Abstract
The development of cost-effective technologies able to comprehensively assess DNA, RNA, protein, and metabolites in patient tumors has fueled efforts to tailor medical care. Indeed validated molecular tests assessing tumor tissue or patient germline DNA already drive therapeutic decision making. However, many theoretical and regulatory challenges must still be overcome before fully realizing the promise of personalized molecular medicine. The masses of data generated by high-throughput technologies are challenging to manage, visualize, and convert to the knowledge required to improve patient outcomes. Systems biology integrates engineering, physics, and mathematical approaches with biologic and medical insights in an iterative process to visualize the interconnected events within a cell that determine how inputs from the environment and the network rewiring that occurs due to the genomic aberrations acquired by patient tumors determines cellular behavior and patient outcomes. A cross-disciplinary systems biology effort will be necessary to convert the information contained in multidimensional data sets into useful biomarkers that can classify patient tumors by prognosis and response to therapeutic modalities and to identify the drivers of tumor behavior that are optimal targets for therapy. An understanding of the effects of targeted therapeutics on signaling networks and homeostatic regulatory loops will be necessary to prevent inadvertent effects as well as to develop rational combinatorial therapies. Systems biology approaches identifying molecular drivers and biomarkers will lead to the implementation of smaller, shorter, cheaper, and individualized clinical trials that will increase the success rate and hasten the implementation of effective therapies into the clinical armamentarium.
INTRODUCTION
Traditionally, much of medical practice is based on standards of care. These interventions are based on knowledge from different levels of evidence generated by epidemiological and clinical studies or evidence-based medicine. However, large randomized studies, our best level of evidence, are designed to determine the best approach for the average populations and not for specific individuals. The development of molecular profiling technologies to assess DNA, RNA, protein, and metabolites provides the potential to tailor medical care, both at tumor and patient levels. These approaches have the potential to fulfill the promise of delivering the right dose for the right indication to the right patient at the right time. Importantly, personalized therapy offers the opportunity to increase therapeutic efficacy by targeting the genomic aberrations driving tumor behavior while at the same time decreasing inadvertent toxicity due to altered drug metabolism encoded by the patients' genetic background. Despite clear and important examples demonstrating the potential of the concept, a plethora of technical and regulatory challenges remain to be resolved before wide-spread implementation of personalized therapy.
Several validated molecular tests performed in tumor tissue or assessing the patients' genome are now part of standard therapeutic decision making in breast, colorectal, and lung cancers. However, the field of personalized medicine raises many challenges including the unexpected high failure rate of molecular targeted therapeutics, difficulties identifying, and validating molecular markers, homeostatic feedback loops, and molecular crosstalk and bypass mechanisms that can lead to unexpected effects on patient outcomes.1–6 It is thus critically important to develop an improved understanding of the pathways and networks to target as well as of the homeostatic loops induced by the interventions.
SYSTEMS BIOLOGY TO APPROACH PERSONALIZED CANCER THERAPY
Complex genomic aberrations targeting multiple genes through mutation, changes in copy number, and methylation occur in most epithelial tumors resulting in marked rewiring of the signaling networks that determine the behavior of the cancer cell and patient outcomes.7–14 High throughput technologies generate incredible masses of data showing many potential aberrations and connections and allowing visualization and integration of the data into testable hypotheses. However, much of the data used to explore the structure of signaling networks are contextual and not generalizable to a cancer cell in its microenvironment. Thus, the avalanche of data generated from efforts to map genetic aberrations in tumors including Genome Wide Associations Studies (GWAS) and the Cancer Genome Atlas (TCGA) as well as international genomics and proteomics efforts underlines the challenge of understanding the effects of the aberrations in patient tumors both alone and when integrated into cellular function. Indeed, the development of high throughput omics technologies that interrogate tumors at the DNA, RNA, protein, and metabolomic level has not been paralleled by improvements in cell biology approaches to understand the consequences of these changes on cellular, organ, and organismal outputs.
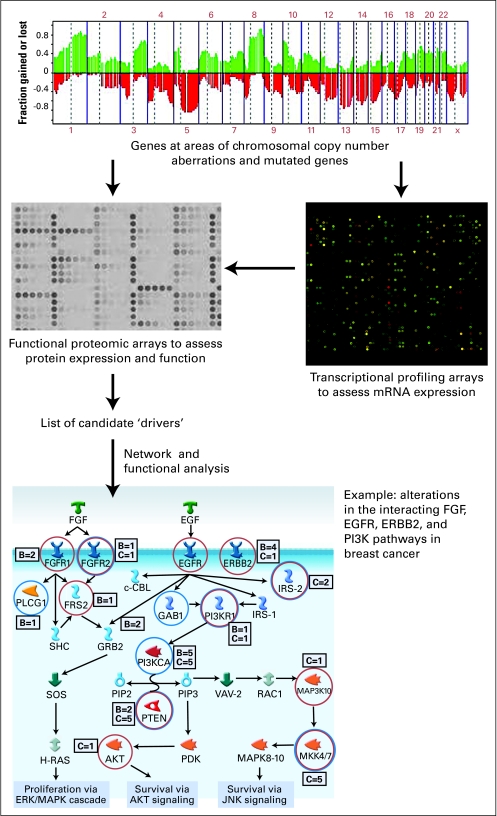
Systems biology uses engineering, computational, and physics approaches combined with biologic and medical inputs in an iterative manner to develop representations of the network of interactions within a cell that regulate cellular, organ, and organismal behavior. As opposed to the traditional reductionist approaches focusing on the manipulation of one gene or protein (eg, tumor suppressor or oncogene), systems biology attempts to integrate important information from reductionist approaches with multidimensional data into a comprehensive map of the way components of a biologic system integrate with external inputs to optimally predict the behavior of the system and how it regulates (Fig 115). This map will ultimately describe the behavior of the cancer cell and thus predict the natural course of the cancer and its response to specific treatments.16
Fig 1.
A systems approach integrating genomic and functional proteomic data to identify molecular strategies for the treatment of breast cancer. A comprehensive list of aberrant genes identified by copy number and sequence studies is filtered using expression arrays and functional proteomics. The found targets most likely represent important drivers of oncogenesis and can be mapped using network and functional analysis approaches into interconnecting molecular pathways. This pathway analysis can then be used to design rational therapeutic approaches in individual patients. Bottom panel reproduced with permission from Leary et al.15 Copyright (2008) National Academy of Sciences, U.S.A. FGF, fibroblast growth factor; EGFR, epidermal growth factor receptor; PI3K, phosphatidylinositol-3-OH kinase.
Thus, a multimodal approach including assessment of DNA, RNA, proteomics, and metabolomics has the potential to interrogate the patient tumor letting the experiment of nature teach us what is important to the initiation and progression of cancer. The discovery of these coordinate events will be a critical event in the process of the development of targeted therapeutics capitalizing on the Achilles' heel of oncogene addiction and synthetic lethality.17,18 High throughput biology approaches must be able to rapidly assess functional outcomes and must be sufficiently flexible to allow simultaneous manipulation of the multiple candidates that arise from omics approaches in a single cell.
TCGA
TCGA, a joint program of the National Cancer Institute (NCI) and the National Human Genome Research Institute, is an effort to accelerate the understanding of the molecular basis of cancer through genome analysis, including large-scale genome sequencing, analysis of DNA copy number, methylation, transcriptional profiling, and assessment of splicing aberrations. The goal of TCGA is to provide the molecular and physical map of cancer aberrations to improve our ability to diagnose, treat, and prevent cancer.19 TCGA is paralleled and extended by other collaborative international efforts. In the initial project, TCGA described the discovery of new genetic mutations and other DNA alterations with potential implications for the diagnosis and treatment of glioblastoma (GBM). The key observation from the GBM project was the demonstration that the genomic aberrations present in GBM integrate into limited pathways in which no single aberration is present at a sufficient level to act as a beacon to change patient management. Rather, the integrated analysis of multiple components of the pathway is necessary to reflect activation state of the pathway and potentially alter patient therapy.20 The challenge is to determine the role of each aberration as biomarkers and as therapeutic targets and the optimal methods to interrogate a pathway.
High Throughput Technologies for the Study of the Pathogenesis of Cancer
DNA copy number.
DNA copy number aberrations (CNA) alter the amount and organization of genomic material, which can increase or decrease the transcriptional activity of critical genes or regulatory RNAs. CNAs can be small altering function of a single gene or potentially affect a large chromosomal region. CNAs are inherited or caused by somatic such as deletions, duplications, inversions, or translocations. High throughput technologies including comparative genomic hybridization, digital karyotyping, representational oligonucleotide microarray (ROMA), single nucleotide polymorphism arrays, molecular inversion probes, and next generation sequencing are now capable of rapidly and efficiently profiling copy number changes across entire cancer genomes.16,21,22
DNA mutation detection.
The strongest predictors of risk of developing cancer and of response to therapy appear to be at the DNA level. Whether this reflects a readily assessed dichotomous variable (presence or absence of a mutation) versus changes in levels of RNA or protein, or reflects a greater effect on underlying biology remains to be determined. When identifying indicators of increased cancer risk, efforts have moved from positional cloning of rare, high-risk alleles such as BRCA1 and BRCA2 to identification of common low-risk variants through candidate gene analysis or GWAS. High throughput DNA sequencing offers a new approach to gene resequencing. However, most approaches incorporate error prone amplification steps requiring deep sequencing and statistical approaches to validate the results.23–25 Single molecule sequencing has the potential to bypass this challenge, but high costs currently restrict the technology to in depth studies of specific questions. A mass spectroscopy–based (MassARRAY) approach (Sequenom Inc, San Diego, CA) aimed at evaluating SNPs can facilitate rapid, high-throughput and cost-effective detection of hot-spot gene mutations (ie, in PIK3CA, AKT1, KRAS).26,27 However, it is not applicable to genes targeted by nonhot-spot mutations in tumor suppressors (ie, TP53, PTEN). SNP detection either through array approach or MassARRAY can be also applied to assess germline changes in metabolizing enzymes that could alter therapy efficacy (ie, CYP-2D6).
Epigenetic profiling.
A number of genome- or methylome-wide approaches to assess methylation state of important genes are under development. As with other technologies, next generation sequencing approaches are in the processes of supplanting current technologies. The challenge remains in validating the observations with orthologous technologies and developing high throughput biology approaches to determine the functional relevance of methylation changes. Some of these epigenetic phenomena responsible for silencing important tumor suppressor genes may be targetable.28
Gene-expression profiling.
The ability to measure thousands of mRNA transcripts in a single experiment has resulted in a rapid increase in our understanding of tumor pathophysiology as well as in identifying tumor classifiers that identify lineages, establish prognosis, and predict therapy response. A number of genomic tests that have been approved or are under evaluation provide better predictions of clinical outcome than traditional clinicopathologic standards, allow prediction of effects of therapies, and explore the activity of signaling pathways with targetable components.29–31 One of the most striking uses of this technology are predictors to identify women with early-stage breast cancer who do not benefit from chemotherapy.32
Table 1.
High Throughput Technologies for the Study of the Pathogenesis of Cancer
| Parameter | Available Technology | Target for Detection | Tissue Requirement | Advantage | Disadvantage |
|---|---|---|---|---|---|
| DNA copy number | Comparative genomic hybridization | DNA copy number aberrations | Blood (cDNA or germline DNA) | High throughput | Most platforms still require high quality frozen material |
| Digital karyotyping | Polymorphisms | Fresh/frozen | Comprehensive/whole genome | Limited dynamic range | |
| ROMA | Genomic rearrangements | Paraffin embedded | Expensive | ||
| Single nucleotide polymorphism arrays | |||||
| Molecular inversion probes deep sequencing | |||||
| DNA mutation detection | Candidate gene analysis | Identification of common low risk variants | Blood (cDNA or germline DNA) | High throughput | Expensive |
| GWAS | Presence or absence of a mutation | Fresh/frozen | Comprehensive | Many platforms require high quality frozen material | |
| High throughput DNA sequencing | Germline changes in metabolizing enzymes | Paraffin embedded | |||
| Single molecule sequencing | |||||
| Mass spectroscopy for single nucleotide polymorphisms | |||||
| Epigenetic profiling | Methylation arrays | Methylation | Blood (cDNA or germline DNA) | High throughput | Most platforms still require high quality frozen material |
| Acethylation arrays | Acethylation | Fresh/frozen | Comprehensive/whole genome | ||
| Paraffin embedded | |||||
| Gene expression profiling | Transcriptional profiles | Messenger RNA | Fresh/frozen | High throughput | Most platforms still require high quality frozen material |
| RT-PCR | Paraffin embedded | Comprehensive/whole genome | Limited dynamic range (eg, in comparison with qPCR) | ||
| Expensive | |||||
| Detection of splicing RNA forms | Exon junction arrays | Exome | Fresh/frozen | High throughput | Requires high quality frozen material |
| Genome tiling arrays | Particularly comprehensive/whole exome | Limited dynamic range | |||
| Most appropriate applications and analysis approaches still to be defined | |||||
| Expensive | |||||
| Functional proteomics | Reverse phase protein arrays | Protein expression and activation | Fresh/frozen | High throughput | Limited to proteins with available high-quality antibodies |
| Mass spectroscopy | Paraffin embedded | Allows simultaneous study of several hundred proteins | Primarily discovery tool | ||
| Applicable to both frozen and formalin fixed paraffin embedded tumor samples | Platforms still not robust and difficult to compare across centers | ||||
| Useful to study specific candidates | |||||
| Inexpensive |
Abbreviations: ROMA, representational oligonucleotide microarray; GWAS, Genome Wide Associations Studies; qPCR, quantitative polymerase chain reaction.
Detection of splicing RNA forms.
It is predicted that each gene is alternately or aberrantly spliced in cancer resulting in five to 50 different proteins from each gene. Tumor-specific or aberrant splicing that alters the function of target proteins appears to be a common event during tumorigenesis. Exon junction arrays use probes specific to the expected or potential splice sites of predicted exons for each gene in the genome. They are designed to detect each individual exon and to detect different splicing isoforms. Genome tiling arrays consist of overlapping probes designed to densely represent a genomic region of interest to detect alternatively spliced forms that may not have been previously known or predicted.33
Protein arrays.
Traditional protein assay techniques like western blotting or enzyme-linked immunosorbent assay can assess expression and phosphorylation of only a limited number of proteins, and they cannot map intracellular signal transduction or apoptotic pathways. Forward phase and reverse phase proteins arrays (RPPA) have the potential to screen large numbers of candidates. Forward phase arrays offer convenience, but are difficult to quantify. RPPA constitutes a low-cost, sensitive, high-throughput platform for marker screening, pathophysiology studies, identification of novel targets important in cancer growth, and therapeutic monitoring. Because most potential molecular markers and targets are proteins, proteomic profiling is expected to yield more direct answers to functional and pharmacologic questions than transcriptional profiling.34,35 RPPA can concurrently evaluate activation, proliferation, apoptosis, or any process for which high quality antibodies exist. Clinical applicability and potential benefits of RPPA have already been demonstrated.36–40
APPLICATION OF SYSTEMS BIOLOGY TO PERSONALIZED CANCER THERAPY
Driver Abnormalities Can Be Identified by the Integration of Multidimensional Data Sets
Multidimensional data sets permit the identification of multimodality aberrations that occur when a critical signaling protein is targeted in multiple ways in different patients. For example, the epidermal growth factor receptor (EGFR) tyrosine kinase can be mutated, amplified, or targeted by other signaling molecules, such as increased ligand expression, formation of heterodimers, or constitutive activation of the pathway. Multimodality aberrations that converge on a single molecule point to this molecule as being a driver or key node in the pathway which may represent an optimal marker or therapeutic target. Further, a signaling protein could be constitutively activated by epigenetic modification, amplification, or a point mutation. Comparing information obtained through different platforms, one will potentially be able to parse passenger aberrations that are usually noise. Conversely, by comparing these multidimensional data sets in different patients, one will potentially be able to identify converging aberrations that target the same driver molecule by different mechanisms and that result in the same functional outcome.20,34
Characterization of Tumors With Unusual Response to Therapies
In early clinical trials, a small but real number of patients demonstrate remarkable responses. They offer the opportunity to serve as beacons that teach important lessons if we are able to deconvolute the underlying mechanisms. Novel high throughput technologies could be used to fully characterize these tumors and to define the molecular aberration(s) that explains this unique therapy responsiveness. Studies of very small numbers of patients with this concept lead to the demonstration that EGFR mutations, but not EGFR protein levels identify a population of patients with lung cancer likely to benefit from EGFR-targeted therapy.41 A more recent study demonstrated that KIT mutations are functionally relevant targets in melanoma.42 This emphasizes the need to obtain biopsies from patients in clinical trials and underpins a major effort to identify and validate new targets.
Interspecies Comparative Genomics May Provide a Novel Approach to Identify Drivers
Evolutionary conservation of genomic aberrations across species points to an important role of those conserved regions in tumorigenesis. Dysregulated pathways involved in oncogenesis are conserved by evolution in different species. Such comparative genomic approaches can reveal important driving mutations in cancer. For example, comparison of gene expression changes in lung cancer in mice and lung cancer in humans uncovered gene expression signatures that demonstrate activation of the K-Ras oncogene.43 Comparisons of point mutations and DNA copy number alterations in different species are now underway to unravel crucial aberrations in tumor formation.
Cell-Line Models
Cell lines provide a first step for validation of putative driver aberrations and for establishing their mechanisms of action at the molecular level. High-throughput small interfering RNA, short hairpin RNA, and microRNA screening assays were developed to facilitate a comprehensive evaluation the role of genes in cellular functions and potential synthetic lethality.34 Similar approaches using overexpression of open reading frames or regulatory RNA can elucidate the function of increased levels of genes. These approaches have been most often applied to cell lines to identify candidate oncogenes. Application to model organisms is allowing comprehensive in vivo screens. For example, high-throughput screens using RNA interference in cell lines can identify and validate candidate oncogenes. High throughput RNAi screens in human cell lines in vitro led to the identification of PIK3CA mutations and resistance to trastuzumab in breast cancer.34
Drug Screens
High-throughput in vitro proliferation/viability drug screening assays have been developed to facilitate a comprehensive evaluation of the antitumor efficacy of drugs across multiple cell lines that are representative of different cancer types, distinct subtypes of specific cancers, and of the various genomic and proteomic aberrations that are present in cancer.44
Using the Unit of Function: The Cancer Cell in Its Microenvironment
Tumor initiation and progression as well as response to therapy depend on the interplay between the cancer and its host—the microenvironment. The success of our attempts at treating cancer will depend on a thorough understanding of these interactions at a molecular level. Therefore, the use of three-dimensional modeling systems45 to study the microenvironment as well as the direct study of the effects of the stroma in tumors cannot be excluded from the systems biology approach.
Future of Clinical Trials: Smaller, Shorter, Cheaper, Individualized
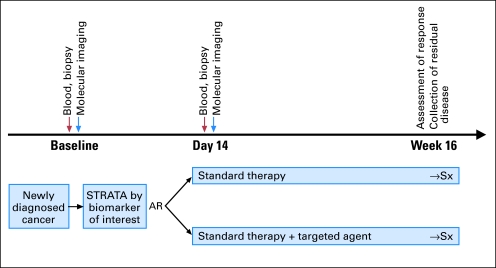
Neoadjuvant trials represent an emerging opportunity for the rapid validation of biomarkers in cancer. Ideally, biomarker-driven trials should require mandated tumor biopsies, testing patient tumors for the presence of specific genetic lesions in specific pathways, to deliver rationally designed therapy that targets the underlying aberrations in tumors as well as the potential bypass mechanisms. This approach must clearly be compared with the standard of care. Patients should be rebiopsied during therapy to monitor pharmacodynamic markers and at recurrence or progression to understand mechanisms of resistance. Figure 2 shows and explains the characteristics and of a generic example of a biomarker driven randomized phase II neoadjuvant study. 46
Fig 2.
Schema of a neoadjuvant phase II molecular marker–driven randomized study. Patients have a baseline biopsy and molecular imaging, and are stratified (STRATA) according to the marker, then adaptively randomly assigned (AR) to standard therapy versus standard therapy plus a targeted therapy directed to the marker of interest. A second biopsy and molecular imaging are performed at 2 weeks of treatment to study pharmacodynamic markers in both arms and correlate the pharmacodynamic changes and marker status with response at the time of surgery. This is a modified “Marker by Treatment Interaction Design”46 in which there is an assumption that a hypothesized marker splits the population into groups in which the efficacy of a particular treatment will differ. This design can be viewed as a classical randomized clinical trial with upfront stratification for the marker. Importantly, patients with and without the marker have the same chance of being randomly assigned to either therapy until the adaptive part of the study begins. Once enough information is gathered (safety or efficacy according with the end points of the study), the group with the advantage is enriched. Strict early stopping rules are built in. The main goal of these studies is to generate a better hypothesis to be tested in an adequately powered clinical trial. Sx, surgery.
CHALLENGES IN SYSTEMS BIOLOGY AND PERSONALIZED CANCER THERAPY
The use of a systems biology approach and the design and execution of biomarker-driven clinical trials remains a challenging task.
Segregating Driving Aberrations From Noise: Finding the Needle in the Haystack
Solid tumors evolve through the accumulation of genetic abnormalities through a process of natural selection of mutations and CNAs that give a proliferative or survival advantage called driver and contributing aberrations. A parallel process leads to the accumulation of passenger mutations (noise) through the induction of genetic instability. A major challenge that researchers face is the parsing of drivers versus noise. The multiple comparisons problem represents an insidious challenge. In most cases, large amounts of data are acquired on a limited number of samples resulting on over-training results and identifying spurious associations. Many approaches to limit this challenge have been proposed, but the most effective is the rigorous use of independent training, test and validation sets. This parallels the concept of the need for confirmatory clinical trials. The success at identifying causal genetic lesions for the aim of therapeutic targeting will involve the use of comparative technologies. Common themes will emerge, and conserved aberrations will have a higher probability of being drivers. Validation of driver genes can be facilitated by the demonstration of alterations at the DNA, RNA, and protein levels to be tested as potential targets for novel therapies.
Regulatory Implications
The progress toward personalized therapy also has important regulatory implications. A large number of exploratory biomarkers can be studied during clinical trials, but to make therapeutic decisions such us stratification for randomized studies, the used test should be done in a Clinical Laboratory Improvement Amendments–certified environment.47 Further, new clinical trial designs to identify and validate biomarkers and targeted therapeutics require education of regulatory committees in institutions and at the US Food and Drug Administration to ensure that effective approaches reach patients efficiently without compromising their safety.
CONCLUSION AND FUTURE DIRECTIONS
A systems biology approach to apply new high throughput technologies will be required to efficiently fulfill the promise of personalized molecular medicine. New clinical trial designs are needed to rapidly evaluate the hundreds of targeted therapeutics and potential biomarkers that are under preclinical evaluation. At our institution, we are implementing an effort designated Project T9 (10,000 therapies, 10,000 tests, and 10,000 treatments). We will characterize aberrations in 10,000 patients for all of the genes shown to be mutationally activated in 5% of any major cancers. Information will be used to determine the frequency of mutation and comutation events, to correlate mutations with patient outcomes, and to direct patients to targeted therapy trials aiming the aberrations present in their tumors. This is designed to develop paradigms and approaches to bypass the challenges associated with wide spread implementation of biomarker-driven personalized molecular medicine.
Footnotes
Supported in part by the Kleberg Center for Molecular Markers at M. D. Anderson Cancer Center, American Society of Clinical Oncology Career Development Award, Grant No. 1K23CA121994-01 from the National Cancer Institute (A.M.G.-A.), Susan G. Komen Foundation Grant No. FAS0703849 (A.M.G.-A., B.T.J.H., G.B.M.), Grant No. P50 CA083639 from the National Cancer Institute (G.B.M.), and Cancer Center Support Grant No. P30CA16672.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The author(s) indicated no potential conflicts of interest.
AUTHOR CONTRIBUTIONS
Conception and design: Ana Maria Gonzalez-Angulo, Bryan T.J. Hennessy, Gordon B. Mills
Financial support: Ana Maria Gonzalez-Angulo, Bryan T.J. Hennessy, Gordon B. Mills
Administrative support: Ana Maria Gonzalez-Angulo, Bryan T.J. Hennessy, Gordon B. Mills
Manuscript writing: Ana Maria Gonzalez-Angulo, Bryan T.J. Hennessy, Gordon B. Mills
Final approval of manuscript: Ana Maria Gonzalez-Angulo, Bryan T.J. Hennessy, Gordon B. Mills
REFERENCES
- 1.Engelman JA, Chen L, Tan X, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008;14:1351–1356. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Massarelli E, Varella-Garcia M, Tang X, et al. KRAS mutation is an important predictor of resistance to therapy with epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. Clin Cancer Res. 2007;13:2890–2896. doi: 10.1158/1078-0432.CCR-06-3043. [DOI] [PubMed] [Google Scholar]
- 3.Hennessy BT, Smith DL, Ram PT, et al. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat Rev Drug Discov. 2005;4:988–1004. doi: 10.1038/nrd1902. [DOI] [PubMed] [Google Scholar]
- 4.O'Reilly KE, Rojo F, She QB, et al. MTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66:1500–1508. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ihle NT, Lemos R, Jr, Wipf P, et al. Mutations in the phosphatidylinositol-3-kinase pathway predict for antitumor activity of the inhibitor PX-866 whereas oncogenic Ras is a dominant predictor for resistance. Cancer Res. 2009;69:143–150. doi: 10.1158/0008-5472.CAN-07-6656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jimeno A, Messersmith WA, Hirsch FR, et al. KRAS mutations and sensitivity to epidermal growth factor receptor inhibitors in colorectal cancer: Practical application of patient selection. J Clin Oncol. 2009;27:1130–1136. doi: 10.1200/JCO.2008.19.8168. [DOI] [PubMed] [Google Scholar]
- 7.Bergamaschi A, Kim YH, Wang P, et al. Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes Chromosomes Cancer. 2006;45:1033–1040. doi: 10.1002/gcc.20366. [DOI] [PubMed] [Google Scholar]
- 8.Chin K, DeVries S, Fridlyand J, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 9.Cheng KW, Lahad JP, Kuo WL, et al. The Rab 25 small GTPase determines aggressiveness of ovarian and breast cancers. Nat Med. 2004;10:1251–1256. doi: 10.1038/nm1125. [DOI] [PubMed] [Google Scholar]
- 10.Hughes-Davies L, Huntsman D, Ruas M, et al. EMSY links the BRCA2 pathway to sporadic breast and ovarian cancer. Cell. 2003;115:523–535. doi: 10.1016/s0092-8674(03)00930-9. [DOI] [PubMed] [Google Scholar]
- 11.Bostner J, Ahnström Waltersson M, Fornander T, et al. Amplification of CCND1 and PAK1 as predictors of recurrence and tamoxifen resistance in postmenopausal breast cancer. Oncogene. 2007;26:6997–7005. doi: 10.1038/sj.onc.1210506. [DOI] [PubMed] [Google Scholar]
- 12.Bellacosa A, de Feo D, Godwin AK, et al. Molecular alterations of the AKT2 oncogene in ovarian and breast carcinomas. Int J Cancer. 1995;64:280–285. doi: 10.1002/ijc.2910640412. [DOI] [PubMed] [Google Scholar]
- 13.Bautista S, Vallès H, Walker RL, et al. In breast cancer, amplification of the steroid receptor coactivator gene AIB1 is correlated with estrogen and progesterone receptor positivity. Clin Cancer Res. 1998;4:2925–2929. [PubMed] [Google Scholar]
- 14.Slamon DJ, Godolphin W, Jones LA, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244:707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 15.Leary RJ, Lin JC, Cummins J, et al. Integrated analysis of homozygous deletions, focal amplifications and sequence alterations in breast and colorectal cancers. Proc Natl Acad Sci U S A. 2008;105:16224–16229. doi: 10.1073/pnas.0808041105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hennessy BT, Gonzalez-Angulo AM, Carey MS, et al. A systems approach to analysis of molecular complexity in breast cancer. Clin Cancer Res. 2009;15:417–419. doi: 10.1158/1078-0432.CCR-08-2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Felsher DW. Tumor dormancy and oncogene addiction. APIMS. 2008;116:629–637. doi: 10.1111/j.1600-0463.2008.01037.x. [DOI] [PubMed] [Google Scholar]
- 18.Moeller BJ, Pasqualini R, Arap W. Targeting cancer-specific synthetic lethality in double-strand DNA break repair. Cell Cycle. 2009;8:1872–1876. doi: 10.4161/cc.8.12.8743. [DOI] [PubMed] [Google Scholar]
- 19.The Cancer Genome Atlas. http://cancergenome.nih.gov/about/index.asp.
- 20.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang TL, Maierhofer C, Speicher MR, et al. Digital karyotyping. Proc Natl Acad Sci U S A. 2002;99:16156–16161. doi: 10.1073/pnas.202610899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shih IeM, Sheu JJ, Santillan A, et al. Amplification of a chromatin remodeling gene, Rsf-1/HBXAP, in ovarian carcinoma. Proc Natl Acad Sci U S A. 2005;102:14004–14009. doi: 10.1073/pnas.0504195102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drew Y, Calvert H. The potential of PARP inhibitors in genetic breast and ovarian cancers. Ann N Y Acad Sci. 2008;1138:136–145. doi: 10.1196/annals.1414.020. [DOI] [PubMed] [Google Scholar]
- 24.Tartaglia M, Pennacchio LA, Zhao C, et al. Gain-of function SOS1 mutations cause a distinctive form of Noonan syndrome. Nat Genet. 2007;39:75–79. doi: 10.1038/ng1939. [DOI] [PubMed] [Google Scholar]
- 25.Thomas RK, Baker AC, Debiasi RM, et al. High throughput oncogene mutation profiling in human cancer. Nat Genet. 2007;39:347–351. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- 26.Jurinke C, van den Boom D, Cantor CR, et al. The use of MassARRAY technology for high throughput genotyping. Adv Biochem Eng Biotechnol. 2002;77:57–74. doi: 10.1007/3-540-45713-5_4. [DOI] [PubMed] [Google Scholar]
- 27.Stemke-Hale K, Gonzalez-Angulo AM, Lluch A, et al. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res. 2008;68:6084–6091. doi: 10.1158/0008-5472.CAN-07-6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dehan P, Kustermans G, Guenin S, et al. DNA methylation and cancer diagnosis: New methods and applications. Expert Rev Mol Diagn. 2009;9:651–657. doi: 10.1586/erm.09.53. [DOI] [PubMed] [Google Scholar]
- 29.Sotiriou C, Pusztai L. Gene-expression signatures in breast cancer. N Engl J Med. 2009;360:790–800. doi: 10.1056/NEJMra0801289. [DOI] [PubMed] [Google Scholar]
- 30.Bild AH, Yao G, Chang JT, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 31.Chang JC, Wooten EC, Tsimelzon A, et al. Patterns of resistance and incomplete response to docetaxel by gene expression profiling in breast cancer patients. J Clin Oncol. 2009;23:1169–1177. doi: 10.1200/JCO.2005.03.156. [DOI] [PubMed] [Google Scholar]
- 32.Paik S, Tang G, Shak S, et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J Clin Oncol. 2006;24:3726–3734. doi: 10.1200/JCO.2005.04.7985. [DOI] [PubMed] [Google Scholar]
- 33.Xi L, Feber A, Gupta V, et al. Whole genome exon arrays identify differential expression of alternatively spliced, cancer-related genes in lung cancer. Nucleic Acids Res. 2008;36:6535–6547. doi: 10.1093/nar/gkn697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berns K, Horlings H, Hennessy BT, et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12:395–402. doi: 10.1016/j.ccr.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 35.Charboneau L, Tory H, Chen T, et al. Utility of reverse phase protein arrays: Applications to signaling pathways and human body arrays. Brief Funct Genomic Proteomic. 2002;1:305–315. doi: 10.1093/bfgp/1.3.305. [DOI] [PubMed] [Google Scholar]
- 36.Nishizuka S, Charboneau L, Young L, et al. Proteomic profiling of the NCI-60 cancer cell lines using new high-density reverse-phase lysate microarrays. Proc Natl Acad Sci U S A. 2003;100:14229–14234. doi: 10.1073/pnas.2331323100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wulfkuhle JD, Aquino JA, Calvert VS, et al. Signal pathway profiling of ovarian cancer from human tissue specimens using reverse-phase protein microarrays. Proteomics. 2003;3:2085–2090. doi: 10.1002/pmic.200300591. [DOI] [PubMed] [Google Scholar]
- 38.Grubb RL, Calvert VS, Wulkuhle JD, et al. Signal pathway profiling of prostate cancer using reverse phase protein arrays. Proteomics. 2003;3:2142–2146. doi: 10.1002/pmic.200300598. [DOI] [PubMed] [Google Scholar]
- 39.Zha H, Raffeld M, Charboneau L, et al. Similarities of prosurvival signals in Bcl-2-positive and Bcl-2-negative follicular lymphomas identified by reverse phase protein microarray. Lab Invest. 2004;8:235–244. doi: 10.1038/labinvest.3700051. [DOI] [PubMed] [Google Scholar]
- 40.Hennessy BT, Gonzalez-Angulo AM, Stemke-Hale KA, et al. Characterization of a naturally occurring breast cancer subset enriched in EMT and stem cell characteristics. Cancer Res. 2009;69:4116–4124. doi: 10.1158/0008-5472.CAN-08-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 42.Woodman SE, Trent JC, Stemke-Hale K, et al. Activity of dasatinib against L576P KIT mutant melanoma: Molecular, cellular, and clinical correlates. Mol Cancer Ther. 2009;8:2079–2085. doi: 10.1158/1535-7163.MCT-09-0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sweet-Cordero A, Mukherjee S, Subramanian A, et al. An oncogenic KRAS2 expression signature identified by cross-species gene-expression analysis. Nat Genet. 2005;37:48–55. doi: 10.1038/ng1490. [DOI] [PubMed] [Google Scholar]
- 44.Monks A, Scudiero D, Skehan P, et al. Feasibility of a high-flux anticancer drug screen using a diverse panel of cultured human tumor cell lines. J Natl Cancer Inst. 1991;83:757–766. doi: 10.1093/jnci/83.11.757. [DOI] [PubMed] [Google Scholar]
- 45.Fischbach C, Chen R, Matsumoto T, et al. Engineering tumors with 3D scaffolds. Nature Methods. 2007;4:855–860. doi: 10.1038/nmeth1085. [DOI] [PubMed] [Google Scholar]
- 46.Sargent DJ, Conley BA, Allegra C, et al. Clinical trial designs for predictive marker validation in cancer treatment trials. J Clin Oncol. 2005;23:2020–2027. doi: 10.1200/JCO.2005.01.112. [DOI] [PubMed] [Google Scholar]
- 47.US Department of Health and Human Services. Centers for Medicare and Medicaid Services. http://www.cms.hhs.gov/CLIA/