Abstract
Carbamazepine (CBZ) is a widely prescribed anticonvulsant whose use is often associated with idiosyncratic hypersensitivity. Sera of CBZ-hypersensitive patients often contain anti-CYP3A antibodies, including those to a CYP3A23 K-helix peptide that is also modified during peroxidative CYP3A4 heme-fragmentation. We explored the possibility that cytochromes P450 (P450s) such as CYP3A4 bioactivate CBZ to reactive metabolite(s) that irreversibly modify the P450 protein. Such CBZ-P450 adducts, if stable in vivo, could engender corresponding serum P450 autoantibodies. Incubation with CBZ not only failed to inactivate functionally reconstituted, purified recombinant CYP3A4 or CYP3A4 Supersomes in a time-dependent manner, but the inclusion of CBZ (0–1 mM) also afforded a concentration-dependent protection to CYP3A4 from inactivation by NADPH-induced oxidative uncoupling. Incubation of CYP3A4 Supersomes with 3H-CBZ resulted in its irreversible binding to CYP3A4 protein with a stoichiometry of 1.58 ± 0.15 pmol 3H-CBZ bound/pmol CYP3A4. Inclusion of glutathione (1.5 mM) in the incubation reduced this level to 1.09. Similar binding (1.0 ± 0.4 pmol 3H-CBZ bound/pmol CYP3A4) was observed after 3H-CBZ incubation with functionally reconstituted, purified recombinant CYP3A4(His)6. The CBZ-modified CYP3A4 retained its functional activity albeit at a reduced level, but its testosterone 6β-hydroxylase kinetics were altered from sigmoidal (a characteristic profile of substrate cooperativity) to near-hyperbolic (Michaelis-Menten) type, suggesting that CBZ may have modified CYP3A4 within its active site.
Carbamazepine (CBZ), one of the most widely used anticonvulsants, has been associated with a series of idiosyncratic adverse reactions, such as skin rash, blood disorders, hepatotoxicity, and hematological disorders (Hart and Easton, 1982; Shear et al., 1988; Leeder, 1998; Kalapos, 2002; Romero Maldonado et al., 2002). These clinical symptoms are consistent with an immune etiology. Although the mechanism of CBZ-induced adverse reactions has not been well characterized, it is widely believed that biotransformation of CBZ into reactive metabolites and their subsequent covalent binding to cellular proteins is responsible for the immune response (Park et al., 1987).
Bioactivation of CBZ has been found to result in covalent adducts of its metabolites with microsomal proteins (Pirmohamed et al., 1992; Lillibridge et al., 1996). In vitro studies with recombinant human P450s expressed in yeast have demonstrated that P450s 1A, 2C, and 3A are responsible for the covalent binding of CBZ to human liver microsomal proteins with 65 and 31% covalent adduct formation attributed to CYP3A4 and CYP1A2, respectively, after normalization of their relative hepatic abundance (Wolkenstein et al., 1998). However, preincubation of human and rat liver microsomes with CBZ in the presence of NADPH with subsequent assay of residual P450 monooxygenation activities provided no evidence for mechanism-based inactivation of rat CYP2C11, CYP3A, CYP1A1/2, or CYP2B1/2 or that of human CYP2C8, CYP2C9, or CYP3A4, whereas time-dependent inactivation of rat CYP2D6 and human CYP1A2 was observed (Masubuchi et al., 2001).
Sera of CBZ-hypersensitive patients often contain anti-CYP3A antibodies including those specific to the CYP3A23 K-helix peptide (EYLDMLNETLRL) (Leeder et al., 1996), a region also modified during peroxidative CYP3A4 heme-fragmentation (He et al., 1998). We explored the possibility that P450s such as CYP3A4 bioactivate CBZ to reactive metabolite(s) that irreversibly modify the P450 protein. Such CBZ-P450 adducts, if stable in vivo, could engender corresponding serum P450 autoantibodies if processed and presented to the immune system in an appropriate immunologic context.
Unusual substrate kinetic interactions involving CYP3A4 have been reported, which are proposed to result from the simultaneous binding of multiple substrate molecules and consequent substrate cooperativity within the CYP3A4 active site (Shou et al., 1994; Ueng et al., 1997; Harlow and Halpert, 1998). Covalent modification of the CYP3A4 active site has been shown to alter the kinetics of CYP3A4 catalysis, indicating the existence of multiple binding sites (Schrag and Wienkers, 2001; Khan et al., 2002). The recently reported crystal structure of CYP3A4 complexed with two molecules of ketoconazole suggested that the atypical kinetics displayed by CYP3A4 could be the result of multiple coexistent binding modes at the active site (Williams et al., 2004; Yano et al., 2004; Ekroos and Sjogren, 2006). The finding that CBZ-modified CYP3A4 still retains considerable functional activity led us to investigate the altered enzyme kinetic profile of CBZ-modified CYP3A4.
Materials and Methods
Materials
CBZ, testosterone, 6β-hydroxytestosterone, GSH, NADPH, EDTA, Hepes, sodium cholate, catalase, magnesium chloride (MgCl2), and ethyl acetate (EtOAc) were purchased from Sigma-Aldrich (St. Louis, MO). [3H]CBZ was purchased from Amersham Biosciences (Piscataway, NJ). Erythromycin was purchased from Calbiochem (San Diego, CA), and the N-desmethylerythromycin standard was synthesized by Pfizer Inc. (San Diego, CA). l-α-Dilauroylphosphatidylcholine, l-α-dioleyl-sn-glycerophosphatidyl-choline, and phosphatidyl-serine were purchased from Doosan Serdary Research Laboratories (Englewood Cliffs, NJ). Microsomes (Supersomes; BD Gentest, Woburn, MA) from baculovirus-infected insect cells expressing CYP3A4 along with coexpressed human NADPH-cytochrome P450 reductase (P450 reductase) and human cytochrome b5 (b5) were purchased from BD Gentest. Ni2+-NTA resin was purchased from Qiagen (Valencia, CA). The recombinant CYP3A4(His)6 was expressed in Escherichia coli DH5αF′ cells and purified exactly as described previously (Wang et al., 1998; Xue et al., 2001). Rat liver b5 and P450 reductase were purified from phenobarbital-pretreated male rat liver microsomes as previously reported (Licad-Coles et al., 1997).
Determination of CBZ Covalent Binding to Functionally Reconstituted Recombinant CYP3A4
Purified recombinant CYP3A4(His)6, assayed spectrophotometrically (Wang et al., 1998), was functionally reconstituted with P450 reductase and cytochrome b5 at a previously determined optimal molar ratio of CYP3A4/P450 reductase/b5 of 1:4:2 using testosterone 6β-hydroxylase as the functional probe as described (Wang et al., 1998). The reagents were added at 4°C in the following order in a final volume of 1 ml: lipid mixture (l-α-dilauroylphosphatidylcholine/l-α-dioleyl-sn-glycerophosphati-dylcholine/phosphatidylserine, 1:1:1, w/w, 20 µg), sodium cholate (200 µg), cytochrome b5 (500 pmol), P450 reductase (1000 pmol), CYP3A4 (250 pmol), and GSH (1.5 µmol). The mixture was reconstituted at room temperature for 10 min and then placed back on ice. The following reagents were then added in order to a final volume of 1 ml: Hepes (50 mM, pH 7.85), 20% glycerol, catalase (200 U), EDTA (1 mM), water, MgCl2 (30 mM), and CBZ with [3H]CBZ (1 mM and 1.25 µCi/ml final, dissolved in methanol). After a 3-min preincubation at 37°C, NADPH (1 mM final) was added to initiate the reaction. Controls for nonspecific binding were prepared as described except that the incubations contained no NADPH. Carrier rat liver microsomal protein (10 mg) was added after a 30-min incubation, and the incubation was immediately quenched with EtOAc. The protein precipitate was then washed three times with methanol containing 5% H2SO4, followed by three washes with a mixture of ethanol and ethyl ether (3:1, v/v) and two washes with 80% aqueous methanol. The pellet was dissolved in 1 N NaOH. An aliquot was neutralized with 1 N HCl, and liquid scintillation fluid was added for radioactivity determination. The protein concentration of another aliquot was also determined to calculate the protein recovery. The stoichiometry of covalent binding was determined on the basis of the recovered residual radioactivity irreversibly bound to the precipitated protein pellet. Higher concentrations of GSH up to 4 mM were also included in the incubation mixture to study the effect of GSH on the covalent binding.
Determination of CBZ-Covalent Binding to CYP3A4 Supersomes
A mixture of CYP3A4 Supersomes (100 pmol), isocitric acid (5 mM), isocitric acid dehydrogenase (0.5 U/ml), potassium phosphate buffer (100 mM, pH 7.4), CBZ with [3H]CBZ (1 mM and 1.25 µCi/ml final, dissolved in methanol), and EDTA (1 mM) was preincubated for 3 min at 37°C. NADPH (1 mM, final) was added to start the reaction (final volume, 0.5 ml). Controls for nonspecific binding consisted of exactly similar incubations without NADPH. Carrier microsomal protein (10 mg) was added after a 30-min incubation, and the reaction was terminated with EtOAc. The protein pellet was washed as detailed above. The stoichiometry of covalent binding was calculated on the basis of the recovered residual radioactivity irreversibly bound to the protein pellet as described above. When the effect of GSH on the CBZ covalent binding to CYP3A4 was assessed, GSH up to 4 mM was also included in the incubation mixture.
Reisolation of [3H]CBZ-CYP3A4(His)6 Adduct
After a 30-min incubation of the functionally reconstituted recombinant CYP3A4(His)6 with [3H]CBZ as described above, the incubation mixture was mixed with pre-equilibrated Ni2+-NTA resin and then loaded onto a column. After removal of free [3H]CBZ by extensive washing with Hepes buffer (50 mM, pH 7.85) containing 20 mM imidazole, the [3H]CBZ-CYP3A4(His)6 adduct was eluted out of the column with the same buffer containing 200 mM imidazole. The purified [3H]CBZ-CYP3A4(His)6 adduct was subjected to SDS-PAGE analyses and the protein quantified by densitometry relative to the content of the purified CYP3A4 used as the starting material and electrophoresed in parallel. The stoichiometry of covalent binding was calculated on the basis of the residual radioactivity irreversibly bound to the recombinant CYP3A4 protein.
Testosterone 6β-Hydroxylase Activity of the CBZ-Modified CYP3A4
After a 30-min incubation of CBZ with either CYP3A4 Supersomes or the functionally reconstituted purified recombinant CYP3A4 as described above, an aliquot of CYP3A4 (10 pmol) was added to a secondary reaction mixture (1 ml final volume) containing testosterone (1.9–500 µM) and other reagents as described above. The reaction was initiated by the addition of NADPH (1 mM), allowed to run at 37°C for 20 min, and then quenched with EtOAc. Progesterone was included as an internal control. The metabolites were extracted twice with EtOAc, dried down under N2, and assayed by high-performance liquid chromatography with peak area quantification for 6β-hydroxytestosterone, the major product, as described previously (Xue et al., 2001).
Kinetic analysis of CYP3A4 6β-hydroxylation of testosterone was performed using the Hill equation:
where υ is the enzyme velocity, C is the substrate concentration, Km is the substrate concentration at the half-maximum velocity, n is the Hill coefficient, and Vmax is the maximal velocity. The nonlinear, least-squares curve fitting was carried out with IGOR Pro 4.01 (WaveMetrics Inc., Lake Oswego, OR).
Time-Dependent Inhibition of CYP3A4 Testosterone Hydroxylase Activity
Functionally reconstituted, purified recombinant CYP3A4 was incubated with various concentrations of CBZ (0–1000 µM) at 37°C as described above. Aliquots of the mixtures (40 µl containing 10 pmol CYP3A4) were taken at various time intervals (0–20 min) and added to a secondary reaction mixture (1 ml final volume) containing 250 µM testosterone, Hepes (50 mM, pH 7.85), 20% glycerol, catalase (200 U), EDTA (1 mM), GSH (1.5 mM), and MgCl2 (30 mM) for the determination of the residual testosterone 6β-hydroxylase activity as described above. CYP3A4 Supersomes were incubated with 150 µM and 1000 µM of CBZ at 37°C as described above. Aliquots of the mixtures (100 µd containing 10 pmol CYP3A4) were taken at various time intervals (0–30 min) and added to a secondary 1-ml reaction mixture containing 250 µM testosterone, isocitric acid (5 mM), isocitric acid dehydrogenase (0.5 U/ml), potassium phosphate buffer (100 mM, pH 7.4), EDTA (1 mM), and GSH (1.5 mM) for the determination of the residual testosterone 6β-hydroxylase activity as described above.
Erythromycin N-Demethylase Activity of the CBZ-Modified CYP3A4
After a 30-min incubation of CBZ with CYP3A4 Supersomes as described above, an aliquot of CYP3A4 (1 pmol) was added to a secondary reaction mixture (0.2 ml, final volume) containing erythromycin (1–350 µM) and other reagents as described above. The reaction was initiated by the addition of NADPH (1 mM), allowed to run at 37°C for 15 min, and then quenched with 0.2 ml acetonitrile. A 10-µl aliquot was subjected to liquid chromatography-mass spectrometric analysis for metabolite quantification. N-Desmethylerythromycin was quantified by Shimadzu LC-10AD VP binary pumps (Shimadzu, Columbia, MD) coupled with a Q-Trap 4000 (Applied Biosystems/MDS Sciex, Foster City, CA). The metabolite was separated by a Synergi Fusion-RP column (4 µm, 100 × 2.0 mm; Phenomenex, Torrance, CA) at a flow rate of 0.2 ml/min. A gradient of (A) water with 0.1% formic acid and (B) acetonitrile with 0.1% formic acid was applied as follows: initiated with 20% B for 1 min and then increased in a linear manner to 65% at 2.3 min and to 100% at 4.5 min. The gradient was maintained at 100% B for 5 min and then decreased to 20% at 5.5 min. The column was allowed to equilibrate at 20% solvent B for 0.5 min before the next injection. The high-performance liquid chromatography effluent going to the mass spectrometer was directed to waste through a divert valve for the initial 1 min after sample injection. The Q-Trap 4000 Electrospray-MS was operated in the positive ionization mode by applying to the capillary a voltage (ion spray) of 4.5 kV. Nitrogen was used as curtain gas, as well as nebulizing (GS1) and turbo spray gas (GS2, heated at 450°C), with the optimum values set, respectively, at 36, 50, and 40 (arbitrary values). Collision-activated dissociation was performed at 6 (arbitrary value) with nitrogen as collision gas. Declustering potential was set at 41 V, whereas entrance potential was set at 10 V; collision energy was optimized at 51 eV. The multiple reaction monitoring transitions used were 720→144 for N-desmethylerythromycin. The concentrations were determined by means of a standard curve, wherein N-desmethylerythromycin was dissolved in the same incubation and quench buffer to prepare standards with concentrations ranging from 0.1 to 100 µg/ml. Kinetic data analyses of CYP3A4-dependent erythromycin N-demethylase were performed as described above for testosterone 6β-hydroxylase.
Flexible Docking of CBZ, Testosterone, and Erythromycin in CYP3A4 Crystal Structure
Initial flexible docking of testosterone into the active site of CYP3A4 [Protein Data Bank accession code 1w0g (Williams et al., 2004)] was carried out with FlexX, version 1.7.6, implemented in the SYBYL, version 7.0 (Tripos Inc., St. Louis, MO.). The docking solutions were ranked according to CScore. Visual inspection identified a low energy binding mode compatible with 6β-hydroxylation of testosterone, and this protein-ligand complex was used as a starting point for further flexible docking experiments with testosterone or CBZ using the Pfizer in-house docking and scoring software (Gehlhaar et al., 1995, 1999; Marrone et al., 1999). Restrained docking of CBZ into the active site of the previously identified complex anchoring its 2, 3, 10, or 11 carbon near the sulfur of Cys239 was performed and followed by protein-ligand minimization to yield a low energy binding mode. In separate experiments, a second testosterone was docked into the previously identified 3A4-testosterone complex using a method that allowed the protein to move throughout the docking simulation via discrete minimization. Erythromycin was also docked into the 1w0g structure using a restrained docking method that anchored the site of metabolism near the heme iron and was followed by a docking simulation that allowed protein flexibility.
Results
Time-Dependent Inactivation of CYP3A4 Testosterone 6β-Hydroxylase Activity by CBZ
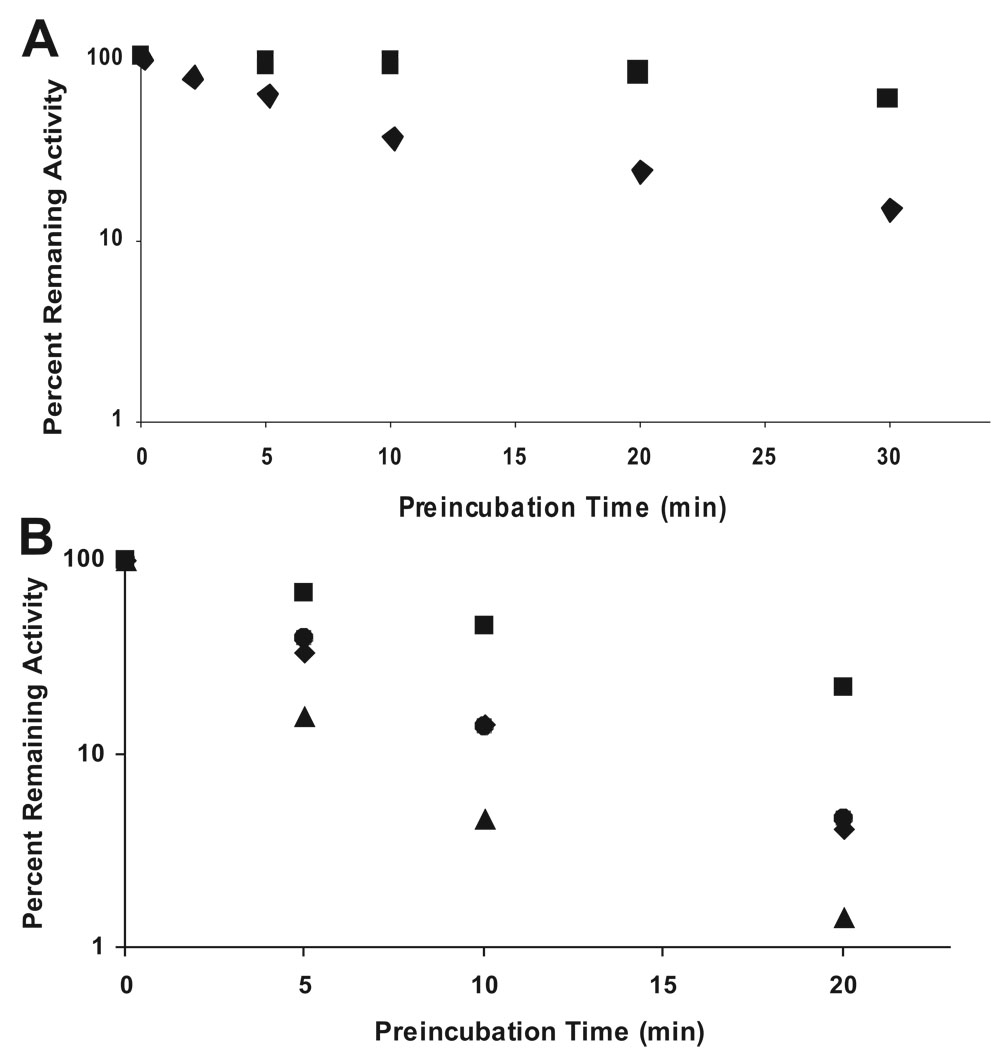
The testosterone 6β-hydroxylase activity of both CYP3A4 Supersomes and functionally reconstituted purified recombinant CYP3A4(His)6 was reduced with increasing preincubation time with CBZ (Fig. 1, A and B). However, the time-dependent functional inactivation of CYP3A4 was not exacerbated by increasing concentrations of CBZ. Instead, the greatest inactivation of enzyme activity, particularly with the functionally reconstituted purified recombinant CYP3A4, was observed in the absence of CBZ (Fig. 1B), and inclusion of CBZ increasingly protected the enzyme from time-dependent inactivation. Functionally reconstituted recombinant CYP3A4(His)6 incubated with NADPH alone also showed a significant reduction in its characteristic reduced CO-binding 450 nm-absorption spectrum (data not shown). These findings do not support the mechanism-based inactivation of CYP3A4 by CBZ; however, they are consistent with significant oxidative uncoupling associated with these CYP3A4 systems containing a 1:4 M excess of P450 reductase in the absence of substrates.
FIG. 1.
Time-dependent inactivation of CYP3A4-dependent testosterone 6β-hydroxylase activity and concentration-dependent protection by CBZ. CYP3A4 Supersomes or functionally reconstituted purified recombinant CYP3A4 was incubated with various concentrations of CBZ (0–1000 µM) and NADPH at 37°C. Aliquots of the mixture were taken at various time intervals (0–30 min) and added to a secondary reaction mixture for the determination of the residual testosterone 6β-hydroxylase activity. A, CYP3A4 Supersomes, CBZ concentration: ♦, 150 µM; ■, 1000 µM. B, functionally reconstituted purified recombinant CYP3A4(His)6, CBZ concentration: ▲, 0 µM; ♦, 10 µM; ●, 100 µM; ■, 1000 µM.
Determination of CBZ-Covalent Binding to CYP3A4 Protein
Incubation of CYP3A4 Supersomes with [3H]CBZ resulted in covalently bound CBZ to CYP3A4. After extensive organic solvent washes to remove free nonspecifically bound [3H]CBZ, the residual radioactivity associated with the protein pellet was monitored. The stoichiometry (molar ratio) of CBZ/CYP3A4 protein was calculated to be 1.58:1. Inclusion of GSH (1.5 mM) in the incubation reduced this molar ratio to 1.09. Similar covalent binding analyses with the recombinant CYP3A4, functionally reconstituted in the presence of GSH (1.5 mM) and using the same protein precipitation method, yielded a stoichiometry of CBZ-CYP3A4 adduct of 0.79:1.
Determination of the Covalent Binding by Reisolation of the CBZ-CYP3A4(His)6 Adduct
The stoichiometry of the adduct was also determined by isolating the CBZ-modified CYP3A4(His)6 protein after incubation of the functionally reconstituted recombinant CYP3A4(His)6 with [3H]CBZ by Ni2+-NTA affinity chromatography. Unbound CBZ and other constituents of the incubation mixture such as P450 reductase and b5 were removed by extensive washing, as monitored by SDS-PAGE analyses, which showed that the residual radioactivity was associated solely with the eluted CYP3A4(His)6 and that the CBZ-CYP3A4(His)6 adduct was clearly separated from P450 reductase and b5. The stoichiometry of the isolated CBZ-CYP3A4(His)6 adduct was calculated to be 1 ± 0.4 pmol CBZ/pmol CYP3A4 on the basis of its residual radioactivity. Together, these findings indicated that the bioactivated CBZ covalently modified CYP3A4, possibly within the active site of this enzyme, to form a CBZ-CYP3A4 protein adduct.
Enzyme Kinetics of CBZ-Modified CYP3A4
Influence on testosterone 6β-hydroxylation
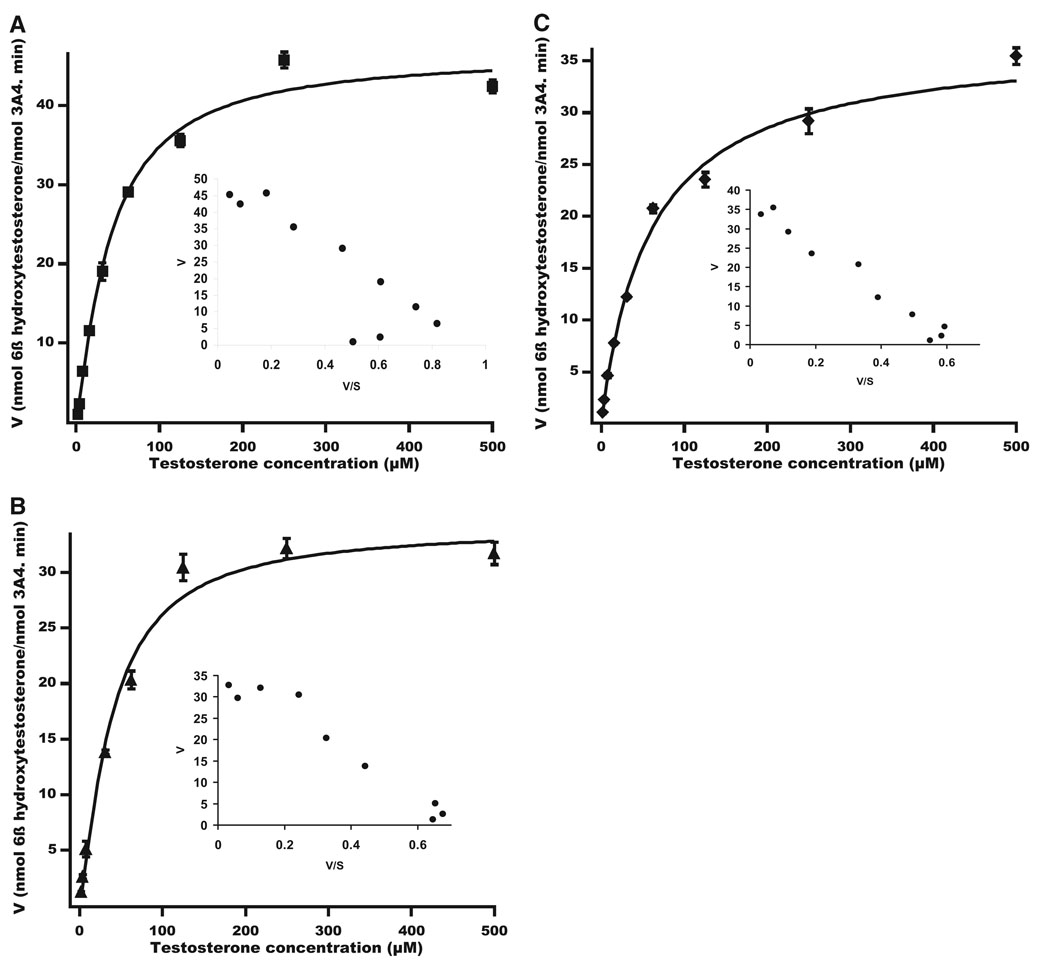
To explore the possible site of CBZ adduction, its effect on the cooperativity with a known CYP3A4 substrate testosterone was probed. Incubation of CYP3A4 Supersomes with testosterone yielded the previously reported sigmoidal kinetics as evidenced by the curved Eadie-Hofstee plot (Fig. 2A; n = 1.2). The kinetic values obtained after the data were fitted to the Hill equation are shown in Table 1. CYP3A4-dependent testosterone 6β-hydroxylase activity was inhibited when CYP3A4 was preincubated with CBZ in the presence or absence of NADPH. When preincubated with CBZ in the absence of NADPH, CYP3A4 still retained its sigmoidal kinetics (Fig. 2B; n = 1.3). In contrast, after preincubation with NADPH, which, as described above, results in a covalent 1:1 molar CBZ-adduction of CYP3A4, the CBZ-modified enzyme displayed an almost hyperbolic kinetic profile of testosterone 6β-hydroxylation (Fig. 2C; n = 1.0). The Km for testosterone was unaffected when CYP3A4 was preincubated with CBZ without NADPH. A larger increase in Km was observed when CYP3A4 was covalently modified by CBZ in the presence of NADPH. However, the Vmax values, albeit lower, were not substantially different after preincubation with CBZ in the presence or absence of NADPH (Table 1).
FIG. 2.
Kinetic profiles of testosterone 6β-hydroxylase activity of CYP3A4 Supersomes before or after CBZ-modification. A, kinetic profile with enzyme preincubation in the absence of both CBZ and NADPH. B, kinetic profile with enzyme preincubation with CBZ in the absence of NADPH. C, kinetic profile with enzyme preincubation with CBZ and NADPH. Data shown are the average mean ± S.D. of three independent experiments.
TABLE 1.
Kinetic parameters of testosterone 6β-hydroxylation by CYP3A4 Supersomes and functionally reconstituted purified recombinant CYP3A4
| CYP3A4 | Vmax | Km | Hill Coefficient (n) |
|---|---|---|---|
| nmol/min/nmol | µM | ||
| Supersomes | |||
| NADPH−, CBZ− | 46.5 ± 1.6 | 40.6 ± 4.2 | 1.2 ± 0.1 |
| NADPH−, CBZ+ | 33.3 ± 1.5 | 36.7 ± 5.3 | 1.3 ± 0.2 |
| NADPH+, CBZ+ | 37.1 ± 1.9 | 59.3 ± 9.7 | 1.0 ± 0.1 |
| Purified CYP3A4 | |||
| NADPH−, CBZ− | 12.2 ± 0.3 | 81.5 ± 4.3 | 1.4 ± 0.1 |
| NADPH−, CBZ+ | 11.2 ± 0.4 | 102.3 ± 8.6 | 1.2 ± 0.1 |
| NADPH+, CBZ+ | 7.2 ± 0.6 | 132.4 ± 26.1 | 1.1 ± 0.1 |
NADPH−, CBZ−: preincubation of CYP3A4 in the absence of NADPH and CBZ followed by assay of testosterone 6β-hydroxylase activity; NADPH−, CBZ+: preincubation of CYP3A4 with CBZ but without NADPH followed by assay of testosterone 6β-hydroxylase activity; NADPH+, CBZ +: preincubation of CYP3A4 in the presence of both NADPH and CBZ followed by assay of testosterone 6β-hydroxylase activity.
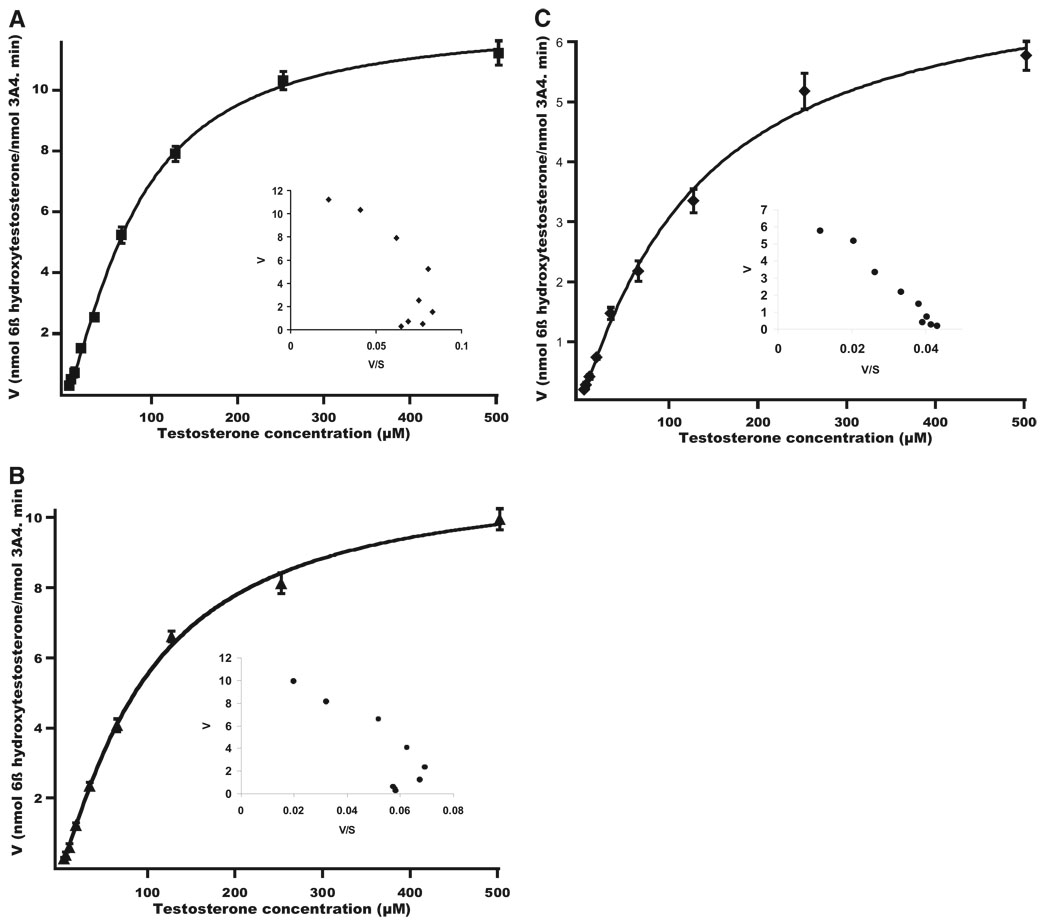
A similar sigmoidal kinetic profile, characterized by a curved Eadie-Hofstee plot (Fig. 3A; n = 1.4), was also observed for the testosterone 6β-hydroxylase activity of functionally reconstituted recombinant CYP3A4. The rate of 6β-hydroxytestosterone formation was inhibited in the presence of CBZ, even in the absence of NADPH. Eadie-Hofstee analyses of the enzyme activity in the presence of CBZ but without preincubation with NADPH revealed a similar sigmoidal profile, although the presence of CBZ lowered the Hill coefficient (Fig. 3B; n = 1.2). The Km of testosterone increased in the presence of CBZ, thereby suggesting CBZ competition at the testosterone-binding site. After covalent modification of CYP3A4 by preincubation with CBZ in the presence of NADPH, its residual testosterone 6β-hydroxylase activity yielded an almost hyperbolic profile with a linear Eadie-Hofstee plot (Fig. 3C). The Hill coefficient n also decreased to 1.1 (Table 1). CBZ-modified CYP3A4 also displayed a lower binding affinity toward testosterone as evidenced by the increased Km value, whereas the Vmax value only slightly decreased when NADPH was included in the preincubation (Table 1).
FIG. 3.
Kinetic profiles of testosterone 6β-hydroxylase activity of purified functionally reconstituted CYP3A4(His)6 before or after CBZ-modification. A, kinetic profile with enzyme preincubation in the absence of both CBZ and NADPH. B, kinetic profile with enzyme preincubation with CBZ in the absence of NADPH. C, kinetic profile with enzyme preincubation with CBZ and NADPH. Data shown are the average mean ± S.D. of three independent experiments.
Influence on erythromycin N-demethylation
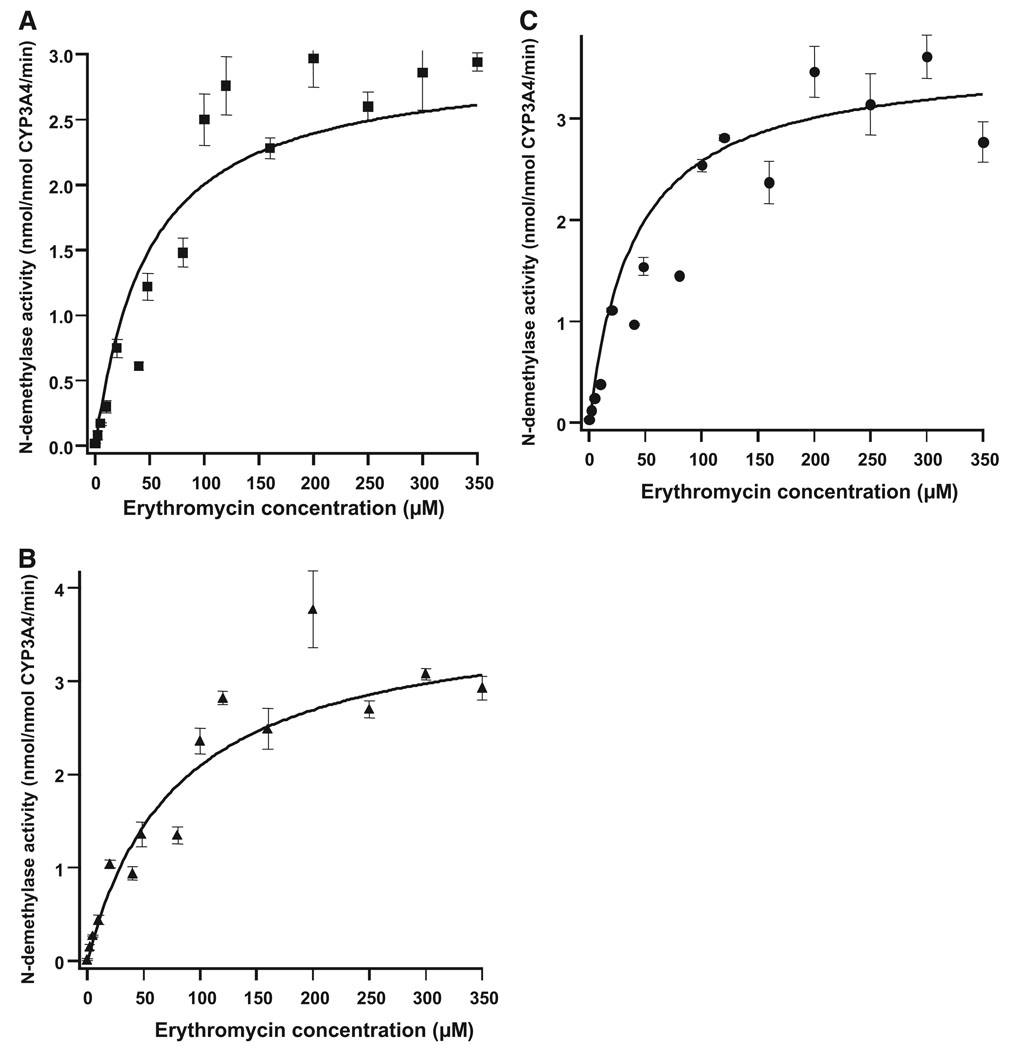
To further assess the effect of covalent CBZ-induced modification on CYP3A4-dependent function, the N-demethylation of erythromycin, another prototypic CYP3A4 substrate with an affinity comparable to that of testosterone and thus a competitive substrate inhibitor of CYP3A4-dependent testosterone 6β-hydroxylase activity (Wang et al., 1997), was examined. However, the kinetic data obtained for erythromycin N-demethylation after CBZ-induced modification of CYP3A4 Supersomes were scattered and failed to provide a good fit to either the Hill or Michaelis-Menten equation. Nevertheless, curve fitting yielded comparable Vmax values for CYP3A4-dependent erythromycin N-demethylation with (3.61 ± 0.22 nmol/nmol CYP3A4/min) or without (3.60 ± 0.58 nmol/nmol CYP3A4/min) CBZ-induced covalent modification of the enzyme (Fig. 4, A, B, and C; Table 2). These results suggested that, contrary to the results obtained with testosterone as a substrate, CBZ-induced covalent modification of CYP3A4 had no significant effect on erythromycin N-demethylation.
FIG. 4.
Kinetic profiles of erythromycin N-demethylase activity of CYP3A4 Supersomes before or after CBZ-modification. A, kinetic profile with enzyme preincubation in the absence of both CBZ and NADPH. B, kinetic profile with enzyme preincubation with CBZ in the absence of NADPH. C, kinetic profile with enzyme preincubation with CBZ and NADPH. Data shown are the average mean ± S.D. of three independent experiments.
TABLE 2.
Kinetic parameters of erythromycin N-demethylation by CYP3A4 Supersomes before and after CBZ-modification
| CYP3A4 Supersomes | Vmax | Km |
|---|---|---|
| nmol/min/nmol | µM | |
| NADPH−, CBZ− | 3.27 ± 0.08 | 44.0 ± 4.0 |
| NADPH−, CBZ+ | 3.60 ± 0.58 | 80.0 ± 0.00 |
| NADPH+, CBZ+ | 3.61 ± 0.22 | 44.0 ± 4.0 |
NADPH−, CBZ−: preincubation of CYP3A4 in the absence of NADPH and CBZ followed by assay of erythromycin N-demethylase; NADPH−, CBZ+: preincubation of CYP3A4 with CBZ but without NADPH followed by assay of erythromycin N-demethylase; NADPH+, CBZ+: preincubation of CYP3A4 in the presence of both NADPH and CBZ followed by assay of erythromycin N-demethylase.
Flexible Docking of CBZ, Testosterone, and Erythromycin in CYP3A4 Crystal Structure
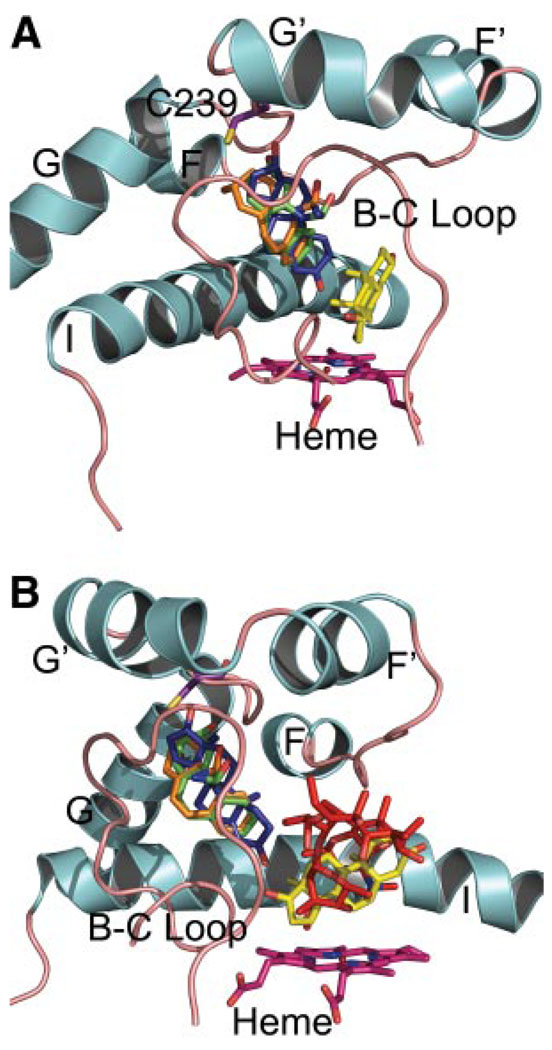
Initial dockings with testosterone into the active site of apo CYP3A4 identified a low energy binding pose of testosterone near the heme that is consistent with its 6β-hydroxylation. This complex was used as the starting point for further docking experiments. When a second molecule of testosterone is docked into the active site using an induced fit method, which allows minimal protein flexibility during docking, one of the low energy binding modes places the second testosterone partly in the active site but extending toward the surface of the protein in a channel that is defined by the helix G′ to helix G loop, the B–C loop, and the I helix. This docking did not require any repositioning of the first testosterone bound (Fig. 5A).
FIG. 5.
Docking of CBZ, testosterone (TS), and erythromycin into the active site of CYP3A4 [Protein Data Bank accession code 1w0g (Williams et al., 2004)]. Only the helix F-G, helix I, and B–C loop regions are displayed for clarity. Helices are depicted in cyan and loops in tan. A, TS molecules are shown in yellow and dark blue stick representations. CBZ molecules bound for carbon 2-adduct shown in green stick and bound for carbon 3-adduct in orange stick. The side chain of Cys239 is shown in purple. The heme group is shown in magenta. The linearity of the CYP3A4-dependent CBZ 10,11-epoxidation with time and our recent findings of the superiority of radiolabeled 3-OHCBZ versus CBZ in CYP3A4 covalent binding after bioactivation lead us to exclude CBZ 10,11-epoxide as the primary reactive intermediate (R. E. Pearce, W. Lu, Y.-Q. Wang, J. P. Uetrecht, M. A. Correia, J. S. Leeder, unpublished observations). B, same as in (A) except that erythromycin, docked in a pose compatible with its N-demethylation, is also shown in a red stick representation. Please note that although these particular dockings are consistent with our in vitro kinetic data, they represent an in silico attempt at capturing a very dynamic system. Other potential low energy binding modes for the second TS molecule in the CYP3A4 active site outside of the access channel are also possible.
Restrained docking of CBZ yielded poses that would be compatible with adduct formation with Cys239 by carbon 2 or 3 of CBZ (Fig. 5A). Furthermore, low energy binding modes were also observed when the 10 or 11 carbon of CBZ was restrained toward Cys239, suggesting that adduct formation arising from the 10,11-epoxide intermediate is also possible (data not shown). All of the restrained docking binding poses observed for CBZ, regardless of which carbon was anchored, are compatible with testosterone binding near the heme but would cause steric clashes with a second molecule of testosterone bound in the active site as described previously (Fig. 5A). On the other hand, restrained docking of erythromycin in the apo active site of 1w0g yielded a low energy binding pose that allowed approach of its dimethylamino nitrogen consistent with the known N-demethylation of erythromycin by CYP3A4 (Fig. 5B).
Discussion
In vitro studies with recombinant human P450s expressed in yeast have documented the covalent binding of CBZ to proteins belonging to the P450 subfamilies 1A, 2C, and 3A (Wolkenstein et al., 1998). However, no mechanism-based inactivation of the covalently modified P450s was detected. Similarly, no evidence for the functional inactivation of human CYP2C8, CYP2C9, or CYP3A4 could be found after preincubation of human liver microsomes with CBZ in the presence of NADPH (Masubuchi et al., 2001). In the present study, we show that CYP3A4 Supersomes formed an adduct with [3H]CBZ with a molar stoichiometry of 1.58:1 (CBZ/CYP3A4). This molar ratio was reduced to approximately 1:1 by inclusion of GSH (1.5 mM) in the incubation; however, this molar ratio was not further affected by higher concentrations of GSH (up to 4 mM). Because both CYP3A4 Supersomes and yeast microsomes contain various other proteins, the possibility existed that proteins other than CYP3A4 were covalently modified by CBZ and thus either were acid coprecipitated with CYP3A4 or comigrated with it after SDS-PAGE. We therefore examined a simpler system consisting of purified functionally reconstituted CYP3A4, wherein after incubation with CBZ, the CBZ-modified recombinant CYP3A4(His)6 was readily separated from P450 reductase and b5 by Ni2+-NTA affinity chromatography. A similar 1:1 molar ratio was observed in this CBZ-modified CYP3A4(His)6 system, thus suggesting that CYP3A4 expressed in baculovirus (CYP3A4 Supersomes) and yeast (Wolkenstein et al., 1998) was most likely the only protein that was covalently modified by CBZ. The fact that GSH (1.5 mM), normally included in CYP3A4 functional reconstitution systems, failed to reduce this 1:1 molar ratio of CBZ/CYP3A4 suggested that CBZ was covalently bound within the CYP3A4 active site, thereby making it somewhat inaccessible for nucleophilic trapping by GSH.
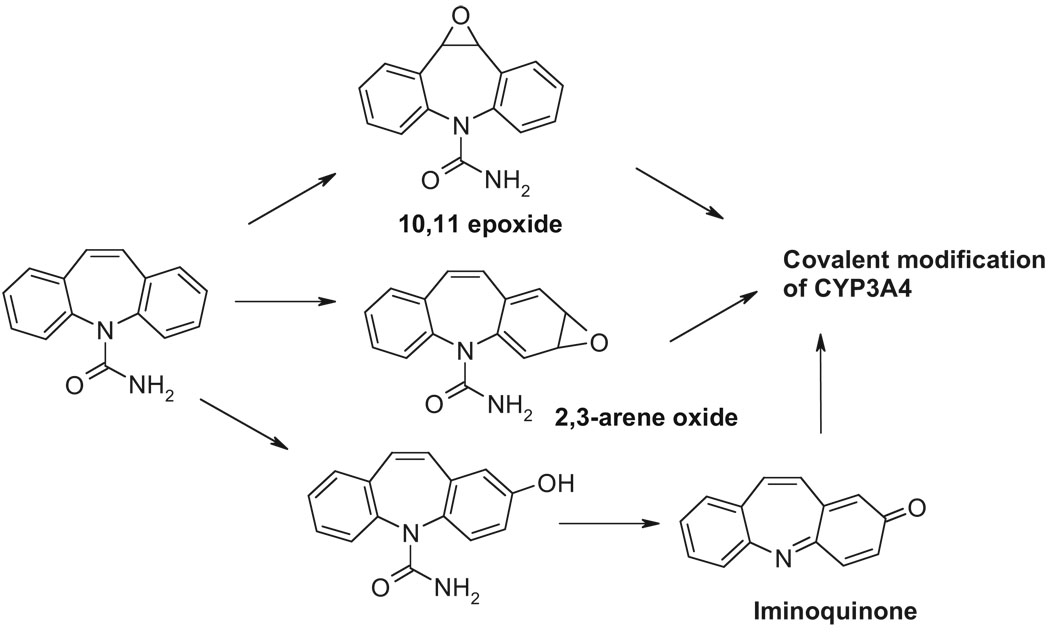
We find it noteworthy, however, that a considerably greater extent of CBZ-induced covalent binding occurs in the absence of GSH. This suggests that individuals with low hepatic GSH content, which may result from pharmacological depletion by coadministered drugs, dietary deprivation, inadequate repletion of GSH-precursors, or even defective GSH synthesis, may be at a higher risk for enhanced covalent modification of P450s. Subsequent proteolytic processing of such CBZ-modified P450s for antigenic presentation may contribute to the pathogenesis of the hypersensitivity/idiosyncratic reactions. Although the mechanism of CBZ bioactivation and covalent binding has yet to be precisely elucidated, previous studies have suggested that CBZ-derived arene oxide, quinone, iminoquinone, catechol, quinone methide, and/or epoxide could be viable reactive metabolites that chemically modify P450s (Madden et al., 1996; Pearce et al., 2002, 2005; Bu et al., 2005) (Scheme 1). Furthermore, although the precise site of this binding remains to be identified, conceivably Cys239 could be a plausible target as in raloxifene-mediated CYP3A4 adduction (Baer et al., 2007). Nevertheless, our finding that CBZ covalently modifies CYP3A4 (possibly at the active site) is intriguing in view of the fact that it maims but does not inactivate the enzyme, consistent with the report that CBZ is not a mechanism-based CYP3A4 inactivator (Masubuchi et al., 2001).
SCHEME 1.
Proposed reactive metabolites and the covalent modification of CYP3A4 active site. The iminoquinone is reportedly derived from 2-hydroxycarbam-azepine (Pearce et al., 2005). Other reactive metabolites include 2,3 arene oxide, 2,3 catechol, dihydroquinone, and the 3-quinone methide.
The unusual sigmoidal kinetic interactions exhibited by CYP3A4 have been rationalized by the binding of multiple substrate molecules at more than one site within the CYP3A4 active site (Shou et al., 1994; Ueng et al., 1997; Harlow and Halpert, 1998; Korzekwa et al., 1998). Similar sigmoidal kinetics of CYP3A4-catalyzed CBZ metabolism imply multiple CBZ-binding sites within the CYP3A4 active site (Nakamura et al., 2003). However, the observed 1:1 molar stoichiometry of the CBZ-CYP3A4 adduct suggests that only one substrate binding site of CYP3A4 was covalently modified by CBZ, with the other CBZ-binding sites remaining unscathed. Furthermore, not only was the CYP3A4-characteristic testosterone 6β-hydroxylase activity only slightly reduced when CYP3A4 was covalently modified by CBZ, but such modification also failed to affect CYP3A4-dependent erythromycin N-demethylase activity. CBZ also fails to functionally inactivate CYP3A4 in a mechanism-based manner in human liver microsomes (Masubuchi et al., 2001). Indeed, the presence of endogenous steroids, such as testosterone in the incubation, drastically increases CYP3A4-dependent CBZ 10,11-epoxidase activity and alters its kinetic profile from sigmoidal to hyperbolic (Michaelis-Menten type) (Nakamura et al., 2003). Such substrate cooperativity at the CYP3A4 active site was taken as evidence for a two-site substrate-binding model. Consistent with this model, our study showed that a complete block of one CYP3A4 substrate-binding site by CBZ covalent modification altered the kinetic profile of CYP3A4-dependent testosterone 6β-hydroxylation from sigmoidal to near-hyperbolic (Michaelis-Menten type). Due to the higher CYP3A4 binding affinity of testosterone (Km, 30–100 µM) than that of CBZ (Km, 250 µM) (Pelkonen et al., 2001), CBZ does not compete effectively for all the CYP3A4-testosterone substrate binding sites, thus leading to the retention of its CYP3A4 sigmoidal kinetic profile. Interestingly, CBZ inclusion led to a greater reduction of the Hill coefficient for the functionally reconstituted purified CYP3A4(His)6 (n = 1.4 to n = 1.2) but an increase for CYP3A4 Supersomes (from n = 1.2 to n = 1.3), which may be explained by the higher binding affinity of testosterone to CYP3A4 Supersomes (Km = 41.0 µM) than to the functionally reconstituted purified CYP3A4(His)6 (Km = 81.5 µM).
In silico docking studies were undertaken in an attempt to rationalize the effect of CBZ-modification of CYP3A4 on its atypical testosterone 6β-hydroxylation kinetics. Although the presence of multiple binding sites and consequent substrate cooperativity within the CYP3A4 active site is supported by the simultaneous docking of both α-naphthoflavone and testosterone into the apo crystal structure, the recently reported CYP3A4 crystal structure with two complexed ketoconazole molecules confirms that the CYP3A4 active site can simultaneously accommodate even relatively large substrate ligands (Yano et al., 2004; Ekroos et al., 2006). In a similar manner, our in silico experiments suggest that at least two molecules of testosterone can simultaneously bind in the CYP3A4 active site. One testosterone is bound near the heme in an orientation compatible with its 6β-hydroxylation and a second extends toward the surface of the protein via an access channel, which also contains Cys239 located on the loop between helix G′ and helix G (Fig. 5). In the absence of any large conformational alterations upon adduct formation, CBZ bound at Cys239 would sterically hinder testosterone binding at this second site but would not affect its catalytic binding near the heme. The persistence of a relatively robust testosterone 6β-hydroxylase activity after CBZ-modification of the CYP3A4 active site is thus consistent not only with the successful simultaneous docking of both CBZ and testosterone therein but also the plausible existence of multiple binding sites within the capacious CYP3A4 active site. Indeed, this particular CYP3A4 feature is further underscored by our findings that the N-demethylation of erythromycin is largely unaffected by CBZ-induced CYP3A4 covalent modification. Our docking analyses reveal that this relatively large macrolide apparently assumes a slightly different orientation within the CYP3A4 active site, which would be unaffected by the CBZ modification in the access channel, thereby precluding any steric/kinetic effects such as those observed with testosterone 6β-hydroxylation (Fig. 5B).
Intriguingly, however, such CBZ-mediated alteration of testosterone hydroxylation kinetic profile from sigmoidal to near hyperbolic following its CYP3A4 active site modification may rationalize the reported failure of midazolam (MDZ) hydroxylation to exhibit any detectable homotropic cooperativity (Khan et al., 2002), in spite of its putative binding at multiple sites within the CYP3A4 active site. The covalent modification of one of these sites during its mechanism-based inactivation via 1′-OH MDZ formation may similarly conceal its sigmoidal kinetic profile. Moreover, such MDZ-mediated covalent modification is reported to differentially inhibit CYP3A4-catalyzed oxidations of testosterone and triazolam, thereby providing further evidence for the possible existence of multiple spatially distinct binding sites or binding orientations for each of these substrates (Schrag and Wienkers, 2001; Khan et al., 2002). Additional evidence for plausible MDZ binding at two distinct albeit overlapping CYP3A4 active sites may be derived from the findings of its regioselective hydroxylation to either 1′-OH MDZ or 4-OH MDZ by CYP3A4 and several of its active site-directed mutants (Khan et al., 2002).
Lastly, it is to be noted that significant time-dependent enzyme inactivation of both CYP3A4 Supersomes and functionally reconstituted purified recombinant CYP3A4(His)6 was observed upon incubation with NADPH alone, consistent with marked oxidative uncoupling associated with these CYP3A4 systems in the absence of substrates (Zangar et al., 2004; Denisov et al., 2006). Interestingly, CBZ inclusion in these incubations protected CYP3A4 from this time-dependent inactivation, and this protection was magnified by increasing CBZ concentrations, thereby precluding the detection of any CBZ-induced mechanism-based inactivation of CYP3A4 testosterone 6β-hydroxylase activity.
In summary, CYP3A4-mediated bioactivation of CBZ generated a covalent CBZ-CYP3A4 adduct with a molar ratio of 1:1 (CBZ/CYP3A4). The formation of this drug-protein adduct could not be abolished by GSH inclusion. However, the CBZ-modified CYP3A4 was still functionally active, albeit at a reduced functional level with testosterone as the substrate. This CYP3A4 covalent modification by CBZ altered its testosterone 6β-hydroxylase kinetic profile from sigmoidal to near-hyperbolic Michaelis-Menten type, thereby suggesting that CBZ-mediated modification occurs within the CYP3A4 active site.
Acknowledgments
We thank Dr. Xin He, Incyte Corp., for valuable discussions and assistance with the initial flexible docking analysis and Prof. Paul Ortiz de Montellano, UCSF, for the use of his Silicon Graphics station and FlexX software. We thank Dr. Bernard Murray, Gilead Sciences Inc., for bringing the Baer et al. findings to our attention. We also acknowledge the UCSF Liver Core Center Facilities on Molecular Analyses (Spectrophotometry) supported by P30DK26743.
This work was supported by National Institutes of Health (NIH) Grant GM58883 to J.S.L. and M.A.C., NIH Grants DK26506 and GM44037 to M.A.C., and NIH Grant P30DK26743.
ABBREVIATIONS
- CBZ
carbamazepine
- P450
cytochrome P450
- GSH
glutathione
- EtOAc
ethyl acetate
- b5
cytochrome b5
- PAGE
polyacryl-amide gel electrophoresis
- MDZ
midazolam
- P450 reductase
NADPH-cytochrome P450 reductase
- Ni2+-NTA
nickel-nitrilotriacetic acid.
References
- Baer BR, Wienkers LC, Rock DA. Time-dependent inactivation of P450 3A4 by raloxifene: identification of Cys239 as the site of apoprotein alkylation. Chem Res Toxicol. 2007;20:954–964. doi: 10.1021/tx700037e. [DOI] [PubMed] [Google Scholar]
- Bu HZ, Kang P, Deese AJ, Zhao P, Pool WF. Human in vitro glutathionyl and protein adducts of carbamazepine-10,11-epoxide, a stable and pharmacologically active metabolite of carbamazepine. Drug Metab Dispos. 2005;33:1920–1924. doi: 10.1124/dmd.105.006866. [DOI] [PubMed] [Google Scholar]
- Denisov IG, Grinkova YV, Baas BJ, Sligar SG. The ferrous-dioxygen intermediate in human cytochrome P450 3A4. Substrate dependence of formation and decay kinetics. J Biol Chem. 2006;281:23313–23318. doi: 10.1074/jbc.M605511200. [DOI] [PubMed] [Google Scholar]
- Ekroos M, Sjogren T. Structural basis for ligand promiscuity in cytochrome P450 3A4. Proc Natl Acad Sci U S A. 2006;103:13682–136876. doi: 10.1073/pnas.0603236103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehlhaar DK, Verkhivker GM, Rejto PA, Sherman CJ, Fogel DB, Fogel LJ, Freer ST. Molecular recognition of the inhibitor AG-1343 by HIV-1 protease: conformationally flexible docking by evolutionary programming. Chem Biol. 1995;2:317–324. doi: 10.1016/1074-5521(95)90050-0. [DOI] [PubMed] [Google Scholar]
- Gehlhaar D, Bouzida D, Rejto PA. Reduced dimensionality in ligand-protein structure prediction: covalent inhibitors of serine proteases and design of site-directed combinatorial libraries. In: Parrill AL, Reddy MR, editors. ACS Symposium Series 719: Rational Drug Design. Washington, DC: ACS Press; 1999. pp. 292–311. [Google Scholar]
- Harlow GR, Halpert JR. Analysis of human cytochrome P450 3A4 cooperativity: construction and characterization of a site-directed mutant that displays hyperbolic steroid hydroxylation kinetics. Proc Natl Acad Sci U S A. 1998;95 doi: 10.1073/pnas.95.12.6636. 6636-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart RG, Easton JD. Carbamazepine and hematological monitoring. Ann Neurol. 1982;11:309–312. doi: 10.1002/ana.410110312. [DOI] [PubMed] [Google Scholar]
- He K, Bornheim LM, Falick AM, Maltby D, Yin H, Correia MA. Identification of the heme-modified peptides from cumene hydroperoxide-inactivated cytochrome P450 3A4. Biochemistry. 1998;37:17448–17457. doi: 10.1021/bi9808464. [DOI] [PubMed] [Google Scholar]
- Kalapos MP. Carbamazepine-provoked hepatotoxicity and possible aetiopathological role of glutathione in the events. Retrospective review of old data and call for new investigation. Adverse Drug React Toxicol Rev. 2002;21:123–141. doi: 10.1007/BF03256188. [DOI] [PubMed] [Google Scholar]
- Khan KK, He YQ, Domanski TL, Halpert JR. Midazolam oxidation by cytochrome P450 3A4 and active-site mutants: an evaluation of multiple binding sites and of the metabolic pathway that leads to enzyme inactivation. Mol Pharmacol. 2002;61:495–5062. doi: 10.1124/mol.61.3.495. [DOI] [PubMed] [Google Scholar]
- Korzekwa KR, Krishnamachary N, Shou M, Ogai A, Parise RA, Rettie AE, Gonzalez FJ, Tracy TS. Evaluation of atypical cytochrome P450 kinetics with two-substrate models: evidence that multiple substrates can simultaneously bind to cytochrome P450 active sites. Biochemistry. 1998;37:4137–4147. doi: 10.1021/bi9715627. [DOI] [PubMed] [Google Scholar]
- Leeder JS. Mechanisms of idiosyncratic hypersensitivity reactions to antiepileptic drugs. Epilepsia. 1998;39 suppl 7:S8–S16. doi: 10.1111/j.1528-1157.1998.tb01679.x. [DOI] [PubMed] [Google Scholar]
- Leeder JS, Gaedigk A, Lu X, Cook VA. Epitope mapping studies with human anti-cytochrome P450 3A antibodies. Mol Pharmacol. 1996;49:234–243. [PubMed] [Google Scholar]
- Licad-Coles E, He K, Yin H, Correia MA. Cytochrome P450 2C11: Escherichia coli expression, purification, functional characterization, and mechanism-based inactivation of the enzyme. Arch Biochem Biophys. 1997;338:35–42. doi: 10.1006/abbi.1996.9795. [DOI] [PubMed] [Google Scholar]
- Lillibridge JH, Amore BM, Slattery JT, Kalhorn TF, Nelson SD, Finnell RH, Bennett GD. Protein-reactive metabolites of carbamazepine in mouse liver microsomes. Drug Metab Dispos. 1996;24:509–514. [PubMed] [Google Scholar]
- Madden S, Maggs JL, Park BK. Bioactivation of carbamazepine in the rat in vivo. Evidence for the formation of reactive arene oxide(s) Drug Metab Dispos. 1996;24:469–479. [PubMed] [Google Scholar]
- Marrone TJ, Luty BA, Rose PW. Proceedings of the Workshop “New Approaches in Drug Design and Discovery,” special topic “Virtual Screening;”. Germany: Springer Netherlands, Schloâ Rauischholzhausen; 1999. Mar 15–18, Discovering high-affinity ligands from the computationally predicted structures and affinities of small molecules bound to a target: a virtual screening approach. Virtual screening: an alternative or complement to high throughput screening? pp. 209–230. [Google Scholar]
- Masubuchi Y, Nakano T, Ose A, Horie T. Differential selectivity in carbamazepine-induced inactivation of cytochrome P450 enzymes in rat and human liver. Arch Toxicol. 2001;75:538–543. doi: 10.1007/s002040100270. [DOI] [PubMed] [Google Scholar]
- Nakamura H, Torimoto N, Ishii I, Ariyoshi N, Nakasa H, Ohmori S, Kitada M. CYP3A4 and CYP3A7-mediated carbamazepine 10,11-epoxidation are activated by differential endogenous steroids. Drug Metab Dispos. 2003;31:432–438. doi: 10.1124/dmd.31.4.432. [DOI] [PubMed] [Google Scholar]
- Park BK, Coleman JW, Kitteringham NR. Drug disposition and drug hypersensitivity. Biochem Pharmacol. 1987;36:581–590. [PubMed] [Google Scholar]
- Pearce RE, Vakkalagadda GR, Leeder JS. Pathways of carbamazepine bioactivation in vitro I. Characterization of human cytochromes P450 responsible for the formation of 2- and 3-hydroxylated metabolites. Drug Metab Dispos. 2002;30:1170–1179. doi: 10.1124/dmd.30.11.1170. [DOI] [PubMed] [Google Scholar]
- Pearce RE, Uetrecht JP, Leeder JS. Pathways of carbamazepine bioactivation in vitro: II. The role of human cytochrome P450 enzymes in the formation of 2-hydroxyiminostilbene. Drug Metab Dispos. 2005;33:1819–1826. doi: 10.1124/dmd.105.004861. [DOI] [PubMed] [Google Scholar]
- Pelkonen O, Myllynen P, Taavitsainen P, Boobis AR, Watts P, Lake BG, Price RJ, Renwick AB, Gomez-Lechon MJ, Castell JV, et al. Carbamazepine: a “blind” assessment of CVP-associated metabolism and interactions in human liver-derived in vitro systems. Xenobiotica. 2001;31:321–343. doi: 10.1080/00498250110055479. [DOI] [PubMed] [Google Scholar]
- Pirmohamed M, Kitteringham NR, Breckenridge AM, Park BK. The effect of enzyme induction on the cytochrome P450-mediated bioactivation of carbamazepine by mouse liver microsomes. Biochem Pharmacol. 1992;44:2307–2314. doi: 10.1016/0006-2952(92)90674-8. [DOI] [PubMed] [Google Scholar]
- Romero Maldonado N, Sendra Tello J, Raboso Garcia-Baquero E, Harto Castano A. Anticonvulsant hypersensitivity syndrome with fatal outcome. Eur J Dermatol. 2002;12:503–505. [PubMed] [Google Scholar]
- Schrag ML, Wienkers LC. Covalent alteration of the CYP3A4 active site: evidence for multiple substrate binding domains. Arch Biochem Biophys. 2001;391:49–55. doi: 10.1006/abbi.2001.2401. [DOI] [PubMed] [Google Scholar]
- Shear NH, Spielberg SP, Cannon M, Miller M. Anticonvulsant hypersensitivity syndrome. In vitro risk assessment. J Clin Invest. 1988;82:1826–1832. doi: 10.1172/JCI113798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou M, Grogan J, Mancewicz JA, Krausz KW, Gonzalez FJ, Gelboin HV, Korzekwa KR. Activation of CYP3A4: evidence for the simultaneous binding of two substrates in a cytochrome P450 active site. Biochemistry. 1994;33:6450–64554. doi: 10.1021/bi00187a009. [DOI] [PubMed] [Google Scholar]
- Ueng YF, Kuwabara T, Chun YJ, Guengerich FP. Cooperativity in oxidations catalyzed by cytochrome P450 3A4. Biochemistry. 1997;36:370–381. doi: 10.1021/bi962359z. [DOI] [PubMed] [Google Scholar]
- Wang RW, Newton DJ, Scheri TD, Lu AY. Human cytochrome P450 3A4-catalyzed testosterone 6 beta-hydroxylation and erythromycin N-demethylation. Competition during catalysis. Drug Metab Dispos. 1997;25:502–507. [PubMed] [Google Scholar]
- Wang H, Dick R, Yin H, Licad-Coles E, Kroetz DL, Szklarz G, Harlow G, Halpert JR, Correia MA. Structure-function relationships of human liver cytochromes P450 3A: aflatoxin B1 metabolism as a probe. Biochemistry. 1998;37:12536–12545. doi: 10.1021/bi980895g. [DOI] [PubMed] [Google Scholar]
- Williams PA, Cosme J, Vinkovic DM, Ward A, Angove HC, Day PJ, Vonrhein C, Tickle IJ, Jhoti H. Crystal structures of human cytochrome P450 3A4 bound to metyrapone and progesterone. Science. 2004;305:683–686. doi: 10.1126/science.1099736. [DOI] [PubMed] [Google Scholar]
- Wolkenstein P, Tan C, Lecoeur S, Wechsler J, Garcia-Martin N, Charue D, Bagot M, Beaune P. Covalent binding of carbamazepine reactive metabolites to P450 isoforms present in the skin. Chem Biol Interact. 1998;113:39–50. doi: 10.1016/s0009-2797(98)00021-0. [DOI] [PubMed] [Google Scholar]
- Xue L, Wang HF, Wang Q, Szklarz GD, Domanski TL, Halpert JR, Correia MA. Influence of P450 3A4 SRS-2 residues on cooperativity and/or regioselectivity of aflatoxin B(1) oxidation. Chem Res Toxicol. 2001;14:483–491. doi: 10.1021/tx000218z. [DOI] [PubMed] [Google Scholar]
- Yano JK, Wester MR, Schoch GA, Griffin KJ, Stout CD, Johnson EF. The structure of human microsomal cytochrome P450 3A4 determined by X-ray crystallography to 2.05-A resolution. J Biol Chem. 2004;279:38091–38094. doi: 10.1074/jbc.C400293200. [DOI] [PubMed] [Google Scholar]
- Zangar RC, Davydov DR, Verma S. Mechanisms that regulate production of reactive oxygen species by cytochrome P450. Toxicol Appl Pharmacol. 2004;199:316–331. doi: 10.1016/j.taap.2004.01.018. [DOI] [PubMed] [Google Scholar]