Abstract
We interrogate the ability of free-space fluorescence tomography to image small animals in vivo using charge-coupled device (CCD) camera measurements over 360-deg noncontact projections. We demonstrate the performance of normalized dual-wavelength measurements that are essential for in-vivo use, as they account for the heterogeneous distribution of photons in tissue. In-vivo imaging is then showcased on mouse lung and brain tumors cross-validated by x-ray microcomputed tomography and histology.
Keywords: fluorescence, tomography, in vivo, molecular imaging, cancer, tumor, fluorescent proteins
Optical tomography of tissues has evolved as a method that three-dimensionally resolves optical contrast in vivo, with applications in small animal research1 and clinical diagnostics.2, 3, 4 The method typically combines theoretical models of photon propagation in diffusive media and boundary measurements of light propagating through tissue utilizing multiple source-detector pairs using a mathematical inversion scheme. Inversion yields the spatial distribution of a number of different possible optical parameters. These parameters may include a combination of the optical absorption coefficient, the reduced scattering coefficient,5 fluorescence concentration,6 or bioluminescence strength.7
Optical detection is particularly important in several small animal imaging applications, especially at areas where several air-tissue interfaces or the utilization of wave-propagation fluids make the application of optical measurements challenging. To increase the tomographic performance over prototype implementations that utilized a relatively small number of source-detector pairs by bringing fiber pairs in contact with tissue, a number of methodologies have been developed to enable imaging using direct charge-coupled device (CCD) camera imaging of free tissue surfaces.8, 9 These techniques moved away from the use of fibers for photon detection, or the use of matching fluids, and were more recently combined in a tomographic scheme for fluorescence imaging, which can collect data over 360-deg projections.9, 10 This approach offers a high quality dataset, as it enables high spatial sampling of photon fields propagating through tissues at any preferred combination of imaging angles. While the performance of this technique has been demonstrated with phantom measurements, there has been no confirmation that this approach can work in vivo. Here, we applied this 360-deg projection approach to imaging lung and brain tumors in murine mouse models.
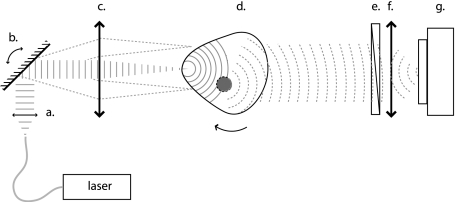
Experimental measurements were performed in a custom-built system (Fig. 1), which is described in detail in Ref. 10. The illumination system and filters have been configured to work both with NIR fluorescent probes and with red-shifted fluorescent proteins. The mouse is illuminated from one side with either a 30-mW laser beam at 750 nm (B&W Tek., Inc. Newark, Delaware), or a 40-mW diode-pumped solid state laser beam (Dream Lasers, Shanghai, China) at 593 nm focused at a 0.5-mm spot on its surface. The transmitted photon fields reaching the opposite side are imaged with the use of a lens (50 mm, f∕1.2, Nikon Corporation, Japan) and a −70 °C cooled CCD camera (Princeton Instruments Incorporated, Trenton, New Jersey). The mouse was imaged in free space, without the use of surrounding matching fluid, and was suspended on a step-motor-controlled rotating stage (Newport Corporation, Irvine California) to acquire images from P=36 different projections. For each projection, the body of the mouse is scanned with a rectangular source pattern created by translating the scanning beam with a set of two galvano-controlled mirrors and a telecentric lens (Nutfield Technology, Incorporated, Windham, New Hampshire). The pattern consisted of S=3×7 sources covering a 4×8.5-mm2 area centered along the center of rotation to reduce the stray light coming from the sides of the animal.
Figure 1.
Schematic of the experimental setup: (a) collimating lens, (b) scanning mirrors, (c) telecentric lens, (d) object, (e) filter, (f) lens, and (g) CCD camera.
Two images were acquired at each projection and each source position employed; one image captured at the emission wavelength (fluorescence image) and one image at the excitation wavelength (normalization image). Excitation and emission data were acquired using 745- to 755-nm and 780- to 820-nm bandpass interference filters, respectively (Andover Corporation Salem, New Hampshire) when imaging probes using the Alexa750 fluorescent dye. Correspondingly, when imaging the mCherry fluorescent protein, the fluorescence images were acquired using a 625- to 635-nm bandpass filter, and the normalization images were acquired using a second diode laser at 650 nm and a 645- to 655-nm bandpass filter, since the excitation wavelength (593 nm) is highly absorbed by the tissue yielding low signal-to-noise ratio. The mouse surface was extracted by capturing the silhouette of the mouse at 72 projections (5-deg step) against a white background. Surface reconstruction was then based on a volume carving algorithm11 developed for this setup.
Each acquired image is sampled by D=11×12 virtual detectors spanning an area 10×11 mm2 by binning 4×4 pixels. To normalize for the inhomogeneity of the tissue absorption and illumination,6 the ratio of the fluorescence over the excitation is calculated and used in the inversion code, i.e.:
| (1) |
where (s,d,p) is the source, detector, and projection indices. For imaging purposes, the mouse body is descretized in a rectangular mesh of M=24×14×22 nodes, and the normalized intensity for each source-detector pair is related to the unknown fluorescence value in each of the nodes via the normalized Born approximation.12 The unknown fluorescence values are then calculated by inverting the resulting weight matrix with an algebraic reconstruction technique (ART),12 accessing randomly the sources and the detectors and sequentially the different projections. A 5% low threshold is applied to both the Ufl and Uex data to exclude signals of low value from the reconstruction. Computation time was approximately 10 min for 50 ART iterations on an Intel Core 2 Duo processor with 4 GB of memory.
Two mouse tumor models were used. In the first one, two nude mice were implanted with 1×106 Lewis lung carcinoma (LLC) cells administered intrercostally into the right lung parenchyma. The tumor had grown for 10 days and the mice were injected via tail vein with 2 nM of AngioSence750 (Visen Medical, Woburn Massachusetts), a fluorescent probe that remains in areas of high vasculature to target angiogenesis in tumors. Mice were imaged by FMT under general anesthesia two days after probe injection for ensuring sufficient clearance from the circulation, allowing presence of a probe mostly at the tumor interstitium. Following FMT imaging, mice received x-ray micro-CT imaging (Gamma Medica, Northridge, California) to anatomically locate the tumor. In the second model, five mice were stereotacticaly implanted in the brain with 105 glioma cells expressing mCherry fluorescent protein under the cytomegalovirus promoter, and were imaged in FMT after 7 days. The mice were then euthanized and dissected for correlative histology. The complete FMT measurement takes 20 to 30 mins depending on the camera exposure time.
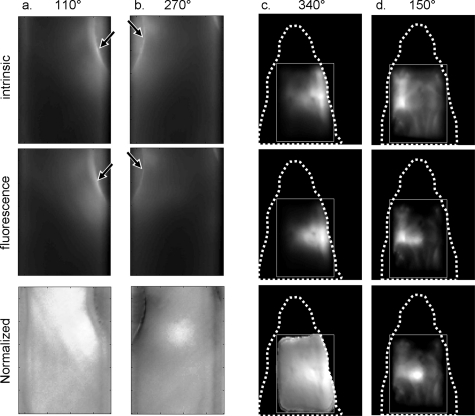
In-vivo imaging requires that fluorescence signals are normalized for the highly heterogeneous attenuation of tissue. This is particularly evident in 360-deg-projection free-space geometries, where a large dynamic range also exists between intensities acquired close to the boundary of the animal versus measurements acquired through the center of the animal’s body. Figure 2 depicts the effects of optical heterogeneity on raw signals and the effects of the subsequent data normalization afforded by Eq. 1 for two representative experimental sets. The first row in Fig. 2 depicts transillumination images from the torso and head of the mice, captured at different projections, and calculated as the sum of the excitation images , i.e., , where S is the number of point sources scanned at the back of the animal at each projection. In the second row of Fig. 2, the corresponding fluorescence images , i.e., are depicted. As shown, the areas of the highest light intensity are at the side edges of the mouse, due to: 1. the shorter propagation distance that light has to travel when penetrating through the mouse side, and 2. the lower overall absorption of the skin, compared to other more vascular structures. However, calculating the sum of the ratios of fluorescence images over the excitation for each source, i.e., , which is shown in the third row of Fig. 2, accounts for the tissue and illumination heterogeneity, and enhances the contrast of true fluorescence biodistribution, in this case fluorescence coming from the tumor. At the 110- and 340-deg projections, the tumors are closer to the sources, so their normalized images appear more diffusive, since fluorescence light has to travel for a longer distance. Conversely, at the 270- and 150-deg projections, the tumors are closer to the detector and are more accurately localized on these raw images.
Figure 2.
Raw transillumination images of mouse tumors at the intrinsic (first raw) and the fluorescence wavelengths (second row), and the normalized fluorescence image (third raw). (a) and (b) Lung tumor at different projections (110 and at 270 deg). (c) and (d) Brain tumor at different projections (340 and 150 deg). Arrows show intensity “hot spots” due to light waveguiding at the edges of the body. Dotted lines describe the outline of the mouse head, and the rectangle is the region of interest.
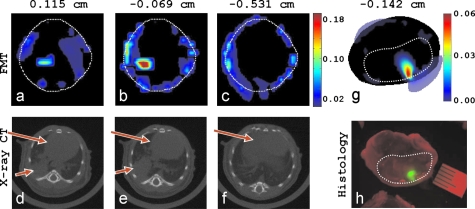
The fluorescence from both tumor models was positively identified with noncontact FMT in all imaging experiments, and it was congruent with the tumor location. Correspondingly, typical results of the tomographic reconstructions using the normalized data at multiple projections are shown in Fig. 3. Three different FMT axial slices of the lung tumor model [Figs. 3a, 3b, 3c] show that a volume of high fluorochrome concentration is reconstructed in the right lung (anterior direction of view), which is congruent with the tumor location seen on the x-ray CT images [Figs. 3d, 3e, 3f]. The tumor volume estimated from FMT and CT is 38 and 40 mm3, respectively. The average thorax movement in anesthetized animals was recently measured in this setup13 and was found to be of the order of 100 to 200 μm. With motion averaging due to high CCD exposure times relative to the animals breathing cycle, thorax movement has negligible effects in the corresponding FMT images and has not been found to affect imaging performance in the current implementation. In Fig. 3g an FMT axial slice of the brain is presented, the high FMT signal at the left hemisphere of the brain that corresponds to the location of the mCherry brain tumor in the histological head section in Fig. 3h. The tumor volumes measured from FMT and histology are 16 and 13 mm3, respectively. There are a few artifacts reconstructed in the periphery of the FMT images that are typical in this type of reconstruction due to boundary theoretical discrepancies. Comparatively, imaging in the near-infrared (i.e., using fluorescent probes) yields a higher number of photons and better signal-to-noise ratios. Contrary to the fluorescent probes, the fluorescent proteins produce lower signals because tissue is more absorbing. However, they exhibit unparalled specificity, since mCherry fluorescence is emerging only from the tumor, and consequently image quality is comparable to those acquired with near-IR probes.
Figure 3.
Tomographic reconstructions of lung and brain tumor in mice (as viewed toward the anterior direction). (a), (b), and (c) FMT axial slices of the Angioscence 750 concentration (arb. units) at three different y positions. The white dotted line is the mouse boundary. (e) and (f) The corresponding x-ray CT axial slices; small arrows are the tumor, and big arrows are the liver and heart. (g) FMT of the mCherry brain tumor, and (f) histology fluorescence image of the corresponding axial head section; dotted line is the brain boundary.
Overall, in-vivo tomographic imaging in free-space 360-deg-projection geometry has been shown to be feasible in this study. We have further showcased the necessity for fluorescence dataset normalization as means to correct for dynamic range variation in the resulting illumination through the tissue and tissue absorption inhomogeneity. The advantages of this particular geometrical implementation are two-fold: it is a more natural and less stressful procedure for the mouse (no matching fluids, no compression, ability to image the head), and it enables the high spatial frequency acquisition of data over multiple projections. Although the total acquisition time has increased compared to the planar matching-fluid acquisition method due to the larger dataset, the spatial resolution and accuracy in the tomographic reconstructions have improved dramatically. The benefits of this implementation on tomographic quality were recently demonstrated over other more conventional tomographic approaches that use fibers or limited-angle projections, using singular value decomposition analysis.14
Acknowledgment
The authors would like to thank Tobias Lasser and Damon Hyde for helpful discussions. This research was supported by a Marie Curie Intra-European Fellowship within the 7th European Community Framework Programme, and by National Institutes of Health grants RO1 EB004382-01, RO1 EB000750-01, and R43-ES012360.
References
- Ntziachristos V., Ripoll J., Wang L. H. V., and Weissleder R., “Looking and listening to light: the evolution of whole-body photonic imaging,” Nat. Biotechnol. 23(3), 313–320 (2005). 10.1038/nbt1074 [DOI] [PubMed] [Google Scholar]
- Brooksby B., Pogue B. W., Jiang S. D., Dehghani H., Srinivasan S., Kogel C., Tosteson T. D., Weaver J., Poplack S. P., and Paulsen K. D., “Imaging breast adipose and fibroglandular tissue molecular signatures by using hybrid MRI-guided near-infrared spectral tomography,” Proc. Natl. Acad. Sci. U.S.A. 103(23), 8828–8833 (2006). 10.1073/pnas.0509636103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corlu A., Choe R., Durduran T., Rosen M. A., Schweiger M., Arridge S. R., Schnall M. D., and Yodh A. G., “Three-dimensional in vivo fluorescence diffuse optical tomography of breast cancer in humans,” Opt. Express 15(11), 6696–6716 (2007). 10.1364/OE.15.006696 [DOI] [PubMed] [Google Scholar]
- Godavarty A., Thompson A. B., Roy R., Gurfinkel M., Eppstein M. J., Zhang C., and Sevick-Muraca E. M., “Diagnostic imaging of breast cancer using fluorescence-enhanced optical tomography: phantom studies,” J. Biomed. Opt. 9(3), 488–496 (2004). 10.1117/1.1691027 [DOI] [PubMed] [Google Scholar]
- Xu Y., Iftimia N., Jiang H. B., Key L. L., and Bolster M. B., “Three-dimensional diffuse optical tomography of bones and joints,” J. Biomed. Opt. 7(1), 88–92 (2002). 10.1117/1.1427336 [DOI] [PubMed] [Google Scholar]
- Ntziachristos V. and Weissleder R., “Experimental three-dimensional fluorescence reconstruction of diffuse media by use of a normalized Born approximation,” Opt. Lett. 26(12), 893–895 (2001). 10.1364/OL.26.000893 [DOI] [PubMed] [Google Scholar]
- Cong W. X., Wang G., Kumar D., Liu Y., Jiang M., Wang L. V., Hoffman E. A., McLennan G., McCray P. B., Zabner J., and Cong A., “Practical reconstruction method for bioluminescence tomography,” Opt. Express 13(18), 6756–6771 (2005). 10.1364/OPEX.13.006756 [DOI] [PubMed] [Google Scholar]
- Schulz R. B., Ripoll J., and Ntziachristos V., “Experimental fluorescence tomography of tissues with noncontact measurements,” IEEE Trans. Med. Imaging 23(4), 492–500 (2004). 10.1109/TMI.2004.825633 [DOI] [PubMed] [Google Scholar]
- Meyer H., Garofaiakis A., Zacharakis G., Psycharakis S., Mamalaki C., Kioussis D., Economou E. N., Ntziachristos V., and Ripoll J., “Noncontact optical imaging in mice with full angular coverage and automatic surface extraction,” Appl. Opt. 46(17), 3617–3627 (2007). 10.1364/AO.46.003617 [DOI] [PubMed] [Google Scholar]
- Deliolanis N. C., Lasser T., Hyde D., Soubret A., Ripoll J., and Ntziachristos V., “Free-space fluorescence molecular tomography utilizing 360° geometry projections,” Opt. Lett. 32(4), 382–384 (2007). 10.1364/OL.32.000382 [DOI] [PubMed] [Google Scholar]
- Potmesil M., “Generating octree models of 3d objects from their silhouettes in a sequence of images,” Comput. Vis. Graph. Image Process. 40(1), 1–29 (1987). 10.1016/0734-189X(87)90053-3 [DOI] [Google Scholar]
- Kak A. C. and Slaney M., Principles of Computerized Tomographic Imaging, IEEE Press, New York, 1999. [Google Scholar]
- Lasser T., Soubret A., Ripoll J., and Ntziachristos V., “Surface reconstruction for free-space 360 degrees fluorescence molecular tomography and the effects of animal motion,” IEEE Trans. Med. Imaging 27(2), 188–194 (2008). 10.1109/TMI.2007.904662 [DOI] [PubMed] [Google Scholar]
- Lasser T. and Ntziachristos V., “Optimization of 360 degrees projection fluorescence molecular tomography,” Med. Image Anal. 11(4), 389–399 (2007). 10.1016/j.media.2007.04.003 [DOI] [PubMed] [Google Scholar]