Abstract
Pannexin 3 (Panx3) is a new member of the gap junction pannexin family, but its expression profiles and physiological function are not yet clear. We demonstrate in this study that Panx3 is expressed in cartilage and regulates chondrocyte proliferation and differentiation. Panx3 mRNA was expressed in the prehypertrophic zone in the developing growth plate and was induced during the differentiation of chondrogenic ATDC5 and N1511 cells. Panx3-transfected ATDC5 and N1511 cells promoted chondrogenic differentiation, but the suppression of endogenous Panx3 inhibited differentiation of ATDC5 cells and primary chondrocytes. Panx3-transfected ATDC5 cells reduced parathyroid hormone-induced cell proliferation and promoted the release of ATP into the extracellular space, possibly by action of Panx3 as a hemichannel. Panx3 expression in ATDC5 cells reduced intracellular cAMP levels and the activation of cAMP-response element-binding, a protein kinase A downstream effector. These Panx3 activities were blocked by anti-Panx3 antibody. Our results suggest that Panx3 functions to switch the chondrocyte cell fate from proliferation to differentiation by regulating the intracellular ATP/cAMP levels.
Keywords: Cell Differentiation, Cyclic AMP (cAMP), Gap Junctions, Gene Expression, Protein Kinase A (PKA)
Introduction
Cartilage plays an important role in mechanical load resistance and in skeletal structure support. It also serves as the skeletal template for endochondral ossification by which most bones in the body, such as long bones, are formed. In endochondral ossification, cartilage development is initiated by mesenchymal cell condensation, followed by a series of proliferation and differentiation processes. Cells undergoing condensation differentiate into chondrocytes, which then proliferate, produce type II collagen and form the proliferative zone of the cartilage molds. As development proceeds, chondrocytes in the center of the cartilage molds (prehypertrophic zone) cease proliferating and differentiate into type X collagen-producing hypertrophic chondrocytes to form the hypertrophic zone. Terminally differentiated hypertrophic chondrocytes mineralize the surrounding matrix. Eventually these cells die by apoptosis and are replaced by osteoblasts that form trabecular bone.
The regulation of chondrocyte proliferation and differentiation must be tightly coordinated to allow formation of properly sized cartilage and bone (1). Parathyroid hormone-related peptide (PTHrP)2 and parathyroid hormone (PTH) sustain chondrocyte proliferation and delay differentiation of the growth plate (2). PTHrP is expressed by perichondrial cells and chondrocytes in the upper region of growing cartilage. Mutant mice that are deficient in PTHrP (3), PTH (4), or its receptor (5) have short proliferative zones and accelerated chondrocyte differentiation, which results in abnormal endochondral bone formation. In contrast, mice that overexpress PTHrP have enlarged proliferative zones and delayed chondrocyte terminal differentiation (6). Humans with an activating mutation in the PTH/PTHrP receptor develop Jansen metaphyseal chondrodysplasia, characterized by disorganization of the growth plates and delayed chondrocyte terminal differentiation (7). These results suggest that PTH/PTHrP signaling regulates skeletal development by promoting cell proliferation and inhibiting hypertrophic differentiation of chondrocytes.
The binding of PTH/PTHrP to its receptor activates both Gs and Gq family heterotrimeric G proteins (8, 9). The activation of Gs is necessary for cAMP production and protein kinase A (PKA) activation, which leads to phosphorylation of the cAMP-response element-binding (CREB) family of transcription factors. CREB then induces genes such as the cyclin D1 and cyclin A genes. The activated cyclin/cyclin-dependent kinases in turn phosphorylate the retinoblastoma protein and its relative factors, which then dissociates the E2F transcription factor and subsequently activates the target genes necessary for DNA replication and cell cycle progression. Thus, CREB is a direct target of PKA and a downstream target of PTH/PTHrP/cAMP signaling and is required for chondrocyte proliferation (10, 11). How proliferation signaling is down-regulated in the prehypertrophic zone to stop proliferation and allow the switch to the postmitotic state is not well understood.
Pannexins, relatives of innexins that had been considered as exclusively invertebrate gap junction proteins, were recently discovered as candidates for a second family of gap junction proteins in vertebrates (12). Although there are some similarities in domain structures between pannexins and connexins, which are well characterized as vertebrate gap junction proteins, these two protein families have no sequence homology (13). The pannexin family has three members as follows: pannexin 1 (Panx1), pannexin 2 (Panx2), and pannexin 3 (Panx3). These proteins were originally identified in the human and mouse genomes (12). The expression of Panx1 is observed in many organs, such as the eye, thyroid, prostate, kidney, and liver, but its expression is especially strong in both the developing and mature central nervous system (12–14). Panx2 is preferentially expressed in the central nervous system. Recently, it was reported that Panx3 is expressed in skin, cartilage, and cochlea (15–17).
In Xenopus oocytes, Panx1 forms both nonjunctional hemichannels and intercellular channels and interacts with Panx2 (18). Panx1 hemichannels are stress-sensitive conduits for ATP (19). Panx1 is a Ca2+-permeable ion channel that is localized on both the ER and plasma membrane and participates in ER Ca2+ leakage and intercellular Ca2+ movement (20). Both Panx1 and Panx3 are glycoproteins, and N-glycosylation of these pannexins plays a role in intracellular trafficking and functional channel function (15, 16). The physiological function of pannexins in cell differentiation has not yet been characterized however.
In this study, we report that Panx3 regulates the proliferation and differentiation of chondrocytes. Panx3 mRNA was expressed in prehypertrophic chondrocytes and induced during the differentiation of chondrogenic ATDC5 cells. The transfection of Panx3 into ATDC5 cells promoted ATDC5 cell differentiation, whereas the inhibition of endogenous Panx3 by shRNA blocked differentiation. Panx3 promoted ATP release into the extracellular space and inhibited PTH-mediated cell proliferation, intracellular levels of cAMP, and phosphorylation of CREB. Thus, our results suggest that Panx3 regulates the transition of proliferation to differentiation in chondrocytes.
EXPERIMENTAL PROCEDURES
Reagents
Insulin/transferrin/sodium selenite (ITS) was obtained from Sigma. Recombinant rat PTH(1–34) was purchased from Bachem. Recombinant human BMP2 was purchased from Humanzyme.
In Situ Hybridization
Digoxigenin-11-UTP-labeled, single-stranded antisense RNA probes for Panx3, Ihh, Col2a1, Col10a1, and Hist1h4c were prepared using the DIG RNA labeling kit (Roche Applied Science) according to the manufacturer's instructions. In situ hybridization of the tissue sections was performed essentially according to the protocol provided with Link-Label ISH core kit (Biogenex). Frozen tissue sections from growth plates (E16.5) were generated and placed on RNase-free glass slides. After drying the frozen sections for 10 min at room temperature and incubating at 37 °C for 30 min, the sections were treated with 10 μg/ml proteinase K at 37 °C for 30 min. Hybridization was performed at 37 °C for 16 h, and washes were carried out with 2× SSC at 50 °C for 15 min and 2× SSC containing 50% formamide at 37 °C for 15 min. The slides were then subjected to digestion with 10 μg/ml RNase A in 10 mm Tris-HCl (pH 7.6), 500 mm NaCl, and 1 mm EDTA at 37 °C for 15 min and then washed. The sections were treated with 2.4 mg/ml Levamisole (Sigma) to inactivate endogenous alkaline phosphatase.
Antibody for Panx3
A rabbit polyclonal antibody to a peptide (amino acid residues, HHTQDKAGQYKVKSLWPH) from the first extracellular loop of the mouse Panx3 protein was prepared. The antiserum was purified by the peptide affinity column. This purified antibody reacts specifically to Panx3 and was used for immunohistochemistry, Western blotting, and functional blocking assay. For inhibition by the Panx3 antibody, the cells were incubated with 10 ng/ml affinity-purified antibody or control IgG for 30 min before the experiments. To abrogate the blocking activity with Panx3 antibody, the Panx3 peptide was preincubated with the Panx3 antibody. A peptide with a scrambled sequence (WHTKYQVGLDPQHKASHK) of the Panx3 peptide was used as a control.
Immunohistochemistry
Frozen tissue sections were fixed with acetone at −20 °C for 2 min and treated with liberate antibody binding solution for 15 min at 37 °C. For cultured cell staining, the cells were fixed with acetone at −20 °C for 2 min. Immunohistochemistry was performed on sections that were incubated with Universal Blocking Reagent (Biogenex) for 7 min at room temperature before incubation with the primary antibody. The primary antibodies were detected by Cy-3- or Cy-5-conjugated secondary antibodies (Jackson ImmunoResearch). Nuclear staining was performed with Hoechst dye (Sigma). A fluorescence microscope (Axiovert 200; Carl Zeiss MicroImaging, Inc.) and 510 Meta confocal microscope (Zeiss) were used for immunofluorescent image analysis. For green fluorescent protein and ER-Tracker Red (Invitrogen) staining, ATDC5 cells were transfected for 2 days and then stained with ER-Tracker Red as described in the manufacturer's instructions. The images were prepared with AxioVision and Photoshop (Adobe Systems, Inc.).
Cell Culture
ATDC5 cells (21) were grown in Dulbecco's modified Eagle's medium/F-12 (Invitrogen) containing 5% fetal bovine serum (HyClone) and under 5% CO2. In proliferation conditions, cells were maintained under confluency, and the media were replaced every other day. In differentiation conditions, cells were plated in confluency and incubated in the same medium plus 10 μg/ml insulin, 10 μg/ml transferrin, and 10 μg of selenium. N1511 cells (22) were cultured with α-minimal essential medium (Invitrogen) containing 2% fetal bovine serum (HyClone) under 5% CO2. As differentiation conditions, 1 μm insulin, 100 ng/ml rhBMP2, and 50 μg/ml ascorbic acid were added in the culture medium.
Mouse primary chondrocytes were isolated from neonatal ICR mice as described previously (23). Distal cartilaginous ends of femurs and humeri were digested with 0.25% trypsin, 0.01% EDTA for 15 min, followed by digestion with 2 mg/ml collagenase type I (Worthington) in Dulbecco's modified Eagle's medium/F-12 overnight. Neonatal mouse cartilage tissue was dispersed by pipetting, and cells were filtered through 100-μm cell strainers (Falcon). Single cells were inoculated onto type I collagen-coated multiwell dishes maintained in 10% fetal bovine serum in α-minimal essential medium.
RT-PCR and Real Time PCR
Total RNA was extracted from cells using the TRIzol reagent kit (Invitrogen). Two μg of total RNA was used for reverse transcription to generate cDNA, which was used as a template for PCR with gene-specific primers. Each cDNA was amplified with an initial denaturation at 95 °C for 3 min; then 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s for 25 cycles; and a final elongation step at 72 °C for 5 min and then separated on agarose gels. Real time PCR was performed with SYBR Green PCR master mix and the TaqMan 7700 sequencer detection system (Applied Biosystems). PCR was performed for 40 cycles as follows: 95 °C for 1 min, 60 °C for 1 min, and 72 °C for 1 min. Gene expression was normalized to the housekeeping gene S29. The reactions were run in triplicate and repeated three times, and the results were combined to generate the graphs. The following primer sequences were used: Panx3, 5′-GCCCCTGGATAAGATGGTCAAG-3′ and 5′-GCGGATGGAACGGTTGTAAGA-3′; Panx1, 5′-TTTGGACCTAAGAGACGGACCTG-3′ and 5′-CGGGAATCAGCAGAGCATACAC-3′; Panx2, 5′-ACAAGGGCAGTGGAGGTGATTC-3′ and 5′-CGATGAGGATAGCGTGCTGATG-3′; Col2a1, 5′-GAAAAACTGGTGGAGCAGCAAGAGC-3′ and 5′-CAATAATGGGAAGGCGGGAGGTC-3′; Agc1, 5′-TGGAGCATGCTAGAACCCTCG-3′ and 5′-GCGACAAGAAGACACCATGTG-3′; Col10a1, 5′-AGCCCCAAGACACAATACTTCATC-3′ and 5′-TTTCCCCTTTCCGCCCATTCACAC-3′; PPR, 5′-ACTACTACTGGATTCTGGTGGAGGG-3′ and 5′-CTGGAAGGAGTTGAAGAGCATCTC-3′; PTHrP, 5′-CAGACGATGAGGGCAGATACCTAAC-3′ and 5′-CAGTTTCCTGGGGAGACAGTTTG-3′; and Gapdh, 5′-ACCACAGTCCATGCCATCAC-3′ and 5′-TCCACCACCCTGTTGCTGTA-3′. The predicted size of each fragment is 373, 380, 442, 392, 325, 463, 470, 558, 470, 372, and 452 bp.
Western Blotting
The cells were washed three times with phosphate-buffered saline containing 1 mm sodium vanadate (Na3VO4) and then solubilized in 100 μl of lysis buffer (10 mm Tris-HCl (pH 7.4), 150 mm NaCl, 10 mm MgCl2, 0.5% Nonidet P-40, 1 mm phenylmethylsulfonyl fluoride, and 20 units/ml aprotinin). Lysed cells were centrifuged at 14,000 rpm for 30 min, and the protein concentration of each sample was measured with Micro-BCA assay reagent (Pierce). The samples were denatured in SDS sample buffer and loaded onto a 12% SDS-polyacrylamide gel. Ten μg of lysate protein was applied to each lane. After SDS-PAGE, the proteins were transferred onto a polyvinylidene difluoride membrane and immunoblotted with anti-CREB, anti-phospho-CREB (Cell Signaling Technology, Inc.), anti-MAPK, and anti-phospho-MAPK (New England Biolabs) and then visualized using an ECL kit (Amersham Biosciences). For CREB and ERK1/2 experiments, the cells were pretreated as follows. The cells (3 × 104 cell/cm2) were plated in a 60-mm dish and cultured with 10 μg/ml insulin, 10 μg/ml transferrin, and 10 μg of selenium for 7 days. They were incubated with serum-free 0.1% albumin-containing Dulbecco's modified Eagle's medium/F-12 medium for 8–12 h and then exposed to 100 nm rPTH(1–34) for the appropriate times.
Plasmid Construction and Transfection
The coding sequence of mouse Panx3 cDNA was subcloned into the pEF1/V5-His vector (pEF1/Panx3) and pcDNA3.1-GFP-TOPO (Panx3-pcDNA-GFP) (Invitrogen). As a control, an empty vector of pEF1 or pcDNA3.1-GFP-TOPO was used. ATDC5 cells or N1511 cells were transiently transfected using Nucleofector (Amaxa). Briefly, cells were resuspended in 100 μl of Nucleofector solution T and transfected with 5 μg of DNA using Program T-20. Transfection efficiency using these conditions was shown to be >70%. For stable transfection of ATDC5 cells, selection was initiated 24 h after transfection using G418 (Invitrogen) at a concentration of 600 μg/ml for 2 weeks. Pools of transfected cells were collected and cultured as the parental ATDC5 cells, in the continued presence of 60 μg/ml G418. All experiments were performed three times, and representative experiments are shown. For transient transfection, Panx3-pcDNA-GFP was transfected into ATDC5 cells under the same conditions as used for stable transfection, except that G418 was omitted.
Alcian Blue Staining
The cells were first rinsed with phosphate-buffered saline three times and then fixed with 100% methanol for 10 min at −20 °C. Staining was accomplished by applying a solution of 0.1% Alcian blue 8 GX in 0.1 m HCl to the cells for 2 h at room temperature. To quantify the intensity of the staining, the stained culture plates were rinsed with phosphate-buffered saline three times, and the well was extracted with 6 m guanidine HCl for 8 h at room temperature. The absorbance of the extracted dye was measured at 650 nm.
Short Hairpin RNA Experiments
Panx3-specific knockdowns were performed with the expression of shRNA using a pSM2 vector (Open Biosystems). The shRNA construct contains the 3′-untranslated region of Panx3 (GGCAGGGTAGAACAATTTA). A nonsilencing shRNA construct from Open Biosystems, whose sequence has been verified to contain no homology to known mammalian genes, was used as a control. The cells were transfected with shRNA plasmids using Nucleofection (Amaxa). Selection was performed 24 h after transfection with puromycin at a concentration of 4 μg/ml for 5 days.
siRNA Experiments
siRNAs targeting mouse Panx3 (NM_172454) (Panx3 siRNA-1, 5′-UAAUAAGGAUGUCCACGUA-3′ and Panx3 siRNA-2, 5′-GGGCUCAGAUUAUGGACUA-3′) were purchased from Dharmacon (siGenome On-Target Plus; Dharmacon). Negative control siRNA duplex (Stealth RNAi negative control; Invitrogen) was used as control. Forward transfection for ATDC5 cells and reverse transfection for primary chondrocytes were carried out using Lipofectamine RNAiMAX reagent (Invitrogen), according to the manufacturer's protocol. In forward transfection, ATDC5 cells were transfected with 50 nm siRNA duplex in regular serum-free culture medium without antibiotics, using Lipofectamine RNAiMAX reagent. After 8 h of incubation with the transfection reagent mixture, the medium was changed to the differentiation medium with BMP-2 and incubated for 8 days. In reverse transfection, freshly isolated chondrocytes were plated at an initial density of 5 × 104/cm2, into the plates that had been coated with Panx3 siRNA-1, siRNA-2, or Stealth RNAi negative control in the presence of Lipofectamine RNAiMAX reagent. After 8 h of incubation with the transfection reagent mix, the medium was changed to the differentiation medium with BMP-2 and incubated for 2 days. Expressions of Panx3, Col2a1, and Col10a1 were analyzed by real time PCR methods.
Measurement of Intracellular cAMP
The cells were seeded at 1 × 104 cells/well in a 96-well plate and cultured for 7 days with Dulbecco's modified Eagle's medium/F-12 in the presence of 10 μg/ml of ITS. They were then incubated with serum-free 0.1% albumin containing Dulbecco's modified Eagle's medium/F-12 medium for 8–12 h, followed by exposure to 100 nm rPTH(1–34) for 10 min. The level of cAMP was determined with a Bridge-It cAMP Designer fluorescence assay kit (Mediomics). Briefly, the cells were incubated with 50 μl of 1× Krebs-Ringer/isobutylmethylxanthine buffer for 15 min at room temperature and with 50 μl of forskolin for 15 min at room temperature. After the solution was removed, the cells were incubated with 100 μl of the cAMP designer assay solution for 30 min while covered with aluminum foil to avoid exposure to light. The supernatant was collected, and the fluorescence intensity was measured with a plate reader (excitation, ∼480–485 nm; emission, ∼520–535 nm).
Measurement of ATP Flux
ATP flux was determined by luminometry. To open the pannexin channels, the cells were depolarized by incubation in KGlu solution (140 mm KGlu, 10 mm KCl, and 5.0 mm TES, pH 7.5) for 10 min. The supernatant was collected and assayed with luciferase/luciferin (Promega).
RESULTS
Expression of Panx3 in Cartilage
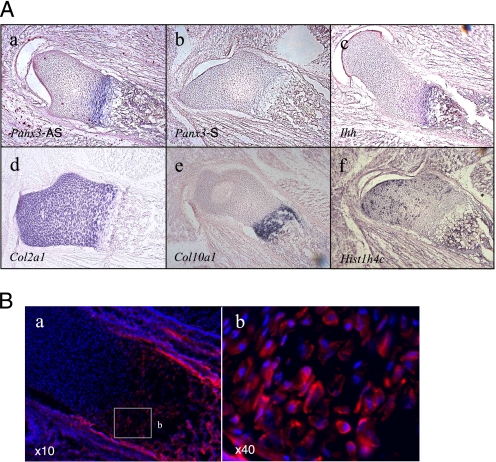
While searching for an expression profile of gap junction genes in skeletal tissues through the EST data base, we found that Panx3 was expressed in limbs and cartilage. Because of its unique expression profile and its potential function in skeletal development, we characterized the expression and function of Panx3 in cartilage. In situ hybridization of the embryonic day (E) 16.5 growth plates revealed that Panx3 mRNA was strongly expressed in the prehypertrophic zone (Fig. 1A, panel a). A control sense probe for Panx3 showed no signal (Fig. 1A, panel b). Indian hedgehog (Ihh) mRNA was expressed in prehypertrophic and hypertrophic chondrocytes at this stage (Fig. 1A, panel c). mRNA for type II collagen (Col2a1), a major collagen in cartilage, was expressed in the resting, proliferative, and prehypertrophic zones (Fig. 1A, panel d). Type X collagen (Col10a1) mRNA was expressed in the prehypertrophic and hypertrophic chondrocyte chondrocytes (Fig. 1A, panel e). Histone H4 (Hist1h4c), a marker for cell proliferation, was expressed in proliferating chondrocytes (Fig. 1A, panel f). Immunostaining with a Panx3 antibody showed that Panx3 protein was expressed in prehypertrophic and hypertrophic chondrocytes and was localized on the surface of these cells (Fig. 1B). Panx3 was also expressed in perichondrial cells and osteoblasts.
FIGURE 1.
Expression of Panx3 in E16.5 growth plates. A, in situ hybridization. Panx3 mRNA was expressed in prehypertrophic chondrocytes, perichondrium, and osteoblasts. The following are shown: antisense Panx3 (panel a); sense Panx3 (panel b); Ihh (panel c); Col2a1 (panel d); Col10a1 (panel e); and Hist1h4c (panel f). B, immunostaining with anti-Panx3 antibody (red) and Hoechst nuclear staining (blue). A magnified view of the areas (panel a) is marked by the square in panel b.
Expression of Panx3 in Differentiating Chondrogenic Cells
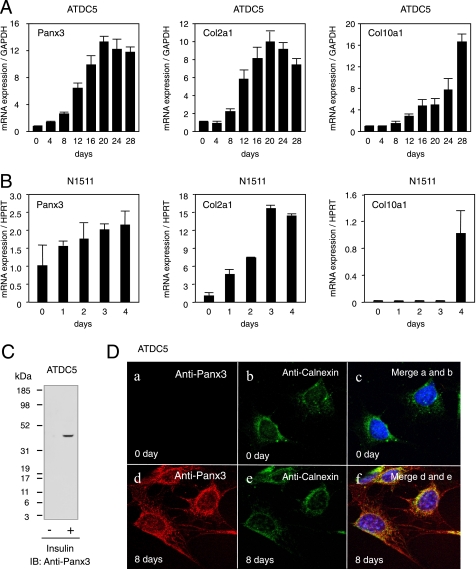
The expression of Panx3 mRNA in a transitional stage between proliferative and hypertrophic chondrocytes suggests that Panx3 regulates chondrocyte proliferation and differentiation. To assess the role of Panx3 in chondrocyte differentiation, we used the murine chondrogenic cell lines ATDC5 and N1511, which are used to study multistep processes of chondrocyte differentiation (21, 22). ATDC5 cells proliferate until confluency and then differentiate into chondrocytic phenotypes in a prolonged culture in the presence of insulin (21). Factors such as BMP-2, GDF-5, and transforming growth factor β accelerate the chondrogenic differentiation of ATDC5 cells (24–26). We first examined the expression of Panx3 mRNA during differentiation of the ATDC5 cells in the presence of insulin using real time RT-PCR (Fig. 2A). Panx3 mRNA expression was induced during ATDC5 cell differentiation and reached its highest level after 20 days of culture (Fig. 2A). The expression of Col2a1 mRNA was increased in a similar manner to that of Panx3 mRNA; Col10a1 mRNA was induced at later stages of differentiation. Without insulin, the induction of Panx3, Col2a1, and Col10a1 was low even in a prolonged culture, indicating that Panx3 expression was clearly linked to the differentiation of ATDC5 cells. We observed similar expression patterns in those genes during BMP-2-induced ATDC5 differentiation, except that BMP-2 promoted differentiation much faster than insulin (data not shown). In another chondrogenic cell line N1511, BMP-2- and insulin-induced expressions of Panx3, Col2a1, and Col10a1 were seen (Fig. 2B). Western blot analysis demonstrated that Panx3 protein with a molecular mass of 45 kDa was induced in differentiated ATDC5 cells (Fig. 2C). These results indicate that Panx3 was induced during chondrogenic differentiation of ATDC5 and N1511 cells.
FIGURE 2.
Expression of Panx3 in differentiating ATDC5 and N1511 cells. A, mRNA expression in differentiating ATDC5 cells. ATDC5 cells were cultured with 10 μg/ml insulin. Total RNA was extracted from cells on the indicated days after insulin treatment and analyzed with real time RT-PCR. In differentiating ATDC5 cells, Panx3, Col2a1, and Col10a1 were strongly induced. GAPDH, glyceraldehyde-3-phosphate dehydrogenase, was used as a control. B, mRNA expression in differentiating N1511 cells. N1511 cells were cultured with 1 μm insulin, 100 ng/ml rhBMP-2, and 50 μg/ml ascorbic acid for differentiation. Panx3 was also progressively induced in differentiating N1511 cells. HPRT, hypoxanthine phosphoribosyltransferase, was used as a control. C, expression of Panx3 protein in undifferentiated (1st lane) and differentiated ATDC5 cells (2nd lane). ATDC5 cells were treated with or without insulin for 20 days, and cell extracts were analyzed by Western blotting using Panx3 antibody. Panx3 protein was induced in differentiated ATDC5 cells. IB, immunoblot. D, immunostaining with anti-Panx3 antibody (red), ER marker (green), calnexin, and Hoechst nuclear staining (blue). In differentiating ATDC5 cells, endogenous Panx3 was observed in the cell membrane, cell processes, and ER (panels d–f) but not in undifferentiated cells (panels a and c).
Membrane Localization of Panx3 in ATDC5 Cells
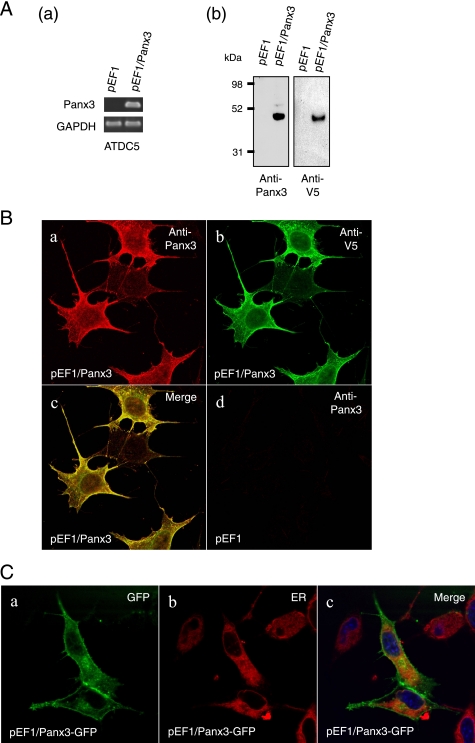
To examine cellular localizations of Panx3, we immunostained differentiated ATDC5 cells using anti-Panx3 antibody (Fig. 2D). Panx3 was expressed in the plasma membrane, cell extensions, and in organelles most likely found in the ER and the Golgi of ATDC5 cells 8 days after differentiation induction. Panx3 was co-localized with calnexin, an ER marker, indicating that the localization of Panx3 was most likely in the ER (Fig. 2D, panels d and e). There was no staining signal for Panx3 in undifferentiated ATDC5 cells (Fig. 2D, panel a). We also examined the expression and localization of Panx3 in pooled, stable Panx3-transfected ATDC5 cells (Fig. 3, A and B). In these cells, Panx3 mRNA and protein were strongly expressed in an undifferentiated condition. Both anti-Panx3 and anti-V5 antibodies detected the recombinant protein as a single band of about 49 kDa, corresponding to the predicted molecular weight of the Panx3-V5-His fusion protein (Fig. 3A). The immunohistochemistry of Panx3-transfected ATDC5 cells showed that both anti-Panx3 and anti-V5 antibodies strongly stained the plasma membranes (Fig. 3B). Some Panx3 signals were also observed in the ER because ER-Tracker Red, which had been transiently transfected with the Panx3 expression vector containing a green fluorescent protein tag, revealed that Panx3 was co-localized with an ER marker. This indicates that the localization of Panx3 was most likely in the ER (Fig. 3C).
FIGURE 3.
Panx3 expression in Panx3-transfected ATDC5 cells. ATDC5 cells were stably transfected with the control empty vector (pEF1) or the Panx3 expression vector (pEF1/Panx3). A, expression of Panx3 mRNA and protein. Pooled transfectants were analyzed by RT-PCR (panel a) and Western blotting (panel b) using anti-Panx3 and anti-V5 antibodies. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. B, immunostaining of Panx3-transfected ATDC5 cells using anti-Panx3 (red) and anti-V5 (blue) antibodies. Fluorescent confocal images showed that the staining signals of Panx3 and V5 antibodies were co-localized in the cell membrane, cell-cell junction, and organelles. No staining of either Panx3 or V5 antibodies was observed in control pEF1-transfected ATDC5 cells. C, co-localization of Panx3-GFP and ER-Tracker Red. ATDC5 cells were transiently transfected with Panx3-pcDNA-GFP or pcDNA3.1-GFP-TOPO (control) for 2 days. Panx3-GFP was co-localized with ER-Tracker Red, indicating the presence of Panx3 in ER. GFP, green fluorescent protein.
Panx3 Promotes ATDC5 and N1511 Cell Differentiation
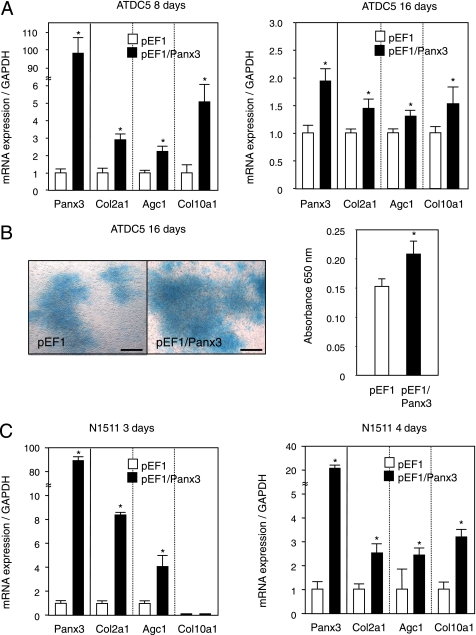
We next examined whether the overexpression of Panx3 affects ATDC5 cell differentiation. Panx3-transfected ATDC5 cells were cultured under the differentiation condition in the presence of insulin. The expression of chondrocyte marker genes for Col2a1, aggrecan (Agc1), and Col10a1 increased by 2.8-, 2.2-, and 5.1-fold, respectively, compared with control pEF1-transfected cells 8 days after induction (Fig. 4A, left panel). The Panx3 mRNA levels in the Panx3-transfected cells were ∼100-fold higher than those in the control pEF1-ATDC5 cells at day 8 of induction. At day 16, after induction, the expression of both Agc1 and Col10a1 in Panx3-transfected cells was still higher than that of the control cells (Fig. 4A). Panx3 mRNA in the transfected cells at day 16 was increased by about 2-fold, when its level was normalized with the level of control pEF1-transfected cells. The large decrease in the relative ratio of Panx3 mRNA levels at day 16 compared with day 8 is due to the endogenous Panx3 mRNA levels increasing substantially from day 8 to 16 as shown in Fig. 2A. The actual Panx3 mRNA levels did not decrease in Panx3-transfected ATDC5 cells during differentiation. Alcian blue staining, often used to stain proteoglycans in cartilage, was also increased in nodules of Panx3-transfected cells compared with the control cells (Fig. 4B). We also examined the expression of chondrogenic marker genes in transiently Panx3-transfected N1511 cells during differentiation in the presence of insulin and BMP-2 (Fig. 4C). At day 3, the expression of Col2a1 and Agc1 mRNA was stimulated in the transfected cells compared with the control pEF1-transfected cells, whereas the Col10a1 mRNA levels were very low in both control pEF1- and Panx3-transfected cells. In day 4, Col10a1 mRNA was induced in control cells, and its expression levels were promoted in Panx3-transfected cells. The total Panx3 mRNA levels, including exogenous and endogenous Panx3 mRNA, did not change at day 3 or 4. Taken together, these results indicate that Panx3 expression promoted chondrogenic cell differentiation.
FIGURE 4.
Panx3 promotes chondrogenic differentiation of ATDC5 and N1511 cells. A, differentiation of ATDC 5 cells. Pooled ATDC5 cells stably transfected with either control pEF1 or pEF1/Panx3 were cultured with 10 μg/ml insulin. Total RNA was extracted at 8 and 16 days after insulin treatment and analyzed by real time RT-PCR. The expression of chondrogenic marker genes for Col2a1, Agc1, and Col10a1 was stimulated in Panx3-transfected ATDC5 cells compared with that in control cells. The expression level of an individual gene in control pEF1-transfected cells was set as 1.0, and we compared it with the level of each gene in Panx3-transfected cells for each day 8 and 18. The exogenous Panx3 levels were the same but endogenous Panx3 levels were strongly increased from day 8 to 16. As results, the ratio at day 16 was less than that in day 8. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. B, Alcian blue staining of ATDC5 cells. After 16 days of culture, Alcian blue staining was performed. Alcian blue staining was increased in Panx3-transfected ATDC5 cells compared with that in pEF1-transfected ATDC5 cells. Scale bar, 200 μm. C, differentiation of N1511 cells. N1511 cells were transfected with either control pEF1 or pEF1/Panx3, were cultured with 1 μm insulin, 100 ng/ml rhBMP-2, and 50 μg/ml ascorbic acid for 3 s and 4 days. Similar to the results of ATDC5 cells, chondrogenic maker genes expression was stimulated by Panx3. Statistical analysis was performed using analysis of variance (*, p < 0.01).
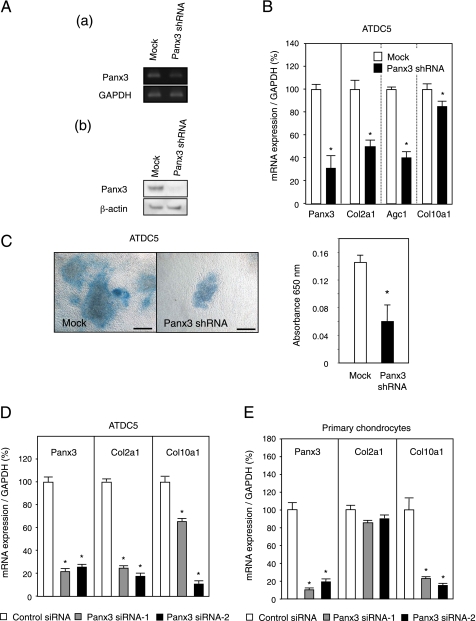
Suppression of Endogenous Panx3 Expression Inhibits Cell Differentiation
To analyze the endogenous Panx3 function in ATDC5 cell differentiation, we knocked down Panx3 expression using Panx3 shRNA. We transfected the Panx3 shRNA expression vector into ATDC5 cells. The resulting stably transfected cells were pooled and induced to differentiate in the presence of insulin. After 8 days in culture, the expression of Panx3 was substantially reduced at both the RNA and protein levels, compared with empty vector-transfected cells (Fig. 5A). Panx3 mRNA was found to be down-regulated by ∼70% in Panx3 shRNA-transfected cells compared with the controls. In addition, the expression of mRNA for Col2a1, Agc1, and Col10a1 in the shRNA-transfected cells was reduced by 50, 64, and 15%, respectively (Fig. 5B). Similarly, Alcian blue staining was reduced to 40% of the control cell level at 16 days in culture (Fig. 5C). Similarly, N1511 cell differentiation was inhibited by Panx3 shRNA transfection (data not shown).
FIGURE 5.
Inhibition of ATDC5 cell differentiation by Panx3 shRNA. Pooled ATDC5 cells stably transfected with either control vector (Mock) or Panx3 shRNA vector were cultured with 10 μg/ml insulin. A, reduced expression of endogenous Panx3. Total RNA and protein were prepared from cells after 8 days of culture and analyzed through RT-PCR (panel a) and Western blotting (panel b), using anti-Panx3 antibody. Panx3 expression was reduced in Panx3 shRNA-transfected ATDC5 cells. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. B, reduced expression of chondrogenic marker genes. Total RNA was prepared from cells after 8 days of culture and analyzed by real time RT-PCR. Expression of Col2a1, Agc1, and Col10a1 was reduced in Panx3 shRNA-transfected ATDC5 cells. C, Alcian blue staining. After 16 days of culture, Alcian blue staining was performed. Alcian blue staining was reduced in Panx3 shRNA-transfected cells. Scale bar, 200 μm. Statistical analysis was performed using analysis of variance (*, p < 0.01). D, reduced expression of Col2a1 and Col10a1 by Panx3 siRNA in ATDC5 cells. ATDC5 cells transfected with control siRNA, Panx3 siRNA-1, or Panx3 siRNA-2 were cultured with 100 ng/ml BMP-2 for 8 days. Expressions of Panx3, Col2a1, and Col10a1 were analyzed by real time PCR methods. Expression of Col2a1 and Col10a1 was reduced in Panx3 siRNA-transfected ATDC5 cells. E, reduced expression of Col10a1 but not Col2a1 by Panx3 siRNA in primary chondrocytes. Primary chondrocytes transfected with control siRNA, Panx3 siRNA-1, or Panx3 siRNA-2 were cultured with 100 ng/ml BMP-2 for 2 days. Expressions of Panx3, Col2a1, and Col10a1 were analyzed by real time PCR methods. Expression of Panx3 and Col10a1, but not Col2a1, was reduced in Panx3 siRNA-transfected primary chondrocytes. Statistical analysis was performed using analysis of variance (*, p < 0.01).
To eliminate the possibility of off-target inhibitory effects of shPanx3 on cell differentiation, we used two different types of siRNA for Panx3. Both Panx3 siRNA-1 and -2 inhibited the expression of Col2a1 and Col10a1 in ATDC5 cells (Fig. 5D). We also tested these siRNAs in primary chondrocytes prepared from cartilage of neonatal mice. Panx3 siRNA-1 and -2 inhibited the expression of Col10a1 but not Col2a1 (Fig. 5E). This may be because primary chondrocytes are a mixture of different chondrocytes in varying stages of differentiation. The suppression of endogenous Panx3 did not affect Col2a-expressing chondrocytes but inhibited differentiation to hypertrophic Col10a1-expressing chondrocytes. These results indicate that the suppression of endogenous Panx3 expression inhibited chondrocyte differentiation.
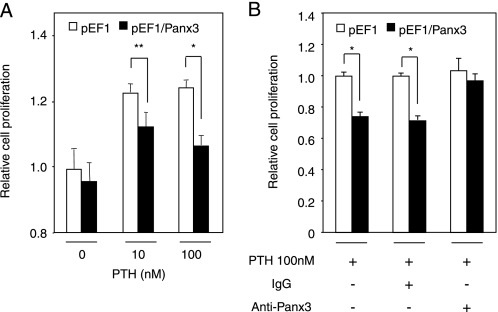
Panx3 Inhibits PTH-induced Cell Proliferation
It has been reported that the expression of connexins such as Cx43, Cx32, and Cx26 inhibits tumor cell growth (27, 28). Panx3 may have similar cell growth inhibitory activity, thereby promoting ATDC5 cell differentiation. Because PTH/PTHrP promotes chondrocyte proliferation, we examined Panx3 activity in PTH-mediated ATDC5 cell proliferation. Three days after the addition of PTH, we found that the number of control ATDC5 cells had increased in a dose-responsive manner and that the maximum cell number was reached at 10 nm PTH (Fig. 6). However, the number of Panx3-transfected ATDC5 cells was reduced in response to increasing amounts of PTH compared with the control cells (Fig. 6A). The endogenous PTHrP mRNA levels remained the same in Panx3- and control pEF1-transfected ATDC5 cells, under either proliferation or differentiation conditions (supplemental Fig. 1). PTH/PTHrP receptor was induced during differentiation, but its expression levels were similar in pEF- and Panx3-transfected cells (supplemental Fig. 1). These results suggest that endogenous PTH/PTHrP receptor and PTHrP did not affect the proliferation and differentiation results. This reduced proliferation activity in Panx3-transfected cells with PTH was blocked by anti-Panx3 antibody but not control IgG (Fig. 6B). These results suggest that Panx3 inhibits PTH-mediated cell proliferation.
FIGURE 6.
Inhibition of PTH-promoted cell proliferation by Panx3. A, Panx3- and pEF1-transfected cells were incubated in the presence of various amounts of PTH. Cell numbers were counted after 3 days of culture. The number of Panx3-transfected cells was reduced compared with the control cells. B, Panx3 antibody, but not IgG, restored PTH-promoted cell proliferation in Panx3-transfected cells. Statistical analysis was performed using analysis of variance (**, p < 0.02; *, p < 0.01).
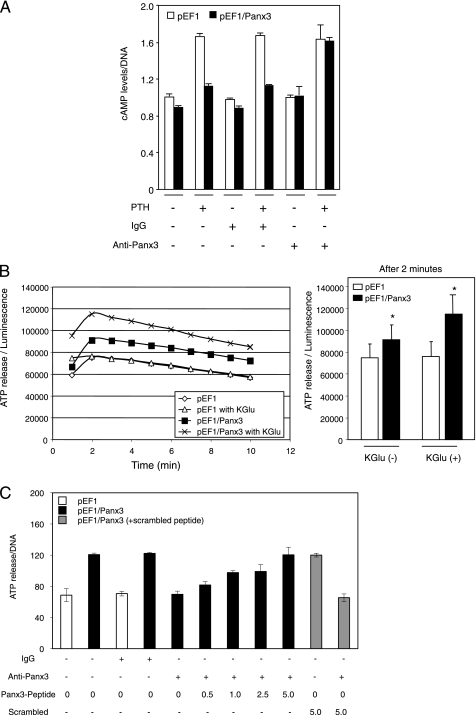
Panx3 Reduces Intracellular cAMP and ATP Levels
Because PTH/PTHrP stimulates the proliferation of chondrocytes through activation of the cAMP pathway (2), we examined the intracellular level of cAMP in Panx3-transfected ATDC5 cells under proliferation conditions (Fig. 7A). In the absence of PTH, the intracellular cAMP level was ∼10% less in Panx3-transfected cells than in control pEF1-transfected cells. The addition of PTH increased the cAMP level within 10 min by 1.7-fold in pEF1-transfected cells. In contrast, Panx3-transfected cells demonstrated only a 1.2-fold induction by PTH. This reduced induction of the cAMP levels in Panx3-transfected cells was reversed to normal levels by addition of anti-Panx3 antibody but not by control IgG (Fig. 7A). These data suggest that Panx3 inhibited PTH-mediated proliferation of ATDC5 cells by reducing intracellular cAMP levels.
FIGURE 7.
Reduced cAMP levels and increased ATP efflux in Panx3-transfected ATDC5 cells. A, intracellular cAMP level. Panx3- and pEF1-transfected ATDC5 cells were cultured with 10 μg/ml insulin for 1 week. The cells were incubated with anti-Panx3, IgG, or without them for 30 min and then exposed to PTH at 100 nm for 10 min, and we analyzed the intracellular cAMP levels. PTH promoted the intracellular cAMP level in control pEF1-transfected cells, whereas this PTH effect was reduced in Panx3-transfected cells. This reduction was blocked by anti-Panx3 antibody but not IgG. B, release of ATP. Cells were plated at ∼50% confluency in the absence or presence of potassium (KGlu), and ATP levels in the media were measured. ATP release to the extracellular space was increased in Panx3-transfected cells. Left panel, time course of ATP release after addition of KGlu. Right panel, data at 2 min after addition of KGlu in the right panel are shown in bar graphs. Statistical analysis was performed using analysis of variance (*, p < 0.01). C, inhibition of ATP release by Panx3 antibody. Cells were incubated with anti-Panx3 antibody, Panx3 peptide, or IgG for 30 min, and ATP release was measured. The Panx3 antibody inhibited ATP release in Panx3-transfected cells. This inhibition was blocked by various concentrations (0.5 to 5.0 ng/ml) of the Panx3 peptide but not its scrambled peptide (5.0 ng/ml).
This result may be due to Panx3 functioning as a hemichannel, releasing ATP to the extracellular space, and thus decreasing the intracellular cAMP level. To examine the hemichannel activity of Panx3, pEF1- and Panx3-transfected ATDC5 cells were treated with PTH, and the release of ATP into the culture medium was then measured (Fig. 7B). Panx3-transfected cells exhibited an elevated ATP release that reached a maximum level at 2 min and then gradually decreased with time. A similar release was not observed in control pEF1-transfected cells. In the presence of a high concentration of potassium glutamate (KGlu), which depolarizes the cell membrane, ATP was released from Panx3-transfected ATDC5 cells (Fig. 7B). A function-blocking antibody specific to the extracellular domain of Panx3 inhibited this ATP efflux (Fig. 7C). This antibody inhibition was blocked by the Panx3 peptide, which was used to raise the Panx3 antibody as an antigen in a dose-dependent manner, whereas its scrambled peptide and control IgG did not affect the antibody inhibition. These results suggest that the Panx3 hemichannel is one of the major ATP release channels.
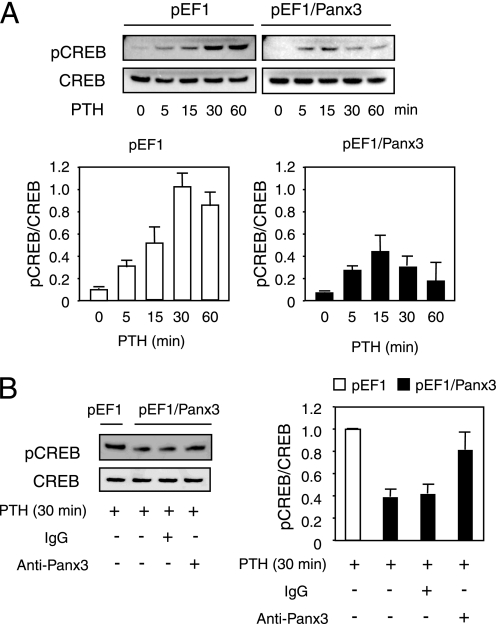
Panx3 Inhibits PTH-induced CREB Phosphorylation
We next examined the activation of CREB, which is a downstream molecule of the PTH/PTHrP-cAMP-PKA pathway in chondrocytes (10, 29). CREB reached its maximum phosphorylation level at 30 min after PTH treatment in the control pEF1-transfected ATDC5 cells. In Panx3-transfected cells, CREB phosphorylation was significantly reduced (Fig. 8A). The reduced CREB phosphorylation in Panx3-transfected cells was blocked by anti-Panx3 antibody but not by IgG (Fig. 8B). Taken together, these results suggest that Panx3 inhibits PTH/PTHrP-cAMP-PKA signaling in ATDC5 cells.
FIGURE 8.
Decrease in phosphorylation of CREB by Panx3. A, time course of CREB phosphorylation. ATDC5 cells were cultured with 10 μg/ml insulin for 1 week and then treated with PTH (100 nm) for the time indicated. Protein extracts were analyzed by Western blotting using anti-phospho-CREB and anti-CREB antibodies. In control pEF1-transfected cells, the phosphorylation of CREB was strongly induced, and in Panx3-transfected cells the phosphorylation levels of CREB were reduced. B, restoration of the CREB phosphorylation levels by Panx3 antibody. Cells were preincubated with Panx3 antibody or IgG for 30 min before the stimulation with 100 nm PTH, and then Western blotting using anti-phospho-CREB and anti-CREB antibodies was performed. Panx3 antibody inhibited the reduction of the phosphorylation of CREB in Panx3-transfected cells. ImageJ 1.33u was used to quantify the protein bands.
DISCUSSION
In this study, we utilized a bioinformatics approach to search for a gap junction protein involved in cartilage development. We found that Panx3 mRNA was preferentially expressed in a transitional stage between proliferative chondrocytes and terminally differentiated hypertrophic chondrocytes in developing growth plates. This suggests that Panx3 plays a role in the switch from proliferation to differentiation in these chondrocytes. To assess Panx3 action in chondrocyte differentiation, we used chondrogenic cell lines ATDC5 and N1511, which can be induced to differentiate into chondrocyte phenotypes. Panx3 mRNA expression was induced during differentiation of ATDC5 and N1511 cells (Fig. 2). We demonstrated that the expression of Panx3 promoted differentiation of ATDC5 and N1511 cells (Fig. 4). In contrast, the inhibition of endogenous Panx3 expression through Panx3 shRNA and siRNA reduced differentiation of these cells and primary chondrocytes (Fig. 5 and data not shown) These results suggest that Panx3 regulates chondrogenic cell differentiation.
PTH/PTHrP functions to keep chondrocytes proliferating and to delay hypertrophic chondrocyte differentiation. Both PTHrP-deficient mice and PTH/PTHrP receptor-deficient mice have similar growth plate abnormalities, i.e. reduced numbers of chondrocytes and premature hypertrophic differentiation, indicating that PTHrP signals primarily through the PTH/PTHrP receptor in the growth plate (3, 5). We found that PTH increased proliferation of ATDC5 cells in culture, but this PTH activity was reduced in Panx3-transfected ATDC5 cells (Fig. 6). There was no significant difference in cell proliferation activity between untransfected and Panx3-transfected ATDC5 cells in the absence of PTH, indicating that the inhibitory activity of Panx3 for cell proliferation is dependent on PTH. The interaction of PTH/PTHrP with PTH/PTHrP receptor promotes the activation of multiple heteromeric G proteins, including Gs, which can activate adenylyl cyclase, Gq, which can activate phospholipase C/protein kinase C, and Gq, whose action occurs opposite the Gs pathway (30, 31). The PTH-induced activation of Gs leads to the cascade activation of downstream molecules, specifically the activation of adenylyl cyclase, an increase in cAMP levels, the activation of PKA, and the phosphorylation of CREB. The activation of CREB induces the expression of genes required for cell proliferation (Fig. 9).
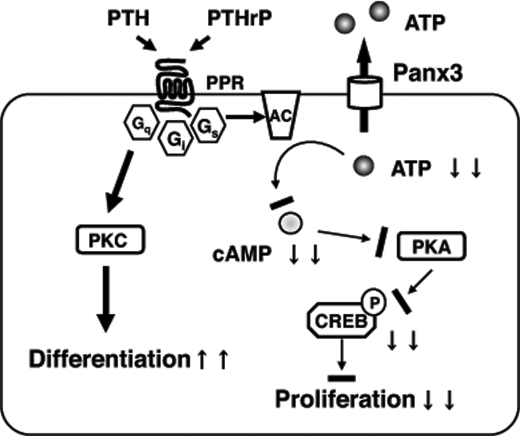
FIGURE 9.
Role of Panx3 in chondrogenic differentiation. The PTH/PTHrP receptor activates multimeric G proteins. The activation of the Gs subunit leads to the activation of adenylyl cyclase (AC) for cAMP generation from ATP and the subsequent activation of PKA. PKA phosphorylates CREB, which promotes the expression of genes for cell proliferation. Panx3 is expressed in prehypertrophic chondrocytes, and it promotes the release of ATP into the extracellular space, which results in a reduction of the intracellular cAMP level and subsequent inhibition of PKA/CREB signaling for cell proliferation. The PTH/PTHrP receptor also activates the Gq subunit and subsequent downstream signaling, such as protein kinase C (PKC), to promote differentiation.
We found that Panx3 expression in ATDC5 cells promoted ATP release from the cytoplasm to the extracellular space, and this ATP release was inhibited by a function-blocking anti-Panx3 antibody, suggesting that Panx3 plays a specific role in the release of ATP (Fig. 7, B and C). The activity of Panx3 as a hemichannel for ATP release would explain the reduced intracellular cAMP levels in Panx3-expressing ATDC5 cells treated with PTH, because ATP is converted to cAMP by adenylyl cyclase (32). Thus, it is conceivable that Panx3-promoted ATP release to the extracellular space results in the reduction of intracellular cAMP, leading to the inhibition of PTH-induced cell proliferation. Panx3 activity in ATDC5 cells as an ATP-release channel is consistent with recent reports that Panx1 can form a hemichannel for stress-sensitive ATP permeability in oocytes and erythrocytes (19, 33).
ATP secreted into the pericellular environment affects a variety of cellular processes. Recently, up-regulation of Panx1 has been reported in macrophages stimulated by endotoxin lipopolysaccharide. This treatment mediates large pore formation of the ATP-gated P2X7Rs receptor, which leads to interleukin-1 release from the activated macrophage (34). The interaction of ATP with P2 receptors reportedly increases the concentration of intracellular Ca2+ in chondrocytes through Gq (35). ATP released into the extracellular space through Panx3 hemichannels at the early differentiation stage may re-enter the cytoplasm through purinergic receptors such as P2Y and P2X and promote differentiation. It is also possible that Panx3 may function as a Ca2+ channel in the ER and increase intracellular Ca2+ levels for chondrocyte maturation.
Although Panx3 mRNA is expressed strongly by prehypertrophic chondrocytes, the Panx3 protein is found in both prehypertrophic and hypertrophic chondrocytes (Fig. 1). We did not detect Panx1 or Panx2 in developing growth plates with in situ hybridization (data not shown). Panx1, but not Panx2, was expressed very weakly in ATDC5 cells but was not induced during ATDC5 cell differentiation (data not shown). Connexin43 is expressed in condensing mesenchymal cells in the primordial cartilage, and also in articular chondrocytes and osteoblasts. Connexin43 has been shown to form functional gap junctions capable of sustaining the propagation of intercellular Ca2+ waves in articular chondrocyte culture (36) and to regulate BMP-2-mediated chondrogenic differentiation in chick mesenchyme micromass culture (37). Many mutations in the connexin43 gene have been identified in association with oculodentodigital dysplasia (38, 39), which is characterized by syndactyly of the hands and feet, hypoplasia of the middle phalanges, and abnormal craniofacial elements. However, connexin-deficient mice do not exhibit any of the gross abnormalities of chondrocyte differentiation, but they do have mineralization defects (40). Immunostaining of developing growth plates revealed that connexin43 was not expressed in prehypertrophic and hypertrophic chondrocytes, but it was expressed in osteoblasts (data not shown). These connexin43 expression patterns are consistent with the skeletal phenotype of connexin43-deficient mice. Thus, Panx3 is likely the major gap junction protein in cartilage.
In conclusion, we demonstrated that Panx3 is uniquely expressed in prehypertrophic and hypertrophic chondrocytes and that it promotes the chondrogenic differentiation of ATDC5 cells, at least in part through its hemichannel activity. Panx3 inhibits PTH-induced ATDC5 cell proliferation and mediates intracellular ATP release into the extracellular space, which in turn leads to a reduction in cAMP/PKA signaling, resulting in decreased proliferation and increased differentiation. Our findings suggest that Panx3 is a regulator of the switching mechanism behind the transition of chondrocytes from a proliferative state to a postmitotic state and that it is required for chondrocyte differentiation.
Supplementary Material
Acknowledgments
We thank Dr. Masahiro Iwamoto for plasmid Hist1h4c and Dr. Motomi Iwamoto and Dr. Hideto Watanabe for N1511 cells.
This work was supported, in whole or in part, by a National Institutes of Health grant from Intramural Research Program of NIDCR (to Y. Y.). This work was also supported by Grant-in-aid for Research Fellows 20791583 from the Japan Society for the Promotion of Science (to T. I.) and Grant-in-aid from the Ministry of Education, Science, and Culture of Japan 20679006 (to S. F.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- PTHrP
- parathyroid hormone-related peptide
- PTH
- parathyroid hormone
- PKA
- protein kinase A
- CREB
- cAMP-response element-binding protein
- ER
- endoplasmic reticulum
- RT
- reverse transcription
- shRNA
- short hairpin RNA
- siRNA
- short interfering RNA
- MAPK
- mitogen-activated protein kinase
- TES
- 2-{[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]amino}ethanesulfonic acid.
REFERENCES
- 1.Kronenberg H. M. (2003) Nature 423, 332–336 [DOI] [PubMed] [Google Scholar]
- 2.Kronenberg H. M. (2006) Ann. N.Y. Acad. Sci. 1068, 1–13 [DOI] [PubMed] [Google Scholar]
- 3.Karaplis A. C., Luz A., Glowacki J., Bronson R. T., Tybulewicz V. L., Kronenberg H. M., Mulligan R. C. (1994) Genes Dev. 8, 277–289 [DOI] [PubMed] [Google Scholar]
- 4.Miao D., He B., Karaplis A. C., Goltzman D. (2002) J. Clin. Invest. 109, 1173–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lanske B., Karaplis A. C., Lee K., Luz A., Vortkamp A., Pirro A., Karperien M., Defize L. H., Ho C., Mulligan R. C., Abou-Samra A. B., Jüppner H., Segre G. V., Kronenberg H. M. (1996) Science 273, 663–666 [DOI] [PubMed] [Google Scholar]
- 6.Weir E. C., Philbrick W. M., Amling M., Neff L. A., Baron R., Broadus A. E. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 10240–10245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schipani E., Lanske B., Hunzelman J., Luz A., Kovacs C. S., Lee K., Pirro A., Kronenberg H. M., Jüppner H. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 13689–13694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abou-Samra A. B., Jüppner H., Force T., Freeman M. W., Kong X. F., Schipani E., Urena P., Richards J., Bonventre J. V., Potts J. T., Jr. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 2732–2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh A. T., Gilchrist A., Voyno-Yasenetskaya T., Radeff-Huang J. M., Stern P. H. (2005) Endocrinology 146, 2171–2175 [DOI] [PubMed] [Google Scholar]
- 10.Beier F., Ali Z., Mok D., Taylor A. C., Leask T., Albanese C., Pestell R. G., LuValle P. (2001) Mol. Biol. Cell 12, 3852–3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beier F., LuValle P. (2002) Mol. Endocrinol. 16, 2163–2173 [DOI] [PubMed] [Google Scholar]
- 12.Panchin Y., Kelmanson I., Matz M., Lukyanov K., Usman N., Lukyanov S. (2000) Curr. Biol. 10, R473–R474 [DOI] [PubMed] [Google Scholar]
- 13.Baranova A., Ivanov D., Petrash N., Pestova A., Skoblov M., Kelmanson I., Shagin D., Nazarenko S., Geraymovych E., Litvin O., Tiunova A., Born T. L., Usman N., Staroverov D., Lukyanov S., Panchin Y. (2004) Genomics 83, 706–716 [DOI] [PubMed] [Google Scholar]
- 14.Vogt A., Hormuzdi S. G., Monyer H. (2005) Brain Res. Mol. Brain Res. 141, 113–120 [DOI] [PubMed] [Google Scholar]
- 15.Penuela S., Bhalla R., Gong X. Q., Cowan K. N., Celetti S. J., Cowan B. J., Bai D., Shao Q., Laird D. W. (2007) J. Cell Sci. 120, 3772–3783 [DOI] [PubMed] [Google Scholar]
- 16.Penuela S., Celetti S. J., Bhalla R., Shao Q., Laird D. W. (2008) Cell Commun. Adhes. 15, 133–142 [DOI] [PubMed] [Google Scholar]
- 17.Wang X. H., Streeter M., Liu Y. P., Zhao H. B. (2009) J. Comp. Neurol. 512, 336–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bruzzone R., Hormuzdi S. G., Barbe M. T., Herb A., Monyer H. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 13644–13649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bao L., Locovei S., Dahl G. (2004) FEBS Lett. 572, 65–68 [DOI] [PubMed] [Google Scholar]
- 20.Vanden Abeele F., Bidaux G., Gordienko D., Beck B., Panchin Y. V., Baranova A. V., Ivanov D. V., Skryma R., Prevarskaya N. (2006) J. Cell Biol. 174, 535–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Atsumi T., Miwa Y., Kimata K., Ikawa Y. (1990) Cell Differ. Dev. 30, 109–116 [DOI] [PubMed] [Google Scholar]
- 22.Kamiya N., Jikko A., Kimata K., Damsky C., Shimizu K., Watanabe H. (2002) J. Bone Miner. Res. 17, 1832–1842 [DOI] [PubMed] [Google Scholar]
- 23.Williams J. A., Kondo N., Okabe T., Takeshita N., Pilchak D. M., Koyama E., Ochiai T., Jensen D., Chu M. L., Kane M. A., Napoli J. L., Enomoto-Iwamoto M., Ghyselinck N., Chambon P., Pacifici M., Iwamoto M. (2009) Dev. Biol. 328, 315–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shukunami C., Ohta Y., Sakuda M., Hiraki Y. (1998) Exp. Cell Res. 241, 1–11 [DOI] [PubMed] [Google Scholar]
- 25.Nakamura K., Shirai T., Morishita S., Uchida S., Saeki-Miura K., Makishima F. (1999) Exp. Cell Res. 250, 351–363 [DOI] [PubMed] [Google Scholar]
- 26.Watanabe H., de Caestecker M. P., Yamada Y. (2001) J. Biol. Chem. 276, 14466–14473 [DOI] [PubMed] [Google Scholar]
- 27.Kumar N. M., Gilula N. B. (1996) Cell 84, 381–388 [DOI] [PubMed] [Google Scholar]
- 28.Yamasaki H., Naus C. C. (1996) Carcinogenesis 17, 1199–1213 [DOI] [PubMed] [Google Scholar]
- 29.Ionescu A. M., Schwarz E. M., Vinson C., Puzas J. E., Rosier R., Reynolds P. R., O'Keefe R. J. (2001) J. Biol. Chem. 276, 11639–11647 [DOI] [PubMed] [Google Scholar]
- 30.Bringhurst F. R., Juppner H., Guo J., Urena P., Potts J. T., Jr., Kronenberg H. M., Abou-Samra A. B., Segre G. V. (1993) Endocrinology 132, 2090–2098 [DOI] [PubMed] [Google Scholar]
- 31.Schwindinger W. F., Fredericks J., Watkins L., Robinson H., Bathon J. M., Pines M., Suva L. J., Levine M. A. (1998) Endocrine 8, 201–209 [DOI] [PubMed] [Google Scholar]
- 32.Cooper D. M. (2003) Biochem. J. 375, 517–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Locovei S., Wang J., Dahl G. (2006) FEBS Lett. 580, 239–244 [DOI] [PubMed] [Google Scholar]
- 34.Pelegrin P., Surprenant A. (2006) EMBO J. 25, 5071–5082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaplan A. D., Kilkenny D. M., Hill D. J., Dixon S. J. (1996) Endocrinology 137, 4757–4766 [DOI] [PubMed] [Google Scholar]
- 36.Tonon R., D'Andrea P. (2000) J. Bone Miner. Res. 15, 1669–1677 [DOI] [PubMed] [Google Scholar]
- 37.Zhang W., Green C., Stott N. S. (2002) J. Cell. Physiol. 193, 233–243 [DOI] [PubMed] [Google Scholar]
- 38.Richardson R., Donnai D., Meire F., Dixon M. J. (2004) J. Med. Genet. 41, 60–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kjaer K. W., Hansen L., Eiberg H., Leicht P., Opitz J. M., Tommerup N. (2004) Am. J. Med. Genet. 127A, 152–157 [DOI] [PubMed] [Google Scholar]
- 40.Lecanda F., Warlow P. M., Sheikh S., Furlan F., Steinberg T. H., Civitelli R. (2000) J. Cell Biol. 151, 931–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.