Summary
The RecQ-helicase family is widespread, highly conserved, and includes human orthologues that suppress genomic instability and cancer. In vivo, some RecQ homologues promote reduction of steady-state levels of bimolecular recombination intermediates (BRIs), which block chromosome segregation if not resolved. We find that in vivo, E. coli RecQ can promote the opposite: the net accumulation of BRIs. We report that cells lacking Ruv and UvrD BRI-resolution and -prevention proteins die and display failed chromosome segregation attributable to accumulation of BRIs. Death and segregation failure require RecA and RecF strand-exchange proteins. FISH data show that replication is completed during chromosome-segregation failure/death of ruv uvrD recA(Ts) cells. Surprisingly, RecQ (and RecJ) promote this death. The data imply that RecQ promotes the net accumulation of BRIs in vivo, indicating a second paradigm for the in-vivo effect of RecQ-like proteins. The E. coli RecQ paradigm may provide a useful model for some human RecQ homologues.
Introduction
The RecQ family of DNA helicases is widespread and highly conserved. Homologues in Escherichia coli, budding and fission yeasts, Drosophila, and human WRN, BLM, and RECQ4 are the best characterized, and two additional human homologues, RECQ1 and RECQ5, are known (reviewed, Bachrati and Hickson, 2003; Wu and Hickson, 2006). Loss of function of some RecQ homologues results in chromosomal aberrations and genomic instability, including hyper-recombination, genome rearrangements, and in humans, cancer proneness (Myung et al., 2001; Saintigny et al., 2002; Stewart et al., 1997; and reviewed, Bachrati and Hickson, 2003). Loss of BLM, WRN, and RECQ4 functions results in Bloom, Werner, and Rothmund-Thomson cancer-predisposition syndromes. In contrast, genome instability is not prominent in E. coli recQ mutants. Instability was reported in E. coli recQ cells expressing phage λ proteins (Hanada et al., 1997), but not observed in the absence of λ (Mashimo et al., 2003).
Despite the importance of RecQ-like proteins, their precise cellular role(s) remain uncertain. RecQ-like helicases unwind multiple substrates in vitro, and the most relevant for each in vivo are unclear. In vitro, 3′–5′ unwinding activity is reported on B-form DNA, Holliday junctions (HJ), forked structures (e.g., BLM, WRN, yeast Sgs1, RecQ), and G4 (tetraplex) DNA (e.g., BLM, Sgs1, RecQ) (reviewed, Bachrati and Hickson, 2003; and see Hishida et al., 2004). In vivo, possible roles in preventing or repairing stalled replication forks are suggested for RecQ and others (Bjergbaek et al., 2005; Courcelle and Hanawalt, 1999; Doe et al., 2000; Florés et al., 2005; Hishida et al., 2004; Wu and Hickson, 2006), though the precise role(s) in such processes is uncertain.
Nevertheless, for some RecQ-like proteins an overall “paradigm” to explain in vivo effects has emerged; several RecQ-like proteins reduce steady-state levels of bimolecular intermediates in homologous recombination (BRIs). BRIs can be toxic if not removed. For example, loss of WRN leads to reduced viability (and decreased mitotic recombination) that can be rescued either by expressing a bacterial HJ resolvase or by loss of RAD51, which catalyzes BRI formation (Saintigny et al., 2002). Accumulated BRIs can block chromosome segregation, reducing viability. Viability is presumably restored by processing the BRIs by resolvase or by the loss of RAD51. Drosophila DmBLM appears to process BRIs in vivo (McVey et al., 2004). Human BLM unwinds model BRIs in vitro (Wu and Hickson, 2003). S. pombe rqh1 mutants display chromosome-segregation defects that are suppressed by expressing a bacterial HJ resolvase (Doe et al., 2000). Both in budding yeast and S. pombe, sgs1 srs2 (rqh1 srs2 in S. pombe) double mutants are inviable only if strand-exchange proteins Rad51, Rad55, and Rad57 are present, implying death by unresolved BRIs and failure of chromosome segregation (Doe and Whitby, 2004; Gangloff et al., 2000; Liberi et al., 2005). srs2, homologue of E. coli uvrD discussed below, encodes a helicase that removes Rad51 from DNA, causing an apparent build-up of BRIs in its absence (Krejci et al., 2003; Veaute et al., 2003). The net cellular levels of BRIs could be reduced by these RecQ-like helicases in either of two ways: they might unwind problematic DNA structures that, if left, require recombinational DNA repair; or they might directly promote unwinding (reversal) or dissolution/resolution of BRIs formed in spontaneous DNA damage-repair events (Bachrati and Hickson, 2003; Wu and Hickson, 2006). Either way, net levels of BRIs are reduced. This in vivo paradigm does not exclude other possible roles for these proteins, or even that they might act at multiple stages in recombination. Here, we address whether E. coli RecQ affects the overall outcome of recombination in vivo similarly.
E. coli RecQ has been hypothesized to act in various roles in DNA metabolism. RecQ promotes re-start of stalled replication forks in vivo (Courcelle and Hanawalt, 1999), and in vitro unwinds the lagging-strand equivalent in model forks (Hishida et al., 2004). Its role in replication re-start might be via recombination, via non-recombinational direct assistance at the replication fork (Courcelle and Hanawalt, 1999), or indirectly via promotion of an SOS DNA damage response (Hishida et al., 2004). In model partial reactions of recombination in vitro, RecQ can promote RecA-mediated strand-exchange (Harmon and Kowalczykowski, 1998). Although this suggests a role in increasing net levels of BRIs in vivo, RecQ also catalyzes reactions compatible with reduction of BRI levels. RecQ stimulates strand-passage activity of Topoisomerase (Topo) III, leading to either catenation or decatenation of duplex DNA molecules (Harmon et al., 1999). The decatenation activity resembles that of BLM with Topo III in resolution of model BRIs (Wu and Hickson, 2003). This would suggest a role in net reduction of BRIs, also suggested on the basis of in vivo data (Lopez et al., 2005, discussed below). Similarly, decatenation by RecQ and Topo III could serve additionally to decatenate and resolve interlinked bacterial chromosomes after recombination and HJ resolution (Harmon et al., 1999), which would also support RecQ acting via the yeast/fly/WRN paradigm of net reduction of levels of BRIs in vivo. Alteration of DNA topology with Topo III might alternatively aid DNA replication-fork progression, not recombination. Finally, RecQ might promote both BRI formation and resolution in vivo. Which of these, if either, is its predominant role in vivo is examined here.
Here we present evidence that E. coli RecQ can promote the net accumulation of BRIs in vivo, the opposite of the paradigm shown for yeast, fly, and two human RecQ homologues. We suggest that this exemplifies a second paradigm for the net effect of a RecQ-family protein in vivo. This paradigm for E. coli RecQ might pertain to human RecQ homologues, perhaps other than BLM or WRN, for which the cellular roles are poorly defined.
Results
Inviability of ΔruvΔuvrD Mutants
We discovered an inability to construct strains carrying both Δruv and ΔuvrD mutations, and hypothesized that the combination might result in inviability (also called “synthetically lethality”). This appears to contradict a previous report of experiments with ruvA uvrD cells (Bierne et al., 1997). A later paper discusses possible inviability of ruv uvrD mutants (Florés et al., 2004); however the authors neither present data nor state how possible inviability was determined. Because rigorous demonstrations of synthetic lethality in E. coli are non-trivial (below), and because of the apparent conflict in the literature, we show by a few means that cells carrying ruvA, ruvB, or ruvC and uvrD null mutations are inviable, then use this fact, and its cause, to probe the role of E. coli RecQ in vivo.
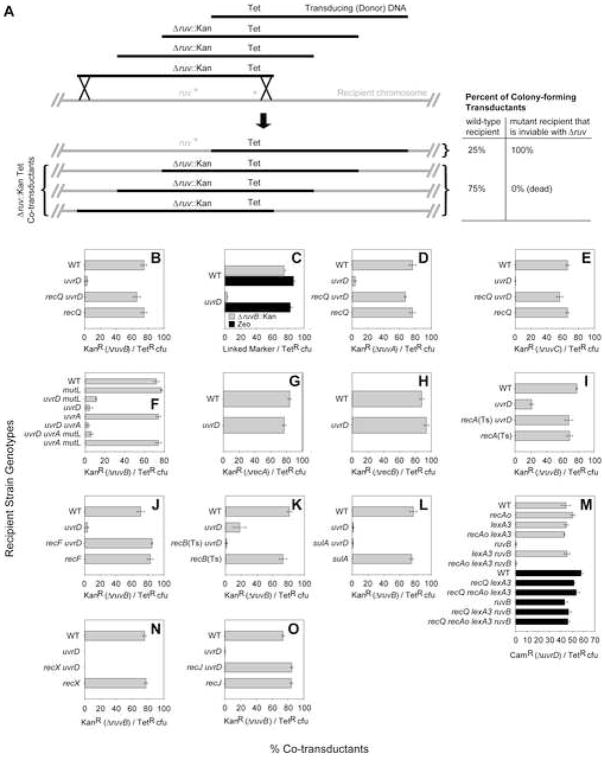
To demonstrate possible inviability of ΔuvrD Δruv mutants, we first used a genetic-linkage/phage P1 co-transduction assay (e.g., Washburn and Kushner, 1991). The strategy is illustrated in Figure 1A. Phage P1 are grown on a donor strain carrying a Δruv::Kan mutation (a ruv deletion/replacement with a kanamycin-resistance cassette). The Δruv::Kan allele in the donor strain is partially genetically linked to a nearby Tet marker (Figure 1A). Transducing particles (phage carrying bacterial DNA) grown on this donor strain will carry DNA fragments, some with the Tet marker, and others with both Tet and Kan, which can therefore “co-transduce” both simultaneously into a recipient strain (Figure 1A). The transducing phage suspension is applied to “wild-type” and isogenic ΔuvrD recipient strains, tetracycline-resistant (TetR) transductants selected, and the frequency of KanR (Δruv::Kan) co-transductants among TetR transductants determined. If the combination is inviable, TetR transductants of the ΔuvrD strain will be obtained, but Δruv::Kan TetR co-transductants will not. As a positive control, both classes should be observable when transducing the wild-type recipient, which stays viable after ruv deletion (Figure 1A).
Figure 1.
RecA-, RecF-, and RecQ-Mediated Inviability of Δruv ΔuvrD Cells
(A) Phage P1 co-transduction assay for synthetic lethality. Transducing DNA fragments from phage grown on a donor strain marked with Tet near a mutation of interest (Δruv::Kan) are used to transduce “wild-type” and isogenic mutant recipients. The frequency of co-transduction of Δruv::Kan with Tet depends on the distance between the two markers. E.g., when there is 75% linkage, the two markers are co-transduced in 75% of Tet-resistant (TetR) transductants. Recipient mutants inviable in combination with Δruv fail to produce colonies that carry both Tet marker and the linked Δruv::Kan, thereby reducing the co-transductant frequency.
(B–O) Co-transduction data. Means ± SEM, n ≥ 3 independent determinations each. WT, wild-type. Inefficient recovery of (B) ΔruvB::Kan zea-3::Tn10, (D) ΔruvA::Kan eda-51::Tn10, or (E) ΔruvC::Kan eda-51::Tn10 co-transductants of ΔuvrD, rescued by ΔrecQ (Tn10 carries the Tet element). (C) ΔuvrD does not alter linkage/co-transductant frequencies generally (black bars, donor has Zeo marker near ruv+), but is specific to transduction of ruv mutations into ΔuvrD strains (gray bars, donor: ΔruvB::Kan zea-3::Tn10). (F) Neither MMR nor NER defects cause inviability with ΔruvB. (G) Loss of HR at the strand-exchange stage (ΔrecA) is viable with ΔuvrD. (H) ΔrecB is viable with ΔuvrD. (I) Loss of RecA rescues the ΔruvB ΔuvrD inviability. (J) ΔrecF rescues the ΔruvB ΔuvrD inviability. (K) Loss of RecB does not rescue ΔruvB ΔuvrD inviability. (L) Loss of SulA does not rescue ΔruvB ΔuvrD inviability. (M) RecQ promotes death of recombination-proficient/SOS-deficient cells. SOS-deficient [lexA3(Ind−)] ΔruvB ΔuvrD cells are dead when RecA levels are not limiting due to a recA operator-constitutive allele, recAo281 [gray bars, recAo281 lexA3(Ind−) ΔruvB compared with lexA3(Ind−) ΔruvB and ΔruvB]. ΔrecQ rescues recAo281 lexA3(Ind−) ΔruvB ΔuvrD inviability (black bars; P1 donor also contains ΔrecQ, which cotransduces normally with ΔuvrD. See Supplemental Data.). (N) Loss of RecX does not rescue ΔruvB ΔuvrD inviability. (O) ΔrecJ rescues the ΔruvB ΔuvrD inviability.
ruvB encodes the helicase component of the RuvABC Holliday-junction (HJ)-processing complex (Sharples et al., 1999). We show that ΔruvB::Kan is co-transduced in ~75% of TetR transductants of wild-type recipients, reflecting ~75% linkage of the two markers (Figure 1A, B, top). In Figure 1B (top), we see >25-fold reduction of co-transduction of ΔruvB::Kan with Tet in the ΔuvrD recipient strain, implying that ΔruvB ΔuvrD mutants are inviable.
A few apparent ΔruvB ΔuvrD transductant colonies were obtained (Figure 1B, top). If the combination is truly inviable, these rare apparent ΔruvB ΔuvrD strains could have formed colonies, first, as a result of transductions into cells that acquired, or acquire during subsequent residual growth, a mutation that suppresses the inviability. Second, ~10−3 of cells carry spontaneous tandem duplications of segments of the bacterial chromosome, and so are partially diploid (Roth et al., 1996). Transduction of the deletion allele replacing only one of the ruv copies would produce viable ΔruvB::Kan/ruvB+ duplication heterozygotes. Though duplications are normally infrequent (~10−3), we found that nine of 46 (~20%) of the rare ΔruvB ΔuvrD colonies obtained were ΔruvB::Kan/ruvB+ duplication heterozygotes (PCR assay, Supplemental Data). This strongly supports the conclusion that ruvB is essential in ΔuvrD cells.
In a further control, reduced co-transductant frequency of ΔruvB::Kan with Tet in ΔuvrD recipient cells is not caused by changes in co-transduction efficiency/genetic-linkage generally by ΔuvrD, but is specific to recovery of co-transductants of a ruv mutation in ΔuvrD cells. Figure 1C shows that co-transduction of a zeocin-resistance (Zeo) marker near ruv+ with the Tet was the same in the ΔurvD and wild-type control strains (black bars), whereas ΔruvB::Kan co-transductants with Tet were reduced greatly (gray bars). Thus reduced frequencies of co-transductants of ΔruvB::Kan with Tet in ΔuvrD recipients is not caused by changes in genetic linkage relationships in ΔuvrD cells, further supporting the conclusion of inviability of Δruv ΔuvrD double mutants.
Inviability with ΔuvrD is also observed with ΔruvA and ΔruvC mutations (Figure 1D,E). ruvA and ruvC encode a HJ-binding protein and endonuclease, that work with RuvB to resolve BRIs (Sharples et al., 1999). Thus strains lacking the UvrD helicase appear to require a functional RuvABC HJ-resolution system, not just the RuvB helicase, for viability.
Neither MMR nor NER Is the UvrD Essential Function
UvrD is required for completion of MMR and NER (Friedberg et al., 2005), and also removes RecA strand-exchange protein from single-strand (ss)DNA, reducing BRIs (Veaute et al., 2005). We show that neither MMR nor NER is required for viability in Δruv cells. In co-transduction experiments, ΔruvB was introduced efficiently into strains carrying null alleles of mutL (MMR) and/or uvrA (NER) genes, indicating that neither deficiency causes the inviability of ΔuvrD withΔruv (Figure 1F).
In addition, both MutL and UvrA act early in repair, producing repair intermediates, whereas UvrD removes those intermediates (Friedberg et al., 2005). We show also that loss of MutL, UvrA, or both simultaneously did not rescue the inviability of Δruv ΔuvrD mutants (Figure 1F). Thus, MMR and/or NER intermediates, which might accumulate in cells lacking UvrD, are also not lethal when RuvB is lost. These data suggest that reduction of BRIs by UvrD, for example via its RecA-removal activity, could be the function that is essential in Δruv cells (supported further, and alternatives discussed, below).
BRI Formation Not Essential in ΔuvrD Cells
The Ruv-dependent essential process in ΔuvrD cells could be homologous recombination (HR) as a whole, or completion of HR by resolution of BRIs specifically. We find that deletions inactivating either of two proteins that promote BRIs do not cause inviability with ΔuvrD (Figure 1G,H): RecA strand-exchange protein required for all HR, and RecBCD double-strand-end (DSE)-processing enzyme, required prior to strand exchange in DSE-repair (Kuzminov, 1999). This extends previous conclusions of viability of uvrD recA and uvrD recB non-deletion strains (reviewed, Kuzminov, 1999). These data rule out models in which, e.g., ΔuvrD mutation results in chronic DNA damage that must be repaired using HR for viability, and support models in which failure to resolve BRIs specifically is toxic in cells lacking UvrD.
Δruv ΔuvrD Inviability Requires RecA and RecF, not RecB
We hypothesized that the inviability of Δruv with ΔuvrD results from accumulation of toxic BRIs, preventing segregation of sister chromosomes after initiation of recombinational repair of spontaneous DNA damage. In support of this idea, we found that loss of either RecA (temperature-sensitive allele at restrictive temperature: 42°C) or RecF strand-exchange/strand-exchange-accessory proteins allowed viability of Δruv ΔuvrD mutants (Figure 1I,J). Similar conclusions were made by Florés et al. (2004, 2005) based on experimental methods and data not reported. Our data support the validity of those conclusions.
Loss of RecB did not restore viability to Δruv ΔuvrD cells (Figure 1K, temperature-sensitive allele at 42°C). Because RecB function is highly specific to initiation of BRIs from DSEs (Kuzminov, 1999), these data imply that the toxic HR that kills Δruv ΔuvrD cells is not mainly from repair of DSEs, but rather suggests ssDNA lesions. Similarly, RecF is implicated mainly in HR-repair of ssDNA gaps (reviewed, Kuzminov, 1999). The data support a model whereby RecF and RecA initiate HR from a substrate other than a DSE to create toxic BRIs in Δruv ΔuvrD cells.
Chromosome-Segregation Defects During Death of Δruv ΔuvrD Mutants
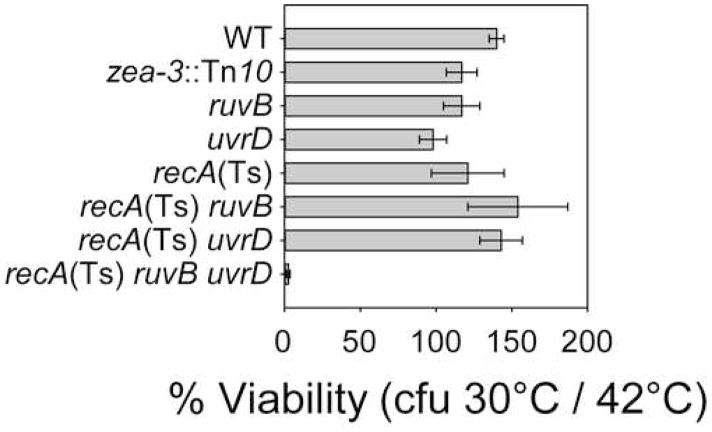
Unresolved BRIs between sister chromosomes can block chromosome segregation (e.g., Ishioka et al., 1998). We examined chromosome segregation by growing cultures of Δruv ΔuvrD cells carrying a temperature-sensitive recA(Ts) allele at restrictive temperature (42°C, RecA− phenotype), then shifting to permissive temperature (30°C, RecA+ phenotype). First, we show that death occurs en masse upon temperature shift (Figure 2). This confirms, by independent means, the inviability of Δruv ΔuvrD mutants and its dependence on RecA+.
Figure 2.
Temperature-shift assay for viability of ΔruvB ΔuvrD recA200(Ts) cells. Inviability of ΔruvB ΔuvrD recA200(Ts) cells shifted to permissive temperature (RecA+). zea-3::Tn10 is an incidental marker present in all strains shown. Means ± SEM, n = 3 independent determinations.
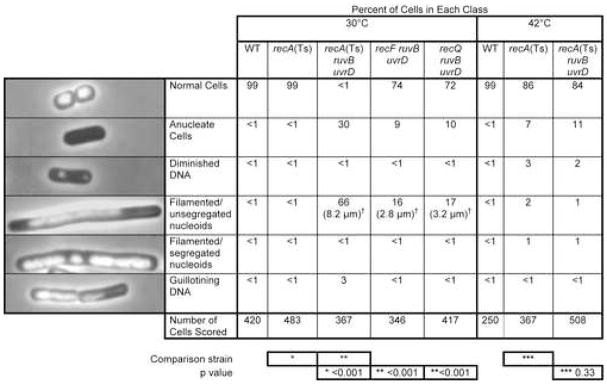
Second, DNA DAPI staining and fluorescence microscopy show dramatic chromosome-segregation defects after the shift (Figure 3). Saturated 42°C cultures were diluted into fresh medium and grown to late-log phase at either the restrictive or permissive temperature. When re-grown at the restrictive temperature (RecA−), essentially all (84%) of ΔruvB ΔuvrD recA(Ts) cells appeared normal, compared with recA(Ts) cells (86% normal), containing distinct bacterial chromosomes or nucleoids (Figure 3). Of those that are abnormal in the recA(Ts) genetic background, most have reduced or no DNA content, probably reflecting the known DNA-degradation phenotype in recA cells (Clark et al., 1966). Importantly, when re-grown at the permissive temperature (30°, RecA+), most (66%) of Δruv ΔuvrD recA(Ts) cells were present as long “filaments” many cell bodies long containing unsegregated nucleoids (Figure 3). Filaments with properly-segregated nucleoids, which occur when cell division is blocked but chromosome partitioning is normal (Chen and Beckwith, 2001), were an insignificant <1% of cells (Figure 3). Most of the filaments have tips devoid of visible DNA (Figure 3), as expected if cell elongation continues without chromosome segregation. Moreover, 30% of Δruv ΔuvrD recA(Ts) cells were anucleate after shift to permissive temperature (Figure 3), implying that cell division occurred in regions of cells that had not received a nucleoid. An additional 3% of cells displayed DNA stretched across the septum of a dividing cell (“guillotining” effect, Figure 3), which has been attributed to the inability of cells to segregate unresolved chromosomes (Hendricks et al., 2000). These are all classic chromosome-segregation-defective phenotypes as in par (“chromosome-partition”) mutants of E. coli and Salmonella typhimurium (Britton and Lupski, 1998). Similar, but less severe phenotypes were observed in UV-irradiated recG and ruvC E. coli cells (Ishioka et al., 1998), which are defective at resolving BRIs, and were also suppressed by loss of RecA, supporting the idea that in that study and here, a toxic build-up of BRIs is responsible.
Figure 3.
Chromosome-Segregation Failure Accompanies Death of Δruv ΔuvrD Cells
Cultures were grown from common 42°C-grown saturated cultures, split, diluted and grown to log-phase at 30°C or 42°, harvested, prepared for DAPI DNA-fluorescence microscopy, and scored blind. p values calculated from Chi-squared analyses indicate significant differences in the distributions between the indicated strains (* versus *, ** versus **, etc.) (SigmaStat 3.1 from SPSS, Inc). Table S3 for comparisons of numbers of normal cells in all relevant single- and double-mutant combinations.
†Median cell lengths. ~2.7-fold reductions by ΔrecF and ΔrecQ compared with the recA(Ts)ΔruvB ΔuvrD cells. The median length of 100 wild-type (normal) cells was 1.5 μm.
Further support for the idea that the chromosome-segregation failure is caused by accumulated BRIs comes from our finding that loss of either RecA or RecF strand-exchange proteins rescued the chromosome-segregation defects of Δruv ΔuvrD mutants (Figure 3). Absence of RecA resulted in normal low frequencies of filamented (1%) and anucleate (11%) cells comparable to those in recA(Ts) single mutants (Figure 3, 2% and 7%, respectively at 42°C). Similarly, loss of RecF resulted in a substantially higher frequency of 74% normal cells and lower length and frequency of filaments with unsegregated nucleoids (Figure 3, only two cell bodies long and 16% filamented at 30°C). All viable single- and double-mutant combinations were comparable to wild type (at 30°C and 42°C), or recA(Ts) at 42°C when strains contain recA(Ts) (Table S3). Thus, the death of Δruv ΔuvrD cells is accompanied by RecA- and RecF-dependent failure of chromosome segregation, as expected for death by toxic accumulation of BRIs.
Apart from recA, SOS-Gene Induction Not Required for Inviability of Δruv ΔuvrD Cells
RecA and RecF strand-exchange proteins could promote failure of chromosome segregation and death of Δruv ΔuvrD cells either directly, via formation of BRIs, or indirectly via their roles in activation of SOS DNA-damage-response genes, or both. The SOS response checkpoint is activated when RecA binds ssDNA, which can form at stalled replication forks or during DNA break repair. Activated RecA facilitates the proteolytic cleavage of the LexA transcriptional repressor, upregulating the expression of >40 DNA-repair and damage-response genes, including recA, and also the sulA cell-division inhibitor which causes filamentation (Friedberg et al., 2005). We tested whether SOS induction causes chromosome-segregation failure and death first by examining sulA. Loss of SulA did not restore viability to Δruv ΔuvrD cells (Figure 1L). Thus, sulA-mediated cell-division arrest is not responsible for the death.
However, SOS-induced levels of RecA are required for death of Δruv ΔuvrD cells as follows. First, blocking upregulation of SOS genes by using a special uncleavable mutant LexA repressor [lexA3(Ind−)] restored viability to Δruv ΔuvrD cells (Figure 1M, gray bars, lexA3 ruvB). This indicates that induction of one or more LexA-repressed genes is required for the death of Δruv ΔuvrD cells. Though recA is expressed constitutively, recA is also repressed by LexA such that RecA levels increase during SOS and are reduced in lexA3(Ind−) cells. This reduction in RecA could result in fewer strand-exchange events. We find that inviability requires increased expression only of recA, and not of other LexA-repressed gene(s); we see that lexA3(Ind−) does not block death of Δruv ΔuvrD when recA is overexpressed due to an operator-constitutive mutation (recAo281), which elevates levels of RecA in lexA3 cells (Volkert et al., 1981) (Figure 1M, gray bars, recAo lexA3 ruvB). Thus, upregulation of no SOS gene other than recA appears to be required for death of Δruv ΔuvrD cells.
recX is a gene downstream of and co-transcribed with recA (reviewed, Lusetti et al., 2004). The possibility that upregulation of recX, and not recA, restored inviability to recAo281 lexA3 Δruv ΔuvrD cells (Figure 1M) is argued against as follows. First, recX encodes a negative regulator of RecA activity (Lusetti et al., 2004; Renzette et al., 2007). Thus RecX should not promote inviability; it should have the opposite effect from RecA. Second, we show that unlike RecA (Figures 1I, 2), RecX is not required for inviability of Δruv ΔuvrD (Figure 1N).
Thus, SOS-induced levels of RecA, and no other SOS gene, are required for the inviability of Δruv ΔuvrD cells. This indicates roles for RecA and RecF in the chromosome-segregation failure and death other than, or in addition to, upregulation of recA.
Completion of Chromosome Replication During Death in Δruv ΔuvrD Mutants
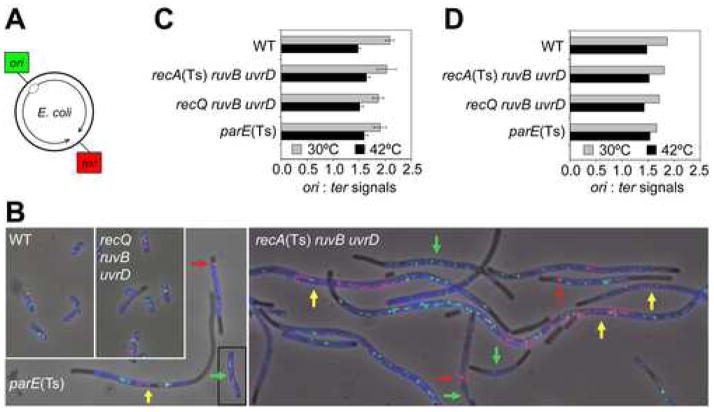
The chromosome-segregation failure/death of Δruv ΔuvrD cells requires RecA and RecF strand-exchange proteins (Figures 1I,J, 2, 3), supporting the idea that accumulated BRIs are responsible. However chromosome-segregation failure can result also from incomplete replication of chromosomes, a process in which RecF has been implicated (Courcelle et al., 1997). FISH analyses show that incomplete replication does not accompany chromosome-segregation failure as follows (Figure 4). Loci near the origin (“ori”) and terminus (“ter”, near dif locus) of replication (Figure 4A) were probed simultaneously to examine the number of each in individual recA(Ts) ΔruvB ΔuvrD cells shifted to permissive temperature, at which death occurs (Figure 2) and chromosome segregation fails (Figure 3). If replication were incomplete, multiple rounds of initiation should produce more copies of ori-containing DNA relative to DNA near the terminus.
Figure 4.
Completion of Chromosome Replication During Defective Chromosome Segregation/Death of Δruv ΔuvrD Cells
(A) Chromosomal positions of ori and ter.
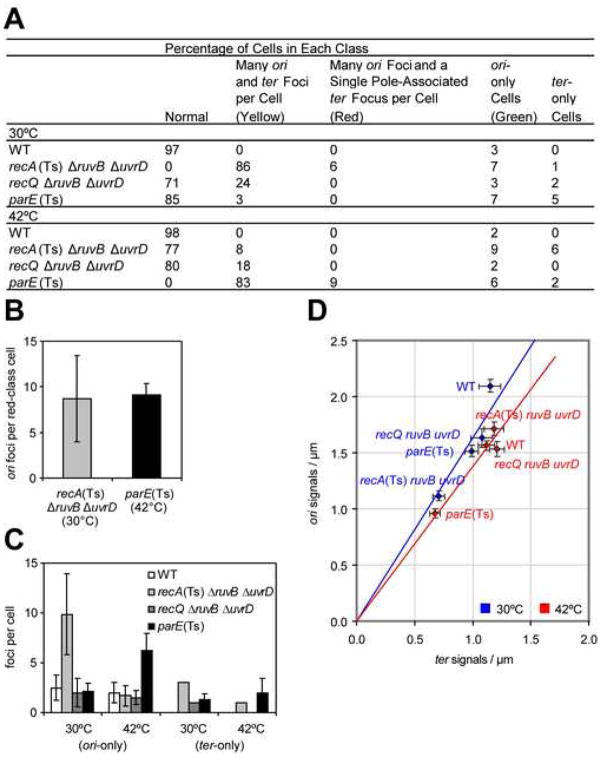
(B) Representative images of selected genotypes at 30°C using two-color FISH to loci shown in (A). Images are overlays of phase-contrast, DAPI (blue), green (ori-hybridization) and red (ter-hybridization) exposures. Common 42°C-grown [30°C for parE(Ts)] saturated cultures were diluted and grown to late-log phase at 30°C [42°C for parE(Ts)], at which each strain dies, prepared and analyzed. Colored arrows: yellow (“yellow class cells”), most ter signals near mid-cell with ori-signals near the cell poles; green, ori-only cells; red, ter signals clustered at one cell pole/to one side of a septum. Figure 5 for quantification of these classes.
(C, D) Completion of replication during chromosome-segregation failure in ΔruvB ΔuvrD recA(Ts) cells. Ratios of ori:ter signals per cell (means ± SEM) from--(C) normal and yellow-class cells (102–128 cells/genotype), and (D) all classes (104–144 cells/genotype). The ori:ter ratios do not differ between genotypes (p = 0.739 and 0.938 for 30°C and 42°C, Chi-squared analysis). All resemble wild-type [shown previously for control parE(Ts) cells (Khodursky et al., 2000)].
First, we find that recA(Ts) ΔruvB ΔuvrD cells at permissive temperature (RecA+) have more of both ori and ter signals per cell than wild-type cells (Figure 4B) but that the signals increase proportionally. The ori- to ter-signal ratio is unchanged from that of wild-type cells and of parE(Ts) cells (Figure 4C,D), a control for a chromosome segregation-defective strain in which chromosome replication is completed (Khodursky et al., 2000). This demonstrates completion of chromosome replication at the gross, whole-chromosome level.
Second, we describe specific aberrant patterns of distribution of ori and ter signals for both recA(Ts) ΔruvB ΔuvrD and parE(Ts) mutants as each dies after temperature shift, and show that the two are essentially identical [Figure 5, compare recA(Ts) ΔruvB ΔuvrD at 30° with parE(Ts) at 42°]. During death of most recA(Ts) ΔruvB ΔuvrD and parE(Ts) cells, most of the ter signals cluster near mid-cell with the ori signals near the cell poles (Figure 4B, yellow arrows, Figure 5A), as in dividing cells just prior to septation (Bates and Kleckner, 2005; Wang et al., 2006). Seven and 6% of the two dying strains’ cells had several ori but no ter signals (Figure 4B, green arrows, Figure 5A). These “ori-only cells” appear to result from inappropriate septation following incomplete chromosome segregation, in that similar numbers of cells had ter-containing DNA in only one of two daughter cells (e.g., Figure 4B, bottom red arrow, Figure 5), or as a single ter focus at one end of the cell with many ori foci throughout (Figure 4B, red arrows). The numbers in these latter two classes were not significantly different (p=1.000 for difference by z-test) for both genotypes during death (Figure 5A). Additional similarities in their FISH profiles are outlined in Figure 5B-D. parE10(Ts) cells are defective in a subunit of Topo IV at restrictive temperature, and par mutants are characterized by apparently replicated chromosomes (Khodursky et al., 2000) that accumulate mid-cell of a filament, but cannot be segregated (Britton and Lupski, 1998). Thus, chromosome-segregation failure of recA(Ts) Δruv ΔuvrD cells mimics failure to disentangle complete chromosomes as in parE(Ts) cells. This rules out replication failure and provides further support for unresolved BRIs as the cause.
Figure 5.
ori- and ter- Focus Distributions in Various Mutants
(A) Percentages of cells in five classes of distributions of ori and ter foci (104–144 cells/genotype). Colors refer to arrows in Figure 4B, showing examples of each class. “Normal” defined per Figure 3 and with ori and ter signal numbers within the range observed in wild-type (WT) cells at the same temperature.
(B) Numbers of ori foci per cell in cells with many ori and a single ter focus at a cell pole (red class). Comparable numbers in dying recA(Ts) ΔruvB ΔuvrD cells and parE(Ts) chromosome-segregation-defective control. Means ± SD.
(C) Numbers of foci in ori-only and ter-only cells in various mutants. In most genotypes, the average numbers were approximately 3±1 (e.g., ori signal for WT at 30°C). Imperfect probe labeling/hybridization and 3-D constraints on cell visualization should yield rare cells with only ori or ter foci. Both dying recA(Ts) ΔruvB ΔuvrD cells (30°C) and parE(Ts) (42°C) had ori-only cells (green class) with significantly more foci than this apparent background, suggesting similar chromosome-segregation defects in these strains. ori-only cells are thought to form from inappropriate septation after failed chromosome segregation. Means ± SD.
(D) Numbers of ori and ter foci per unit of cell length are reduced (proportionately) in ΔruvB ΔuvrD recA(Ts) cells (30°C) and parE(Ts) cells (42°C) during chromosome-segregation failure (yellow-class population of C). This might indicate loss of the coordinate regulation of cell growth and chromosome replication. Blue and red lines, best-fit regressions passing through zero for strains grown at 30°C or 42°C. Means ± SEM. Supplemental Data for strains.
RecQ Promotes Inviability and Chromosome-Segregation Failure of Δruv ΔuvrD Cells
Surprisingly, we find that deletion of recQ allows viability of Δruv ΔuvrD cells (Figure 1B,D,E). Thus, RecQ is required for the inviability of Δruv ΔuvrD mutants, shown above to result from accumulation of unresolved BRIs.
Similarly, RecQ is also required for chromosome-segregation defects of ΔruvB ΔuvrD mutants (Figures 3, 4B and 5):
First, loss of RecQ mimics loss of RecF strand-exchange-accessory protein in Δruv ΔuvrD cells conferring a strong partial suppression of chromosome-segregation failure (Figure 3); 72% of cells showed normal segregation and only 10% and 17% were anucleate or in short filaments with unsegregated nucleoids, respectively.
Second, FISH analyses showed nearly normal (small) numbers of ori and ter signals per cell in a ratio similar to wild-type (Figures 4B-D and 5A,C,D).
Third, like RecA and RecF, RecQ plays a role in SOS induction (Hishida et al., 2004). However, RecQ does not appear to promote accumulation of unresolved BRIs via indirect effects on SOS induction. We see that ΔrecQ rescues recA-constitutive/SOS-uninducible recAo lexA3 Δruv ΔuvrD cells (Figure 1M). These cells have enough RecA to initiate strand-exchange but cannot induce increased expression of SOS genes, and will die without the ΔrecQ deletion. Thus, RecQ promotes the chromosome-segregation failure and death independently of possible effects on upregulation of SOS genes (Figure 1M).
These data imply that, in vivo, RecQ promotes the net accumulation of toxic BRIs that lead to failed chromosome segregation in cells lacking the UvrD anti-recombination helicase and Ruv BRI-resolution proteins.
The promotion of the net accumulation of BRIs by RecQ contrasts with the net cellular effects of budding yeast Sgs1, S. pombe Rqh1, and human WRN, which prevent the net accumulation of BRIs in vivo (reviewed in Introduction). This appears to represent a second net in-vivo effect for a RecQ-family protein in recombination. Further evidence for this difference between E. coli RecQ and the two yeast homologues is also shown in Figure 1. On the one hand, loss of Sgs1 or Rqh1 causes death by toxic recombination in cells lacking Srs2 helicase (Doe and Whitby, 2004; Gangloff et al., 2000), the homologue of UvrD (Aboussekhra et al., 1989), which reduces BRIs by stripping Rad51 from DNA (Krejci et al., 2003; Veaute et al., 2003). In contrast, cells lacking E. coli RecQ are viable in combination with loss of the Srs2 homologue, UvrD (Figure 1B,D,E).
In possible contrast with our conclusions, data not shown led previous authors to conclude that loss of RecF, but not RecQ, rescued inviability of ruvABC uvrD mutants that also carry mutations in other genes [dnaE recBC or dnaN recBC (Florés et al., 2005)]. We cannot evaluate whether a true discrepancy exists, or its possible cause(s) if so, because we have seen neither the data nor the means of testing of inviability employed (both unpublished).
Inviability of Δruv ΔuvrD Cells Requires RecJ
RecJ is a 5′-to-3′ single-strand exonuclease that functions in several DNA repair processes. recJ and recQ mutations confer similar phenotypes in some assays/repair processes, prompting models in which the two act together in these processes (discussed below). We find that, like RecQ, RecJ is required for (promotes) the death by toxic BRI accumulation in Δruv ΔuvrD cells; loss of RecJ restored viability of Δruv ΔuvrD mutants (Figure 1O). This extends a previous mention of data not shown that cells carrying recBC(Ts) ruvABC uvrD recJ and dnaN(Ts) or dnaE(Ts) mutations are viable (Florés et al., 2005). At least two possible, non-exclusive roles of RecJ, could promote net accumulation of BRIs in vivo. RecJ could help expose ssDNA used by RecA in recombinational synapsis of sister chromosomes, for example at stalled replication forks (per Courcelle and Hanawalt, 1999; Florés et al., 2005; Hishida et al., 2004; and Figure 6B), and/or RecJ could act post-synaptically to stabilize BRIs (Corette-Bennett and Lovett, 1995).
Figure 6.
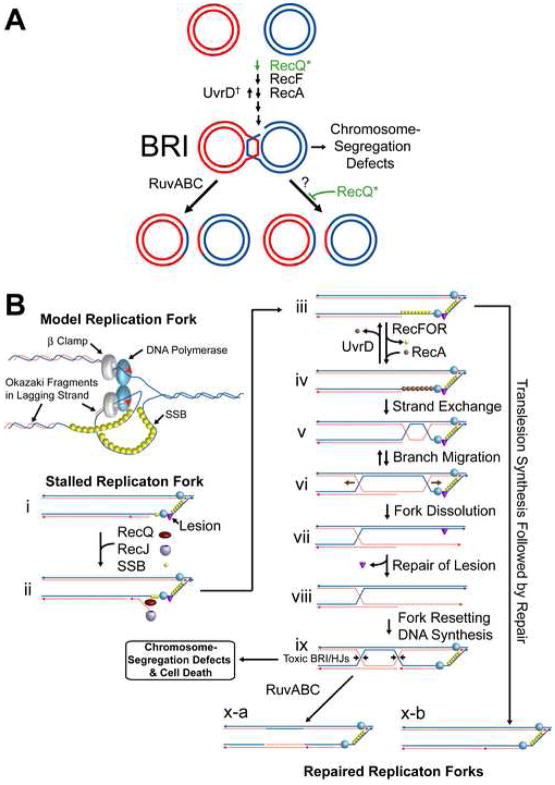
Models for RecQ-Promoted Net Accumulation of Bimolecular Recombination Intermediates (BRIs)
(A) RecQ might either promote strand exchange (top) or block/slow a Ruv-independent BRI-resolution pathway (bottom). Alternatives, “*”. BRIs accumulate in cells lacking RuvABC HJ-resolution activity and UvrD causing chromosome-segregation failure and inviability. Blocking net accumulation of BRIs by loss of RecQ, RecF, or RecA suppresses these phenotypes.
†UvrD could keep BRIs at survivable levels in ruv cells via its RecA-removal/anti-BRI activity (Veaute et al., 2005) (shown), but might in principle promote Ruv-independent BRI resolution [not shown or demonstrated, but similarly to a proposal for UvrD homologue Srs2 (Ira et al., 2003)]. Either way, loss of UvrD and Ruv lead to BRI accumulation.
(B) Model: RecQ-Promoted BRI Formation for Lagging-Strand-Template Repair. (i) A lesion in the lagging-strand template stalls the replication fork. (ii) RecQ unwinds the nascent lagging strand (Courcelle and Hanawalt, 1999; Hishida et al., 2004), which is degraded by RecJ (Courcelle and Hanawalt, 1999). (iii) SSB binds the ssDNA. (iv) RecFOR promotes exchange of SSB for RecA (Morimatsu and Kowalczykowski, 2003); UvrD counteracts (somewhat), removing RecA (Veaute et al., 2005), and loss of UvrD increases BRI formation. (v) RecA-promoted strand exchange creating a BRI. (vi) Branch migration produces a Holliday junction (HJ). (vii) Repair of the DNA lesion and (viii, ix) resetting of the fork, which now precedes two HJs, linking the sister chromosomes until (ix, arrows, to x-a) RuvABC resolve the HJs. In the absence of Ruv, replication proceeds, but the entangled chromosomes (BRI) cause chromosome-segregation failure and death. Alternative pathway (arrow from iii to x-b) operating when RecA is counteracted by UvrD. Fork regression and repair (not shown) or translesion synthesis allowing replication restart (x-b).
Discussion
E. coli RecQ In Vivo Paradigm
The data presented imply that in cells defective for HJ resolution and anti-recombinase activities of the RuvABC system and UvrD helicase, respectively, E. coli RecQ promotes death via toxic recombination (Figures 1–5, alternative role for UvrD discussed Figure 6A, legend). The inviability requires RecA and RecF strand-exchange proteins, and RecQ and RecJ, and is accompanied by a dramatic failure to segregate chromosomes (Figures 3–5). The data imply that sister chromosomes joined in unresolved BRIs during acts of recombinational-repair of spontaneous DNA damage cause death, and that RecQ (and RecJ) promote the net accumulation of BRIs in these cells.
Although interpreted differently, the results of a previous study support the conclusion that RecQ promotes net accumulation of BRIs in vivo. Lopez et al. (2005) found a role for E. coli Topo III in a HJ-resolution pathway alternative to RuvABC. They also found death with failed chromosome segregation in cells lacking Topo III and Topo IV, and showed that RecA and RecQ were required for that death. This implies, we now suggest, that RecA and RecQ promoted net BRI accumulation leading to chromosome-segregation failure.
RecQ could promote a net accumulation of BRIs in vivo in either of two general ways (Figure 6A). It could act, like RecA and RecF, to facilitate BRI formation (Figure 6A, top, and a specific model in Figure 6B, discussed below). Alternatively, RecQ might stabilize BRIs after their formation, for example by blocking an as-yet unidentified pathway for BRI resolution/removal that operates in the absence of the Ruv system (Figure 6A, lower right).
Other, non-recombinational roles for RecQ have been proposed, including in promoting the regression of replication forks into “chicken-foot” configuration which stops replication (Florés et al., 2005). However, our data showing completion of chromosome replication during RecQ-promoted death by recombination (Figures 4, 5) appear to rule out models relying on blocked replication as the cause of death.
In either of the two general models in Figure 6A (promoting BRI formation or blocking Ruv-independent resolution), the net effect is that RecQ promotes accumulation of BRIs in vivo, the opposite of the net effect of some RecQ homologues (Introduction and below). This indicates a different net effect or paradigm for the function of a RecQ-family protein in recombination in vivo. Because several RecQ homologues (including RecQ) show biochemical and/or in vivo activities compatible with both initiation, and dissolution/resolution of BRIs (reviewed, Introduction), we suggest that considering the overall effect on recombination/BRIs in vivo is useful for understanding their predominant functions in vivo.
Model for RecQ in Lagging-Strand-Template Repair
A possible specific model in which RecQ promotes BRI formation is shown (Figure 6B). In it, RecQ is part of a recombinational pathway that allows otherwise irreparable DNA lesions to be repaired. Figure 6Bi shows a replication fork stalled due to a replication-blocking lesion (triangle) in the lagging-strand template. The lesion is refractory to repair because it is in single-stranded DNA (Friedberg et al., 2005). RecQ might act with RecJ to remove the nascent lagging strand, as proposed (Courcelle and Hanawalt, 1999; Florés et al., 2005; Hishida et al., 2004), revealing ssDNA capable of being bound by RecA (Figure 6Bi–iii). RecA binding is reversed by UvrD (Veaute et al., 2005) (Figure 6Biii-iv). When UvrD fails to remove RecA [sometimes in wild-type cells; always in ΔuvrD cells (iv)], RecA-promoted strand exchange with a sister chromosome (v) leads to formation of a HJ(s) (vi). Further branch migration/strand exchange (vi–vii) incorporates the lesion into double-stranded DNA, where it can be repaired (vii–viii). In Ruv+ cells, fork re-setting and Ruv-catalyzed resolution of the BRI (ix, at arrows) completes the process (x-a). In Δruv cells, although replication is completed (Figures 4, 5), the HJs trailing the replication fork (Figure 6Bix) persist, blocking chromosome segregation and causing death.
An alternative, Ruv-independent, repair pathway is shown by the arrow from step iii to x-b. It can explain why UvrD+ Δruv cells survive lagging-strand template lesions. In it, the lesion could be bypassed by a trans-lesion polymerase, then repaired without producing potentially toxic BRIs.
Though attractive, many important aspects of this model remain to be tested.
RecQ Homologues
Whereas we have shown that E. coli RecQ can promote accumulation of toxic BRIs in vivo, yeast Sgs1, Rqh1, human WRN and Drosophila DmBLM do the opposite (reviewed, Introduction). BLM possesses biochemical activities compatible with this role (Wu and Hickson, 2003), possibly linking BLM with the yeast/fly/WRN group. These yeast, fly, and human RecQ homologues could reduce BRIs in either of two general ways. They could unwind BRIs, directly promoting their resolution or dissolution (Fabre et al., 2002; Ira et al., 2003; Liberi et al., 2005; Saintigny et al., 2002; Wu and Hickson, 2003). Similarly, they could act as anti-recombinases, controlling the fidelity of recombination by aborting inappropriate BRIs such as those containing mismatches (Bachrati and Hickson, 2003; Myung et al., 2001; Spell and Jinks-Robertson, 2004). Alternatively, these helicases could act pre-recombinationally to unwind aberrant DNA structures, such as G4 (tetraplex) DNA, which might otherwise require recombinational repair, or be processed to structures such as breaks that require repair (Fabre et al., 2002; Huber et al., 2002). Either way, these proteins have the net effect of reduction of toxic BRIs in cells, constituting the first described phenotypic “paradigm” for the net effect of RecQ-like proteins on recombination in vivo, and contrasting with the E. coli RecQ paradigm described here.
E. coli RecQ provides a second paradigm for the overall effect of RecQ-like proteins in recombination in vivo, a paradigm that might pertain to some of the less understood human proteins, and the defect(s) that result from their loss. Not all human RecQ homologues are known to fall into the yeast/fly/WRN paradigm for RecQ-like proteins: reducing net levels of BRIs in vivo. Humans possess three RecQ homologues in addition to BLM and WRN: RECQ4, which is deficient in some cases of Rothmund-Thompson syndrome, and RECQ5 and RECQ1 (reviewed, Bachrati and Hickson, 2003). Of these, RECQ4 might be a candidate for similarity to the E. coli RecQ paradigm in that its Xenopus orthologue is implicated in creation/stabilization of ssDNA used to load ssDNA-binding replication proteins (SSBs) at origins (Sangrithi et al., 2005). Loading SSBs is also an obligatory first step in strand-exchange (Friedberg et al., 2005) that promotes BRI formation. If BLM falls within the yeast/fly/WRN group, then RECQ5 might also in that RECQ5 functions in a non-redundant pathway with BLM, perhaps similarly to BLM (Hu et al., 2005). RECQ1 is important for maintenance of genome stability (Sharma et al., 2006) but poorly understood, and thus might fit with either (or neither) of these two general paradigms for in vivo effect of RecQ homologues on recombination and genome stability.
Experimental Procedures
See Supplemental Data for strains, new alleles, media and growth conditions.
Quantitative Phage P1-Mediated Co-transduction Assays
P1 transductions (as Miller, 1992, with the following modifications) were performed at 32°C, immediately plated on LBH plates with tetracycline (Tet) and sodium citrate, and incubated 4–5 days at 32°C. Examination of plates on subsequent days showed that apparent decreases in transductants were not caused by slow growth of colonies. 100 colonies/genotype were purified by streaking, grown at 32°C for 1–3 days, and single colonies patched sequentially to an LBH-Kan-citrate, or LBH-Cam-citrate, or LBH-Zeo-citrate plate, then to LBH-Tet-citrate. Co-transductant frequencies are numbers of KanR/TetR, CamR/TetR, or ZeoR/TetR patches. Genotypes were verified by antibiotic resistance, and when relevant, PCR of the deletion allele and UV sensitivity. For recA(Ts) and recB(Ts) recipients, transductions were performed at 30°C and all subsequent incubations at 42°C with pre-warmed plates. ΔrecB::Kan experiments were incubated at 32°C.
Temperature-Shift Assays
Cfu were measured from saturated cultures grown at 42°C, plated on pre-warmed LBH plates then incubated at 30°C or 42°C and numbers of colonies scored. The inviable genotype (Figure 2) was scored again after ≥4 days.
Microscopy
For chromosome-segregation analyses (Figure 3), cells were fixed and stained (Addinall et al., 1996, with minor modifications). Cultures were diluted 1/500, grown to the indicated growth phase, and fixed by resuspension of pellets in equal volumes of PBS with 4% paraformaldehyde for 30 minutes at room temperature, then washed 3 times in PBS and stored in GTE (pH 7.5). Appropriate numbers of cells embedded in 0.7% agar on slides were stained with 4′, 6-diamidino-2-phenylindole (DAPI; 1 μg/ml). Samples were photographed with an Olympus BX51 microscope equipped with an Uplan Fluorite 100X oil objective, DAPI filter (U-N31000, Olympus), and an Olympus MagnaFire CCD digital camera. Merges of phase-contrast and DAPI-fluorescence images were made with Magnafire software (Optronics). Blind scoring was performed on captured images.
Fluorescence in situ Hybridization (FISH)
FISH probes were 6 kb DNA fragments PCR-amplified (Phusion DNA polymerase, NEB) from MG1655 DNA. Primers for the origin (ori) and the terminus (ter) probes were as (Bates and Kleckner, 2005). The PCR fragments were gel purified and ligated (Quick Ligation Kit, NEB). ori and ter probes were labeled with SpectrumGreen and SpectrumOrange, respectively (Vysis Nick Translation Kit). For each slide, 200 ng of each probe was ethanol precipitated and resuspended individually in 10 μl hybridization buffer (1.4X SSC, 35% formamide, 7% dextran sulfate), then mixed together, denatured at 73°C for 5 minutes and placed on ice immediately before adding to the slides.
To prepare slides for FISH, cells were washed as above before the following steps. Slides were dehydrated by soaking in a series of 70%, 90%, and 100% ethanol for 2 min. each at room temperature, air dried, then incubated in 75°C denaturation solution [70% formamide (Sigma), 2X SSC, pH 7.2] for 5 min. and immediately dehydrated in a −20°C ethanol series of 70%, 90%, and 100% for 2 min. each. Slides were dried by gently blowing compressed air over the top of each slide. Denatured probes were added, glass cover slips applied and sealed with rubber cement before overnight incubation in the dark in a 37°C humidity chamber. After removing the cover slip, slides were washed for 2 sec. in 73°C 0.4X SSC, 5 sec. in room temperature 2X SSC/0.05% Tween-20 (Sigma), air dried in darkness, then mounted using VECTASHIELD Mounting Medium (Vector Laboratories) with 1 μg/ml DAPI. Probes were visualized using GFP (41017 EN-GFP, Chroma Tech, Inc.) or OR (41003 HQ:R&B Phy, Chroma Tech, Inc.) filters and images analyzed and prepared with ImageJ (Abramoff et al., 2004) and Adobe Photoshop.
Supplementary Material
Acknowledgments
We thank R Kolodner, EL Zechiedrich, R Cirz and F Romesberg for strains, D Bates and C Herman for helpful suggestions, and F Fabre, R Galhardo, J Gibson, J Halliday, PJ Hastings, G Ira, R Monnat, P Opresko, S Plon, and H Shinagawa for comments. Supported by a DOD Breast Cancer Research Program Predoctoral Fellowship (JMP), Baylor College of Medicine Mental Retardation Research Center (HD 2406407, JAL and JRL), and NIH grant R01-CA85777.
Footnotes
Supplemental Data including Supplemental Results, Experimental Procedures, and three tables can be found at http://www.molecule.org.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aboussekhra A, Chanet R, Zgaga Z, Cassier-Chauvat C, Heude M, Fabre F. RADH, a gene of Saccharomyces cerevisiae encoding a putative DNA helicase involved in DNA repair. Characteristics of radH mutants and sequence of the gene. Nucleic Acids Res. 1989;17:7211–7219. doi: 10.1093/nar/17.18.7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abramoff MD, Magalhães PJ, Ram SJ. Image processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- Addinall SG, Bi E, Lutkenhaus J. FtsZ ring formation in fts mutants. J Bacteriol. 1996;178:3877–3884. doi: 10.1128/jb.178.13.3877-3884.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachrati CZ, Hickson ID. RecQ helicases: suppressors of tumorigenesis and premature aging. Biochem J. 2003;374:577–606. doi: 10.1042/BJ20030491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D, Kleckner N. Chromosome and replisome dynamics in E. coli: loss of sister cohesion triggers global chromosome movement and mediates chromosome segregation. Cell. 2005;121:899–911. doi: 10.1016/j.cell.2005.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne H, Seigneur M, Ehrlich SD, Michel B. uvrD mutations enhance tandem repeat deletion in the Escherichia coli chromosome via SOS induction of the RecF recombination pathway. Mol Microbiol. 1997;26:557–567. doi: 10.1046/j.1365-2958.1997.6011973.x. [DOI] [PubMed] [Google Scholar]
- Bjergbaek L, Cobb JA, Tsai-Pflugfelder M, Gasser SM. Mechanistically distinct roles for Sgs1p in checkpoint activation and replication fork maintenance. EMBO J. 2005;24:405–417. doi: 10.1038/sj.emboj.7600511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton RA, Lupski JR. Partitioning and segregation of the bacterial chromosome. In: de Bruijn FJ, Lupski JR, Weinstock GM, editors. Bacterial Genomes, Physical Structure and Analysis. New York, New York: Chapman and Hall; 1998. pp. 103–111. [Google Scholar]
- Chen JC, Beckwith J. FtsQ, FtsL and FtsI require FtsK, but not FtsN, for co-localization with FtsZ during Escherichia coli cell division. Mol Microbiol. 2001;42:395–413. doi: 10.1046/j.1365-2958.2001.02640.x. [DOI] [PubMed] [Google Scholar]
- Clark AJ, Chamberlin M, Boyce RP, Howard-Flanders P. Abnormal metabolic response to ultraviolet light of a recombination-deficient mutant of Escherichia coli K12. J Mol Biol. 1966;19:442–454. doi: 10.1016/s0022-2836(66)80015-3. [DOI] [PubMed] [Google Scholar]
- Corette-Bennett S, Lovett ST. Enhancement of RecA strand transfer activity by the RecJ exonuclease of Escherichia coli. J Biol Chem. 1995;270:6881–6885. doi: 10.1074/jbc.270.12.6881. [DOI] [PubMed] [Google Scholar]
- Courcelle J, Carswell-Crumpton C, Hanawalt PC. RecF and RecR are required for resumption of replication at DNA replication forks in Escherichia coli. Proc Natl Acad Sci USA. 1997;94:3714–3719. doi: 10.1073/pnas.94.8.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courcelle J, Hanawalt PC. RecQ and RecJ process blocked replication forks prior to the resumption of replication in UV-irradiated Escherichia coli. Mol Gen Genet. 1999;262:543–551. doi: 10.1007/s004380051116. [DOI] [PubMed] [Google Scholar]
- Doe CL, Dixon J, Osman F, Whitby MC. Partial suppression of the fission yeast rqh1− phenotype by expression of a bacterial Holliday junction resolvase. EMBO J. 2000;19:2751–2762. doi: 10.1093/emboj/19.11.2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe CL, Whitby MC. The involvement of Srs2 in post-replication repair and homologous recombination in fission yeast. Nucleic Acids Res. 2004;32:1480–1491. doi: 10.1093/nar/gkh317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre F, Chan A, Heyer WD, Gangloff S. Alternate pathways involving Sgs1/Top3, Mus81/Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc Natl Acad Sci USA. 2002;99:16887–16892. doi: 10.1073/pnas.252652399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florés MJ, Bidnenko V, Michel B. The DNA repair helicase UvrD is essential for replication fork reversal in replication mutants. EMBO Rep. 2004;5:983–988. doi: 10.1038/sj.embor.7400262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florés MJ, Sanchez N, Michel B. A fork-clearing role for UvrD. Mol Microbiol. 2005;57:1664–1675. doi: 10.1111/j.1365-2958.2005.04753.x. [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2. Washington, DC: ASM Press; 2005. [Google Scholar]
- Gangloff S, Soustelle C, Fabre F. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat Genet. 2000;25:192–194. doi: 10.1038/76055. [DOI] [PubMed] [Google Scholar]
- Hanada K, Ukita T, Kohno Y, Saito K, Kato J, Ikeda H. RecQ DNA helicase is a suppressor of illegitimate recombination in Escherichia coli. Proc Natl Acad Sci USA. 1997;94:3860–3865. doi: 10.1073/pnas.94.8.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon FG, DiGate RJ, Kowalczykowski SC. RecQ helicase and topoisomerase III comprise a novel DNA strand passage function: a conserved mechanism for control of DNA recombination. Mol Cell. 1999;3:611–620. doi: 10.1016/s1097-2765(00)80354-8. [DOI] [PubMed] [Google Scholar]
- Harmon FG, Kowalczykowski SC. RecQ helicase, in concert with RecA and SSB proteins, initiates and disrupts DNA recombination. Genes Dev. 1998;12:1134–1144. doi: 10.1101/gad.12.8.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks EC, Szerlong H, Hill T, Kuempel P. Cell division, guillotining of dimer chromosomes and SOS induction in resolution mutants (dif, xerC and xerD) of Escherichia coli. Mol Microbiol. 2000;36:973–981. doi: 10.1046/j.1365-2958.2000.01920.x. [DOI] [PubMed] [Google Scholar]
- Hishida T, Han YW, Shibata T, Kubota Y, Ishino Y, Iwasaki H, Shinagawa H. Role of the Escherichia coli RecQ DNA helicase in SOS signaling and genome stabilization at stalled replication forks. Genes Dev. 2004;18:1886–1897. doi: 10.1101/gad.1223804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Lu X, Barnes E, Yan M, Lou H, Luo G. Recql5 and Blm RecQ DNA helicases have nonredundant roles in suppressing crossovers. Mol Cell Biol. 2005;25:3431–3442. doi: 10.1128/MCB.25.9.3431-3442.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber MD, Lee DC, Maizels N. G4 DNA unwinding by BLM and Sgs1p: substrate specificity and substrate-specific inhibition. Nucleic Acids Res. 2002;30:3954–3961. doi: 10.1093/nar/gkf530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira G, Malkova A, Liberi G, Foiani M, Haber JE. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishioka K, Fukuoh A, Iwasaki H, Nakata A, Shinagawa H. Abortive recombination in Escherichia coli ruv mutants blocks chromosome partitioning. Genes Cells. 1998;3:209–220. doi: 10.1046/j.1365-2443.1998.00185.x. [DOI] [PubMed] [Google Scholar]
- Khodursky AB, Peter BJ, Schmid MB, DeRisi J, Botstein D, Brown PO, Cozzarelli NR. Analysis of topoisomerase function in bacterial replication fork movement: use of DNA microarrays. Proc Natl Acad Sci USA. 2000;97:9419–9424. doi: 10.1073/pnas.97.17.9419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejci L, Van Komen S, Li Y, Villemain J, Reddy MS, Klein H, Ellenberger T, Sung P. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003;423:305–309. doi: 10.1038/nature01577. [DOI] [PubMed] [Google Scholar]
- Kuzminov A. Recombinational repair of DNA damage in Escherichia coli and bacteriophage λ. Microbiol Mol Biol Rev. 1999;63:751–813. doi: 10.1128/mmbr.63.4.751-813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberi G, Maffioletti G, Lucca C, Chiolo I, Baryshnikova A, Cotta-Ramusino C, Lopes M, Pellicioli A, Haber JE, Foiani M. Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes Dev. 2005;19:339–350. doi: 10.1101/gad.322605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez CR, Yang S, Deibler RW, Ray SA, Pennington JM, Digate RJ, Hastings PJ, Rosenberg SM, Zechiedrich EL. A role for topoisomerase III in a recombination pathway alternative to RuvABC. Mol Microbiol. 2005;58:80–101. doi: 10.1111/j.1365-2958.2005.04812.x. [DOI] [PubMed] [Google Scholar]
- Lusetti SL, Drees JC, Stohl EA, Seifert HS, Cox MM. The DinI and RecX proteins are competing modulators of RecA function. J Biol Chem. 2004;279:55073–55079. doi: 10.1074/jbc.M410371200. [DOI] [PubMed] [Google Scholar]
- Mashimo K, Kawata M, Yamamoto K. Roles of the RecJ and RecQ proteins in spontaneous formation of deletion mutations in the Escherichia coli K12 endogenous tonB gene. Mutagenesis. 2003;18:355–363. doi: 10.1093/mutage/geg004. [DOI] [PubMed] [Google Scholar]
- McVey M, Larocque JR, Adams MD, Sekelsky JJ. Formation of deletions during double-strand break repair in Drosophila DmBlm mutants occurs after strand invasion. Proc Natl Acad Sci USA. 2004;101:15694–15699. doi: 10.1073/pnas.0406157101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. A short course in bacterial genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- Morimatsu K, Kowalczykowski SC. RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: a universal step of recombinational repair. Mol Cell. 2003;11:1337–1347. doi: 10.1016/s1097-2765(03)00188-6. [DOI] [PubMed] [Google Scholar]
- Myung K, Datta A, Chen C, Kolodner RD. SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination. Nat Genet. 2001;27:113–116. doi: 10.1038/83673. [DOI] [PubMed] [Google Scholar]
- Renzette N, Gumlaw N, Sandler SJ. DinI and RecX modulate RecA-DNA structures in Escherichia coli K-12. Mol Microbiol. 2007;63:103–115. doi: 10.1111/j.1365-2958.2006.05496.x. [DOI] [PubMed] [Google Scholar]
- Roth JR, Benson N, Galitski T, Haack K, Lawrence JG, Meisel L. Rearrangements of the bacterial chromosome: formation and applications. In: Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella Cellular and Molecular Biology. Washington, D. C: ASM Press; 1996. pp. 2256–2276. [Google Scholar]
- Saintigny Y, Makienko K, Swanson C, Emond MJ, Monnat RJ., Jr Homologous recombination resolution defect in Werner syndrome. Mol Cell Biol. 2002;22:6971–6978. doi: 10.1128/MCB.22.20.6971-6978.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangrithi MN, Bernal JA, Madine M, Philpott A, Lee J, Dunphy WG, Venkitaraman AR. Initiation of DNA replication requires the RECQL4 protein mutated in Rothmund-Thomson syndrome. Cell. 2005;121:887–898. doi: 10.1016/j.cell.2005.05.015. [DOI] [PubMed] [Google Scholar]
- Sharma S, Stumpo DJ, Balajee AS, Bock CB, Lansdorp PM, Brosh RM, Jr, Blackshear PJ. RECQL, a Member of the RecQ family of DNA Helicases, suppresses chromosomal instability. Mol Cell Biol. 2006 doi: 10.1128/MCB.01620-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharples GJ, Ingleston SM, Lloyd RG. Holliday junction processing in bacteria: insights from the evolutionary conservation of RuvABC, RecG, and RusA. J Bacteriol. 1999;181:5543–5550. doi: 10.1128/jb.181.18.5543-5550.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spell RM, Jinks-Robertson S. Examination of the roles of Sgs1 and Srs2 helicases in the enforcement of recombination fidelity in Saccharomyces cerevisiae. Genetics. 2004;168:1855–1865. doi: 10.1534/genetics.104.032771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart E, Chapman CR, Al-Khodairy F, Carr AM, Enoch T. rqh1+, a fission yeast gene related to the Bloom’s and Werner’s syndrome genes, is required for reversible S-phase arrest. EMBO J. 1997;16:2682–2692. doi: 10.1093/emboj/16.10.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veaute X, Delmas S, Selva M, Jeusset J, Le Cam E, Matic I, Fabre F, Petit MA. UvrD helicase, unlike Rep helicase, dismantles RecA nucleoprotein filaments in Escherichia coli. EMBO J. 2005;24:180–189. doi: 10.1038/sj.emboj.7600485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veaute X, Jeusset J, Soustelle C, Kowalczykowski SC, Le Cam E, Fabre F. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 2003;423:309–312. doi: 10.1038/nature01585. [DOI] [PubMed] [Google Scholar]
- Volkert MR, Margossian LJ, Clark AJ. Evidence that rnmB is the operator of the Escherichia coli recA gene. Proc Natl Acad Sci USA. 1981;78:1786–1790. doi: 10.1073/pnas.78.3.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Liu X, Possoz C, Sherratt DJ. The two Escherichia coli chromosome arms locate to separate cell halves. Genes Dev. 2006;20:1727–1731. doi: 10.1101/gad.388406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn BK, Kushner SR. Construction and analysis of deletions in the structural gene (uvrD) for DNA helicase II of Escherichia coli. J Bacteriol. 1991;173:2569–2575. doi: 10.1128/jb.173.8.2569-2575.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Hickson ID. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- Wu L, Hickson ID. DNA helicases required for homologous recombination and repair of damaged replication forks. Annu Rev Genet. 2006;40:279–306. doi: 10.1146/annurev.genet.40.110405.090636. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.