Abstract
Calnexin is a molecular chaperone and a component of the quality control of the secretory pathway. We have generated calnexin gene-deficient mice (cnx−/−) and showed that calnexin deficiency leads to myelinopathy. Calnexin-deficient mice were viable with no discernible effects on other systems, including immune function, and instead they demonstrated dysmyelination as documented by reduced conductive velocity of nerve fibers and electron microscopy analysis of sciatic nerve and spinal cord. Myelin of the peripheral and central nervous systems of cnx−/− mice was disorganized and decompacted. There were no abnormalities in neuronal growth, no loss of neuronal fibers, and no change in fictive locomotor pattern in the absence of calnexin. This work reveals a previously unrecognized and important function of calnexin in myelination and provides new insights into the mechanisms responsible for myelin diseases.
Keywords: Endoplasmic Reticulum (ER), Membrane Proteins, Mouse, Neuroscience, Protein Folding, Calnexin, Myelin, Neuropathy
Introduction
The endoplasmic reticulum (ER)5 is the first compartment in the secretory pathway responsible for protein synthesis, post-translational modification, and correct folding. The resident molecular chaperones ensure that only correctly folded proteins leave the ER. Calnexin is a type I ER membrane protein, a major component in assuring the quality control of the secretory pathway, and together with calreticulin and the oxidoreductase ERp57, it promotes the correct folding of newly synthesized glycoproteins (2). Calnexin and calreticulin bind monoglucosylated carbohydrate on newly synthesized glycoproteins, whereas ERp57 catalyzes rearrangements of disulfide bonds within the calnexin/calreticulin substrate proteins (2). Despite its ubiquitous expression, the absence of calnexin has a different effect in different organisms. Calnexin deficiency is lethal in Schizosaccharomyces pombe but not in Saccharomyces cerevisiae (3), Dictyostelium (4, 5), or Caenorhabditis elegans (6, 7). The loss of calnexin affects phagocytosis in Dictyostelium (4, 5) and promotes necrotic cell death in C. elegans (7). It has been reported that deletion of the calnexin gene in a mouse results in early postnatal death (1), and thus the molecular consequences of calnexin deficiency could not be studied.
Here we show that calnexin deficiency in the mouse did not result in early postnatal death (1). These animals developed myelinopathy with no discernible effects on other systems, including immune function. The phenotype was linked to slow nerve conduction velocities in the absence of calnexin with evidence of peripheral axon dysmyelination. The dysmyelinating phenotype described here underscores the emerging importance of calnexin and ER-associated pathways as contributors to these severe neurological disorders.
EXPERIMENTAL PROCEDURES
Generation of Calnexin-deficient Mice
Gene trapping with the trap vector pGT1TMpfs was used to generate the calnexin gene disrupted embryonic stem cells, designated KST286. The cell line KST286 was from the Gene Trap Resource (BayGenomics, University of San Francisco, San Francisco, California). The KST286ES cell line was generated from the 129P2 (formerly 129/Ola) embryonic stem cell line, the E14Tg2A.4 subclone. Parental cell lines (CGR8 and E14Tg2A) were established from delayed blastocysts. Embryonic stem cells were microinjected into 3.5-day-old C57BL/6J blastocysts to generate chimeric mice (8). Chimeric males were analyzed for germ line transmission by mating with C57BL/6J females, and the progeny was identified by PCR analysis, β-galactosidase staining, and Western blot analysis. All of the animal experimental procedures were approved by the Animal Welfare Program at the Research Ethics Office of the University of Alberta and conformed to the guidelines set forth by the Canadian Council on Animal Care.
Genotype Analysis of Calnexin-deficient Mice
Genomic DNA was isolated from mouse tails by lysis with a buffer containing 10 mm Tris, pH 8.0, 150 mm NaCl, 10 mm EDTA, 0.5% SDS, and proteinase K digestion followed by phenol-chloroform extraction. An inverse PCR technique was used to identify the gene trap insertion site in the calnexin gene. Briefly, genomic DNA was first digested with BfaI restriction enzyme that cleaves at frequent intervals and digests the gene trap vector near the 3′-terminal end. The resulting DNA fragments were ligated under conditions that favor intramolecular circularization of single fragments. The nucleotide sequence located at the 3′-terminal end of the gene trap vector was then selectively amplified using inverse DNA primers (INVF1, 5′-TCAAGGCGAGTTACATGATCCC-3′; and INVR1, 5′-AAGCCATACCAAACGACGAGCG-3′) derived from the nucleotide sequence of the gene trap vector. The resulting PCR product was amplified a second time using nested DNA primers (F2, 5′-TCAAGGCGAGTTACATGATCCC-3′; and R2, 5′-CGAGCGTGACACCACGATGC-3′), purified, and sequenced. The PCR product obtained corresponded to the gene trap vector and extension into the genomic sequence that resides immediately downstream. This allowed determination of the precise point of the vector integration in the calnexin gene. Once the integration site was identified, it was possible to design a protocol for genotyping wild-type, heterozygote, and homozygote calnexin-deficient mice (see Fig. 1A). DNA primers that flank the integration site allow detection of the wild-type allele (Primers F1 (exon 7), 5′-GGCCAGATGCAGATCTGAAGACC-3′; and R3 (intron 7–8), 5′-CACACAGGGTATGGGGCTGTTTCAG-3′), whereas DNA primer F2 within the insertion vector sequence with primer R3 was used to detect the gene trap allele insertion (see Fig. 1A).
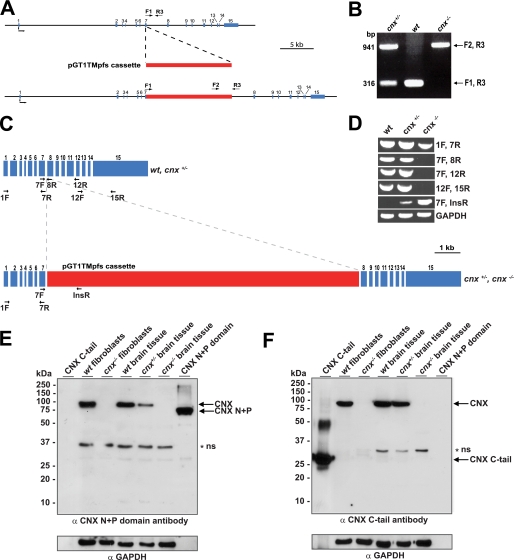
FIGURE 1.
Generation of a calnexin-deficient mouse. A, random gene trapping was used to generate the calnexin-deficient mice. An interruption cassette (pGT1TMpfs cassette, in red) containing β-galactosidase and neomycin genes was inserted into the calnexin gene. The numbers indicate the locations of calnexin gene exons (in blue). Forward (F1 and F2) and reverse (R3) primers are indicated with arrows. B, PCR analysis of genomic DNA isolated from wild-type (wt), heterozygote (cnx+/−), and homozygote (cnx−/−) calnexin-deficient mice. Forward (F1 and F2) and reverse (R3) primers were used as indicated for A. A DNA product of 941 pairs (bp amplified with primers F2 and R3 identifies successful cassette insertion and interruption of the calnexin gene (cnx−/−), whereas a DNA product of 316 bp amplified with primers F1 and R3 indicates the presence of the wild-type allele (wt). The presence of both 941- and 316-bp DNA products identifies heterozygotes. C, schematic representation of calnexin mRNA. The location of the insertion cassette (in red) is shown. The location of specific primers used for RT-PCR analysis in D is indicated in the figure. D, RT-PCR was carried out using wild-type (wt), heterozygote (cnx+/−), and calnexin-deficient (cnx−/−) RNA isolated from brain tissue. Pairs of specific DNA primers used for the analysis are indicated. E and F, Western blot analysis of wild-type, heterozygote, and calnexin-deficient tissues and wild-type and calnexin-deficient fibroblasts with anti-calnexin antibodies. The location of molecular weight markers is indicated to the left of the gel. In E, anti-N terminus (N+P domain) calnexin antibodies were used. In F, the blot was probed with anti-C terminus calnexin antibodies. N+P, N+P domain of calnexin; C-tail, cytoplasmic C-terminal domain of calnexin. The asterisk and ns designate the nonspecific reactive protein band. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
To determine that no alternative splicing around the interruption cassette took place, RNA was isolated from wild-type, heterozygote, and calnexin-deficient brain tissue using TRIzol reagent followed by RT-PCR analysis (8). The following DNA primers (see Fig. 1) were used: primers upstream of the insertion (1F, 5′-GAAGGTGAGGGAGCCGCCAGTG-3′; and 7R, 5′-GTAGTCCTCTCCACACTTATCTGG 3′), primers flanking the site of insertion (7F, 5′-CCAGATAAGTGTGGAGAGGACTAC-3′; and 8R, 5′-CACGTGAAGGGTTTACAGGAGGAG-3′; and 12R, 5′-GACGCTCTTCAGCTGCCTCCAG-3′), primers within the insertion (InsR, 5′-CCTTCTCTGCCTTCCATCTCAACTC-3′), and primers following the insertion (12F, 5′-CTGGAGGCAGCTGAAGAGCGTC-3′; and 15R, 5′-GTCTCTAGGGCAACAGAACACTGC-3′). RT-PCR analysis of mRNA encoding glyceraldehyde-3-phosphate dehydrogenase was used as a loading control using the following DNA primers (forward primer, 5′-TTCACCACCATGGAGAAGGC-3′; and reverse primer, 5′-GGCATGGACTGTGGTCATGA-3′).
Escherichia coli Expression and Purification of Calnexin Domains
cDNA encoding calnexin C-tail (amino acid residues 486–573) was synthesized by PCR-driven amplification using the following primers: forward primer, 5′-CATGCCATGGCTGGAAAGAAACAGTCAAG-3′; and reverse primer, 5′-GCTCTAGACACTCTCTTCGTGGCTTTC-3′. cDNA was cloned into pBAD His tag vector using NcoI and XbaI restriction enzymes. cDNA encoding the N+P domain of calnexin (amino acid residues 1–461) was amplified by a PCR-driven reaction using the following primers: forward primer, 5′-CATGCCATGGATCATGAAGGACATGATGAT-3′; and reverse primer, 5′-GCTCTAGAGGGCGCTCCTCAGCTGCCTC-3′. cDNA was cloned into pBAD plasmid using NcoI and XbaI restriction sites. The proteins were expressed in Top 10 F′ E. coli according to the pBAD expression system using 0.02% l-arabinose induction for 4 h. His-tagged protein purification was carried out as previously described (9).
Western Blot Analysis
Two distinct polyclonal rabbit anti-calnexin antibodies were used: SPA-860 (Stressgen Biotechnologies) raised against a synthetic peptide corresponding to the C terminus of calnexin (amino acid residues 575–593) and SPA-865 (Stressgen Biotechnologies) raised against a synthetic peptide near the N terminus. Antibodies were used at 1:1000 and 1:500 dilutions, respectively. Preparation of cell extracts, Western blot analysis, and immunostaining of wild-type and calnexin-deficient cells were carried out as described previously (21). Twenty μg of cell and brain tissue extracts and 200 ng of purified recombinant protein (C-tail and N+P domain) was loaded for analysis of calnexin protein expression. The membranes were stripped with a buffer containing 1% SDS, 100 mm β-mercaptoethanol, and 50 mm Tris-HCl, pH 6.8. Anti-glyceraldehyde-3-phosphate dehydrogenase antibodies (1:500; Abcam) were used to normalize for protein loading.
Electrophysiology Measurements
Newborn, 1-day-old, and 2-day-old mice were used for the electrophysiological experiments (10). The spinal cord was pinned ventral side up in a recording chamber and perfused with oxygenated Ringer's solution containing 111 mm NaCl, 3.08 mm KCl, 11 mm glucose, 25 mm NaHCO3, 1.18 mm KH2PO4, 1.25 mm MgSO4, and 2.52 mm CaCl2 at room temperature. Electroneurogram recordings were made by placing bipolar suction electrodes on a combination of the second and fifth lumbar ventral roots (lL2-rL2 or lL2-lL5) (10). The second lumbar ventral roots consist of primarily flexor motor axons, and the L5 ventral roots consist of primarily extensor motor axons; therefore fictive locomotion involves alternation between lL2 and lL5 as well as alternation between lL2 and rL2. Electroneurogram signals were amplified, bandpass-filtered (100 Hz-1 kHz), digitized, and collected using Axoscope software (Axon Instruments). Rhythmic fictive locomotor activity was induced by the addition of 5 μm 5-hydroxytryptamine and 10 μm N-methyl-d-aspartic acid to the Ringer's solution (10).
Multifiber Motor and Sensory Conduction
Multifiber motor and sensory conduction studies were carried out in mice briefly anesthetized with isoflurane, using protocols previously reported (11). In brief, sciatic-tibial motor fibers were supramaximally stimulated at the sciatic notch and knee, and a compound muscle action potential was recorded (base line-peak amplitude) from the motor end plate of tibial innervated dorsal interosseous foot muscles. Motor conduction velocity was calculated for the notch to knee segment. For sensory conduction, digital hind paw nerves were supramaximally stimulated, and the sciatic-tibial sensory nerve action potential (base line-peak amplitude) was recorded from the knee after averaging (5–10 times). Stimulation and recording were carried out using E2 subdermal platinum electrodes (Grass/Astromed). All of the recordings were carried out with near nerve temperatures maintained at 37.0 ± 0.5 °C.
Histological and Electron Microscopy Analyses
The mice were decapitated, and the brains were rapidly removed and flash frozen in 2-methylbutane on solid carbon dioxide (12). Serial coronal sections of 20 μm were obtained, and the sections were thaw mounted on charged slides. To visualize neuronal cell bodies and astrocytes, immunoassays were performed using antibodies recognizing the neuron-specific marker mouse anti-neuronal nuclei (1:1,000; NeuN, Chemicon) and glial fibrillary acidic protein (1:750; Dako), respectively. The fresh frozen sections were brought to room temperature, postfixed in buffered formalin, and taken through graded ethanol washes. The sections were incubated in a humidifying chamber with 1% hydrogen peroxide to quench endogenous peroxidase enzyme activity and were subsequently blocked with universal blocking serum (Dako) containing 0.2% Triton X-100. The sections were washed with phosphate-buffered saline (PBS) and incubated with the primary antibodies for 1 h at room temperature followed by the secondary antibody (rabbit anti-mouse, 1:200; Dako) for 30 min. Then the sections were washed with PBS and incubated for 30 min with an avidin-biotin complex (1:100; Vector Laboratories) and washed three times, and immunoreactivity was visualized with 3,3′-diaminobenzedine tetrahydrochloride. The sections were then rinsed, dehydrated in a series of ethanol washes, and mounted with Permount.
To assess myelinated axons, Weil's stain for myelin was used. Fresh frozen sections were postfixed in buffered formalin, rinsed with water, and dehydrated. The sections were then incubated for 45 min at 55 °C in Weil's staining solution containing 10% hematoxylin and 4% ferric ammonium sulfate solution preheated to 55 °C. The sections were then washed with tap water and differentiated macroscopically with 4% ferric ammonium sulfate and microscopically with Weigert's differentiator (potassium ferricyanide with borax).
For electron microscopy analysis, the mice were euthanized by decapitation, and their brains and spinal cord tissues were removed. The following regions of the brain were dissected: rostral spinal cord, medulla, cerebellum, diencephalon, fornix, striatum, internal capsule, corpus callosum, and motor cortex. Primary fixation was carried out at 4 °C for 4 h in a freshly prepared solution containing 2.5% glutaraldehyde and 2% paraformaldehyde in 100 mm cacodylate, pH 7.2 (13). Spinal cord and sciatic nerve samples were obtained following fixation by perfusion or by euthanasia by cervical displacement and dissection, followed by fixation by immersion. Identical results were obtained for samples fixed by tissue perfusion or by fixation by immersion of the tissue in fixative. In both cases, the fixative used was 2.5% glutaraldehyde and 0.1 m sodium cacodylate, pH 7.0. The samples were processed for electron microscopy and examined with a Hitachi Transmission Electron Microscope H-7000.
For morphological analysis of a comprehensive range of tissues, the organs were obtained following euthanasia by cervical displacement and dissection. The samples were fixed in Zinc-Formal Fixx (Fisher) overnight, then processed, and embedded in paraffin blocks, after which 5-μm sections were cut and placed onto Histobond slides (Fisher). The sections were rehydrated to water and stained with Harris hematoxylin and alcoholic eosin Y (Electron Microscopy Sciences) as per standard histology protocols, followed by mounting with Entellan medium (Electron Microscopy Sciences).
Flow Cytometry
The thymus, lymph nodes, and spleen were harvested from the mice following euthanasia. Single-cell suspensions were generated, and 2 × 106 cells were aliquoted into the wells of a 96-well plate for antibody staining. All of the antibody incubations were carried out for 30 min on ice in a fluorescence-activated cell sorting (FACS) buffer (PBS containing 1% fetal calf serum and 0.02% sodium azide), and the cells were washed twice with FACS buffer following antibody incubations. The cell events were collected with a BD FACS Canto II flow cytometer and analyzed with FlowJo software (Treestar).
Retinal Analysis
For retinal analysis of calnexin-deficient mice, the following antibodies were employed: the 1D4 monoclonal anti-rhodopsin antibodies raised against a synthetic peptide located at the C terminus of bovine rhodopsin (amino acid residues TETSQVAPA) (from R. Molday, University of British Columbia, Vancouver, Canada) (14), polyclonal rabbit anti-M-opsin antibodies (a gift from C. Craft, University of Southern California, Los Angeles, CA) (15), and polyclonal rabbit anti-melanopsin antibodies raised against a synthetic peptide located at the N terminus of mouse melanopsin (amino acid residues QTLSSLVRPGSPSDM) (a gift from I. Provencio, University of Virginia, Charlottesville, VA) (16). Whole retinas from both wild-type and calnexin-deficient mice were sonicated in Laemmli buffer, and 2.5 μg of protein were loaded per lane for detection of rhodopsin using the 1D4 antibody. For detection of calnexin, M-opsin, and melanopsin, 7.5 μg of protein was loaded per lane. The proteins were separated by electrophoresis in SDS-PAGE (12% acrylamide) and electroblotted onto nitrocellulose membranes. The immunoreactive proteins were visualized using horseradish peroxide-conjugated goat anti-mouse or anti-rabbit IgG (Invitrogen) followed by ECL detection (Amersham Biosciences).
For histological analysis of eye tissue, calnexin-deficient and wild-type mice were subjected to intracardiac perfusion with a modified Karnovsky's fixative containing 2% paraformaldehyde and 2% glutaraldehyde in 100 mm phosphate buffer. Eye cups were fixed and processed as previously described (17). 500-nm tissue sections were stained with a 0.25% azure II, 0.25% methylene blue stain containing 0.25% sodium borate in water.
Miscellaneous Procedures
Axon cultures (Campenot cultures) were set up and maintained as previously described (18). Protein concentration was estimated using a Bio-Rad DC protein assay (19). To determine the number of neurons, wild-type and cnx−/− mice were anesthetized and fixed by sequential intracardiac perfusion of PBS and 4% paraformaldehyde in PBS. Brain and whole spinal cord were dissected out, postfixed in the same fixative overnight, processed for paraffin embedding, and cut in 10-μm-thick coronal sections. After paraffin removal in xylol and graded alcohols, the sections were counterstained with cresyl violet. Spinal cord motoneurons exhibiting clear nucleus/nucleolus in layer IX were counted every 10 sections. Similarly, the number of brain stem facial motoneurons was counted every three sections. Ca2+ measurements were carried out using 1 mm Fura 2-acetoxymethyl ester as described previously (20). Ca2+ measurements were analyzed in response to 1 μm thapsigargin and 600 nm bradykinin using a Photon International Technology fluorometer at excitation of 340 nm and emission of 380 nm. For electrocardiogram analysis, ECG Leads I, II, III, AVR, and AVL of nonsedated transgenic and control mice were simultaneously recorded using E for M ECG amplifiers (PPG Biomedical Systems Inc., Pleasantville, NY) (21).
RESULTS
Calnexin-deficient Mice
Fig. 1A summarizes the gene targeting strategy used to generate the calnexin gene knock-out mice. The calnexin gene was disrupted by random gene trapping using a cassette containing the β-galactosidase-neomycin genes. Using specific primers F1, F2, and R3 (Fig. 1A), we determined the site of insertion to be preceding the first nucleotide of intron 7–8 (Fig. 1A). DNA sequence analysis confirmed that the interruption cassette was inserted directly following exon 7. Primers F1 and R3 correspond to the calnexin gene, whereas primer F2 corresponds to a sequence within the β-galactosidase gene in the insertion cassette. We used these primers for PCR-driven amplification of genomic DNA to genotype the mice. As expected, analysis of DNA isolated from wild-type mice showed only a 316-bp DNA product with F1/R3 primers (Fig. 1B) and no DNA product when F2/R3 primers were used (Fig. 1B). Analysis of genomic DNA from cnx−/− mice showed amplification of a 941-bp DNA product with the use of F2/R3 primers and no DNA product with primers F1/R3, indicating that both alleles of the calnexin gene were interrupted by the insertion cassette (Fig. 1B). In contrast, PCR analysis of genomic DNA from heterozygote mice with F1/R3 and F2/R3 primers produced both 316- and 941-bp DNA fragments corresponding to the presence of both wild-type and calnexin gene interrupted alleles, respectively (Fig. 1B). To determine that there was no mRNA alternative splicing around the interruption cassette, RT-PCR analysis was carried out of RNA isolated from wild-type, heterozygote, and calnexin-deficient brain tissue using a specific set of primers (Fig. 1C). Fig. 1D shows that mRNA encoded by exons 1–7, prior to the interruption cassette, was transcribed with no detectable alternative splicing near the interruption cassette. No RT-PCR product was detected with primers covering exons 8–15 of the calnexin gene (Fig. 1, C and D). Similarly, no RT-PCR product was seen when primers 7F (exon 7) and primers 8R and 12R (exon 8 and 12, respectively) were used (Fig. 1, C and D). As expected, RT-PCR analysis of cnx+/− and cnx−/− RNA, but not wild-type RNA, with primers covering exon 7 and insertion cassette, resulted in a specific DNA product (Fig. 1D). Western blot analysis revealed that there was no detectable expression of calnexin protein when both alleles of the gene were interrupted (Fig. 1, E and F). Identical results were obtained with a calnexin antibody specific for either the N-terminal ER luminal portion (Fig. 1E) or an antibody that recognizes the C-terminal cytoplasmic domain (Fig. 1F). We concluded that the expression of calnexin protein was fully inactivated.
Absence of Calnexin Results in Impaired Nerve Conduction Velocity
The heterozygote mice had a normal phenotype, being viable and fertile. Intercrossing of heterozygote females and males was carried out to generate homozygote calnexin-deficient mice. In stark contrast to the results of Denzel et al. (1), we did not observe early postnatal death in mice with the complete loss of calnexin. Instead, newborn calnexin-deficient mice were indistinguishable from wild-type and heterozygote littermates with respect to their size, weight, and external appearance. However, a size difference between wild-type and calnexin-deficient (cnx−/−) mice became apparent as early as 7 days after birth, and a marked size discrepancy was evident 14–16 days following birth, resulting in cnx−/− mice that are 30–50% smaller than their wild-type littermates. Calnexin-deficient mice showed neurological abnormalities manifested by a gait disturbance with instability, splaying of the hind limbs, ataxia, tremors, lower limb motor defects, and a rolling walk (Fig. 2A and supplemental video).
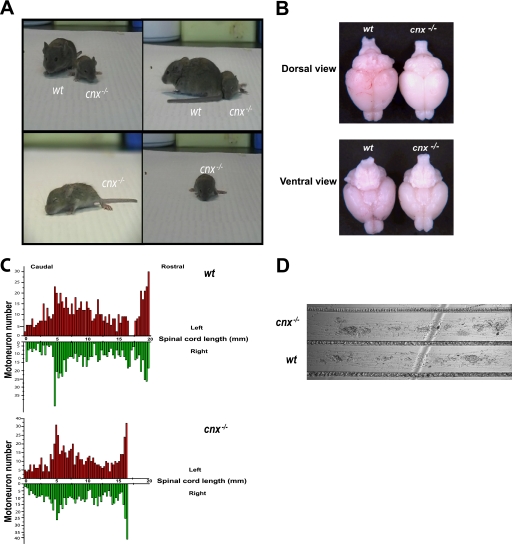
FIGURE 2.
Calnexin in neuronal tissue. A, video clips of wild-type (wt) and calnexin-deficient sibling mice. The full-length video is available as supplemental material. Calnexin-deficient mice are smaller than their wild-type counterparts, with splaying of the hind limbs and gait instability. B, cnx−/− and wt brains isolated from 21-day-old mice, viewed from the dorsal and ventral side. There were no morphological differences in the anatomical structure between cnx−/− and wt brains as examined in multiple animals. C, analysis of motor neuron number in wild-type (wt) and calnexin-deficient (cnx−/−) mice. The spinal cord length is shorter in cnx−/− mice, but the spinal motoneuron distribution and number were similar to that in wild-type mice. In red are the neurons counted on the left side of the spinal cord; in green are the neurons counted on the right side of the spinal cord. Motoneurons were counted in two sets of wild-type and calnexin-deficient mice. D, sympathetic neurons from day 1 neonate cnx−/− and wild-type (wt) mice were grown in compartmentalized cultures (Campenot cultures). Axonal growth was examined in three sets of wild-type and calnexin-deficient cultures. The absence of calnexin had no effect on axonal growth.
To evaluate neuronal status, we carried out morphological analysis of the brain tissue, counted motoneurons of the spinal cord, and examined neuronal growth and function in the absence of calnexin. We did not observe any significant changes in the gross morphology of the brain in calnexin-deficient mice (Fig. 2B). Examination of the motoneuron distribution in the spinal cord indicated that although the spinal cord was shorter in the absence of calnexin (consistent with their smaller size), the motoneuron distribution was comparable with that of wild type (Fig. 2C). Furthermore, a careful count of motoneurons in wild-type and calnexin-deficient mice revealed no difference in the number of motoneurons (Fig. 2C). Similarly, amplitudes of the compound muscle action potentials, indices of motor axon innervations, were not altered in cnx−/− mice. Neuronal growth in cnx−/− neurons was investigated by culturing sympathetic neurons in compartmentalized cultures (18). We observed no difference in neuronal growth in the absence of calnexin (Fig. 2D).
To assess neuron status in calnexin-deficient mice, we examined pharmacologically induced lower limb walking movements in the isolated spinal cord. To evoke fictive locomotion (characterized by the oscillatory bursting of motor neurons in a step cycle period of 2–4 s), we applied 5 μm 5-hydroxytryptamine (serotonin) and 10 μm N-methyl-d-aspartic acid (Fig. 3A) to isolated spinal cords (10) taken from wild-type and cnx−/− neonatal mice. Electroneurograms were recorded from the second and fifth lumbar ventral root on the left side (i.e. IL2, IL5) and the second lumbar ventral root on the left and right side (i.e. IL2, rL2). Appropriate alternation between bursts was noted in wild-type and calnexin-deficient preparations (Fig. 3A), indicating that the fictive locomotor pattern was undisturbed in the cnx−/− mouse.
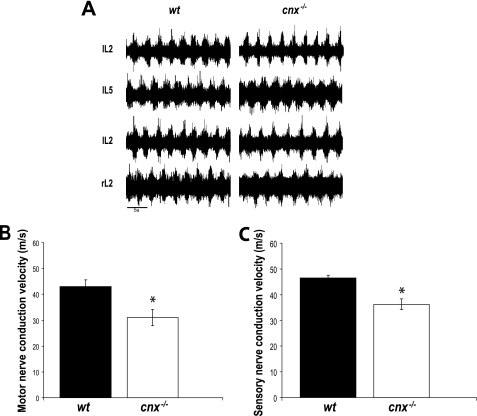
FIGURE 3.
Functional analysis of calnexin in neuronal tissue. A, the fictive locomotor pattern is undisturbed in the cnx−/− mouse. Electroneurograms were recorded from the second and fifth lumbar ventral root on the left side (i.e. IL2, IL5) and the second lumbar ventral root on the left and right side (i.e. IL2, rL2) of the spinal cord in the wild-type (wt, left traces) and cnx−/− (right traces) mice. Fictive locomotion was evoked with 5 μm serotonin (5-hydroxytryptamine) and 10 μm N-methyl-d-aspartic acid. The spinal cords used were obtained from newborn, 1-day-old, and 2-day-old mice (n = 4). Note the appropriate alternation between bursts in both cases. B, motor nerve conductive analysis of wild-type (wt) and calnexin-deficient (cnx−/−) mice. Motor nerve conduction velocity was significantly slower in the absence of calnexin. C, sensory nerve conductive analysis of wild-type (wt) and calnexin-deficient (cnx−/−) mice. In the absence of calnexin, there was reduced sensory nerve conduction velocity. Six sex-matched sets of wild-type and calnexin-deficient mice of 4–5 months of age were examined for motor and sensory nerve conduction. The asterisks indicate significant differences (p = 0.01).
Next, we tested for electrophysiological parameters of motor and sensory neurons in calnexin-deficient and wild-type mice. Fig. 3B shows that motor nerve conduction velocities were significantly slowed in the absence of calnexin. There was a significant difference between wild-type and calnexin-deficient motor conduction velocities at values of 43.1 ± 2.5 and 31.0 ± 3.2 m/s, respectively (n = 4; p = 0.01) (Fig. 3B). Wild-type and calnexin-deficient mouse compound muscle action potential amplitudes were preserved at 9.2 and 10.0 mV, respectively. Sensory nerve conduction velocities were also reduced in the cnx−/− mice (Fig. 3C). The sensory nerve conduction velocity in wild-type mice was 46.1 m/s with a significant decrease in sensory nerve conduction velocity (38.2 m/s) in the absence of calnexin (Fig. 3C). The amplitude of the sensory nerve action potentials in the wild-type mice was also comparable between the groups: 14.1 μV in controls compared with 10.9 μV in the calnexin-deficient animals. The sensory nerve action potential amplitude reflects the number of excitable myelinated axons that can be recruited by stimulation. Taken together, these results indicate that neuronal growth and neuron number were not altered in the absence of calnexin, but there was a significant decrease in the nerve conduction velocity in cnx−/− mice.
Dysmyelination in the Calnexin-deficient Mouse
Myelin surrounds axons and allows for rapid nerve conduction that is essential to nervous system function. Loss of myelin leads to reduced nerve conduction velocity, and therefore we tested whether myelination was affected in the absence of calnexin. First, we carried out electron microscopic analysis of spinal cord and sciatic nerve in calnexin-deficient mice to examine, at a higher resolution, whether myelin formation was impaired in the absence of calnexin. Calnexin-deficient spinal cords had a thinner, wavy, and decompacted myelin in the absence of calnexin (Fig. 4, A–D), indicating that the absence of calnexin affects myelination of the spinal cord. A different kind of myelination defect was apparent in the sciatic nerve. Electron microscopic analysis of calnexin-deficient sciatic nerve revealed, in addition to wavy and decompacted myelin, a hypermyelination that appeared to invade the neuronal areas (Fig. 4, E–H). The findings resembled “G fibers” or tomaculae that are described in human hereditary neuropathy with sensitivity to pressure palsy or focally folded myelin described in Charcot-Marie-Tooth disease 4B. To test whether calnexin deficiency resulted in a reduced amount of myelin in nervous tissue (hypomyelination), the g ratio was calculated for calnexin-deficient and wild-type spinal cord and sciatic nerve. The g ratio is defined as the ratio of the axonal diameter divided by the diameter of the axon plus the thickness of its myelin sheath. Calculation of the g ratio revealed that calnexin deficiency resulted in modest hypomyelination in the spinal cord and did not affect myelin sheath thickness in the sciatic nerve (cnx−/− spinal cord g ratio was 0.78 ± 0.05 (n = 40) compared with wild-type spinal cord at 0.71 ± 0.05 (n = 40); cnx−/− sciatic nerve g ratio was 0.73 ± 0.09 (n = 10) compared with wild-type sciatic nerve at 0.71 ± 0.07 (n = 10); n represents the number of neurons measured from representative electron micrographs). We concluded that in the absence of calnexin there was no significant reduction in myelin but defective formation and compaction of myelin sheaths. These findings indicate significant changes in the peripheral and central nervous systems of calnexin-deficient animals and help to explain the neuronal phenotype and decreased nerve conduction velocity in cnx−/− mice.
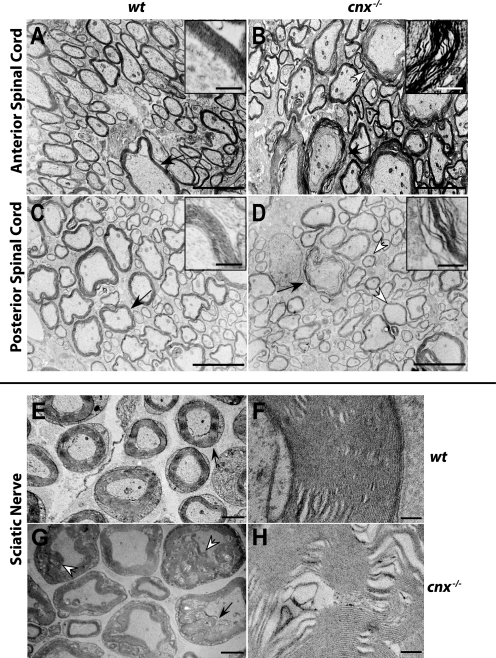
FIGURE 4.
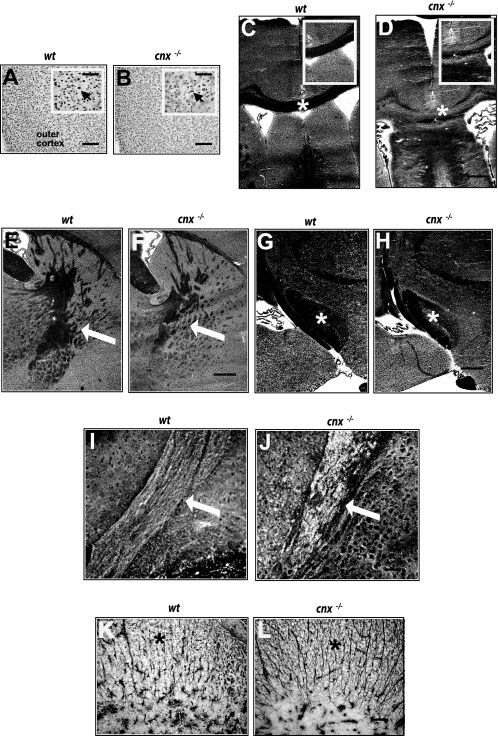
Dysmyelination in the spinal cord and sciatic nerve in the absence of calnexin. Electron micrographs demonstrate dysmyelination. Representative electron microscopy micrographs shown here depict spinal cords and sciatic nerves from 21-day old mice, although dysmyelination was observed in mice from 3 weeks to 2 years of age. Four sets of 21-day-old sex-matched mice were examined, using either tissue fixation by immersion or perfusion. Areas of wavy, decompacted myelin and hypomyelination appear in anterior (A and B) and posterior (C and D) regions of the spinal cord of cnx−/− mice. The black arrows indicate areas of myelin shown magnified in the insets. Myelin abnormalities also appeared in the sciatic nerve (E and G). Redundant myelin folds are noted. F and H are higher magnifications of the regions indicated by black arrows in E and G, respectively. The white arrows indicate the areas of wavy myelination (B), hypomyelination (D), and aberrant myelination (G). A–D, scale bar, 5 μm; inset, scale bar, 500 nm; E and G, scale bar, 2 μm; F and H, scale bar, 200 nm. wt, wild type; cnx−/−, calnexin-deficient.
Histological analysis of the brain tissue from wild-type and cnx−/− animals was carried out to evaluate the consequences of calnexin deficiency on various regions of the central nervous system. In all of the brain regions examined, neuronal cell bodies, visualized using anti-neuronal specific nuclear protein (NeuN) antibodies, appeared normal and healthy in both the wild-type (Fig. 5A) and calnexin-deficient (Fig. 5B) mice. However, large white matter tracts were variably affected. In calnexin-deficient animals the rostral corpus callosum was thinner and, particularly at the medial rise, displayed areas of patchy and irregular myelination or dysmyelination (Fig. 5, C and D). However, the axons appeared to be spared. In the absence of calnexin, the internal capsule (Fig. 5F) displayed less branching and was narrower than that observed in the wild type (Fig. 5E), suggesting fewer myelinated fibers traveling between the periphery and cerebral cortex. Patchy areas of myelination were evident in the cerebral peduncle of the calnexin-deficient mouse (Fig. 5, G and H). The cerebellar peduncles of the cnx−/− mouse were also characterized by a patchy, loose myelination pattern (Fig. 5, I and J). At low resolution, white matter tracts of the spinal cord did not show obvious dysmyelination, and the cell bodies of the horns appeared normal and healthy. An increased number of glial fibrillary acid protein-positive astrocytic fibers was observed in the absence of calnexin (Fig. 5L) compared with the wild type (Fig. 5K). The perpendicular organization of the glial fibers in the calnexin knock-out mice was similar to that observed early in central nervous system development, suggesting that in the absence of calnexin, spinal cord development was altered.
FIGURE 5.
Dysmyelination in calnexin-deficient mice. A and B, antibodies to neuronal specific nuclear protein (NeuN) were used to visualize neuronal cell bodies in all brain regions. A and B, representative photomicrographs of the premotor cortex are shown for wt (A) and cnx−/− (B). Normal cortical cytoarchitecture was observed, and neurons appeared normal and healthy (arrows point to NeuN positive neurons). Scale bar, 50 μm. Inset, scale bar, 10 μm. C and D, examination of large white matter tracts showed that the rostral corpus callosum was thinner and contained patchy areas of myelination in the absence of calnexin (D). The asterisks in C and D identify rostral corpus callosum and areas magnified for insets. Scale bar, 50 μm. Inset, scale bar, 20 μm. E and F, compared with wild type (E), the overall amount of myelin in the internal capsule (identified with arrows) was reduced in cnx−/− mice (F). Scale bar, 50 μm. G and H, the cerebral peduncle (identified with asterisks) in the cnx−/− mouse (H) contained areas of patchy myelin compared with wild type (G). Scale bar, 50 μm. I and J, the cerebellar peduncles (identified with arrows) in the cnx−/− mouse (J) showed a patchy, loose myelination pattern as compared with wild-type animals (I). Scale bar, 20 μm. K and L, in the white matter of the spinal cord, an increased number of glial fibrillary acid protein-positive astrocytic fibers (identified with asterisks) was observed in the absence of calnexin (L) compared with the wild type (K). Scale bar, 10 μm. Three sets of 6–8-week-old wild-type and calnexin-deficient mice were examined. wt, wild type; cnx−/−, calnexin-deficient.
Calnexin Deficiency Specifically Affects Myelination
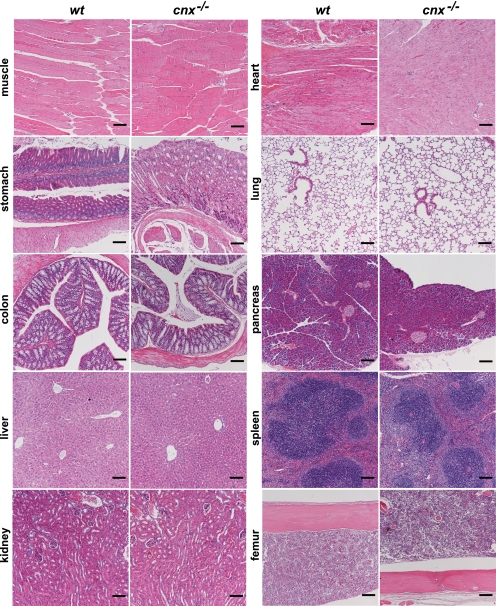
Considering that calnexin is a ubiquitously expressed ER-associated protein, both the specificity of the neurological phenotype and the effect on myelination in cnx−/− mice were surprising. We expected that calnexin deficiency may also affect other tissues, and this may have been masked by the predominant neurological phenotype described above. However, we found no gross histological abnormalities in heart, lung, pancreas, spleen, femur, skeletal muscle, colon, liver, kidney, or stomach in the absence of calnexin (Fig. 6).
FIGURE 6.
Histological analysis of calnexin-deficient tissues. Wild-type (wt) and calnexin-deficient (cnx−/−) tissues were stained with hematoxylin and eosin. No significant morphological changes were observed between wild-type and calnexin-deficient tissues. Representative pictures are shown from the three sets of 21-day-old wt and cnx−/− mice examined. Scale bar, 100 μm.
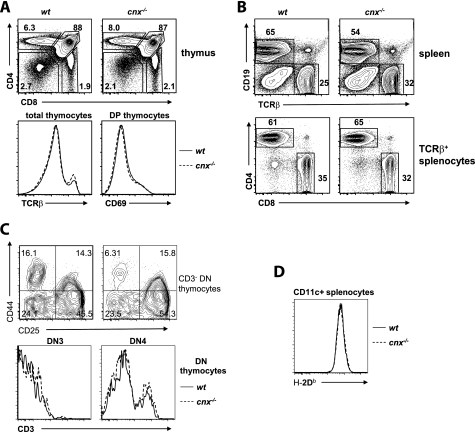
Given the role that calnexin plays in the early events of MHC Class I protein folding (22) and the fact that calnexin is able to associate with cell surface CD3 complexes (23) and regulate T cell receptor (TCR) assembly (24), we anticipated that the elimination of calnexin would lead to aberration of these processes. Unexpectedly, calnexin deficiency had no impact on the immune system (Fig. 7). The CD4/CD8 profiles of wild-type and cnx−/− thymocytes were indistinguishable (Fig. 7A), and the number of thymocytes recovered from each strain was similar. We did not detect any differences in CD25/CD44 expression within the CD4−CD8− double negative compartment or changes in the expression of MHC Class I in any thymocyte subpopulation in cnx−/− mice (Fig. 7C). The expression of TCRβ on bulk thymocytes and CD69 on CD4+CD8+ double positive thymocytes was also identical (Fig. 7A), suggesting that positive selection occurred normally in the absence of calnexin. Maturation of CD8 single positive thymocytes as evaluated by CD24 down-regulation occurred normally in cnx−/− mice. The splenic lymphocyte population also appeared grossly normal in cnx−/− mice (Fig. 7B). We did notice a decrease in splenic cellularity in cnx−/− mice; however, we attributed this to the reduced size and weight of cnx−/− animals. The percentage of T cells in the spleen as well as the CD4/CD8 profile of the T cell population was unaffected by calnexin deficiency (Fig. 7B). Again, no dramatic change in cell surface marker expression (CD44, CD62L, CD25, TCRβ, and MHC Class I) was observed in cnx−/− T cells (not shown and Fig. 7B). Finally, we examined MHC Class I expression on dendritic cells from cnx−/− mice and found no change when compared with the wild-type mice (Fig. 7D). Overall, the thymus and peripheral T cell populations appeared normal in cnx−/− mice. These results demonstrate that the absence of calnexin had no significant impact on the immune system, indicating that this protein is not essential for the formation of the immune system in mice.
FIGURE 7.
Calnexin deficiency does not affect the immune system. A, thymocytes from wild-type and cnx−/− mice were probed with anti-CD4, anti-CD8, anti-CD69, and anti-TCRβ antibodies. The cells were analyzed by flow cytometry. The CD4/CD8 profile of bulk thymocytes (top row), TCRβ expression on bulk thymocytes, and CD69 expression on CD4+CD8+ thymocytes from wild-type (wt, solid line) or cnx−/− (broken line) mice are shown (bottom row). B, splenocytes from wild-type and cnx−/− mice were stained with anti-CD19, anti-TCRβ, anti-CD4, and anti-CD8 antibodies followed by flow cytometry analysis. The CD19/TCRβ profile of bulk splenocytes (top row) and the CD4/CD8 profile of TCRβ+ cells are depicted (bottom row). C, cells were probed with anti-CD25 and anti-CD44 followed by FACS analysis. There are neither detectable differences in CD25/CD44 expression within the CD4−CD8− double negative compartment nor changes in the expression of MHC Class I in any thymocyte subpopulation in calnexin-deficient mice. D, analysis of H-2Db expression in wild-type (wt) and calnexin-deficient (cnx−/−) cells. There is no change in MHC Class I expression on dendritic cells from cnx−/− mice when compared with the wild-type mice.
DISCUSSION
Calnexin is a molecular lectin-like chaperone, and together with calreticulin, the protein promotes folding of glycosylated proteins (2). In this study we showed that the loss of calnexin has a specific and detrimental effect on myelin formation. We have carried out a systematic analysis of the impact of calnexin deficiency in mice. We found that a bona fide calnexin deficiency results in a neurological disorder resulting not from a loss of neuronal fibers (1) but instead from myelin defects in both the central and peripheral nervous systems. We did not observe any abnormalities in neuronal growth or fictive locomotor pattern in the absence of calnexin. However, cnx−/− mice exhibit dysmyelination as documented by reduced conductive velocity of nerve fibers and electron microscopy analysis of sciatic nerve and spinal cord. Most importantly, nerve conduction abnormalities and decompacted myelin similar to that observed in calnexin-deficient mice are the hallmark of Charcot-Marie-Tooth diseases and other demyelinating/dysmyelinating neuropathies (26).
Previously, Denzel et al. (1) reported that their strain of mice with a disrupted calnexin gene exhibited early postnatal death. In stark contrast to that report, we did not observe postnatal lethality in the cnx−/− mice produced in our laboratory (this study). The calnexin-deficient mice generated in the present study did not have a reduced number of neurons, they were fertile, and they had a normal lifespan. The discrepancy between this study and that of Denzel et al. (1) may represent differences in the genetic background of the animals and/or the difference between only partial inactivation (25) and full disruption of the calnexin gene (this study). Regrettably, the cause of early postnatal death was not investigated (1), and the calnexin gene-disrupted mouse strain generated by Denzel et al. is no longer available (25). The early postnatal death of the calnexin gene-disrupted mice reported by Denzel et al. (1) precludes the meaningful analysis of the molecular consequences of calnexin deficiency. Nevertheless, three of their calnexin gene-disrupted mice lived up to 3 months (1), and these mice, which represented less than 5% of the calnexin gene-disrupted mouse population, were subjected to further analysis (1). These mice were reported to have a reduction in the number of large myelinated nerve fibers, although unfortunately the definition of large myelinated fibers was not provided. Furthermore, statistical analysis of the findings was not carried out, and the authors attributed the large variation in their numbers to the age and sex differences of two sets of one wild-type and one calnexin-deficient mouse. It is unlikely that the data reported in the earlier study (1) represents an accurate assessment of the phenotype of mice with a fully documented deficiency of calnexin.
Considering that calnexin is a ubiquitously expressed ER-associated protein, both the specificity of the neurological phenotype and the effect on myelination in cnx−/− mice were surprising. We expected that calnexin deficiency may also affect other tissues, and this may have been masked by the predominant neurological phenotype described above. We show that adult cnx−/− mice have no discernible abnormalities in the immune system, indicating that the role of calnexin in the development of the immune system is dispensable. This was unexpected considering the importance of calnexin and ER quality control in MHC Class I protein folding (22) and its association with CD3 complexes (23) and TCR (24).
Calnexin and calreticulin are structurally and functionally similar lectin-like chaperones of the ER (27). Calreticulin-deficient mice are embryonically lethal because of a defect in cardiac development (8). Despite the structural and functional similarities between calnexin and calreticulin, the complete loss of calnexin did not have any effect on cardiac development and function or on cellular Ca2+ homeostasis (supplemental Fig. S1). These findings further support our conclusions that calnexin plays a critical and specific role during myelination. The molecular chaperone function of calnexin is essential for proper formation of the myelin sheaths, for which there are no compensatory mechanisms provided by other ER chaperones, including homologous calreticulin. Yet, evidently, there must be compensatory redundancy in the other tissues of calnexin-deficient mice, including in the immune system.
Mutations in the Drosophila homolog of the calnexin gene (calnexin 99A) lead to severe defects in rhodopsin expression (28), suggesting that calnexin deficiency may also affect visual pigments. However, the complete lack of calnexin did not affect expression of rhodopsin, M-opsin, or melanopsin in mouse retinas. Consistent with Western blot analysis, the photoreceptor outer segments did not display significant defects in the cnx−/− mice (supplemental Fig. S2). However, there was an increased number of nuclei in the outer and inner nuclear layers, and the nuclei were disorganized. There was also vacuolization in the retinal pigment epithelial layer, indicating that calnexin was required for proper function of the retinal pigment epithelial layer and the retina.
The systematic and comprehensive analysis described here reveals that calnexin plays an important role in the pathogenesis of peripheral neuropathies. The specificity of calnexin deficiency toward myelin proteins and myelination is intriguing and supports early observations of transient association of myelin glycoprotein PMP22 with calnexin (29). PMP22 accounts only for a small fraction of peripheral nervous system myelin, whereas P0 is a major peripheral nervous system myelin glycoprotein (30). Both myelin proteins are involved in the compaction and maintenance of myelin (30). In the absence of calnexin, it is possible that PMP22 folding and function are modified, leading to dysmyelination. Our findings have identified a previously unknown role for calnexin in myelination and myelin diseases and as a novel contributor to the diversity of neurological disorders.
Supplementary Material
Acknowledgments
We thank S. Aldred, M. Dabrowska, P. Gajda, and A. Thorne for superb technical support. We thank R. Campenot for culturing neurons and C. Craft, I. Provencio, R. Molday and L. Notterpek for antibodies. We thank K. Reue and L. Vergnes for the inverse PCR protocol. We thank N. Nation for help with histology and gross tissue analysis. We thank S. Steiner for help with the initial electron microscopy analysis. We thank B. Lemire and R. C. Bleackley for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants EYEY008768 (to N. J. C.) and F31AG032176 (to E. E. R.). This work was also supported by Grants MOP-15291 (to M. M.), MOP-8659 (to T. A. B.), and MOP-86470 (to S. G.) and other grants (to D. Z. and K. T.) from the Canadian Institutes of Health Research and grants from the Swiss National Science Foundation (to K.-H. K.), the Retina Research Foundation (to N. J. C.), Alberta Innovates-Health Solutions (to S. G.), the March of Dimes (to S. G.), and the Davey Fund for Brain Research (to K. T.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and movie.
- ER
- endoplasmic reticulum
- MHC
- major histocompatibility complex
- RT
- reverse transcription
- PBS
- phosphate-buffered saline
- FACS
- fluorescence-activated cell sorting
- TCR
- T cell receptor.
REFERENCES
- 1.Denzel A., Molinari M., Trigueros C., Martin J. E., Velmurgan S., Brown S., Stamp G., Owen M. J. (2002) Mol. Cell. Biol. 22, 7398–7404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hebert D. N., Molinari M. (2007) Physiol. Rev. 87, 1377–1408 [DOI] [PubMed] [Google Scholar]
- 3.Parlati F., Dignard D., Bergeron J. J., Thomas D. Y. (1995) EMBO J. 14, 3064–3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Müller-Taubenberger A., Lupas A. N., Li H., Ecke M., Simmeth E., Gerisch G. (2001) EMBO J. 20, 6772–6782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fajardo M., Schleicher M., Noegel A., Bozzaro S., Killinger S., Heuner K., Hacker J., Steinert M. (2004) Microbiology 150, 2825–2835 [DOI] [PubMed] [Google Scholar]
- 6.Lee W., Kim K. R., Singaravelu G., Park B. J., Kim D. H., Ahnn J., Yoo Y. J. (2006) Proteomics 6, 1329–1339 [DOI] [PubMed] [Google Scholar]
- 7.Xu K., Tavernarakis N., Driscoll M. (2001) Neuron 31, 957–971 [DOI] [PubMed] [Google Scholar]
- 8.Mesaeli N., Nakamura K., Zvaritch E., Dickie P., Dziak E., Krause K. H., Opas M., MacLennan D. H., Michalak M. (1999) J. Cell Biol. 144, 857–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin V., Groenendyk J., Steiner S. S., Guo L., Dabrowska M., Parker J. M., Müller-Esterl W., Opas M., Michalak M. (2006) J. Biol. Chem. 281, 2338–2346 [DOI] [PubMed] [Google Scholar]
- 10.Gosgnach S., Lanuza G. M., Butt S. J., Saueressig H., Zhang Y., Velasquez T., Riethmacher D., Callaway E. M., Kiehn O., Goulding M. (2006) Nature 440, 215–219 [DOI] [PubMed] [Google Scholar]
- 11.Toth C., Martinez J. A., Liu W. Q., Diggle J., Guo G. F., Ramji N., Mi R., Hoke A., Zochodne D. W. (2008) Neuroscience 154, 767–783 [DOI] [PubMed] [Google Scholar]
- 12.Jantzie L. L., Cheung P. Y., Todd K. G. (2005) J. Cereb. Blood Flow Metab. 25, 314–324 [DOI] [PubMed] [Google Scholar]
- 13.Lozyk M. D., Papp S., Zhang X., Nakamura K., Michalak M., Opas M. (2006) BMC Dev. Biol. 6, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.MacKenzie D., Arendt A., Hargrave P., McDowell J. H., Molday R. S. (1984) Biochemistry 23, 6544–6549 [DOI] [PubMed] [Google Scholar]
- 15.Zhu X., Brown B., Li A., Mears A. J., Swaroop A., Craft C. M. (2003) J. Neurosci. 23, 6152–6160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Provencio I., Jiang G., De Grip W. J., Hayes W. P., Rollag M. D. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 340–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colley N. J., Cassill J. A., Baker E. K., Zuker C. S. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 3070–3074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacInnis B. L., Campenot R. B. (2002) Science 295, 1536–1539 [DOI] [PubMed] [Google Scholar]
- 19.Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 20.Mery L., Mesaeli N., Michalak M., Opas M., Lew D. P., Krause K. H. (1996) J. Biol. Chem. 271, 9332–9339 [DOI] [PubMed] [Google Scholar]
- 21.Nakamura K., Robertson M., Liu G., Dickie P., Nakamura K., Guo J. Q., Duff H. J., Opas M., Kavanagh K., Michalak M. (2001) J. Clin. Invest. 107, 1245–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wearsch P. A., Cresswell P. (2008) Curr. Opin. Cell Biol. 20, 624–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wiest D. L., Burgess W. H., McKean D., Kearse K. P., Singer A. (1995) EMBO J. 14, 3425–3433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bennett M. J., Van Leeuwen J. E., Kearse K. P. (1998) J. Biol. Chem. 273, 23674–23680 [DOI] [PubMed] [Google Scholar]
- 25.Kosmaoglou M., Cheetham M. E. (2008) Mol. Vis. 14, 2466–2474 [PMC free article] [PubMed] [Google Scholar]
- 26.Berger P., Niemann A., Suter U. (2006) Glia 54, 243–257 [DOI] [PubMed] [Google Scholar]
- 27.Michalak M., Groenendyk J., Szabo E., Gold L. I., Opas M. (2009) Biochem. J. 417, 651–666 [DOI] [PubMed] [Google Scholar]
- 28.Rosenbaum E. E., Hardie R. C., Colley N. J. (2006) Neuron 49, 229–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dickson K. M., Bergeron J. J., Shames I., Colby J., Nguyen D. T., Chevet E., Thomas D. Y., Snipes G. J. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 9852–9857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quarles R. H. (2007) J. Neurochem. 100, 1431–1448 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.