Abstract
An important event in cholesterol metabolism is the efflux of cellular cholesterol by apolipoprotein A-I (apoA-I), the major protein of high density lipoproteins (HDL). Lipid-free apoA-I is the preferred substrate for ATP-binding cassette A1, which promotes cholesterol efflux from macrophage foam cells in the arterial wall. However, the vast majority of apoA-I in plasma is associated with HDL, and the mechanisms for the generation of lipid-free apoA-I remain poorly understood. In the current study, we used fluorescently labeled apoA-I that exhibits a distinct fluorescence emission spectrum when in different states of lipid association to establish the kinetics of apoA-I transition between the lipid-associated and lipid-free states. This approach characterized the spontaneous and rapid exchange of apoA-I between the lipid-associated and lipid-free states. In contrast, the kinetics of apoA-I exchange were significantly reduced when apoA-I on HDL was cross-linked with a bi-functional reagent or oxidized by myeloperoxidase. Our observations support the hypothesis that oxidative damage to apoA-I by myeloperoxidase limits the ability of apoA-I to be liberated in a lipid-free form from HDL. This impairment of apoA-I exchange reaction may be a trait of dysfunctional HDL contributing to reduced ATP-binding cassette A1-mediated cholesterol efflux and atherosclerosis.
Keywords: Apolipoproteins, Cholesterol, Cholesterol Metabolism, High Density Lipoprotein (HDL), Lipid Transport, Apolipoprotein A-I, Exchangeable Apolipoproteins, Oxidation
Introduction
Apolipoprotein A-I (apoA-I),4 the primary protein constituent of HDL, inhibits atherosclerosis in hypercholesterolemic mice (1–3). Moreover, increasing evidence indicates that HDL is cardioprotective in humans (4). The ability of HDL to promote cholesterol efflux from cholesteryl ester-laden macrophage foam cells is of central importance to the initiation and progression of atherosclerosis (5–7).
In plasma, the vast majority of apoA-I associates with spherical HDL, a complex of apolipoproteins, phospholipids, triglycerides, free cholesterol, and cholesterol esters (8). However, the primary acceptor of cholesterol and phospholipids from macrophages is lipid-free or lipid-poor apoA-I (containing up to four phospholipid molecules (9)), which is the preferred substrate of the plasma membrane transporter ATP-binding cassette A1 (ABCA1) (10–15). Circulating HDL is remodeled by the action of proteins and enzymes (16) such as cholesteryl ester transfer protein (17), lechitin:cholesterol acyltransferase (LCAT) (18, 19), phospholipid transfer protein (20, 21), and hepatic lipase (22). Plasma HDL remodeling can result in the destabilization of HDL and the release of lipid-free/lipid-poor apoA-I. Furthermore, the selective uptake of lipids from HDL through scavenger receptor B, type 1 can yield lipid-poor apoA-I (23).
We hypothesized that the production of lipid-free/lipid-poor apoA-I from mature HDL and relipidation by ABCA1 is a dynamic process in the arterial wall, which is critical in protecting macrophages from cholesteryl ester accumulation. Previously, we showed that lipid-free/lipid-poor apoA-I is released from reconstituted HDL (rHDL) by long term (3–7 days) incubation at 37 °C (24), a time frame consistent with the in vivo half-life of HDL (25).
In the current study, we have employed a fluorescent apoA-I variant to demonstrate that apoA-I spontaneously exchanges between rHDL-associated and lipid-free states. Furthermore, using fluorescence resonance energy transfer spectroscopy, we quantified the rate of apoA-I exchange.
Förster (or fluorescence) resonance energy transfer (FRET) spectroscopy is the distance-dependent transfer of energy from the excited state of a donor to an acceptor fluorophore. Energy transfer is exhibited by a reduction in donor fluorophore fluorescence intensity and a concomitant increase in acceptor fluorophore fluorescence intensity. By quantifying these changes, it is possible to detect nanoscale separations in donor-acceptor distance (10–100 Å). FRET has been successfully applied as a means of estimating intramolecular and intermolecular distances in macromolecular systems, especially proteins (26–28) and has been exploited in an array of techniques ranging from the assay of interaction of antigen with an antibody in vitro to the real time imaging of protein folding in vivo (29, 30), as reviewed in Refs. 31–33.
By strategically positioning donor-acceptor fluorophores, we generated an apoA-I variant that exhibits lipidation state-specific FRET. We used this fluorescent apoA-I variant to examine the parameters governing the exchange of apoA-I between rHDL-associated and lipid-free states. Furthermore, we demonstrated that chemical cross-linking and oxidation by myeloperoxidase (MPO) severely impair the ability of apoA-I to exchange between the lipid-associated and lipid-free forms. These apoA-I modifying reactions replicate oxidative events that occur in human atherosclerotic lesions (34–43), suggesting that processes that impair the rate of apoA-I exchange reduce the cholesterol mobilizing capacity of HDL, thereby contributing to foam cell formation and atherogenesis (44).
EXPERIMENTAL PROCEDURES
Production of Recombinant ApoA-I Proteins
Mutations within human apoA-I cDNA were generated by primer-directed PCR mutagenesis as previously described (45). DNA sequences were verified by dideoxy automated fluorescent sequencing and confirmed by MS analysis of the protein. Wild-type and variant apoA-I proteins were expressed in Escherichia coli using the pET-20b bacterial expression vector (Novagen, Inc., Madison, WI) and purified as described (46).
Rationale for the Design of Fluorescent ApoA-I Variants
Glu at position 136 was mutated to Cys to generate apoA-I:A350, apoA-I:W19-AED136, and apoA-I:W19-A350–136 (Table 1). According to the current knowledge of lipid-free apoA-I structure derived from different biophysical methods (47, 48), position 136 resides in an unstructured and flexible central region of the protein. This residue was thus selected to minimize the impact of amino acid substitution and fluorescent labels on structure and lipid binding properties of the protein.
TABLE 1.
List of protein variants used in this study sorted as they appear in the text
| Protein variant name | Amino acid substitutions | FRET | Extrinsic fluorophore |
|---|---|---|---|
| ApoA-I:A350 | E136C | ALEXA350 | |
| ApoA-I:W19-AED136 | W8F, W50F, W72F, W108F, V19W, E136C | Donor-acceptor | AEDANS |
| ApoA-I:W19-A350–136 | W8F, W50F, W72F, W108F, V19W, E136C | ALEXA350 | |
| ApoA-I:ΔW | W8F, W50F, W72F, W108F | Null | |
| ApoA-I:ΔW-W19 | W8F, W50F, W72F, W108F, V19W | Donor-only | |
| ApoA-I:ΔW-AED136 | W8F, W50F, W72F, W108F, E136C | Acceptor-only | AEDANS |
| ApoE3-NT:ΔW | N-terminal (1–183) W39F, W74F, W118F, W162F | Null |
Similarly, to minimize the impact of the mutation on the N-terminal helix bundle of apoA-I, position 19 was selected to be substituted with a Trp in apoA-I:W19-AED136 and apoA-I:W19-A350–136. Residue 19 either lies at the beginning of the first α-helical segment at the protein N terminus (47) or in a dynamic secondary structure ensemble (48). Flexible structural regions proximal to Val-19 and Glu-136 tolerate mutations and bulky substituents without affecting critical structural constraints in apoA-I.
FRET Donor-Acceptor ApoA-I:W19-AED136 Variant
To generate an apoA-I variant with lipidation state-specific FRET properties (apoA-I:W19-AED136), six amino acid substitutions were performed (Table 1). Endogenous Trp fluorescence was eliminated by substituting the four native Trp residues with Phe (residues 8, 50, 72, and 108). Additionally, Val-19 was substituted with a Trp (the FRET donor), and Glu-136 was substituted with a Cys residue. The Cys in position 136 was reacted with I-AEDANS (the FRET acceptor) to generate apoA-I:W19-AED136 (supplemental Fig. S1A).
Trp to Phe substitution in apoA-I does not significantly affect protein structure (49) and function, as demonstrated by comparable ABCA1-mediated cholesterol efflux activity of wild-type and variant apoA-I (50). To further demonstrate that the six amino acid substitutions introduced in apoA-I:W19-AED136 do not significantly affect the HDL binding properties of the protein, we generated an ALEXA350-labeled variant (apoA-I:W19-A350–136) equivalent to apoA-I:W19-AED136 (Table 1). ApoA-I:W19-A350–136 was incubated at 37 °C in the presence of a 5-fold molar excess of wild-type apoA-I 7.8-nm rHDL particles. Fluorescent and total protein staining imaging of NDGGE gels showed a time-dependent increase of fluorescence in the rHDL fraction without rHDL remodeling (supplemental Fig. S1B). Displacement of rHDL-associated wild-type apoA-I by apoA-I:W19-A350–136 clearly demonstrates that the six mutations introduced in apoA-I do not significantly reduce the lipid binding capacity of the protein.
Fluorescent Labeling of Proteins
AEDANS and ALEXA350 fluorophores were covalently linked to Cys-136 as described (51). Briefly, the cysteine disulfide bond was reduced by overnight incubation in the presence of tris(2-carboxyethyl)phosphine at a final molar ratio of 1:10 apoA-I:tris(2-carboxyethyl)phosphine. The reduced protein (8 mg) was immobilized on a nickel-chelating column (1 ml of Hi-Trap Chelating HP columns; Pharmacia Biotech) and prewashed with 40 mm imidazole and 3 m guanidine in phosphate-buffered saline, pH 7.4. The column was incubated with the labeling reagent at a final molar ratio apoA-I:reagent 1:5 in 3 m guanidine HCl (to eliminate conformation-specific labeling) for 3 h at 37 °C. Labeled protein was eluted by 0.5 m imidazole and dialyzed extensively against Tris-buffered saline (8.2 mm Tris, 150 mm NaCl, 1 mm EDTA, pH 7.4) to remove guanidine and unreacted label. Similar labeling techniques have been employed with apoA-I (49, 51) and other proteins (52, 53), wherein such photolabeling and equivalent amino acid substitutions did not affect the structure and stability of the proteins.
Preparation and Characterization of Reconstituted HDL
Palmitoyl oleoyl phosphatidyl choline and free cholesterol containing HDL particles 7.8, 8.4, and 9.6 nm in diameter were prepared by the sodium deoxycholate dialysis method and purified as described (24). rHDL particles containing apoA-I variants were equivalent to control particles containing wild-type apoA-I in size (by NDGGE and size exclusion chromatography) (supplemental Fig. S2), lipid and cholesterol composition, and number of apoA-I molecules/particle (by cross-linking). rHDL particles containing apoA-I variants also retained their size-dependent ability to activate LCAT. These measures indicate that the mutations introduced did not significantly affect the physicochemical and physiological properties of rHDL.
Fluorescence Spectroscopy
Steady-state fluorescence spectra were collected on a Horiba Jobin-Yvon fluoromax-4 spectrofluorometer using a 1.5-nm slit width for both the excitation and emission monochronomators. The samples were in Tris-buffered saline, and all of the spectra were collected at 20 °C. For kinetic analysis of apoA-I exchange reaction, at each time point, the exchange reaction mixture was rapidly transferred from 37 °C incubation conditions to a quartz cuvette. Fluorescence spectra were recorded in less than 5 min, and then the exchange reaction mixture was promptly returned to 37 °C.
FRET Analysis
To determine structural changes in apoA-I caused by lipidation, nonradiative energy transfer from Trp-19 (donor) to AEDANS (acceptor) bound to Cys-136 was used. The absorption spectrum of AEDANS overlaps with the fluorescence spectrum of the intrinsic Trp residues of apoA-I (data not shown), making nonradiative resonance energy transfer from Trp to protein-conjugated AEDANS possible, upon excitation of Trp. This process causes a decrease in Trp fluorescence intensity and leads to the appearance of sensitized fluorescence of AEDANS (see Fig. 4, A and D).
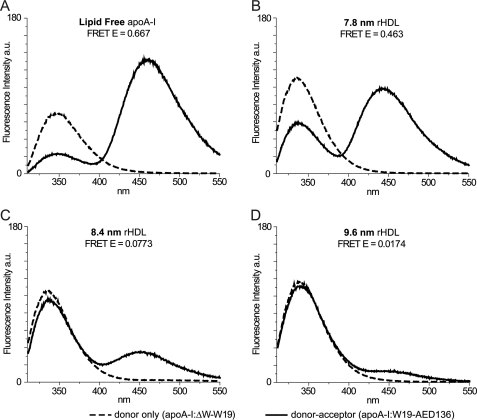
FIGURE 4.
Fluorescence spectra of protein variants. Sample concentrations were 0.25 mg/ml; excitation wavelength was 280 nm; and emission was from 310 to 550 nm. The bandwidth of excitation and emission monochromators was 1.5 nm. Emission of the single Trp at position 19 of apoA-I:ΔW-W19 (donor-only) is reported as a dashed line for lipid-free (A) and each lipidation state (7.8-nm(B), 8.4-nm (C), and 9.6-nm (D) rHDL). Background fluorescence from Tyr was eliminated by subtracting the emission spectrum of apoA-I:ΔW (Trp-null background) from the emission spectrum of apoA-I:ΔW-W19 (donor-only) in the same lipidation state. The solid lines are emission spectra of Trp and AEDANS in apoA-I:W19-AED136 (donor-acceptor). Acceptor fluorescence is due to energy transfer alone; Tyr background and direct excitation of AEDANS were eliminated by subtracting the emission spectra of apoA-I:ΔW-AED136 (acceptor-only in Trp-null background) from the emission spectra of apoA-I:W19-AED136 in the same lipidation state. Energy transfer efficiency was calculated from background-corrected spectra by comparing Trp fluorescence intensities (integration over 310–395 nm) of apoA-I:ΔW-W19 (donor-only) and apoA-I:W19-AED136 (donor-acceptor).
For FRET analysis we used 0.25 mg/ml of fluorescently labeled protein. The samples were excited at 280 nm, and the emission spectra were recorded from 310 to 550 nm.
Even though fluorescence is a sensitive technique, interference from background emissions can affect data quality and lead to misinterpretation. We successfully overcame this problem using the controls described below, correcting for two sources of interfering fluorescence emission: (i) weak emission from the 10 Tyr residues of apoA-I and (ii) residual emission by direct excitation of AEDANS. The controls employed are: (i) a Trp-null apoA-I variant (apoA-I:ΔW; Table 1), wherein the four native Trp residues were substituted with Phe; apoA-I:ΔW was used to eliminate the background noise from Tyr residues (51); (ii) a donor-only apoA-I variant (apoA-I:ΔW-W19; Table 1), wherein the four native Trp residues were substituted with Phe and Val-19 was substituted with Trp; this provided us a control for monitoring the intrinsic fluorescence of apoA-I; and (iii) an acceptor-only apoA-I variant (apoA-I:ΔW-AED136; Table 1), wherein the four native Trp residues were substituted with Phe, and Glu-136 was substituted with a Cys residue, labeled with AEDANS. The emission spectrum of apoA-I:ΔW-AED136 was used to eliminate the interference from direct excitation of AEDANS.
Efficiency of energy transfer was calculated by comparison of the integrated emissions (310–395 nm) of background-corrected apoA-I:ΔW-W19 (donor-only) and apoA-I:W19-AED136 (donor-acceptor) fluorescence spectra, for each lipidation state (as described under “Results” and in Fig. 4).
ApoE3-NT and Plasma-purified ApoA-I
A variant of the N-terminal fragment (residues 1–183) of apoE3 where all of the Trp residues were mutated to Phe was generously provided by Dr. Taichi Yamamoto (54, 55). Dr. Trudy M. Forte supplied the plasma apoA-I used in this study, which had been purified as described (56).
Cross-linking of rHDL
7.8 rHDL (apoA-I:W19-AED136) was incubated for 2 h at room temperature with a 200-fold molar excess (relative to apoA-I) of ((bis)sulfosuccinimidyl) suberate (BS3) in phosphate-buffered saline (20 mm phosphate, 500 mm NaCl, pH 7.4). The reaction was terminated by the addition of 500 mm Tris-HCl. The cross-linking mixtures were dialyzed overnight versus 4 liters of Tris-buffered saline with two buffer changes and analyzed via 4–20% gradient SDS-PAGE and NDGGE (Invitrogen) (supplemental Fig. S2).
Oxidation Reactions and Liquid Chromatography-Electrospray Ionization-MS Analysis of Modified Met and Tyr Residues
ApoA-I:ΔW was oxidized by ONOO− or the MPO-H2O2-nitrite system as described (57). Briefly, the reactions were carried out at 37 °C for 1 h in phosphate-buffered saline (with 100 μm diethylenetriaminepentaacetic acid, pH 7.4) containing 20 μm apoA-I. ONOO− was synthesized from nitrite and H2O2 (58), and 500 μm ONOO− was used for the reaction. For the MPO-H2O2-nitrite system, the reaction mixture was supplemented with 50 nm MPO, 150 μm NaNO2, and 200 μm H2O2. Native or oxidized apoA-I:ΔW was digested with sequencing grade modified trypsin (Promega, Madison, WI) or sequencing grade endoproteinase Glu-C (Roche Applied Science). Liquid chromatography-MS and tandem MS analyses of tryptic or Glu-C digest peptides were performed in the positive ion mode with a Thermo-Finnigan LCQ Deca XP Plus instrument (San Jose, CA) interfaced with an Agilent 1100 series high pressure liquid chromatography system (Santa Clara, CA) as described (59). Product yield of oxidized peptides was determined with reconstructed ion chromatograms of product and precursor peptides, calculated as: product yield (%) = [(product ion peak area)/(precursor ion peak area + product ion peak area)] × 100 (59).
RESULTS
ApoA-I Is Freely Exchangeable between rHDL-associated and Lipid-free States
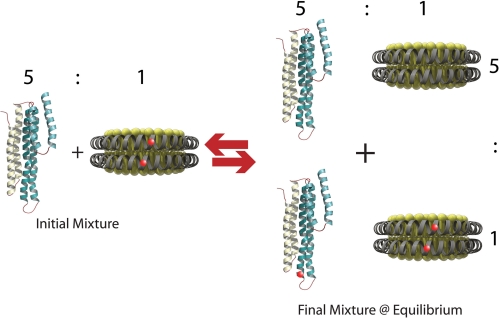
To determine whether apoA-I can exchange between rHDL-associated and lipid-free states without the need for HDL remodeling enzymes, we labeled an apoA-I variant, bearing a Glu to Cys substitution at position 136, with ALEXA350 (apoA-I:A350; Table 1 and “Experimental Procedures”). We hypothesized that under physiological conditions there is an equilibrium between rHDL-associated and lipid-free apoA-I. To test this hypothesis we reconstituted apoA-I:A350, palmitoyl oleoyl phosphatidyl choline, and cholesterol into HDL and purified 7.8-nm-diameter particles as previously described for wild-type apoA-I (24). No differences were detectable between rHDL particles containing apoA-I:A350 or wild-type apoA-I by NDGGE and particle composition analysis. rHDL (apoA-I:A350) was incubated at 37 °C in the presence of a 5-fold molar excess of unlabeled plasma-purified lipid-free apoA-I and assessed by NDGGE after 5 h and at 24-h intervals thereafter. According to our hypothesis, the addition of an excess of lipid-free apoA-I would re-establish an equilibrium state between lipid-free and lipid-associated protein by displacement of apoA-I:A350 from the rHDL particle (Fig. 1). At equilibrium, if all of the apoA-I species have an equal affinity for rHDL, approximately five-sixths (∼83%) of apoA-I:A350 should be in the lipid-free protein pool, whereas unlabeled apoA-I should compose approximately five-sixths of the rHDL-associated apoA-I.
FIGURE 1.
Schematic of the apoA-I exchange assay. Fluorescently labeled apoA-I-containing rHDL is incubated at 37 °C with a 5-fold molar excess of unlabeled lipid-free apoA-I. At equilibrium, five-sixths (∼83%) of the original rHDL-associated fluorescent apoA-I are displaced by unlabeled protein into the lipid-free pool. By this scheme, at equilibrium, one-sixth of the lipid-free apoA-I pool will be fluorescent.
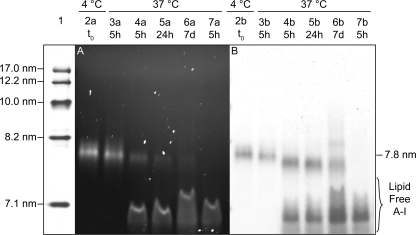
Fluorescent imaging of NDGGE gels (Fig. 2A) showed that ∼70% of apoA-I:A350 was in the lipid-free pool of apoA-I after 5 h of incubation at 37 °C. At 24 h, >80% of apoA-I:A350 was detected in the apoA-I lipid-free pool. No further changes were detectable after longer incubation times (up to 7 days). Protein staining of these NDGGE gels (Fig. 2B) revealed that no significant change in rHDL size (7.8 nm) occurred within 5 h (Fig. 2B, lane 4b), and additional rHDL species were barely detectable after 7 days of incubation (Fig. 2B, lane 6b). This result confirms our proposition that excess lipid-free apoA-I can efficiently displace HDL-associated apoA-I.
FIGURE 2.
NDGGE analysis of apoA-I exchange. rHDL (apoA-I:A350) particles of 7.8-nm diameter were incubated at 37 °C in the presence (lanes 4a–6a) or absence (lane 3a) of a 5-fold molar excess of plasma-purified unlabeled lipid-free apoA-I. Equivalent amounts of rHDL-associated protein (1.5 μg) were separated from the lipid-free protein by electrophoresis on 4–20% nondenaturing Tris-glycine polyacrylamide gels. Fluorescently labeled protein was visualized using an UV trans-illuminator (excitation light, 365 nm; emission blue filter, 440–480 nm) (A). Total protein was detected with GelCode Blue staining (Pierce) (B). Lane 1, molecular weight markers (high molecular weight calibration kit from GE Healthcare). Lanes 2 and 3, rHDL (apoA-I:A350) stored at 4 °C or incubated at 37 °C for 5 h, respectively. Lanes 4–6, incubation mixtures of rHDL (apoA-I:A350) and a 5-fold molar excess of plasma-purified lipid-free apoA-I at 5 h, 24 h, and 7 days, respectively. Lanes 7, lipid-free apoA-I:A350 incubated with a 5-fold excess of lipid-free plasma-purified unlabeled lipid-free apoA-I and incubated at 37 °C for 5 h. The relative band intensities were estimated by densitometry (Imagequant 5.0 software; Amersham Biosciences).
Stability of rHDL
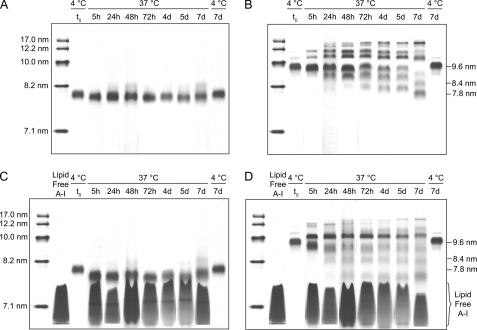
To establish a suitable model system for kinetic analysis of apoA-I exchange, we assessed the stability of rHDL of different sizes during prolonged incubation at 37 °C. rHDL (wild-type apoA-I) of 7.8- and 9.6-nm diameter were incubated at 37 °C up to 7 days in the presence and absence of a 5-fold molar excess of plasma-purified lipid-free apoA-I. Remodeling of rHDL was evaluated by daily NDGGE analysis. No additional HDL subclasses were detectable in the 7.8-nm rHDL sample for either incubation condition (Fig. 3, A and C, and supplemental Fig. S3). In contrast (Fig. 3, B and D), extensive remodeling of 9.6-nm rHDL had occurred after 5 h of incubation in the presence and absence of lipid-free apoA-I, leading to the formation of a significant number of additional rHDL species. By day 7, 90% of 9.6-nm rHDL remodeled preferentially toward the 7.8-nm rHDL subclass. Because of the absence of spontaneous rHDL particle remodeling, 7.8-nm rHDL is the ideal rHDL to monitor the kinetics of protein exchange between lipid-associated and lipid-free states.
FIGURE 3.
Remodeling of rHDL. NDGGE (4–20% Tris-glycine polyacrylamide gel) of 7.8-nm (A and C) and 9.6-nm (B and D) wild-type apoA-I rHDL incubated at 37 °C for 7 days in the presence (C and D) or absence (A and B) of a 5-fold molar excess of lipid-free plasma-purified apoA-I is shown. Incubation of 7.8- and 9.6-nm rHDL at 4 °C is reported in the last lane of each gel. The samples were analyzed as described for Fig. 2. The molecular weight markers were from the high molecular weight calibration kit from GE Healthcare. The figure represents composite gels in which lanes from the electrophoretic runs at each time point were combined by alignment of the molecular weight markers. See supplemental Fig. S3, for a representative original gel (24-h time point).
Fluorescently Labeled ApoA-I Reports the Molecule Lipidation State
To measure the kinetics of apoA-I exchange reaction between rHDL-associated and lipid-free states, we created a fluorescent apoA-I variant with donor and acceptor FRET fluorophores (apoA-I:W19-AED136) (see “Experimental Procedures” and Fig. 4). The fluorescence spectra of donor-only (apoA-I:ΔW-W19) and donor-acceptor (apoA-I:W19-AED136) apoA-I variants in different lipidation states are shown in Fig. 4 and summarized in Table 2.
TABLE 2.
Maximal fluorescence emission wavelengths in different lipidation states
The excitation wavelength was 280 nm, with instrument settings as described under “Experimental Procedures.” The values are from background corrected spectra as described for Fig. 4. Energy transfer efficiency was calculated from background corrected spectra by comparing Trp fluorescence intensities (integration over 310–395 nm) of apoA-I:ΔW-W19 (donor-only) and apoA-I:W19-AED136 (donor-acceptor).
| Lipidation state | λmax (Trp) of donor-only | Donor-acceptor |
||
|---|---|---|---|---|
| λmax (Trp) | λmax (AEDAN) | Energy transfer efficiency | ||
| nm | nm | nm | ||
| Lipid-free | 347 | 347 | 461 | 0.667 |
| Lipid-free control samplea | 343 | 460 | ||
| 7.8-nm rHDL | 336 | 337 | 442 | 0.463 |
| 8.4-nm rHDL | 335 | 336 | 451 | 0.0773 |
| 9.6-nm rHDL | 337 | 339 | 454 | 0.0174 |
a From the spectrum of a control sample composed of lipid-free donor-acceptor (apoA-I:W19-AED136) and nonfluorescent (apoA-I:ΔW; Trp-null background) apoA-I variants in a ratio that is predicted from our equilibrium reaction model and incubated at 37 °C for 72 h (lipid-free control sample) (see Fig. 5A, red line). Tyr background and direct excitation of AEDANS were eliminated by subtracting the emission spectra of apoA-I:ΔW (Trp-null background) and apoA-I:ΔW-AED136 (acceptor-only in Trp-null background) in the appropriate concentrations from the emission spectrum of the lipid-free control sample.
For lipid-free apoA-I, fluorescence emission λmax values for donor Trp and acceptor AEDANS indicate that both fluorophores were largely solvent-exposed (Table 2; Trp λmax > 336 (60)). The spectra reported in Fig. 4 were recorded at a protein concentration of 0.25 mg/ml. Lipid-free apoA-I is prone to self-associate at concentrations greater than 0.1 mg/ml (47, 61); thus, lipid-free apoA-I spectra in Fig. 4A might be from a mixture of monomeric and high molecular weight oligomeric species. As a consequence, the estimated energy transfer efficiency (66.7%) is the weighted average of intramolecular and intermolecular energy transfer contributions favored by close proximity of multiple apoA-I molecules within oligomeric complexes.
In lipid-associated states, the emissions of Trp and AEDANS were blue-shifted (∼10 and 10–20 nm, respectively) compared with lipid-free apoA-I. This may be attributed to the altered protein-protein or newly established lipid-protein interactions. In the lipid-bound state, apoA-I:W19-AED136 showed an inverse correlation between the size of the rHDL particles and energy transfer efficiency (46.3% (7.8 nm), 7.7% (8.4 nm), and 1.7% (9.6 nm). This may be due to the progressive separation of the N-terminal (Trp-19, donor) from the central region (Cys-136, acceptor) triggered by lipid enrichment in the center of rHDL, from smaller (7.8 nm) to larger (9.6 nm) particles.
To further validate the conformational dependence of energy transfer in apoA-I:W19-AED136, we examined the fluorescence spectrum of lipid-free apoA-I:W19-AED136 in 1% SDS (supplemental Fig. S4A). Disruption of lipid-free apoA-I:W19-AED136 native tertiary structure impaired AEDANS fluorescence and recovered >90% of the Trp florescence emission observed for apoA-I:ΔW-W19 (FRET donor-only). The Trp emission was blue-shifted by ∼25 nm (both apoA-I:W19-AED136 and apoA-I:ΔW-W19 spectra) because of the effect of SDS on the polarity of the local environment around the Trp. Furthermore, after SDS treatment, the emission of acceptor-only apoA-I:ΔW-AED136 was similar (for emission λ>425 nm) to that of the SDS-treated donor-acceptor apoA-I:W19-AED136, showing that the disruption of the native tertiary structure of the protein abolished a majority of FRET-induced acceptor fluorescence. Taken together, our data suggest that the fluorescence properties of double-labeled apoA-I can be used as a “molecular marker” to distinguish different rHDL subclasses and to monitor the proportion of apoA-I in different states of lipidation (lipid-free/rHDL subclasses).
Lipid-free ApoA-I Promotes the Rapid Exchange of ApoA-I between the rHDL-associated and Lipid-free States
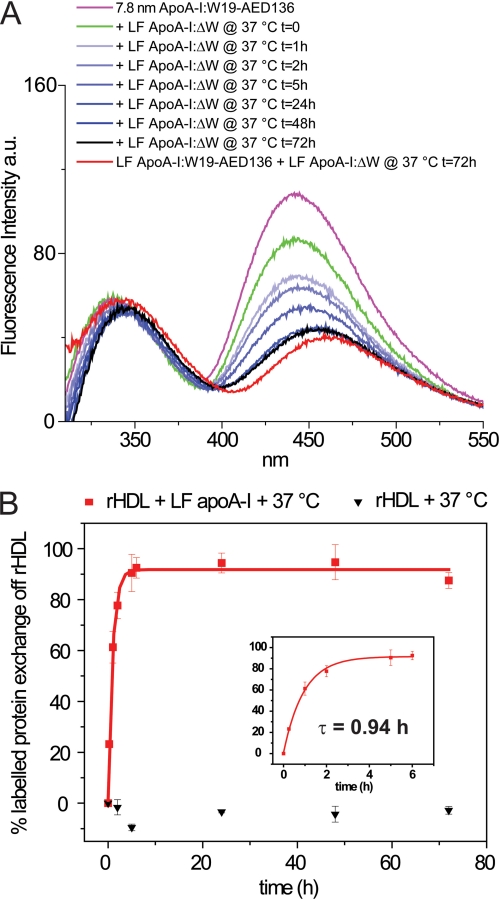
To perform kinetic measurements of apoA-I exchange between rHDL-associated and lipid-free states, we employed 7.8-nm rHDL (apoA-I:W19-AED136) and examined the apoA-I:W19-AED136 rate of exchange off rHDL. A 5-fold molar excess of lipid-free Trp-null apoA-I:ΔW was added to 7.8-nm rHDL (apoA-I:W19-AED136) and incubated at 37 °C (exchange reaction mixture). Fluorescence spectra were recorded at regular time intervals (Fig. 5A). As a control, acceptor-only 7.8-nm rHDL (apoA-I:ΔW-AED136) was mixed with a 5-fold molar excess of lipid-free Trp-null apoA-I:ΔW (control reaction mixture) and incubated at 37 °C. To eliminate Tyr and residual AEDANS direct excitation fluorescence background contributions, the fluorescence emission of the control reaction mixture (acceptor-only + Trp-null) was subtracted from the fluorescence emission spectra of the exchange reaction mixture (donor-acceptor + Trp-null) at each time point.
FIGURE 5.
Kinetic analysis of the exchange reaction between rHDL-associated and lipid-free apoA-I. A, rHDL (apoA-I:W19-AED136) particles (7.8-nm diameter; 0.25 mg of protein/ml) were incubated at 37 °C for 72 h in the presence of a 5-fold molar excess of lipid-free apoA-I:ΔW (1.25 mg of protein/ml) (exchange reaction mixture). The fluorescence spectra were recorded following the addition of lipid-free apoA-I:ΔW. The instrument setting was as described in Fig. 4. A control was produced in which acceptor-only 7.8-nm rHDL (apoA-I:ΔW-AED136) was mixed with a 5-fold molar excess of lipid-free Trp-null apoA-I:ΔW and incubated at 37 °C (control reaction mixture). To eliminate Tyr and direct excitation of AEDANS backgrounds, the exchange reaction mixture spectra shown in A were obtained by subtracting the fluorescence emission of the control reaction mixture (acceptor-only + Trp-null) from the fluorescence emission spectra of the exchange reaction mixture (donor-acceptor + Trp-null) at each time point. The spectrum of a lipid-free control sample composed of lipid-free donor-acceptor (apoA-I:W19-AED136) and nonfluorescent (apoA-I:ΔW) apoA-I variants in a ratio that is predicted from our equilibrium reaction model and incubated at 37 °C for 72 h is shown in red (see “Results”). B, kinetic analysis. The data points are the average values and standard deviations of at least six experiments. Red squares, 7.8-nm rHDL + lipid-free apoA-I:ΔW. Black triangles, 7.8-nm rHDL alone (see also supplemental Fig. S4B). The percentage of apoA-I exchanged from rHDL was calculated by the intensity of the fluorescence emission of AEDANS at λmax at each time point relative to the average intensity of AEDANS fluorescence emission at λmax at equilibrium (72 h) from six experiments. The solid lines show least squares fitting of the data by single exponential. The exponential relaxation time (τ) for apoA-I exchange reaction as calculated by fitting of the first 6 h of data points is indicated (inset).
Upon the addition of lipid-free apoA-I:ΔW, AEDANS fluorescence was rapidly reduced by ∼ 19% without affecting λmax (442 nm; Fig. 5A, magenta line versus green line). We attribute this initial drop in FRET to the disruption of interparticle interactions that contribute to energy transfer.
Incubation at 37 °C led to a time-dependent change in the emission spectra of the exchange reaction mixture. We propose that these spectral changes were due to the transition of apoA-I:W19-AED136 from the lipid-associated to the lipid-free state. Because the initial ratio of rHDL-associated apoA-I:W19-AED136 to lipid-free apoA-I:ΔW is 1:5, our reaction model predicts that when the exchange reaction mixture is at equilibrium, five-sixths of fluorescent apoA-I is liberated into the lipid-free protein pool and mixed with a 5-fold excess of lipid-free apoA-I:ΔW (Fig. 1). To confirm that the predicted equilibrium state matches the observed equilibrium mixture, lipid-free apoA-I:W19-AED136 (0.208 mg/ml) and lipid-free apoA-I:ΔW (1.042 mg/ml) were mixed in the proportions predicted in the final equilibrium state (5:1) and incubated at 37 °C for 72 h (lipid-free control sample). Upon incubation at 37 °C, the λmax of Trp and AEDANS of the exchange reaction mixture deviated from t = 0 values (337 and 442 nm, respectively) matching the spectral features of apoA-I:W19-AED136 in 7.8-nm rHDL and shifted toward values (at 72 h, 344 and 457 nm) similar to those observed in the lipid-free control sample (343 and 460 nm) (Fig. 5A, black and red lines, respectively). Similarly, the quantum yields of Trp and AEDANS emissions in the exchange reaction mixture at equilibrium were comparable with those of the lipid-free control sample. This correlation confirms that, at equilibrium, the composition of the exchange reaction mixture matches the predicted composition. Consistent with this observation, when lower ratios of lipid-free to lipid-associated apoA-I were used, the fluorescence emission of AEDANS in the exchange reaction mixture at equilibrium was significantly higher than in the lipid-free control sample. Similarly, λmax of AEDANS emission was not as red-shifted, indicating that less of apoA-I:W19-AED136 exchanged off 7.8-nm rHDL (data not shown).
The rate of apoA-I exchange was derived from the relative intensity of the fluorescence emission of AEDANS at λmax. The emission spectrum of the exchange reaction mixture at 0 h was used as a reference for 0% exchange (all fluorescently labeled protein in 7.8-nm rHDL), and the spectrum at 72 h was used as the final equilibrium state (100% exchange; average of six experiments) (Fig. 5B). Mono-exponential data fitting analysis yielded an exponential relaxation time (time at which 50% of the exchange occurred, τ) equal to 0.94 h, a measure consistent with the qualitative evaluation of apoA-I (apoA-I:A350) exchange observed by NDGGE (Fig. 2). The fluorescence emission spectra of 7.8-nm rHDL (apoA-I:W19-AED136) alone showed no changes in intensity or λmax during the same time period (supplemental Fig. S4B). Thus, changes in the fluorescence emission spectra of 7.8-nm rHDL (apoA-I:W19-AED136) in the exchange reaction mixture are not a result of incubation at 37 °C.
Apolipoprotein Exchange Rates Are Dependent on Relative Lipid Affinity
We compared the relative rate of exchange for a variant of the N-terminal fragment (residues 1–183) of apoE3 wherein the endogenous Trp residues were substituted with Phe (apoE3-NT:ΔW; Table 1) to that of apoA-I. Because of the very low lipid binding affinity of the N-terminal domain of apoE3 (62), it was anticipated that a lower rate of exchange would be observed.
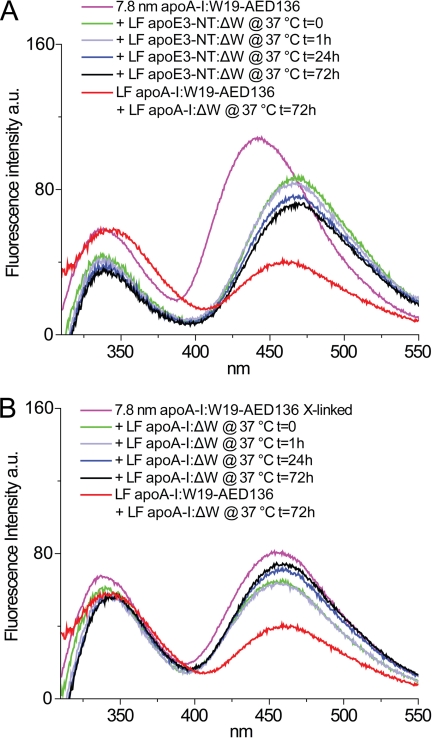
A 5-fold molar excess of lipid-free Trp-null apoE3-NT:ΔW was added to 7.8-nm rHDL (apoA-I:W19-AED136) and incubated at 37 °C (exchange reaction mixture). Background corrections were performed as described for Fig. 6A, eliminating Tyr and AEDANS direct excitation fluorescence. Upon the addition of lipid-free apoE3-NT:ΔW, the λmax of the fluorescence emission of AEDANS was red-shifted by 25 nm, and its fluorescence intensity was immediately diminished by ∼19% (Fig. 6A). This reduction in fluorescence intensity was equal to that observed when inter-HDL particle interactions were disrupted by the addition of lipid-free apoA-I (Fig. 5A). Incubation at 37 °C produced very modest changes in AEDANS fluorescence, even after long incubation times (up to 7 days), demonstrating that apoE3-NT:ΔW was unable to displace apoA-I from the rHDL particle. Our observations provide further validation that this fluorescence-based method presents an accurate assessment of the relative exchange rate and lipid affinity.
FIGURE 6.
ApoA-I exchange between rHDL-associated apoA-I and lipid-free apoE3-NT:ΔW (A) or BS3 cross-linked rHDL-associated apoA-I and lipid-free apoA-I:ΔW (B). In both panels, 7.8-nm rHDL and lipid-free protein were mixed at final concentrations of 0.25 and 1.25 mg/ml, respectively. Collection of fluorescence spectra of exchange reaction mixtures was initiated immediately upon the addition of lipid-free protein, with instrument setting and controls as described in Figs. 4 and 5. The spectrum of a lipid-free control sample representing the predicted equilibrium mixture is shown in red (see Fig. 5 and “Results”). A, a control was produced in which acceptor-only 7.8-nm rHDL (apoA-I:ΔW-AED136) was mixed with a 5-fold molar excess of lipid-free Trp-null apoE3-NT:ΔW and incubated at 37 °C (control reaction mixture). To eliminate Tyr and direct excitation of AEDANS backgrounds, the exchange reaction mixture spectra were obtained by subtracting the fluorescence emission of the control reaction mixture (acceptor-only + apoE3-NT Trp-null) from the fluorescence emission spectra of the exchange reaction mixture (donor-acceptor + apoE3-NT Trp-null) at each time point. B, rHDL particles of 7.8-nm diameter were reconstituted using apoA-I:W19-AED136 and cross-linked by BS3 as described under “Experimental Procedures.” To eliminate Tyr and direct excitation of AEDANS backgrounds, the exchange reaction mixture spectra were obtained by subtracting the fluorescence emission of the control reaction mixture (acceptor-only + Trp-null) from the fluorescence emission spectra of the exchange reaction mixture (cross-linked donor-acceptor + Trp-null) at each time point.
Cross-linking of rHDL-associated ApoA-I Impairs the Exchange of ApoA-I
To further test the hypothesis that apoA-I exchange occurs through desorption of a single apoA-I molecule from the surface of HDL, independent of rHDL remodeling, the two molecules of apoA-I that compose rHDL were fixed in a dimeric state by chemical cross-linking. Treatment of 7.8-nm rHDL (apoA-I:W19-AED136) particles with a molar excess of BS3 did not lead to sample degradation. The cross-linked rHDL migrated with higher electrophoretic mobility than untreated rHDL (supplemental Fig. S2). This change in electrophoretic mobility was due to the addition of negatively charged sulfonic groups from the BS3 cross-linking agent. The majority (>90%) of apoA-I in the sample was cross-linked with an apparent molecular weight equal to dimeric apoA-I, as determined by SDS-PAGE. AEDANS fluorescence emission of cross-linked rHDL (apoA-I:W19-AED136) was reduced in intensity (∼ 25%) and red-shifted (13 nm) relative to noncross-linked rHDL (apoA-I:W19-AED136) (Fig. 6, A and B).
A 5-fold molar excess of lipid-free Trp-null apoA-I:ΔW was added to cross-linked 7.8-nm rHDL (apoA-I:W19-AED136) and incubated at 37 °C (exchange reaction mixture). Fluorescence background corrections were performed as described in Fig. 6B. Incubation at 37 °C did not significantly change the fluorescence emission spectrum of cross-linked rHDL (Fig. 6B). This result indicates that cross-linking of apoA-I impairs its ability to freely exchange off the rHDL particle and supports the notion that apoA-I exchanges on and off HDL as a monomer.
MPO-mediated Oxidation of Lipid-free ApoA-I Reduces Its Rate of Exchange
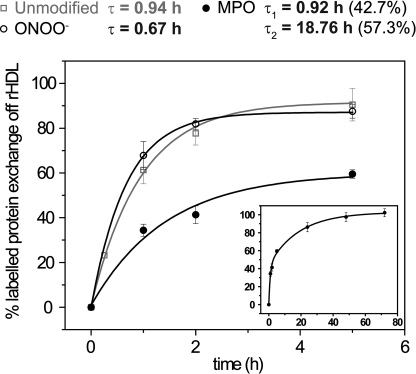
We hypothesized that MPO-mediated modifications of apoA-I decrease its conformational adaptability, thereby diminishing the ability of the protein to exchange into a lipid-free (ABCA1 efflux capable) state. We measured the exchange rate of lipid-free apoA-I:ΔW exposed to the complete MPO-H2O2-nitrite system, a potent source of NO2 radicals (63). The oxidation conditions were optimized to produce levels of nitrotyrosine in apoA-I comparable with those reported for HDL from human atherosclerotic lesion material (∼9%) (42). We also measured the exchange rate of apoA-I:ΔW exposed to peroxynitrite (ONOO−), another potent reactive nitrogen species generated by activated macrophages.
The rate of exchange of ONOO−-oxidized apoA-I:ΔW was similar to that of control apoA-I:ΔW and fit a single exponential model (Fig. 7). The exponential relaxation time (τ = 0.67 h) was also similar to that of the unmodified protein (τ = 0.94 h). MPO-oxidized apoA-I:ΔW exchange kinetics were complex. The simplest model was a bi-exponential fitting with two distinct relaxation times. A faster kinetic component, which accounted for 43% of the exchange rate, was similar to untreated apoA-I (τ = 0.92 h). In contrast, a slower component (57% of the exchange rate) showed a significantly reduced rate with a relaxation time of 18.8 h.
FIGURE 7.
Kinetic analysis of the exchange reactions between rHDL-associated apoA-I and unmodified (open squares), ONOO−-oxidized (open circles), or MPO-oxidized (solid circles) lipid-free apoA-I. Lipid-free apoA-I:ΔW was oxidized either with ONOO− generated from nitrite and H2O2 or with the MPO-H2O2-nitrite system. Exchange reactions were monitored as described for Fig. 5. For kinetics analysis, the data points were the average values and standard deviations of at least three experiments. The percentage of apoA-I exchanged off rHDL was calculated by taking the intensity of the fluorescence emission of AEDANS at λmax at each time point relative to the average intensity of AEDANS fluorescence emission at λmax at equilibrium (72 h) from at least three independent experiments. The solid lines are the fitting of the experimental data by single exponential for unmodified and ONOO−-oxidized apoA-I and by bi-exponential for MPO-oxidized apoA-I. The exponential relaxation times (τ) are indicated. Inset, fitting of data points up to the exchange equilibrium for MPO-oxidized apoA-I (48–72 h).
Previous studies have implicated specific Tyr and Met residues in the loss of LCAT and ABCA1 activity in MPO oxidized apoA-I (64, 65). To derive potential mechanisms for the difference in τ values of oxidized apoA-I, we used liquid chromatography-electrospray ionization-MS analysis (supplemental Figs. S5 and S6) to determine the product yields of methionine sulfoxide [Met+16] and 3-nitrotyrosine residues [Tyr+45] in apoA-I:ΔW exposed to ONOO− or the MPO-H2O2-nitrite system. At a 25:1 molar ratio of ONOO− to apoA-I:ΔW, 20% of Met-86, 65% of Met-112, and 40% of Met-148 were oxidized. Except for Tyr-192, the other six Tyr residues were nitrated by ONOO− with product yields of 15–30% (supplemental Fig. S6).
The yields of methionine sulfoxide and 3-nitrotyrosine were significantly lower when we exposed apoA-I:ΔW to the MPO-H2O2-nitrite system supplemented with a 10:1 molar excess of H2O2 (H2O2/apoA-I:ΔW, mol/mol). Thus, the overall product yields for methionine sulfoxide (6% of Met-86, 23% of Met-112, and 2% of Met-148) and 3-nitrotyrosine (5–15%) were approximately one-third and one-half of those observed with the ONOO− system, respectively (supplemental Fig. S6).
These observations suggest that the ability of apoA-I to exchange with HDL is impaired by MPO-dependent apoA-I modifications that are distinct from Met or Tyr oxidation. SDS-PAGE analysis of MPO-oxidized samples showed an array of apoA-I oligomers together with significant cleavage of apoA-I (supplemental Fig. S1A). Thus, oligomeric apoA-I and/or a truncated form of the protein produced by MPO oxidation may be responsible for the observed slow exchange kinetic component.
DISCUSSION
A central element of lipid metabolism is the exchange of apolipoproteins between lipoprotein particles. Lipid-poor apolipoproteins have been proposed as intermediates in this process (66). However, an understanding of how apolipoproteins exchange between or off lipoproteins remains elusive because of the difficulties in discriminating between lipid-free and lipid-associated apolipoproteins in a relevant time frame. The steady-state measurements of HDL proteins and lipids mostly relied on ultracentrifugation to separate the various HDL subclasses. Because ultracentrifugation techniques require at least 3 h and apoA-I exchange is at near equilibrium in 5 h, as demonstrated in this study, ultracentrifugation is not suitable for measuring the rate of apoA-I exchange. Ultracentrifugation also subjects HDL particles to a high concentration of chaotropic salts, which may alter the distribution of apolipoproteins. Similar time and resolution limitations exist for other techniques, such as size exclusion chromatography, sequential immunoprecipitation (67), and proton nuclear magnetic resonance (68).
Our FRET-based approach provides a solution to overcome this problem. Here we demonstrated that the fluorescent apoA-I variant, apoA-I:W19-AED136, shows distinct fluorescence properties in different lipidation states (Table 2 and Fig. 4). The positions of donor and acceptor fluorophores were based on the observation that apoA-I undergoes significant structural changes upon lipidation.
In the absence of lipids, apoA-I can assume a compact four-helix bundle (69–71). Upon lipidation, the amphipathic α-helices substitute protein-protein contact for protein-lipid interaction. The partitioning of the hydrophobic face of amphipathic α-helices into the lipid phase corresponds with an opening of the helical bundle into an extended belt-like α-helix, which wraps around the perimeter of the nascent HDL particle (72, 73).
On the smallest observed lipid-associated rHDL particle (7.8-nm diameter), apoA-I has been predicted to assume a compact structure resulting from the small surface area of the lipid component, imparting a saddle-like shape to the particle (74). This compact particle geometry positions the N-terminal and central domains proximally. In agreement with this prediction, apoA-I:W19-AED136 exhibited high levels of FRET when on 7.8-nm rHDL.
Increases in the rHDL lipid:protein ratio have been shown to expand the surface area of the lipid portion of the rHDL, transforming the bilayer into a more planar configuration and driving N-terminal and central domains of apoA-I to more distal positions (75). The degree of observed FRET decreased as the lipid to protein ratios increased, consistent with the progressive unfurling of the apoA-I molecule and the attendant separation of N-terminal and central domains. As a result 9.6-nm rHDL (apoA-I:W19-AED136) exhibited very little FRET in comparison with 7.8-nm rHDL.
Upon displacement of apoA-I:W19-AED136 from 7.8-nm rHDL, by an excess of unlabeled lipid-free apoA-I, there was a reduction of FRET (Fig. 5A, red line versus magenta line). This is likely the result of a loss of tertiary contact because any apoA-I released from rHDL would likely associate with the excess of unlabeled lipid-free apoA-I, whereas at t = 0, the two apoA-I molecules on 7.8-nm rHDL are both apoA-I:W19-AED136.
This fluorescent approach to monitoring the apoA-I lipidation state has allowed us to measure the kinetic parameters governing the transition of apoA-I from the lipid-associated to the lipid-free state. This kinetic analysis revealed that the exchange of rHDL-associated apoA-I with a pool of lipid-free apoA-I can reach equilibrium within 5 h.
Although apolipoprotein exchange can describe the movement of an apolipoprotein from one lipoprotein particle to another, in the case of apoA-I, exchange also implies spontaneous and reversible adsorption and desorption of lipid-free apoA-I on and off an HDL particle. Interestingly, Liang et al. (18, 19) found that unless LCAT was present, no change in HDL size, chemical composition, or level of incorporation of exogenous lipid-free apoA-I (0.5:1 mol:mol; lipid-free A-I:HDL A-I) was observed for 7.6-nm spherical rHDL or 7.9-nm discoidal rHDL incubated at 37 °C (24 h). Consistent with these results, we observed no significant size changes for 7.8-nm rHDL upon incubation at 37 °C (up to 7 days), independent of the presence of a 5-fold molar excess of lipid free apoA-I. However, despite the apparent stability of the 7.8-nm rHDL subclass, 100% of apoA-I exchange occurred in ∼5 h.
Reijngoud and Phillips (76) also observed partial incorporation of exogenous lipid-free apoA-I into rHDL upon incubation at 24 °C for 24 h of rHDL and radioactively labeled lipid-free apoA-I at a 1:1 molar ratio. Recently, Lund-Katz et al. (77) confirmed by surface plasmon resonance that the lipid-free apoA-I can bind to plasma-purified spherical HDL but with low affinity (Kd ≈ 8 μm). Quantitative differences between our results and these previous reports may be due to the significantly higher molar ratio of lipid-free apoA-I used in our system versus the one employed by Reijngoud and Phillips (5:1 versus 1:1 mol:mol; lipid-free A-I:HDL A-I) or the higher incubation temperature (37 °C versus 24 °C). In fact, we observed that the rate of apoA-I exchange at 24 °C was ∼50% that at 37 °C.
Mehta et al. demonstrated that chemical denaturation of HDL is a kinetically controlled process. This encompasses two phases: the fast phase (τ ≈ 0.55 h) comprises rapid protein unfolding, partial dissociation of apoA-I, and HDL particle fusion, and the slow phase (τ ≈ 5.5 h) involves further protein dissociation and particle rupture (78). Although apoA-I has been shown to exist in a meta-stable state, the dissociation of protein from HDL was observed so far only during enzymatic (17–22, 79), chemical, or physical (76, 78, 80–82) perturbations. These conditions led to extensive remodeling of HDL particles.
Here, we were able to demonstrate that apoA-I spontaneously exchanges between rHDL-associated and lipid-free protein pools through the addition of excess lipid-free apoA-I under physiological conditions. Protein exchange occurred in the absence of particle disruption, in contrast to chemical, physical, or enzymatic approaches.
Relevance of ApoA-I Exchange to HDL Function and Metabolism
Cholesterol efflux from cholesterol-laden macrophages is dependent upon two main factors: 1) the presentation of cholesterol by an energy-dependent process (ABCA1) and 2) the availability of lipoprotein recipients/carriers of cholesterol (83). In the arterial wall, desorption of lipid-free/lipid-poor apoA-I from HDL is the likely source of the cholesterol acceptor required to initiate de novo ABCA1-mediated cholesterol efflux from macrophages. Because the availability of lipid-free/lipid-poor apoA-I is likely a limiting factor in cholesterol mobilization (15, 84), the relative rate of apoA-I exchange off rHDL may be a measure of HDL functionality and of cholesterol efflux efficiency. Furthermore, the requirement for lipid-free/lipid-poor apoA-I by ABCA1-mediated cholesterol efflux may be met via apoA-I displacement from HDL by apolipoproteins with greater lipid affinity. That the N-terminal portion of apoE3, a protein fragment with low lipid affinity, cannot displace rHDL-associated apoA-I validates the conclusion that the observed rate of spontaneous apolipoprotein exchange with apoA-I on rHDL is a function of the relative lipid affinity of that apolipoprotein.
When the conformational flexibility of the two apoA-I molecules on rHDL was impaired by intermolecular cross-linking, the modified form of apoA-I was unable to desorb from the particles. This suggests that processes that lead to apoA-I cross-linking in vivo, such as oxidation and carbonyl stress (39–41, 65), might impair lipoprotein function by limiting the ability of apoA-I to adopt different conformations.
Similarly, MPO, a heme protein expressed by macrophages in human atherosclerotic lesions (85) may affect the conformational adaptability of apoA-I. Elevated levels of chlorotyrosine and nitrotyrosine, two characteristic products of MPO, have been detected in HDL from human atherosclerotic lesions (42, 43, 86, 87). Moreover, when MPO is expressed in macrophages, atherosclerosis increases in LDL receptor-deficient mice (88), raising the possibility that the heme enzyme promotes atherosclerosis by targeting apoA-I for oxidative damage.
Oxidation of apoA-I by MPO impairs its ability to activate ABCA1 and LCAT, key steps in reverse cholesterol transport and HDL maturation (64, 65, 87). We found that MPO-mediated oxidation also severely limited the ability of apoA-I to exchange between the HDL-associated and lipid-free/lipid-poor states. This in turn could limit the availability of lipid-free/lipid-poor apoA-I to promote ABCA1-mediated cholesterol mobilization.
ABCA1 and NADPH oxidase, an important source of the peroxide used by MPO, are located in the plasma membrane of macrophages, consistent with the hypothesis that pericellular oxidation of apoA-I and HDL could be important for producing macrophage foam cells in the human arterial wall (89). Furthermore, our in vitro studies used apoA-I nitrated to a level similar to that detected in the artery wall (42), supporting the physiological relevance of our observations.
A New Analytical Approach to Test HDL Functionality
As we gain a better understanding of the anti-atherosclerotic nature of HDL, it is becoming evident that a greater focus should be placed on HDL functionality (4, 90–94). Through the presented fluorescence-based approach to monitor the apoA-I lipidation state, we have furthered our understanding of factors that influence the rate of apoA-I exchange. Because lipid-free apoA-I in the arterial wall may be the primary cholesterol acceptor in cholesterol efflux from macrophages, a measure of apoA-I exchange rate in patients could provide insight into the antiatherosclerotic quality of their HDL. Thus, apoA-I exchange rates combined with plasma HDL-cholesterol levels may serve as a more accurate predictor of risk for cardiovascular disease.
Supplementary Material
Acknowledgments
We thank Dr. Taichi Yamamoto for providing the Trp-null apo-E3 and Dr. Trudy M. Forte for plasma-purified apoA-I and useful suggestions. We also thank Dr. Shobini Jayaraman for useful insights on fluorescence data interpretation and Dr. Mariana F. Tioni for generous suggestions about kinetic analysis.
This work was supported, in whole or in part, by National Institutes of Health Grants HL077268, HL086708, and HL030086. This work was also supported by Tobacco-related Disease Research Program of California Grant 16FT-0163.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
- apoA-I
- apolipoprotein A-I
- ABCA-1
- ATP-binding cassette transporter A-1
- BS3
- ((bis)sulfosuccinimidyl) suberate
- FRET
- fluorescence resonance energy transfer
- HDL
- high density lipoproteins
- I-AEDANS
- N-iodoacetyl-N′-(5-sulfo-1-napthyl)ethylenediamine
- LCAT
- lechitin:cholesterol acyltransferase
- MPO
- myeloperoxidase
- NDGGE
- nondenaturing gradient gel electrophoresis
- rHDL
- reconstituted high density lipoproteins
- MS
- mass spectrometry.
REFERENCES
- 1.Pászty C., Maeda N., Verstuyft J., Rubin E. M. (1994) J. Clin. Invest. 94, 899–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Plump A. S., Scott C. J., Breslow J. L. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 9607–9611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kawashiri M. A., Zhang Y., Puré E., Rader D. J. (2002) Atherosclerosis 165, 15–22 [DOI] [PubMed] [Google Scholar]
- 4.Rader D. J. (2006) J. Clin. Invest. 116, 3090–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marcel Y. L., Ouimet M., Wang M. D. (2008) Curr. Opin. Lipidol. 19, 455–461 [DOI] [PubMed] [Google Scholar]
- 6.Jessup W., Gelissen I. C., Gaus K., Kritharides L. (2006) Curr. Opin. Lipidol. 17, 247–257 [DOI] [PubMed] [Google Scholar]
- 7.Cuchel M., Rader D. J. (2006) Circulation 113, 2548–2555 [DOI] [PubMed] [Google Scholar]
- 8.Rye K. A., Barter P. J. (2004) Arterioscler. Thromb. Vasc. Biol. 24, 421–428 [DOI] [PubMed] [Google Scholar]
- 9.Duong P. T., Weibel G. L., Lund-Katz S., Rothblat G. H., Phillips M. C. (2008) J. Lipid Res. 49, 1006–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X., Rader D. J. (2007) Curr. Opin. Cardiol. 22, 368–372 [DOI] [PubMed] [Google Scholar]
- 11.Van Eck M., Singaraja R. R., Ye D., Hildebrand R. B., James E. R., Hayden M. R., Van Berkel T. J. (2006) Arterioscler. Thromb. Vasc. Biol. 26, 929–934 [DOI] [PubMed] [Google Scholar]
- 12.Adorni M. P., Zimetti F., Billheimer J. T., Wang N., Rader D. J., Phillips M. C., Rothblat G. H. (2007) J. Lipid Res. 48, 2453–2462 [DOI] [PubMed] [Google Scholar]
- 13.Wang N., Silver D. L., Costet P., Tall A. R. (2000) J. Biol. Chem. 275, 33053–33058 [DOI] [PubMed] [Google Scholar]
- 14.Mulya A., Lee J. Y., Gebre A. K., Thomas M. J., Colvin P. L., Parks J. S. (2007) Arterioscler. Thromb. Vasc. Biol. 27, 1828–1836 [DOI] [PubMed] [Google Scholar]
- 15.Sacks F. M., Rudel L. L., Conner A., Akeefe H., Kostner G., Baki T., Rothblat G., de la Llera-Moya M., Asztalos B., Perlman T., Zheng C., Alaupovic P., Maltais J. A., Brewer H. B. (2009) J. Lipid Res. 50, 894–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rye K. A., Clay M. A., Barter P. J. (1999) Atherosclerosis 145, 227–238 [DOI] [PubMed] [Google Scholar]
- 17.Rye K. A., Hime N. J., Barter P. J. (1997) J. Biol. Chem. 272, 3953–3960 [DOI] [PubMed] [Google Scholar]
- 18.Liang H. Q., Rye K. A., Barter P. J. (1995) Biochim. Biophys. Acta 1257, 31–37 [DOI] [PubMed] [Google Scholar]
- 19.Liang H. Q., Rye K. A., Barter P. J. (1996) J. Lipid Res. 37, 1962–1970 [PubMed] [Google Scholar]
- 20.Lusa S., Jauhiainen M., Metso J., Somerharju P., Ehnholm C. (1996) Biochem. J. 313, 275–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ryan R. O., Yokoyama S., Liu H., Czarnecka H., Oikawa K., Kay C. M. (1992) Biochemistry 31, 4509–4514 [DOI] [PubMed] [Google Scholar]
- 22.Clay M. A., Newnham H. H., Forte T. M., Barter P. I. (1992) Biochim. Biophys. Acta 1124, 52–58 [DOI] [PubMed] [Google Scholar]
- 23.Acton S., Rigotti A., Landschulz K. T., Xu S., Hobbs H. H., Krieger M. (1996) Science 271, 518–520 [DOI] [PubMed] [Google Scholar]
- 24.Cavigiolio G., Shao B., Geier E. G., Ren G., Heinecke J. W., Oda M. N. (2008) Biochemistry 47, 4770–4779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malmendier C. L., Delcroix C., Ameryckx J. P. (1983) Clin. Chim. Acta 131, 201–210 [DOI] [PubMed] [Google Scholar]
- 26.Lakowicz J. R. (2006) Principles of Fluorescence Spectroscopy, 3rd Ed., pp. 367–394, Springer, New York [Google Scholar]
- 27.Förster T. (1965) in Modern Quantum Chemistry (Sinanoglu O. ed) Academic Press Inc., New York [Google Scholar]
- 28.Stryer L. (1978) Annu. Rev. Biochem. 47, 819–846 [DOI] [PubMed] [Google Scholar]
- 29.Schobel U., Egelhaaf H. J., Brecht A., Oelkrug D., Gauglitz G. (1999) Bioconjug Chem. 10, 1107–1114 [DOI] [PubMed] [Google Scholar]
- 30.Lillo M. P., Szpikowska B. K., Mas M. T., Sutin J. D., Beechem J. M. (1997) Biochemistry 36, 11273–11281 [DOI] [PubMed] [Google Scholar]
- 31.Selvin P. R. (2000) Nat. Struct. Biol. 7, 730–734 [DOI] [PubMed] [Google Scholar]
- 32.Jares-Erijman E. A., Jovin T. M. (2003) Nat. Biotechnol. 21, 1387–1395 [DOI] [PubMed] [Google Scholar]
- 33.Miyawaki A., Sawano A., Kogure T. (2003) Nat. Cell Biol. (suppl.) S1–S7 [PubMed] [Google Scholar]
- 34.Heinecke J. W. (2003) Am. J. Cardiol. 91, 12A–16A [DOI] [PubMed] [Google Scholar]
- 35.Bernhard D., Csordas A., Henderson B., Rossmann A., Kind M., Wick G. (2005) FASEB J. 19, 1096–1107 [DOI] [PubMed] [Google Scholar]
- 36.Ambrose J. A., Barua R. S. (2004) J. Am. Coll Cardiol. 43, 1731–1737 [DOI] [PubMed] [Google Scholar]
- 37.Bielicki J. K., McCall M. R., van den Berg J. J., Kuypers F. A., Forte T. M. (1995) J. Lipid Res. 36, 322–331 [PubMed] [Google Scholar]
- 38.Wang Y., Rosen H., Madtes D. K., Shao B., Martin T. R., Heinecke J. W., Fu X. (2007) J. Biol. Chem. 282, 31826–31834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng L., Nukuna B., Brennan M. L., Sun M., Goormastic M., Settle M., Schmitt D., Fu X., Thomson L., Fox P. L., Ischiropoulos H., Smith J. D., Kinter M., Hazen S. L. (2004) J. Clin. Invest. 114, 529–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Artola R. L., Conde C. B., Bagatolli L., Pécora R. P., Fidelio G. D., Kivatinitz S. C. (1997) Biochem. Biophys. Res. Commun. 239, 570–574 [DOI] [PubMed] [Google Scholar]
- 41.Malle E., Marsche G., Panzenboeck U., Sattler W. (2006) Arch. Biochem. Biophys. 445, 245–255 [DOI] [PubMed] [Google Scholar]
- 42.Zheng L., Settle M., Brubaker G., Schmitt D., Hazen S. L., Smith J. D., Kinter M. (2005) J. Biol. Chem. 280, 38–47 [DOI] [PubMed] [Google Scholar]
- 43.Pennathur S., Bergt C., Shao B., Byun J., Kassim S. Y., Singh P., Green P. S., McDonald T. O., Brunzell J., Chait A., Oram J. F., O'brien K., Geary R. L., Heinecke J. W. (2004) J. Biol. Chem. 279, 42977–42983 [DOI] [PubMed] [Google Scholar]
- 44.Ansell B. J., Fonarow G. C., Fogelman A. M. (2007) Curr. Opin. Lipidol. 18, 427–434 [DOI] [PubMed] [Google Scholar]
- 45.Oda M. N., Bielicki J. K., Berger T., Forte T. M. (2001) Biochemistry 40, 1710–1718 [DOI] [PubMed] [Google Scholar]
- 46.Ryan R. O., Forte T. M., Oda M. N. (2003) Protein Expr. Purif. 27, 98–103 [DOI] [PubMed] [Google Scholar]
- 47.Chetty P. S., Phillips M. C., Mayne L., Lund-Katz S., Stranz D., Englander W. S. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 19005–19010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lagerstedt J. O., Budamagunta M. S., Oda M. N., Voss J. C. (2007) J. Biol. Chem. 282, 9143–9149 [DOI] [PubMed] [Google Scholar]
- 49.Brouillette C. G., Dong W. J., Yang Z. W., Ray M. J., Protasevich, Cheung H. C., Engler J. A. (2005) Biochemistry 44, 16413–16425 [DOI] [PubMed] [Google Scholar]
- 50.Peng D. Q., Brubaker G., Wu Z., Zheng L., Willard B., Kinter M., Hazen S. L., Smith J. D. (2008) Arterioscler. Thromb. Vasc. Biol. 28, 2063–2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martin D. D., Budamagunta M. S., Ryan R. O., Voss J. C., Oda M. N. (2006) J. Biol. Chem. 281, 20418–20426 [DOI] [PubMed] [Google Scholar]
- 52.Phu M. J., Hawbecker S. K., Narayanaswami V. (2005) J. Neurosci. Res. 80, 877–886 [DOI] [PubMed] [Google Scholar]
- 53.Fisher C. A., Ryan R. O. (1999) J. Lipid Res. 40, 93–99 [PubMed] [Google Scholar]
- 54.Yamamoto T., Lamoureux J., Ryan R. O. (2006) J. Lipid Res. 47, 1091–1096 [DOI] [PubMed] [Google Scholar]
- 55.Fisher C. A., Narayanaswami V., Ryan R. O. (2000) J. Biol. Chem. 275, 33601–33606 [DOI] [PubMed] [Google Scholar]
- 56.Forte T. M., Bielicki J. K., Knoff L., McCall M. R. (1996) J. Lipid Res. 37, 1076–1085 [PubMed] [Google Scholar]
- 57.Shao B., Bergt C., Fu X., Green P., Voss J. C., Oda M. N., Oram J. F., Heinecke J. W. (2005) J. Biol. Chem. 280, 5983–5993 [DOI] [PubMed] [Google Scholar]
- 58.Beckman J. S., Chen J., Ischiropoulos H., Crow J. P. (1994) Methods Enzymol. 233, 229–240 [DOI] [PubMed] [Google Scholar]
- 59.Shao B., Heinecke J. W. (2008) Methods Enzymol. 440, 33–63 [DOI] [PubMed] [Google Scholar]
- 60.Maiorano J. N., Jandacek R. J., Horace E. M., Davidson W. S. (2004) Biochemistry 43, 11717–11726 [DOI] [PubMed] [Google Scholar]
- 61.Massey J. B., Gotto A. M., Jr., Pownall H. J. (1981) Biochem. Biophys. Res. Commun. 99, 466–474 [DOI] [PubMed] [Google Scholar]
- 62.De Pauw M., Vanloo B., Weisgraber K., Rosseneu M. (1995) Biochemistry 34, 10953–10966 [DOI] [PubMed] [Google Scholar]
- 63.Gaut J. P., Byun J., Tran H. D., Lauber W. M., Carroll J. A., Hotchkiss R. S., Belaaouaj A., Heinecke J. W. (2002) J. Clin. Invest. 109, 1311–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shao B., Cavigiolio G., Brot N., Oda M. N., Heinecke J. W. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 12224–12229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shao B., Oda M. N., Bergt C., Fu X., Green P. S., Brot N., Oram J. F., Heinecke J. W. (2006) J. Biol. Chem. 281, 9001–9004 [DOI] [PubMed] [Google Scholar]
- 66.Gursky O. (2005) Curr. Opin. Lipidol. 16, 287–294 [DOI] [PubMed] [Google Scholar]
- 67.Dallinga-Thie G. M., Schneijderberg V. L., van Tol A. (1986) J. Lipid Res. 27, 1035–1043 [PubMed] [Google Scholar]
- 68.Petersen M., Dyrby M., Toubro S., Engelsen S. B., Nørgaard L., Pedersen H. T., Dyerberg J. (2005) Clin. Chem. 51, 1457–1461 [DOI] [PubMed] [Google Scholar]
- 69.Roberts L. M., Ray M. J., Shih T. W., Hayden E., Reader M. M., Brouillette C. G. (1997) Biochemistry 36, 7615–7624 [DOI] [PubMed] [Google Scholar]
- 70.Rogers D. P., Roberts L. M., Lebowitz J., Datta G., Anantharamaiah G. M., Engler J. A., Brouillette C. G. (1998) Biochemistry 37, 11714–11725 [DOI] [PubMed] [Google Scholar]
- 71.Silva R. A., Hilliard G. M., Fang J., Macha S., Davidson W. S. (2005) Biochemistry 44, 2759–2769 [DOI] [PubMed] [Google Scholar]
- 72.Borhani D. W., Rogers D. P., Engler J. A., Brouillette C. G. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 12291–12296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Silva R. A., Hilliard G. M., Li L., Segrest J. P., Davidson W. S. (2005) Biochemistry 44, 8600–8607 [DOI] [PubMed] [Google Scholar]
- 74.Miyazaki M., Nakano M., Fukuda M., Handa T. (2009) Biochemistry 48, 7756–7763 [DOI] [PubMed] [Google Scholar]
- 75.Gu F., Jones M. K., Chen J., Patterson J. C., Catte A., Jerome W. G., Li L., Segrest J. P. (2010) J. Biol. Chem. 285, 4652–4665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Reijngoud D. J., Phillips M. C. (1982) Biochemistry 21, 2969–2976 [DOI] [PubMed] [Google Scholar]
- 77.Lund-Katz S., Nguyen D., Dhanasekaran P., Kono M., Nickel M., Saito H., Phillips M. C. (2010) J. Lipid Res. 51, 606–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mehta R., Gantz D. L., Gursky O. (2003) J. Mol. Biol. 328, 183–192 [DOI] [PubMed] [Google Scholar]
- 79.Gillard B. K., Courtney H. S., Massey J. B., Pownall H. J. (2007) Biochemistry 46, 12968–12978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sparks D. L., Lund-Katz S., Phillips M. C. (1992) J. Biol. Chem. 267, 25839–25847 [PubMed] [Google Scholar]
- 81.Pownall H. J. (2005) Biochemistry 44, 9714–9722 [DOI] [PubMed] [Google Scholar]
- 82.Pownall H. J., Hosken B. D., Gillard B. K., Higgins C. L., Lin H. Y., Massey J. B. (2007) Biochemistry 46, 7449–7459 [DOI] [PubMed] [Google Scholar]
- 83.Curtiss L. K., Valenta D. T., Hime N. J., Rye K. A. (2006) Arterioscler. Thromb. Vasc. Biol. 26, 12–19 [DOI] [PubMed] [Google Scholar]
- 84.Okuhira K., Tsujita M., Yamauchi Y., Abe-Dohmae S., Kato K., Handa T., Yokoyama S. (2004) J. Lipid Res. 45, 645–652 [DOI] [PubMed] [Google Scholar]
- 85.Daugherty A., Dunn J. L., Rateri D. L., Heinecke J. W. (1994) J. Clin. Invest. 94, 437–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Leeuwenburgh C., Rasmussen J. E., Hsu F. F., Mueller D. M., Pennathur S., Heinecke J. W. (1997) J. Biol. Chem. 272, 3520–3526 [DOI] [PubMed] [Google Scholar]
- 87.Bergt C., Pennathur S., Fu X., Byun J., O'Brien K., McDonald T. O., Singh P., Anantharamaiah G. M., Chait A., Brunzell J., Geary R. L., Oram J. F., Heinecke J. W. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 13032–13037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.McMillen T. S., Heinecke J. W., LeBoeuf R. C. (2005) Circulation 111, 2798–2804 [DOI] [PubMed] [Google Scholar]
- 89.Shao B., Oda M. N., Oram J. F., Heinecke J. W. (2010) Chem. Res. Toxicol. 23, 447–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Barter P. J., Caulfield M., Eriksson M., Grundy S. M., Kastelein J. J., Komajda M., Lopez-Sendon J., Mosca L., Tardif J. C., Waters D. D., Shear C. L., Revkin J. H., Buhr K. A., Fisher M. R., Tall A. R., Brewer B. (2007) N. Engl. J. Med. 357, 2109–2122 [DOI] [PubMed] [Google Scholar]
- 91.Nissen S. E., Tardif J. C., Nicholls S. J., Revkin J. H., Shear C. L., Duggan W. T., Ruzyllo W., Bachinsky W. B., Lasala G. P., Tuzcu E. M. (2007) N. Engl. J. Med. 356, 1304–1316 [DOI] [PubMed] [Google Scholar]
- 92.Pennathur S., Heinecke J. W. (2007) Curr. Diab. Rep. 7, 257–264 [DOI] [PubMed] [Google Scholar]
- 93.Navab M., Yu R., Gharavi N., Huang W., Ezra N., Lotfizadeh A., Anantharamaiah G. M., Alipour N., Van Lenten B. J., Reddy S. T., Marelli D. (2007) Curr. Atheroscler Rep. 9, 244–248 [DOI] [PubMed] [Google Scholar]
- 94.Kontush A., Chapman M. J. (2006) Pharmacol. Rev. 58, 342–374 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.