To the Editor
Inherited hearing impairment is a highly heterogeneous genetic trait. To date, 22 and 27 genes have been associated with autosomal dominant and recessive non-syndromic hearing loss, respectively (ADNHSL, ARNSHL) (1). Mutations in seven of these genes – GJB2, GJB3, GJB6, MYO6, MYO7A, COL11A2 and TMC1 – cause both ADNSHL and ARNSHL.
TMC1 belongs to a large TMC (Trans Membrane Channel-like) gene family. Its eight members encode transmembrane proteins with intracellular amino- and carboxyl- termini and multiple transmembrane domains. In the inner ear, murine Tmc1 is expressed in outer and inner hair cells after P3. Tmc1 deficiency in the recessive deafness mouse mutant dn leads to complete deafness and hair cell degeneration, while the Tmc1 p.M412K missense mutation in the semi-dominant Bth mutant causes progressive hearing loss and hair cell degeneration in heterozygotes and early-onset deafness in homozygotes (2, 3). These data suggest that Tmc1 is necessary for maturation and survival of hair cells in the murine cochlea (4).
Twenty-two recessive mutations of TMC1 are associated with ARNSHL at the DFNB7/11 locus in 39 families worldwide (2, 5–10), however only two dominant mutations have been reported. These two mutations - p.D572N and p.D572H - affect the same amino acid (2, 11, 12) and were found in three unrelated North American families with progressive ADNSHL at the DFNA36 locus.
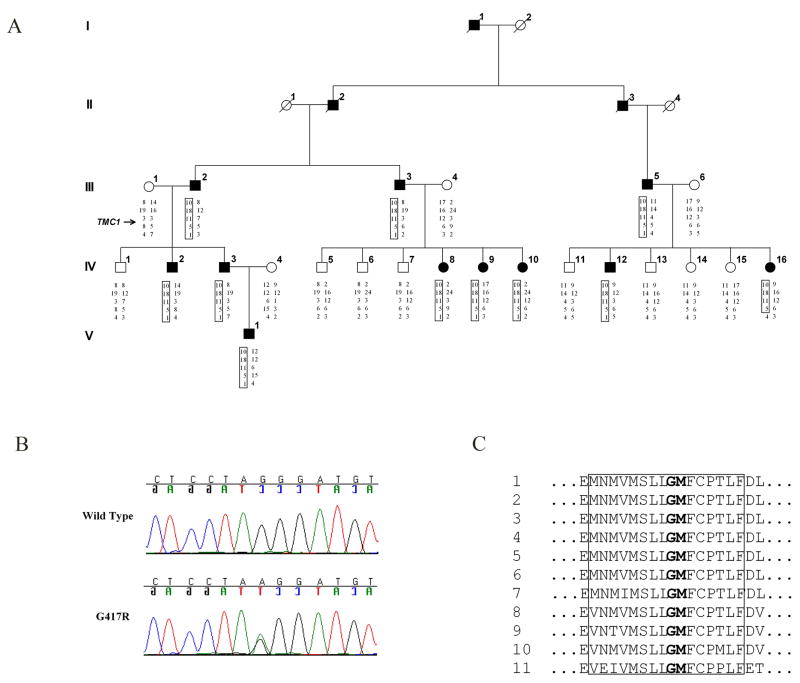
We have identified a novel dominant mutation, p.G417R, and a novel recessive mutation, p.N50KfsX26, in TMC1 in a large Iranian DFNA36 family (Family L1754) and a consanguineous Iranian DFNB7/11 family (Family L787), respectively (Fig 1A). These families were ascertained through the Research Centre of Ear, Nose, Throat and Head and Neck Surgery, Iran University of Medical Sciences, Tehran, Iran, as part of a study approved by the Human Research Institutional Review Boards at the participating institutions. Written informed consent was obtained from all study subjects.
Figure 1.
Family L1754 carrying the G417R mutation in TMC1 segregates ADNSHL at the DFNA36 locus. (A) Pedigree and haplotype segregation of STRP markers D9S1817, D9S273, D9S175, D9S167 and D9S283 (top to bottom). Note the recombination event in the paternal chromosome inherited by IV-16. (B) Sequencing result showing the heterozygous p.G417R mutation in affected individuals. (C) Amino acids p.G417 and p.M418 (bolded) of TMC1 are conserved across species: 1 Homo sapiens, 2 Pan troglodytes, 3 Canis familiaris, 4 Bos taurus, 5 Equus caballus, 6 Mus musculus, 7 Monodelphis domestica, 8 Gallus gallus, 9 Taeniopygia guttata, 10 Danio rerio and 11 human TMC2). The predicted transmembrane domain is boxed.
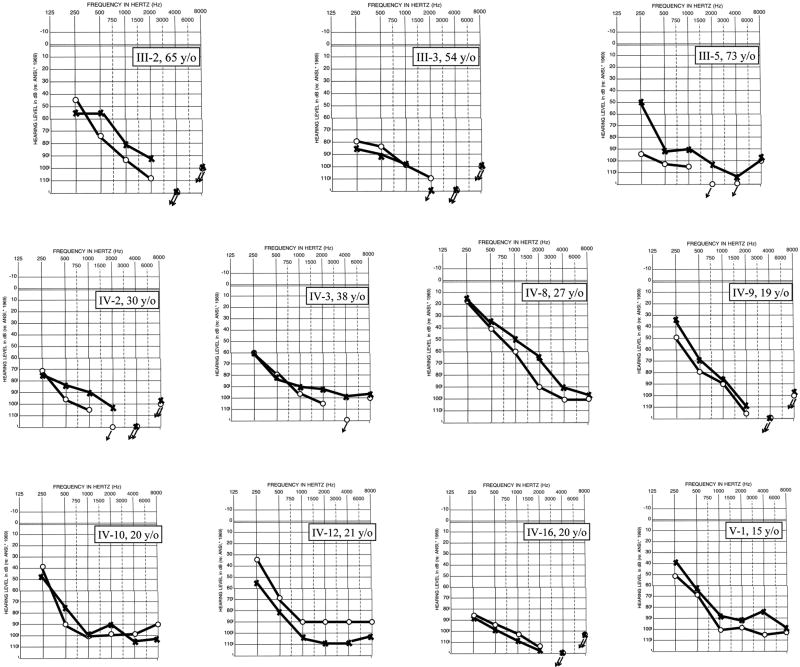
Consistent with the phenotypes of the DFNA36 and DFNB7/11 families as previously reported, hearing impairment in Family L1754 is rapidly progressive, starting in the higher frequencies and ultimately affecting all frequencies, while hearing impairment in Family L787 is congenital and profound (Fig 2). Haplotype analysis in both families was consistent with linkage to DFNA36 (LOD score >6.02) and DFNB7/11 (LOD score >2.5 – the maximum possible for this family), and sequence analysis of TMC1 revealed a p.G417R (c.1249G>A) mutation in Family L1754 and a p.N50KfsX26 (c.150delT) mutation leading to a premature stop in Family L787. Neither mutation was seen in 500 control chromosomes.
Figure 2.
Audiograms from all affected persons shown in Figure 1A (○, right ear; x, left ear; arrows, no response).
As in many other genes involved in both dominant and recessive non-syndromic hearing loss, the dominant mutations of TMC1 mostly likely cause disease by a dominant-negative or gain-of-function mechanism rather than haploinsufficiency. Studying of the pathogenic roles of these specific mutations, therefore, may provide us important insights into the function of TMC1 and its interacting proteins. It is noteworthy that our report is the first to document a dominant mutation of TMC1 from a region other than North America. The p.G417R mutation in Family L1754 is the second amino acid position of TMC1 associated with ADNSHL (Fig 1B). The adjacent amino acid, p.M418, is orthologous to murine p.M412, which is replaced by lysine in the Bth mouse mutant (Fig 1C). Both p.G417 and p.M418 lie within a predicted transmembrane (13) and the substitution of these hydrophobic amino acids with highly charged, hydrophilic amino acids is likely to lead to abnormal protein folding or abnormal protein-protein interactions. Consistent with the positional proximity and substitution similarity of p.G417R and p.M412K mutations, the phenotypes in both Family L1754 and the Bth mouse are characterized by rapidly progressive hearing loss. We hypothesize that these two mutations therefore share pathogenic mechanisms making the Bth mouse an excellent animal model to study TMC1 p.G417R-associated ADNSHL.
Acknowledgments
This project was supported in part by: The Science and Technology Commission of Shanghai Municipality, China (09DJ1400604 to TY); The National Institutes of Health-National Institute on Deafness and Other Communication Disorders (Grants DC02842 and DC03544 to RJHS); and The Iranian National Science Foundation (Grants 85073/23 and 85033/10 to HN). RJHS is the Sterba Hearing Research Professor, University of Iowa College of Medicine.
References
- 1.Van Campm G, Smith RJH. Hereditary Hearing Loss Homepage. 2009 http://webhostuaacbe/hhh/
- 2.Kurima K, Peters LM, Yang Y, et al. Dominant and recessive deafness caused by mutations of a novel gene, TMC1, required for cochlear hair-cell function. Nat Genet. 2002;30:277–284. doi: 10.1038/ng842. [DOI] [PubMed] [Google Scholar]
- 3.Vreugde S, Erven A, Kros CJ, et al. Beethoven, a mouse model for dominant, progressive hearing loss DFNA36. Nat Genet. 2002;30:257–258. doi: 10.1038/ng848. [DOI] [PubMed] [Google Scholar]
- 4.Marcotti W, Erven A, Johnson SL, et al. Tmc1 is necessary for normal functional maturation and survival of inner and outer hair cells in the mouse cochlea. J Physiol. 2006;574:677–698. doi: 10.1113/jphysiol.2005.095661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hilgert N, Alasti F, Dieltjens N, et al. Mutation analysis of TMC1 identifies four new mutations and suggests an additional deafness gene at loci DFNA36 and DFNB7/11. Clin Genet. 2008;74:223–232. doi: 10.1111/j.1399-0004.2008.01053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalay E, Karaguzel A, Caylan R, et al. Four novel TMC1 (DFNB7/DFNB11) mutations in Turkish patients with congenital autosomal recessive nonsyndromic hearing loss. Hum Mutat. 2005;26:591. doi: 10.1002/humu.9384. [DOI] [PubMed] [Google Scholar]
- 7.Kitajiri SI, McNamara R, Makishima T, et al. Identities, frequencies and origins of TMC1 mutations causing DFNB7/B11 deafness in Pakistan. Clin Genet. 2007;72:546–550. doi: 10.1111/j.1399-0004.2007.00895.x. [DOI] [PubMed] [Google Scholar]
- 8.Meyer CG, Gasmelseed NM, Mergani A, et al. Novel TMC1 structural and splice variants associated with congenital nonsyndromic deafness in a Sudanese pedigree. Hum Mutat. 2005;25:100. doi: 10.1002/humu.9302. [DOI] [PubMed] [Google Scholar]
- 9.Santos RL, Wajid M, Khan MN, et al. Novel sequence variants in the TMC1 gene in Pakistani families with autosomal recessive hearing impairment. Hum Mutat. 2005;26:396. doi: 10.1002/humu.9374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tlili A, Rebeh IB, Aifa-Hmani M, et al. TMC1 but not TMC2 is responsible for autosomal recessive nonsyndromic hearing impairment in Tunisian families. Audiol Neurootol. 2008;13:213–218. doi: 10.1159/000115430. [DOI] [PubMed] [Google Scholar]
- 11.Hilgert N, Monahan K, Kurima K, et al. Amino acid 572 in TMC1: hot spot or critical functional residue for dominant mutations causing hearing impairment. J Hum Genet. 2009;54:188–190. doi: 10.1038/jhg.2009.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kitajiri S, Makishima T, Friedman TB, et al. A novel mutation at the DFNA36 hearing loss locus reveals a critical function and potential genotype-phenotype correlation for amino acid-572 of TMC1. Clin Genet. 2007;71:148–152. doi: 10.1111/j.1399-0004.2007.00739.x. [DOI] [PubMed] [Google Scholar]
- 13.Keresztes G, Mutai H, Heller S. TMC and EVER genes belong to a larger novel family, the TMC gene family encoding transmembrane proteins. BMC Genomics. 2003;4:24. doi: 10.1186/1471-2164-4-24. [DOI] [PMC free article] [PubMed] [Google Scholar]