Abstract
We report, for the first time, on the preparation, characterization and in vitro testing of poly (D,L-lactide-co-glycolide) (PLGA) microparticles loaded with polyamidoamine (PAMAM)-plasmid DNA (pDNA) dendriplexes. Loading of pDNA into the PLGA microparticles increased by 150% when pDNA was first complexed with PAMAM dendrimers relative to loading of pDNA alone. Scanning electron microscopy (SEM) showed that the presence of PAMAM dendrimers in the PLGA microparticles created porous features and indentations on the surface of the microparticles. Loading PLGA microparticles with PAMAM-pDNA dendriplexes lowered the average PLGA microparticle size and changed the surface charge of the microparticles from negative to positive when compared to PLGA microparticles loaded with pDNA alone. The zetapotential and buffering capacity of the microparticles increased as the generation of the PAMAM dendrimer loaded in the PLGA microparticles increased. Gel electrophoresis assays showed that all the PLGA microparticle formulations were able to entrap the pDNA within the PLGA matrix. There was no significant difference in the cytotoxicity of PLGA microparticles loaded with PAMAM-pDNA dendriplexes when compared to PLGA microparticles loaded with pDNA alone. Furthermore, and in contrast to PAMAM dendrimers alone, the generation of the PAMAM dendrimer loaded in the PLGA microparticles had no significant impact on cytotoxicity or transfection efficiencies in human embryonic kidney (HEK293) or Monkey African green kidney fibroblast-like (COS7) cells. The transfection efficiency of PLGA microparticles loaded with generation 3 (G3) PAMAM-pDNA dendriplexes was significantly higher than PLGA microparticles loaded with pDNA alone in HEK293 and COS7 cells. PLGA microparticles loaded with G3 PAMAM-pDNA dendriplexes generated equivalent transfection efficiencies as (G3 to G6) PAMAM-pDNA dendriplexes alone in COS7 cells when the transfection was carried out in serum containing media. The delivery system developed in this report has low toxicity, high pDNA loading efficiencies and high transfection efficiencies that are not reduced in the presence of serum. A delivery system with these characteristics is expected to have significant potential for translational applications.
Keywords: Polyamidoamine (PAMAM); poly (D,L-lactide-co-glycolide) (PLGA); Non-viral Gene Delivery; Vaccine; Plasmid DNA; Formulation; Dendriplex; Microparticle
1. Introduction
Biodegradable microparticles can be used to provide sustained delivery of plasmid DNA (pDNA) and have shown significant potential in vaccination applications1–3. A common approach to loading pDNA into biodegradable microparticles is to use the double emulsion solvent evaporation method4–6. A challenge with preparing pDNA loaded biodegradable microparticles using the double emulsion solvent evaporation method is that the manufacturing process can cause damage to the pDNA thereby denaturing or inactivating it1,5. This occurs because the process of entrapping the pDNA into the microparticles exposes the pDNA to organic solvents, high shear forces and high interfacial tensions at the oil/water interface1,5. These processes can nick the DNA or lead to breakage in the strands of DNA. In addition, the hydrophilic characteristics of pDNA can lead to poor loading levels in hydrophobic microparticles1,5. Plasmid DNA could alternatively be bound to the surface of cationic microparticles thereby avoiding exposing the pDNA to processes that could degrade the pDNA7–11. However, this approach does not allow for pDNA to be released over a sustained period of time. Other approaches to protecting plasmid DNA include entrapping the pDNA in blends of biodegradable polymers and cationic polymers4,12,13.
We have previously shown that the most efficient transfection efficiencies generated by biodegradable polymeric microparticles are achieved by first complexing the pDNA with a cationic polymer prior to entrapping the pDNA into the microparticles4. Examples of cationic polymers and excipients that have been used for complexation with pDNA prior to entrapment in microparticles include polyethylenimine (PEI), polylysine (PL) and chitosan4,14–16.
A promising class of cationic polymers that efficiently complex with pDNA is the polyamidoamine (PAMAM) dendrimers (figure 1)17–20. PAMAM dendrimers have a number of advantages over linear cationic polymers such as PEI20,21. In comparison to linear cationic polymers such as PEI, PAMAM dendrimers are spherical, monodisperse polymers with a reduced structural density in the intramolecular core20.
Figure 1.
Structure of poly (lactic-co-glycolic acid) (PLGA) and PAMAM dendrimers with ethylenediamine core.
In this study, we show for the first time, that loading PAMAM-pDNA dendriplexes into biodegradable microparticles results in significantly higher transfection efficiencies than loading pDNA alone. We also show that loading PAMAM-pDNA dendriplexes into PLGA microparticles significantly lowers the toxicity associated with the PAMAM dendrimers. Finally, we show that loading PAMAM-pDNA dendriplexes into biodegradable microparticles produces a delivery system that can generate transgene expression levels that are as high as the PAMAM-pDNA dendriplexes alone when the transfections are carried out in serum-containing media. PLGA microparticles loaded with PAMAM-pDNA dendriplexes have low toxicity and high transfection efficiencies that are not reduced in the presence of serum and is therefore expected to have significant potential for translational applications in vitro and in vivo.
2. Materials and Methods
2.1 Materials
D, L-Lactide/Glycolide copolymer (PLGA, molar ratio: 50:50, inherent viscosity 0.54dL/g) was purchased from Absorbable Polymers International (Pelham, AL USA). PAMAM dendrimers from generation 3 to 6 (G3, G4, G5 and G6, ethylenediamine core) and poly (vinyl alcohol) (PVA, MW 30–70kDa) were purchased from Sigma-Aldrich (St. Louis, MO). The Bicinchoninic acid (BCA) protein assay kit was purchased from Pierce Biotechnology Inc. (Rockford, IL). Dulbecco’s Modified Eagle’s Medium (DMEM) was obtained from Gibco BRL (Grand Island, NY). The luciferase assay system was purchased from Promega (Madison, WI). For cellular uptake studies, FITC (Fluorescein Isothiocyanate, Sigma-Aldrich) labeled dendrimers G3-G6 were prepared by reaction of FITC and PAMAM dendrimers in 0.1M sodium carbonate buffer (pH 9) at 4°C in darkness overnight. FITC-conjugated dendrimers were purified by dialysis (MWCO 7000, Pierce Biotechnology Inc., Rockford, IL) and then lyophilized (Labconco FreeZone 4.5, Kansas City, Missouri). The level of fluorescein in the dendrimers was determined by measuring their absorbance at 498nm using standard curves of FITC. Spectrofluorometric analysis (Spectramax M5 Microplate reader, Molecular Device) found that approximately 3% of the dendrimer amino groups were attached with FITC.
2.2 Cell Culture
Human embryonic kidney cells (HEK293) and Monkey African green kidney fibroblast-like cell line (COS7) were obtained from the American Type Culture Collection (ATCC, Rockville, MD). The cells were maintained in DMEM supplemented with 10% FBS, streptomycin at 100μg/ml, penicillin at 100U/ml, and 4mM L-glutamine at 37°C in a humidified 5% CO2-containing atmosphere.
2.3 Amplification and Purification of Plasmid DNA
VR1255 plasmid is a 6.4-kb cDNA encoding firefly luciferase driven by the cytomegalovirus (CMV) promoter/enhancer. The plasmid was transformed in Escherichia coli DH5α and amplified in Terrific Broth media at 37°C overnight with a shaking speed of 300rpm. The plasmid was purified by an endotoxin-free QIAGEN Giga plasmid purification kit (QIAGEN, Valencia, CA) according to the manufacturer’s protocol. Purified pDNA was dissolved in saline, and its purity and concentration were determined by UV absorbance at 260 and 280nm.
2.4 Preparation of pDNA loaded PLGA Microparticles and PAMAM-pDNA dendriplex loaded PLGA Microparticles
2.4.1 PLGA microparticles loaded with pDNA
PLGA microparticles were prepared using a w/o/w double emulsion, solvent evaporation technique. Briefly, 200 mg of 50:50 PLGA was dissolved in 5ml of dichloromethane (DCM). VR1255 pDNA in 0.5% (w/v) PVA solution was prepared at a concentration of 4mg/ml. Using a microtip probe sonicator set at level 2 (Sonic Dismembrator Model 100, Fisher Scientific, Pittsburgh, PA), 500μl of the PVA solution containing 2mg of VR1255 pDNA was mixed with the PLGA/DCM solution for 20 seconds to form the first emulsion. This emulsion was then rapidly added to 50ml of 0.5% (w/v) PVA solution with stirring at 13,500 rpm for 30 seconds using an IKA Ultra-Turrax T25 basic homogenizer (IKA, Wilmington, NC). The second emulsion was stirred overnight during which time the DCM solvent was evaporated. The supernatant was collected and analyzed spectrophotometrically at 260nm using a SpectraMax Plus384 Microplate Spectrophotometer (Molecular device) for pDNA content. The microparticles were then washed 3 times with deionized water and then lyophilized (Labconco FreeZone 4.5, Kansas City, Missouri). Plasmid DNA entrapped in the PLGA microparticles was calculated by subtracting the pDNA content in the supernatant from the initial concentration of pDNA added. Microparticles were stored at −20°C until use. Empty microparticles in acetonitrile were spiked with a known amount of pDNA. Following the procedure, recovery of the extracted pDNA was found to be complete. Microparticles containing pDNA were also dissolved in acetonitrile and the pDNA was extracted in TE buffer (10 mM, pH 8.3). The amount of pDNA was determined based on absorbance at 260 nm. For in vitro microparticle uptake studies, PLGA microparticles loaded with Rhodamine 123 were prepared using a single emulsion evaporation methodology. Briefly, 200 mg of 50:50 PLGA and 2 mg Rhodamine 123 (Sigma) was dissolved in 5mL DCM. This was then rapidly added to 50mL of 0.5% (w/v) PVA in deionized water with stirring at 13500 rpm for 30s. After evaporation overnight, the rhodamine labeled microparticles were washed and collected as described above.
2.4.2 PLGA Microparticles loaded with PAMAM-pDNA dendriplexes
PAMAM dendrimer (G3-G6)-pDNA dendriplexes at N/P ratio of 5 were prepared by mixing 150 μg of VR1255 with 97.5μg of PAMAM dendrimer. The mixture was vortexed for 20 s, and incubated for 30 min at room temperature. Then 500 μl of PAMAM-pDNA complexes solution was mixed with 5 ml of DCM containing 200 mg of PLGA 50:50 using the microtip probe sonicator set at level 2 for 30 seconds to form the first emulsion. This emulsion was then rapidly added to 50ml of 0.5% (w/v) PVA solution and homogenized at 13,500 rpm for 30 seconds. The mixture was stirred overnight during which time the DCM solvent evaporated. The resulting particles, which were thereafter named as PG3, PG4, PG5, PG6 were then washed 4 times with deionized water and lyophilized. Particles were stored at −20°C until use. Microparticles containing dendriplexes were dissolved in acetonitrile and the dendriplexes were extracted in TE buffer (10 mM, pH 8.3). Loading of dendriplexes in the PLGA microparticles was analyzed spectrophotometrically at 480nm/520nm (Ex/Em) using the PicoGreen Reagent and a SpectraMax Plus384 Microplate Spectrophotometer (Molecular device) after it was found that complexing the pDNA with PAMAM dendrimers interfered with absorbance readings at 260 nm. The level of dendriplexes loaded into the microparticles was then determined using a linear calibration curve prepared using known concentrations of PAMAM-pDNA dendriplexes and the PicoGreen Reagent.
2.5 Microparticle Size and Surface Morphology
Microparticle size and zetapotential measurements were conducted using the Zetasizer Nano ZS (Malvern, Southborough, MA). Briefly, the microparticles were suspended in deionized water at a concentration of 1mg/ml. The size measurements were performed at 25°C at a 173° scattering angle. The mean hydrodynamic diameter was determined by cumulative analysis. The zetapotential determinations were based on electrophoretic mobility of the microparticles in the aqueous medium, which were performed using folded capillary cells in automatic mode. Microparticle morphology was assessed by scanning electron microscopy (SEM, Hitachi S-4000). Briefly, air-dried microparticles were placed on adhesive carbon tabs mounted on SEM specimen stubs. The specimen stubs were coated with approximately 5nm of gold by ion beam evaporation before examination in the SEM operated at 5kV accelerating voltage.
2.6 In vitro release of PAMAM-pDNA dendriplexes
PLGA microparticles (200 mg) loaded with PAMAM-pDNA dendriplexes were suspended in phosphate buffered saline (PBS, 10 ml, 50 mM, pH 7.4) for 2 months. The suspension was gently shaken in a water bath at 37°C. At various time intervals, the supernatant was removed after centrifugation and replaced with fresh medium. PAMAM-pDNA dendriplex containing microparticles were incubated in TE buffer and the released fraction were estimated spectrophotometrically at 480nm/520nm (Ex/Em) using the PicoGreen Reagent and a SpectraMax Plus384 Microplate Spectrophotometer (Molecular device).
2.7 Buffering Capacity of pDNA loaded PLGA Microparticles and PAMAM-pDNA dendriplex loaded PLGA Microparticles
The ability of PLGA microparticles loaded with PAMAM-pDNA dendriplexes (PG3 to PG6) to resist acidification was tested using the acid titration assay as described by Tang et al 22. Briefly, 10 mg/ml PG3, PG4, PG5 or PG6 microparticles were suspended in 150mM NaCl, respectively. The pH was first adjusted to ~9.0 and then titrated in small increments with 0.1 N HCl until a pH of 3.0 was reached. Multiple pH readings were taken to ensure that equilibrium had been reached. The slope of the pH versus HCl added graph provides an indication of the intrinsic buffering capacity of the delivery vehicles.
2.8 Cytotoxicity Evaluation using the MTT assay
The cytotoxicity of the pDNA loaded PLGA microparticles and the PAMAM-pDNA dendriplex loaded PLGA microparticles was evaluated using the MTT (3-[4,5-dimethylthiazol-2-yl]-2, 5-diphenyl tetrazolium bromide) assay.23 COS7 and HEK293 cells were seeded in a 96-well plate at a density of 10,000 cells/well. Twenty-four hours later, cells were incubated with 200 μl of complete DMEM containing PLGA microparticles loaded with pDNA or PLGA microparticles loaded with PAMAM-pDNA dendriplexes at various concentrations. After 4h of incubation, the medium in each well was replaced with 100 μl of fresh complete medium. 25μl of 5mg/ml MTT solution in PBS was added to each well and incubated with cells for additional 2 h. Cells were lysed with 100 μl of the extraction buffer (20% SDS in 50% DMF, pH4.7) overnight. The optical density of the lysate was measured at 550 nm using a Spectramax plus384 Microplate Spectrophotometer (Molecular Device). Values were expressed as a percentage of the control to which no microparticles were added.
2.9 Cellular uptake of pDNA loaded PLGA Microparticles, PAMAM-pDNA dendriplexes and PAMAM-pDNA dendriplex loaded PLGA Microparticles
PAMAM-pDNA dendriplexes (G3, G4, G5, and G6) and PAMAM-pDNA dendriplex loaded rhodamine labeled PLGA Microparticles (PG3, PG4, PG5 and PG6) were prepared using FITC-labeled dendrimers. HEK293 cells were seeded into 12-well plate at a density of 1×106/well 24h before transfection. After 24h incubation, the medium was replaced with fresh DMEM containing 10% FBS. 0.5mg/well PLGA/Rhodamine 123, FITC labeled G3, G4, G5, G6, PG3, PG4, PG5 or PG6 microparticles were incubated with HEK293 cells for 16 h. Then the cells were washed with PBS three times to remove free fluorescent labeled particles and the particles adsorbed on the cells surface. Samples were then assessed using flow cytometry (Becton Dickinson). Dot plots were gated on FSC/SSC properties of HEK293 cells to exclude free fluorescent labeled particles. Data was analyzed using Cell-QuestPro software. All samples were tested in triplicate.
2.10 Evaluation of Luciferase Expression in COS7 and HEK293 cells
COS7 and HEK293 cells were seeded into 24-well plates at a density of 8×104/well 24 h before transfection. 0.2 mg/well PLGA microparticles loaded with pDNA and PLGA microparticles loaded with PAMAM-pDNA dendriplexes were added to the cells in transfection medium (serum-free) and incubated for 4 h at 37 °C, followed by further incubation in serum containing medium for 44 h. Transfection experiments were also carried out in serum (10%) containing medium in COS7 cells with additional PAMAM-pDNA dendriplexes alone included as control groups. The PAMAM-pDNA dendriplexes were prepared and tested using PAMAM dendrimers of generation 3, 4, 5 and 6 (denoted as G3/DNA, G4/DNA, G5/DNA and G6/DNA respectively). The concentration of the microparticles was selected based on a target pDNA loading of 5μg/mg microparticles and an equivalent pDNA dose of 1 μg/well. After the incubation, cells were treated with 200 μl of lysis buffer (Promega). The lysate was subjected to two cycles of freezing and thawing, then transferred into tubes and centrifuged at 13200 rpm for 5 min. Twenty microliters of supernatant was added to 100 μl of luciferase assay reagent (Promega) and samples were measured on a luminometer for 10 s (Lumat LB 9507, EG&G Berthold, Bad Wildbad, Germany). The relative light units (RLU) were normalized against protein concentration in the cell extracts, measured by a BCA protein assay kit (Pierce). Luciferase activity was expressed as relative light units (RLU/mg protein in the cell lysate). The data were reported as mean ± standard deviation for triplicate samples. Every transfection experiment was repeated at least twice.
2.11 Statistical Analysis
Group data were reported as mean+/−SD. Differences between groups were analyzed by one way analysis of variance with a Tukey post-test analysis. Levels of significance were accepted at the P<0.05 level. Statistical analyses were performed using Prism 5 software (Graphpad Software, Inc., San Diego, CA.)
3. Results
3.1 Formation of PAMAM-pDNA dendriplexes and loading into PLGA Microparticles using Double Emulsion Solvent Evaporation
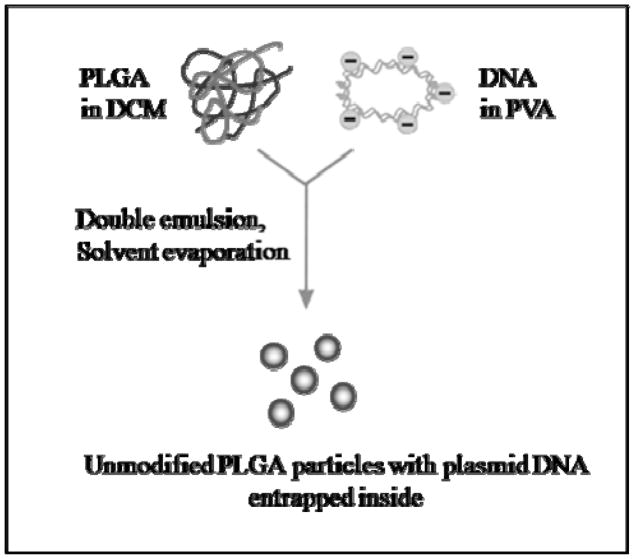
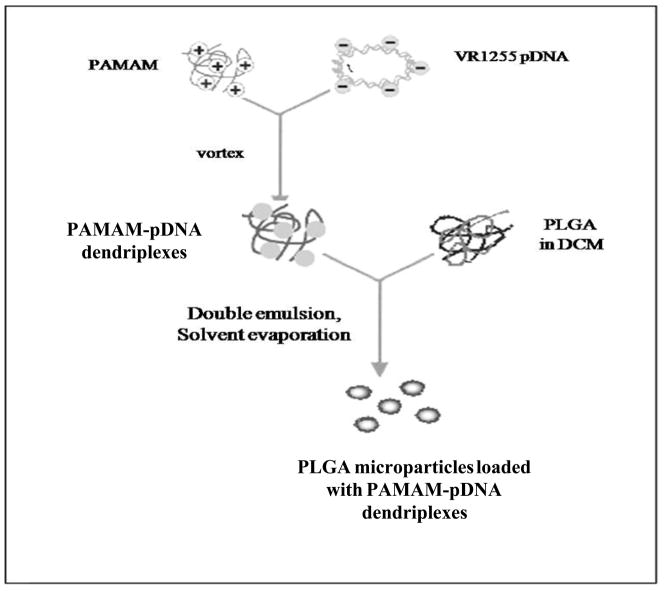
Figure 2 schematically shows our approach for preparing PLGA particles loaded with pDNA and figure 3 schematically shows our approach for preparing PLGA particles loaded with PAMAM-pDNA dendriplexes. PLGA particles loaded with pDNA were prepared using a w/o/w double emulsion solvent evaporation technique. 0.5% (w/v) PVA solution was selected to prepare microparticles based on a previous optimization study on microparticle fabrication. To load dendriplexes into the PLGA microparticles, PAMAM dendrimers and pDNA were complexed at an N:P ratio of 5 and then entrapped into the PLGA microparticles using the same w/o/w double emulsion solvent evaporation technique as the loading of pDNA alone.
Figure 2.
Schematic of preparation of PLGA microparticles loaded with plasmid DNA alone.
Figure 3.
Schematic of preparation of PLGA microparticles loaded with PAMAM-pDNA dendriplexes.
3.2 Microparticles Size, Zetapotential and Morphology of Microparticles
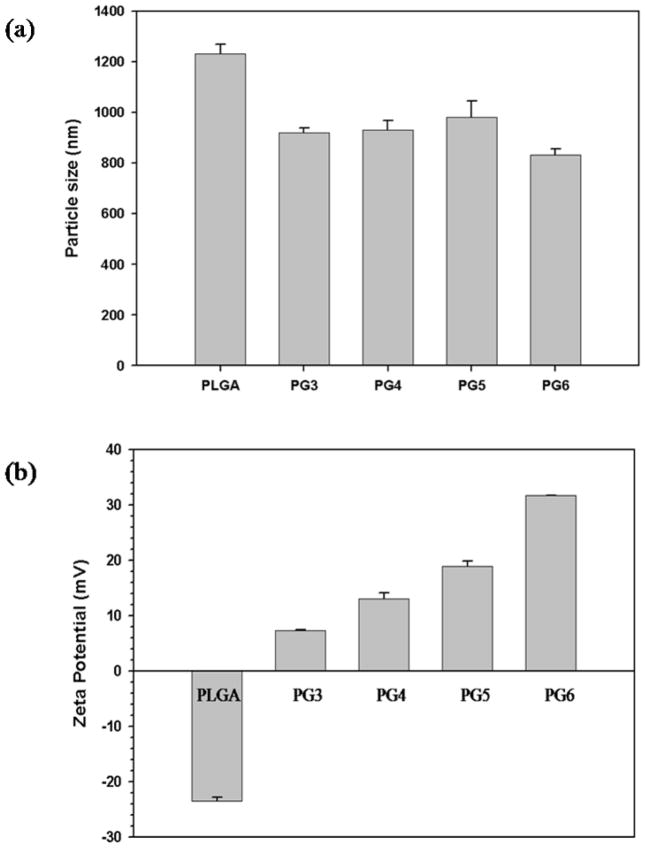
Figure 4a shows the microparticle size of the PLGA microparticles loaded with pDNA alone and PLGA microparticles loaded with PAMAM-pDNA dendriplexes prepared with varying generation levels of PAMAM dendrimers. PLGA microparticles loaded with pDNA alone had an average size of 1.2 μm. PLGA microparticles loaded with PAMAM-pDNA dendriplexes had an average size, ranging from 800 to 1000 nm, that was significantly lower than PLGA microparticles loaded with pDNA alone (P<0.001). The generation of the PAMAM dendrimer used did not have a significant impact on the average size of the PLGA microparticles loaded with PAMAM-pDNA dendriplexes (P>0.05).
Figure 4.
Particle size and zeta potential of PLGA microparticles loaded with PAMAM-pDNA dendriplexes (PG3 – PG6). Particle size (a) and zeta potential (b) of PLGA microparticles loaded with PAMAM-pDNA dendriplexes. Data is represented as the mean ± standard deviation (n=3).
Figure 4b shows the zetapotential of PLGA microparticles loaded with pDNA alone or with PAMAM-pDNA dendriplexes prepared with varying generations of PAMAM dendrimers. PLGA microparticles loaded with pDNA alone showed a net negative zetapotential of −28 mV. In contrast, the zetapotential of all PLGA particles loaded with PAMAM-pDNA dendriplexes (PG3 to PG6) was positive and the value of zetapotential increased as the PAMAM dendrimer generation used for complexing with pDNA increased (Figure 4b).
The surface morphology of PLGA microparticles loaded with pDNA alone and PLGA microparticles loaded with PAMAM-pDNA dendriplexes (PG3 to PG6) was examined by scanning electron microscopy (SEM). Particle morphology was influenced by the presence of PAMAM-pDNA dendriplexes in the inner aqueous phase. PLGA microparticles containing pDNA alone were round spheres with a smooth surface. In contrast, many of the PLGA microparticles loaded with PAMAM-pDNA had indentations and porous features on the surface (Figure 5).
Figure 5.
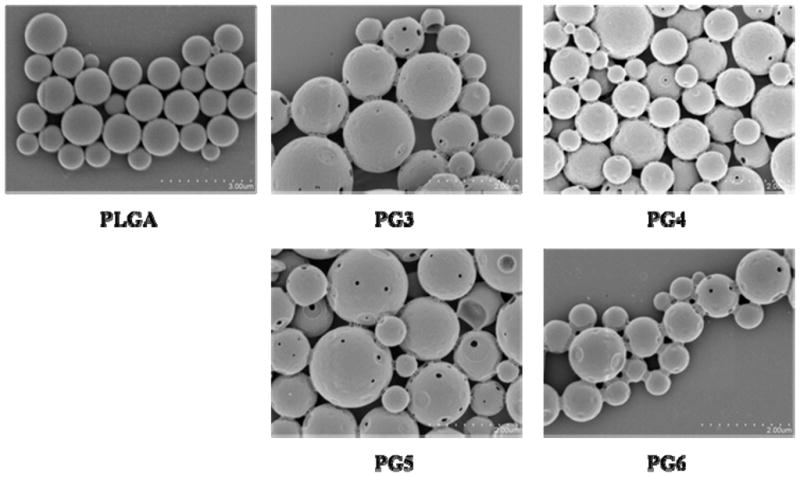
SEM images show the morphology of all the PLGA microparticles loaded with PAMAM-pDNA dendriplexes is smooth and spherical in appearance with porous features intermittently spread throughout the surface.
3.3 Release of PAMAM-pDNA Dendriplexes from PLGA Microparticles
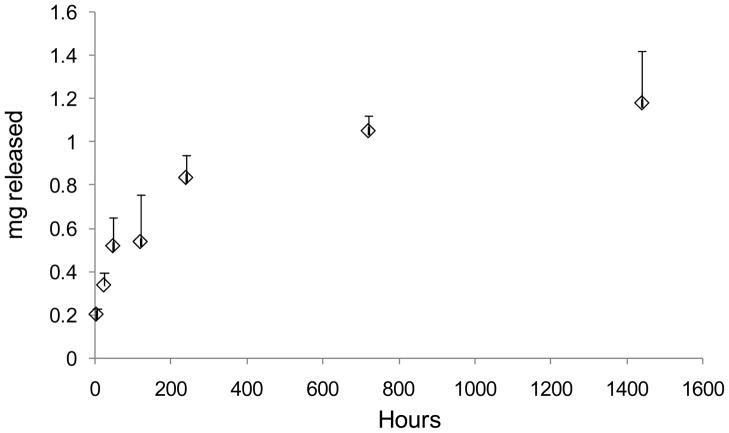
Release of PAMAM-pDNA Dendriplexes (Figure 6) was initially fast with with 0.2 mg dendriplexes (pDNA conc) released by 4 hrs. This was followed by a more sustained cumulative release of 1.05 mg of pDNA by 720 hrs.
Figure 6.
PAMAM (G3)-pDNA dendriplex release profiles from PLGA microparticles determined using UV spectrophotometry and the picogreen assay reagent (averages representative of 8 measurements + SD).
3.4 Increases in Buffering Capacity of PAMAM-pDNA dendriplex loaded PLGA Microparticles are correlated with the Generation of the PAMAM Dendrimer Used
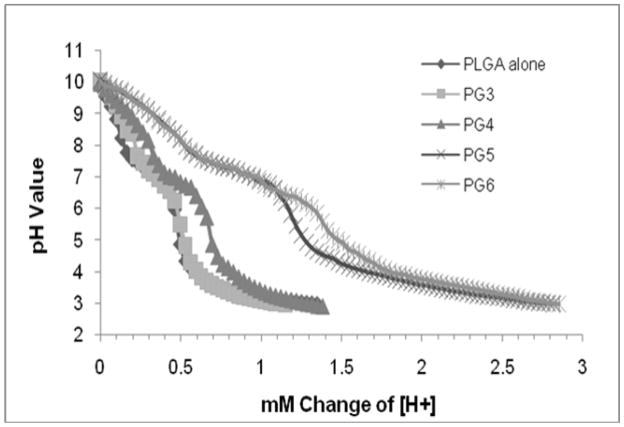
The buffering capacity of the PLGA microparticles loaded with pDNA alone and PLGA microparticles loaded with PAMAM-pDNA dendriplexes was assessed by measuring the change in pH of a particle suspension (10mg/ml) upon addition of increasing amounts of 0.1 N HCL. As shown in figure 7, the buffering capacity of PLGA microparticles loaded with PAMAM-pDNA dendriplexes (PG3 to PG6) is correlated with the generation level. The higher the generation level of the PAMAM dendrimers encapsulated in the PLGA microparticles, the more effective it is at buffering against pH changes. Similar results have been previously reported for PAMAM dendrimers alone 11,22,24,25. This is indicated by the shift and a decrease in the slope of the titration curves. The most significant shifts in buffering capacity were observed in PLGA microparticles loaded with PAMAM-pDNA dendriplexes prepared using generation 5 and generation 6 (P<0.001).
Figure 7.
Acid titration experiments with 0.1 N HCL to demonstrate the buffering ability of PLGA microparticles loaded with pDNA alone and PLGA microparticles loaded with PAMAM-pDNA dendriplexes. The data shows significantly increased buffering of PLGA microparticles loaded with PAMAM-pDNA dendriplexes when compared to PLGA microparticles loaded with pDNA alone.
3.5 Loading PAMAM-pDNA Dendriplexes into PLGA Microparticles Results in Higher pDNA Loading in Comparison to PLGA Microparticles Loaded with pDNA Alone
The loading efficiency of PLGA microparticles loaded with pDNA alone and PLGA microparticles loaded with PAMAM-pDNA dendriplexes were determined by dissolving 200 mg of lyophilized particles in 2mL of acetonitrile. The pDNA and PAMAM-pDNA dendriplexes were extracted in TE buffer (10 mM, pH 8.3). The amount of pDNA was evaluated based on absorbance measurements using standard calibration curves. The actual loading of PLGA microparticles loaded with pDNA alone was 3.2 μg pDNA/mg particles (Table 1). Complex formation of PAMAM dendrimer and pDNA before encapsulation into PLGA particles leads to an approximate 150% increase in pDNA loading relative to the pDNA alone loading in PLGA microparticles (P<0.001). The actual loading of pDNA in PG3 to PG6 microparticles is approximately 8.1 μg pDNA/mg particles. The generation of the PAMAM dendrimer used to complex the pDNA prior to loading in PLGA microparticles had no significant effect on the level of pDNA loaded (P>0.05).
Table 1.
pDNA loading in PLGA microparticles (Averages representative of 3 measurements ± standard deviation).
| pDNA actual loading (μg DNA/mg particles) | |
|---|---|
| PLGA/pDNA | 3.2±0.3 |
| PG3 | 8.0±0.2 |
| PG4 | 8.0±0.1 |
| PG5 | 8.2±0.3 |
| PG6 | 8.2±0.2 |
3.6 Evaluation of the ability of PLGA microparticles to entrap pDNA
PLGA microparticles loaded with pDNA alone or PAMAM-pDNA dendriplexes (PG3 to PG6) were prepared as described in the materials and method section. The ability of the PLGA microparticles to entrap pDNA was analyzed by agarose gel electrophoresis as shown in figure 8. All the PLGA microparticle formulations completely inhibit pDNA migration on the gel, suggesting a strong pDNA entrapment capacity and/or that the entrapment of the dendriplexes in PLGA microparticles did not cause the dendriplexes to disassociate.
Figure 8.
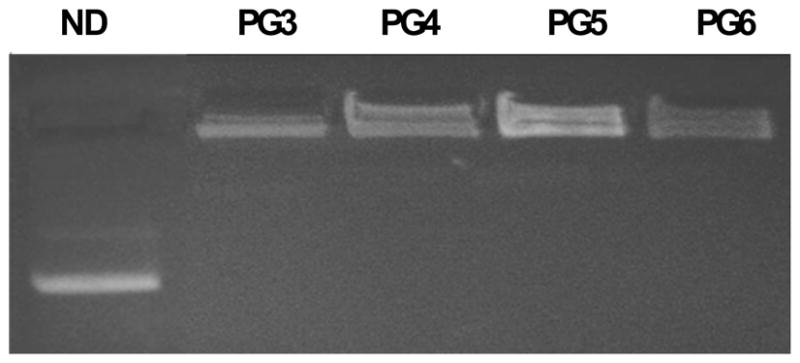
The ability of PLGA microparticles loaded with PAMAM-pDNA dendriplexes to prevent pDNA migration was analyzed on agarose gel stained with ethidium bromide.
3.7 Loading PAMAM-pDNA dendriplexes into PLGA microparticles reduces the cytotoxicity associated with PAMAM dendrimers
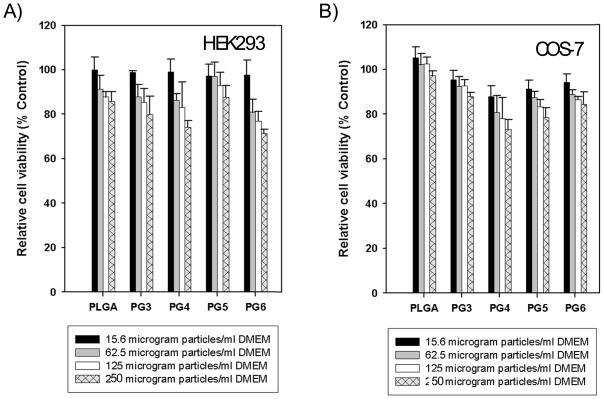
In vitro cytotoxicity was evaluated in COS7 and HEK293 cells with increasing doses of PLGA microparticles loaded with pDNA and PLGA microparticles loaded with PAMAM-pDNA dendriplexes. The concentrations tested ranged from 15.6 to 250 μg of PLGA particles per milliliter of DMEM. As shown in figure 9a and 9b, PLGA microparticles loaded with PAMAM-pDNA dendriplexes (PG3 to PG6) had similar cytotoxicity profiles to PLGA microparticles loaded with pDNA alone in both HEK293 and COS7 cells. No significant difference in toxicity was observed when the generation of the PAMAM used to complex with pDNA prior to entrapment in the PLGA microparticles was increased (P>0.05). Similar differences in toxicity between the groups tested were observed at varying time-points (data not shown).
Figure 9.
A) Cytotoxicity study of PLGA PAMAM (PG3 to PG6) pDNA particles in HEK293 cell lines. Cell viabilities were evaluated by MTT assay as described in the experimental section. Data is represented as the mean ± standard deviation (n=6). B) Cytotoxicity study of PLGA PAMAM (PG3 to PG6) pDNA particles in COS-7 cell lines. Cell viabilities were evaluated by MTT assay as described in the experimental section. Data is represented as the mean ± standard deviation (n=6).
3.8 PLGA microparticles loaded with PAMAM-pDNA dendriplexes can be efficiently taken up by HEK293 cells
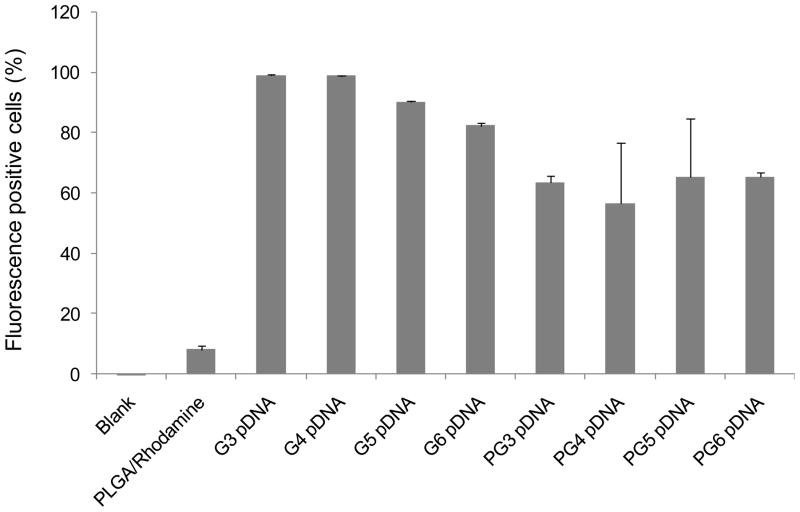
Figure 10 shows cellular uptake data of fluorescence-labeled microparticles incubated with HEK293 cells. PLGA microparticles loaded with PAMAM-pDNA dendriplexes displayed 8 fold higher uptake in cells than unmodified PLGA microparticles (P<0.001). No significant difference in the cell uptake of PLGA microparticles loaded with PAMAM-pDNA dendriplexes was observed for increasing generation levels of the PAMAM dendrimer complexed with pDNA.
Figure 10.
Flow cytometry data of fluorescence-labeled particles that have been incubated with HEK293 cells. The cellular uptake of the PAMAM –pDNA dendrimers alone or PLGA microparticles loaded with PAMAM dendriplexes are presented as percentage of fluorescence-labeled cells (n=3 per group).
3.9 Loading PAMAM-pDNA dendriplexes into PLGA microparticles significantly increases the transfection in COS7 and HEK293 cells when compared to PLGA microparticles loaded with pDNA alone
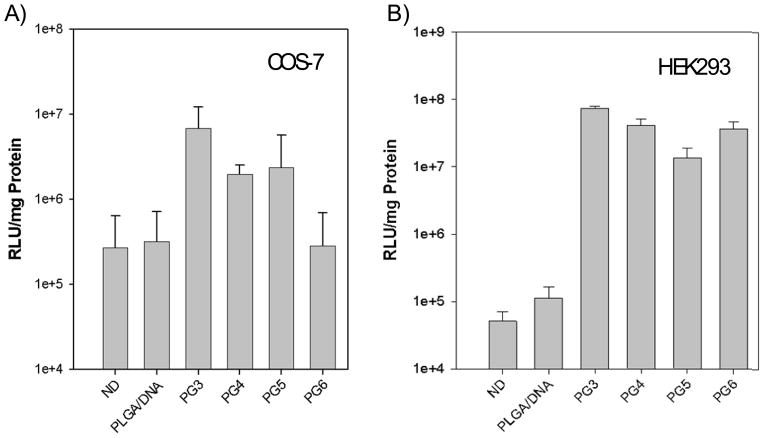
Gene transfection mediated by PLGA microparticles loaded with pDNA alone and PLGA microparticles loaded with PAMAM-pDNA dendriplexes was evaluated in COS7 and HEK293 cells (Figure 11a and 11b). All of the PLGA microparticles loaded with PAMAM-pDNA poyplexes showed significantly higher transgene expression than PLGA microparticles loaded with pDNA alone. The only exception was PG6, which in COS7 cells, generated transgene expressions that were similar to pDNA alone (P>0.05). The transfection efficiency of PLGA microparticles loaded with PAMAM-pDNA dendriplexes was not dependent on the generation of PAMAM dendrimer incorporated. Among the PLGA microparticles loaded with PAMAM-pDNA dendriplexes, the PG3 particles mediated the highest luciferase expression in both cell lines, which was 60-fold or 27-fold higher than PLGA microparticles loaded with pDNA alone in HEK293 or COS7 cells, respectively. PLGA microparticles loaded with PAMAM-pDNA dendriplexes generated higher transfection efficiencies in HEK293 cells than COS7 cells.
Figure 11.
A) PLGA pDNA and PLGA PAMAM-pDNA (PG3 to PG6) particles mediated gene transfection in COS7 cells. Particles were incubated with cells at a concentration of 0.2mg/well with a target pDNA dose of 1μg/well. Cell harvesting and luciferase assays were performed 48h after transfection as described in the materials and methods section. Data is represented as mean ± standard deviation (n=3). All the PLGA particles loaded with PAMAM-pDNA polyplexes exhibited efficient gene delivery in COS-7 cells. B) PLGA pDNA and PLGA PAMAM pDNA (PG3 to PG6) particles mediated gene transfection in HEK293 cells. Particles were incubated with cells at a concentration of 0.2mg/well with a target pDNA dose of 1μg/well. Cell harvesting and luciferase assays were performed 48h after transfection as described in the materials and methods section. Data is represented as mean ± standard deviation (n=3). All the PLGA PAMAM pDNA particles exhibited efficient gene delivery in HEK293 cells.
3.10 PLGA microparticles loaded with PAMAM-pDNA dendriplexes retain high transfection efficiencies when tested in serum containing media
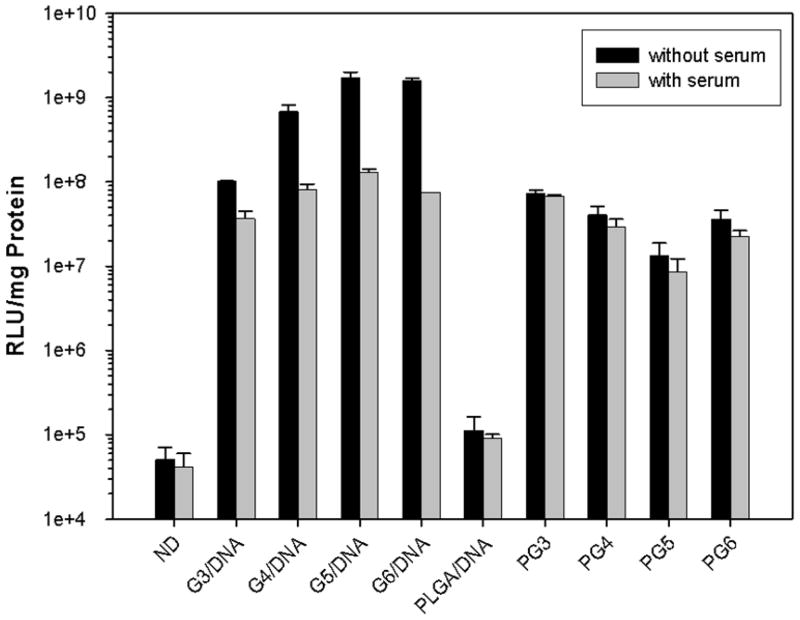
Gene transfection mediated by PLGA microparticles loaded with pDNA alone, PLGA microparticles loaded with PAMAM-pDNA dendriplexes and PAMAM-pDNA dendriplexes alone were evaluated in COS7 cells (Figure 12). The results showed that transgene expression generated by PAMAM-pDNA dendriplexes in serum containing media was significantly lower than PAMAM-pDNA dendriplexes tested in serum-free media (P<0.05). The presence of serum in the media reduced the transfection efficiency of PAMAM (G3)-pDNA and PAMAM (G4)-pDNA dendriplexes by 279% and 838% in COS7 cells, respectively. The transgene expression generated by PAMAM (G5)-pDNA and PAMAM (G6)-pDNA dendriplexes was reduced by 1318% and 2127%, respectively, when transfections were carried out in serum containing media. In contrast, no significant reduction in transfection efficiency was observed for any of the PLGA microparticles loaded with PAMAM-pDNA dendriplexes (PG3-PG6) when transfections were carried out in serum-containing media relative to transfections carried out in serum-free media (P>0.05).
Figure 12.
Effect of serum on transgene expression of PLGA microparticles loaded with pDNA alone and PLGA microparticles loaded with PAMAM-pDNA (PG3 to PG6) dendriplexes in COS7 cells in serum-free media (Black bar) or media containing 10% serum (Gray bar). Microparticles were incubated with cells at a concentration of 0.2mg/well with a target pDNA dose of 1μg/well. Cell harvesting and luciferase assays were performed 48h after transfection as described in the materials and methods section. Data is represented as mean ± standard deviation (n=3).
4. Discussion
When pDNA is delivered using liposomes or cationic polymer vectors such as polyethylenimine, the expression of the genes is transient and the delivery systems are frequently associated with high toxicity4,26. A potential approach to overcoming transient gene expression and high toxicity is to utilize PLGA polymers for sustained release of pDNA1. For example, sustained pDNA delivery using PLGA microparticles has shown significant potential in transfecting human arterial smooth muscles27,28 and airway epithelia29. PLGA microparticles are biodegradable, biocompatible, have the capacity to protect pDNA from nuclease degradation and have FDA approval for several biomedical and drug delivery applications.
A significant challenge in the development of PLGA delivery systems has been the difficulty of entrapping hydrophilic DNA type molecules within the hydrophobic PLGA1. Additionally the DNA can become damaged during the encapsulation procedure or become inactivated as a result of the acidic environment created by accumulation of oligomers within the particles1.
A promising approach to overcoming these limitations is to utilize cationic polymers or excipients1,4,8,10–12,15,16,30–36. Cationic polymers can be incorporated into PLGA based microparticles using a number of approaches. PLGA and cationic polymers can be blended and pDNA entrapped into the blended polymer mixture4,12,13. Cationic polymers can be attached onto the surface of the PLGA microparticles and pDNA then immobilized onto the surface of the PLGA microparticles using electrostatic interactions1,8,9,11,37. The advantage to this approach is that pDNA is never exposed to any of the harsh manufacturing conditions for preparation of the microparticles including exposure to organic solvents and shear forces. A drawback to this approach is that the pDNA is not released over a sustained period of time. Finally, pDNA can be complexed with cationic polymers and the complexes can then be loaded into PLGA microparticles1,4. This approach compacts the pDNA providing significant protection to the pDNA during the process of entrapment into PLGA microparticles1. We have previously shown, using PEI as the cationic polymer, that complexing pDNA with PEI and then loading the PEI-pDNA polyplexes into PLGA microparticles resulted in a delivery system that generated higher transgene activity in HEK293 and COS7 cells than either conjugating PEI to the surface of the microparticles or blending PLGA and PEI before loading pDNA into the microparticles4.
In comparison to linear cationic polymers such as PEI or PL, PAMAM dendrimers have several unique and advantageous features11,38,39. PAMAM dendrimers, similar to polymers such as PEI, have amine groups capable of protonation at physiological conditions20,39. PAMAM dendrimers are thus able to generate significant buffering capacity that allow the dendriplexes to escape from the endosome and into the cytoplasm using the “proton sponge mechanism”39. In comparison to polymers such as PEI, however, dendrimers are spherical, monodisperse polymers with lower structural density in the intramolecular core39. Similar to PEI, however, using PAMAM dendrimers as a standalone delivery system has a number of drawbacks. Cationic polymers such as PAMAM tend to aggregate in the presence of serum and this significantly reduces transfection efficiencies and therefore, the potential for in vivo applications 26,40–42. Additionally, as the generation of the PAMAM dendrimer increases, so does the cytotoxicity43–45. Given our other recent observations that complexing the cationic polymer PEI with pDNA prior to entrapping the pDNA in the microparticles generates stronger transgene activity in cells than conjugation of PEI to the surface of PLGA microparticles or blending of PEI/PLGA4, we decided, for the first time, to evaluate the potential of PLGA microparticles loaded with PAMAM-pDNA dendriplexes for applications in non-viral gene delivery.
Negatively charged pDNA condenses and complexes with PAMAM to form a dendriplex. The extent of dendriplex formation and condensation is a function of the charge balance in the pDNA/dendrimer complex and commonly referred to as the nitrogen/phosphate or N/P ratio. The negative charge on pDNA arises from the presence of an extensive phosphate (P) backbone. For the purposes of calculating the N/P ratio, we use the average molecular weight of the amine residue of the repeating unit in the PAMAM dendrimer to be the corresponding number of “N” moieties per molecule.
In this approach, PAMAM-pDNA dendriplexes were prepared at an N:P ratio of 5. This N:P ratio was selected based on previous optimization studies showing that it generated the strongest transgene expressions in HEK293 cells and COS7 cells. The dendriplexes were then loaded into PLGA microparticles using a double emulsion solvent evaporation technique. Complex formation between PAMAM dendrimer and pDNA before entrapment into PLGA particles leads to an approximate 150% increase in pDNA loading relative to loading of pDNA alone in PLGA microparticles (P<0.001). We presume that the increased loading is primarily due to the reduced hydrophilicity of the pDNA when complexed with the PAMAM dendrimers that then allows for better distribution throughout the hydrophobic PLGA matrix. Similar results have been reported for increasing loading efficiency of pDNA in PLGA particles using poly-L-lysine46 or in PLA particles using chitosan47. SEM analysis showed that PLGA microparticles loaded with pDNA alone were smooth and spherical in morphology. In contrast, when PLGA microparticles were loaded with PAMAM-pDNA dendriplexes, porous features and indentations appeared on the surface of the microparticles. Previous studies have shown that porous particles are formed when PEI/oligonucleotide complexes are encapsulated within PLGA microparticles48. Higher osmotic pressure within particles may contribute to increased surface porosity49. For example, the encapsulation of osmotically active PAMAM dendrimers, may lead to a difference in osmotic pressure between the inner and outer aqueous phases that, in turn may lead to an influx of water to the inner aqueous phase during the particle hardening process. An increased inner aqueous phase volume would be expected to result in increased porosity48,50.
Encapsulating PAMAM-pDNA dendriplexes into PLGA microparticles resulted in significant particle size reduction (P<0.001) in comparison to PLGA particles loaded with pDNA alone. This may have resulted from the decrease in pDNA size after condensation of pDNA by PAMAM dendrimers. Previous studies have showed that pDNA complexed with cationic polymers such as PEI display reduced sizes in the range of 20–200 nm51. The generation of the PAMAM dendrimer used did not significantly change the size of the PLGA microparticles loaded with PAMAM-pDNA dendriplexes.
The zetapotential of PLGA particles loaded with PAMAM-pDNA dendriplexes was positive in phosphate buffered saline. In contrast, the zetapotential of PLGA microparticles loaded with pDNA alone showed a net negative zetapotential value of −23.48 mV. This suggests that some of the PAMAM dendrimer moieties are stably associated with the surface of the PLGA microparticles. The zetapotential of PLGA microparticles loaded with PAMAM-pDNA dendriplexes showed an increasing correlation with increasing generation of the PAMAM dendrimer. An increase in dendrimer generation from G3 to G6, resulted in a zetapotential increase from +7.29 mV to +31.65 mV. This change in zetapotential is associated with an increase in higher positive charge density as the generation of the dendrimer increases. We have previously shown that changing the charge of the surface of microparticles from negative to positive significantly increases cell uptake and the incorporation of the PAMAM-pDNA dendriplexes into PLGA microparticles is expected to have the same impact4,11. Release profiles of PAMAM (G3)-pDNA dendriplexes clearly showed that the microparticles are capable of providing an initially higher dose of dendriplexes followed by lower doses released over a sustained period of time. The sustained release of the dendriplexes over the time tested confirms that dendriplexes are distributed throughout the PLGA microparticle matrix.
The presence of primary and tertiary amines on the PAMAM dendrimer loaded in the PLGA microparticles allowed for the microparticles to resist acidification in acid titration experiments. This buffering capacity against pH changes is purported to lead to endophagosomal escape into the cytoplasm through a “proton sponge mechanism”22,39. Increases in the generation level of the PAMAM incorporated into the PLGA particles was correlated to increases in buffering capacity. The most significant differences in buffering capacity were observed when PAMAM dendrimers of generation 5 and 6 were used to complex pDNA prior to entrapment in PLGA microparticles (P<0.001 in comparison to PLGA microparticles loaded with pDNA alone). As with our previous studies on the development of cationic microparticles, zetapotential and buffering capacity were broadly correlated4,11. Confirmation that pDNA or PAMAM-pDNA dendriplexes loaded into the PLGA microparticles remained entrapped in the PLGA microparticles and that dendriplexes remained intact after fabrication was observed through the lack of mobility of pDNA in gel electrophoresis assays.
We used the MTT assay to make dose dependent toxicity measurements of the PLGA microparticles loaded with pDNA alone or PAMAM-pDNA dendriplexes. PLGA microparticles loaded with PAMAM-pDNA dendriplexes were incubated at levels significantly above the levels used for transfection experiments (250 μg of particles per ml of DMEM). In both HEK293 and COS7 cells, no significant difference in toxicity was observed in comparison to the biocompatible PLGA microparticles without any PAMAM dendrimer incorporated.
A number of studies have shown that increasing the generation and concentration of the PAMAM dendrimer increases cytotoxicity to levels similar to that of PEI 24,43–45,52,53. In this study, no significant differences in cytotoxicity were observed from PLGA microparticles loaded with PAMAM-pDNA dendriplexes relative to PLGA microparticles loaded with pDNA alone. We have previously shown that these levels of toxicity are significantly lower than those generated by cationic polymers such as PEI 4,11. Furthermore, incorporation of PAMAM dendrimers into biodegradable PLGA microparticles removed generation dependent cytotoxicity of PAMAM dendrimers. We have observed similar results when PAMAM dendrimers were conjugated to the surface of PLGA microparticles. This result suggests that combining PLGA and PAMAM dendrimers provides an effective way to reduce PAMAM dendrimer toxicity. A comparison of the toxicity data from our previous studies on PLGA microparticles with PAMAM dendrimers conjugated to the surface versus PLGA microparticles loaded with PAMAM-pDNA dendriplexes in this study reveals that loading dendriplexes into PLGA microparticles results in a significant reduction in toxicity. This is likely to be due to the more sustained release of PAMAM-pDNA dendriplexes from the PLGA microparticles reducing the concentration of PAMAM dendrimer exposed to the cells at any one time. Flow cytometry studies showed that loading PAMAM-pDNA dendriplexes into PLGA microparticles significantly enhanced cell uptake by 8 fold in comparison to unmodified PLGA microparticles (P<0.001). A drop in particle size from 1200 nm to approximately 800 nm was observed when comparing unmodified PLGA microparticles with PLGA microparticles loaded with dendriplexes that could be potentially contributing to this increased uptake. However, previous studies have shown that its likely to be the change in zetapotential from negative to positive when incorporating dendriplexes into the PLGA microparticles that has the strongest impact on increasing cellular uptake11. This is because positively charged microparticles can demonstrate an enhanced interaction with the negatively charged cellular membranes. However, increasing the generation of the PAMAM dendrimer conjugated to the PLGA microparticles had no effect on increasing cell uptake further.
Gene transfection experiments were carried out on HEK293 and COS7 cells, which are two model cell lines commonly used to evaluate transfection efficiency26,54. PLGA microparticles loaded with pDNA-PAMAM dendriplexes significantly enhanced luciferase expression in comparison to PLGA microparticles loaded with pDNA alone in both COS7 and HEK293 cell lines. PLGA microparticles loaded with G3 PAMAM dendrimer-pDNA dendriplexes generated the highest transgene luciferase expression. In contrast to PAMAM dendrimers alone, however, increasing the generation of the PAMAM dendrimer complexed with pDNA and loaded into the PLGA microparticles did not increase luciferase transgene expression. This result was observed in both HEK293 and COS7 cell lines. Lower generation PAMAM dendrimers are cheaper, easier to synthesize, less toxic and have longer circulation times38,43–45,55. Thus, the ability to generate the highest transfection efficiencies using G3 PAMAM dendrimers complexed with pDNA and loaded into PLGA microparticles will have significant translational benefits. These results are also consistent with ours and others previous studies in which zetapotential and buffering capacity of PAMAM dendrimers do not necessarily correlate with transfection efficiencies11,18,38,56. The increase in transfection efficiency generated by PLGA microparticles loaded with PAMAM-pDNA dendriplexes in comparison to PLGA microparticles loaded with pDNA alone is therefore related to increased pDNA loading, protection of pDNA during loading, and sustained release of pDNA-PAMAM dendriplexes. The positively charged PLGA microparticles are also likely to have increased interactions with the cell surface. Although buffering capacity may enhance transfection to some degree, our results indicate that it is not the dominant parameter in enhancing transfection efficiency with this delivery system.
Another parameter that can significantly impair the transfection efficiency of cationic polymer-pDNA dendriplexes is the presence of serum. Serum can cause aggregation of cationic polymer-pDNA dendriplexes and reduced uptake by endocytosis. This results in reduced transfection efficiencies and is a major limitation for adapting cationic polymer based non-viral delivery systems for in vivo applications57,58. We tested the transfection efficiency of PLGA microparticles loaded with PAMAM-pDNA dendriplexes and PAMAM-pDNA dendriplexes alone in serum free media and serum containing media. The transgene expression generated by PAMAM-pDNA dendriplexes alone in serum-free media was significantly higher than the transgene expression generated when transfections were carried out in serum-containing media. In contrast, there is no significant difference in the transgene expression generated by PLGA microparticles loaded with PAMAM-pDNA dendriplexes in serum-free or serum containing medium (P>0.05). These results indicated that loading PAMAM-pDNA dendriplexes into PLGA microparticles can reduce the direct interaction between anionic components in serum and cationic PAMAM-pDNA dendriplexes. The ability of a non-viral gene delivery vector to generate equivalent transfection efficiencies in serum free and serum containing media increases the potential for translational applications in vivo.
5. Conclusions
PLGA particles have significant potential for generating sustained transgene expression. Loading pDNA into the PLGA microparticles generates non-viral vectors with poor transfection efficiencies. Cationic polymers such as PAMAM dendrimer can generate high but transient transgene expression. However, higher generation PAMAM dendrimers alone are toxic and all PAMAM dendrimers display reduced transfection efficiencies in the presence of serum. Loading PAMAM-pDNA dendriplexes into PLGA microparticles generates a delivery system that has several advantages over either PLGA microparticles alone or PAMAM dendrimers alone. PLGA microparticles loaded with PAMAM-pDNA dendriplexes have desirable cytotoxicity profiles similar to PLGA microparticles alone, have higher cell uptake than PLGA microparticles alone, provide sustained release of PAMAM-pDNA dendriplexes, have higher loading efficiencies of pDNA and generate strong transfection efficiencies in serum free or serum containing media. PLGA microparticles loaded with PAMAM-pDNA dendriplexes are therefore likely to have significant translational potential for applications in RNA, DNA and oligonucleotide delivery.
Acknowledgments
We gratefully acknowledge support from the American Cancer Society (RSG-09-015-01-CDD), the National Cancer Institute at the National Institutes of Health (1R21CA13345-01/1R21CA128414-01A2), and the Pharmaceutical Research and Manufacturers of America Foundation. J. Intra acknowledges support from the Parenteral Drug Association for a predoctoral fellowship.
References
- 1.Abbas AO, Donovan MD, Salem AK. Formulating poly(lactide-co-glycolide) particles for plasmid DNA delivery. Journal of Pharmaceutical Sciences. 2008;97(7):2448–2461. doi: 10.1002/jps.21215. [DOI] [PubMed] [Google Scholar]
- 2.Eldridge JH, Staas JK, Meulbroek JA, Tice TR, Gilley RM. Biodegradable and Biocompatible Poly(Dl-Lactide-Co-Glycolide) Microspheres as an Adjuvant for Staphylococcal Enterotoxin-B Toxoid Which Enhances the Level of Toxin-Neutralizing Antibodies. Infection and Immunity. 1991;59(9):2978–2986. doi: 10.1128/iai.59.9.2978-2986.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Hagan DT, Singh M, Ulmer JB. Microparticles for the delivery of DNA vaccines. Immunological Reviews. 2004;199(1):191–200. doi: 10.1111/j.0105-2896.2004.00153.x. [DOI] [PubMed] [Google Scholar]
- 4.Zhang XQ, Intra J, Salem AK. Comparative study of poly (lactic-co-glycolic acid)-poly ethyleneimine-plasmid DNA microparticles prepared using double emulsion methods. Journal of Microencapsulation. 2008;25:1–12. doi: 10.1080/02652040701659347. [DOI] [PubMed] [Google Scholar]
- 5.Walter E, Moelling K, Pavlovic J, Merkle HP. Microencapsulation of DNA using poly(DL-lactide-co-glycolide): stability issues and release characteristics. Journal of Controlled Release. 1999;61(3):361–374. doi: 10.1016/s0168-3659(99)00151-0. [DOI] [PubMed] [Google Scholar]
- 6.Tinsley-Bown AM, Fretwell R, Dowsett AB, Davis SL, Farrar GH. Formulation of poly(D,L-lactic-co-glycolic acid) microparticles for rapid plasmid DNA delivery. Journal of Controlled Release. 2000;66(2–3):229–241. doi: 10.1016/s0168-3659(99)00275-8. [DOI] [PubMed] [Google Scholar]
- 7.Kasturi SP, Qin H, Thomson KS, El-Bereir S, Cha SC, Neelapu S, Kwak LW, Roy K. Prophylactic anti-tumor effects in a B cell lymphoma model with DNA vaccines delivered on polyethylenimine (PEI) functionalized PLGA microparticles. Journal of Controlled Release. 2006;113(3):261–270. doi: 10.1016/j.jconrel.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 8.Kasturi SP, Sachaphibulkij K, Roy K. Covalent conjugation of polyethyleneimine on biodegradable microparticles for delivery of plasmid DNA vaccines. Biomaterials. 2005;26(32):6375–6385. doi: 10.1016/j.biomaterials.2005.03.043. [DOI] [PubMed] [Google Scholar]
- 9.Manuel WS, Zheng J, Hornsby PJ. Transfection by polyethyleneimine-coated microspheres. Journal of Drug Targeting. 2001;9(1):15. doi: 10.3109/10611860108995629. [DOI] [PubMed] [Google Scholar]
- 10.Singh M, Briones M, Ott G, O’Hagan D. Cationic microparticles: A potent delivery system for DNA vaccines. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(2):811–816. doi: 10.1073/pnas.97.2.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang XQ, Intra J, Salem AK. Conjugation of polyamidoamine dendrimers on biodegradable microparticles for nonviral gene delivery. Bioconjugate Chemistry. 2007;18:2068–2076. doi: 10.1021/bc070116l. [DOI] [PubMed] [Google Scholar]
- 12.Sutton D, Durand R, Shuai XT, Gao JM. Poly(D, L-lactide-co-glycolide)/poly(ethylenimine) blend matrix system for pH sensitive drug delivery. Journal of Applied Polymer Science. 2006;100(1):89–96. [Google Scholar]
- 13.Oster CG, Kissel T. Comparative study of DNA encapsulation into PLGA microparticles using modified double emulsion methods and spray drying techniques. Journal of Microencapsulation. 2005;22(3):235–244. doi: 10.1080/02652040500100295. [DOI] [PubMed] [Google Scholar]
- 14.Yun YH, Jiang HL, Chan R, Chen WL. Sustained release of PEG-g-chitosan complexed DNA from poly(lactide-co-glycolide) Journal of Biomaterials Science-Polymer Edition. 2005;16(11):1359–1378. doi: 10.1163/156856205774472281. [DOI] [PubMed] [Google Scholar]
- 15.Kim IS, Lee SK, Park YM, Lee YB, Shin SC, Lee KC, Oh IJ. Physicochemical characterization of poly(L-lactic acid) and poly(D,L-lactide-co-glycolide) nanoparticles with polyethylenimine as gene delivery carrier. International Journal of Pharmaceutics. 2005;298(1):255–262. doi: 10.1016/j.ijpharm.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 16.Oster CG, Kim N, Grode L, Barbu-Tudoran L, Schaper AK, Kaufmann SHE, Kissel T. Cationic microparticles consisting of poly(lactide-co-glycolide) and polyethylenimine as carriers systems for parental DNA vaccination. Journal of Controlled Release. 2005;104(2):359–377. doi: 10.1016/j.jconrel.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 17.Bielinska AU, Chen CL, Johnson J, Baker JR. DNA complexing with polyamidoamine dendrimers: Implications for transfection. Bioconjugate Chemistry. 1999;10(5):843–850. doi: 10.1021/bc990036k. [DOI] [PubMed] [Google Scholar]
- 18.Braun CS, Vetro JA, Tomalia DA, Koe GS, Koe JG, Middaugh CR. Structure/function relationships of polyamidoamine/DNA dendrimers as gene delivery vehicles. Journal of Pharmaceutical Sciences. 2005;94(2):423–436. doi: 10.1002/jps.20251. [DOI] [PubMed] [Google Scholar]
- 19.Cheng YY, Xu ZH, Ma ML, Xu TW. Dendrimers as drug carriers: Applications in different routes of drug administration. Journal of Pharmaceutical Sciences. 2008;97(1):123–143. doi: 10.1002/jps.21079. [DOI] [PubMed] [Google Scholar]
- 20.Esfand R, Tomalia DA. Poly(amidoamine) (PAMAM) dendrimers: from biomimicry to drug delivery and biomedical applications. Drug Discovery Today. 2001;6(8):427–436. doi: 10.1016/s1359-6446(01)01757-3. [DOI] [PubMed] [Google Scholar]
- 21.Zhou JH, Wu JY, Hafdi N, Behr JP, Erbacher P, Peng L. PAMAM dendrimers for efficient siRNA delivery and potent gene silencing. Chemical Communications. 2006;(22):2362–2364. doi: 10.1039/b601381c. [DOI] [PubMed] [Google Scholar]
- 22.Tang MX, Szoka FC. The influence of polymer structure on the interactions of cationic polymers with DNA and morphology of the resulting complexes. Gene Therapy. 1997;4(8):823–832. doi: 10.1038/sj.gt.3300454. [DOI] [PubMed] [Google Scholar]
- 23.Hansen MB, Nielsen SE, Berg K. Re-Examination and Further Development of a Precise and Rapid Dye Method for Measuring Cell-Growth Cell Kill. Journal of Immunological Methods. 1989;119(2):203–210. doi: 10.1016/0022-1759(89)90397-9. [DOI] [PubMed] [Google Scholar]
- 24.Nam HY, Nam K, Hahn HJ, Kim BH, Lim HJ, Kim HJ, Choi JS, Park JS. Biodegradable PAMAM ester for enhanced transfection efficiency with low cytotoxicity. Biomaterials. 2009;30(4):665–673. doi: 10.1016/j.biomaterials.2008.10.013. [DOI] [PubMed] [Google Scholar]
- 25.Tang MX, Redemann CT, Szoka FC. In vitro gene delivery by degraded polyamidoamine dendrimers. Bioconjugate Chemistry. 1996;7(6):703–714. doi: 10.1021/bc9600630. [DOI] [PubMed] [Google Scholar]
- 26.Intra J, Salem AK. Characterization of the transgene expression generated by branched and linear polyethylenimine-plasmid DNA nanoparticles in vitro and after intraperitoneal injection in vivo. Journal of Controlled Release. 2008;130(2):129–138. doi: 10.1016/j.jconrel.2008.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Panyam J, Labhasetwar V. Biodegradable nanoparticles for drug and gene delivery to cells and tissue. Advanced Drug Delivery Reviews. 2003;55(3):329–347. doi: 10.1016/s0169-409x(02)00228-4. [DOI] [PubMed] [Google Scholar]
- 28.Prabha S, Labhasetwar V. Critical determinants in PLGA/PLA nanoparticle-mediated gene expression. Pharmaceutical Research. 2004;21(2):354–364. doi: 10.1023/b:pham.0000016250.56402.99. [DOI] [PubMed] [Google Scholar]
- 29.Stern M, Ulrich K, Geddes DM, Alton EWFW. Poly (D, L-lactide-co-glycolide)/DNA microspheres to facilitate prolonged transgene expression in airway epithelium in vitro, ex vivo and in vivo. Gene Therapy. 2003;10(16):1282–1288. doi: 10.1038/sj.gt.3301994. [DOI] [PubMed] [Google Scholar]
- 30.Bivas-Benita M, Romeijn S, Junginger HE, Borchard G. PLGA-PEI nanoparticles for gene delivery to pulmonary epithelium. European Journal of Pharmaceutics and Biopharmaceutics. 2004;58(1):1–6. doi: 10.1016/j.ejpb.2004.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Rosa G, Bochot A, Quaglia F, Besnard M, Fattal E. A new delivery system for antisense therapy: PLGA microspheres encapsulating oligonucleotide/polyethyleneimine solid complexes. International Journal of Pharmaceutics. 2003;254(1):89–93. doi: 10.1016/s0378-5173(02)00689-0. [DOI] [PubMed] [Google Scholar]
- 32.De Smedt SC, Demeester J, Hennink WE. Cationic polymer based gene delivery systems. Pharmaceutical Research. 2000;17(2):113–126. doi: 10.1023/a:1007548826495. [DOI] [PubMed] [Google Scholar]
- 33.Fu HL, Cheng SX, Zhang XZ, Zhuo RX. Dendrimer/DNA complexes encapsulated functional biodegradable polymer for substrate-mediated gene delivery. Journal of Gene Medicine. 2008;10(12):1334–1342. doi: 10.1002/jgm.1258. [DOI] [PubMed] [Google Scholar]
- 34.Ribeiro S, Hussain N, Florence AT. Release of DNA from dendriplexes encapsulated in PLGA nanoparticles. International Journal of Pharmaceutics. 2005;298(2):354–360. doi: 10.1016/j.ijpharm.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 35.Takashima Y, Saito R, Nakajima A, Oda M, Kimura A, Kanazawa T, Okada H. Spray-drying preparation of microparticles containing cationic PLGA nanospheres as gene carriers for avoiding aggregation of nanospheres. International Journal of Pharmaceutics. 2007;343:262–269. doi: 10.1016/j.ijpharm.2007.05.042. [DOI] [PubMed] [Google Scholar]
- 36.Ungaro F, De Rosa G, Quaglia F, Fattal E, La Rotonda MI. Controlled release of oligonucleotide/polyethyleneimine complexes from PLGA-based microspheres: potential of spray-drying technique. Journal of Drug Delivery Science and Technology. 2005;15(2):137–143. [Google Scholar]
- 37.Zhang XQ, Dahle CE, Weiner GJ, Salem AK. A comparative study of the antigen-specific immune response induced by co-delivery of CpG ODN and antigen using fusion molecules or biodegradable microparticles. Journal of Pharmaceutical Sciences. 2007;96:3283–3292. doi: 10.1002/jps.20978. [DOI] [PubMed] [Google Scholar]
- 38.Radu DR, Lai CY, Jeftinija K, Rowe EW, Jeftinija S, Lin VSY. A polyamidoamine dendrimer-capped mesoporous silica nanosphere-based gene transfection reagent. Journal of the American Chemical Society. 2004;126(41):13216–13217. doi: 10.1021/ja046275m. [DOI] [PubMed] [Google Scholar]
- 39.Lee CC, MacKay JA, Frechet JMJ, Szoka FC. Designing dendrimers for biological applications. Nature Biotechnology. 2005;23(12):1517–1526. doi: 10.1038/nbt1171. [DOI] [PubMed] [Google Scholar]
- 40.Verbaan FJ, Bos GW, Oussoren C, Woodle MC, Hennink WE, Storm G. A comparative study of different cationic transfection agents for in vivo gene delivery after intravenous administration. Journal of Drug Delivery Science and Technology. 2004;14(2):105–111. [Google Scholar]
- 41.Verbaan FJ, Oussoren C, van Dam IM, Takakura Y, Hashida M, Crommelin DJA, Hennink WE, Storm G. The fate of poly(2-dimethyl amino ethyl)methacrylate-based polyplexes after intravenous administration. International Journal of Pharmaceutics. 2001;214(1–2):99–101. doi: 10.1016/s0378-5173(00)00642-6. [DOI] [PubMed] [Google Scholar]
- 42.Li S, Tseng WC, Stolz DB, Wu SP, Watkins SC, Huang L. Dynamic changes in the characteristics of cationic lipidic vectors after exposure to mouse serum: implications for intravenous lipofection. Gene Therapy. 1999;6(4):585–594. doi: 10.1038/sj.gt.3300865. [DOI] [PubMed] [Google Scholar]
- 43.Duncan R, Izzo L. Dendrimer biocompatibility and toxicity. Advanced Drug Delivery Reviews. 2005;57(15):2215–2237. doi: 10.1016/j.addr.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 44.Kuo JHS, Jan MS, Chu HW. Mechanism of cell death induced by cationic dendrimers in RAW 264.7 murine macrophage-like cells. Journal of Pharmacy and Pharmacology. 2005;57(4):489–495. doi: 10.1211/0022357055803. [DOI] [PubMed] [Google Scholar]
- 45.Malik N, Wiwattanapatapee R, Klopsch R, Lorenz K, Frey H, Weener JW, Meijer EW, Paulus W, Duncan R. Dendrimers: Relationship between structure and biocompatibility in vitro, and preliminary studies on the biodistribution of I-125-labelled polyamidoamine dendrimers in vivo. Journal of Controlled Release. 2000;65(1–2):133–148. doi: 10.1016/s0168-3659(99)00246-1. [DOI] [PubMed] [Google Scholar]
- 46.Capan Y, Woo BH, Gebrekidan S, Ahmed S, DeLuca PP. Preparation and characterization of poly (D,L-lactide-co-glycolide) microspheres for controlled release of poly(L-lysine) complexed plasmid DNA. Pharmaceutical Research. 1999;16(4):509–513. doi: 10.1023/a:1018862827426. [DOI] [PubMed] [Google Scholar]
- 47.Munier S, Messai I, Delair T, Verrier B, Ataman-Onal Y. Cationic PLA nanoparticles for DNA delivery: Comparison of three surface polycations for DNA binding, protection and transfection properties. Colloid Surf B-Biointerfaces. 2005;43(3–4):163–173. doi: 10.1016/j.colsurfb.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 48.De Rosa G, Quaglia F, La Rotonda MI, Appel M, Alphandary H, Fattal E. Poly(lactide-co-glycolide) microspheres for the controlled release of oligonucleotide/polyethylenimine complexes. Journal of Pharmaceutical Sciences. 2002;91(3):790–799. doi: 10.1002/jps.10063. [DOI] [PubMed] [Google Scholar]
- 49.Pistel KF, Kissel T. Effects of salt addition on the microencapsulation of proteins using W/O/W double emulsion technique. J Microencapsul. 2000;17(4):467–483. doi: 10.1080/026520400405723. [DOI] [PubMed] [Google Scholar]
- 50.Crotts G, Park TG. Preparation of Porous and Nonporous Biodegradable Polymeric Hollow Microspheres. Journal of Controlled Release. 1995;35(2–3):91–105. [Google Scholar]
- 51.Thomas M, Klibanov AM. Non-viral gene therapy: polycation-mediated DNA delivery. Appl Microbiol Biotechnol. 2003;62(1):27–34. doi: 10.1007/s00253-003-1321-8. [DOI] [PubMed] [Google Scholar]
- 52.Jevprasesphant R, Penny J, Jalal R, Attwood D, McKeown NB, D’Emanuele A. The influence of surface modification on the cytotoxicity of PAMAM dendrimers. International Journal of Pharmaceutics. 2003;252(1–2) doi: 10.1016/s0378-5173(02)00623-3. PII S0378–5173(0302)00623–00623. [DOI] [PubMed] [Google Scholar]
- 53.Jevprasesphant R, Penny J, Attwood D, McKeown NB, D’Emanuele A. Engineering of dendrimer surfaces to enhance transepithelial transport and reduce cytotoxicity. Pharmaceutical Research. 2003;20(10):1543–1550. doi: 10.1023/a:1026166729873. [DOI] [PubMed] [Google Scholar]
- 54.Intra J, Glasgow JA, Mai HQ, Salem AK. Pulsatile release of biomolecules from polydimethylsiloxane (PDMS) chips with hydrolytically degradable seals. Journal of Controlled Release. 2008;127(3):280–287. doi: 10.1016/j.jconrel.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 55.Zhang XQ, Wang XL, Huang SW, Zhuo RX, Liu ZL, Mao HQ, Leong KW. In vitro gene delivery using polyamidoamine dendrimers with a trimesyl core. Biomacromolecules. 2005;6(1):341–350. doi: 10.1021/bm040060n. [DOI] [PubMed] [Google Scholar]
- 56.Zinselmeyer BH, Mackay SP, Schatzlein AG, Uchegbu IF. The lower-generation polypropylenimine dendrimers are effective gene-transfer agents. Pharmaceutical Research. 2002;19(7):960–967. doi: 10.1023/a:1016458104359. [DOI] [PubMed] [Google Scholar]
- 57.Ogris M, Brunner S, Schuller S, Kircheis R, Wagner E. PEGylated DNA/transferrin-PEI complexes: reduced interaction with blood components, extended circulation in blood and potential for systemic gene delivery. Gene Therapy. 1999;6(4):595–605. doi: 10.1038/sj.gt.3300900. [DOI] [PubMed] [Google Scholar]
- 58.Dash PR, Read ML, Barrett LB, Wolfert M, Seymour LW. Factors affecting blood clearance and in vivo distribution of polyelectrolyte complexes for gene delivery. Gene Therapy. 1999;6(4):643–650. doi: 10.1038/sj.gt.3300843. [DOI] [PubMed] [Google Scholar]