Abstract
Fanconi anemia (FA) and dyskeratosis congenita (DC) are rare inherited syndromes that cause head and neck squamous cell cancer (HNSCC). Prior studies of inherited forms of cancer have been extremely important in elucidating tumor suppressor genes inactivated in sporadic tumors. Here, we studied whether sporadic tumors have epigenetic silencing of the genes causing the inherited forms of HNSCC. Using bisulfite sequencing, we investigated the incidence of promoter hypermethylation of the 17 Fanconi- and DC-associated genes in sporadic HNSCC. Genes that only showed methylation in the tumor patients were chosen for quantitative methylation-specific PCR (qMSP) in a set of 45 tumor and 16 normal patients. Three gene promoters showed differences in methylation: FancB (FAAP95, FA core complex), FancJ (BRIP1, DNA Helicase/ATPase), and DKC1 (dyskeratin). Bisulfite sequencing revealed that only FancB and DKC1 showed no methylation in normal patients, yet the presence of promoter hypermethylation in tumor patients. On qMSP, 1/16 (6.25%) of the normal mucosal samples from non-cancer patients and 14/45 (31.1%) of the tumor patients demonstrated hypermethylation of the FancB locus (p < 0.05). These results suggest that inactivation of FancB may play a role in the pathogenesis of sporadic HNSCC.
Key Words: Hypermethylation, Fanconi anemia, Dyskeratosis congenita, Leukoplakia, Head and neck cancer
Introduction
Head and Neck Squamous Cell Carcinoma (HNSCC) is the fifth most common cancer, and more than 500,000 cases annually are diagnosed worldwide. Exposure to tobacco and alcohol are the most strongly associated clinical factors for the development of sporadic disease. Many of the genetic and epigenetic changes causing sporadic head and neck cancer, including p53, VEGF, and cyclin D1 [1], have been described; however, much remains to be learned. Head and neck cancer continues to be seen as a clonal progression of genetic and epigenetic changes yielding a cancer phenotype [2]. Previous work on inherited forms of cancer susceptibility have led to understanding of the sporadic forms of disease; limited examples include p53 (Li-Fraumeni syndrome) [3] and APC (familial adenomatous polyposis) [4]. Inactivation, as proposed by Knudsen's two-hit hypothesis, has been expanded to include epigenetic mechanisms – including promoter methylation.
In head and neck cancer, 2 syndromes (both with documented genetic alterations) exist: Fanconi anemia (FA) and dyskeratosis congenita (DC). Both are inherited syndromes that cause bone marrow failure, and are associated with early development of HNSCC in the absence of tobacco and alcohol exposure. FA is a rare autosomal recessive disorder characterized by progressive pancytopenia, requiring bone marrow transplant, and associated congenital abnormalities, such as microcephaly, short stature, skeletal, cardiac, and renal malformations. Mortality is associated with bone marrow failure and leukemia in the first 2 decades of life, with the majority of deaths occurring within 5 years after the onset of anemia [5,6]. The disease is caused by mutations within a variety of genes associated with the FA pathway. Many genes have been linked in the pathogenesis of FA, and at least 12 genes, including a 9-gene FA nuclear core complex with associated downstream proteins, all function to promote genetic stability via repair of DNA following damage [7]. Germline mutations of these genes are associated with dysregulation of cell cycle and apoptosis [8,9] and chromosomal instability [8]. It has been postulated that disruption of the FA complex and associated pathways are associated with chromosomal and telomere instability characterized by impaired DNA repair and susceptibility to double-strand DNA breaks leading to the inactivation of tumor suppressor genes and activation of oncogenes [10]. The exact mechanisms by which mutation of the FA protein pathway promotes carcinogenesis are being actively investigated.
FA patients, if they survive past bone marrow failure, have an extremely high rate of premalignant leukoplakia of the oral mucosa [11]. Most lesions arise from the upper aerodigestive tract in areas of mucosal or mucocutaneous squamous epithelium. Patients with FA have a HNSCC incidence 700 times greater than the general population [12], and tend to develop the disease at a younger age [13].
DC is also a rare inherited bone marrow failure disorder with a triad of lacey reticular pigmentation of the skin, nail dystrophy, and mucosal leukoplakia [14]. Like FA, mortality is associated with bone marrow failure, which happens in 90% of DC patients, mostly in the first decade of life [15]. X-linked recessive, autosomal dominant, and autosomal recessive forms of the disorder have been identified clinically, with 75% of cases attributable to the X-linked recessive form [15]. DC appears to be a disorder of telomere maintenance, with gene products of both the XLR (DKC1) and AD (TERC) forms associated with the telomerase complex, and a characteristic short telomere phenotype in all patients with DC [16]. There are multiple mutations in the DKC1 and TERC genes associated with DC, which lead to heterogenic presentations of varying phenotypic severity [17]. The exact mechanisms by which dysfunction of the DKC1 and TERC gene products contribute to carcinogenesis in DC patients is poorly understood. Recent studies have demonstrated the role of the mutant DKC1 gene in alterations of messenger RNA transcription [18]. DKC1 resides on the end of the X chromosome in an area surrounded by members of the cancer-testis gene family, whose expression is controlled by imprinting (methylation silencing) [19].
Oral leukoplakia is present in 70–80% of DC patients with almost uniform involvement of the dorsum of the tongue [20]. These patients are at high risk for the development of squamous cell carcinoma from these lesions [21]. Due to the rarity of DC (1:1,000,000 individuals), quantification of the cancer risk is difficult, but reports have shown 9% of male patients develop malignancies [22] and multiple case reports have demonstrated the association of DC with oral cancer [23,24,25,26,27,28,29,30,31].
Transcriptional inactivation of tumor suppressor genes by methylation of CpG islands is well described in HNSCC [32,33,34,35,36]. By far the most studied has been promoter hypermethylation of tumor suppressor genes, including: Cyclin A1, MGMT, DCC, and p16[37]. Methylation of cytosine-guanine dinucleotides by the enzyme class of DNA methyltransferases transfers a methyl group from S-adenosyl-methionine, and is associated with gene silencing [38]. In this study, we examine the presence of promoter hypermethylation as a means of tumor suppressor gene silencing in head and neck cancer.
One fascinating study implicated methylation of the Fanconi pathway in the pathogenesis of ovarian cancer and the presence of cisplatin resistance. D’Andrea [39] found FANCF to be transcriptionally silenced in sporadic ovarian tumors by methylation, and more importantly this was correlated with cisplatin sensitivity. Restoration of this pathway was found to be associated with demethylation of FANCF, leading to acquired cisplatinum resistance. This was followed up by work in advanced-stage invasive cervical cancer indicating that the FANCF gene is disrupted by either promoter hypermethylation and/or deregulated gene expression in the majority of cervical cancers, and that cervical cancer cell lines also exhibit a chromosomal hypersensitivity phenotype after exposure to an alkylating agent, a characteristic of FA patients [40]. Indeed, some researchers have already looked at the methylation status of sporadic head and neck cancer. In that study, they investigated epigenetic alterations in a limited part of the FANC-BRCA pathway in HNSCC and non-small-cell lung cancers using methylation-specific PCR (MSP), and found that promoter methylation of FANCF occurred in 15% (13/89) of HNSCCs [41]. This study, while extremely promising, acknowledges that FA can be caused by inactivation in any of the 12 genes involved in the complex or downstream, but just looked at FANCF and did not consider genes causing DC.
Materials and Methods
Histopathology
Samples were taken from surgical specimens of patients with head and neck cancer and biopsies from normal mucosa (taken from uvulopalatopharyngoplasty procedures for sleep apnea) treated at the Johns Hopkins Hospital between 1998 and 2005. Samples were collected in an IRB-approved protocol. All samples were analyzed in the Pathology Department at Johns Hopkins Hospital. Normal samples were microdissected, and DNA was prepared from the mucosa. Tumor samples were confirmed to be head and neck squamous, and were subsequently microdissected to separate the tumor from the stromal elements.
DNA Extraction
Samples were centrifuged and digested in a solution of detergent (sodium dodecylsulfate) and proteinase K, for the removal of proteins bound to the DNA. Samples were first purified and desalted with phenol/chloroform extraction. The digested sample was subjected to ethanol precipitation, twice, and subsequently resuspended in 50 μl LoTE (EDTA 2.5 mM and Tris-HCl 10 mM) and stored at −80°C.
Bisulfite Treatment
DNA from salivary rinses was subjected to bisulfite treatment, as described previously [42]. In short, 2 μg genomic DNA was denatured in 0.2 M NaOH for 30 min at 50°C. This denatured DNA was then diluted into 500 μl of a solution of 10 mM hydroquinone and 3 M sodium bisulfite. This was incubated for 3 h at 70°C. Afterwards, the DNA sample was purified with a Sepharose column (Wizard DNA Clean-Up System; Promega, Madison, Wisc., USA). Eluted DNA was treated with 0.3 M NaOH for 10 min at room temperature, and precipitated with ethanol. This bisulfite-modified DNA was subsequently resuspended in 120 μl LoTE (EDTA 2.5 mM and Tris-HCl 10 mM) and stored at −80°C.
Bisulfite Sequencing
Sequencing reactions were conducted with initial PCR of the gene promoter of interest, utilizing 20 ng of bisulfite-treated DNA. Primers (available upon request) were designed using MethPrimer (www.urogene.org/methprimer) in areas without CpG islands. Reactions were conducted in 25 μl total volume, with template DNA, forward and reverse primers (0.5 nM each), 0.5 μl Platinum Taq (Invitrogen), 10% DMSO, 200 μM each of DNTPs, 16.6 mM ammonium sulfate, 67 mM Trizma (Sigma, St Louis, Mo., USA); 6.7 mM MgCl2, and 10 mM mercaptoethanol. Products were run on to test specific product amplification, gel bands at predicted lengths were cut under UV light, and bands were extracted using the QIAquick Gel Extraction Kit (Qiagen, Valencia, Calif., USA) per protocol. Products were tested for adequate concentration, and sequenced by cycle sequencing (Applied Biosystems, Norwalk, Conn., USA) using big dye terminator.
Methylation Analysis and Quantitative MSP
The bisulfite-modified DNA was used as a template for fluorescence-based real-time PCR, as described previously [43]. Primers and probes were designed to amplify the bisulfite-converted promoters of DKC1, FANCB. All probes had 5′ FAM and 3′ TAMRA moieties. The ratios between the values of the gene of interest and β-actin, an internal reference gene, were obtained by quantitative MSP (qMSP) analysis, and were used as a measure for representing the relative level of methylation in a particular sample. Fluorogenic PCRs were carried out in a 20-μl reaction volume consisting of 600 nM of each primer, 200 nM probe, 0.75 U Platinum Taq DNA polymerase (Invitrogen, Carlsbad, Calif. USA), 200 μM each of DNTPs, 16.6 mM ammonium sulfate, 67 mM Trizma (Sigma), 6.7 mM MgCl2 (2.5 mM for p16), 10 mM mercaptoethanol, and 0.1% dimethylsulfoxide. Three microliters of treated DNA solution were used in each qMSP reaction. Reactions were carried out in 384-well plates in an ABI 7900 real-time PCR system (Perkin-Elmer Applied Biosystems, Norwalk, Conn., USA). Each plate consisted of triplicate patient samples, as well as positive and negative controls. Reaction conditions consisted of an initial denaturation for 2 min at 95°C, followed by cycles of denaturation for 15 s at 95°C, and annealing for 1 min at 60°C. Leukocytes from a healthy individual were methylated in vitro with SssI methyltransferase (New England Biolabs Inc, Beverly, Mass., USA) to generate completely methylated DNA, and serial dilutions of this DNA were used for constructing the calibration curves on each plate.
Statistical Analysis
Statistical analyses were performed using Stata 9.0 (Statacorp LP; College Station, Tex., USA), and p values were calculated using Pearson's χ2 test. If the observed frequencies were below 5, then Fisher's exact correlation was performed. All statistical tests were two-sided. All differences were considered statistically significant when p ≤ 0.05.
Results
Bisulfite Sequencing
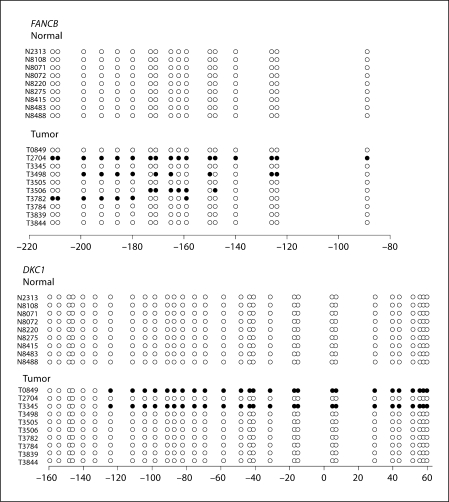
Ten tumor and 10 normal samples were bisulfite-sequenced at each of the gene promoters listed in table 1. Results from FANCB and DKC1 only are shown in figure 1. Four of 10 tumors (40%) showed some degree of promoter methylation in HSNCC without evidence of promoter methylation in normal samples (0/10). In DKC1, 20% (2/10) of the tumors tested showed promoter hypermethylation, which was again absent in normal samples (0/10). There was no overlap in the samples demonstrating promoter hypermethylation of the DKC1 or FANCB loci.
Table 1.
Genes bisulfite-sequenced in this study
| Genes linked to inherited HNSCC | Chromosomal location | CpG island |
|---|---|---|
| FANCA – FA core complex | 16q24.3 | yes |
| FANCB (FAAP95) – FA core complex | Xp22.31 | yes |
| FANCC – FA core complex, cytoplasmic functions | 9q22.3 | yes |
| FANCD1/BRCA2 – RAD51 recruitment | 13q12–13 | yes |
| FANCD2 – Monoubiquitinated | 3p25.3 | yes |
| FANCE – FA core complex, direct binding | 6p21–22 | yes |
| FANCF – FA core complex | 11p15 | yes |
| FANCG/XRCC9 – FA core complex | 9p13 | yes |
| FancI – not identified, rare | ? | NA |
| FANCJ/BACH1/BRIP1 – DNA helicase/ATPase, binding to BRCA1 | 17q22–q24 | yes |
| FANCL/PHF9/POG (FAAP43) – FA core complex, ubiquitin ligase | 2p16.1 | yes |
| FANCM/Hef (FAAP250) – FA core complex, ATPase | 14q21.3 | yes |
| RAD51 | 15q15.1 | no |
| ATM | 11q22.3 | yes |
| NBS1 | 8q21.3 | yes |
| DKC1 | Xq28 | yes |
| TERC | 3q26.2 | yes |
Fig. 1.
Bisulfite sequencing was done in order to screen targets for promoter hypermethylation and gene silencing: 2/10(20%) of the DKC1 tumors were methylated; 4/10 (40%) of the FANCB promoters showed some degree of methylation. All other targets did not show promoter hypermethylation in the tumors. In the case of FANCJ, there was methylation in both tumor and normal samples.
Every other gene, including FANCA, FANCB, FANCC, FANCD1/BRCA2, FANCD2, FANCE, FANCF, FANCG, FANCJ/BRIP1, FANCL/PHF9/POG, FANCM, RAD51, ATM, and NBS1, were all bisulfite-sequenced on sporadic tumors and normal tissues, and did not show promoter hypermethylation specific to tumors. Bisulfite sequencing was used to screen possible gene targets for subsequent studies evaluating promoter methylation status. In this study, methylation was not detected in FANCA, FANCC, FANCD1/BRCA2, FANCD2, FANCE, FANCF, FANCG, FANCL/PHF9/POG, FANCM, RAD51, ATM, or NBS1 with bisulfite sequencing (data not shown).
Quantitative Methylation-Specific PCR
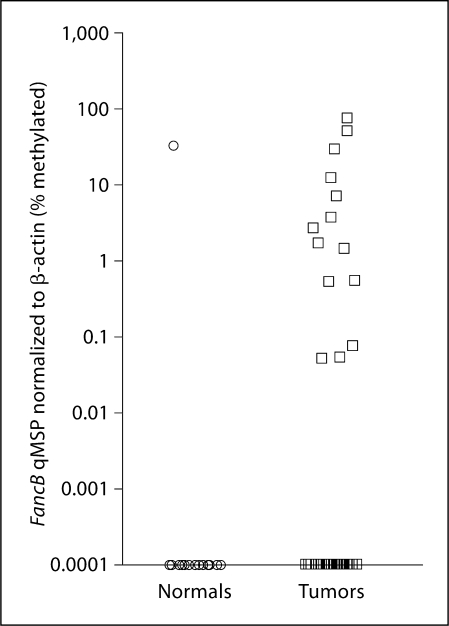
The demographics of the 2 groups of patients studied by qMSP are shown in table 2. The normal patients had a slightly lower median age (41 years; 20–76 years) compared to HNSCC patients (56 years; 39–78 years). Two genes, DKC1 and FANCB, showed promising differences in promoter methylation between tumors and normal tissues. Figure 2 shows the qMSP differences in promoter methylation of the FANCB locus. On qMSP, 14/45 (31.1%) of the tumor patients compared to 1/16 (6.25%) of the normal patients demonstrated hypermethylation of the FANCB locus. This difference was significant and associated with a value of p < 0.05 (χ2).
Table 2.
Demographics of study patients
| Head and neck cancer (n = 45) | Normal patients (n = 16) | ||
|---|---|---|---|
| Median age, years (range) | 56 (39–78) | 41 (20–76) | |
| Sex | Male | 35 (78) | 11 (69) |
| Female | 10 (22) | 5 (31) | |
| Race | Caucasian | 34 (76) | 15 (94) |
| Black | 6 (13) | 1 (6.3) | |
| Other | 5 (11) | 0 (0) | |
| Smoking status | Never | 14 (31) | 10 (63) |
| Former | 18 (40) | 3 (19) | |
| Current | 13 (29) | 3 (19) | |
| Stage | I | 4 (8.9) | – |
| II | 5 (11) | – | |
| III | 6 (13) | – | |
| IV | 30 (67) | – | |
| Site | Nose/sinus | 1 (2.2) | – |
| Oral cavity | 12 (27) | – | |
| Oropharynx | 21 (47) | – | |
| Hypopharynx | 2 (4.4) | – | |
| Larynx | 4 (8.9) | – | |
| Neck/other | 5 (11) | – | |
Figures in parentheses are percentages.
Fig. 2.
Using qMSP, the FancB promoter was shown to be hypermethylated in HN-SCC patients: 14/45 (31.1%) of the tumor patients demonstrated hypermethylation of the FANCB locus compared to 1/16 (6.25%) of the normal patients (χ2; p < 0.05).
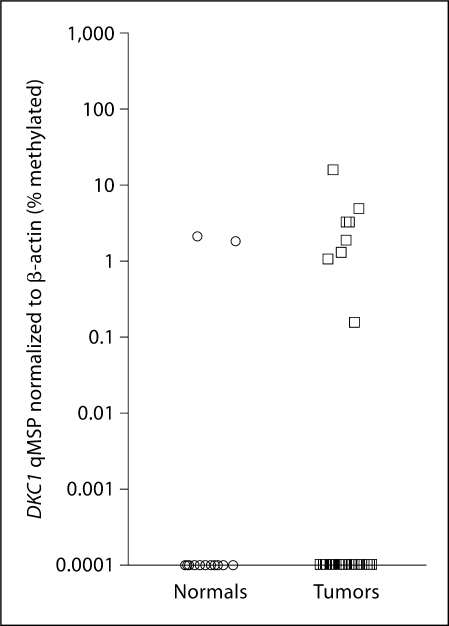
In qMSP from DKC1, 8/45 (17.8%) of the tumor patients and 2/16 (12.5%) of the normal patients showed promoter hypermethylation and inactivation of the DKC1 locus (fig. 3); however, the p value was not significant.
Fig. 3.
The DKC1 promoter demonstrates increased methylation in HNSCC patients on qMSP [8/45 (17.8%) compared to 2/16 (12.5%)of the normal patients].
Discussion
FA and DC are cancer susceptibly syndromes with many well-characterized genetic mutations. The inactivation of these targets in sporadic head and neck cancer has not been well studied. To date, no studies looking at mutational inactivation of all of these targets (except BRCA2) have taken place. Other studies have implicated these FA and DC targets for silencing via loss of heterozygosity, and the most comprehensive high-definition comparative genomic hybridization study conducted on oral squamous cell carcinoma [44] found only 21 targets that were most commonly deleted in head and neck cancer, 2 of which were part of these inherited syndromes (FANCD1 on 13q13.3, 38% of tumors, and FANCD2 on 3p25.3, 33% of tumors – clearly implicating these genes as areas silenced in sporadic head and neck cancers). Epigenetic inactivation has been increasingly studied in cancer pathogenesis for many reasons, which include: heritability, the ability for it to be switched off/on, and its usability in screening studies that take advantage of the stability of DNA in body fluids. We hypothesized that epigenetic inactivation (promoter hypermethylation) was a cause of inactivation of the tumor suppressor genes associated with FA and DC.
To test this hypothesis, we employed bisulfite sequencing of sporadic tumors versus normal controls and qMSP of the genes responsible for FA and DC. We were able to demonstrate promoter methylation specific to tumors in both the DKC1 and FANCB genes. qMSP showed that only FANCB demonstrated gene inactivation by promoter hypermethylation. This target (FANCB), as well as the other targets shown in table 1, was not previously studied in prior reports on epigenetic inactivation of the FANC/BRCA gene family.
This paper is the first comprehensive study of epigenetic silencing of all the tumor suppressors involved in the 2 inherited syndromes of head and neck cancer susceptibility. FANCB shows promoter methylation in tumors compared to normal tissues. This target should prove to be useful in screening studies of the saliva or serum. Further work will include considering other ways in which these genes can be inactivated, including mutation and loss of heterozygosity.
Acknowledgements
J.A.C. is supported by a Clinical Innovator Award from the Flight Attendant Medical Research Institute and the National Institute of Dental and Craniofacial Research (1R01DE015939-01). I.M.S. is supported by an NIH grant (T32DC00027). This work was supported by an NIDCR grant (5R37DE012588).
References
- 1.Kim MM, Califano JA. Molecular pathology of head-and-neck cancer. Int J Cancer. 2004;112:545–553. doi: 10.1002/ijc.20379. [DOI] [PubMed] [Google Scholar]
- 2.Califano J, van der Riet P, Westra W, Nawroz H, Clayman G, Piantadosi S, Corio R, Lee D, Greenberg B, Koch W, Sidransky D. Genetic progression model for head and neck cancer: implications for field cancerization. Cancer Res. 1996;56:2488–2492. [PubMed] [Google Scholar]
- 3.Li FP, Fraumeni JF., Jr Soft-tissue sarcomas, breast cancer, and other neoplasms: a familial syndrome?*. Ann Intern Med. 1969;71:747–752. doi: 10.7326/0003-4819-71-4-747. [DOI] [PubMed] [Google Scholar]
- 4.Gardner EJ. A genetic and clinical study of intestinal polyposis, a predisposing factor for carcinoma of the colon and rectum. Am J Hum Genet. 1951;3:167–176. [PMC free article] [PubMed] [Google Scholar]
- 5.Kennedy AW, Hart WR. Multiple squamous-cell carcinomas in Fanconi's anemia. Cancer. 1982;50:811–814. doi: 10.1002/1097-0142(19820815)50:4<811::aid-cncr2820500432>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 6.Alter BP. Cancer in Fanconi anemia 1927–2001. Cancer. 2003;97:425–440. doi: 10.1002/cncr.11046. [DOI] [PubMed] [Google Scholar]
- 7.Collins N, Kupfer GM. Molecular pathogenesis of Fanconi anemia. Int J Hematol. 2005;82:176–183. doi: 10.1532/IJH97.05108. [DOI] [PubMed] [Google Scholar]
- 8.Bagby GC., Jr Genetic basis of Fanconi anemia. Curr Opin Hematol. 2003;10:68–76. doi: 10.1097/00062752-200301000-00011. [DOI] [PubMed] [Google Scholar]
- 9.Joenje H, Patel KJ. The emerging genetic and molecular basis of Fanconi anaemia. Nat Rev Genet. 2001;2:446–457. doi: 10.1038/35076590. [DOI] [PubMed] [Google Scholar]
- 10.D'Andrea AD. The Fanconi road to cancer. Genes Dev. 2003;17:1933–1936. doi: 10.1101/gad.1128303. [DOI] [PubMed] [Google Scholar]
- 11.Van Waes C. Head and neck squamous cell carcinoma in patients with Fanconi anemia. Arch Otolaryngol Head Neck Surg. 2005;131:640–641. doi: 10.1001/archotol.131.7.640. [DOI] [PubMed] [Google Scholar]
- 12.Rosenberg PS, Greene MH, Alter BP. Cancer incidence in persons with Fanconi anemia. Blood. 2003;101:822–826. doi: 10.1182/blood-2002-05-1498. [DOI] [PubMed] [Google Scholar]
- 13.Alter BP, Joenje H, Oostra AB, Pals G. Fanconi anemia: adult head and neck cancer and hematopoietic mosaicism. Arch Otolaryngol Head Neck Surg. 2005;131:635–639. doi: 10.1001/archotol.131.7.635. [DOI] [PubMed] [Google Scholar]
- 14.Sirinavin C, Trowbridge AA. Dyskeratosis congenita: clinical features and genetic aspects: report of a family and review of the literature. J Med Genet. 1975;12:339–354. doi: 10.1136/jmg.12.4.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dokal I. Dyskeratosis congenita in all its forms. Br J Haematol. 2000;110:768–779. doi: 10.1046/j.1365-2141.2000.02109.x. [DOI] [PubMed] [Google Scholar]
- 16.Walne AJ, Marrone A, Dokal I. Dyskeratosis congenita: a disorder of defective telomere maintenance?*. Int J Hematol. 2005;82:184–189. doi: 10.1532/IJH97.05067. [DOI] [PubMed] [Google Scholar]
- 17.Vulliamy TJ, Marrone A, Knight SW, Walne A, Mason PJ, Dokal I. Mutations in dyskeratosis congenita: their impact on telomere length and the diversity of clinical presentation. Blood. 2006;107:2680–2685. doi: 10.1182/blood-2005-07-2622. [DOI] [PubMed] [Google Scholar]
- 18.Yoon A, Peng G, Brandenburger Y, Zollo O, Xu W, Rego E, Ruggero D. Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science. 2006;312:902–906. doi: 10.1126/science.1123835. [DOI] [PubMed] [Google Scholar]
- 19.Loukinov DI, Pugacheva E, Vatolin S, Pack SD, Moon H, Chernukhin I, Mannan P, Larsson E, Kanduri C, Vostrov AA, Cui H, Niemitz EL, Rasko JE, Docquier FM, Kistler M, Breen JJ, Zhuang Z, Quitschke WW, Renkawitz R, Klenova EM, Feinberg AP, Ohlsson R, Morse HC, 3rd, Lobanenkov VV. BORIS, a novel male germ-line-specific protein associated with epigenetic reprogramming events, shares the same 11-zinc-finger domain with CTCF, the insulator protein involved in reading imprinting marks in the soma. Proc Natl Acad Sci USA. 2002;99:6806–6811. doi: 10.1073/pnas.092123699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wald C, Diner H. Dyskeratosis congenita with associated periodontal disease. Oral Surg Oral Med Oral Pathol. 1974;37:736–744. doi: 10.1016/0030-4220(74)90139-x. [DOI] [PubMed] [Google Scholar]
- 21.Treister N, Lehmann LE, Cherrick I, Guinan EC, Woo SB. Dyskeratosis congenita vs. chronic graft versus host disease: report of a case and a review of the literature. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2004;98:566–571. doi: 10.1016/j.tripleo.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 22.Knight S, Vulliamy T, Copplestone A, Gluckman E, Mason P, Dokal I. Dyskeratosis Congenita (DC) Registry: identification of new features of DC. Br J Haematol. 1998;103:990–996. doi: 10.1046/j.1365-2141.1998.01103.x. [DOI] [PubMed] [Google Scholar]
- 23.Addison M, Rice MS. The association of dyskeratosis congenita and Fanconi's Anaemia. Med J Aust. 1965;18:797–799. doi: 10.5694/j.1326-5377.1965.tb72218.x. [DOI] [PubMed] [Google Scholar]
- 24.Anil S, Beena VT, Raji MA, Remani P, Ankathil R, Vijayakumar T. Oral squamous cell carcinoma in a case of dyskeratosis congenita. Ann Dent. 1994;53:15–18. [PubMed] [Google Scholar]
- 25.Benoit S, Kraemer D, Brocker EB, Goebeler M. Dyskeratosis congenita in a 40-year-old patient (in German) Hautarzt. 2006;57:313–316. doi: 10.1007/s00105-005-0937-2. [DOI] [PubMed] [Google Scholar]
- 26.Cannell H. Dyskeratosis congenita. Br J Oral Surg. 1971;9:8–20. doi: 10.1016/s0007-117x(71)80003-3. [DOI] [PubMed] [Google Scholar]
- 27.Cole HN, Rauschkolb J, Toomey J. Dyskeratosis congenita with pigmentation, dystrophia unguium, and leucokeratosis oris: review of the known cases reported to date and discussion of the disease from various aspects. AMA Arch Derm. 1955;71:451–456. doi: 10.1001/archderm.1955.01540280027005. [DOI] [PubMed] [Google Scholar]
- 28.Garb J. Dyskeratosis congenita with pigmentation, dystrophia unguium, and leukoplakia oris; a follow-up report of two brothers. AMA Arch Derm. 1958;77:704–712. doi: 10.1001/archderm.1958.01560060070012. [DOI] [PubMed] [Google Scholar]
- 29.Hyodo M, Sadamoto A, Hinohira Y, Yumoto E. Tongue cancer as a complication of dyskeratosis congenita in a woman. Am J Otolaryngol. 1999;20:405–407. doi: 10.1016/s0196-0709(99)90082-0. [DOI] [PubMed] [Google Scholar]
- 30.Llistosella E, Moreno A, de Moragas JM. Dyskeratosis congenita with macular cutaneous amyloid deposits. Arch Dermatol. 1984;120:1381–1382. [PubMed] [Google Scholar]
- 31.Moretti S, Spallanzani A, Chiarugi A, Muscarella G, Battini ML. Oral carcinoma in a young man: a case of dyskeratosis congenita. J Eur Acad Dermatol Venereol. 2000;14:123–125. doi: 10.1046/j.1468-3083.2000.00029.x. [DOI] [PubMed] [Google Scholar]
- 32.Dong SM, Sun DI, Benoit NE, Kuzmin I, Lerman MI, Sidransky D. Epigenetic inactivation of RASSF1A in head and neck cancer. Clin Cancer Res. 2003;9:3635–3640. [PubMed] [Google Scholar]
- 33.Sanchez-Cespedes M, Esteller M, Wu L, Nawroz-Danish H, Yoo GH, Koch WM, Jen J, Herman JG, Sidransky D. Gene promoter hypermethylation in tumors and serum of head and neck cancer patients. Cancer Res. 2000;60:892–895. [PubMed] [Google Scholar]
- 34.Reed AL, Califano J, Cairns P, Westra WH, Jones RM, Koch W, Ahrendt S, Eby Y, Sewell D, Nawroz H, Bartek J, Sidransky D. High frequency of p16 (CDKN2/MTS-1/INK4A) inactivation in head and neck squamous cell carcinoma. Cancer Res. 1996;56:3630–3633. [PubMed] [Google Scholar]
- 35.Merlo A, Herman JG, Mao L, Lee DJ, Gabrielson E, Burger PC, Baylin SB, Sidransky D. 5′ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat Med. 1995;1:686–692. doi: 10.1038/nm0795-686. [DOI] [PubMed] [Google Scholar]
- 36.Herman JG, Merlo A, Mao L, Lapidus RG, Issa JP, Davidson NE, Sidransky D, Baylin SB. Inactivation of the CDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res. 1995;55:4525–4530. [PubMed] [Google Scholar]
- 37.Ha PK, Califano JA. Promoter methylation and inactivation of tumour-suppressor genes in oral squamous-cell carcinoma. Lancet Oncol. 2006;7:77–82. doi: 10.1016/S1470-2045(05)70540-4. [DOI] [PubMed] [Google Scholar]
- 38.Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 39.D';Andrea AD. The Fanconi anemia/BRCA signaling pathway: disruption in cisplatin-sensitive ovarian cancers. Cell Cycle. 2003;2:290–292. [PubMed] [Google Scholar]
- 40.Narayan G, Arias-Pulido H, Nandula SV, Basso K, Sugirtharaj DD, Vargas H, Mansukhani M, Villella J, Meyer L, Schneider A, Gissmann L, Durst M, Pothuri B, Murty VV. Promoter hypermethylation of FANCF: disruption of Fanconi anemia-BRCA pathway in cervical cancer. Cancer Res. 2004;64:2994–2997. doi: 10.1158/0008-5472.can-04-0245. [DOI] [PubMed] [Google Scholar]
- 41.Marsit CJ, Liu M, Nelson HH, Posner M, Suzuki M, Kelsey KT. Inactivation of the Fanconi anemia/BRCA pathway in lung and oral cancers: implications for treatment and survival. Oncogene. 2004;23:1000–1004. doi: 10.1038/sj.onc.1207256. [DOI] [PubMed] [Google Scholar]
- 42.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harden SV, Tokumaru Y, Westra WH, Goodman S, Ahrendt SA, Yang SC, Sidransky D. Gene promoter hypermethylation in tumors and lymph nodes of stage I lung cancer patients. Clin Cancer Res. 2003;9:1370–1375. [PubMed] [Google Scholar]
- 44.Sparano A, Quesnelle KM, Kumar MS, Wang Y, Sylvester AJ, Feldman M, Sewell DA, Weinstein GS, Brose MS. Genome-wide profiling of oral squamous cell carcinoma by array-based comparative genomic hybridization. Laryngoscope. 2006;116:735–741. doi: 10.1097/01.mlg.0000205141.54471.7f. [DOI] [PubMed] [Google Scholar]