (supplemental Fig. S4). Furthermore, there was an absence of drug-induced beclin-1 expression upon drug treatment in the presence of tiron as seen by immunoblotting methods (Fig. 3C, ii). Taken together, these data suggested ROS-mediated induction of autophagy in PC-3 cells.
Red-Br-nos Induces ROS-dependent Apoptosis That Is Caspase-independent
Several tubulin-binding drugs recruit mitochondrially mediated ROS signaling to induce apoptotic cell death (41). To investigate the involvement of ROS in Red-Br-nos-induced cell death, we monitored the drug-induced sub-G1 population (an indicator of cell death) in the absence or presence of tiron using flow cytometry. Fig. 3C, iii, is a three-dimensional representation of cell-cycle profiles of PC-3 cells that were treated with drug in the absence or presence of tiron. Quantitation data showed that there was a decline in the drug-induced sub-G1 population by ∼2.7-fold when ROS was inhibited by tiron, suggesting that ROS played a crucial role in induction of cell death (Fig. 3C, iv). Because caspase activation is a hallmark of classical apoptosis, cleaved caspase-3 expression was examined by immunoblotting (Fig. 3C, v). There was an absence of cleaved caspase-3 expression upon drug treatment over time (Fig. 3C, v). Furthermore, we also measured caspase-3/7 activity at 24 h of drug treatment (Fig. 3C, vi). Docetaxel was included as a positive control. We found that there were no changes in caspase-3/7 activity patterns in drug-treated cells compared with controls (Fig. 3C, vi). However, 2 nm docetaxel increased caspase-3 activity (Fig. 3C, vi). In addition, there was an absence of activated caspase-3, caspase-7, caspase-8, caspase-9, and caspase-2 upon a 48-h Red-Br-nos treatment (supplemental Fig. S5). Docetaxel treatment, however, showed an increase of activated caspase-3, caspase-7, caspase-8, and caspase-9 compared with controls (supplemental Fig. S5). Drug treatment in the presence of the pan-caspase inhibitor (Z-VAD-fmk) showed no significant difference in the sub-G1 population compared with drug alone (supplemental Fig. S6). This suggested that ROS-mediated cell death was indeed caspase-independent.
Red-Br-nos Induces AIF and Endo-G Nuclear Translocation
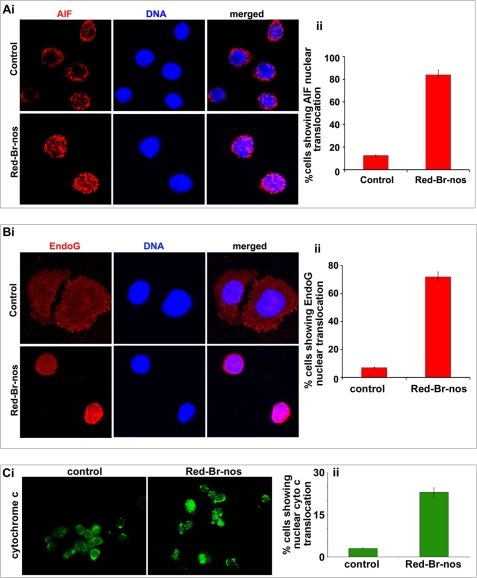
AIF and Endo-G are mitochondrial proteins that potentially contribute to the induction of apoptotic cell death via a caspase-independent pathway (42). Upon release into the cytosol, both AIF and Endo-G shuttle to the nucleus and trigger nuclear chromatin condensation and high molecular weight (50 kb) DNA loss (43). Thus, we next examined whether AIF and/or Endo-G were implicated in Red-Br-nos-induced cell death. Using immunofluorescence microscopy, analysis of subcellular localization clearly showed that Endo-G and AIF were released from the mitochondria into the cytosol translocated to the nucleus following a 48-h treatment with Red-Br-nos in PC-3 cells (Fig. 4, A, i, and B, i). Quantitation of a total of 200 cells from random image fields showed that there was a ∼6.7-fold increase in cells with nuclear AIF upon drug treatment compared with controls (Fig. 4A, ii). Red-Br-nos treatment also increased the number of nuclear Endo-G containing cells by ∼10-fold compared with controls (Fig. 4B, ii).
FIGURE 4.
Red-Br-nos treatment induces caspase-independent cell death in PC-3 cells. Micrographs show immunocytochemical staining of 48 h Red-Br-nos-treated PC-3 cells for AIF (panel A, i), Endo-G (panel B, i), and cytochrome c (panel C, i). Bar graph representation of the quantitation of percent cells showing nuclear AIF translocation (panel A, ii), nuclear Endo-G translocation (panel B, ii) and nuclear cytochrome c translocation (panel C, ii) in control and drug-treated PC-3 cells from 6 to 8 random image fields totaling 200 cells and reported as mean ± S.D. (p < 0.05, compared with controls).
Red-Br-nos Induces Nuclear Translocation of Cytochrome c
The release of cytochrome c from the mitochondria marks a major event during apoptosis (44, 45). Once released into the cytosol, cytochrome c usually complexes with Apaf-1, which then activates the caspase cascade leading to DNA fragmentation. However, several reports have demonstrated a caspase-independent cell death despite release of cytochrome c (46). The nuclear translocation of cytochrome c in caspase-independent nuclear apoptosis has been recently reported (47). Consistent with this notion, our immunofluorescence micrographs depict nuclear translocation of cytochrome c indicating that cytochrome c induces cell death by nuclear DNA fragmentation in a caspase-3 independent manner (Fig. 4C, i). Quantitation of a total of ∼200 cells from random image fields showed a ∼7.5-fold increase in cells with nuclear cytochrome c upon drug treatment for 48 h compared with controls (Fig. 4C, ii).
Red-Br-nos-induced Autophagy Is Protective
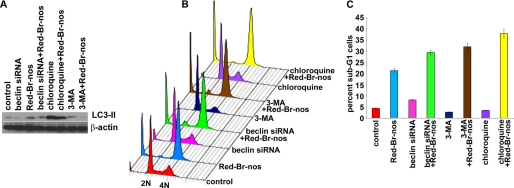
Red-Br-nos-triggered ROS generation mediated a caspase-independent apoptosis. We next asked if autophagy affected cell death. To address this question, multiple strategies were adopted to inhibit drug-induced autophagy by employing pharmacologic (small molecule inhibitor) and genetic approaches (siRNA knockdown). Among the pharmacologic autophagy inhibitors, we employed 3-MA (early phase inhibitor) and chloroquine (late phase inhibitor). The inhibitors were added 1 h prior to the addition of Red-Br-nos. This was essential because the timing of the autophagic process is just as important as detecting autophagy. Class III PI3K plays a critical role in the early stages of autophagosome formation in mammals through formation of an essential complex with beclin-1. Hence, inhibition of its activity (using a nucleotide derivative that blocks class III PI3K activity, 3-MA, or beclin-1 siRNA) inhibits the early autophagic process (48, 49). Although early autophagy inhibition blocks class III PI3K activity, late phase inhibitors such as chloroquine appear to block the fusion of autophagosomes and lysosomes. Fig. 5A shows expression of LC3-II upon treatment with 3-MA, beclin-1 siRNA, or chloroquine alone or in the presence of Red-Br-nos using immunoblotting methods. Because chloroquine essentially interrupts at the end stage when autophagosomes fuse with lysosome to execute the degradative process, we did not expect to see decreased LC3-II levels upon co-treatment of chloroquine and Red-Br-nos. Paradoxically, chloroquine alone increased LC3-II expression levels and acted as a autophagy inducer in PC-3 cells as shown by immunoblot analysis (Fig. 5A). Flow cytometric data showed that inhibition of the early autophagic process using beclin-1 siRNA duplexes and 3-MA increased the sub-G1 population to ∼32 and ∼37%, respectively (Fig. 5, B and C), compared with Red-Br-nos alone (∼22%). This indicated that autophagy is a protective response of drug-induced apoptosis. Although analysis of the sub-G1 population gives an estimate of the extent of apoptosis, it might not be a true representation of total cell death. To this end, we determined total cell death upon 25 μm Red-Br-nos treatment by trypan blue staining (supplemental Fig. S7). We found that total cell death was comparable with the sub-G1 cell population indicating negligible necrotic cell death. Co-treatment of Red-Br-nos and chloroquine significantly increased the sub-G1 population (Fig. 5, B and C), suggesting that inhibitory intervention at the late stage of autophagy is apoptosis promoting when enhanced autophagy has perhaps failed in its mission to render protection and has given way to apoptosis.
FIGURE 5.
Red-Br-nos induced autophagy protects PC-3 cells from cell death. Panel A, immunoblotting analysis of LC3-II in cells treated with early phase autophagy inhibitors (3-MA and beclin-1 siRNA) and widely claimed late phase autophagy inhibitor (chloroquine). Panel B, a representative depiction of flow histograms to show subdiploid fraction in PC-3 cultures treated for 24 h with DMSO (control) or 25 μm Red-Br-nos in the presence or absence of 3-MA, or beclin-1 siRNA or chloroquine. Panel C, quantitation of the sub-G1 population shows that abrogating autophagy increases the sub-G1 population suggesting a protective role of autophagy induction. Columns, mean ± S.D. (*, p < 0.05, compared with controls).
Having identified that autophagy inhibited the extent of apoptosis, we next asked if inhibition of drug-induced autophagy would allow the caspase-dependent cell death program. Our immunoblotting data for caspase-3 in the presence of drug and autophagy inhibitor, 3-MA, showed the absence of any immunoreactive bands suggesting that inhibition of autophagy did not trigger caspase-dependent cell death in PC-3 cells (supplemental Fig. S8).
Red-Br-nos Affects Mitochondrial Integrity
Mitochondria are central organelles that integrate apoptosis and autophagy (50, 51). Mitochondria generate apoptotic signals and at the same time autophagy is responsible for removing damaged mitochondria. Because mitochondria represent a nexus at which the two pathways may cross-talk, we next asked if Red-Br-nos directly affected the mitochondria.
Perturbation of Mitochondrial Architecture
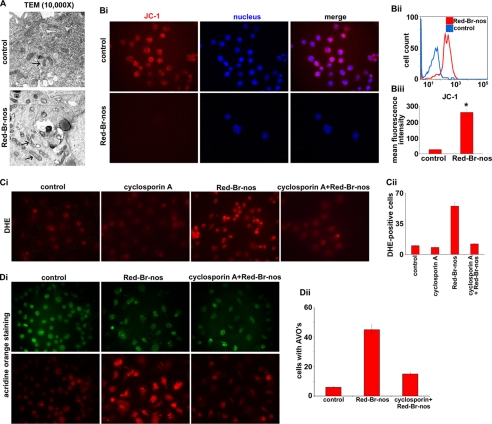
We first examined mitochondrial ultrastructure by classical electron microscopy in control and 24-h drug-treated cells. Although control cells showed a complement of healthy looking mitochondria with intact cristae structure, the mitochondria in drug-treated cells had an intact outer membrane but disorderly cristae structure (Fig. 6A). The mitochondria of Red-Br-nos-treated cells resembled type II mitochondria, characterized by a serpentine electron transparent intracristal compartment interrupted by “sausage”-shaped electron dense matrix spaces as described by Scorrano et al. (52).
FIGURE 6.
Red-Br-nos directly affects mitochondria. Panel A, representative transmission electron micrographs of PC-3 cell following a 24-h treatment with DMSO (control) or 25 μm Red-Br-nos (magnification, ×10,000). Control PC-3 cells displayed healthy looking mitochondria with intact cristae structure. However, Red-Br-nos treatment caused disruption of mitochondrial cristae structure (arrow), and a large fraction of mitochondria in Red-Br-nos-treated cells resembled type III mitochondria (52). Panel B, i, fluorescence micrographs of JC-1-stained PC-3 cells showing red JC-1 aggregates in controls, whereas there was a significant reduction in the number of red fluorescent cells indicating disruption of mitochondrial transmembrane potential upon drug treatment. Panel ii, flow cytometric histogram profiles showing percentage of cells with cytosolic monomeric JC-1-associated green fluorescence (indicating collapse of mitochondrial membrane potential) in PC-3 cultures treated with DMSO (control, blue profile) or 25 μm Red-Br-nos (red profile) for 24 h. Representative data from a single experiment, which was repeated three times with similar results, are shown. Panel iii, quantitation of the increase in mean fluorescence intensity (i.e. the percentage of green JC-1-stained cells) in PC-3 cultures treated with DMSO (control) or 25 μm Red-Br-nos for 24 h. Preserving mitochondrial integrity by cyclosporin A pretreatment remarkably attenuates ROS generation as well as autophagy. Panel C, i, cyclosporin A pretreatment was followed by drug treatment and microsopic evaluation of drug-induced ROS generation using DHE staining. Panel ii, quantitation of DHE-stained fluorescent micrographs showed that there was a significant reduction in ethidium bromide fluorescence upon drug treatment after cyclosporin A pretreatment. Cyclosporin A pretreatment yielded attenuated autophagic responses. Panel D, i, immunofluorescence microscopy of acridine orange-stained PC-3 cells pretreated with cyclosporin A for 4 h before treatment with 25 μm Red-Br-nos for 24 h. A significant decrease in the number of cells with acridine orange accumulating acidic vesicular organelles (orange-red fluorescence) in Red-Br-nos-treated cells was seen. Panel ii, quantitation of cells with AVOs from 6 to 8 random image fields totaling 200 cells and reported as mean ± S.D. (p < 0.05, compared with controls).
Dissipation of Mitochondrial Transmembrane Potential
Next, we examined the effect of Red-Br-nos on mitochondrial transmembrane potential following staining with JC-1, a cationic dye that exhibits a potentially dependent accumulation in mitochondria (53). Microscopic inspection of drug-treated cells showed high red fluorescence intensity of JC-1 aggregates in mitochondria of living control cells that was significantly diminished in 24-h Red-Br-nos-treated cells (Fig. 6B, i). Mitochondrial depolarization was indicated by a decrease in the red to green fluorescence intensity ratio. The increase in green JC-1 monomeric form indicative of collapse of transmembrane potential was quantitatively determined using flow cytometry. As can be seen in Fig. 6B, ii, the treated PC-3 cells predominantly showed a right-shift in the mean fluorescence intensity of green JC-1 monomers compared with controls (blue profile). Quantitation of FACS data indicated a ∼90% increase in the mean fluorescence intensity of drug-treated JC-1-stained cells compared with controls (Fig. 6B, iii). In addition, Red-Br-nos treatment caused a time-dependent and statistically significant increase in the percentage of cells with green fluorescence, indicating collapse of the mitochondrial membrane potential, up to 48 h (data not shown).
Mitochondrial Disruption Generates ROS
We next asked if mitochondria were the major source of ROS generation that was responsible for the induction of two opposing pathways, death-inhibiting autophagy and death-inducing apoptosis. We approached this question by preserving mitochondrial integrity by cyclosporin A pretreatment followed by evaluation of drug-induced ROS generation using DHE staining (Fig. 6C, i). There was a significant reduction in drug-induced EtBr-positive cells by ∼78% upon a 4-h cyclosporin A pretreatment compared with Red-Br-nos alone (Fig. 6C, ii). This indicated that mitochondrial disruption was essential for production of ROS. In addition, pretreatment of cyclosporin A also prevented beclin-1 up-regulation (supplemental Fig. S9A) as well as nuclear translocation of AIF (supplemental Fig. S9B).
Induction of Autophagy Is Downstream of Mitochondrial Disruption and ROS Generation
Autophagy is also known to trigger ROS (54). Thus, we examined if induction of autophagy was upstream or downstream of ROS generation. Because induction of autophagy upon drug exposure was observable as early as 12 h, we determined if mitochondrial disruption and ROS production was evident at even earlier time points. Our data showed significant DCF staining suggesting ROS generation, upon the 6-h Red-Br-nos treatment (supplemental Fig. S10A). In addition, we observed a ∼39% increase in the mean fluorescence intensity of the 6-h drug-treated JC-1-stained cells compared with controls, using flow cytometry (supplemental Fig. S10B). Having identified the attenuation of ROS upon cyclosporin A treatment, we asked if reduction of ROS levels caused a dampening of the drug-induced autophagic response (Fig. 6D, i and ii). Cells with AVOs that indicate acidic compartments reflecting autophagic vacuoles were significantly decreased (∼66%) upon drug treatment of cells pretreated with cyclosporin A for 4 h (Fig. 6D, i and ii). In addition, inhibition of autophagy by 3-MA did not have any effects on ROS generation as quantitated by DCF staining using flow cytometry (data not shown). Taken together, these data strongly suggested that mitochondrial ROS production was essential for autophagy and was upstream of autophagy induction.
DISCUSSION
The present study uses multiple strategies to detect autophagy including (a) electron microscopy, (b) conversion of LC3-I to LC3-II, (c) quantification of GFP-LC3-tagged compartments, (d) AVO quantifying, and (e) up-regulation of beclin-1 expression. These methods have provided strong evidence for a complete autophagy process upon Red-Br-nos treatment. Increased autophagy offers a distinct advantage in various physiological and stress conditions suggesting that autophagy represents an adaptive mechanism to rescue cells from death (3). Despite the fact that cells rely on autophagy to recycle their components to suppress tumor development, recent data suggest that autophagy can even protect cancer cells from chemotherapeutic regimes and foster tumor development (55). Consistent with its protective role during stress response, autophagy has been considered as protective mechanism against apoptosis (10). Thus, the presence of autophagic vesicles in dying cells may reflect an adaptive response that eventually failed to maintain cell survival under stress conditions rather than a reflection of death induced by autophagy (56). However, it is still debatable whether apoptotic and autophagic cell death are mutually exclusive or can be considered as a continuum situation (57). It is also likely that the final outcome of death depends on the severity of the response, the influence of cellular constituents and/or the influence of other signaling pathways.
Autophagy and apoptosis are closely interrelated because major players of both pathways cross-talk to each other (57). In several studies, autophagy induction has been shown to promote survival of tumor cells, thereby counteracting or limiting the efficacy of apoptosis induction by chemotherapeutic agents (58). In agreement with the notion, our data are in conformity of the protective role of autophagy upon Red-Br-nos treatment in PC-3 cells. On the contrary, induction of autophagy has been shown to enhance cell death in several studies indicating a death-inducing role of autophagy. Nevertheless, most evidence linking autophagy to cell death is circumstantial. Whether autophagy enables cells to survive or enhances their death is context driven, depending on the type of stimuli, nutrient availability, and organism (59). Given the paradoxical scenarios in which autophagy has been linked either to survival or death, it is reasonable to speculate that autophagy may protect tumor cells from undergoing apoptosis in response to treatment with anticancer agents, but may be a mechanism of cell death in tumor cells with defective apoptotic machinery. Our data helps us to propose that Red-Br-nos-induced cell death can be enhanced by manipulating autophagy. The autophagy inhibitor 3-MA augmented the Red-Br-nos-induced sub-G1 apoptotic population indicating that autophagy is a protective mechanism in the context of PC-3 cells that perhaps allowed cells to escape from apoptosis. This finding also indicated that Red-Br-nos-induced autophagic protection is partially PI3K dependent.
Mitochondrial damage plays an important role in both autophagy and apoptosis. Mitochondrial outer membrane permeabilization accompanied by collapse of the mitochondrial transmembrane potential is known to cause release of a battery of destructive molecules into the cytosol. These molecules exert pleiotropic effects ranging from caspase activation, DNA strand breakage, and ROS generation all of which set the stage for cellular demise. On the other hand, depolarized mitochondria, a feature of apoptosis, are known to be rapidly eliminated by autophagy (4). Eliminating damaged mitochondria prevents release of pro-apoptotic molecules from mitochondria, preventing apoptosis. In addition, ROS generation can elicit autophagy as well as apoptotic cell death. Our data show that Red-Br-nos-triggered ROS generation, which induced death-inhibiting autophagy and death-inducing apoptosis. Perhaps, the initial ROS release triggered autophagy, as a protective defense mechanism of cancer cells to deal with chemotherapeutic stress, but the temporal order of protective autophagy followed by apoptosis makes us wonder if autophagy sets the stage for apoptosis to kill prostate cancer cells after a futile attempt to rescue them from death.
Caspase-dependent apoptosis is most often recognized as the preferred type of physiological cell death. However, situations involving imbalance of ROS generation and detoxification, limited energy metabolism, or lack of proper protein synthesis might restrict the ability of cells to activate caspase-driven apoptosis. Under such circumstances, cells might choose to die through alternative non-caspase-dependent apoptotic pathways that rely on a different set of players of mitochondrial origin. Although previous reports have shown that Red-Br-nos induces caspase-3-driven apoptosis in ovarian cancer cells, our present study in prostate cancer cells shows the activation of mitochondrially mediated ROS-dependent death cascade, which is caspase-independent. Existence of other cell death options suggest that there exists plasticity in the choice of cell death executioner programs, which is tissue-type specific, despite treatment with the same drug. Several reports indicate that cells can survive apoptotic cytochrome c release (60). This suggests that cytochrome c release might not always be an apoptogenic event; rather cytochrome c can intensify ROS generation. It is likely that release of cytochrome c upon Red-Br-nos treatment is due to a “mitochondrial catastrophe,” a consequence of loss of mitochondrial function that ultimately leads to cell death in a caspase-independent manner. Our data demonstrate that Red-Br-nos exposure showed an absence of cytochrome c-mediated caspase-3 activation. We believe that in such conditions, other mitochondrial intermembrane space proteins that are released along with cytochrome c, such as Endo-G and AIF. Our immunofluorescence microscopy data showed that Red-Br-nos caused the nuclear translocation of AIF and Endo-G, both of which are known to cause DNA fragmentation.
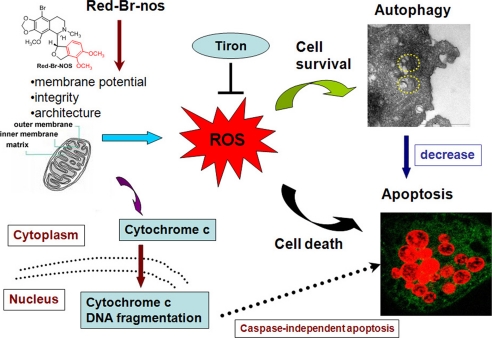
The role of mitochondrial damage in autophagy is intricate and complex. Based on our data, we hypothesize that cells perhaps react to mitochondrial damage in a graded manner: damage to a few mitochondria stimulates autophagy and mitochondria are degraded; damage to several mitochondria provokes apoptosis and cells die (28). We have found that Red-Br-nos directly affects the mitochondrial morphology resulting in loss of transmembrane potential. Using cyclosporin A that preserves mitochondrial integrity, there is an attenuation of ROS levels and a significantly lesser protective response from autophagy. These data allow us to conclude that drug-induced ROS generation is upstream of autophagy induction. In addition, our data underscore that a cross-talk between autophagy and apoptosis does exist, with early autophagy trying to rescue cancer cells by recycling damaged organelles, however, when the mitochondrial damage is large scale and ROS generation is robust, apoptosis is induced and cells die. Based on our results, we propose the following model, as illustrated in Fig. 7.
FIGURE 7.
Schematic diagram illustrates a proposed model depicting the interrelationships of events in drug-induced activation of ROS and autophagy that determine the extent of apoptosis.
Based upon our data, it is clearly conceivable that autophagy offers protection and enables tumor cell survival. Thus, combination regimens of chemotherapeutic drugs with autophagy inhibitors might be essential to achieve near complete cancer eradication. In summary, our data using a tubulin-binding agent, Red-Br-nos, sheds light on the role of autophagy in cancer chemotherapy and on the potential of suppressing autophagy as a novel strategy to enhance therapeutic efficiency.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant 1K99CA131489 from the NCI and Department of Defense Grant PC073104 (to R. A.) and a grant from the Georgia Research Alliance.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S10.
- PI3K
- phosphatidylinositol 3-OH kinase
- Red-Br-nos
- (R)-9-bromo-5-((S)-4,5-dimethoxy-1,3-dihydro-isobenzofuran-1-yl)-4-methoxy-6-methyl-5,6,7,8-tetrahydro-[1,3]di-oxolo[4,5-g]isoquinoline
- AVO
- acidic vesicular organelles
- AO
- acridine orange
- DCFDA
- 2′,7′-dichlorofluorescein diacetate
- 3-MA
- 3-methyladenine
- DHE
- dihydroethidium
- JC-1
- 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolyl carbocyanine iodide
- Z-VAD-fmk
- benzyloxycarbonyl-Val-Ala-Asp (OMe) fluoromethyl ketone
- ROS
- reactive oxygen species
- siRNA
- small interfering RNA
- DMSO
- dimethyl sulfoxide
- PBS
- phosphate-buffered saline
- GFP
- green fluorescent protein
- Endo-G
- endonuclease G
- LC3
- light chain 3.
REFERENCES
- 1.Peng X. H., Karna P., O'Regan R. M., Liu X., Naithani R., Moriarty R. M., Wood W. C., Lee H. Y., Yang L. (2007) Mol. Pharmacol. 71, 101–111 [DOI] [PubMed] [Google Scholar]
- 2.Yu L., Wan F., Dutta S., Welsh S., Liu Z., Freundt E., Baehrecke E. H., Lenardo M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 4952–4957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levine B. (2005) Cell 120, 159–162 [DOI] [PubMed] [Google Scholar]
- 4.Levine B., Yuan J. (2005) J. Clin. Invest. 115, 2679–2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levine B., Klionsky D. J. (2004) Dev. Cell 6, 463–477 [DOI] [PubMed] [Google Scholar]
- 6.Pattingre S., Levine B. (2006) Cancer Res. 66, 2885–2888 [DOI] [PubMed] [Google Scholar]
- 7.Pattingre S., Tassa A., Qu X., Garuti R., Liang X. H., Mizushima N., Packer M., Schneider M. D., Levine B. (2005) Cell 122, 927–939 [DOI] [PubMed] [Google Scholar]
- 8.Levine B., Abrams J. (2008) Nat. Cell. Biol. 10, 637–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosenbluth J. M., Pietenpol J. A. (2009) Autophagy 5, 114–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levine B., Kroemer G. (2009) Cell Death Differ. 16, 1–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kroemer G., Levine B. (2008) Nat. Rev. Mol. Cell Biol. 9, 1004–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Høyer-Hansen M., Bastholm L., Mathiasen I. S., Elling F., Jäättelä M. (2005) Cell Death Differ. 12, 1297–1309 [DOI] [PubMed] [Google Scholar]
- 13.Lee S. B., Tong S. Y., Kim J. J., Um S. J., Park J. S. (2007) DNA Cell Biol. 26, 713–720 [DOI] [PubMed] [Google Scholar]
- 14.Shao Y., Gao Z., Marks P. A., Jiang X. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 18030–18035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanzawa T., Germano I. M., Komata T., Ito H., Kondo Y., Kondo S. (2004) Cell Death Differ. 11, 448–457 [DOI] [PubMed] [Google Scholar]
- 16.Kanzawa T., Kondo Y., Ito H., Kondo S., Germano I. (2003) Cancer Res. 63, 2103–2108 [PubMed] [Google Scholar]
- 17.Bursch W., Ellinger A., Kienzl H., Török L., Pandey S., Sikorska M., Walker R., Hermann R. S. (1996) Carcinogenesis 17, 1595–1607 [DOI] [PubMed] [Google Scholar]
- 18.Aneja R., Lopus M., Zhou J., Vangapandu S. N., Ghaleb A., Yao J., Nettles J. H., Zhou B., Gupta M., Panda D., Chandra R., Joshi H. C. (2006) Cancer Res. 66, 3782–3791 [DOI] [PubMed] [Google Scholar]
- 19.Aneja R., Zhou J., Vangapandu S. N., Zhou B., Chandra R., Joshi H. C. (2006) Blood 107, 2486–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aneja R., Liu M., Yates C., Gao J., Dong X., Zhou B., Vangapandu S. N., Zhou J., Joshi H. C. (2008) Cancer Res. 68, 1495–1503 [DOI] [PubMed] [Google Scholar]
- 21.Aneja R., Zhou J., Zhou B., Chandra R., Joshi H. C. (2006) Mol. Cancer Ther. 5, 2366–2377 [DOI] [PubMed] [Google Scholar]
- 22.Zhou J., Gupta K., Aggarwal S., Aneja R., Chandra R., Panda D., Joshi H. C. (2003) Mol. Pharmacol. 63, 799–807 [DOI] [PubMed] [Google Scholar]
- 23.Zhou J., Panda D., Landen J. W., Wilson L., Joshi H. C. (2002) J. Biol. Chem. 277, 17200–17208 [DOI] [PubMed] [Google Scholar]
- 24.Heidemann S. (2006) Blood 107, 2216–2217 [Google Scholar]
- 25.Zhou J., Liu M., Luthra R., Jones J., Aneja R., Chandra R., Tekmal R. R., Joshi H. C. (2005) Cancer Chemother. Pharmacol. 55, 461–465 [DOI] [PubMed] [Google Scholar]
- 26.Zhou J., Liu M., Aneja R., Chandra R., Joshi H. C. (2004) Biochem. Pharmacol. 68, 2435–2441 [DOI] [PubMed] [Google Scholar]
- 27.Millot C., Millot J. M., Morjani H., Desplaces A., Manfait M. (1997) J. Histochem. Cytochem. 45, 1255–1264 [DOI] [PubMed] [Google Scholar]
- 28.Kondo Y., Kondo S. (2006) Autophagy 2, 85–90 [DOI] [PubMed] [Google Scholar]
- 29.Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. (2000) EMBO J. 19, 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kirisako T., Baba M., Ishihara N., Miyazawa K., Ohsumi M., Yoshimori T., Noda T., Ohsumi Y. (1999) J. Cell Biol. 147, 435–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang X. H., Jackson S., Seaman M., Brown K., Kempkes B., Hibshoosh H., Levine B. (1999) Nature 402, 672–676 [DOI] [PubMed] [Google Scholar]
- 32.Daido S., Kanzawa T., Yamamoto A., Takeuchi H., Kondo Y., Kondo S. (2004) Cancer Res. 64, 4286–4293 [DOI] [PubMed] [Google Scholar]
- 33.Paglin S., Hollister T., Delohery T., Hackett N., McMahill M., Sphicas E., Domingo D., Yahalom J. (2001) Cancer Res. 61, 439–444 [PubMed] [Google Scholar]
- 34.Xu Y., Kim S. O., Li Y., Han J. (2006) J. Biol. Chem. 281, 19179–19187 [DOI] [PubMed] [Google Scholar]
- 35.Cathcart R., Schwiers E., Ames B. N. (1983) Anal. Biochem. 134, 111–116 [DOI] [PubMed] [Google Scholar]
- 36.Vanden Hoek T. L., Li C., Shao Z., Schumacker P. T., Becker L. B. (1997) J. Mol. Cell Cardiol. 29, 2571–2583 [DOI] [PubMed] [Google Scholar]
- 37.Scherz-Shouval R., Shvets E., Fass E., Shorer H., Gil L., Elazar Z. (2007) EMBO J. 26, 1749–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Azad M. B., Chen Y., Gibson S. B. (2009) Antioxid. Redox Signal 11, 777–790 [DOI] [PubMed] [Google Scholar]
- 39.Wu W. S. (2006) Cancer Metastasis Rev. 25, 695–705 [DOI] [PubMed] [Google Scholar]
- 40.Scherz-Shouval R., Elazar Z. (2007) Trends Cell Biol. 17, 422–427 [DOI] [PubMed] [Google Scholar]
- 41.Mizumachi T., Suzuki S., Naito A., Carcel-Trullols J., Evans T. T., Spring P. M., Oridate N., Furuta Y., Fukuda S., Higuchi M. (2008) Oncogene 27, 831–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chipuk J. E., Green D. R. (2005) Nat. Rev. Mol. Cell Biol. 6, 268–275 [DOI] [PubMed] [Google Scholar]
- 43.Schneiders U. M., Schyschka L., Rudy A., Vollmar A. M. (2009) Mol. Cancer Ther. 8, 2914–2925 [DOI] [PubMed] [Google Scholar]
- 44.Ow Y. P., Green D. R., Hao Z., Mak T. W. (2008) Nat. Rev. Mol. Cell Biol. 9, 532–542 [DOI] [PubMed] [Google Scholar]
- 45.Petronilli V., Penzo D., Scorrano L., Bernardi P., Di Lisa F. (2001) J. Biol. Chem. 276, 12030–12034 [DOI] [PubMed] [Google Scholar]
- 46.Perchellet E. M., Wang Y., Weber R. L., Lou K., Hua D. H., Perchellet J. P. (2004) Anticancer Drugs 15, 929–946 [DOI] [PubMed] [Google Scholar]
- 47.Nur-E-Kamal A., Gross S. R., Pan Z., Balklava Z., Ma J., Liu L. F. (2004) J. Biol. Chem. 279, 24911–24914 [DOI] [PubMed] [Google Scholar]
- 48.Petiot A., Ogier-Denis E., Blommaart E. F., Meijer A. J., Codogno P. (2000) J. Biol. Chem. 275, 992–998 [DOI] [PubMed] [Google Scholar]
- 49.Tassa A., Roux M. P., Attaix D., Bechet D. M. (2003) Biochem. J. 376, 577–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen Y., Gibson S. B. (2008) Autophagy 4, 246–248 [DOI] [PubMed] [Google Scholar]
- 51.Elmore S. P., Qian T., Grissom S. F., Lemasters J. J. (2001) FASEB J. 15, 2286–2287 [DOI] [PubMed] [Google Scholar]
- 52.Scorrano L., Ashiya M., Buttle K., Weiler S., Oakes S. A., Mannella C. A., Korsmeyer S. J. (2002) Dev. Cell 2, 55–67 [DOI] [PubMed] [Google Scholar]
- 53.Cossarizza A., Baccarani-Contri M., Kalashnikova G., Franceschi C. (1993) Biochem. Biophys. Res. Commun. 197, 40–45 [DOI] [PubMed] [Google Scholar]
- 54.Matsui Y., Kyoi S., Takagi H., Hsu C. P., Hariharan N., Ago T., Vatner S. F., Sadoshima J. (2008) Autophagy 4, 409–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mathew R., Kongara S., Beaudoin B., Karp C. M., Bray K., Degenhardt K., Chen G., Jin S., White E. (2007) Genes Dev. 21, 1367–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thorburn A. (2008) Apoptosis 13, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maiuri M. C., Zalckvar E., Kimchi A., Kroemer G. (2007) Nat. Rev. Mol. Cell Biol. 8, 741–752 [DOI] [PubMed] [Google Scholar]
- 58.Sinha S., Levine B. (2008) Oncogene 27, Suppl. 1, S137–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim R. H., Coates J. M., Bowles T. L., McNerney G. P., Sutcliffe J., Jung J. U., Gandour-Edwards R., Chuang F. Y., Bold R. J., Kung H. J. (2009) Cancer Res. 69, 700–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Colell A., Ricci J. E., Tait S., Milasta S., Maurer U., Bouchier-Hayes L., Fitzgerald P., Guio-Carrion A., Waterhouse N. J., Li C. W., Mari B., Barbry P., Newmeyer D. D., Beere H. M., Green D. R. (2007) Cell 129, 983–997 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.