Abstract
The cohesin complex, discovered for its role in sister chromatid cohesion, also plays roles in gene expression and development in organisms from yeast to man. This review highlights what has been learned about the gene control and developmental functions of cohesin and the Nipped-B (NIPBL/Scc2) cohesin loading factor in Drosophila. The Drosophila studies have provided unique insights into the etiology of Cornelia de Lange syndrome (CdLS), which is caused by mutations affecting sister chromatid cohesion proteins in humans. In vivo experiments with Drosophila show that cohesin and Nipped-B have dosage-sensitive effects on the functions of many evolutionarily conserved genes and developmental pathways. Genome-wide studies with Drosophila cultured cells show that Nipped-B and cohesin co-localize on chromosomes, and bind preferentially, but not exclusively, to many actively-transcribed genes and their regulatory sequences, including many of the proposed in vivo target genes. In contrast, the cohesion factors are largely excluded from genes silenced by Polycomb group (PcG) proteins. Combined, the in vivo genetic data and the binding patterns of cohesin and Nipped-B in cultured cells are consistent with the hypothesis that they control the action of gene regulatory sequences, including transcriptional enhancers and insulators, and suggest that they might also help define active chromatin domains and influence transcriptional elongation.
Keywords: cut gene, bithorax complex, ecdysone receptor, trithorax group, Polycomb group, insulator, CTCF, Cornelia de Lange syndrome
Introduction
The subunits of the cohesin complex (Figure 1) were discovered in genetic screens in fungi for mutations that cause defects in sister chromatid cohesion (Guacci et al. 1997; Michaelis et al. 1997). The structure, regulation and diverse functions of cohesin are reviewed in other articles in this issue (Austin et al., Gartenberg, MacNairn and Gerton, Sakuno and Watanabe, Wendt and Peters). This article focuses on the roles of cohesin and its regulatory factors in gene expression and development in the fruitfly, Drosophila melanogaster.
Figure 1.
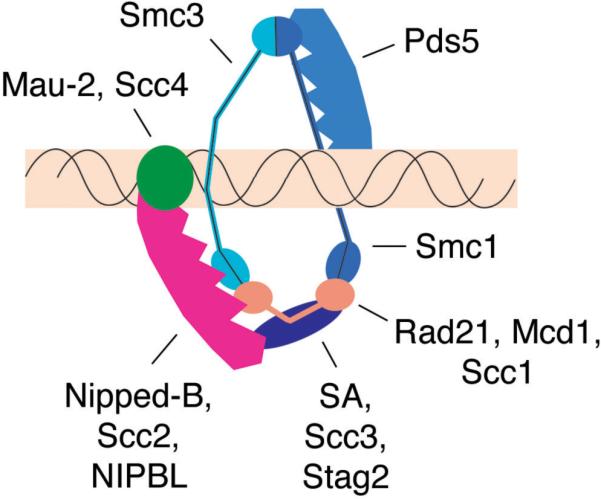
The cohesin complex and regulatory factors. Proteins are labeled with the Drosophila names, followed by the S. cerevisiae and human names, if different. The core cohesin complex forms a ring-like structure containing a heterodimer of the Smc1 and Smc3 proteins, Rad21 (Mcd1/Scc1) and Stromalin (SA/Scc3) (Anderson et al. 2002, Gruber et al. 2003) that can encircle a chromosome. The Nipped-B-Mau-2 (Scc2-Scc4) complex is required for cohesin to bind to chromosomes (Ciosk et al. 2000, Tomonaga et al. 2000), and Pds5 is required to establish and maintain sister chromatid cohesion (Hartman et al. 2000, Panizza et al. 2000, Dorsett et al. 2005). The depicted interactions between cohesin and the Nipped-B-Mau-2 complex, and with Pds5 are speculative, and based on the findings that Mau-2 interacts with the N terminal region of Nipped-B (Seitan et al. 2006, Takahashi et al. 2008), that the N terminal portion of Nipped-B and Mau-2 bind chromatin (Takahashi et al. 2008), that Nipped-B complexes with cohesin subunits (Arumugam et al. 2003, Gause et al. 2008b, Takahashi et al. 2008), and that Nipped-B and cohesin co-localize genome-wide (Misulovin et al. 2008). The Pds5 protein co-localizes with cohesin on chromosomes (Hartman et al. 2000, Panizza et al. 2000, Gause et al. 2008b), interacts with cohesin (Sumara et al. 2000), and requires cohesin to bind to chromosomes (Hartman et al. 2000, Panizza et al. 2000). Förster resonance energy transfer experiments indicate that Pds5 interacts with the hinge region where Smc1 and Smc3 dimerize (Mc Intyre et al. 2007).
Similar to other SMC (Structural Maintenance of Chromosome) complexes, cohesin contains a heterodimer of two SMC proteins and other factors. In cohesin these are the Smc1–Smc3 heterodimer, the Rad21 kleisin protein, and Stromalin (Figure 1). Cohesin is structurally distinct from other SMC complexes in that a kink in the arm of Smc3 creates an open ring-like structure, with Rad21 linking the Smc1 and Smc3 ATPase head domains (Figure 1). There is substantial evidence that the cohesin ring encircles chromosomes (Ivanov and Nasmyth 2005, Ivanov and Nasmyth 2007, Haering et al. 2008), supporting the ideas that cohesin mediates sister chromatid cohesion by encircling both sisters, or by interactions between two cohesin rings encircling different sisters.
The Nipped-B protein and its Mau-2 partner (Figure 1) are required for cohesin to associate with chromosomes, and the Eco1 and Pds5 proteins are needed to establish and/or maintain sister chromatid cohesion. Cohesin is removed from chromosome arms during prophase, and from centromeres at the metaphase-to-anaphase transition, to permit sister separation and cell division.
In most organisms, cohesin rebinds to chromosomes immediately after cell division in telophase. Thus cohesin binds extensively along chromosomes throughout the entire interphase of the cell cycle. Thus it is not altogether surprising that it has been discovered that cohesin regulates gene expression. As described in other articles in this issue, cohesin influences the function of silencing proteins in yeast (Gartenberg), and helps mediate CTCF protein insulator activity in mouse and human cells (Wendt and Peters).
Studies in Drosophila have revealed that cohesin, Nipped-B, and Pds5 regulate gene expression, and that this regulation is important for organismal development. Based on the Drosophila studies described below, it seems likely that multiple changes in gene expression underlie the many developmental deficits, such as slow growth, mental retardation, autism, limb and organ structural defects, that occur in Cornelia de Lange syndrome (CdLS, Dorsett 2007, Liu and Krantz 2008, Dorsett and Krantz 2009). CdLS is caused by heterozygous loss-of-function mutations in the human NIPBL ortholog of Nipped-B and Scc2 in at least half the cases (Krantz et al. 2004, Tonkin et al. 2004). In some 5% of cases, CdLS is caused by amino acid changes in the Smc1 or Smc3 cohesin subunits, indicating that the role of NIPBL in human development involves regulation of cohesin function (Musio et al. 2006, Deardorff et al. 2007). This article first reviews several examples of genetic evidence showing that sister chromatid cohesion factors control segmental identity, and development of limbs and nervous system in Drosophila. This is followed by examination of the molecular data supporting the idea that the diverse in vivo effects on development are through direct regulation of gene expression, and discussion of some of the potential mechanisms by which such regulation might occur.
In vivo effects of sister chromatid cohesion factors on Drosophila gene expression and development
The cut gene and wing margin development
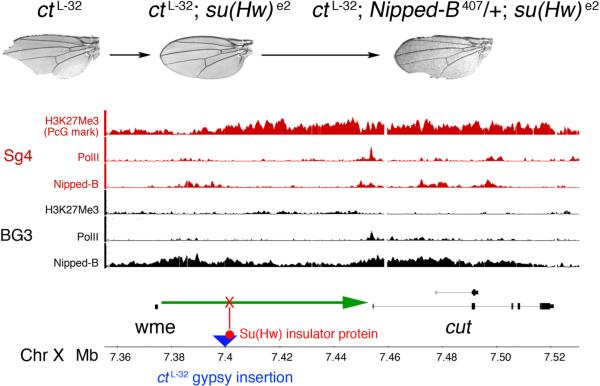
The first evidence that cohesion factors regulate gene expression and development in eumetazoa arose from studies on the Drosophila cut gene (Figure 2, Rollins et al. 1999). cut encodes a homeobox protein involved in many developmental processes, and has human orthologs with similar functions (Nepveu 2001, Sansregret and Nepveu 2008). During early wing development, the Notch receptor signaling pathway activates cut in a thin strip of cells located between the cells that will eventually form the dorsal and ventral surfaces of the adult wing. The cut-expressing cells give rise to the bristle-forming cells around adult wing margin, and loss of cut expression results in loss of these margin cells (Figure 2).
Figure 2.
The cut locus and Nipped-B/cohesin binding patterns in Sg4 and BG3 cells. A diagram of the cut gene is at the bottom. cut is activated (green arrow) in wing margin precursor cells by the wing margin enhancer (wme) located more than 80 kbp upstream of the transcription start site (Jack et al. 1991). The ctL-32 gypsy insertion (blue triangle) into the region between the wing margin enhancer and the cut gene binds the Su(Hw) insulator protein (red) that blocks (red X) enhancer-promoter communication, causing loss of adult wing margin cells (top left, Jack et al. 1991). Reduction of Su(Hw) activity by the weak su(Hw)e2 mutation partially restores cut expression and suppresses the cut wing phenotype (top middle). Reduction of Nipped-B dosage in ctL-32; su(Hw)e2 flies decreases cut expression, causing partial loss of the adult wing margin (top right, Rollins et al. 1999). The red (Sg4 cells) and black (BG3 cells) tracks show chromatin immunoprecipitation (ChIP-chip) results for histone H3 lysine 27 trimethylation (H3K27Me3, a mark of PcG silencing), RNA polymerase II (PolII) and Nipped-B, on a log2 IP/control scale of −0.5 to 3. The cohesin (Smc1) ChIP pattern (not shown) is virtually indistinguishable from the Nipped-B pattern (Misulovin et al. 2008). In the Sg4 cell line of embryonic origin, cut is a Polycomb group (PcG) silencing target, showing high H3K27Me3 PcG silencing mark (Schwartz et al. 2006), and lower Nipped-B and cohesin binding.
Activation of cut in the developing wing margin in response to Notch receptor signaling is mediated by a transcriptional enhancer located more than 80 kbp upstream of the cut transcription start site (Figure 2, Jack et al. 1991). In one of the first descriptions of insulators, it was shown that the ability of this distant enhancer to activate cut is impeded by several independent natural insertions of the gypsy retrovirus into the region between the enhancer and promoter (Figure 2, Jack et al. 1991). Gypsy binds the Suppressor of Hairy-wing [Su(Hw)] insulator protein, and long-range enhancer-promoter communication and the wing margin phenotype of cut gypsy insertion alleles vary quantitatively with the in vivo activity of Su(Hw) over a several hundred-fold range (Dorsett 1993).
The quantitative nature of Su(Hw) insulation was exploited to identify genes that control long-range transcriptional activation of cut. A gypsy insertion allele of cut was partially suppressed by a weak su(Hw) mutation, and mutations that dominantly decrease cut activation (or increase insulator activity) were isolated by virtue of the increased loss of adult wing margin (Figure 2, Morcillo et al. 1996, Rollins et al. 1999). These screens identified several loss-of-function alleles of Nipped-B, named for the gaps in the adult wing margin (Figure 2). Although the Nipped-B mutations were isolated in a screen using a gypsy insertion, they also reduce expression of wild-type cut alleles, indicating that Nipped-B normally regulates cut. Based on several lines of genetic evidence, it was postulated that Nipped-B is particularly critical for long-range enhancer-promoter communication (Rollins et al. 1999).
Nipped-B encodes a homolog of the S. cerevisiae Scc2 and S. pombe Mis4 proteins, which are required for sister chromatid cohesion and binding of cohesin to chromosomes (Ciosk et al. 2000, Tomonaga et al. 2000). Heterozygous Nipped-B mutants, which have reduced cut expression in the wing margin, are viable and do not show chromatid cohesion defects. In contrast, homozygous Nipped-B mutants survive only a few days into development and display significantly reduced sister chromatid cohesion prior to death, confirming that Nipped-B is a true Scc2/Mis4 ortholog (Rollins et al. 2004, Gause et al. 2008b).
Identification of Nipped-B as a sister chromatid cohesion protein raised the question of whether cohesin also participates in long-range activation of cut. Because Nipped-B facilitates both long-range activation and cohesin binding to chromosomes, it was expected that cohesin would have the same effect as Nipped-B. In contrast to this expectation, however, non-lethal in vivo reduction of the Rad21, SA or Smc1 cohesin subunit dosage by RNAi or mutation increases expression of cut in the developing wing (Rollins et al. 2004, Dorsett et al. 2005).
Based on these results, it was hypothesized that cohesin functions as an insulator, inhibiting enhancer-promoter communication in cut, and that Nipped-B facilitates enhancer-promoter interaction by dynamically altering cohesin binding (Dorsett 2004). As described by Wendt and Peters in this issue, this hypothesis is supported by recent studies showing that cohesin contributes to the insulator activity of the CTCF (CCCTC-binding factor) zinc finger protein in mouse and human cells (Parelho et al. 2008, Wendt et al. 2008). It is also consistent with the findings described later, that Nipped-B and cohesin bind to the regulatory and transcribed regions of cut in cultured cells, and that the binding pattern varies with transcription (Misulovin et al. 2008).
Although the eco and separation anxiety (san) Drosophila homologs of Eco1 are required for centromeric cohesion (Williams et al. 2003), unlike the cohesin subunits, they do not have dosage-sensitive effects on cut expression (Dorsett et al. 2005). More recently, it has been suggested that the Scc2 yeast ortholog of Nipped-B is involved in the chromosomal loading and/or positioning of the condensin (D'Ambrosio et al. 2008) and Smc5/6 DNA repair (Lindroos et al. 2006) SMC complexes. Condensin-like complexes regulate gene expression in C. elegans (Ercan and Lieb, this issue; Meyer 2005), and the Barren condensin subunit has been implicated in gene silencing in Drosophila (Lupo et al. 2001). Mutations affecting the Smc5/6 proteins are not available, but mutations in the Drosophila barren and gluon genes encoding condensin subunits, do not have dominant effects on cut expression (R.A. Rollins and D.D., unpublished).
Other genetic evidence supports the idea that Nipped-B regulates cut expression through control of cohesin. The Pds5 protein was identified in genetic screens in S. cerevisiae for defects in sister chromatid cohesion (Hartman et al. 2000, Panizza et al. 2000), and like cohesin and Nipped-B, it is also conserved from yeast to man. Similar to its orthologs in other organisms, Drosophila Pds5 is required for sister chromatid cohesion, but not for binding of cohesin to chromosomes, and it co-localizes with cohesin on chromosomes (Dorsett et al. 2005, Gause et al. 2008b). A null pds5 allele dominantly decreases cut expression in the developing wing margin to a slight extent (Dorsett et al. 2005). In contrast to the null allele, an allele predicted to encode a Pds5 protein lacking the N terminus increases cut expression. This truncation allele also diminishes cohesin binding to salivary gland polytene chromosomes when homozygous, suggesting that the mutant protein may destabilize or block binding of cohesin to chromosomes. The Pds5 data thus further suggest that cohesin interferes with cut expression in the developing wing margin.
Recently it was shown that homozygous mutations in Pds5B, one of the two mouse Pds5 genes, cause perinatal lethality and several developmental abnormalities reminiscent of some seen in Cornelia de Lange syndrome (Zhang et al. 2007). No defects in sister chromatid cohesion were detected, and thus these findings indicate that Pds5 proteins also likely regulate genes in mammals.
Ultrabithorax (Ubx), the bithorax complex (BX-C), and segmental identity
The bithorax complex (BX-C) contains three homeobox (HOX) genes, Ubx, abdominal-A (abd-A) and Abdominal-B (Abd-B) that control segmental identity, and limb and organ development (Maeda and Karch 2006). The BX-C genes, and the transcriptional enhancers that activate them, occupy nearly 320 kbp and are arranged along the chromosome in the anterior-to-posterior order of the thoracic and abdominal segments in which they are expressed, and whose identity they determine (Figure 3). This gene order is conserved in the mammalian HOX gene complexes.
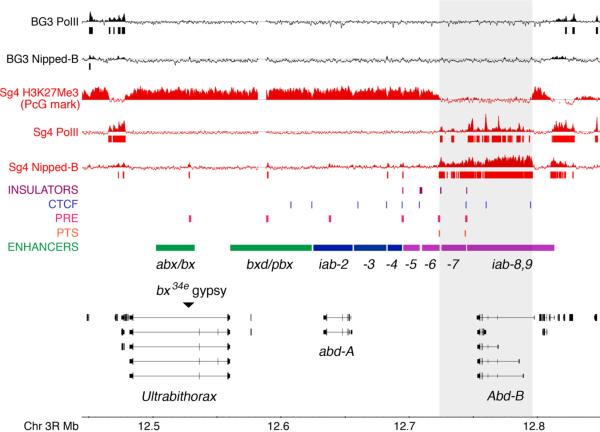
Figure 3.
The bithorax HOX gene complex (BX-C) and Nipped-B/cohesin binding patterns in Sg4 and BG3 cells. The bottom shows a diagram of the BX-C, which contains the Ultrabithorax (Ubx), abdominal-A(abd-A) and Abdominal-B (Abd-B) genes (transcribed from right to left). Ubx is activated by the abx/bx and bxd/pbx region transcriptional enhancers (green), abd-A is activated by the iab-2,3,4 enhancers (blue), and Abd-B is activated by the iab-5,6,7,8,9 enhancers (violet), shown above the gene diagrams. The genes and enhancers are in the order of the body segments in which they function: Ubx is expressed in parasegments 5–12, abd-A in parasegment 7 to parasegment 12, and Abd-B in parasegment 10 to parasegment 14. The actions of the enhancers are controlled by several insulator elements (violet), some of which bind CTCF (light blue, Holohan et al. 2007). In Abd-B, the iab-6 and iab-7 enhancers bypass the intervening insulators separating them from the transcription start sites with the aid of Promoter Targeting Sequences (PTS, orange). Several Polycomb Response Elements (PRE, rose) bind the PcG silencing complexes and some activating trithorax group (trxG) proteins. The bx34e gypsy insertion mutation (black triangle) blocks activation of Ubx by the downstream abx/bx enhancers, causing abdominal to thoracic transformations. Nipped-B mutations magnify the effect of the bx34e gypsy, increasing the severity of the segmental transformation (Figure 4). The tracks at the top show chromatin immunoprecipitation (ChIP-chip) data for BG3 (black) and Sg4 (red) cells. Bars immediately underneath the PolII and Nipped-B ChIP-chip profiles indicate where binding is detected with a p value of 10−4 or less (G. Sahota, G. Stormo, D.D., unpublished). In Sg4 cells, Abd-B is highly transcribed, and shows reduced levels of the H3K27Me3 PcG silencing mark over a region starting at a CTCF binding site near the distal transcription start site, extending down to the Fab-7 insulator located between the iab-6 and iab-7 enhancers (Schwartz et al. 2006). PolII, Nipped-B and cohesin bind to the region with reduced H3K27Me3.
Segmentation genes control expression of the BX-C genes during early embryogenesis, and beginning in mid-embryogenesis, they are regulated positively by trithorax group (trxG) genes, and negatively by Polycomb group (PcG) genes. PcG proteins, and some trxG proteins, bind to Polycomb Response Elements (PRE), or “maintenance elements” located in multiple locations in the BX-C (Figure 3).
In addition to the enhancers and PREs, several insulators, or boundary elements, are required for proper regulation of the BX-C, and some are tightly linked to PREs (Figure 3). The insulators in the BX-C require different proteins, including GAGA factor encoded by the Trithorax-like (Trl) gene and CTCF. In general, these insulators prevent inappropriate activation of individual BX-C genes by enhancers for the neighboring gene. Some of the enhancers that activate Abd-B are separated from the Abd-B transcription start sites by insulators, but in these cases, the insulators attenuate activation, and there are Promoter Targeting Sequences (PTS) that enable the enhancers to bypass the insulators and target the promoters in an epigenetic manner (Figure 3, Zhou and Levine 1999, Lin et al. 2004, Chen et al. 2005, Lin et al. 2007).
Reduced Ubx expression in parasegment 5 causes abdominal to thoracic transformations. Ubx expression in this parasegment is regulated by the abx/bx enhancers located several kilobases downstream of the transcription start site (Simon et al. 1990, Qian et al. 1991), and the bx34e gypsy retrovirus insertion (Peifer and Bender 1986) reduces activation of Ubx by these enhancers (Figure 3). The segmental transformation caused by the bx34e gypsy insertion is reversed by reduced Su(Hw) insulator protein activity. Reducing Nipped-B dosage or activity, however, strongly magnifies the effect of the weakened insulator, and increases the segmental transformation (Figure 4, Rollins et al. 1999, Gause et al. 2008b). Thus Nipped-B also facilitates long-range gene activation in the BX-C. As described below, Nipped-B and cohesin bind to the Abd-B gene and its 3' regulatory domain in cultured cells in which Abd-B is highly transcribed, but are absent from Abd-B and other BX-C genes, including Ubx, when they are silenced by PcG proteins (Misulovin et al. 2008).
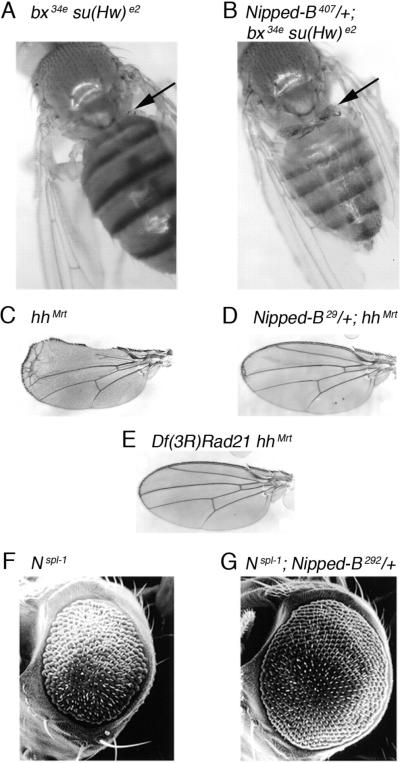
Figure 4.
Dominant effects of Nipped-B and Rad21 (verthande) mutations on mutant developmental phenotypes. (A) Dorsal view of adult fly with the bx34e gypsy transposon insertion in the Ultrabithorax (Ubx) gene (Figure 3) partially suppressed by a weak su(Hw) mutation. The arrow points to bristles showing a weak abdominal to thoracic transformation. The fly shown also has a heterozygous Nipped-A mutation that does not affect the phenotype (Rollins et al. 1999). (B) Suppressed bx34e in the presence of a heterozygous loss-of-function Nipped-B mutation. The arrow points to additional cuticle and bristles caused by an increased abdominal to thoracic transformation. (C) hhMrt gain-of-function mutant wing showing transformation and overgrowth of anterior compartment. (D) Suppression of hhMrt wing phenotype by heterozygous Nipped-B mutation. (E) Suppression of hhMrt wing phenotype by heterozygous deletion of Rad21 (vtd). (F) Nspl-1 mutant eye, showing small rough phenotype. (G) Nspl-1 eye with heterozygous loss-of-function Nipped-B mutation. Panels A,B,F, and G are taken from Rollins et al. (1999) Genetics 152: 577–593 with permission, and panels C–E are from Hallson et al. (2008) Proc Natl Acad Sci USA 105: 12405–12410 with permission.
Mutations in the PcG genes that repress BX-C gene expression often cause dominant mild segmental transformations in adults. For example, Polycomb (Pc) loss-of-function alleles cause extra sets of sex comb bristles that are normally specific for the anterior set of legs to form on the middle and posterior legs in males. Loss-of-function mutations in the verthande (vtd) gene were isolated in a screen for trxG mutations that dominantly suppress the dominant extra sex comb phenotypes of Pc mutations (Kennison and Tamkun 1988). Recently it was shown that the vtd trxG gene encodes the Rad21 subunit of the cohesin complex, and that Nipped-B mutations also dominantly suppress Pc dominant phenotypes (Hallson et al. 2008). These data indicate that Nipped-B and Rad21 both positively regulate BX-C expression in developing legs. This contrasts with cut, where Nipped-B and Rad21 have opposing effects, and suggest that the effect of cohesin on gene expression is context-dependent.
Rad21 (vtd) mutations even have opposite effects on the different phenotypes displayed by a single PcG gene mutation. The Enhancer of zeste [E(z)] PcG gene encodes the methyltransferase responsible for trimethylation of the lysine 27 residue of histone H3 (H3K27Me3) that coats PcG-silenced loci (Müller et al. 2002, Kahn et al. 2006, Schwartz et al. 2006). The E(z) trithorax mimic allele [E(z)Trm] is a gain-of-function missense mutation that causes E(z) to target BX-C genes when they should not be silenced (Bajusz et al. 2001). The phenotypes caused by E(z)Trm thus include thoracic and abdominal segmental transformations, recognized by bristle and pigmentation changes. The vtd2 loss-of-function allele of Rad21 dominantly increases the penetrance of the abdominal transformation, as do several other trxG mutations, but unlike most trxG mutations, reduces the frequency the thoracic transformations (Bajusz et al. 2001). These results further indicate that cohesin regulates the BX-C and segmental identity. If as the evidence suggests, cohesin directly regulates the BX-C, then its effects on expression are more context-dependent than those of most trxG proteins.
Rad21 also has context-dependent effects on gene expression in vertebrates. In early zebrafish embryogenesis, the runx1 gene is involved in haematopoietic and neural development. Homozygous rad21 mutations reduce runx1 expression in haematopoietic precursor cells, but not in most of the mechanosensory neurons (Horsfield et al. 2007). Expression of the runx3 gene, which is also expressed in the mechanosensory neurons, is lost in rad21 mutants, indicating that Rad21 still regulates other genes in these cells. Thus, runx1 expression is regulated by cohesin in some cells and not others. Expression of runx1 is also reduced in heterozygous rad21 mutants, indicating that expression of some genes is sensitive to cohesin dosage as in Drosophila.
Hedgehog (Hh) and wing development
The hedgehog protein (Hh) is a highly conserved signaling molecule that regulates several developmental processes. Hh is normally expressed in the posterior compartment of the developing wing, but the Moonrat gain-of-function hh mutation (hhMrt) causes expression in the anterior compartment (Felsenfeld and Kennison 1995). This results in variable overgrowth of the anterior compartment in heterozygous hhMrt mutants (Figure 4). Both Rad21 (vtd) and Nipped-B loss-of-function mutations dominantly suppress the hhMrt mutant wing phenotype (Figure 4, Schulze et al. 2001, Hallson et al. 2008), indicating that hh, or other genes in the hh signaling pathway, are regulated by cohesin and Nipped-B.
Enhancer of split gene complex [E(spl)-C] and eye development
Effects of Nipped-B on Drosophila eye development have been observed, and likely involve regulation of the Enhancer of split gene complex. The Notch receptor plays early and late roles in the development of photoreceptors in the eye (Baker et al. 1996, Baker and Yu 1997). First, Notch activates basic helix-loop-helix (bHLH) DNA-binding proneural genes, creating groups of neural-competent cells that have the potential to become photoreceptors. Later, similar to its role in “lateral inhibition” during bristle development, Notch directly activates repressor-encoding genes in the E(spl)-C (Nelleson et al. 1999), which repress proneural genes, such that only one cell in the cluster retains the neural fate. Many of the E(spl)-C genes encode bHLH proteins that can interact directly with proneural proteins.
The Nspl-1 mutation in the Notch receptor gene causes small rough adult eyes by reducing the number of photoreceptors (Nagel and Preiss 1999). E(spl)D is a dominant gain-of-function mutation in one of the E(spl)-C genes that strongly enhances the eye phenotype, making Nspl-1 dominant (Welshons 1956, Nagel and Preiss 1999). Reducing E(spl)-C dosage diminishes the effect of E(spl)D, and reducing proneural gene dosage increases its effect (Nagel and Preiss 1999). E(spl)D encodes a hyperactive protein that over-represses proneural activity and reduces photoreceptors, possibly through direct interactions with proneural proteins (Nagel et al. 1999, Nagel and Preiss 1999). Thus the leading ideas are that Nspl-1 reduces eye size through reduced early activation of proneural genes, and/or by hyperactivation of the E(spl)-C.
Nipped-B mutations dominantly suppress Nspl-1, increasing the size of the adult eye (Figure 4, Rollins et al. 1999). The simplest explanation is that Nipped-B mutations reduce expression of genes in the E(spl)-C, although it is also possible that they could increase proneural gene expression. It is unlikely that Nipped-B mutations alter expression of Notch, because they do not affect other sensitive Notch mutant phenotypes (Rollins et al. 1999). Rad21 (vtd) mutations have the opposite effect as Nipped-B mutations, and reduce the size of Nspl-1 mutant eyes (C.A. Schaaf and D.D., unpublished), providing another case where Nipped-B and cohesin may have opposing effects on gene expression. As described below, Nipped-B and cohesin coat the entire E(spl)-C in cells derived from larval nervous system (Figure 5, Misulovin et al. 2008), supporting the idea that they directly regulate the E(spl)-C.
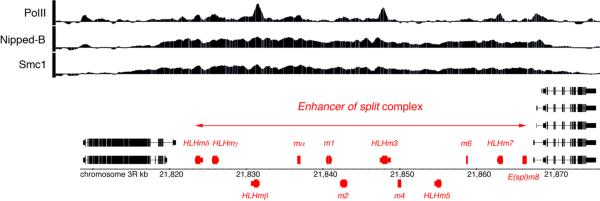
Figure 5.
The Enhancer of split gene complex [E(spl)-C] and Nipped-B/cohesin binding patterns in BG3 cells. The E(spl)-C contains twelve genes (red) that encode bHLH or bearded family proteins involved in suppressing neural fates in developing nervous system. The ChIP-chip profiles show that Nipped-B and cohesin bind to the full 50 kbp length of the active E(spl)-C in BG3 cells derived from central nervous system (Misulovin et al. 2008). Decreased expression of the E(spl)-C might cause the suppression of the Nspl-1 mutant eye phenotype by Nipped-B mutations as depicted in Figure 4.
Trans-generational effects on the Irregular facets allele of Krüppel (KrIf-1) and eye development
The Krüppel (Kr) zinc finger protein regulates development of many tissues, beginning with its role as a gap gene during early embryogenesis. KrIf-1 is a dominant gain-of-function allele that causes ectopic expression in the developing eye, reducing its size (Carrera et al. 1998, Abrell et al. 2000). Loss-of-function mutations in certain trxG genes, including Rad21 (vtd), cause ectopic outgrowths in KrIf-1 mutant eyes (Sollars et al. 2003). Of all the mutations tested, Rad21 (vtd3) has by far the strongest effect, causing eye outgrowths in over half the progeny. The most intriguing observation is that the vtd3 (Rad21) mutation has an epigenetic effect on KrIf-1 expression (Sollars et al. 2003). The eye outgrowths are seen in the next generation, even in many of the progeny that do not inherit the vtd3 mutation. By breeding progeny that have outgrowths but lack the vtd3 mutation, it was shown that this effect continues for at least five generations. Although these experiments need to be performed with mutations affecting other cohesin subunits, they raise the intriguing likelihood that cohesin has epigenetic effects on gene expression and development.
Ecdysone receptor (EcR) and axon pruning in post-mitotic neurons
The mushroom body is a structure in the Drosophila brain that mediates olfactory learning and memory. Like many other neurons, the γ neuron of the mushroom body extends excess axons, and then prunes them back. A genetic screen for mutations that block axon pruning in the γ neuron recovered loss-of-function mutations in the Smc1 and Stromalin (SA) cohesin subunit genes (Schuldiner et al. 2008). Neurons homozygous mutant for the cohesin subunit genes generated by mitotic recombination fail to prune their axons. Axon pruning was rescued by expressing Smc1 specifically in postmitotic neurons, demonstrating that the loss of pruning does not result from reduced cell proliferation (Schuldiner et al. 2008).
The effects of cohesin on axon pruning in the mushroom body γ neuron were confirmed using an engineered Tobacco Etch mosaic Virus (TEV) protease-sensitive Rad21 cohesin subunit (Rad21TEV, Pauli et al. 2008). A Rad21 (vtd) mutation was fully rescued by a transgene encoding Rad21TEV. Ubiquitous expression of TEV protease, which has no obvious effect on wild-type flies, is lethal and causes release of cohesin from chromosomes in Rad21TEV-rescued flies. Expression of TEV protease specifically in post-mitotic neurons is also lethal in Rad21TEV flies, and blocks pruning of the γ neuron axons (Pauli et al. 2008). Combined, the studies on the mushroom body γ neuron show definitively that cohesin plays an important role in the morphogenesis of a non-dividing cell, and thus is unlikely to involve its role in chromosome segregation.
The role of cohesin in axon pruning in the mushroom body γ neuron likely involves regulation of the EcR (ecdysone receptor) gene, which encodes a homolog of mammalian nuclear steroid hormone and retinoic acid receptors (Schuldiner et al. 2008). EcR is expressed in the mushroom body γ neuron, and is required for axon pruning (Lee et al. 2000). Strikingly, EcR levels are reduced in Smc1 mutant γ neurons, and targeted expression of EcR in post-mitotic neurons rescues the pruning defect (Schuldiner et al. 2008). As described below, cohesin and Nipped-B bind to the active EcR gene in cultured cells, and thus it is predicted that the effect of Smc1 mutations on EcR-B1 protein levels in the γ neurons results from reduced transcription.
Developmental effects of cohesin are also seen in other Drosophila neurons, in addition to the mushroom body γ neurons and eye photoreceptors, although candidate target genes responsible for these effects have yet to be identified. Smc1 and SA mutations were also isolated in screens for mutations that cause defects in dendrite targeting by olfactory projection neurons (Schuldiner et al. 2008), and targeted expression of TEV protease in cholinergic neurons of Rad21TEV-rescued flies causes defects in larval locomotion (Pauli et al. 2008).
Do the effects of cohesion proteins on Drosophila development involve disruption of sister chromatid cohesion or changes in cell proliferation?
Several lines of evidence argue that the in vivo effects of cohesin, Nipped-B, and Pds5 on Drosophila development detailed above result from effects on gene expression, and that the effects on gene expression are unlikely to result from defects in sister cohesion or cell proliferation. In the cases of EcR expression, axon pruning, and dendrite formation in the mushroom body, although the cohesin subunit mutations are homozygous, the cells are non-dividing, and thus the chromosome segregation function is not involved. Most convincingly, expression of the missing cohesin subunit specifically in postmitotic neurons reverses the pruning defect (Pauli et al. 2008, Schuldiner et al. 2008). It is unknown, however, if the postmitotic neurons go through S phase before they differentiate (Liqun Luo, personal communication), and thus it is possible that sister chromatid interactions could be important for gene expression.
In the cases of cut, Ubx, Pc, E(z)Trm, hhMrt, KrIf-1 and Nspl-1, changes in Nipped-B and/or cohesin dosage that are too small to cause overt defects in sister chromatid cohesion (Rollins et al. 2004, Hallson et al. 2008) alter expression. Many of these changes in dosage are even smaller than might be expected - heterozygous Nipped-B null mutations cause only a 25 to 30% reduction in Nipped-B mRNA (Rollins et al. 2004). Reduction of Drosophila Nipped-B mRNA by 50% using in vivo RNAi is lethal, also without causing visible cohesion defects (Rollins et al. 2004).
Intriguingly, the partial dosage compensation of Nipped-B seen in Drosophila is also seen in tissues from heterozygous NIPBL mutant mice (Arthur Lander and Anne Calof, personal communication) and cell lines derived from CdLS patients with NIPBL mutations (Jinglan Liu and Ian Krantz, personal communication). All cases show a maximum reduction of 30% in NIPBL mRNA. This suggests that there is a conserved compensation mechanism to maintain Nipped-B/NIPBL levels.
Although it is possible that there are subtle effects on cohesion that might alter gene expression and development with reduced Nipped-B and cohesin subunit dosage, certain observations argue against the notion that minor changes in sister cohesion underlie the effects on gene expression. If they have a subtle effect, reducing Nipped-B or cohesin dosage would both be expected to slightly reduce sister cohesion, but as described above, they have opposing effects on development in some cases. Also, two pds5 gene mutations that cause equivalent recessive defects in sister chromatid cohesion have opposite dominant effects on cut expression (Dorsett et al. 2005). Thus it seems likely that the effects of cohesion factors on gene expression also occur in G1, and not just in the S or G2 phases of the cell cycle, when cohesion occurs.
Although the effects of Nipped-B and cohesin dosage on gene expression are unlikely to involve changes in sister chromatid cohesion, the role of Nipped-B in gene expression appears to involve the same molecular activity that is required for its role in cohesion. This idea is based upon the finding that missense Nipped-B mutations show both milder dominant effects on gene expression and weaker homozygous effects on cohesion compared to null or truncation alleles (Gause et al. 2008b).
Nipped-B and cohesin associate with active genes and are absent from Polycomb group-silenced genes
The in vivo data described above indicate that Nipped-B and cohesin regulate diverse and conserved developmental pathways in Drosophila, and that most of these developmental roles likely involve regulation of gene expression. The finding that small reductions in Nipped-B or cohesin dosage have particularly strong effects on specific mutant alleles of proposed target genes, such as those documented with various cut alleles, argues strongly that Nipped-B and cohesin directly regulate some of the proposed gene targets. It is essential, however, to obtain molecular data to determine if the regulation is direct, and also to gain insights into the mechanisms by which cohesion factors regulate genes.
A first step towards this goal was the genome-wide mapping of Nipped-B and cohesin in Drosophila cell lines by genome-wide chromatin immunoprecipitation using tiled microarrays (ChIP-chip), and comparison of their binding patterns to those of RNA polymerase II (PolII) and PcG silencing (Misulovin et al. 2008). One of the cell lines, Sg4, is embryonic in origin, and another, MLDmBG3 (BG3), is derived from larval central nervous system. There are differences in gene expression between these lines, which as described below, provide key insights.
Contrary to what is seen in yeast, where cohesin loads at Scc2 (Nipped-B) binding sites and translocates away (Lengronne et al. 2004), Nipped-B and cohesin co-localize virtually completely throughout the non-repetitive genome (Misulovin et al. 2008), consistent with the notion that Nipped-B could dynamically regulate cohesin chromosome-binding. Also contrary to what occurs in yeast, where cohesin binds mostly between convergently transcribed genes (Lengronne et al. 2004, Glynn et al. 2004), Nipped-B and cohesin bind to transcribed regions (Misulovin et al. 2008). This is similar, however, to what is seen in mouse and human cells, where cohesin also binds transcribed regions (Parelho et al. 2008, Wendt et al. 2008).
The reasons for the differences between yeast and higher eukaryotes, in the co-localization of the Nipped-B/Scc2 cohesin loader, and the binding of cohesin to transcribed genes, are unknown. The answers may lie in differences in chromatin structure, or genome organization. Transcription might facilitate cohesin binding by unwinding higher order chromatin structures in higher organisms, while this is probably unnecessary in yeast, where chromosomes likely have a more open chromatin configuration. Yeast also have little repetitive DNA and intergenic sequences, and few long-range enhancers or introns. Thus the positions of cohesin binding regions between genes might have occurred by selection to reduce transcriptional interference.
Drosophila Nipped-B and cohesin prefer transcription units, binding to intron sequences about 30 to 45% more than expected if binding was random, and to 5' UTRs some 6 to 8-fold more than expected (Misulovin et al. 2008). Although Nipped-B and cohesin bind preferentially to transcription units, they also bind intergenic regions, but about 20 to 30% less than expected if binding were random. This is similar to human cells, where cohesin binds some 14% less to intergenic regions than expected on a random basis (Wendt et al. 2008). Although ChIP-chip studies in mouse suggest that binding might be equally distributed between genes and intergenic regions, these studies covered only 3% of the genome, and many intergenic cohesin binding regions are close to genes (Parelho et al. 2008).
Drosophila Nipped-B and cohesin bind preferentially to actively-transcribed genes. There is a strong genome-wide correlation and overlap between Nipped-B/cohesin and RNA polymerase II (PolII) binding, although most of actual peaks of binding for PolII and Nipped-B/cohesin differ (Misulovin et al. 2008). When Nipped-B/cohesin binding to genes in the region spanning from 10 kbp upstream to 10 kbp downstream of genes is averaged genome-wide, the peak is at the transcription start site (Misulovin et al. 2008), similar to what is seen with cohesin at start sites in mouse cells (Parelho et al. 2008).
Importantly, differences in gene expression between Drosophila cell lines also correlate with differences in cohesin binding. In the vast majority of the hundred or so genes that were found to bind PolII in one cell type and not another, cohesin binds to the gene in those cells in which it also binds PolII (Misulovin et al. 2008).
Although Nipped-B and cohesin associate preferentially with active genes, they do not bind all active genes. Only some 20% to 35% of transcription units that bind PolII in BG3 and Sg4 cells also bind cohesin (Gurmukh Sahota, Gary Stormo, DD, unpublished). The factors that determine which active genes bind Nipped-B and cohesin are currently unknown.
In addition to preferential association of cohesin and Nipped-B with a subset of active genes, they are virtually absent from genes silenced by PcG proteins. PcG silenced genes were identified in Sg4 cells by ChIP-chip for PcG proteins, and for the histone H3 lysine 27 trimethylation (H3K27Me3) associated with PcG silencing (Kahn et al. 2006, Schwartz et al. 2006). Genome-wide there is a negative correlation between H3K27Me3 and Nipped-B/cohesin binding, indicating that PcG silencing is generally incompatible with cohesin binding (Misulovin et al. 2008).
Nipped-B and cohesin bind putative in vivo target genes in cultured cells
Some of the key examples of cohesin binding to active genes, and absence from PcG-silenced genes, involve genes for which there is in vivo evidence for regulation by cohesin. For example, Nipped-B and cohesin bind to the upstream and transcribed regions of the cut locus in Sg4 and BG3 cells (Figure 2, Misulovin et al. 2008). Importantly, there is substantially more binding in BG3 cells. In Sg4 cells, cut is a PcG target, resulting in the H3K27Me3 modification over most of the regulatory region and transcription unit (Figure 2, Schwartz et al. 2006). In these cells, Nipped-B and cohesin are largely restricted to regions with reduced histone methylation (Figure 2, Misulovin et al. 2008). In contrast, cut is not a PcG target in BG3 cells (Y. Schwartz and V. Pirrotta, personal communication), and it is transcribed at least 200-fold more than in Sg4 cells (Z. Misulovin, C.A. Schaaf, D.D., unpublished). The lack of PcG repression and increased transcription correlates with an expansion of Nipped-B and cohesin binding over a 150 kbp region starting upstream of the wing margin enhancer, and extending past the 3' end of the cut gene (Figure 2, Misulovin et al. 2008).
As described above, the effects of Nipped-B and Rad21 (vtd) mutations on Nspl-1 mutant phenotypes suggests that they likely regulate the E(spl)-C, and the effects of cohesin mutations on the levels of ecdysone receptor protein and axon pruning in the mushroom body γ neuron suggests that cohesin regulates the EcR gene. In BG3 cells, the entire 50 kbp E(spl)-C is bound by Nipped-B and cohesin (Figure 4, Misulovin et al. 2008), and most of the genes in the complex are transcribed (Z. Misulovin, C.A. Schaaf, D.D., unpublished). The EcR gene is transcribed in both Sg4 and BG3 cell lines (Z. Misulovin, C.A. Schaaf, D.D., unpublished) and the entire transcribed region shows significant Nipped-B and cohesin binding in both cell lines (Misulovin et al. 2008, Dorsett 2008). Thus it is probable that Nipped-B and cohesin directly regulate the E(spl)-C and EcR in vivo.
The BX-C provides a particularly instructive example. The Ubx gene, which genetic evidence indicates is regulated by Nipped-B in vivo (Figure 4, Rollins et al. 1999), does not bind Nipped-B or cohesin in either Sg4 or BG3 cells, but is PcG-silenced in both lines (Figure 3, Misulovin et al. 2008). The entire BX-C is silenced in BG3 cells, and there is no detectable Nipped-B or cohesin binding to any of the three genes (Figure 3). Abd-B, however, is highly expressed in Sg4 cells, and Nipped-B and cohesin cover a 75 kbp region extending from the distal promoter region through much of the downstream 3' regulatory region (Figure 3). This region corresponds precisely with a region that has high PolII binding and low H3K27Me3 (Schwartz et al. 2006, Misulovin et al. 2008).
The borders of the Abd-B Nipped-B/cohesin binding region are also intriguing. The upstream border near the distal transcription start site corresponds to a CTCF binding site (Figure 3, Holohan et al. 2007) and the downstream border coincides with a well-characterized insulator (Fab-7), which requires the GAGA (Trithorax-like) protein for activity (Schweinsberg et al. 2004). Combined with the finding that CTCF positions cohesin at CTCF-dependent insulators in mammalian cells (Parelho et al. 2008, Rubio et al. 2008, Stedman et al. 2008, Wendt et al. 2008), this raises the possibilities that diverse types of insulators can help determine cohesin-binding domains, and that cohesin might contribute to the activity of different chromatin boundaries. In particular, the cohesin binding pattern in Abd-B combined with the genetic interactions between the cohesion factors and Ubx, Pc and E(z) methyltransferase mutations described above, suggests that cohesin might help define active chromatin domains and determine the boundaries between PcG-silenced and active chromatin. This is related to a function proposed for cohesin at the boundaries of the HMR silent mating type locus in yeast, where it helps prevent spread of the SIR silencing protein complexes (Donze et al. 1999).
Does transcription facilitate cohesin binding by unwinding chromatin?
One idea that arises from the preferential association of cohesin with transcribed genes, and absence from PcG-silenced genes, is that unwinding of higher order chromatin structures by transcription might facilitate the binding of cohesin. The internal opening of cohesin, which is some 35 by 50 nm, may be too small to encircle higher order chromatin structures of 30 nm fiber and above, but could easily encircle a 10 nm nucleosomal fiber.
Also favoring the idea that unwound chromatin facilitates cohesin binding is the finding that many cohesin binding domains correlate in position with early replication origins, which are also likely to have an unwound structure (McAlpine et al. 2004, D. MacAlpine, D.D., unpublished). Indeed, many replication origins also correlate with transcribed regions (MacAlpine et al. 2004), but the correlation between cohesin binding and replication origins extends to large intergenic regions with little or no transcription (D. MacAlpine, Z. Misulovin, D.D., unpublished). These findings fit nicely with the requirement for replication origin licensing for Nipped-B and cohesin binding in Xenopus egg extracts (Gillespie and Hirano 2004, Takahashi et al. 2004), and interactions of the Nipped-B/Mau-2 complex with origin licensing factors (Takahashi et al. 2008). However, the findings that many transcribed genes do not bind Nipped-B and cohesin, and that many genes that bind cohesin do not overlap replication origins, indicate that there are factors besides open chromatin structure that determine cohesin binding.
Potential molecular effects of the sister chromatid cohesion factors on gene expression
The genetic evidence that Nipped-B and cohesin regulate enhancer-promoter interactions in the cut and Ubx genes, and the ChIP-chip data showing that they bind to the large regulatory domains of the cut and Abd-B genes when they are active, argue that they play a role in controlling gene activation through effects on enhancer-promoter communication and chromatin domain definition, consistent with the finding that cohesin contributes to insulation by the CTCF protein in mammalian cells (Parelho et al. 2008, Wendt et al. 2008, Wendt and Peters, this issue). It tempting to propose that cohesin regulates enhancer-promoter interactions and insulator activity by mediating intra-chromosomal cohesion, similar to way it mediates sister chromatid cohesion (Gause et al. 2008a). One of the current leading ideas, supported by multiple lines of evidence, is that insulators pair with other insulators through protein-mediated interactions to form both intra- and interchromosomal loops (Dorman et al. 2007, Maeda and Karch 2007, Wallace and Felsenfeld 2007). Other long-range looping interactions mediated by cohesin might stabilize long-range enhancer-promoter interactions, and could, for example, contribute to the function of promoter-targeting sequences (PTS) that help enhancers bypass insulators in the BX-C (Figure 3).
Data from Drosophila and yeast also raise the possibility that cohesin could also influence transcriptional elongation. Nipped-B and cohesin bind to the long transcribed regions of cut, Abd-B, and EcR genes, among others. Cohesin binding to transcribed regions could potentially influence transcriptional elongation by acting as a roadblock for PolII. It seems likely that cohesin rings have to be moved to permit passage of RNA polymerase, similar to what occurs in the rare cases where cohesin binds a transcription unit in S. cerevisiae, and the gene is activated (Bausch et al. 2007). This idea is also consistent with the finding that cohesin contributes to transcriptional termination between convergent genes in S. pombe during G2 (Gullerova and Proudfoot 2008). In addition to these negative effects, however, it can also be imagined that cohesin could have positive effects by holding the gene in an unwound state, maintaining an open chromatin conformation more conducive to both transcriptional initiation and elongation. The direction and extent of the effect of reducing Nipped-B or cohesin dosage on gene expression would therefore depend on multiple factors, including the dependence on long-range activation, the length of the transcription unit, and the extent to which cohesin might counteract negative effects of chromatin folding.
One question to be answered is why Nipped-B and cohesin have opposite effects on gene expression in some cases, such as with cut. This puzzle is compounded by the fact that heterozygous Nipped-B null mutations only reduce Nipped-B mRNA levels by 25 to 30%. Thus, gene expression is exquisitely sensitive to Nipped-B dosage. This is also seen for NIPBL in cell lines from CdLS patients, and some CdLS individuals have only a 15% reduction in NIPBL mRNA (Borck et al. 2006). The actual mechanisms by which Nipped-B loads cohesin onto chromosomes, and/or stabilizes its binding are unknown, but likely involve direct Nipped-B-cohesin interactions (Arumugam et al. 2003, Gause et al. 2008b, Takahashi et al. 2008). It is possible that Nipped-B does more than simply load cohesin onto chromosomes, but also determines the binding stability and might, in collaboration with other factors such as Pds5 or Wapl (Ghandi et al. 2006, Kueng et al. 2006), dynamically unload and reload cohesin. Whether or not binding dynamics proves to be the explanation, knowing why small changes in Nipped-B and cohesin dosage have significant effects will be important for understanding the molecular mechanisms by which they regulate gene expression.
Summary
Genetic and molecular studies in Drosophila have uncovered diverse roles for cohesin and Nipped-B in gene expression and development, and have framed key questions regarding the potential mechanisms by which the sister chromatid cohesion apparatus regulate gene transcription. As described in other articles in this issue, some of these proposed mechanisms, such as chromatin domain boundary and insulator functions, also likely occur in yeast and mammalian cells. The findings in Drosophila have already provided useful insights into the molecular etiology of Cornelia de Lange syndrome, which is caused by reduced NIPBL (Nipped-B) activity, or amino acid changes in cohesin subunits. In particular, the discoveries showing that cohesin and Nipped-B regulate multiple conserved developmental pathways, and that cohesin and Nipped-B bind to many of the likely target genes, already helps understand why CdLS individuals display diverse developmental deficits, and have important implications for potential therapeutic strategies. Future work in multiple experimental systems are certain to focus on determining the molecular mechanisms by which cohesin and associated factors regulate gene expression, and the genetic and molecular tools available in Drosophila will continue to make important contributions to these efforts.
Acknowledgements
The author thanks Kirsten Wendt, Katsuhiko Shirahige, Jan-Michael Peters, Matthias Merkenschlager, Brian Calvi, David MacAlpine, Yuri Schwartz, Vince Pirrotta, Arthur Lander, Anne Calof, Ian Krantz, Jinglan Liu, Jim Kennison, Barry Honda, Oren Schuldiner, Liqun Luo, Gurmukh Sahota, Gary Stormo, Cheri Schaaf, Maria Gause, and Ziva Misulovin for sharing data and/or helpful discussions. The author also thanks Christian Haering and two anonymous reviewers for helpful comments on the manuscript. Research in the author's laboratory is supported by grants from the NIH (R01 GM055683, P01 HD052860) and the March of Dimes (#1FY05-103).
References
- Abrell S, Carrera P, Jäckle H. A modifier screen of ectopic Krüppel activity identifies autosomal Drosophila chromosomal sites and genes required for normal eye development. Chromosoma. 2000;109:334–342. doi: 10.1007/s004120000092. [DOI] [PubMed] [Google Scholar]
- Anderson DE, Losada A, Erickson HP, Hirano T. Condensin and cohesin display different arm conformations with characteristic hinge angles. J Cell Biol. 2002;156:419–424. doi: 10.1083/jcb.200111002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam P, Gruber S, Tanaka K, Haering CH, Mechtler K, Nasmyth K. ATP hydrolysis is required for cohesin's association with chromosomes. Curr Biol. 2003;13:1941–1953. doi: 10.1016/j.cub.2003.10.036. [DOI] [PubMed] [Google Scholar]
- Bajusz I, Sipos L, Györgypál Z, Carrington EA, Jones RS, Gausz J, Gyurkovics H. The Trithorax-mimic allele of Enhancer of zeste renders active domains of target genes accessible to polycomb-group-dependent silencing in Drosophila melanogaster. Genetics. 2001;159:1135–1150. doi: 10.1093/genetics/159.3.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NE, Yu S, Han D. Evolution of proneural atonal expression during distinct regulatory phases in the developing Drosophila eye. Curr Biol. 1996;6:1290–1301. doi: 10.1016/s0960-9822(02)70715-x. [DOI] [PubMed] [Google Scholar]
- Baker NE, Yu SY. Proneural function of neurogenic genes in the developing Drosophila eye. Curr Biol. 1997;7:122–132. doi: 10.1016/s0960-9822(06)00056-x. [DOI] [PubMed] [Google Scholar]
- Bausch C, Noone S, Henry JM, Gaudenz K, Sanderson B, Seidel C, Gerton JL. Transcription alters chromosomal locations of cohesin in Saccharomyces cerevisiae. Mol Cell Biol. 2007;27:8522–8532. doi: 10.1128/MCB.01007-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borck G, Zarhrate M, Cluzeau C, Bal E, Bonnefont JP, Munnich A, Cormier-Daire V, Colleaux L. Father-to-daughter transmission of Cornelia de Lange syndrome caused by a mutation in the 5' untranslated region of the NIPBL gene. Hum Mutat. 2006;27:731–735. doi: 10.1002/humu.20380. [DOI] [PubMed] [Google Scholar]
- Carrera P, Abrell S, Kerber B, Walldorf U, Preiss A, Hoch M, Jäckle H. A modifier screen in the eye reveals control genes for Krüppel activity in the Drosophila embryo. Proc Natl Acad Sci USA. 1998;95:10779–10784. doi: 10.1073/pnas.95.18.10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Lin L, Smith S, Lin Q, Zhou J. Multiple Promoter Targeting Sequences exist in Abdominal-B to regulate long-range gene activation. Dev Biol. 2005;286:629–36. doi: 10.1016/j.ydbio.2005.08.025. [DOI] [PubMed] [Google Scholar]
- Ciosk R, Shirayama M, Shevchenko A, Tanaka T, Toth A, Shevchenko A, Nasmyth K. Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell. 2000;5:243–254. doi: 10.1016/s1097-2765(00)80420-7. [DOI] [PubMed] [Google Scholar]
- D'Ambrosio C, Schmidt CK, Katou Y, Kelly G, Itoh T, Shirahige K, Uhlmann F. Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev. 2008;22:2215–2227. doi: 10.1101/gad.1675708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deardorff MA, Kaur M, Yaeger D, Rampuria A, Korolev S, Pie J, Gil-Rodríguez C, Arnedo M, Loeys B, Kline AD, Wilson M, Lillquist K, Siu V, Ramos FJ, Musio A, Jackson LS, Dorsett D, Krantz ID. Mutations in cohesin complex members SMC3 and SMC1A cause a mild variant of Cornelia de Lange syndrome with predominant mental retardation. Am J Hum Genet. 2007;80:485–494. doi: 10.1086/511888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donze D, Adams CR, Rine J, Kamakaka RT. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes Dev. 1999;13:698–708. doi: 10.1101/gad.13.6.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman ER, Bushey AM, Corces VG. The role of insulator elements in large-scale chromatin structure in interphase. Semin Cell Dev Biol. 2007;18:682–690. doi: 10.1016/j.semcdb.2007.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D. Distance-independent inactivation of an enhancer by the suppressor of Hairy-wing DNA-binding protein of Drosophila. Genetics. 1993;134:1135–1144. doi: 10.1093/genetics/134.4.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D. Adherin: key to the cohesin ring and Cornelia de Lange syndrome. Curr Biol. 2004;14:R834–836. doi: 10.1016/j.cub.2004.09.035. [DOI] [PubMed] [Google Scholar]
- Dorsett D. Roles of the sister chromatid cohesion apparatus in gene expression, development, and human syndromes. Chromosoma. 2007;116:1–13. doi: 10.1007/s00412-006-0072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D. Running rings around chromosomes to trim axons and target dendrites. Dev Cell. 2008;14:156–158. doi: 10.1016/j.devcel.2008.01.010. [DOI] [PubMed] [Google Scholar]
- Dorsett D, Eissenberg JC, Misulovin Z, Martens A, Redding B, McKim K. Effects of sister chromatid cohesion proteins on cut gene expression during wing development in Drosophila. Development. 2005;132:4743–4753. doi: 10.1242/dev.02064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D, Krantz ID. On the molecular etiology of Cornelia de Lange syndrome. NY Acad Sci. 2009 doi: 10.1111/j.1749-6632.2008.03450.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenfeld AL, Kennison JA. Positional signaling by hedgehog in Drosophila imaginal disc development. Development. 1995;121:1–10. doi: 10.1242/dev.121.1.1. [DOI] [PubMed] [Google Scholar]
- Gause M, Eissenberg JC, Macrae AF, Dorsett M, Misulovin Z, Dorsett D. Nipped-A, the Tra1/TRRAP subunit of the Drosophila SAGA and Tip60 complexes, has multiple roles in Notch signaling during wing development. Mol Cell Biol. 2006;26:2347–2359. doi: 10.1128/MCB.26.6.2347-2359.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gause M, Schaaf CA, Dorsett D. Cohesin and CTCF: cooperating to control chromosome conformation? Bioessays. 2008a;30:715–718. doi: 10.1002/bies.20787. [DOI] [PubMed] [Google Scholar]
- Gause M, Webber HA, Misulovin Z, Haller G, Rollins RA, Eissenberg JC, Bickel SE, Dorsett D. Functional links between Drosophila Nipped-B and cohesin in somatic and meiotic cells. Chromosoma. 2008b;117:51–66. doi: 10.1007/s00412-007-0125-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi R, Gillespie PJ, Hirano T. Human Wapl is a cohesin-binding protein that promotes sister-chromatid resolution in mitotic prophase. Curr Biol. 2006;16:2406–2417. doi: 10.1016/j.cub.2006.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie PJ, Hirano T. Scc2 couples replication licensing to sister chromatid cohesion in Xenopus egg extracts. Curr Biol. 2004;14:1598–1603. doi: 10.1016/j.cub.2004.07.053. [DOI] [PubMed] [Google Scholar]
- Glynn EF, Megee PC, Yu HG, Mistrot C, Unal E, Koshland DE, DeRisi JL, Gerton JL. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2004;2:E259. doi: 10.1371/journal.pbio.0020259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber S, Haering CH, Nasmyth K. Chromosomal cohesin forms a ring. Cell. 2003;112:765–77. doi: 10.1016/s0092-8674(03)00162-4. [DOI] [PubMed] [Google Scholar]
- Guacci V, Koshland D, Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullerova M, Proudfoot NJ. Cohesin complex promotes transcriptional termination between convergent genes in S. pombe. Cell. 2008;132:983–995. doi: 10.1016/j.cell.2008.02.040. [DOI] [PubMed] [Google Scholar]
- Haering CH, Farcas AM, Arumugam P, Metson J, Nasmyth K. The cohesin ring concatenates sister DNA molecules. Nature. 2008;454:297–301. doi: 10.1038/nature07098. [DOI] [PubMed] [Google Scholar]
- Hallson G, Syrzycka M, Beck SA, Kennison JA, Dorsett D, Keall R, Page SL, Warren WD, Brock HW, Sinclair DAR, Honda BM. The Drosophila cohesin subunit Rad21 is a trithorax group (trxG) protein. Proc Natl Acad Sci USA. 2008;105:12405–12410. doi: 10.1073/pnas.0801698105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman T, Stead K, Koshland D, Guacci V. Pds5p is an essential chromosomal protein required for both sister chromatid cohesion and condensation in Saccharomyces cerevisiae. J Cell Biol. 2000;151:613–626. doi: 10.1083/jcb.151.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holohan EE, Kwong C, Adryan B, Bartkuhn M, Herold M, Renkawitz R, Russell S, White R. CTCF genomic binding sites in Drosophila and the organisation of the bithorax complex. PLoS Genet. 2007;3:e112. doi: 10.1371/journal.pgen.0030112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsfield JA, Anagnostou SH, Hu JK, Cho KH, Geisler R, Lieschke G, Crosier KE, Crosier PS. Cohesin-dependent regulation of Runx genes. Development. 2007;134:2639–2649. doi: 10.1242/dev.002485. [DOI] [PubMed] [Google Scholar]
- Ivanov D, Nasmyth K. A topological interaction between cohesin rings and a circular minichromosome. Cell. 2005;122:849–860. doi: 10.1016/j.cell.2005.07.018. [DOI] [PubMed] [Google Scholar]
- Ivanov D, Nasmyth K. A physical assay for sister chromatid cohesion in vitro. Mol Cell. 2007;27:300–310. doi: 10.1016/j.molcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Jack J, Dorsett D, Delotto Y, Liu S. Expression of the cut locus in the Drosophila wing margin is required for cell type specification and is regulated by a distant enhancer. Development. 1991;113:735–747. doi: 10.1242/dev.113.3.735. [DOI] [PubMed] [Google Scholar]
- Kahn TG, Schwartz YB, Dellino GI, Pirrotta V. Polycomb complexes and the propagation of the methylation mark at the Drosophila Ubx gene. J Biol Chem. 2006;281:29064–29075. doi: 10.1074/jbc.M605430200. [DOI] [PubMed] [Google Scholar]
- Kennison J, Tamkun JW. Dosage-dependent modifiers of Polycomb and Antennapedia mutations in Drosophila. Proc Natl Acad Sci USA. 1988;85:8136–8140. doi: 10.1073/pnas.85.21.8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krantz ID, McCallum J, DeScipio C, Kaur M, Gillis LA, Yaeger D, Jukofsky L, Wasserman N, Bottani A, Morris CA, Nowaczyk MJ, Toriello H, Bamshad MJ, Carey JC, Rappaport E, Kawauchi S, Lander AD, Calof AL, Li HH, Devoto M, Jackson LG. Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat Genet. 2004;36:631–635. doi: 10.1038/ng1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kueng S, Hegemann B, Peters BH, Lipp JJ, Schleiffer A, Mechtler K, Peters JM. Wapl controls the dynamic association of cohesin with chromatin. Cell. 2006;127:955–967. doi: 10.1016/j.cell.2006.09.040. [DOI] [PubMed] [Google Scholar]
- Lee T, Marticke S, Sung C, Robinow S, Luo L. Cell-autonomous requirement of the USP/EcR-B ecdysone receptor for mushroom body neuronal remodeling in Drosophila. Neuron. 2000;28:807–818. doi: 10.1016/s0896-6273(00)00155-0. [DOI] [PubMed] [Google Scholar]
- Lengronne A, Katou Y, Mori S, Yokobayashi S, Kelly GP, Itoh T, Watanabe Y, Shirahige K, Uhlmann F. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature. 2004;430:573–578. doi: 10.1038/nature02742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Chen Q, Lin L, Smith S, Zhou J. Promoter targeting sequence mediates enhancer interference in the Drosophila embryo. Proc Natl Acad Sci USA. 2007;104:3237–3242. doi: 10.1073/pnas.0605730104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Chen Q, Lin L, Zhou J. The Promoter Targeting Sequence mediates epigenetically heritable transcription memory. Genes Dev. 2004;18:2639–2651. doi: 10.1101/gad.1230004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroos HB, Ström L, Itoh T, Katou Y, Shirahige K, Sjögren C. Chromosomal association of the Smc5/6 complex reveals that it functions in differently regulated pathways. Mol Cell. 2006;22:755–767. doi: 10.1016/j.molcel.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Liu J, Krantz ID. Cohesin and human disease. Annu Rev Genomics Hum Genet. 2008;9:303–320. doi: 10.1146/annurev.genom.9.081307.164211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A, Yokochi T, Hirano T. Functional contribution of Pds5 to cohesin-mediated cohesion in human cells and Xenopus egg extracts. J Cell Sci. 2005;118:2133–2141. doi: 10.1242/jcs.02355. [DOI] [PubMed] [Google Scholar]
- Lupo R, Breiling A, Bianchi ME, Orlando V. Drosophila chromosome condensation proteins Topoisomerase II and Barren colocalize with Polycomb and maintain Fab-7 PRE silencing. Mol Cell. 2001;7:127–136. doi: 10.1016/s1097-2765(01)00161-7. [DOI] [PubMed] [Google Scholar]
- Maeda RK, Karch F. The ABC of the BX-C: the bithorax complex explained. Development. 2006;133:1413–1422. doi: 10.1242/dev.02323. [DOI] [PubMed] [Google Scholar]
- Maeda RK, Karch F. Making connections: boundaries and insulators in Drosophila. Curr Opin Genet Dev. 2007;17:394–399. doi: 10.1016/j.gde.2007.08.002. [DOI] [PubMed] [Google Scholar]
- MacAlpine DM, Rodríguez HK, Bell SP. Coordination of replication and transcription along a Drosophila chromosome. Genes Dev. 2004;18:3094–3105. doi: 10.1101/gad.1246404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mc Intyre J, Muller EG, Weitzer S, Snydsman BE, Davis TN, Uhlmann F. In vivo analysis of cohesin architecture using FRET in the budding yeast Saccharomyces cerevisiae. EMBO J. 2007;26:3783–3793. doi: 10.1038/sj.emboj.7601793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer BJ. X-Chromosome dosage compensation. WormBook. 2005 Jun 25;:1–14. doi: 10.1895/wormbook.1.8.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- Misulovin Z, Schwartz YB, Li XY, Kahn TG, Gause M, MacArthur S, Fay JC, Eisen MB, Pirrotta V, Biggin MD, Dorsett D. Association of cohesin and Nipped-B with transcriptionally active regions of the Drosophila melanogaster genome. Chromosoma. 2008;117:89–102. doi: 10.1007/s00412-007-0129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcillo P, Rosen C, Dorsett D. Genes regulating the remote wing margin enhancer in the Drosophila cut locus. Genetics. 1996;144:1143–1154. doi: 10.1093/genetics/144.3.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O'Connor MB, Kingston RE, Simon JA. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- Musio A, Selicorni A, Focarelli ML, Gervasini C, Milani D, Russo S, Vezzoni P, Larizza L. X-linked Cornelia de Lange syndrome owing to SMC1L1 mutations. Nat Genet. 2006;38:528–530. doi: 10.1038/ng1779. [DOI] [PubMed] [Google Scholar]
- Nagel AC, Preiss A. Notchspl is deficient for inductive processes in the eye, and E(spl)D enhances split by interfering with proneural activity. Dev Biol. 1999;208:406–415. doi: 10.1006/dbio.1999.9203. [DOI] [PubMed] [Google Scholar]
- Nagel AC, Yu Y, Preiss A. Enhancer of split [E(spl)(D)] is a gro-independent, hypermorphic mutation in Drosophila. Dev Genet. 1999;25:168–179. doi: 10.1002/(SICI)1520-6408(1999)25:2<168::AID-DVG11>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Nellesen DT, Lai EC, Posakony JW. Discrete enhancer elements mediate selective responsiveness of Enhancer of split complex genes to common transcriptional activators. Dev Biol. 1999;213:33–53. doi: 10.1006/dbio.1999.9324. [DOI] [PubMed] [Google Scholar]
- Nepveu A. Role of the multifunctional CDP/Cut/Cux homeodomain transcription factor in regulating differentiation, cell growth and development. Gene. 2001;270:1–15. doi: 10.1016/s0378-1119(01)00485-1. [DOI] [PubMed] [Google Scholar]
- Panizza S, Tanaka T, Hochwagen A, Eisenhaber F, Nasmyth K. Pds5 cooperates with cohesin in maintaining sister chromatid cohesion. Curr Biol. 2000;10:1557–1564. doi: 10.1016/s0960-9822(00)00854-x. [DOI] [PubMed] [Google Scholar]
- Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, Cobb BS, Yokomori K, Dillon N, Aragon L, Fisher AG, Merkenschlager M. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Pauli A, Althoff F, Oliveira RA, Heidmann S, Schuldiner O, Lehner CF, Dickson BJ, Nasmyth K. Cell-type-specific TEV protease cleavage reveals cohesin functions in Drosophila neurons. Dev Cell. 2008;14:239–251. doi: 10.1016/j.devcel.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peifer M, Bender W. The anterobithorax and bithorax mutations of the bithorax complex. EMBO J. 1986;5:2293–2303. doi: 10.1002/j.1460-2075.1986.tb04497.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian S, Capovilla M, Pirrotta V. The bx region enhancer, a distant cis-control element of the Drosophila Ubx gene and its regulation by hunchback and other segmentation genes. EMBO J. 1991;10:1415–1425. doi: 10.1002/j.1460-2075.1991.tb07662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins RA, Korom M, Aulner N, Martens A, Dorsett D. Drosophila Nipped-B protein supports sister chromatid cohesion and opposes the stromalin/Scc3 cohesion factor to facilitate long-range activation of the cut gene. Mol Cell Biol. 2004;24:3100–3111. doi: 10.1128/MCB.24.8.3100-3111.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins RA, Morcillo P, Dorsett D. Nipped-B, a Drosophila homologue of chromosomal adherins, participates in activation by remote enhancers in the cut and Ultrabithorax genes. Genetics. 1999;152:577–593. doi: 10.1093/genetics/152.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, Baliga NS, Aebersold R, Ranish JA, Krumm A. CTCF physically links cohesin to chromatin. Proc Natl Acad Sci USA. 2008;105:8309–8314. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansregret L, Nepveu A. The multiple roles of CUX1: insights from mouse models and cell-based assays. Gene. 2008;412:84–94. doi: 10.1016/j.gene.2008.01.017. [DOI] [PubMed] [Google Scholar]
- Schuldiner O, Berdnik D, Levy JM, Wu JS, Luginbuhl D, Gontang AC, Luo L. piggyBac-based mosaic screen identifies a postmitotic function for cohesin in regulating developmental axon pruning. Dev Cell. 2008;14:227–238. doi: 10.1016/j.devcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze S, Sinclair DA, Silva E, Fitzpatrick KA, Singh M, Lloyd VK, Morin KA, Kim J, Holm DG, Kennison JA, Honda BM. Essential genes in proximal 3L heterochromatin of Drosophila melanogaster. Molec Gen Genet. 2001;264:782–789. doi: 10.1007/s004380000367. [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, Biggin M, Pirrotta V. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet. 2006;38:700–705. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- Schweinsberg S, Hagstrom K, Gohl D, Schedl P, Kumar RP, Mishra R, Karch F. The enhancer-blocking activity of the Fab-7 boundary from the Drosophila bithorax complex requires GAGA-factor-binding sites. Genetics. 2004;168:1371–1384. doi: 10.1534/genetics.104.029561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitan VC, Banks P, Laval S, Majid NA, Dorsett D, Rana A, Smith J, Bateman A, Krpic S, Hostert A, Rollins RA, Erdjument-Bromage H, Tempst P, Benard CY, Hekimi S, Newbury SF, Strachan T. Metazoan Scc4 homologs link sister chromatid cohesion to cell and axon migration guidance. PLoS Biol. 2006;4:e242. doi: 10.1371/journal.pbio.0040242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J, Peifer M, Bender W, O'Connor M. Regulatory elements of the bithorax complex that control expression along the anterior-posterior axis. EMBO J. 1990;9:3945–3956. doi: 10.1002/j.1460-2075.1990.tb07615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollars V, Lu X, Xiao L, Wang X, Garfinkel MD, Ruden DM. Evidence for an epigenetic mechanism by which Hsp90 acts as a capacitor for morphological evolution. Nat Genet. 2003;33:70–74. doi: 10.1038/ng1067. [DOI] [PubMed] [Google Scholar]
- Stedman W, Kang H, Lin S, Kissil JL, Bartolomei MS, Lieberman PM. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 2008;27:654–666. doi: 10.1038/emboj.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi TS, Basu A, Bermudez V, Hurwitz J, Walter JC. Cdc7-Drf1 kinase links chromosome cohesion to the initiation of DNA replication in Xenopus egg extracts. Genes Dev. 2008;22:1894–1905. doi: 10.1101/gad.1683308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi TS, Yiu P, Chou MF, Gygi S, Walter JC. Recruitment of Xenopus Scc2 and cohesin to chromatin requires the pre-replication complex. Nat Cell Biol. 2004;6:991–996. doi: 10.1038/ncb1177. [DOI] [PubMed] [Google Scholar]
- Tomonaga T, Nagao K, Kawasaki Y, Furuya K, Murakami A, Morishita J, Yuasa T, Sutani T, Kearsey SE, Uhlmann F, Nasmyth K, Yanagida M. Characterization of fission yeast cohesin: essential anaphase proteolysis of Rad21 phosphorylated in the S phase. Genes Dev. 2000;14:2757–2770. doi: 10.1101/gad.832000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkin ET, Wang TJ, Lisgo S, Bamshad MJ, Strachan T. NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat Genet. 2004;36:636–641. doi: 10.1038/ng1363. [DOI] [PubMed] [Google Scholar]
- Wallace JA, Felsenfeld G. We gather together: insulators and genome organization. Curr Opin Genet Dev. 2007;17:400–407. doi: 10.1016/j.gde.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welshons WJ. Dosage experiments with split mutants in the presence of an enhancer of split. DIS. 1956;30:157–158. [Google Scholar]
- Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, Yahata K, Imamoto F, Aburatani H, Nakao M, Imamoto N, Maeshima K, Shirahige K, Peters JM. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- Williams BC, Garrett-Engele CM, Li Z, Williams EV, Rosenman ED, Goldberg ML. Two putative acetyltransferases, san and deco, are required for establishing sister chromatid cohesion in Drosophila. Curr Biol. 2003;13:2025–2036. doi: 10.1016/j.cub.2003.11.018. [DOI] [PubMed] [Google Scholar]
- Zhang B, Jain S, Song H, Fu M, Heuckeroth RO, Erlich JM, Jay PY, Milbrandt J. Mice lacking sister chromatid cohesion protein PDS5B exhibit developmental abnormalities reminiscent of Cornelia de Lange syndrome. Development. 2007;134:3191–3201. doi: 10.1242/dev.005884. [DOI] [PubMed] [Google Scholar]
- Zhou J, Levine M. A novel cis-regulatory element, the PTS, mediates an anti-insulator activity in the Drosophila embryo. Cell. 1999;99:567–575. doi: 10.1016/s0092-8674(00)81546-9. [DOI] [PubMed] [Google Scholar]