Abstract
Kinetoplastid RNA (k-RNA) editing is a complex process in the mitochondria of kinetoplastid protozoa, including Trypanosoma brucei, that involves the guide RNA-directed insertion and deletion of uridines from precursor-mRNAs to produce mature, translatable mRNAs. k-RNA editing is performed by multiprotein complexes called editosomes. Additional non-editosome components termed k-RNA-editing accessory factors affect the extent of editing of specific RNAs or classes of RNAs. The T. brucei p22 protein was identified as one such accessory factor. Here we show that p22 contributes to cell growth in the procyclic form of T. brucei and functions as a cytochrome oxidase subunit II-specific k-RNA-editing accessory factor. To gain insight into its functions, we solved the crystal structure of the T. brucei p22 protein to 2.0-Å resolution. The p22 structure consists of a six-stranded, antiparallel β-sheet flanked by five α-helices. Three p22 subunits combine to form a tight trimer that is primarily stabilized by interactions between helical residues. One side of the trimer is strikingly acidic, while the opposite face is more neutral. Database searches show p22 is structurally similar to human p32, which has a number of functions, including regulation of RNA splicing. p32 interacts with a number of target proteins via its α1 N-terminal helix, which is among the most conserved regions between p22 and p32. Co-immunoprecipitation studies showed that p22 interacts with the editosome and the k-RNA accessory protein, TbRGG2, and α1 of p22 was shown to be important for the p22-TbRGG2 interaction. Thus, these combined studies suggest that p22 mediates its role in k-RNA editing by acting as an adaptor protein.
Keywords: Protein, Protein/Structure, RNA, RNA/Editing, RNA/Modification, RNA/MicroRNA, RNA/Splicing, RNA/Structure
Introduction
The kinetoplastid protozoa, including Trypanosoma brucei, comprise a group of parasitic flagellated organisms that parasitize virtually all animal groups. In humans, T. brucei causes African sleeping sickness (1). T. brucei is transmitted between mammalian hosts by the tse tse fly and undergoes complex changes to facilitate its survival in different hosts (1, 2). In the mammalian bloodstream form, energy generation is dependent on glycolytic reactions that are compartmentalized within specialized organelles of the protozoa termed glycosomes (3). However, the insect or procyclic form (PF)2 does not have the luxury of blood glucose as an abundant energy source and instead contains a highly active and unusual mitochondrion (1, 2). These differentiation events are triggered by changes in specific biological processes. One of these is kinetoplastid RNA (k-RNA) editing, which is a highly complex biological process that is unique to the mitochondria of the kinetoplastid parasites (4–8).
k-RNA editing involves the insertion and deletion of uridine nucleotides into/from precursor-mRNAs to produce mature, translatable mRNAs (9, 10). The accumulation of edited RNAs in T. brucei is developmentally regulated and life cycle-specific (11, 12). For example, RNAs encoding the cytochrome components apocytochrome b (7, 11, 12) and cytochrome oxidase subunit II (COII) (5) are only edited in PF (12), whereas editing of RNAs encoding components of the NADH dehydrogenase complex is highly up-regulated in the bloodstream form. Interestingly, a few RNAs (apocytochrome b, MURF2, and COII) are edited only in small regions near their 3′- or 5′-ends (minimally edited), whereas the remaining nine edited RNAs are edited throughout their lengths (pan-edited).
In the kinetoplastid mitochondria, maxicircle DNA molecules encode the pre-edited mRNAs, whereas smaller mitochondrial DNA molecules, termed minicircles, encode the vast majority of guide RNAs (gRNAs) (13–15). gRNAs are necessary for RNA editing as they contain the sequence information required to direct the editing process (16, 17). These small RNAs, which are typically 50–70 nucleotides long, give rise to >1200 different gRNAs (10). gRNAs are structurally conserved and contain a region at their 5′-end that is complementary with the mRNA sequence just downstream of the editing sites. They also contain a non-encoded poly(U) tail at the 3′-end (13, 18, 19). Several related 20 S editosome complexes or RNA ligase-containing (L-complexes), which perform the insertions and deletions of uridines, contain at least 20 proteins, including gRNA-dependent riboendonucleases that cut the mRNA into fragments at gRNA-mRNA mismatches and a 3′-terminal uridylyl transferase (or KRET2) that adds uridines to the 3′-end of the 5′-mRNA fragment as guided by base-pairing with gRNA (21–31). Additional editosome proteins are exonucleases that remove bulged uridines and RNA ligases that connect RNA fragments (2, 10). Interestingly, the mRNA encoding COII exhibits several unique features with regard to the editing process described above. COII precursor-mRNA is edited by insertion of just four uridine residues (5), and, unlike all other edited RNAs in T. brucei, these insertions are not specified by trans-acting gRNAs. Rather, the COII gRNA is contained within the 3′-end of the COII precursor-mRNA and acts in cis (32). Additionally, of the three editosome subclasses, one is devoted entirely to editing of the COII RNA (24), and this editosome type is defined by the presence of a COII-specific riboendonuclease (KREN3) and adaptor protein (KREPB6).
In addition to the editosome or L-complex, several non-editosome components termed RNA-editing accessory factors regulate the specificity, accuracy, and/or efficiency of editing (33). These include MRP1, MRP2, RBP16, TbRGG1, and TbRGG2 (34–37). The latter two proteins have been reported to associate with the ill-defined MRB1 (or GRBC) complex, several components of which have also been shown to affect RNA editing (38–41). The p22 protein was originally identified as an interaction partner for RBP16 (42), a multifunctional RNA-editing accessory factor (43) that is subject to arginine methylation (44, 45). The p22 open reading frame encodes a 227-residue pre-protein that is transported into the mitochondria via a 46-residue signal peptide and subsequently processed to the mature 181-residue protein (42). The specific roles of p22 in k-RNA editing are still unclear, but its interaction with RBP16 suggests it could be involved in regulation of RNA editing (42). To gain insight into p22 protein structure and function, we carried out cellular and structural studies on the T. brucei p22 protein. Our studies show that p22 is a trimeric protein with an asymmetric surface charge distribution. RNA interference data showing the consequences of p22 depletion on different never-edited, minimally edited, and pan-edited RNAs, define p22 as a COII-specific editing accessory factor. Co-immunoprecipitation experiments demonstrate that p22 interacts with core editosomes and editing TbRGG2, and p22 deletion experiments reveal that its N-terminal helix mediates key contacts with TbRGG2.
EXPERIMENTAL PROCEDURES
Expression and Purification
The sequence encoding the T. brucei mature p22 protein (residues 47–227) was cloned into pET-21a (Novagen) to produce a protein with a C-terminal His6 tag. p22 was expressed in Escherichia coli Rosetta 2 (DE3)/pRARE (Novagen). Cells were grown in LB medium in the presence of 100 μg/ml ampicillin and 34 μg/ml chloramphenicol to an A600 of ∼0.6, and protein expression was induced by the addition of 1 mm isopropyl 1-thio-β-d-galactopyranoside for 4 h. After harvesting, the cell pellet was resuspended in buffer A (20 mm Tris HCl, pH 7.5, 300 mm NaCl, 10 mm imidazole, pH 8) and incubated with protease inhibitor mixture tablets, 10 μg/ml RNase, and 10 μg/ml DNase for 30 min. After cell disruption, the resulting supernatant was purified via nickel-nitrilotriacetic acid column chromatography. Pure p22-containing fractions were pooled and concentrated using an Amicon Ultra 10-kDa Filter. The protein was further purified using size-exclusion chromatography (S200 column, Amersham Biosciences) in buffer C (20 mm Tris HCl, pH 7.5, 150 mm NaCl, 5% glycerol). A single peak was obtained from size-exclusion chromatography that corresponded to a p22 trimer. Selenomethionine-substituted p22 was expressed by using the methionine inhibitory pathway, and the protein was purified as for wild type, with the exception that 10 mm β-mercaptoethanol was added to all buffers (46).
Crystallization and Data Collection of p22
For crystallization, p22 was concentrated to 25 mg/ml using an Amicon Ultra 10-kDa Filter. The protein was crystallized via hanging drop, vapor diffusion using 40% polyethylene glycol 400 and 0.1 m imidazole, pH 8, as a crystallization reagent. The crystals took the space group P63 with cell constants a = b = 82.0 Å, c = 51.9 Å, α = β = 90°, and γ = 120° and diffracted to 2.0-Å resolution. The crystals were cryopreserved straight from the drop. Native x-ray intensity data were collected using an R-AXIS IV imaging plate, and x-rays were generated by a Rigaku RU-300HB x-ray generator fitted with osmic mirrors. Data were processed with MOSFLM (47) as implemented in CCP4 (48). Selenomethionine p22 crystals were grown as for wild type except that 10 mm β-mercaptoethanol was added to the reservoir. Multiple wavelength anomalous diffraction data were collected at Beamline 8.2.1 at the Advanced Light Source (Lawrence Berkeley National Laboratory, Berkeley, CA). Data were processed using MOSFLM (47) as implemented in CCP4 (48). The selenomethionine substituted crystals were isomorphous with the wild type p22 crystals and took the space group P63 with cell constants a = b = 82.1 Å, c = 51.8 Å, α = β = 90°, and γ = 120°. Relevant data statistics are shown in Table 1.
TABLE 1.
Selected crystallographic data and statistics
| Selenomethionine-p22 multiple wavelength anomalous diffraction data collection and analysis | |||
|---|---|---|---|
| Wavelength (Å) | 0.97970 | 0.97980 | 0.91840 |
| Cell constants | a = b = 82.2 Å | ||
| c = 51.8 Å | |||
| α = β = 90° | |||
| γ = 120° | |||
| Spacegroup | P63 | ||
| Resolution (Å) | 71.30-2.28 | 71.40-2.28 | 71.40-2.28 |
| High resolution shell (Å) | 2.40-2.28 | 2.40-2.28 | 2.40-2.28 |
| Rsym (%)a | 7.3 (39.2) | 7.4 (41.1) | 8.4 (46.9) |
| Mean I/σ(I) | 32.9 (4.6) | 32.9 (4.4) | 31.0 (4.0) |
| Total reflections (#) | 93,749 | 94,360 | 94,985 |
| Unique reflections (#) | 9,016 | 9,053 | 9,051 |
| Completeness (%) | 100 | 99.7 | 99.8 |
| Selenium sites (#) | 3 | ||
| Overall figure of meritb | 0.46 | ||
| p22 native data collection | |
|---|---|
| Cell constants | a = b = 82.1 Å |
| c = 51.9 Å | |
| α = β = 90° | |
| γ = 120° | |
| Space group | P63 |
| Resolution (Å) | 29.30-2.00 |
| High resolution shell (Å) | 2.11-2.00 |
| Rsym (%)a | 8.3 (13.85) |
| Mean I/σ(I) | 25.8 (1.5) |
| Total reflections (#) | 141,921 |
| Unique reflections (#) | 13,469 |
| Refinement statistics | |
| Completeness (%) | 99.1 |
| Resolution (Å) | 29.30-2.00 |
| Rwork/Rfree (%)c | 22.1/25.9 |
| Ave. B-factor (Å2) | 45.5 |
| Total atoms (#) | 1,501 |
| Water molecules (#) | 54 |
| Root mean square deviation | |
| Bond angles (°) | 1.4 |
| Bond lengths (Å) | 0.007 |
| Ramachandran analysis | |
| Most favored region (%/#) | 93.3/152 |
| Additional allowed region (%/#) | 6.1/10 |
| Generously allowed region (%/#) | 0/0 |
| Disallowed region (%/#) | 0.6/1 |
a Rsym = ΣΣ|Ihkl − Ihkl(j)|/ΣIhkl, where Ihkl(j) is the observed intensity and Ihkl is the final average value of intensity.
b Figure of merit = 〈|ΣP(α)eiα/ΣP(α)|〉, where α is the phase and P(α) is the phase probability distribution.
c Rwork = Σ‖Fobs| − |Fcalc‖/Σ|Fobs| and Rfree = Σ‖Fobs| − |Fcalc‖/Σ|Fobs|, where all reflections belong to a test set of 5% of the data randomly selected and not used in the atomic refinement. Data for the high resolution shell are shown in parentheses.
Structure Determination of p22
The structure of p22 was solved by selenomethionine multiple wavelength anomalous diffraction (MAD) phasing to 2.28-Å resolution using data collected at three wavelengths. Of the four possible selenomethionine residues in the p22 selenomethionine protein, three were located using SOLVE (49, 50). The N-terminal selenomethionine residue was disordered. After density modification a partial structure was built automatically by using RESOLVE (49, 50). Manual fitting of the remaining residues into the electron density map was carried out using COOT (51). The structure was refined using CNS (52), and the model was used in molecular replacement with the high resolution native p22 data. The structure was then refined until convergence using the high resolution data. The final model has an Rwork/Rfree of 22.1%/25.9%, to 2.0-Å resolution and includes residues 46–227 and 54 water molecules. Refinement statistics are shown in Table 1.
RNAi of p22
Primers p22 5′ (GAGGATCCATGCGTCGTGCACTTGTATTCACAGCTTTTG) and p22 3′ (GACTCGAGCGAAACAAATTTGTTAATGCTGCTCAGCC) (underlines indicate restriction sites) were used to amplify the p22 cDNA from oligo(dT)-primed PF T. brucei cDNA. The resultant 697-bp product was subcloned first into the pJET cloning vector (Fermentas) and then ligated into the BamHI-XhoI restriction sites of the RNAi vector p2T7-177 (53), resulting in p2T7-177-p22. p2T7-177-p22 was then transfected by electroporation into the 29-13 cell line, which contains a tetracycline (tc)-inducible integrated T7 polymerase (54). Resultant phleomycin-resistant cell lines were selected by clonal dilution. Cells were seeded at 5 × 105 cells/ml and grown in the absence or presence of 2.5 μg/ml tc. Cell growth was analyzed over 12 days. Using recombinant p22 (42), polyclonal antibodies were raised against p22 (Bethyl Laboratories). RNAi depletion of p22 was monitored by Western blot, during 1 × 106 cell equivalents were analyzed using antibodies against p22. Antibodies against RBP16 (42) were used as a loading control.
Quantitative RT-PCR Analysis of p22 RNAi Cells
p22 RNAi cells were grown in the absence or presence of tc for 3 days, pelleted, and treated with TRIzol (Invitrogen) to extract total RNA. Using the DNA-free DNase kit (Ambion), any residual DNA was subsequently removed. cDNA was then reverse transcribed as detailed in a previous study (34). Using quantitative RT-PCR primers detailed previously (23), multiple mRNAs were analyzed by quantitative RT-PCR using the standard curve method and using both β-tubulin and 18 S rRNA RNAs as separate steady-state standards. Results are represented as the mean change and standard deviation relative to RNA in the absence of tc, and each value is the result of at least six determinations.
Construction of p22 Truncation Mutants
The p22 sequence encoding residues W79-S227, p22(79–227), was amplified via PCR with the primers p22_W79_NdeI_fwd_n (5′-ATAATAATACATATGTGGACAATAGACCGTAAGC-3′) and p22_XhoI_rev (5′-ATAATAATACTCGAGTTATTACGAAACAAATTTGTTAATG-3′) introducing NdeI and XhoI sites (underlined), respectively, and pET-21a-p22 as template. The p22 sequence encoding residues V47-A192, p22(47–192), was amplified using the oligonucleotides p22_NdeI_fwd (5′-ATAATAATACATATGGTATCGGACCAACGA-3′) and p22_A192_XhoI_rev_n (5′-ATAATAATACTCGAGTTATGCACTCAAATAGCTTGTAAA-3′) introducing NdeI and XhoI sites (underlined), respectively, with pET-21a-p22 as the template DNA. The p22 sequence encoding residues W79-A192, p22(79–192), was amplified with the primers p22_W79_NdeI_fwd_n and p22_A192_XhoI_rev_n using pET-21a-p22 as the template DNA. The resulting PCR fragments were cloned via NdeI/XhoI into pET-15b, resulting in pET-15b-p22_W79-S227, pET-15b-p22_V47-A192, and pET-15b-p22_W79-A192, respectively. The mature p22 sequence encoding residues V47-S227, p22(47–227), was amplified via PCR with the primers p22_NdeI_fwd and p22_XhoI_rev introducing NdeI and XhoI sites (underlined), respectively, and pET-21a-p22 as template. The resulting PCR fragment was cloned via NdeI/XhoI into pET-Duet1 giving mature p22 fused to a N-terminal His6 tag. The p22(47–227) and p22(47–192) proteins were both produced in inclusion bodies under all conditions tested. The p22(79–227) protein was expressed and purified as wild type.
Co-immunoprecipitation Studies
Mitochondria were isolated from the 29-13 strain of PF T. brucei as described previously (55). Mitochondria (equivalent of 5 × 109 starting cells) were then lysed in 0.2% Nonidet P-40 as in a previous study (35) except either 40 units of SuperaseIN (Ambion) was added (− nucleases) or a mixture of nucleases (Micrococcal nuclease (Fermentas), Ribonuclease V1 (Amersham Biosciences), Ribonucleases T1, H, and 1 (all Ambion), and DNase I (Invitrogen) were added (+ nucleases) for 20 min at 37 °C. Mitochondrial lysates were then recovered and incubated with 100 μl of Protein A-Sepharose beads (Amersham Biosciences) alone, or beads that had been cross-linked to 200 μg of anti-p22 polyclonal antibodies (42). The slurry was incubated with rocking for 2 h at 4 °C. Beads were centrifuged and washed with a 50-bead volume excess of phosphate-buffered saline. The beads were then boiled in SDS-PAGE loading buffer and analyzed by Western blotting using antibodies against TbRGG2 (34), RBP16 (42), MRP2, MRP1, and KREL1 (the latter two antibodies were provided as generous gifts from Ken Stuart, Seattle BioMed). Recombinant MRP2 was produced as described (56), and polyclonal antibodies were produced by Proteintech, Inc. (Chicago, IL).
GST-pulldown Experiments
GST-His-TbRGG2 was purified as indicated previously (34), and GST alone was purified using a standard GST purification. Ten micrograms of GST-His-TbRGG2 (300 nm) or 41.9 μg of GST (3 μm) was incubated with 300 nm p22 or 300 nm p22(79–227) in 500 μl of binding buffer (1× phosphate-buffered saline with protease inhibitors, 0.1 mg/ml bovine serum albumin, and 0.1% Nonidet P-40) and 100 μl of glutathione-agarose (Invitrogen) for 2 h at 4 °C with rocking. Unbound flow-through was collected, and the beads were washed with a 10-fold excess of binding buffer. Bound proteins were then eluted with binding buffer containing 10 mm reduced glutathione and analyzed by Western blotting. GST protein was detected using anti-GST, GST-His-TbRGG2 was detected using anti-His antibodies (Clontech), and p22 and p22(79–227) were detected using anti-p22 antibodies.
RESULTS AND DISCUSSION
Overall Structure of p22
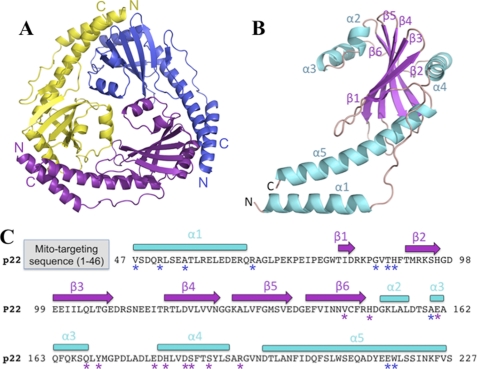
The crystal structure of the T. brucei p22 was solved by multiple wavelength anomalous diffraction phasing using selenomethionine-substituted p22 protein. The structure contains one p22 subunit in the crystallographic asymmetric unit and the final model has an Rwork/Rfree of 22.1%/25.9% to 2.0-Å resolution (Table 1). Size-exclusion chromatography revealed that p22 is a trimer (data not shown). Consistent with this, crystallographic symmetry generates a highly intertwined p22 trimer with a diameter of ∼75 Å and a thickness of ∼35 Å (Fig. 1A). Each subunit of p22 consists of six antiparallel β-strands (β1–β6: β1, residues 80–82; β2, residues 91–96; β3, residues 99–108; β4, residues 117–125; β5, residues 128–137; and β6, residues 140–149) surrounded by a long N-terminal α-helix (α1, residues 47–65) and four C-terminal α-helices (α2–α5: α2, residues 152–156; α3, residues 160–168; α4, residues 180–193; and α5, residues 197–226) (Fig. 1B).
FIGURE 1.
p22 crystal structure. A, ribbon diagram of the p22 trimer. The different subunits are colored yellow, purple, and blue. The N and C termini of each subunit are indicated as N and C according to the color of the subunit, respectively. A and B and Figs. 2, 3, and 4 were made using PyMOL (20). The PDB code is 3JV1. B, ribbon diagram of the p22 monomer. The secondary structure elements are labeled and numbered. α-Helices and β-strands are colored cyan and magenta, respectively, whereas loops are colored salmon. The N and C termini are indicated as N and C, respectively. C, p22 sequence and secondary structure elements. The secondary structure elements of p22 are indicated above the sequence. α-Helices are shown as cyan rectangles and β-strands as magenta arrows. The location and length of mitochondrial signal sequence is indicated (gray). Residues that mediate trimer formation via hydrogen bonds are highlighted with asterisks (purple: subunit 1; blue: subunit 2).
The arrangement of the secondary structural elements of each p22 subunit is shown schematically in Fig. 1C. The β-sheet core is flanked on one side by helices α2 and α3, and the other side by α1, α4, and α5. Helix α5 is notably long and adopts a curved conformation. The arrangement of the three α5 helices from each subunit in the trimer led to its characteristic triangular shape. α4 lies parallel to the β-sheet core, which stabilizes the structure by direct interactions. The structure is further stabilized by the N-terminal part of α5. In contrast, the long helix, α1, is connected via a loop to β1, but does not form any contacts to the β-sheet. α1 lies partially antiparallel to the C-terminal part of α5 and bolsters α5 and its interactions within and between subunits.
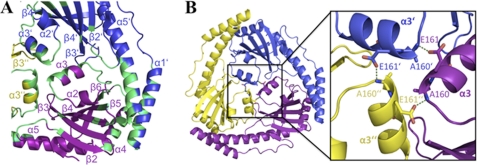
Subunit Interface of p22
The p22 trimer interface buries 1180 Å2 of accessible surface area per subunit. Because the trimer is crystallographic, the interface contacts are identical in all subunits. Although some subunit contacts involve residues from the β-sheet, the interface is formed primarily by interactions from helical residues (Fig. 2A). This highly helical interface is distinct from those found in typical β-barrel and other β-structures, which tend to oligomerize by forming contiguous β-sheets through hydrogen bonding interactions between β-strands in adjacent subunits. Nonetheless, the p22 oligomer interface is rich in hydrogen bonds involving backbone groups as well as side chains (supplemental Fig. S1). Specifically, the carbonyl of Val-146 (β6) and the side chain of His-150 (loop 7) form hydrogen bonds to the side chains of Trp-218′ and Glu-217′of α5′, respectively (where the prime symbol indicates the other p22 subunit). The carbonyl moiety of Gln-168 (α3) contacts the backbone carbonyl groups of Thr-88′ and His-89′ (loop 2′). The amide nitrogen group of Tyr-170 (loop 9) interacts with the carbonyl backbone of Gly-86′ on loop 2′, whereas the side chain of Glu-179 (loop 9) makes ionic interactions with that of Arg-66′ (loop 1′). Multiple contacts are also found between residues on α4 and α1′. These include hydrogen bonds from the His-181 side chain to the carbonyl of Ala-55′, the carbonyl of Asp-184 to the side chain of Arg-51′, the side chain of Ser-185 and the carbonyl of Arg-51′, and finally, two hydrogen bonds between the side chains of Arg-193 and Asp-49′. The central region of the p22 trimer is stabilized by contacts between residues in α3, α3′. and α3″. Especially critical is the hydrogen bond between the side chain of Glu-161 to the backbone amide nitrogen of Ala-160′ (Fig. 2B).
FIGURE 2.
Subunit interface of p22. A, regions involved in the p22 subunit interface. The different p22 subunits are colored as in Fig. 1A. The regions involved in the oligomer interface are colored green, and the secondary structural elements are labeled for reference. B, residues involved in the α3 interface or “α3-triangle.” The left panel shows the location of the α3-triangle in the p22 trimer. Inset (to the right) is a close-up view of the α3-triangle. Residues forming hydrogen bonds are indicated as sticks, and hydrogen bonds are shown as green dashed lines.
p22 Belongs to the Mitochondrial Glycoprotein Mam33-like Family
Database searches show that p22 is a member of the mitochondrial acidic matrix protein (Mam33p) family. Homologous proteins have been identified in several organisms, including human, yeast, mouse, chicken, turkey, Aspergillus nidulans, Vibrio cholerae, and Leishmania major (57–63). Mam33-like proteins contain an unusual structural topology that is shared only by its members. Indeed, homology searches revealed that p22 shares significant structural similarity to only the three Mam33-like proteins present in the protein data base; LMAJ011689, a hypothetical protein from L. major, VC1805, a hypothetical protein from a V. cholerae pathogenicity island (61), and human p32. The root mean squared deviations for comparison of similar Cα atoms for the p22 trimer with the LMAJ011689 and p32 trimers are 1.40 Å and 1.87 Å, respectively.
In contrast to p22, LMAJ011689, and p32, which are trimeric, the VC1805 protein is a monomer. The p22 subunit and the VC1805 monomer superimpose with a root mean squared deviation of 3.05 Å. The conservation between p22 and VC1805 is limited to the β-sheet core and α4, which stabilizes the β-sheet. Notably, the long C-terminal helix, α5, which is critical for trimer formation by p32, LMAJ011689, and p22, is not found in VC1805. In addition, helix α1 is significantly shorter in VC1805 than in the other Mam33-like proteins. Because α5 and α1 are critical for trimer formation by Mam33-like proteins, the lack of α5 and the presence of a significantly shorter α1 in VC1805 appear to explain why it is monomeric. How the oligomeric state affects the function(s) of the Mam33-like proteins is not known. Indeed, remarkably little is known in detail about the specific cellular roles of any of the Mam33-like proteins. Human p32 is the best-characterized member, yet even its specific physiological functions have not been defined. It was first identified as a protein that interacts with the nuclear precursor-mRNA splicing factor SF2/ASF (64). However, a later study suggested that it plays a critical role in maintaining oxidative phosphorylation in the mitochondria (65). To date, many diverse functions of p32 have been proposed that involve interactions with multiple factors in different subcellular localizations such as the nucleus, mitochondria, cytoplasm, and even the extracellular side of the plasma membrane (58–68).
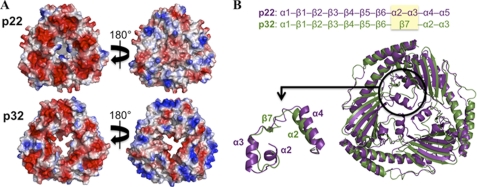
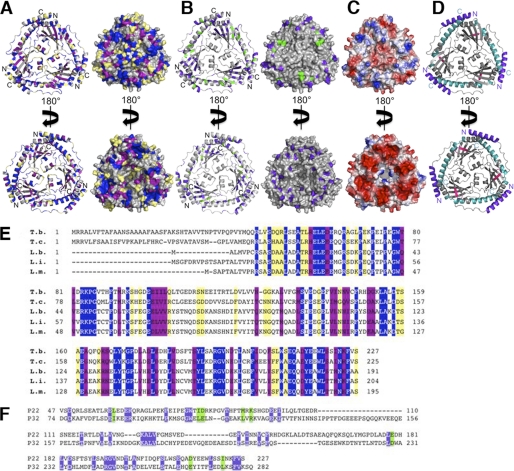
A characteristic feature of the Mam33-like proteins, which is likely important for their functions, is their highly acidic nature. For example, p22 has a pI of 4.4, whereas p32 has a pI of 4.0 (57). Interestingly, however, the electronegative charge distribution on these proteins is localized primarily on one face of each of the Mam33-like structures (Fig. 3A). This localized negative charge led Jiang and coworkers to propose that the p32 protein may associate with the inner mitochondrial membrane in the presence of divalent cations. This idea seemed to be supported by circumstantial evidence that p32 is capable of associating with membranes and led to the theory that, once bound to the membrane, the central channel present in the p32 trimer might function as a pore for molecules on the order of 0.4–3.0 kDa in size (57). However, for the p32 pore to be functional, the loops between β6 and β7, which partially cover the channel, must undergo a conformational change. It was proposed that this structural change might be induced by binding to partner proteins (57). Given this possibility, it is interesting that a major difference between p32 and p22 is the absence of a visible pore in p22 (Fig. 3B). In fact, p22 contains two additional α-helices, α2 and α3, that are not found in p32 (Fig. 3B, upper panel) and that completely covers its central region. Thus, if p22 forms a channel it would appear that a different set of conformational changes compared with p32 would be required to induce its formation (57). In that regard, it is notable that the Leishmania LMAJ011689 trimer also contains helices corresponding to α2 and α3 in p22. This conservation indicates that these helices might have one or more important functions that are specific for kinetoplastid p22 homologues. Indeed, when the conserved residues of known p22 homologues from T. cruzi, L. infantum, L. braziliensus, and L. major are mapped onto the T. brucei p22 structure, α2 and α3 are found to be among the most highly conserved regions in all these proteins (Fig. 4, A–E). Additional regions that show high conservation among these kinetoplastid proteins are α1, α4, and α5. The β-sheet region also contains scattered conserved residues. However, with the exception of β6, these residues are somewhat buried in the protein trimer core and thus likely play a structural role, explaining their conservation. Interestingly, although p32 does not share the conserved α2 and α3 helices like the kinetoplastid proteins, it displays the highest degree of sequence conservation to p22 in regions corresponding to p22 α1, α4, and α5 (Fig. 4, A and B). This overlap in conserved surface-exposed residues between kinetoplastid p22 proteins and p32 may indicate some shared function(s) such as interactions with target proteins.
FIGURE 3.
Comparison of T. brucei p22 to human p32. A, electrostatic surface comparisons of p22 and p32. Electropositive regions are colored blue, and electronegative surfaces are red (color code: −104.1 to 104.1 for p22; −102.7 to 102.7 for p32). p22 and p32 are both notably acidic proteins. However, only one side of each molecule (left) contains a continuous electronegative face. The other side displays a mixed charge in both p22 and p32. B, superimposition of p22 and p32. p22 is shown in purple, and p32 is shown in green. The enlarged view (lower panel) highlights the differences in the central region between p22 and p32. In this region p22 contains two α-helices (α2 and α3) compared with a β-strand (β7) in p32, as indicated in the secondary structure comparison (upper panel).
FIGURE 4.
Conserved regions in p22. A, conserved residues among p22 proteins. Residues conserved in p22 proteins (T. brucei, T. cruzi, L. infantum, L. braziliensis, and L. major) are mapped in blue (fully conserved), purple (highly conserved), and yellow (minimally conserved) onto the T. brucei p22 ribbon diagram (left panel) and surface (right panel) in two different orientations (upper and lower panels). The non-conserved regions are colored gray. N and C termini are indicated on the ribbon diagrams. B, conserved residues in p22 and p32. Conserved residues in p22 and p32 are mapped in blue (fully conserved) and green (highly conserved) onto the T. brucei p22 ribbon diagram (left panel) and surface (right panel) in two different orientations (upper and lower panel). The non-conserved regions are shown in gray. N and C termini are indicated on the ribbon diagrams. C, electrostatic surface representation of p22 (taken from Fig. 3A). D, model for potential p22 interaction regions. The known protein interaction regions of p32 were mapped onto the p22 structure. The three main regions of p32 found to interact with other proteins are colored purple, cyan, and pink, respectively. E, sequence alignment of different p22 proteins. Residues conserved in p22 proteins (T. brucei: T. b., T. cruzi: T. c., L. infantum: L. i., L. braziliensis: L. b., and L. major: L. m.) are shown in blue (fully conserved), purple (highly conserved), and yellow (minimally conserved). F, structure-based sequence alignment of mature p22 and mature p32. Conserved residues in p22 and p32 are shown in blue (fully conserved) and green (highly conserved).
As noted, p32 appears to be a multifunctional protein that has been implicated in several different regulatory pathways and shown to bind to various factors, including viral, bacterial, and cellular proteins (69–94). Thus far, three regions in p32 have been demonstrated to be important for the recognition of binding partners. One region corresponds to the N-terminal helix, α1, residues 74–96. This region binds gC1q and also interacts with vitronectin (68, 92). The second interaction region on p32 maps to β6 residues 196–208, which appear to be involved in the interaction with the HIV-1 Rev protein (76, 80). The third interacting region of p32 is composed of the long C-terminal helix, α3 in p32 (corresponding to α5 in p22). Residues 244–255 in this region bind HIV-1 Tat, whereas residues 260–279 have been proposed to form a complex with TFIIB (75, 93, 94). These data suggest that p32 acts as an adaptor protein to regulate the functions of the proteins it binds. Therefore, it seems plausible that p22 may also bind several proteins or macromolecules. p32 is the only Mam33-like protein in which interacting regions of its structure have been identified. When the p32-interacting regions are mapped onto the p22 structure, we find that these regions cluster primarily on the outside of the trimer on both faces of the molecule (Fig. 4D). These p32-interacting regions correspond to α1, β6, α4, and α5 in p22, which strikingly, as noted, represent the most conserved regions between p22 and p32 (Fig. 4, A, B, E, and F).
p22 Contributes to Cell Growth in PF T. brucei Cells
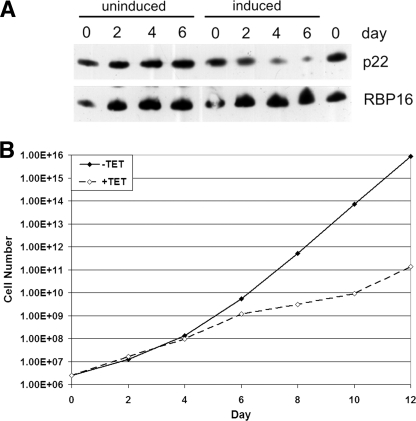
To obtain some insight into the functional importance of p22 in vivo, we down-regulated p22 expression in PF life cycle stages using tc-regulated RNAi. For these experiments, p22 was cloned into p2T7-177 resulting in a tc-controlled p22 expression system. The RNAi vector p2T7-177-p22 was transfected into the PF strain 29-13 of T. brucei. The resulting system allows down-regulation of p22 by addition of tc to the growth medium. The depletion of p22 protein was confirmed by immunoblot (Fig. 5A). Subsequently, we analyzed the T. brucei growth behavior in the presence and absence of tc (Fig. 5B). In the PF life cycle stage, down-regulation of p22 drastically affected the T. brucei growth behavior resulting in a sharp growth arrest after 4–5 days (Fig. 5B). The decrease in cell growth correlates with the decrease in p22 protein level (Fig. 5A). In contrast, tc addition to the parental PF 29-13 cells has no effect on cell growth (43). From these data, we conclude that p22 contributes to cell growth in PF T. brucei cells.
FIGURE 5.
p22 contributes to cell growth in PF T. brucei cells. The RNAi vector p2T7-177-p22 was transfected into the PF strain 29-13 of T. brucei. RNAi was induced by the addition of 2.5 μg/ml tc to the cell media. A, 1 × 106 cells grown either in the absence (uninduced) or presence (induced) of tc were pelleted and immunoblotted for the presence of p22. The abundance of the mitochondrial RBP16 protein was analyzed as a control. B, cells grown either in the absence (−TET, filled diamonds) or presence (+TET, open diamonds) of tc were counted every 2 days, and cumulative cell numbers are indicated.
p22 Is a COII-specific RNA-editing Accessory Factor
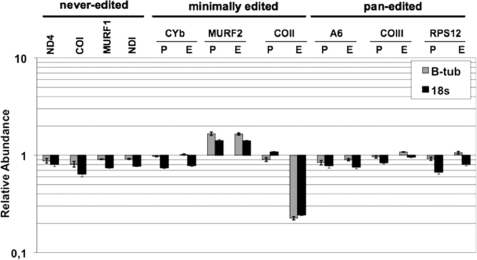
To investigate the role of p22 in k-RNA editing, we extracted total RNA from PF cells either uninduced or induced for p22 RNAi for 3 days. Subsequently, RNA was reverse-transcribed and analyzed by quantitative real-time RT-PCR using primers that have previously been used to analyze mitochondrial RNAs in T. brucei (24). We analyzed three classes of RNA in PF p22 RNAi cells, including never-edited (ND4, COI, MURF1, and NDI), minimally edited (apocytochrome b, MURF2, and COII), and pan-edited (A6, COIII, RPS12, and ND7) RNAs and standardized results to both β-tubulin and 18 S RNAs. The results of quantitative RT-PCR analysis of different RNAs from PF p22 RNAi cells are shown in Fig. 6.
FIGURE 6.
Effect of p22 down-regulation on mitochondrial RNA levels. Quantitative RT-PCR analysis of different RNAs from PF p22 RNAi cells at 3 days post induction. RNA levels represent the mean of at least six determinations for each mRNA set with error bars representing the standard error mean. A value of 1 indicates no change in RNA levels in induced cells relative to uninduced cells. P, pre-edited; E, edited; CYb, apocytochrome b; COI, COII, and COIII, cytochrome oxidase subunits I–III; NDI, ND4, and ND7, NADH dehydrogenase subunits I, 4, and 7; A6, ATPase subunit 6; RPS12, ribosomal protein S12; and MURF1 and -2, mitochondrial unknown reading frames 1 and 2. Mitochondrial RNA levels were standardized to both β-tubulin and 18 S rRNA.
Depletion of p22 had little effect on the levels of most mitochondrial RNAs examined, although both pre-edited and edited MURF2 RNAs were slightly increased (1.5- to 1.8-fold). Unexpectedly, the sole dramatic effect of p22 down-regulation was on edited COII RNA, which was present at ≤15% of wild-type levels in cells with decreased p22. Levels of pre-edited COII RNA were unchanged.
Identification of p22 Interacting Proteins Involved in k-RNA Editing
The specific effect of p22 depletion on COII RNA is of particular interest, because the peculiarities of this RNA make it likely that its editing requires COII-specific accessory factors. For example, such factors may stabilize interaction of the cis-acting gRNA with the edited portion of the mRNA, facilitate interaction of COII RNA with the KREN3-containing editosomes, or modulate association of KREN3 and/or KREPB6 with other editosome components. We have shown that p22 does not interact with RNA (42).3 Thus, based on our structural data showing that p22 is a Mam33-like protein, which bind numerous intracellular targets, we predicted that p22 may interact with k-RNA-editing proteins, such as members of the RNA-editing machinery or proteins that regulate the editing machinery.
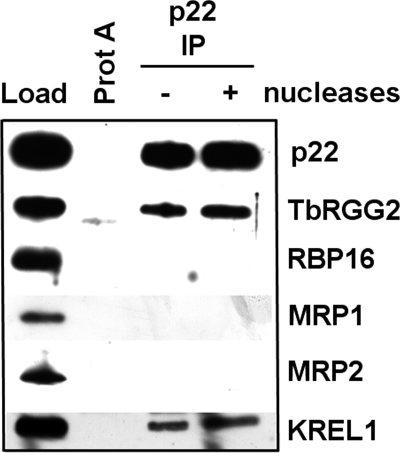
To address this possibility we carried out in vivo co-immunoprecipitation studies (Fig. 7). Mitochondrial extracts were prepared from PF T. brucei and either left untreated or treated with a mixture of nucleases to disrupt interactions mediated by an RNA or DNA bridge. p22 was immunoprecipitated with anti-p22 antibodies, and associated proteins were detected by Western blot. These studies revealed that p22 could be co-immunoprecipitated with core editosomes, as indicated by the presence of KREL1 in p22 immunoprecipitates, and with the k-RNA accessory protein, TbRGG2 (Fig. 7). The accessory factors RBP16, MRP1, and MRP2 did not form stable associations with p22 in this assay. The p22-TbRGG2 and p22-editosome interactions were both refractory to nuclease treatment, indicating that they likely constitute direct protein-protein interactions.
FIGURE 7.
p22 interacts with RNA editing factors in vivo. Mitochondrial lysate (equivalent of 5 × 109 total cells) was either treated with SuperaseIN RNase inhibitor (− nuclease) or a mixture of RNases and DNase (+ nuclease). Extracts were precipitated with either Protein A-Sepharose beads alone (Prot A) or with p22 polyclonal antibody cross-linked to Protein A-Sepharose (p22 IP). Load represents 0.5% of the total starting mitochondrial lysate. The p22 lanes were normalized for the total amount of p22 immunoprecipitated to determine effects of nuclease treatment on analyzed proteins (ProtA − p22 IP nuclease lanes contain 10% total eluate; p22 IP + nuclease lane contains 3% total eluate).
Identification of Interacting Regions of p22 for TbRGG2
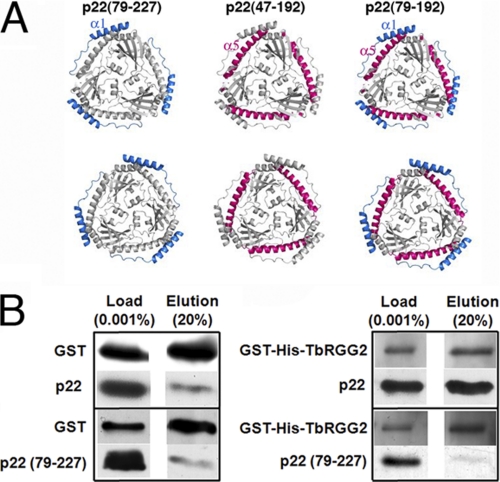
As noted, the main interaction regions of p32 for target proteins have been mapped to regions that correspond to α1, β6, α4, and α5 in p22. In particular, residues in α1 and α5 have been identified as interacting regions for numerous proteins. These surface regions are highly conserved between p22 and p32 suggesting a functional importance. Thus, in efforts to elucidate regions of p22 that are involved in interaction with TbRGG2, we constructed p22 truncation mutants in which α1 and α5 were removed (Fig. 8A). Removal of α5, in p22(47–192) and p22(79–192), led to the production of insoluble and likely unstable proteins (“Experimental Procedures”). By contrast, p22(79–227), in which the N-terminal α1 helix and the loop that follows were removed, produced functional protein that was found to be trimeric by size-exclusion chromatography. We next examined the in vitro binding of p22 and p22(79–227) to GST-TbRGG2 by GST-pulldown assays. Consistent with our in vivo co-immunoprecipitation studies, the wild-type p22 protein was shown to bind avidly to recombinant GST-TbRGG2 (Fig. 8B). However, the p22(79–227) truncation mutant showed marked reduction in GST-TbRGG2 binding compared with wild-type p22 (Fig. 8B). Thus, these data identify the N-terminal α1 helix of p22 as a critical region for interacting with the k-RNA-editing accessory protein TbRGG2.
FIGURE 8.
Recombinant p22 and the accessory factor TbRGG2 directly interact in vitro and p22 residues 47–78 are critical for this interaction. A, p22 structure showing the regions that were removed to examine their roles in TbRGG2 binding by GST-pulldown studies. The leftmost p22 shows the location of residues 47–78 (colored blue), the middle shows the location of the C-terminal residues, 193–227, that were removed (colored red), and the final figure (rightmost) shows the relative location of both truncated regions. Only p22(79–227) was successfully expressed and purified and therefore tested. B, purified GST tag alone (3 μm) or GST-His-TbRGG2 (300 nm) were incubated with 300 nm full-length (p22) or truncated p22 (p22(79–227)) in the presence of glutathione-agarose beads. Left: results using the GST protein alone. The input proteins are shown in the Load lane. Following binding and subsequent washes, bound proteins were eluted from the beads with excess free glutathione. Twenty percent of the total elution volume was analyzed by Western blotting for GST and the p22 proteins. Right: results as in A, except using recombinant GST-His-TbRGG2.
Our data show that p22 is important in k-RNA editing and appears to function primarily as a COII-specific editing factor. We also show that p22 interacts with the core editosome and the recently discovered multifunctional RNA-editing accessory factor, TbRGG2 (34). Interaction of p22 with core editosomes may reflect a role in shuttling of various editing endoribonucleases, facilitating structural rearrangements, or recruitment of COII RNA to the editosomes. However, TbRGG2 affects an entire class of RNAs, namely the pan-edited RNAs; its depletion does not affect editing of COII RNA (34). One possible explanation for these observations is that TbRGG2 is involved in COII RNA editing, but that another mitochondrial protein can substitute for TbRGG2 in this regard. Multiple RNA-binding proteins are present in T. brucei mitochondria, including TbRGG1, which affects editing of most mitochondrial RNAs (39), and another RGG motif-containing protein, Tb927.3.1820, which associates with TbRGG1 and the MRB1 complex (39). There may be functional redundancy between TbRGG2 and these or additional mitochondrial RNA-binding proteins in facilitating COII RNA editing. It is also possible that the p22-TbRGG2 interaction is involved in another, as yet unknown, function of p22. This would not seem unexpected given the finding that p22 is a Mam33-like protein. Indeed, the mammalian Mam33-like p32 protein has been implicated in diverse functions ranging from apoptosis to oxidative phosphorylation and apparently mediates these myriad affects by interacting with a vast number of intracellular targets in multiple cellular compartments. Future studies will examine the role(s) of p22 in T. brucei.
Supplementary Material
Acknowledgments
M. S. is very grateful to Drs. Lisheng Ni and Jinhui Dong for their great support. We thank Dr. Ken Stuart for providing antibodies. We also thank the Advanced Light Source and their support staff.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1 GM074815 (to M. A. S.), RO1 AI061580 (to L. K. R.), and F32 AI07718501 (to J. C. F.). The Advanced Light Source is supported by the Director, Office of science, Office of Basic Energy Sciences, Material Sciences Division, of the US Dept. of Energy under contract DE-AC03-76SF00098 at Lawrence Berkeley National Laboratory.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
The atomic coordinates and structure factors (code 3JV1) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
M. Sprehe, J. C. Fisk, S. M. McEvoy, L. K. Read, and M. A. Schumacher, unpublished data.
- PF
- procyclic form
- RNAi
- RNA interference
- k-RNA
- kinetoplastid RNA
- COII
- cytochrome oxidase subunit II
- gRNA
- guide RNA
- tc
- tetracycline
- RT
- reverse transcription
- GST
- glutathione S-transferase.
REFERENCES
- 1.Simpson A. G., Stevens J. R., Lukes J. (2006) Trends Parasitol. 22, 168–174 [DOI] [PubMed] [Google Scholar]
- 2.Matthews K. R. (2005) J. Cell Sci. 118, 283–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parsons M. (2004) Mol. Microbiol. 53, 717–724 [DOI] [PubMed] [Google Scholar]
- 4.Simpson L., Sbicego S., Aphasizhev R. (2003) RNA 9, 265–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benne R., Van den Burg J., Brakenhoff J. P., Sloof P., Van Boom J. H., Tromp M. C. (1986) Cell 46, 819–826 [DOI] [PubMed] [Google Scholar]
- 6.Feagin J. E., Shaw J. M., Simpson L., Stuart K. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 539–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horváth A., Berry E. A., Maslov D. A. (2000) Science 287, 1639–1640 [DOI] [PubMed] [Google Scholar]
- 8.Shaw J. M., Feagin J. E., Stuart K., Simpson L. (1988) Cell 53, 401–411 [DOI] [PubMed] [Google Scholar]
- 9.Lukes J., Hashimi H., Zíková A. (2005) Curr. Genet 48, 277–299 [DOI] [PubMed] [Google Scholar]
- 10.Stuart K. D., Schnaufer A., Ernst N. L., Panigrahi A. K. (2005) Trends Biochem. Sci. 30, 97–105 [DOI] [PubMed] [Google Scholar]
- 11.Feagin J. E., Jasmer D. P., Stuart K. (1987) Cell 49, 337–345 [DOI] [PubMed] [Google Scholar]
- 12.Feagin J. E., Stuart K. (1988) Mol. Cell. Biol. 8, 1259–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blum B., Bakalara N., Simpson L. (1990) Cell 60, 189–198 [DOI] [PubMed] [Google Scholar]
- 14.Klingbeil M. M., Drew M. E., Liu Y., Morris J. C., Motyka S. A., Saxowsky T. T., Wang Z., Englund P. T. (2001) Protist 152, 255–262 [DOI] [PubMed] [Google Scholar]
- 15.Klingbeil M. M., Englund P. T. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 4333–4334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corell R. A., Feagin J. E., Riley G. R., Strickland T., Guderian J. A., Myler P. J., Stuart K. (1993) Nucleic Acids Res. 21, 4313–4320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seiwert S. D., Stuart K. (1994) Science 266, 114–117 [DOI] [PubMed] [Google Scholar]
- 18.Hermann T., Schmid B., Heumann H., Göringer H. U. (1997) Nucleic Acids Res. 25, 2311–2318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmid B., Read L. K., Stuart K., Göringer H. U. (1996) Eur. J. Biochem. 240, 721–731 [DOI] [PubMed] [Google Scholar]
- 20.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
- 21.Aphasizhev R., Sbicego S., Peris M., Jang S. H., Aphasizheva I., Simpson A. M., Rivlin A., Simpson L. (2002) Cell 108, 637–648 [DOI] [PubMed] [Google Scholar]
- 22.Aphasizheva I., Aphasizhev R., Simpson L. (2004) J. Biol. Chem. 279, 24123–24130 [DOI] [PubMed] [Google Scholar]
- 23.Carnes J., Trotter J. R., Ernst N. L., Steinberg A., Stuart K. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 16614–16619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carnes J., Trotter J. R., Peltan A., Fleck M., Stuart K. (2008) Mol. Cell. Biol. 28, 122–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corell R. A., Read L. K., Riley G. R., Nellissery J. K., Allen T. E., Kable M. L., Wachal M. D., Seiwert S. D., Myler P. J., Stuart K. D. (1996) Mol. Cell. Biol. 16, 1410–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Osato D., Rogers K., Guo Q., Li F., Richmond G., Klug F., Simpson L. (2009) RNA 15, 1338–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Panigrahi A. K., Allen T. E., Stuart K., Haynes P. A., Gygi S. P. (2003) J. Am. Soc. Mass Spectrom 14, 728–735 [DOI] [PubMed] [Google Scholar]
- 28.Panigrahi A. K., Gygi S. P., Ernst N. L., Igo R. P., Jr., Palazzo S. S., Schnaufer A., Weston D. S., Carmean N., Salavati R., Aebersold R., Stuart K. D. (2001) Mol. Cell. Biol. 21, 380–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Panigrahi A. K., Schnaufer A., Carmean N., Igo R. P., Jr., Gygi S. P., Ernst N. L., Palazzo S. S., Weston D. S., Aebersold R., Salavati R., Stuart K. D. (2001) Mol. Cell. Biol. 21, 6833–6840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Panigrahi A. K., Schnaufer A., Ernst N. L., Wang B., Carmean N., Salavati R., Stuart K. (2003) RNA 9, 484–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simpson L., Aphasizhev R., Gao G., Kang X. (2004) RNA 10, 159–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Golden D. E., Hajduk S. L. (2005) RNA 11, 29–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stuart K., Panigrahi A. K. (2002) Mol. Microbiol. 45, 591–596 [DOI] [PubMed] [Google Scholar]
- 34.Fisk J. C., Ammerman M. L., Presnyak V., Read L. K. (2008) J. Biol. Chem. 283, 23016–23025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hayman M. L., Read L. K. (1999) J. Biol. Chem. 274, 12067–12074 [DOI] [PubMed] [Google Scholar]
- 36.Schumacher M. A., Karamooz E., Zíková A., Trantírek L., Lukes J. (2006) Cell 126, 701–711 [DOI] [PubMed] [Google Scholar]
- 37.Vanhamme L., Perez-Morga D., Marchal C., Speijer D., Lambert L., Geuskens M., Alexandre S., Ismaïli N., Göringer U., Benne R., Pays E. (1998) J. Biol. Chem. 273, 21825–21833 [DOI] [PubMed] [Google Scholar]
- 38.Acestor N., Panigrahi A. K., Carnes J., Zíková A., Stuart K. D. (2009) RNA 15, 277–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hashimi H., Zíková A., Panigrahi A. K., Stuart K. D., Lukes J. (2008) RNA 14, 970–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Panigrahi A. K., Zíková A., Dalley R. A., Acestor N., Ogata Y., Anupama A., Myler P. J., Stuart K. D. (2008) Mol. Cell Proteomics 7, 534–545 [DOI] [PubMed] [Google Scholar]
- 41.Weng J., Aphasizheva I., Etheridge R. D., Huang L., Wang X., Falick A. M., Aphasizhev R. (2008) Mol. Cell 32, 198–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hayman M. L., Miller M. M., Chandler D. M., Goulah C. C., Read L. K. (2001) Nucleic Acids Res. 29, 5216–5225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pelletier M., Read L. K. (2003) RNA 9, 457–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goulah C. C., Pelletier M., Read L. K. (2006) RNA 12, 1545–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goulah C. C., Read L. K. (2007) J. Biol. Chem. 282, 7181–7190 [DOI] [PubMed] [Google Scholar]
- 46.Doublié S. (1997) Methods Enzymol. 276, 523–530 [PubMed] [Google Scholar]
- 47.Leslie A. G. (1992) CCP4 + ESF-EAMCB News-letter on Protein Crystallography 26, 27–33 [Google Scholar]
- 48.CCP4 (1994) Acta Crystallogr. D. Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 49.Terwilliger T. (2004) J. Synchrotron Radiat. 11, 49–52 [DOI] [PubMed] [Google Scholar]
- 50.Terwilliger T. C., Berendzen J. (1999) Acta Crystallogr. D. Biol. Crystallogr. 55, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Emsley P., Cowtan K. (2004) Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 52.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D. Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 53.Wickstead B., Ersfeld K., Gull K. (2002) Mol. Biochem. Parasitol. 125, 211–216 [DOI] [PubMed] [Google Scholar]
- 54.Wirtz E., Leal S., Ochatt C., Cross G. A. (1999) Mol. Biochem. Parasitol. 99, 89–101 [DOI] [PubMed] [Google Scholar]
- 55.Harris M. E., Moore D. R., Hadjuk S. L. (1990) J. Biol. Chem. 265, 11368–11376 [PubMed] [Google Scholar]
- 56.Ammerman M. L., Fisk J. C., Read L. K. (2008) RNA 14, 1069–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jiang J., Zhang Y., Krainer A. R., Xu R. M. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 3572–3577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luo Y., Yu H., Peterlin B. M. (1994) J. Virol. 68, 3850–3856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Okagaki T., Nakamura A., Suzuki T., Ohmi K., Kohama K. (2000) J. Cell Biol. 148, 653–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Seytter T., Lottspeich F., Neupert W., Schwarz E. (1998) Yeast 14, 303–310 [DOI] [PubMed] [Google Scholar]
- 61.Sheikh M. A., Potter J. A., Johnson K. A., Sim R. B., Boyd E. F., Taylor G. L. (2008) Proteins 71, 1563–1571 [DOI] [PubMed] [Google Scholar]
- 62.Simos G., Georgatos S. D. (1994) FEBS Lett. 346, 225–228 [DOI] [PubMed] [Google Scholar]
- 63.Van Den Brulle J., Steidl S., Brakhage A. A. (1999) Appl. Environ Microbiol. 65, 5222–5228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Petersen-Mahrt S. K., Estmer C., Ohrmalm C., Matthews D. A., Russell W. C., Akusjärvi G. (1999) EMBO J. 18, 1014–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Muta T., Kang D., Kitajima S., Fujiwara T., Hamasaki N. (1997) J. Biol. Chem. 272, 24363–24370 [DOI] [PubMed] [Google Scholar]
- 66.Ghebrehiwet B., Lim B. L., Peerschke E. I., Willis A. C., Reid K. B. (1994) J. Exp. Med. 179, 1809–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Herwald H., Dedio J., Kellner R., Loos M., Müller-Esterl W. (1996) J. Biol. Chem. 271, 13040–13047 [DOI] [PubMed] [Google Scholar]
- 68.Lim B. L., Holmskov U. (1996) Biochem. Biophys. Res. Commun. 218, 260–266 [DOI] [PubMed] [Google Scholar]
- 69.Beatch M. D., Everitt J. C., Law L. J., Hobman T. C. (2005) J. Virol. 79, 10807–10820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang Y., Finan J. E., Middeldorp J. M., Hayward S. D. (1997) Virology 236, 18–29 [DOI] [PubMed] [Google Scholar]
- 71.Matthews D. A., Russell W. C. (1998) J. Gen Virol. 79, 1677–1685 [DOI] [PubMed] [Google Scholar]
- 72.Marschall M., Marzi A., aus dem Siepen P., Jochmann R., Kalmer M., Auerochs S., Lischka P., Leis M., Stamminger T. (2005) J. Biol. Chem. 280, 33357–33367 [DOI] [PubMed] [Google Scholar]
- 73.Liang X., Shin Y. C., Means R. E., Jung J. U. (2004) J. Virol. 78, 12416–12427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kittlesen D. J., Chianese-Bullock K. A., Yao Z. Q., Braciale T. J., Hahn Y. S. (2000) J. Clin. Invest. 106, 1239–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yu L., Loewenstein P. M., Zhang Z., Green M. (1995) J. Virol. 69, 3017–3023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tange T. O., Jensen T. H., Kjems J. (1996) J. Biol. Chem. 271, 10066–10072 [DOI] [PubMed] [Google Scholar]
- 77.Hall K. T., Giles M. S., Calderwood M. A., Goodwin D. J., Matthews D. A., Whitehouse A. (2002) J. Virol. 76, 11612–11622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bryant H. E., Matthews D. A., Wadd S., Scott J. E., Kean J., Graham S., Russell W. C., Clements J. B. (2000) J. Virol. 74, 11322–11328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bruni R., Roizman B. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 10423–10427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Braun L., Ghebrehiwet B., Cossart P. (2000) EMBO J. 19, 1458–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nguyen T., Ghebrehiwet B., Peerschke E. I. (2000) Infect Immun 68, 2061–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Krainer A. R., Conway G. C., Kozak D. (1990) Genes Dev. 4, 1158–1171 [DOI] [PubMed] [Google Scholar]
- 83.Robles-Flores M., Rendon-Huerta E., Gonzalez-Aguilar H., Mendoza-Hernandez G., Islas S., Mendoza V., Ponce-Castaneda M. V., Gonzalez-Mariscal L., Lopez-Casillas F. (2002) J. Biol. Chem. 277, 5247–5255 [DOI] [PubMed] [Google Scholar]
- 84.Itahana K., Zhang Y. (2008) Cancer Cell 13, 542–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chattopadhyay C., Hawke D., Kobayashi R., Maity S. N. (2004) Nucleic Acids Res. 32, 3632–3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yanagida M., Hayano T., Yamauchi Y., Shinkawa T., Natsume T., Isobe T., Takahashi N. (2004) J. Biol. Chem. 279, 1607–1614 [DOI] [PubMed] [Google Scholar]
- 87.Joseph K., Ghebrehiwet B., Peerschke E. I., Reid K. B., Kaplan A. P. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 8552–8557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Deb T. B., Datta K. (1996) J. Biol. Chem. 271, 2206–2212 [DOI] [PubMed] [Google Scholar]
- 89.Nikolakaki E., Simos G., Georgatos S. D., Giannakouros T. (1996) J. Biol. Chem. 271, 8365–8372 [DOI] [PubMed] [Google Scholar]
- 90.Mallick J., Datta K. (2005) Exp. Cell Res. 309, 250–263 [DOI] [PubMed] [Google Scholar]
- 91.Sunayama J., Ando Y., Itoh N., Tomiyama A., Sakurada K., Sugiyama A., Kang D., Tashiro F., Gotoh Y., Kuchino Y., Kitanaka C. (2004) Cell Death Differ. 11, 771–781 [DOI] [PubMed] [Google Scholar]
- 92.Ghebrehiwet B., Lim B. L., Kumar R., Feng X., Peerschke E. I. (2001) Immunol. Rev. 180, 65–77 [DOI] [PubMed] [Google Scholar]
- 93.Berro R., Kehn K., de la Fuente C., Pumfery A., Adair R., Wade J., Colberg-Poley A. M., Hiscott J., Kashanchi F. (2006) J. Virol. 80, 3189–3204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yu L., Zhang Z., Loewenstein P. M., Desai K., Tang Q., Mao D., Symington J. S., Green M. (1995) J. Virol. 69, 3007–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.