Abstract
PARP-1 is involved in multiple cellular processes, including transcription, DNA repair, and apoptosis. PARP-1 attaches ADP-ribose units to target proteins, including itself as a post-translational modification that can change the biochemical properties of target proteins and mediate recruitment of proteins to sites of poly(ADP-ribose) synthesis. Independent of its catalytic activity, PARP-1 binds to chromatin and promotes compaction affecting RNA polymerase II transcription. PARP-1 has a modular structure composed of six independent domains. Two homologous zinc fingers, Zn1 and Zn2, form the DNA-binding module. Zn1-Zn2 binding to DNA breaks triggers catalytic activity. Recently, we have identified a third zinc binding domain in PARP-1, the Zn3 domain, which is essential for DNA-dependent PARP-1 activity. The crystal structure of the Zn3 domain revealed a novel zinc-ribbon fold and a homodimeric Zn3 structure that formed in the crystal lattice. Structure-guided mutagenesis was used here to investigate the roles of these two features of the Zn3 domain. Our results indicate that the zinc-ribbon fold of the Zn3 domain mediates an interdomain contact crucial to assembly of the DNA-activated conformation of PARP-1. In contrast, residues located at the Zn3 dimer interface are not required for DNA-dependent activation but rather make important contributions to the chromatin compaction activity of PARP-1. Thus, the Zn3 domain has dual roles in regulating the functions of PARP-1.
Keywords: Chromatin Regulation, DNA-binding Protein, DNA Repair, Protein Structure, Transcription Regulation, PARP-1, Poly(ADP-ribose) Polymerases
Introduction
Poly(ADP-ribose) polymerase-1 (PARP-1)4 is a multifunctional enzyme that covalently attaches ADP-ribose to target proteins (1, 2). Using NAD+ as a donor, PARP-1 can polymerize multiple units of ADP-ribose to create long and branched polymers termed poly(ADP-ribose) (PAR). This post-translational modification can change the biochemical properties and cellular location of acceptor proteins. The main substrate of PARP-1 in the cell is PARP-1 itself (automodification activity), although PARP-1 also ADP-ribosylates various substrates, including core histones, histone H1, and transcription regulatory proteins (3–7). PARP-1 can bind to nucleosomes and induce chromatin compaction, an activity that is reversed by automodification of PARP-1 (8, 9). Ecdysone- and heat shock-inducible “puffing” (i.e. chromatin decondensation) of specific loci in Drosophila requires PARP-1 enzymatic activity and is accompanied by the accumulation of PAR in the puffs (10). In vivo PARP-1 localizes to actively transcribed RNA polymerase II promoters (11).
PARP-1 also plays important roles in DNA repair. Following DNA damage, PARP-1 is recruited to sites of DNA lesion by its ability to bind DNA strand breaks. Binding to DNA strand breaks stimulates the automodification activity of PARP-1 and could lead to recruitment of DNA repair machinery to the sites of damage through an interaction with XRCC1 (12, 13). More recent results support an alternative model whereby PARP-1 acts to protect DNA single strand breaks from being converted to more deleterious double strand breaks, until they can be repaired by the base excision repair machinery (14, 15). Importantly, PAR created through PARP-1 automodification serves as a ligand that can recruit PAR-binding factors, such as histone variants and chromatin-remodeling enzymes (16–18). Thus, PARP-1 acts as an early responder to DNA damage that can influence the kinetics of DNA repair by recruiting repair factors and imparting local changes in chromatin structure. PARP-1 involvement in DNA repair has made it a promising drug target in the treatment of cancer (19, 20). Inhibition of PARP-1 by small interfering RNA or the use of small molecule inhibitors specifically kills cells with defects in the breast cancer susceptibility genes BRCA1 or BRCA2 (21, 22).
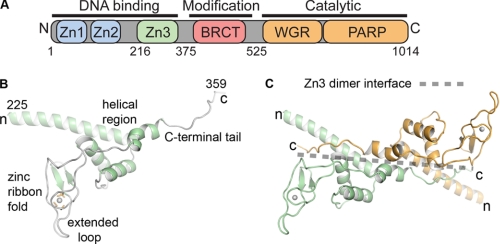
PARP-1 is a modular protein of 114 kDa (human) containing multiple independent domains that perform distinct functions (Fig. 1A). Two homologous zinc fingers, Zn1 and Zn2, at the extreme N terminus of PARP-1 are responsible for binding to DNA strand breaks, which dramatically stimulates PARP-1 activity (23, 24). An internal automodification domain contains a BRCA1 C-terminal fold involved in mediating protein-protein interactions and three lysines that are targeted for automodification (13, 25, 26). The catalytic domain (CAT) is located at the extreme C terminus of PARP-1. DNA binding studies using a Zn1-Zn2 domain fragment indicate that PARP-1 binds to DNA as a dimer (27), and kinetic analysis of PARP-1 activity indicates that PARP-1 is a catalytic dimer (28). Therefore, dimerization of PARP-1 molecules is thought to be a critical step in DNA-dependent PARP-1 activation (29). It is not established how other PARP-1 domains are involved in DNA-dependent activation.
FIGURE 1.
PARP-1 has a modular structure composed of six domains. A, schematic representation of PARP-1 domain structure. B, ribbon representation of the x-ray structure of the Zn3 domain (30). C, crystallographic dimer of the Zn3 domain. One Zn3 monomer is drawn in green, and the second monomer is drawn in orange. The transparent dotted line highlights the Zn3 dimer interface.
We identified and determined the crystal structure of the novel Zn3 zinc binding domain located between the Zn1 and Zn2 zinc fingers and the automodification domain of PARP-1 (Fig. 1, A and B) (30). The Zn3 domain is critically required for DNA-dependent PARP-1 activity (30, 31). The Zn3 domain structure consists of a unique type of zinc-ribbon fold and an α-helical N-terminal region (Fig. 1B). Importantly, the structure is entirely unrelated to the Zn1 and Zn2 domain structures and is therefore expected to have a distinct function. Zinc-ribbon folds frequently mediate protein-protein interactions (32); therefore, this is a likely role for the zinc-ribbon fold of the Zn3 domain. In the crystal structure, the C-terminal tail of the Zn3 domain forms an extensive interface between two Zn3 monomers that are related by 2-fold crystallographic symmetry (Fig. 1C). Although the isolated Zn3 domain is a monomer in solution, conserved residues are involved in forming the Zn3 homodimer interface. This led us to suggest that the Zn3 domain might exist as a dimer in the DNA-activated state of PARP-1. Current models indicate that PARP-1 functions as a catalytic dimer; therefore, it was speculated that the Zn3 dimer might participate in assembling two molecules of PARP-1 for activation. Notably, a recent NMR study reported a monomeric structure of the Zn3 domain in which the C-terminal tail interacts with the N-terminal helical region (31), rather than forming the homodimer contacts observed in the x-ray structure. It is therefore possible that the Zn3 domain can adopt two conformational states, which might have functional relevance in the context of full-length PARP-1.
To better define the role of the PARP-1 Zn3 domain, we have created 23 structure-guided mutations in both the zinc-ribbon fold of the Zn3 domain and the region forming the Zn3 dimer interface in the crystal structure. Biochemical analysis of the PARP-1 mutants indicates that the zinc-ribbon fold of the Zn3 domain is essential for DNA-dependent activity of PARP-1, but it is not required for basal PARP-1 activity in the absence of DNA. Point mutations located on one face of an extended loop of the zinc-ribbon fold drastically affect the catalytic turnover of the DNA-dependent PAR synthesis reaction (kcat), without altering the affinity for NAD+ (Km) or the DNA binding affinity (KD). We suggest that the zinc-ribbon fold of the Zn3 domain is essential for the assembly of an activated enzyme upon PARP-1 binding to DNA. In contrast to mutation of the zinc-ribbon fold, mutation of residues located at the crystallographic Zn3 dimer interface does not affect the catalytic activity of PARP-1, indicating that this interface does not play a role in activating PARP-1. Interestingly, a number of these Zn3 mutations are deficient in their ability to compact chromatin structure, revealing that the Zn3 domain contributes to the ability of PARP-1 to modulate chromatin structure. Thus, two distinct functions for the Zn3 of PARP-1 have been identified and mapped to two different structural regions of the Zn3 domain.
EXPERIMENTAL PROCEDURES
Gene Cloning and Mutagenesis
Full-length wild-type (WT) PARP-1 (residues 1–1014) and mutants and PARP-1 ΔZn3 (deletion of residues 215–374) were cloned in the pET28 expression vector with an N-terminal hexahistidine tag. The WT PARP-1 Zn3 domain (residues 216–366) and the W318R mutant Zn3 domain constructs were cloned in the pET24 expression vector with a C-terminal hexahistidine tag as described previously (30). All mutations and deletions were performed using the QuickChange Protocol (Stratagene) and verified by automated sequencing (Kimmel Cancer Center).
Protein Expression and Purification
PARP-1 proteins were expressed in Escherichia coli strain BL21(DE3)Rosetta2 (Novagen). Cells were grown in LB media containing 100 μm ZnSO4 and induced with 200 μm isopropyl 1-thio-β-d-galactopyranoside at 16 °C for 20 h. Cells were resuspended in 20 mm Hepes, pH 8.0, 500 mm NaCl, 0.5 mm Tris(2-carboxyethyl)phosphine (TCEP), 0.1% Nonidet P-40, 1 mm phenylmethylsulfonyl fluoride (and other protease inhibitors) and lysed using a cell disrupter (Avestin). Cell debris was pelleted for 2 h at 40,000 × g, and the supernatant was then filtered and loaded onto a 5-ml HP-chelating column (GE Healthcare) charged with Ni(II) and preequilibrated in lysis buffer without Nonidet P-40. The column was washed in three consecutive steps with lysis buffer containing the following: 1) 20 mm imidazole, 500 mm NaCl; 2) 20 mm imidazole, 1 m NaCl; and 3) 20 mm imidazole, 500 mm NaCl. Proteins were eluted with lysis buffer containing 400 mm imidazole (500 mm NaCl), diluted to the appropriate salt concentration (see below), and then loaded onto a 5-ml HP heparin column (GE Healthcare). For full-length WT PARP-1 and mutants and ΔZn3 PARP-1, the heparin column was equilibrated with 50 mm Tris-HCl, pH 7.0, 250 mm NaCl, 0.1 mm TCEP, and 1 mm EDTA, and eluted with a gradient from 250 mm to 1 m NaCl. For WT and W318R Zn3 domains, the heparin column was equilibrated with 50 mm NaCl, and the elution gradient was from 50 to 500 mm NaCl. Full-length WT and most mutants (see below for exceptions), ΔZn3 PARP-1, and WT and W318R Zn3 domains were next passed over a Sephacryl S200 gel filtration column (GE Healthcare) in 20 mm Hepes, pH 8.0, 150 mm NaCl, 0.1 mm TCEP, and 0.1 mm EDTA. In the case of mutants Q241L, F357Y, L348D/V350D, and P358G/P359G, proteins were dialyzed in gel filtration buffer after elution from the heparin column.
DNA-dependent Automodification Assay (Nonradioactive)
PARP-1 automodification reactions were performed essentially as described previously (30). WT PARP-1 and mutants (0.62 μm) were first preincubated with 1 μm duplex DNA for 10 min at room temperature (22 °C). 5 mm NAD+ was then added to the reaction, and the mixture was incubated for various times. In the complementation experiments, ΔZn3 PARP-1 (0.62 μm), the Zn3 domain (0.62 μm), full-length E988A (0.31 μm), and W318R (0.31 μm) were preincubated for 10 min at room temperature prior to DNA addition. NAD+ was then added, and the reactions were allowed to proceed for 1- and 10-min time points. In each experiment, reactions were stopped by the addition of SDS-loading buffer containing 0.1 m EDTA. The samples were resolved on SDS-PAGE (gel percentages listed in figure legends) and stained with Imperial protein stain (Pierce).
DNA-dependent Automodification Assay (Radioactive)
WT PARP-1 and mutants (0.5 μm) were preincubated with 1 μm DNA for 10 min at room temperature. 0.2 μm radiolabeled 32P-NAD+ (0.16 μCi) or 0.2 μm unlabeled NAD+ was added for the indicated time points. Reactions were stopped by the addition of SDS-loading buffer containing 0.1 m EDTA. The samples were resolved on 12% SDS-PAGE. The gel was either treated with Imperial protein stain (Pierce) when unlabeled NAD+ was used or exposed on a phosphorimager screen and read on a Typhoon scanner (GE Healthcare) when radiolabeled NAD+ was used.
Fluorescence Polarization DNA Binding Assay
An 18-nucleotide DNA strand (5′-GGGTTGCGGCCGCTTGGG-3′) was annealed to a complementary 18-nucleotide DNA strand that carried a fluorescein derivative (6-carboxyfluorescein) on the 5′ terminus. The binding reactions were performed in 12 mm Hepes, pH 8.0, 60 mm KCl, 0.12 mm EDTA, 5.5 μm β-mercaptoethanol, 8 mm MgCl2, 0.05 mg/ml BSA, and 4% glycerol. Reactions contained 5 nm DNA probe and various concentrations of protein. Reactions were incubated at room temperature for 30 min. Fluorescence polarization data were collected on a Victor3V plate reader (PerkinElmer Life Sciences). The observed binding constant was obtained from a nonlinear least squares fit to the data using a two-state binding model (SigmaPlot).
Colorimetric PARP-1 Automodification Assay
The kinetics of PARP-1 automodification were analyzed using a colorimetric assay that measures the incorporation of biotinylated NAD+ (bNAD+, 6-biotin-17-NAD) into polymers of ADP-ribose (PAR). bNAD+ serves as a substrate for PARP-1 (33); therefore, the amount of biotin in PAR is proportional to the amount of PAR synthesized. All steps of the assay were performed at 22 °C. Histidine-tagged PARP-1 proteins were immobilized on Ni2+-chelating plates (5 PRIME) in a 50-μl reaction volume containing 18 mm Hepes, pH 8.0, 150 mm NaCl, 0.5 mm TCEP, and 10 μg/ml BSA. 20 nm PARP-1 was used in DNA-dependent reactions (40 nm of 18-bp duplex), and 60 nm PARP-1 was used in DNA-independent reactions. Reactions were initiated by the addition of NAD+ at various concentrations (15–1000 μm). The ratio of NAD+ to bNAD+ was 99:1; therefore, 1000 μm NAD+ is 990 μm NAD+ and 10 μm bNAD+ (bNAD+ from Trevigen). Reactions were quenched by the addition of 150 μl of 6 m guanidine hydrochloride (4.5 m final concentration). The reaction wells were washed three times with 200 μl of phosphate-buffered saline (PBS) containing 1% BSA (PBS/BSA). Streptavidin-conjugated horseradish peroxidase (Pierce) was added to each reaction for a 30-min incubation (1:20,000 dilution or 1:12,000 dilution in PBS/BSA, for DNA-dependent and DNA-independent, respectively). Reaction wells were then washed three times with PBS/BSA. 75 μl of the streptavidin-conjugated horseradish peroxidase substrate Ultra-TMB (Pierce) was added to each reaction well, and this reaction was quenched with 75 μl of 2 n sulfuric acid. The absorbance at 450 nm (A450) was read on a Victor3V (PerkinElmer Life Sciences); the absorbance at 550 nm (A550) was subtracted from the A450 reading to account for nonuniform variations in the plastic plates. A biotinylated, histidine-tagged peptide (biotin-Ser-Trp-His-His-His-His-His-His-His-His) of known concentration was immobilized on the Ni2+ plate and served as a standard for converting the A450 reading into a quantity of biotin. The amount of ADP-ribose was then estimated as 100 times the amount of biotin, following the 99:1 ratio of NAD+ to bNAD+. A background A450 reading was performed for each NAD+ concentration in the absence of PARP-1 to account for nonspecific binding of bNAD+ and streptavidin-conjugated horseradish peroxidase to the reaction well. Time points in the linear portion of the reaction profile were measured for each NAD+ concentration to provide initial rates. Initial rates were plotted versus total NAD+ concentration (supplemental Fig. S1), and fitting the Michaelis-Menten model to the data yielded Km (μm) and Vmax values (μmol/min/mg). Vmax was divided by the molecular weight of PARP-1 (115,000 g/mol) to calculate kcat (s−1). The kinetic parameters presented in Table 1 represent the average of three independent experiments.
TABLE 1.
Kinetic analysis of the PARP-1 automodification reaction
Kinetic parameters were obtained by fitting the Michaelis-Menten model to data obtained from a PARP-1 automodification assay that determines the rate of PAR synthesis by measuring the amount of biotinylated-NAD+ incorporated into PAR. The values represent the average of three independent experiments and their standard deviation.
| Km | Vmax | kcat | |
|---|---|---|---|
| μm | (μmol·min−1·mg−1) | s−1 | |
| DNA-dependent PARP-1 automodification activity | |||
| Wild type | 86.5 ± 39.0 | 2.58 ± 0.84 | 4.95 ± 1.61 |
| T316A | 79.1 ± 25.8 | 0.011 ± 0.0037 | 0.022 ± 0.007 |
| W318R | 31.4 ± 17.5 | 0.0018 ± 0.0011 | 0.0035 ± 0.0022 |
| DNA-independent PARP-1 automodification activity | |||
| Wild type | 109.5 ± 31.3 | 0.00148 ± 0.00028 | 0.00284 ± 0.00054 |
| T316A | 51.7 ± 13.3 | 0.000879 ± 0.00018 | 0.00169 ± 0.00034 |
| W318R | 94.8 ± 51.0 | 0.000552 ± 0.00038 | 0.00106 ± 0.00073 |
Assembly of Chromatin Templates
Chromatin assembly was performed using recombinant ACF, as described previously (8, 34). An ∼3.2-kb DNA plasmid (pERE, a plasmid containing four tandem estrogen response elements upstream of the adenovirus E4 core promoter (35)) was assembled into chromatin using purified native Drosophila core histones, Acf-1, ISWI, and human NAP-1, under conditions described previously (8). The assembly reaction was incubated for 2.5 h at 27 °C.
MNase Digestion Assays
ACF-assembled chromatin (30-μl aliquots containing 150 ng of DNA) was incubated without or with PARP-1 or PARP-1 mutant (33, 66, or 132 nm) for 30 min at 27 °C. We then added 5 μl of MNase (5 milliunits; United States Biochemical Corp.) and incubated the samples for 10 min at room temperature. The samples were deproteinized with proteinase K and phenol/chloroform extraction. The deproteinized samples were run on a 1.3% agarose gel and visualized by staining with ethidium bromide. All MNase assays were run a minimum of three times to ensure reproducibility.
In Vitro Transcription
In vitro transcription assays with ACF-assembled chromatin were performed as described previously (8). The chromatin (15-μl aliquots containing 75 ng of DNA) was incubated with or without 40 nm WT PARP-1 or mutant, 300 μm ATP, and 300 μm NAD+ as indicated for 30 min at 27 °C. Next, 10 nm estrogen receptor α (ERα) and 100 nm 17β-estradiol were added as indicated, followed by a 20-min incubation at room temperature. These samples were preincubated for 15 min with HeLa cell nuclear extract as a source of the polymerase II transcriptional machinery and incubated for 30 min at 30 °C after addition of rNTPs to allow transcription to occur. The synthesized RNA was analyzed using primer extension, followed by separation on an 8% urea-polyacrylamide gel with autoradiographic analysis. Quantification was performed using a PhosphorImager (GE Healthcare) with ImageQuant software. All reactions were performed in duplicate, and at least three independent experiments were performed to ensure reproducibility.
RESULTS
Residues Trp-318 and Thr-316 Are Critically Required for DNA-dependent PARP-1 Activation
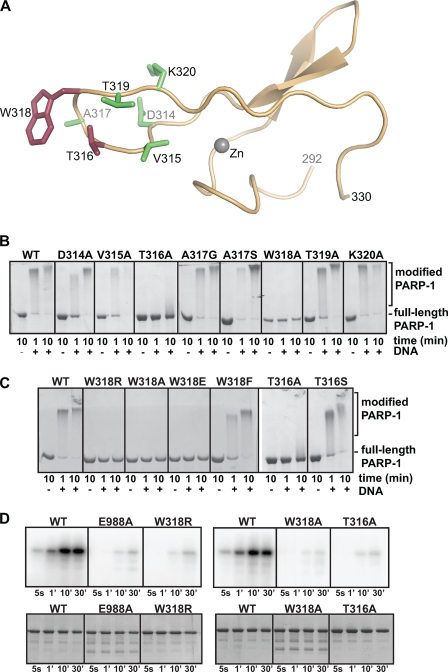
Structure-based mutagenesis was used to study the role of the Zn3 domain of PARP-1. Two defining regions of the Zn3 domain revealed by the x-ray crystal structure were the focus of site-directed mutagenesis experiments of full-length PARP-1, the zinc ribbon fold and the Zn3 dimer interface. The first region of interest (residue 290 to 332) forms a zinc-binding motif reminiscent of a zinc-ribbon fold (Fig. 2A) (30, 31). Zinc-ribbon folds are often involved in mediating protein-protein interactions (32). A unique aspect of the Zn3 zinc-ribbon fold is a large loop that extends from the Zn3 domain structure (amino acid residues 314–320, Fig. 2A; Fig. 1B, “extended loop”).
FIGURE 2.
The zinc-ribbon fold of the Zn3 domain is essential for PARP-1 DNA-dependent activity. A, x-ray structure of the zinc-ribbon fold of the PARP-1 Zn3 domain. Residues mutated in B–D are drawn as sticks. Mutation of residues colored red led to a defect in DNA-dependent activity, although mutation of residues colored green had no affect on DNA-dependent activity. B and C, DNA-dependent automodification activity of WT PARP-1 or mutants (0.62 μm) with 1 μm duplex DNA and 5 mm NAD+. The indicated time points were analyzed by 12% SDS-PAGE. D, radioactive automodification assay. Top, WT PARP-1 and mutants (0.5 μm) were incubated with 1 μm duplex DNA and radiolabeled NAD+ (0.2 μm). Phosphorimage analysis of a 12% SDS-PAGE is shown. Bottom, same experiment was performed in the presence of unlabeled NAD+ as a control for the amount of the enzymes used in the reaction. The SDS-PAGE was treated with Imperial protein stain.
We have previously suggested that the Zn3 domain is important for mediating an interdomain contact that is critical for PARP-1 DNA-dependent activity (30). This was based on the fact that a fragment, including the first three zinc fingers (Zn1-Zn2-Zn3; residues 1–366), could restore DNA-dependent activity of a BRCA1 C-terminal WGR-CAT fragment of PARP-1 (residues 379–1014), whereas a fragment lacking the Zn3 domain could not (Zn1-Zn2; residues 1–234), despite having a robust affinity for DNA. Thus, deletion of the Zn3 domain dramatically compromises PARP-1 activity without compromising DNA binding affinity (30, 31). Previous mutagenesis experiments targeting the Zn3 domain are likely to have disrupted the fold of the Zn3 domain (30, 36) and therefore are comparable with a deletion mutant, which does not provide insights into the importance of particular regions of the Zn3 structure. Here, mutagenesis was focused on the extended loop of the Zn3 domain (residues 314–320) that is formed of well conserved surface-exposed residues that could likely be involved in domain-domain interactions important for PARP-1 activity but are not likely to be important for the fold and stability of the Zn3 domain. Single-point alanine mutants of full-length PARP-1 were created for each residue of the extended loop from Asp-314 to Lys-320; residue Ala-317 was substituted with a glycine and a serine (Fig. 2A).
Each of the mutants was first tested in a DNA-dependent automodification assay (Fig. 2B). In this assay, PARP-1 automodification was assessed by monitoring a shift in mobility on SDS-PAGE upon PAR addition to the PARP-1 polypeptide. The majority of the single-point mutants of the extended loop showed an activity level comparable with WT PARP-1. However, the T316A and the W318A mutants were severely deficient in automodification activity as compared with WT PARP-1. A slight shift in mobility was visible after 10 min of incubation for the T316A mutant, although the W318A mutant had no detectable activity in this automodification assay. Additional substitutions at Thr-316 and Trp-318 indicated that only conservative mutations were tolerated at these two positions on the extended loop (Fig. 2C). For example, substitution of Trp-318 with a charged group (Arg or Glu) inactivated PARP-1 automodification. In contrast, the conservative mutations W318F and T316S showed only slight deficiency compared with WT PARP-1. Importantly, the defects seen in the Thr-316 and Trp-318 mutants were not due to disruption of PARP-1 binding to DNA. A fluorescence polarization DNA binding assay showed that WT PARP-1 and each of the zinc-ribbon fold mutants bind to an 18-bp DNA fragment with similar affinities (supplemental Fig. S2). This is the expected result because all mutations are located distant from the Zn1 and Zn2 domains that mediate DNA binding. Analytical gel filtration confirmed an identical elution profile for WT and W318R PARP-1 suggesting that the mutations did not affect the global conformation of the protein (supplemental Fig. S3). Furthermore, CD spectra analysis of WT and W318R PARP-1 showed no change in the secondary structure, indicating that the overall fold of the protein is preserved (supplemental Fig. S4). Instead, the mutations are expected to have disrupted a specific domain-domain contact between the extended loop of the zinc-ribbon fold and an as yet unidentified region of PARP-1. The two residues that are critical for DNA-dependent activation are both located on the same face of the extended loop, suggesting that the domain-domain contact occurs primarily on one side of the extended loop.
The mutations at position Thr-316 and Trp-318 were further examined to identify which stage of the PARP-1 reaction is deficient. Synthesis of poly(ADP-ribose) consists of three unique chemical steps as follows: (i) initiation on protein side chains; (ii) linear extension of the polymer; and (iii) branched extension of the polymer. The automodification assay above only identifies mutations that prevent formation of long chains of PAR. The mutants T316A and W318R might still be able to initiate PAR synthesis and/or create short chains of PAR. To determine whether PARP-1 that is mutated at position Thr-316 or Trp-318 remains efficient at the initiation step and/or synthesis of short PAR chains, an automodification assay was performed using radiolabeled 32P-NAD+. In this automodification assay, mono-ADP-ribose or short chains of PAR that do not alter PARP-1 electrophoretic mobility could be detected (Fig. 2D). The zinc-ribbon fold mutants W318R, W318A, and T316A all had very weak activity in this assay compared with WT PARP-1. The level of activity of the three zinc-ribbon fold mutants was comparable with that of PARP-1 E988A (Fig. 2D), a mutation located near the NAD+-binding site of the CAT domain that is altered in both its initiation and elongation activities (37). Therefore, we conclude that the Zn3 zinc-ribbon mutants are altered in their ability to catalyze the first steps of the PAR synthesis reaction. Moreover, targeting the Zn3 domain can inhibit PARP-1 activity to a similar level as targeting the CAT domain, suggesting a novel strategy for PARP-1 inhibition.
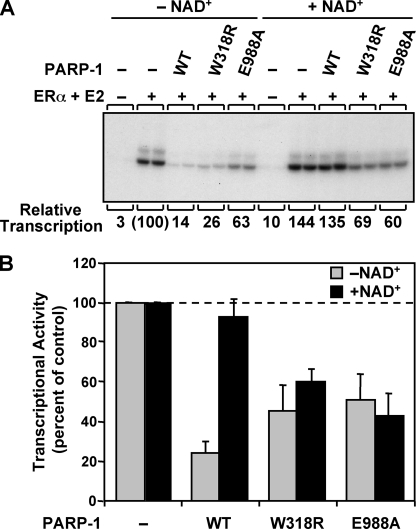
We next explored the relationship between the ability of the zinc ribbon mutants to synthesize PAR and their ability to derepress transcription. We have previously shown that WT PARP-1 can repress transcription with chromatin templates in vitro and that addition of NAD+ reverses the repressive effects (8, 9). We tested the W318R mutant for its ability to derepress transcription in the presence of NAD+ in an ERα-regulated in vitro transcription assay with chromatin templates. ACF-assembled chromatin containing a DNA template with four tandem estrogen response elements upstream of the adenoviral E4 promoter was transcribed in the presence of ERα, its cognate ligand 17β-estradiol, WT and mutant PARP-1 proteins, and HeLa cell nuclear extract as a source of the RNA polymerase II transcription machinery (Fig. 3). As expected, WT PARP-1 repressed ERα-dependent transcription, and the repression was reversed upon the addition of NAD+. The W318R mutant also repressed transcription, but reversal of this repression was not observed in the presence of NAD+, a result similar to that observed with the catalytic active site mutant E988A (Fig. 3) and E988K (9). Taken together, these data indicate that NAD+-dependent transcriptional derepression requires PARP-1 automodification, and the Zn3 domain is an essential contributor to PARP-1 activity in the context of chromatin.
FIGURE 3.
The zinc-ribbon fold of the Zn3 domain of PARP-1 is necessary for NAD+-dependent transcriptional derepression. Results of ERα-dependent in vitro transcription assays with chromatin templates are shown. The pERE plasmid DNA template was assembled into chromatin using ACF and subjected to in vitro transcription under the conditions shown. A, representative gel image of the in vitro transcription assay with WT, W318R, and E988A PARP-1 proteins in the presence or absence of NAD+. E2, 17β-estradiol, B, quantification of three independent in vitro transcription assays. Each bar represents the mean ± S.E.
The Zinc-ribbon Fold Is Required for Efficient Assembly of the DNA-activated Form of PARP-1
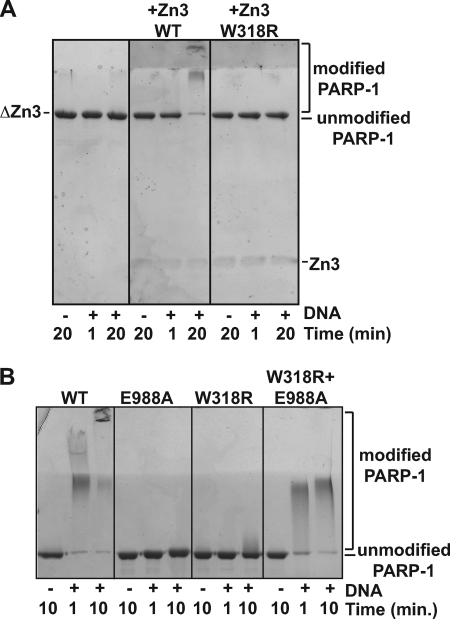
We next tested whether the zinc-ribbon fold is important for mediating contacts between the Zn3 domain and other domains of PARP-1 using a complementation assay with a PARP-1 deletion mutant lacking the Zn3 domain (ΔZn3). As expected, the ΔZn3 mutant is inactive in the automodification assay (Fig. 4A, left panel). Addition of the isolated Zn3 domain in trans restores automodification activity (Fig. 4A, middle panel), indicating that the Zn3 domain interacts with the deletion mutant to help establish the catalytically active form of PARP-1. This result reinforces the view that PARP-1 is an enzyme with several independent modules that can be added separately to create a functional enzyme. In support of this notion, two inactive mutants, W318R and E988A, can restore PARP-1 automodification activity when combined (Fig. 4B). In this case, the Zn3 domain of E988A must communicate with the catalytic domain of W318R to restore activity. This result suggests that the automodification occurs in trans, with the catalytic domain of W318R modifying E988A; however, both PARP-1 proteins are shifted in this reaction indicating that the active catalytic domain of W318R can also modify itself in cis. Importantly, an isolated Zn3 domain bearing the W318R mutation was not able to restore ΔZn3 activity (Fig. 4A, right panel), suggesting that the extended loop of the zinc-ribbon fold is necessary for the Zn3 interdomain contact. All attempts to detect an interaction between the isolated Zn3 domain and any other PARP-1 domain using affinity pulldown experiments were unsuccessful (with or without DNA) (data not shown). We concluded that the interaction is either too weak for detection in our assays or that it only occurs in a robust manner in the context of intact, full-length PARP-1 in the presence of DNA and potentially NAD+.
FIGURE 4.
Zn3 domain added in trans restores activity of a PARP-1 ΔZn3 deletion mutant. A, DNA-dependent automodification activity of PARP-1 ΔZn3 (0.62 μm) is restored by the addition of the WT Zn3 domain (0.62 μm, but not by the W318R mutant of the Zn3 domain (0.62 μm). 1 μm duplex DNA and 5 mm NAD+ were used. The indicated time points were analyzed by SDS-PAGE (15%) stained with Imperial protein stain. B, DNA-dependent automodification activity of WT PARP-1 (0.62 μm), E988A, and W318R mutants (0.62 μm in individual experiments, 0.31 μm each in combination experiment) on 7.5% SDS-PAGE.
Rather than mediating a robust interdomain contact, the extended loop could be crucial for catalytic activity per se, for example by contributing residues to the CAT domain active site to help shape or alter the NAD+-binding pocket. To test this possibility, we developed a catalytic assay that measures the affinity (Km) of WT PARP-1 and mutants for NAD+ (see “Experimental Procedures”) and can therefore identify whether Zn3 domain mutants contribute to the CAT domain active site. Time course experiments at multiple NAD+ concentrations yielded the Vmax, Km, and kcat values for WT PARP-1 and key PARP-1 mutants in the presence and absence of DNA (Table 1). It is noteworthy that the DNA-independent activity of PARP-1 is extremely weak and requires long time points to detect appreciable amounts of PAR (1–5 h). In contrast, DNA-dependent activity is robust for WT PARP-1, resulting in measurable PAR formation within seconds. The inherently weak DNA-independent activity results in a higher error in fitting the kinetic data (supplemental Fig. S1B). However, the measurements are reproducible and the changes in kinetic parameters highlighted below are several orders of magnitude larger than the experimental variability.
For WT PARP-1, the presence of DNA stimulated PARP-1 turnover (kcat) by over 1700-fold with little affect on the Km value for NAD+. In the presence of DNA, the mutants W318R and T316A showed a substantial reduction in their kcat value compared with WT PARP-1 (over 1000- and 200-fold, respectively), but no alteration of their Km value for NAD+. Notably, the W318R mutant has weak PAR synthesis in the presence of DNA that is comparable with its level of DNA-independent activity. In the absence of DNA, the kcat and Km values of the two zinc-ribbon fold mutants were similar to WT PARP-1. Therefore, we conclude that the zinc-ribbon fold plays an essential role in the DNA-dependent activity of PARP-1 but not in the basal catalytic activity that occurs in the absence of DNA. Because mutations of the extended loop of the zinc-ribbon fold have not altered the Km value for NAD+, we conclude that these residues do not contribute important contacts to the NAD+-binding site. We propose that the extended loop of the Zn3 zinc-ribbon fold is involved in an interdomain contact that occurs following DNA binding to form a fully activated conformation of PARP-1. Importantly, we have identified a new class of mutants located outside the conserved catalytic active site and the Zn1/Zn2 domain that is essential for DNA-dependent activity. The extended loop of the Zn3 domain might therefore be a potential target for the design of PARP-1-specific inhibitors, because the Zn3 domain is not found in any of the other PARP family members.
The Crystallographic Zn3 Dimer Is Not Relevant to DNA-dependent PARP-1 Activation
Structure-based mutagenesis was next focused on the region identified in the crystal structure as the Zn3 dimer interface. The Zn3 dimer interface is formed by conserved residues and buries over 2000 Å2 of accessible surface area on each monomer. Two Zn3 monomers are organized such that the N-terminal helix of one molecule is juxtaposed to the C-terminal tail of the second molecule (Fig. 5A). In the context of full-length PARP-1, the Zn3 dimer might therefore bring the DNA binding domain of one PARP-1 molecule in close proximity to the CAT domain of a second PARP-1 molecule (30). To determine the potential functional relevance of the crystallographic Zn3 dimer, we investigated whether the isolated Zn3 domain exists as a dimer in solution. Sedimentation equilibrium data and gel filtration analysis indicate that the Zn3 domain exists as a monomer in solution (data not shown). We reasoned that the Zn3 dimer observed in the crystal structure might only form in solution when PARP-1 binds to DNA as a dimer (27), whereby the high local concentration of PARP-1 molecules could drive the dimerization of two Zn3 domains. Therefore, the Zn3 dimer could serve as a DNA-dependent interaction that organizes the activated conformational state of PARP-1.
FIGURE 5.
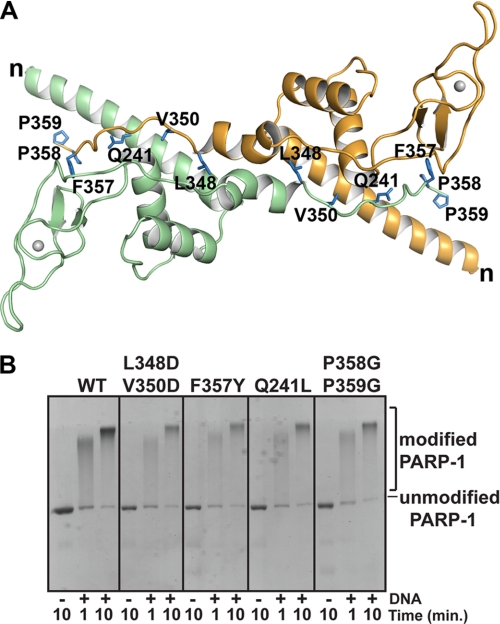
Mutations at the Zn3 dimer interface do not disrupt PARP-1 DNA-dependent automodification activity. A, ribbon representation of the Zn3 dimer with mutated residues drawn as blue sticks and labeled on each monomer. The N terminus is labeled n, and the C terminus is residue Pro-359. B, DNA-dependent automodification activity of WT PARP-1 and Zn3 dimer mutants (0.62 μm) with 1 μm duplex DNA and 5 mm NAD+. See supplemental Fig. S5 for additional Zn3 dimer interface mutants.
Eleven residues involved in contacts at the crystallographic Zn3 dimer interface were targeted for mutagenesis. These residues were substituted with amino acids that are incompatible with the interactions that form the Zn3 dimer. Each PARP-1 mutant was as efficient as WT PARP-1 in the DNA-dependent automodification assay (Fig. 5B and supplemental Fig. S5). On the basis of these results, we conclude that the Zn3 dimer observed in the crystal structure is not important for DNA-dependent activation of PARP-1.
The Zn3 Domain Makes Important Contributions to PARP-1 Compaction of Chromatin
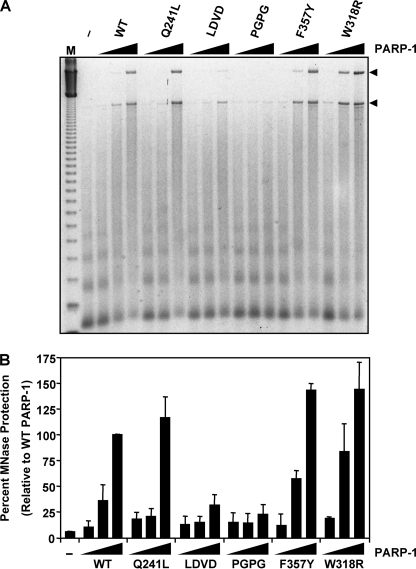
The Zn3 domain mutations at the dimer interface had no affect on DNA-dependent PARP-1 catalytic activity. Fluorescence polarization DNA binding experiments further demonstrated that these Zn3 domain mutants bind to duplex DNA as efficiently as WT PARP-1 (supplemental Fig. S2). Collectively, these results indicate that these mutants have no deficiencies in the DNA-binding zinc fingers, the catalytic domain, or the ability to form the DNA-activated conformation of PARP-1. We sought to determine whether these Zn3 mutations might reveal other roles for the Zn3 domain important for PARP-1 function. PARP-1 was previously shown to compact chromatin in an MNase protection assay (8, 9). In this assay, ACF-assembled chromatin is subjected to limited MNase digestion, and the extent of digestion is analyzed by agarose gel electrophoresis with ethidium bromide staining (9). The addition of increasing amounts of WT PARP-1 compacts the chromatin and thereby protects the linker DNA from MNase digestion, yielding a higher molecular weight DNA species (Fig. 6A). To determine whether the Zn3 domain is an important contributor to chromatin compaction, we analyzed the effects of Zn3 domain mutations on chromatin compaction using the MNase protection assay. The conservative PARP-1 mutant F357Y and the zinc-ribbon fold mutant W318R compacted chromatin in a manner similar to WT PARP-1. In contrast, mutations within the dimer interface of the Zn3 domain, including the double mutants L348D/V350D and P358G/P359G, were defective in chromatin compaction, as shown by a reduction in their ability to generate the high molecular weight protected DNA species in the MNase assay (Fig. 6, A and B). The point mutant Q241L shows a deficiency in chromatin compaction at the intermediate concentration of PARP-1 used in the assay, but this can be overcome at higher concentrations. The results of this assay indicate that the Zn3 domain is an important element for the chromatin compaction capabilities of PARP-1.
FIGURE 6.
Zn3 dimer interface mutations are deficient in the ability to compact chromatin. MNase protection assay with chromatin templates. The pERE plasmid DNA template was assembled into chromatin using ACF and subjected to MNase digestion under the conditions shown. A, representative gel image of the MNase protection assay with WT PARP-1 and five mutant PARP-1 proteins. Shown is a 2-fold serial dilution for each of the PARP-1 proteins at a final concentration of 33, 66, and 132 nm. The control lane indicates the amount of digestion in the absence of PARP-1 protein. The amount of MNase used in the assay was empirically determined. M, 123-bp molecular weight ladder. B, quantification of three MNase protection assays. Arrowheads in A indicate the MNase-resistant bands that were quantified. Percentages determined are relative to WT PARP-1 at the highest concentration used. Each bar represents the mean ± S.E.
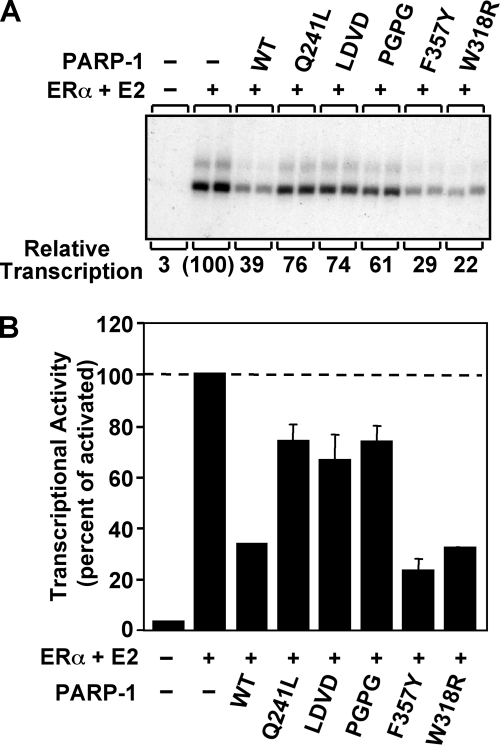
PARP-1 binding to chromatin and compaction of chromatin lead to repression of activator-dependent polymerase II transcription in vitro (Fig. 3 and Fig. 7) (8, 9). The Zn1-Zn2 zinc fingers are the major contributors to PARP-1 binding to chromatin (9), and Zn1-Zn2 binding to chromatin on its own represses transcription to a certain degree, most likely by restricting nucleosome mobility (8). An additional level of repression is mediated through PARP-1 compaction of chromatin structure. The Zn3 domain mutants showed deficiencies in chromatin compaction in the MNase assay; therefore, they were examined for their ability to repress activator-dependent transcription using chromatin templates. The L348D/V350D and P358G/P359G double mutants showed a reduction in their ability to repress transcription compared with WT PARP-1 (Fig. 7), consistent with their chromatin compaction defects. The Q241L mutant also showed a reduction in its ability to repress transcription, consistent with its chromatin compaction deficiencies observed at intermediate concentrations in the MNase assay. A Zn3 domain deletion mutant (ΔZn3) showed a similar level of repression as the Zn3 domain double mutants (data not shown), indicating that these residues constitute major elements of the chromatin compaction contribution made by the Zn3 domain. Each of these mutants shows an intermediate level of transcriptional repression. This is indicative of a PARP-1 mutant that is able to bind to chromatin but unable to compact chromatin as shown previously (8). Therefore, we conclude that these Zn3 domain mutants are able to bind to chromatin and partially repress transcription through their Zn1-Zn2 zinc finger domains. This is consistent with the WT level of activity observed for these mutants in the DNA binding assay (supplemental Fig. S2) and the automodification assay (Fig. 5); both assays require functional Zn1 and Zn2 domains. Thus, the defect in transcriptional repression is due to a deficiency in chromatin compaction activity. In contrast to the double mutants, the PARP-1 mutants F357Y and W318R repressed transcription as effectively as WT PARP-1, consistent with the ability to compact chromatin as assessed by the MNase protection assay (Fig. 6). Collectively, these results indicate that specific residues within the dimer interface of the Zn3 domain are required for PARP-1 regulation of chromatin compaction and activated transcription.
FIGURE 7.
Zn3 mutations in the dimer interface region are unable to fully repress transcription. Results of ERα-dependent in vitro transcription assays with chromatin templates are shown. The pERE plasmid DNA template was assembled into chromatin using ACF and subjected to in vitro transcription under the conditions shown. A, in vitro transcription assay with WT PARP-1 and four Zn3 dimer interface mutants (Q241L, LDVD, PGPG, and F357Y) and one zinc-ribbon fold mutant (W318R). ERα and 17β-estradiol (E2) were used to activate transcription. Quantification of the relative levels of transcription is indicated below the image of the representative gel. B, quantification of three independent in vitro transcription assays. Each bar represents the mean ± S.E.
DISCUSSION
We have identified two functions for the Zn3 domain of PARP-1 and associated them with two distinct structural features of the Zn3 domain. We have demonstrated that the zinc-ribbon fold of the Zn3 domain is essential for DNA-dependent automodification activity (Fig. 2). More precisely, residues Thr-316 and Trp-318 located at the distal end of the extended loop of the zinc-ribbon fold are critical for DNA-dependent activity but are dispensable for basal PAR synthesis activity in the absence of DNA. These mutations in the Zn3 domain zinc-ribbon fold do not affect the Km value for NAD+ and are therefore not expected to make contributions to the NAD+-binding site (Table 1). Instead, these zinc-ribbon fold mutations dramatically reduce the efficiency of PAR synthesis (Table 1; kcat). The catalytic activity of a Zn3 domain deletion of PARP-1 (ΔZn3) is restored when the isolated Zn3 domain is added in trans but not if the Zn3 domain bears a W318R mutation (Fig. 4). These biochemical data indicate a physical interaction between the Zn3 domain and another domain(s) of PARP-1. Thr-316 and Trp-318 are surface-exposed and located on the same face of the extended loop of the zinc-ribbon fold; therefore, we propose that this region of the Zn3 domain mediates a contact essential for PARP-1 activation following DNA binding. Mutations such as T316A and W318R are likely to have weakened the Zn3 interdomain contact and thus lead to an inefficient assembly of the multiple domains of PARP-1 and a dramatic decrease in the efficiency of PAR synthesis. A recent in vitro study using fragments of human PARP-1 suggests that the Zn3 domain mediates an interaction between an N- and C-terminal fragment of PARP-1, consistent with a Zn3 interdomain contact playing a role in organizing PARP-1 domain structure (26).
There are several possible mechanisms by which the Zn3 interdomain contact might regulate PARP-1 activity. The Zn3 domain could bridge an interaction between two PARP-1 domains, bringing them into the appropriate orientation for an association necessary for PAR synthesis. Another possibility is that the Zn3 interdomain contact acts to orient the automodified region of PARP-1 for efficient addition of PAR. Further structural data for PARP-1 domain interactions are required to specifically understand how the Zn3 domain is involved in DNA-dependent activation of PARP-1.
This study has also revealed a class of Zn3 domain mutants that affect the ability of PARP-1 to compact chromatin but have no affect on the PAR synthesis activity of PARP-1. These mutants were designed to disrupt a homodimeric interface observed in the x-ray structure of the Zn3 domain to test its potential role in DNA-dependent PARP-1 activity. However, each of the mutations introduced at the dimer interface demonstrated a level of automodification activity that is comparable with WT PARP-1 (Fig. 5B and supplemental Fig. S5). Thus we have concluded that the Zn3 dimer observed in our crystal structure is not relevant to DNA-dependent PAR synthesis activity. In contrast, the Zn3 dimer interface mutations demonstrated deficiencies in two assays that evaluate the ability of PARP-1 to compact chromatin (Figs. 6 and 7), identifying a new role for the Zn3 domain of PARP-1. Zinc fingers Zn1 and Zn2 are required for PARP-1 to bind to chromatin (9); however, a collaboration of multiple PARP-1 domains is required for PARP-1 to compact chromatin (8). Fluorescence polarization DNA binding experiments demonstrate that the Zn3 domain mutants bind to duplex DNA as efficiently as WT PARP-1 (supplemental Fig. S2), indicating that the DNA binding activity of the Zn1 and Zn2 domains has not been compromised in these mutants, and therefore, the Zn1-Zn2 chromatin binding properties of these Zn3 mutants has not been affected. Indeed, these mutants repress transcription to a moderate level (Fig. 7), which further demonstrates an intact ability to interact with chromatin. Thus the MNase protection and transcriptional repression assays demonstrate that the Zn3 domain is an important element required for chromatin compaction that leads to maximal repression of transcription by PARP-1.
How does the Zn3 domain contribute to chromatin compaction? A potential mechanism is homodimerization of two adjacent nucleosome-bound PARP-1 molecules. Several studies have indicated that the Zn1-Zn2 domains are necessary and sufficient for the nucleosome binding property of PARP-1. The Zn3 dimer observed in the crystal structure is a possible candidate for mediating the homodimerization of PARP-1 on chromatin. The fact that mutation of residues located at the Zn3 dimer interface compromises chromatin compaction is consistent with this hypothesis. However, earlier studies indicated that the Zn1-Zn2-Zn3 domains could not compact chromatin, but when linked to the CAT domain the chromatin compaction capability of PARP-1 was restored (8). These data underscore the possibility that both the Zn3 domain and the CAT domain are intricately associated in the homodimerization of PARP-1. Our model identifies the Zn1-Zn2 domains as necessary for the binding of PARP-1 to chromatin, whereas the Zn3 and CAT domains are collectively required for the dimerization of PARP-1. Studies to examine this model are complicated by the fact that the Zn3 domain does not form detectable dimers in solution, which prevents us from directly measuring the potential dimerization defects of the Zn3 mutants.
Rather than mediating homodimerization of chromatin-bound PARP-1, the Zn3 domain alternatively could be important for positioning other PARP-1 domains for full compaction of chromatin. The DNA binding domain (Zn1-Zn2-Zn3) and the CAT domain of PARP-1 collaborate to induce chromatin compaction (8); therefore, a potential function of the Zn3 domain is to coordinate the activities of the Zn1-Zn2 domains and the CAT domain. The Zn3 domain double mutants L348D/V350D and P358G/P359G each affected the ability of PARP-1 to protect chromatin from MNase digestion and to repress transcription in vitro. These residues are located at the C-terminal tail of the Zn3 domain, which is positioned adjacent to the long N-terminal helix in the dimeric x-ray structure (Fig. 5). The arrangement of the N and C termini of the Zn3 domain could serve to juxtapose the Zn1-Zn2 domains and the CAT domain. The substitution of two prolines with glycines has likely introduced flexibility in the C-terminal tail that could affect the relative orientation of the CAT domain and the Zn1-Zn2 domains. Similarly, the introduction of supplementary negative charges in the case of the L348D/V350D double mutant could perturb the positioning of the C-terminal tail. The two double mutants could therefore be unable to spatially organize the Zn1-Zn2 domains and the CAT domain for compaction of chromatin. The mutation Q241L also showed a deficiency in its ability to compact chromatin in the transcription repression assays. Residue Gln-241 is located on the long N-terminal α-helix of the Zn3 domain, and the side chain of this residue makes direct hydrogen bonding contacts to the main chain of the C-terminal tail. Therefore, the Gln-241 substitution with a nonpolar leucine could disrupt the ability of the Zn1-Zn2 domains and the CAT domain to cooperate to compact chromatin. It is important to emphasize that these potential perturbations in the organization of PARP-1 domains for chromatin compaction have no affect on the DNA-dependent catalytic activity of PARP-1, because each of these mutants is as active as WT PARP-1 in the automodification reaction that requires both DNA binding and catalytic activities.
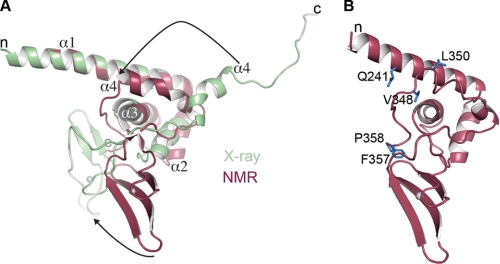
The Zn3 dimer interface mutants map to the N-terminal helical region when viewed in the context of the monomeric NMR structure (31). The crystal structure and the NMR structure of the Zn3 domain are overall very similar (Fig. 8A), but there are two notable differences. The relative orientation of the zinc-ribbon fold and the N-terminal helical region are different in the two structures, with a roughly 60° rotation relating the zinc-ribbon folds when the α-helical regions are aligned (Fig. 8A). There is also a difference in the arrangement of the C terminus in the two Zn3 domain structures. In the NMR structure, the C terminus of the polypeptide folds back onto the main body of the Zn3 domain, contributing to the helical region at the N terminus of the Zn3 domain (Fig. 8, A and B). In contrast, the C terminus in the x-ray structure extends away from the main body of the Zn3 domain to make contacts with a symmetry-related Zn3 domain, forming the crystallographic dimer. The two conformations of the C-terminal tail of the Zn3 domain might represent different functional states of the Zn3 domain (i.e. PARP-1 bound to open chromatin versus PARP-1 bound to compacted chromatin). Despite this uncertainty in the positioning of the C-terminal tail of the Zn3 domain, the mutational analysis has clearly revealed that the Zn3 domain is an important factor contributing to PARP-1 compaction of chromatin.
FIGURE 8.
X-ray and NMR structures of the Zn3 domain demonstrate different positions of the C-terminal tail. A, comparison of the x-ray and NMR models of the Zn3 domain. Structures were superimposed by aligning the first three α-helices of the Zn3 domain. The relative orientation of the zinc-ribbon fold is rotated ∼60° (bottom arrow), and the C terminus of the Zn3 domain is in two different conformations (top arrow). B, Zn3 mutations from Fig. 5 mapped onto the NMR structure.
Our study defines two roles for the Zn3 domain of PARP-1. One role is to contribute to the chromatin compaction activity of PARP-1. We have proposed the following two potential mechanisms for this role of the Zn3 domain: (i) homodimerization of chromatin-bound PARP-1 molecules, or (ii) optimal spatial positioning of the Zn1-Zn2 domains and the CAT domain for maximal chromatin compaction. In the second role, the zinc-ribbon fold of the Zn3 domain is a vital component of DNA-dependent PAR synthesis activity. We propose that the zinc-ribbon fold mediates an interdomain contact that is essential to form the DNA-activated conformation of PARP-1. Importantly, recent clinical trials for the treatment of cancer using PARP-1 inhibitors have shown promising results (38). Current inhibitors target the catalytic domain of PARP-1, which is present in the other PARP family members (39). Because the Zn3 domain is unique to PARP-1, we have identified a particular region and precise residues of the zinc-ribbon fold as potential new targets for the design of more specific PARP-1 inhibitors.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant DK069710 from NIDDK (to W. L. K). This work was also supported by American Cancer Society Grants IRG0806001 and RSG0918301DMC, The Emerald Foundation, Inc. (to J. M. P.), and The Susan G. Komen Breast Cancer Foundation (to D. D. R.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- PARP-1
- poly(ADP-ribose) polymerase-1
- PAR
- poly(ADP-ribose)
- WT
- wild type
- CAT
- catalytic domain
- TCEP
- Tris(2-carboxyethyl)phosphine
- BSA
- bovine serum albumin
- PBS
- phosphate-buffered saline
- MNase
- micrococcal nuclease
- bNAD+
- biotinylated NAD+
- ERα
- estrogen receptor α
- ACF
- ATP-utilizing chromatin assembly and remodeling factor.
REFERENCES
- 1.D'Amours D., Desnoyers S., D'Silva I., Poirier G. G. (1999) Biochem. J. 342, 249–268 [PMC free article] [PubMed] [Google Scholar]
- 2.Hassa P. O., Hottiger M. O. (2008) Front. Biosci. 13, 3046–3082 [DOI] [PubMed] [Google Scholar]
- 3.Poirier G. G., de Murcia G., Jongstra-Bilen J., Niedergang C., Mandel P. (1982) Proc. Natl. Acad. Sci. U.S.A. 79, 3423–3427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huletsky A., de Murcia G., Muller S., Hengartner M., Ménard L., Lamarre D., Poirier G. G. (1989) J. Biol. Chem. 264, 8878–8886 [PubMed] [Google Scholar]
- 5.Kraus W. L. (2008) Curr. Opin. Cell Biol. 20, 294–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu W., Ginjala V., Pant V., Chernukhin I., Whitehead J., Docquier F., Farrar D., Tavoosidana G., Mukhopadhyay R., Kanduri C., Oshimura M., Feinberg A. P., Lobanenkov V., Klenova E., Ohlsson R. (2004) Nat. Genet. 36, 1105–1110 [DOI] [PubMed] [Google Scholar]
- 7.Gamble M. J., Kraus W. L. (2007) Cell 128, 433–434 [DOI] [PubMed] [Google Scholar]
- 8.Wacker D. A., Ruhl D. D., Balagamwala E. H., Hope K. M., Zhang T., Kraus W. L. (2007) Mol. Cell. Biol. 27, 7475–7485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim M. Y., Mauro S., Gévry N., Lis J. T., Kraus W. L. (2004) Cell 119, 803–814 [DOI] [PubMed] [Google Scholar]
- 10.Tulin A., Spradling A. (2003) Science 299, 560–562 [DOI] [PubMed] [Google Scholar]
- 11.Krishnakumar R., Gamble M. J., Frizzell K. M., Berrocal J. G., Kininis M., Kraus W. L. (2008) Science 319, 819–821 [DOI] [PubMed] [Google Scholar]
- 12.Caldecott K. W., Aoufouchi S., Johnson P., Shall S. (1996) Nucleic Acids Res. 24, 4387–4394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masson M., Niedergang C., Schreiber V., Muller S., Menissier-de Murcia J., de Murcia G. (1998) Mol. Cell. Biol. 18, 3563–3571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woodhouse B. C., Dianov G. L. (2008) DNA Repair 7, 1077–1086 [DOI] [PubMed] [Google Scholar]
- 15.Woodhouse B. C., Dianova I. I., Parsons J. L., Dianov G. L. (2008) DNA Repair 7, 932–940 [DOI] [PubMed] [Google Scholar]
- 16.Timinszky G., Till S., Hassa P. O., Hothorn M., Kustatscher G., Nijmeijer B., Colombelli J., Altmeyer M., Stelzer E. H., Scheffzek K., Hottiger M. O., Ladurner A. G. (2009) Nat. Struct. Mol. Biol. 16, 923–929 [DOI] [PubMed] [Google Scholar]
- 17.Ahel D., Horejsí Z., Wiechens N., Polo S. E., Garcia-Wilson E., Ahel I., Flynn H., Skehel M., West S. C., Jackson S. P., Owen-Hughes T., Boulton S. J. (2009) Science 325, 1240–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gottschalk A. J., Timinszky G., Kong S. E., Jin J., Cai Y., Swanson S. K., Washburn M. P., Florens L., Ladurner A. G., Conaway J. W., Conaway R. C. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 13770–13774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Helleday T., Petermann E., Lundin C., Hodgson B., Sharma R. A. (2008) Nat. Rev. Cancer 8, 193–204 [DOI] [PubMed] [Google Scholar]
- 20.Martin S. A., Lord C. J., Ashworth A. (2008) Curr. Opin. Genet. Dev. 18, 80–86 [DOI] [PubMed] [Google Scholar]
- 21.Bryant H. E., Schultz N., Thomas H. D., Parker K. M., Flower D., Lopez E., Kyle S., Meuth M., Curtin N. J., Helleday T. (2005) Nature 434, 913–917 [DOI] [PubMed] [Google Scholar]
- 22.Farmer H., McCabe N., Lord C. J., Tutt A. N., Johnson D. A., Richardson T. B., Santarosa M., Dillon K. J., Hickson I., Knights C., Martin N. M., Jackson S. P., Smith G. C., Ashworth A. (2005) Nature 434, 917–921 [DOI] [PubMed] [Google Scholar]
- 23.Gradwohl G., Ménissier de Murcia J. M., Molinete M., Simonin F., Koken M., Hoeijmakers J. H., de Murcia G. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 2990–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ikejima M., Noguchi S., Yamashita R., Ogura T., Sugimura T., Gill D. M., Miwa M. (1990) J. Biol. Chem. 265, 21907–21913 [PubMed] [Google Scholar]
- 25.Mendoza-Alvarez H., Alvarez-Gonzalez R. (1999) Biochemistry 38, 3948–3953 [DOI] [PubMed] [Google Scholar]
- 26.Altmeyer M., Messner S., Hassa P. O., Fey M., Hottiger M. O. (2009) Nucleic Acids Res. 37, 3723–3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pion E., Ullmann G. M., Amé J. C., Gérard D., de Murcia G., Bombarda E. (2005) Biochemistry 44, 14670–14681 [DOI] [PubMed] [Google Scholar]
- 28.Mendoza-Alvarez H., Alvarez-Gonzalez R. (1993) J. Biol. Chem. 268, 22575–22580 [PubMed] [Google Scholar]
- 29.Bauer P. I., Buki K. G., Hakam A., Kun E. (1990) Biochem. J. 270, 17–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langelier M. F., Servent K. M., Rogers E. E., Pascal J. M. (2008) J. Biol. Chem. 283, 4105–4114 [DOI] [PubMed] [Google Scholar]
- 31.Tao Z., Gao P., Hoffman D. W., Liu H. W. (2008) Biochemistry 47, 5804–5813 [DOI] [PubMed] [Google Scholar]
- 32.Gamsjaeger R., Liew C. K., Loughlin F. E., Crossley M., Mackay J. P. (2007) Trends Biochem. Sci. 32, 63–70 [DOI] [PubMed] [Google Scholar]
- 33.Zhang J. (1997) Methods Enzymol. 280, 255–265 [DOI] [PubMed] [Google Scholar]
- 34.Ito T., Levenstein M. E., Fyodorov D. V., Kutach A. K., Kobayashi R., Kadonaga J. T. (1999) Genes Dev. 13, 1529–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kraus W. L., Kadonaga J. T. (1998) Genes Dev. 12, 331–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trucco C., Flatter E., Fribourg S., de Murcia G., Ménissier-de Murcia J. (1996) FEBS Lett. 399, 313–316 [DOI] [PubMed] [Google Scholar]
- 37.Marsischky G. T., Wilson B. A., Collier R. J. (1995) J. Biol. Chem. 270, 3247–3254 [DOI] [PubMed] [Google Scholar]
- 38.Fong P. C., Boss D. S., Yap T. A., Tutt A., Wu P., Mergui-Roelvink M., Mortimer P., Swaisland H., Lau A., O'Connor M. J., Ashworth A., Carmichael J., Kaye S. B., Schellens J. H., de Bono J. S. (2009) N. Engl. J. Med. 361, 123–134 [DOI] [PubMed] [Google Scholar]
- 39.Rouleau M., Patel A., Hendzel M. J., Kaufmann S. H., Poirier G. G. (2010) Nat. Rev. Cancer 10, 293–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.