Abstract
Molecular dynamics (MD) simulations and quantum mechanical/molecular mechanical (QM/MM) calculations were performed on the prereactive enzyme-substrate complex, transition states, intermediates, and product involved in the process of human butyrylcholinesterase (BChE)-catalyzed hydrolysis of (−)-cocaine. The computational results consistently reveal a unique role of the oxyanion hole (consisting of G116, G117, and A199) in BChE-catalyzed hydrolysis of cocaine, as compared to acetylcholinesterase (AChE)-catalyzed hydrolysis of acetylcholine. During BChE-catalyzed hydrolysis of cocaine, only G117 has a hydrogen bond with the carbonyl oxygen (O31) of the cocaine benzoyl ester in the prereactive BChE-cocaine complex, and the NH groups of G117 and A199 are hydrogen-bonded with O31 of cocaine in all of the transition states and intermediates. Surprisingly, the NH hydrogen of G116 forms an unexpected hydrogen bond with the carboxyl group of E197 side chain and, therefore, is not available to form a hydrogen bond with O31 of cocaine in the acylation. The NH hydrogen of G116 is only partially available to form a weak hydrogen bond with O31 of cocaine in some structures involved in the deacylation. The change of the estimated hydrogen bonding energy between the oxyanion hole and O31 of cocaine during the reaction process demonstrates how the protein environment can affect the energy barrier for each step of the BChE-catalyzed hydrolysis of cocaine. These insights concerning the effects of the oxyanion hole on the energy barriers provide valuable clues on how to rationally design BChE mutants with a higher catalytic activity for the hydrolysis of (−)-cocaine.
Introduction
Cocaine addiction and overdose are a major medical and public health problem that continues to defy treatment.1,2 The disastrous medical and social consequences of cocaine addiction, such as violent crime, loss in individual productivity, illness and death, have made the development of an effective pharmacological treatment a high priority.3,4 However, cocaine mediates its reinforcing and toxic effects by blocking neurotransmitter reuptake and the classical pharmacodynamic approach has failed to yield small-molecule receptor antagonists due to the difficulties inherent in blocking a blocker.1-4
An alternative to receptor-based approaches is to interfere with the delivery of cocaine to its receptors and accelerate its metabolism in the body.4 An ideal molecule for this purpose should be a potent enzyme catalyzing the hydrolysis of cocaine into biologically inactive metabolites. The dominant pathway for cocaine metabolism in primates is butyrylcholinesterase (BChE)-catalyzed hydrolysis at the benzoyl ester group (Chart 1).4,5 Only 5% of the cocaine is deactivated through oxidation by the liver microsomal cytochrome P450 system.6 Cocaine hydrolysis at benzoyl ester group yields ecgonine methyl ester, whereas the oxidation produces norcocaine.7 The metabolite ecgonine methyl ester is biologically inactive metabolite, whereas the metabolite norcocaine is hepatotoxic and a local anesthetic. Clearly, BChE-catalyzed hydrolysis of cocaine at the benzoyl ester is the pathway most suitable for amplification and, therefore, we focus on the improvement of the catalytic activity of the primary cocaine-metabolizing enzyme. BChE is synthesized in the liver and widely distributed in the body, including plasma, brain, and lung.8 Extensive experimental studies in animals and humans demonstrate that enhancement of BChE activity by administration of exogenous enzyme substantially decreases cocaine half-life.9,10,11,12 So, enhancement of cocaine metabolism by administration of BChE has been recognized to be a promising pharmacokinetic approach for treatment of cocaine abuse and dependence.4 However, the catalytic activity of this plasma enzyme is relatively low against the naturally occurring (−)-cocaine. (−)-cocaine has a plasma half-life of ~ 45 −90 min, long enough for manifestation of the effects on central nervous system (CNS) which peak in minutes.13,14 Hence, BChE mutants with high activity against (−)-cocaine are highly desired for use as an exogenous enzyme in humans.
Chart 1.
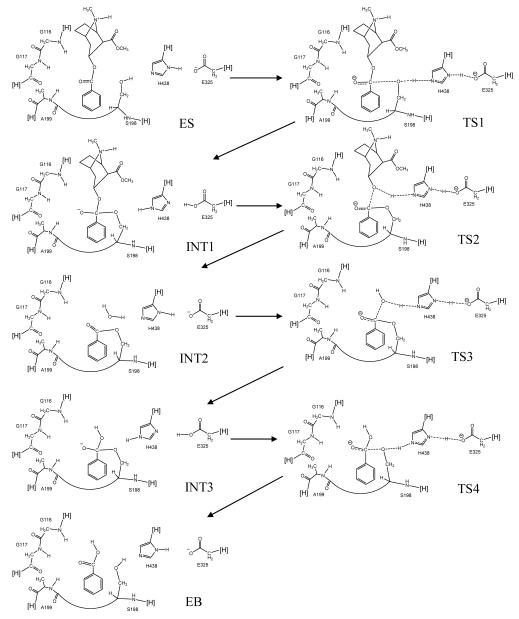
Schematic representation of BChE-catalyzed hydrolysis of (−)-cocaine. Only QM-treated high-layer part of the reaction system in the ONIOM (QM/MM) calculations are drawn. Notation [H] refers to a non-hydrogen atom in the MM-treated low-layer part of the protein and the cut covalent bond with this atom is saturated by a hydrogen atom. The transition covalent bonds existing in all of the transition states are indicated with dash lines.
For the purpose of rational design of high-activity BChE mutants against (−)-cocaine, we first need to understand the detailed reaction mechanism concerning how cocaine is hydrolyzed in human BChE. Previous computational studies15,16,17,18,19 led to understanding how cocaine binds with BChE, although only one of the computational studies18 reported so far dealt with the reaction pathway for BChE-catalyzed cocaine hydrolysis. The MD simulations of the prereactive BChE-cocaine binding and ab initio reaction coordinate calculations with an active site model18 demonstrated that the fundamental reaction pathway for BChE-catalyzed hydrolysis of cocaine consists of acylation and deacylation stages. A total of four individual reaction steps are involved in the acylation and deacylation stages, which is similar to the mechanism for ester hydrolysis catalyzed by other serine hydrolases, and the calculated highest free energy barrier is associated with the third reaction step.18 Based on the computational modeling,18 amino acid residues S198, H438, and E325 clearly form a catalytic triad. The MD simulations of the prereactive BChE-cocaine complexes18 also led to a mechanistic hypothesis that the peptidic NH groups of G116, G117, and A199 could constitute an oxyanion hole forming up to three strong N-H…O hydrogen bonds with the negatively charged carbonyl oxygen of the benzoyl ester group of cocaine in certain transition states and intermediates. The possible oxyanion hole and the hypothetic N-H…O hydrogen bonds would be similar to those found for acetylcholinesterase (AChE)-catalyzed hydrolysis of neurotransmitter acetylcholine (ACh).20
In light of the reported computational studies,18 the key to the rational design of high-activity BChE mutants against (−)-cocaine is to further understand the protein environmental effects on the reaction process based on the fundamental reaction pathway. To understand the protein environmental effects on the reaction process, especially on the hypothetic N-H…O hydrogen bonds with the possible oxyanion hole, is particularly important concerning how to better stabilize the transition states through certain mutations and, therefore, lower the activation free energy for the BChE-catalyzed hydrolysis of (−)-cocaine. However, the hypothetic N-H…O hydrogen bonds existing in the transition states and intermediates with the protein environment have not been simulated. This is because the previous ab initio reaction coordinate calculations18 with an active site model can only account for breaking and formation of covalent bonds during the catalytic reaction process. The used active site model18 includes cocaine and functional groups from the catalytic triad (S198, H438, and E325) and the possible oxyanion hole (G116, G117, and A199), giving only 78 atoms in the gas phase (without any explicit or implicit solvent). In the ab initio reaction coordinate calculations,18 the geometries were optimized at the HF/3-21G level (and also at the B3LYP/6-31+G* level for the rate-determining step) without any geometric constraint, followed by single-point energy calculations at the B3LYP/6-31+G*, B3LYP/6-31++G**, B3LYP/6-311++G**, and MP2/6-31+G* levels; no MD simulation was performed on any of the transition states and intermediates. The calculations with such an active site model cannot account for the complicated protein environmental effects on the reaction process, particularly on the transition states and intermediates binding with the protein environment.
To examine the hypothetic N-H…O hydrogen bonds with the possible oxyanion hole during the reaction process, one needs to simulate the entire BChE-cocaine system in its transition states and intermediates. In the present computational study, we have performed extensive MD simulations and quantum mechanical/molecular mechanical (QM/MM) calculations on the entire BChE-cocaine structures for all of the transition states and intermediates, in addition to the product and prereactive complex. The MD simulations of the transition states were subject to certain constraints on the bond lengths of covalent bonds that are breaking and forming based on the reaction coordinate calculations with the active site model. All of the simulations and calculations consistently support the hypothesis of the oxyanion hole and the corresponding N-H…O hydrogen bonds and further reveal remarkable features of the protein environmental effects on BChE-catalyzed hydrolysis of (−)-cocaine. For comparison, we also simulated prereactive AChE-ACh binding, demonstrating important mechanistic similarity and differences between BChE-catalyzed hydrolysis of (−)-cocaine and AChE-catalyzed hydrolysis of ACh.
Computational methods
Amber7 program package21 was used to perform all MD simulations on the prereactive reactant, transition states, intermediates, and product during BChE-catalyzed hydrolysis of (−)-cocaine. We must address a critical issue before describing how we performed any MD simulation on a transition state. In principle, MD simulation using a classical force field (molecular mechanics) can only simulate a stable structure corresponding to a local minimum on the potential energy surface, whereas a transition state during a reaction process is always associated with a first-order saddle point on the potential energy surface. Hence, MD simulation using a classical force field cannot directly simulate a transition state without any restraint on the geometry of the transition state. Nevertheless, if we can technically remove the freedom of imaginary vibration in the transition state structure, then the number of vibrational freedoms (normal vibration modes) for a nonlinear molecule will decrease from 3N – 6. The transition state structure is associated with a local minimum on the potential energy surface within a subspace of the reduced vibrational freedoms, although it is associated with a first-order saddle point on the potential energy surface with all of the 3N – 6 vibrational freedoms. Theoretically, the vibrational freedom associated with the imaginary vibrational frequency in the transition state structure can be removed by appropriately freezing the reaction coordinate. The reaction coordinate corresponding to the imaginary vibration of the transition state is generally characterized by a combination of some key geometric parameters. These key geometric parameters are bond lengths of the forming and breaking covalent bonds for BChE-catalyzed hydrolysis of cocaine, as seen in Chart 1. Thus, we just need to maintain the bond lengths of the forming and breaking covalent bonds during the MD simulation on each transition state. Technically, we can maintain the bond lengths of the forming and breaking covalent bonds by simply fixing all atoms within the reaction center, by using some constraints on the forming and breaking covalent bonds, or by redefining the forming and breaking covalent bonds. It should be pointed out that the only purpose of performing such type of MD simulation on a transition state is to examine the dynamic change of the protein environment surrounding the reaction center and the interaction between the reaction center and the protein environment. We are only interested in the simulated structures, as the total energies calculated in this way are meaningless.
The starting three-dimensional (3D) structure of human BChE used in our MD simulations came from the X-ray crystal structure22 deposited in the Protein Data Bank (pdb code: 1POP).23 The missing residues (D2, D3, E255, D378, D379, N455, L530, E531, and M532) in the X-ray crystal structure were built using the automated homology modeling tool Modeler24,25/InsightII software (Accelrys, Inc.) with the default parameters. To determine the position of (−)-cocaine, in the prereactive BChE-cocaine complex, the backbone atoms of the human BChE were superimposed with the corresponding atoms in the previously simulated prereactive BChE-substrate complex based on a homology model of BChE. Then, we took the coordinates of the human BChE and cocaine as the initial structure of prereactive BChE-cocaine complex for MD simulation in water. Starting from the simulated prereactive BChE-cocaine complex and the geometries obtained from the previous ab initio reaction coordinate calculations18 on cocaine with a BChE active site model, we were able to construct initial geometries of all transition states, intermediates, and product involved in the BChE-catalyzed hydrolysis of cocaine for performing MD simulations on these structures with the entire BChE structure in water. To construct each of these initial geometries used for MD simulations, the active site atoms in the optimized geometry obtained from the ab initio reaction coordinate calculations were superimposed with the corresponding atoms in the simulated prereactive BChE-cocaine complex. Thus, in these initial geometries used for MD simulations, the geometric parameters within the reaction center are the same as those obtained from our previous ab initio reaction coordinate calculations on the model reaction system.18 The difference is that the initial geometries constructed in this study include the entire protein environment. So, the interaction of the reaction center with its protein environment can be examined through computational studies on the entire reaction system.
We note that S198, H438, and E325 in the catalytic triad are no longer standard residues in the transition states and intermediates. For each transition state or intermediate, the fundamental structure of the reaction center was maintained by redefining the covalent bonds that will break or form during the reaction process. Such a covalent bond is only partially formed or partially broken in a transition state. For convenience, this type of partial covalent bond existing in a transition state structure will be called “transition” covalent bond in our discussion below. The transition covalent bonds are indicated with dash lines in Chart 1 for all transition states. As shown in Chart 1, the transition covalent bonds exist within the catalytic triad and among the catalytic triad, cocaine, and a water molecule. For example, in the first intermediate (INT1 in Chart 1), a bond was defined between the carbonyl carbon of cocaine benzoyl ester group and the Oγ atom of S198, so was a bond between S198 Oγ and H438 Hε2 and a bond between H438 Hδ1 and E325 Oε2. In the first transition state (TS1 in Chart 1), the similar (transition) bonds were also defined, although the corresponding bond lengths needed were all different. In addition, we also needed to redefine the types of atoms involved in the breaking or formation of covalent bonds and to modify/define some bond angles and dihedral angles within the reaction center in order for the MD simulation to correctly maintain the key geometric parameters obtained from the previous ab initio reaction coordinate calculations.18
The partial atomic charges for the non-standard residue atoms, including cocaine atoms, were calculated by using the RESP protocol implemented in the Antechamber module of the Amber7 package following electrostatic potential (ESP) calculations at ab initio HF/6-31G* level. The geometries used in the ESP calculations came from those obtained from the previous ab initio reaction coordinate calculations,18 but the functional groups representing the oxyanion hole were removed. Thus, residues G116, G117, and A199 were the standard residues in our MD simulations.
Each aforementioned initial structure during the reaction process was neutralized by adding chloride counterions and was solvated in a rectangular box of TIP3P water molecules26 with a minimum solute-wall distance of 10 Å. The total numbers of atoms in the solvated protein structures for the MD simulations are nearly 70,000, although the total number of atoms of BChE and cocaine is only 8417. The general procedure for carrying out the MD simulations in water is essentially the same as that used in our previously reported other computational studies.18,27 The MD simulations in this study were performed by using the Sander module of Amber7 package. The solvated systems were carefully equilibrated and fully energy minimized. These systems were gradually heated from T = 10 K to T = 298.15 K in 30 ps before production MD simulation of 1.5 ns or longer, making sure that we obtained a stable MD trajectory for each of the simulated structures. The time step used for the MD simulations was 2 fs. Periodic boundary conditions in the NPT ensemble at T = 298.15 K with Berendsen temperature coupling28 and P = 1 atm with isotropic molecule-based scaling28 were applied. The SHAKE algorithm29 was used to fix all covalent bonds containing hydrogen atoms. The non-bonded pair list was updated every 10 steps. The particle mesh Ewald (PME) method30 was used to treat long-range electrostatic interactions. A residue-based cutoff of 10 Å was utilized to the non-covalent interactions. The coordinates of the simulated systems were collected every 1 ps during the production MD stages.
For each structure examined, after the MD simulation was completed and a stable MD trajectory was obtained, all of the collected snapshots of the simulated structure, excluding those before the trajectory was stabilized, were averaged. The average structure was stripped off the solvent water molecules and re-solvated with TIP3P water molecules in order to get rid of some unexpected close contacts of some side chain atoms due to averaging. The re-solvated average structure was then energy-minimized in a water bath again. To examine the solvent effects on the geometry, we also performed energy minimization on the structure after removing all solvent water molecules from the energy-minimized re-solvated average structure.
The finally energy-minimized average coordinates of each structure simulated were used as an initial geometry to carry out further geometry optimization by using the ONIOM approach31 implemented in the Gaussian03 program.32 Two layers were defined in our ONIOM calculation: the high layer (depicted in Chart 1) was calculated quantum mechanically at the ab initio HF/3-21G level and the low layer was calculated molecular mechanically by using the Amber force field as used in our MD simulations and energy minimizations with Amber7 program. The ONIOM calculations at the HF/3-21G:Amber level in this study are a type of QM/MM calculations.33,34 As depicted in Chart 1, for all of these QM/MM calculations, the same part of the protein was included in the QM-treated high layer. So, the QM-treated high layer included the three residues (G116, G117, and A199) of the possible oxyanion hole, key functional groups from the catalytic triad (S198, H438, and E325), and cocaine, whereas the entire protein structure of BChE was included in the MM-treated low layer. In addition, a water molecule was also included in the QM-treated high layer for the calculations on the structures involved in the deacylation stage of the reaction. We developed a C program to automatically generate the input files for the ONIOM calculations following the MD simulation and subsequent energy minimization in order to make sure that the atom types used for all low-layer atoms are the same as what we used in the Amber7. The lengths of the transition covalent bonds were frozen during the QM/MM geometry optimizations of the transition state structures. For comparison, we also carried out QM/MM geometry optimizations on the transition states with only one transition C-O bond (involving the carbonyl carbon of cocaine) fixed. No bond length, angle, or dihedral angle was frozen during the QM/MM optimization of other structures. To examine whether the HF/3-21G level for the high layer is adequate for the geometry optimization, the geometries of TS3 and INT3 optimized at the HF/3-21G:Amber level were further refined at the more sophisticated B3LYP/6-31+G*:Amber level.
Most of the simulations and calculations were performed in parallel on a supercomputer (HP Superdome, with 4 nodes for 256 shared-memory processors) at the Center for Computational Sciences, University of Kentucky. The other computations were carried out on a 34-processors IBM ×335 Linux cluster and SGI Fuel workstations in our own lab.
Results and discussion
Structures from the MD simulations and QM/MM geometry optimizations
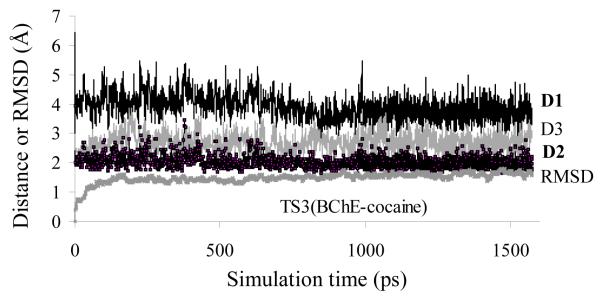
All transition states and intermediates were simulated for at least 1.5 ns to allow the protein structure to have sufficient time to adapt to the fixed reaction center geometries obtained from the ab initio reaction coordinate calculations. The MD trajectories actually became stable quickly, so were the internuclear distances involved in the potential N-H…O hydrogen bonds, i.e. the distances from the carbonyl oxygen (O31) of cocaine benzoyl ester to the NH hydrogen atoms of G116, G117, and A199. Depicted in Figure 1 (and supplementary material) are plots of these important distances in the MD-simulated transition states and intermediates for BChE-catalyzed hydrolysis of (−)-cocaine versus the simulation time, along with root-mean-square deviation (RMSD) of the coordinates of backbone atoms in the simulated structure from those in the initial structure. The numerical results are summarized in Table 1. As seen in Table 1, the RMSD values are all smaller than 2.0 Å for all of the MD trajectories, demonstrating that the backbone of BChE did not dramatically change in going from the prereactive BChE-cocaine complex (ES) to the transition states, intermediates, and product. Some key distances of the cocaine carbonyl oxygen O31 to the NH hydrogen atoms of G116, G117, and A199 are denoted by D1, D2, and D3, respectively, in Figure 1 and Table 1 (see supplementary material for more detailed results).
Figure 1.
Plots of the key internuclear distances (in Å) versus the simulation time in the simulated transition state of the third reaction step (TS3) for BChE-catalyzed hydrolysis of (−)-cocaine. Traces D1, D2, and D3 refer to the distances between the carbonyl oxygen of cocaine benzoyl ester and the NH hydrogen of G116, G117, and A199, respectively. RMSD represents the root-mean-square deviation (in Å) of the simulated positions of BChE backbone atoms from those in the initial structure.
Table 1.
Summary of the MD-simulated and optimized key distances (Å) and the root-mean-square deviation (RMSD) of the simulated structures from the initial structure.
| Structure | Method | Distancea | RMSDb | ||||
|---|---|---|---|---|---|---|---|
| D1 | D2 | D3 | D4 | D5 | |||
| ES | MDc | 3.79 | 2.14 | 4.47 | 3.11 | 4.89 | 1.28 |
| QM/MM(a)d | 4.05 | 2.16 | 4.60 | 3.41 | 5.22 | ||
| TS1 | MD | 4.59 | 2.91 | 1.92 | 2.00 | 3.61 | 1.59 |
| QM/MM(a) | 4.12 | 2.24 | 2.04 | 2.34 | 4.36 | ||
| INT1 | MD | 3.92 | 1.94 | 2.36 | 2.08 | 4.01 | 1.57 |
| QM/MM(a) | 4.09 | 2.03 | 1.93 | 2.06 | 4.04 | ||
| TS2 | MD | 3.49 | 1.98 | 2.22 | 2.26 | 4.13 | 1.53 |
| QM/MM(a) | 3.55 | 1.80 | 1.90 | 2.36 | 4.54 | ||
| INT2 | MD | 3.12 | 2.15 | 2.50 | 3.94 | 3.37 | 1.76 |
| QM/MM(a) | 2.60 | 1.89 | 2.81 | 5.15 | 3.41 | ||
| TS3 | MD | 3.76 | 1.98 | 2.68 | 3.86 | 1.96 | 1.61 |
| QM/MM(a) | 3.76 | 2.14 | 1.87 | 4.04 | 2.07 | ||
| QM/MM(b)d | 4.10 | 2.27 | 1.85 | 3.99 | 2.07 | ||
| INT3 | MD | 3.01 | 2.03 | 1.88 | 4.05 | 5.68 | 1.70 |
| QM/MM(a) | 2.04 | 1.80 | 1.64 | 4.27 | 6.07 | ||
| QM/MM(b) | 2.23 | 1.73 | 1.69 | 4.18 | 6.00 | ||
| TS4 | MD | 3.73 | 2.04 | 1.87 | 3.04 | 4.26 | 1.53 |
| QM/MM(a) | 3.24 | 1.91 | 1.69 | 2.96 | 4.84 | ||
| EB | MD | 4.14 | 3.22 | 4.23 | 3.64 | 2.06 | 1.48 |
| QM/MM(a) | 3.37 | 2.14 | 2.12 | 4.87 | 2.87 | ||
| BChEe | MD | 3.64 | 4.35 | 1.49 | |||
| AChE-ACh f | MD | 2.32 | 1.92 | 3.53 | 4.17 | 5.40 | 1.37 |
D1, D2, and D3 represent the distances between the carbonyl oxygen of cocaine benzoyl ester and the NH hydrogen of G116, G117, and A199, respectively. D4 and D5 respectively refer to the distances between the NH hydrogen of G116 and the oxygen atoms Oε1 and Oε2 of E197 side chain.
Root-mean-square deviation of the coordinates of backbone atoms in the simulated structure from those in the initial structure.
Average distances from the stable trajectory of MD simulation.
Distances in the geometry optimized by performing the QM/MM calculation at the HF/3-21G:Amber level (a) or at the B3LYP/6-31+G*:Amber level (b). The bond lengths for all of the transition bonds were fixed in the geometry optimizations.
Results for the pure protein without cocaine or any other ligand in the active site.
Results for prereactive complex between mouse acetylcholinesterase (AChE) and acetylcholine (ACh). For this system, D1, D2, and D3 represent the distances between the carbonyl oxygen of ACh and the NH hydrogen of G121, G122, and A204, respectively. D4 and D5 respectively refer to the distances between the NH hydrogen of G121 and the oxygen atoms Oε1 and Oε2 of E202 side chain.
As seen in Table 1, in the QM/MM-optimized geometries, the distances optimized for TS3 and INT3 (associated with the rate-determining step of the chemical reaction18) at the B3LYP/6-31+G*:Amber level are all reasonably close to the corresponding distances optimized at the HF/3-21G:Amber level, showing that the HF/3-21G level is adequate for the treatment of high-layer atoms in this study. This is consistent with the conclusion based on the previous ab initio QM calculations18 on the active site model. The previous ab initio QM calculations18 demonstrated that the HF/3-21G level is adequate for the geometry optimizations and that the energy barriers calculated at the B3LYP/6-31+G*//HF/3-21G level are all close to those calculated at the higher levels using the geometries optimized at the B3LYP/6-31G* level. Further, the QM/MM-optimized distances are also close to the corresponding distances in the MM-optimized structures, suggesting that it is reasonable using the classical force field for this kind of enzymatic reaction system.
We note that the significance of the QM/MM calculations described in this report is limited, because the geometries of the transition states were not fully optimized and, thus, the energy barriers cannot be evaluated explicitly.35 The QM/MM results for the transition states summarized in Table 1 refer to the geometries optimized with all the transition bond lengths fixed. For comparison, the QM/MM geometry optimizations were also performed by fixing only the length of one transition C-O bond involving the carbonyl carbon of cocaine. The geometric parameters (provided as supplementary material) in the geometries optimized in this way are very close to the corresponding distances listed in Table 1. For example, the D1, D2, D3, D4, and D5 values (i.e. 3.73, 2.04, 1.86, 4.06, and 2.09 Å, respectively) in the TS3 geometry optimized by fixing only the length of the forming C-O bond (at 1.80 Å)18 are all in good agreement with the corresponding distances (i.e. 3.76, 2.14, 1.87, 4.04, and 2.07 Å, respectively) listed in Table 1.
To examine how the protein environment affects the reaction coordinate, we first checked the calculated force (i.e. the –DE/DX value in the Gaussian03 output) associated with the forming C-O bond length (an internal coordinate) in the TS3 geometry optimized with the forming C-O bond length fixed at 1.80 Å and found that the –DE/DX value for the forming C-O bond length in the optimized TS3 geometry is −0.0321 au. This negative –DE/DX value suggests that the first-order saddle point corresponding to TS3 should have the forming C-O bond length longer than 1.80 Å. Hence, we also carried out the QM/MM geometry optimization on TS3 with this forming C-O bond length fixed at 2.00 Å and found that the –DE/DX value for the forming C-O bond length in the optimized TS3 geometry is 0.0067 au, suggesting that the first-order saddle point corresponding to TS3 should have the forming C-O bond length shorter than 2.00 Å. Clearly, the optimum length of the forming C-O bond in the TS3 geometry corresponding to the first-order saddle point should be between 1.80 and 2.00 Å, with 2.00 Å being the up limit. This means that the protein environmental effects change the forming C-O bond length for < 0.20 Å. The change of < 0.20 Å is significant, but not dramatic. When the forming C-O bond length changes from 1.80 Å to 2.00 Å, the optimized D1, D2, and D3 values change from 3.73, 2.04, and 1.86 Å to 3.57, 2.19, and 1.83 Å, respectively. It follows that the limited geometric change in the reaction center does not considerably change the hydrogen bonding with the oxyanion hole.
The computational results summarized in Table 1 confirm the existence of the oxyanion hole consisting of G116, G117, and A199 during BChE-catalyzed hydrolysis of cocaine, because G116, G117, and A199 all had hydrogen bonding or close interaction with O31 of cocaine in at least one transition state or intermediate. However, the N-H…O hydrogen bonds are mostly between cocaine O31 and the NH hydrogen atoms of G117 and A199. The NH hydrogen of G116 was not hydrogen-bonded to cocaine O31, except very briefly in INT2 and INT3. Only G117 had an N-H…O hydrogen bond with cocaine O31 in the MD-simulated prereactive BChE-cocaine complex ES. The simulated average H…O distance in this N-H…O hydrogen bond is 2.14 Å. After further energy minimization with Amber7 and the QM/MM geometry optimization, this H…O distance became 2.02 and 2.16 Å, respectively, as seen in Table 1 (and supplementary material). Changing from the prereactive complex to the transition states and intermediates, another N-H…O hydrogen bond formed between cocaine O31 and A199 while the N-H…O hydrogen bond with G117 was fully or partially maintained. For example, the N-H…O hydrogen bond between cocaine O31 and G117 existed partially in TS1 and fully in INT1. Thus, compared to ES, the transition states and intermediates were stabilized further by the hydrogen bonding of cocaine O31 with A199.
Hydrogen bonding energies
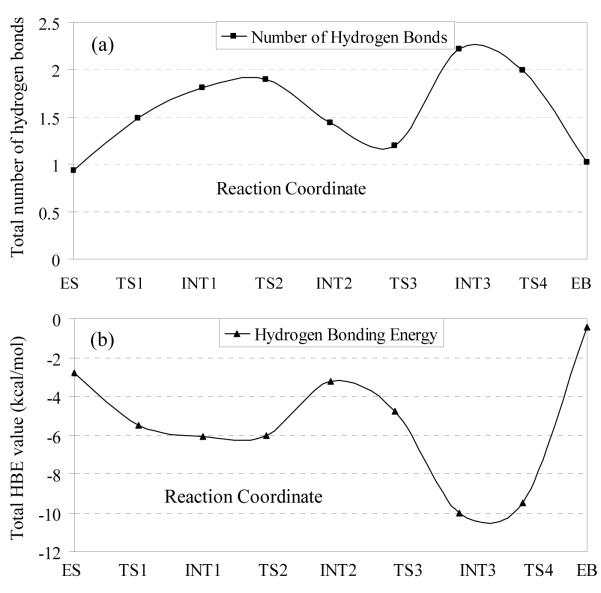
To better represent the overall strength of hydrogen bonding of cocaine O31 with the oxyanion hole in each MD-simulated structure, we theoretically estimated total number of hydrogen bonds with cocaine O31 by using a cutoff value of 2.5 Å for the H…O distances in order to roughly determine whether any hydrogen bond existed in any snapshot of MD simulation. Thus, we were able to calculate the total number of hydrogen bonds with cocaine O31 at each snapshot. The simulated total number of hydrogen bonds with cocaine O31 was estimated as the average over all of the snapshots taken in stable range of the MD trajectory. Figure 2(a) shows the simulated total numbers of hydrogen bonds estimated in this way for all of the structures involved in the reaction process: ES → TS1 → INT1 → TS2 → INT2 → TS3 → INT3 → TS4 → EB. Here, TS’s refer to the transition states, INT’s represent the intermediates, and EB refers to the BChE-benzoate acid complex. The results depicted in Figure 2(a) clearly suggest that the total average number of hydrogen bonds between cocaine O31 and the oxyanion hole gradually increased from ES to TS1, INT1, and TS2, whereas the total average number of hydrogen bonds gradually decreased from TS2 to INT2 and TS3 before a remarkable increase from TS3 to INT3. In going from INT3 to TS4 and EB, the total average number of hydrogen bonds gradually decreased again.
Figure 2.
(a) The total average number of hydrogen bonds between the carbonyl oxygen of cocaine benzoyl ester and the oxyanion hole in the simulated the prereactive BChE-cocaine complex (ES), transition states, intermediates, and product for BChE-catalyzed hydrolysis of (−)-cocaine. (b) The corresponding total hydrogen bonding energy calculated.
As seen in Table 1, the H…O distances optimized at various levels of theory are consistent with the change of the simulated total average number of hydrogen bonds during the reaction process. For example, the optimized distances revealed only one N-H…O hydrogen bond with G117 in ES, a stronger N-H…O hydrogen bond with A199 and a weaker N-H…O hydrogen bond with G117 in TS1, and stronger N-H…O hydrogen bonds with both G117 and A199 in INT1. This supports the conclusion of the gradual increase of the hydrogen bonding for ES → TS1 → INT1.
With each of the simulated H…O distances, we also estimated the hydrogen bonding energy (HBE) by using the general HBE equation implemented in AutoDock 3.0 program suite.36 Specifically, for each hydrogen bond with cocaine O31, a HBE value can be evaluated with each snapshot of the MD-simulated structure. The final HBE of the MD-simulated hydrogen bond is considered to be the average HBE value of all snapshots taken from the stable MD trajectory. The total hydrogen bonding energy between cocaine O31 and the oxyanion hole in each MD-simulated state is depicted in Figure 2(b).
Comparing Figure 2(b) with Figure 2(a), one can clearly see that the calculated total hydrogen bonding energy (HBE) is mostly proportional to the simulated total number of hydrogen bonds, but the exception exists in some cases, particularly for the change from INT2 to TS3. The exception is due to the reason that different hydrogen bonds may have quite different average H…O distances and, therefore, have quite different hydrogen bonding energies. Thus, the total hydrogen bonding energy should be a better indicator of the overall hydrogen bonding between cocaine O31 and the oxyanion hole. In going from ES to TS1, the simulated total number of hydrogen bonds increases from 0.93 to 1.49 and, correspondingly, the calculated total HBE value changes from −2.9 kcal/mol to −5.5 kcal/mol, suggesting that the hydrogen bonding decreases the energy barrier for the first reaction step by ~2.6 kcal/mol. In going from INT1 to TS2, the simulated total number of hydrogen bonds changes from 1.81 to 1.90 and the calculated total HBE value changes from −6.1 kcal/mol to −6.0 kcal/mol, indicating that the hydrogen bonding effects on the energy barrier for the second reaction step are negligible. In going from INT2 to TS3, the simulated total number of hydrogen bonds decreases from 1.44 to 1.20, whereas the calculated total HBE value changes from −3.2 kcal/mol to −4.8 kcal/mol, showing that the average hydrogen bonding strength per bond in TS3 is stronger than that in INT2. These energetic data suggest that the hydrogen bonding effects might decrease the energy barrier for the third reaction step by ~1.6 kcal/mol. In going from INT3 to TS4, the simulated total number of hydrogen bonds decreases from 2.22 to 1.99 and the calculated total HBE value changes from −10.1 kcal/mol to −9.5 kcal/mol, suggesting that the hydrogen bonding may slightly decrease the energy barrier for the fourth reaction step by ~0.6 kcal/mol.
Comparison with acetylcholinesterase-catalyzed hydrolysis of acetylcholine
It is interesting to compare the mechanism of BChE-catalyzed hydrolysis of cocaine with that of acetylcholinesterase (AChE)-catalyzed hydrolysis of neurotransmitter acetylcholine (ACh), because AChE and BChE have very similar active sites including the same type of catalytic triad and the same type of oxyanion hole. In terms of mouse AChE, the catalytic triad consists of S203, H447, and E334 and the oxyanion hole consists of G121, G122, and A204. The only significant difference is that the cavity of BChE active site is larger so that it can accommodate a larger substrate like cocaine. McCammon et al.20,37,38 reported MD simulations and QM/MM calculations on mouse AChE-ACh system. Their MD simulations37,38 were performed on the prereactive AChE-ACh complex, whereas their QM/MM calculations20 were carried out on the initial step of the acylation stage of the AChE-catalyzed ACh hydrolysis. Their QM/MM results indicate that in the AChE-ACh Michaelis complex, two hydrogen bonds exist between the carbonyl oxygen of ACh and the peptidic NH groups of G121 and G122. In going from the AChE-ACh Michaelis complex to the (first) transition state and (first) intermediate, the distance between the carbonyl oxygen of ACh and NH group of A204 becomes shorter, and the third hydrogen bond is formed both in the transition state and in the tetrahedral intermediate.20 Based on the structural similarity of these two closely related cholinesterases, one might easily assume that the N-H…O hydrogen bonding between the carbonyl oxygen of the substrate and the three residues of oxyanion hole in the BChE-cocaine and in the AChE-ACh systems should be very close to each other during the catalytic hydrolysis processes. However, a remarkable difference exists on the role of the first residue, i.e. G121 in AChE or G116 in BChE, of the oxyanion hole. Our MD simulations and QM/MM calculations on BChE-catalyzed cocaine hydrolysis revealed that G116 was never hydrogen-bonded to O31 of cocaine during the acylation stage and was rarely involved in hydrogen bonding with O31 of cocaine during the deacylation stage. Structures ES, TS1, INT1, and TS2 simulated in this study are associated with the acylation stage, in which all of the cocaine atoms were included in the simulations and calculations. The simulated structures INT2, TS3, INT3, and TS4 are all associated with the deacylation stage, in which the free product ecgonine methyl ester had left the active site and cocaine only had benzoate atoms included in the simulations and calculations.
To make sure that this remarkable difference between the structures simulated for these two hydrolysis processes was not an artifact of the possibly different computational strategies used for the two different reaction processes, we also simulated the prereactive complex between ACh and mouse AChE (using X-ray crystal structure 1MAH in the Protein Data Bank23) by using the same MD approach as we used for the BChE-cocaine system. As seen in Table 1, our MD simulation of the prereactive AChE-ACh complex also revealed an N-H…O hydrogen bond between the carbonyl oxygen of ACh and the NH hydrogen of G121, which is qualitatively consistent with the computational results reported by McCammon et al.20,37,38 This consistency supports the remarkable difference between the two catalytic hydrolysis processes.
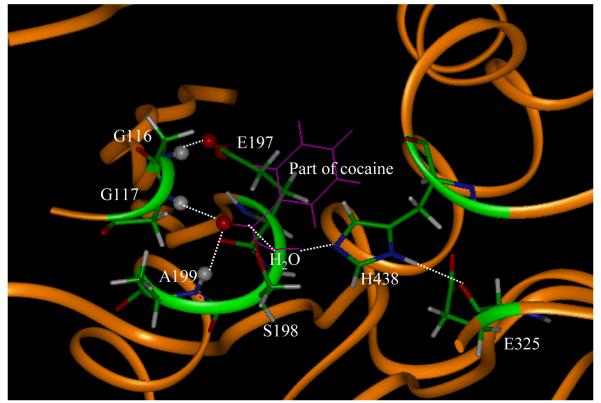
The remaining question is why the role of G116 in human BChE is remarkably different from that of G121 in mouse AChE in terms of the hydrogen bonding with the substrate. Interestingly, our results revealed an unexpected hydrogen bond between the NH hydrogen of G116 and the carboxylate group (with either Oε1 or Oε2 atom) of E197 side chain in all of the simulated structures during the reaction process, except INT3. Part of the QM/MM-optimized TS3 structure is depicted in Figure 3, showing the unexpected hydrogen bond between the NH hydrogen of G116 and the oxygen of E197 side chain. Having a hydrogen bond with the carboxylate group of E197 side chain, the NH hydrogen of G116 was not available to form another hydrogen bond with O31 of cocaine at the same time. We noticed that the NH hydrogen of G116 hydrogen-bonded mostly with Oε1 or Oε2 of E197 during the MD simulations of ES, TS1, INT1, TS2, TS3, and EB structures. Such an N-H…Oε1 or N-H…Oε2 hydrogen bond should be strong. However, in the simulated INT2 and TS4 structures, the relatively weaker N-H…Oε1 and N-H…Oε2 hydrogen bonds were exchanged frequently during the MD simulations. Due to the frequent exchange of the relatively weaker N-H…Oε1 and N-H…Oε2 hydrogen bonds, the difference between the average H…Oε1 and H…Oε2 distances was smaller than those in the above-mentioned structures and the NH hydrogen of G116 was sometimes able to be closer to O31 of cocaine. In the simulated INT3 structure, the NH hydrogen of G116 did not have a hydrogen bond with E197 and, therefore, had a hydrogen bond with O31 of cocaine for significant part of the simulation time.
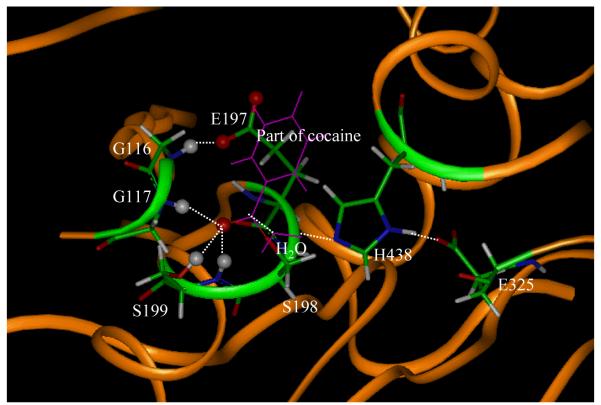
Figure 3.
Part of the MD-simulated TS3 structure for the cocaine hydrolysis catalyzed by wild-type BChE. The geometry was energy-minimized molecular mechanically starting from the MD-simulated average structure. Highlighted as balls are the H atoms (grey balls) and O atoms (red balls) involved in the key hydrogen bonds with the oxyanion hole.
To further examine the possible effect of the bulky ligand (cocaine) on the hydrogen bond between G116 and E197, we also carried out an MD simulation on BChE in a water bath without cocaine or any other ligand. As seen in Table 1, the simulated BChE structure did not show a hydrogen bond between G116 and E197, showing that the ligand has a role in the hydrogen bonding between the NH hydrogen of G116 and the carboxyl group of E197 side chain.
Insights into rational design of BChE mutants
Generally speaking, for rational design of a mutant enzyme with a higher catalytic activity for a given substrate, one needs to find a mutation that can accelerate the rate-determining step of the entire catalytic reaction while the other steps are not slowed down by the mutation. To accelerate a chemical reaction step, one needs to find a mutation that can lower the energy barrier for the reaction step. For this purpose, the knowledge concerning the hydrogen bonding between the carbonyl oxygen (O31) of cocaine benzoyl ester and the oxyanion hole of the wild-type BChE during the reaction process will be valuable for rational design of BChE mutants with the desirable higher catalytic activity for cocaine hydrolysis. The aforementioned MD and QM/MM results indicate that the effects of hydrogen bonding between cocaine O31 and the oxyanion hole of wild-type BChE should be insignificant for the (less-important) second and fourth reaction steps. However, the overall hydrogen bonding effects are expected to lower the energy barriers for the first and third reaction steps of the cocaine hydrolysis catalyzed by the wild-type BChE, although the change of the estimated total hydrogen binding energy from INT2 to TS3 (the rate-determining step of the chemical reaction process) is only ~1.6 kcal/mol for the wild-type BChE. To decrease the energy barrier for the third reaction step, one can try to design a possible mutation that can enhance the hydrogen bonding in TS3 so that the hydrogen bonding in TS3 becomes much stronger than that in INT2. To theoretically test this idea, we also carried out the same type of MD simulations on the INT2 and TS3 structures with a new BChE mutant, A199S/A328W/Y332G, in water. The MD simulations clearly revealed that, in addition to the hydrogen bond between cocaine O31 and the backbone NH of G117, the new residue at position #199, i.e. S199, forms hydrogen bonding with cocaine O31 through both the backbone NH and the hydroxyl group of the S199 side chain in the simulated TS3 structure (see Figure 4). However, the MD simulation on the INT2 structure revealed no hydrogen bonding between cocaine O31 and hydroxyl group of S199 side chain. The total hydrogen bonding energy between cocaine O31 and the oxyanion hole of A199S/A328W/Y332G BChE was calculated as −8.8 kcal/mol for TS3 and −1.4 kcal/mol for INT2. The difference in the total hydrogen bonding energy between INT2 and TS3 becomes ~7.4 kcal/mol for this BChE mutant, thus indicating that the formed new hydrogen bond with S199 can significantly stabilize the transition state TS3. The calculated results predict that mutation A199S/A328W/Y332G can significantly decrease the energy barrier for the third reaction step and, therefore, could significantly increase the catalytic activity of the enzyme for the cocaine hydrolysis.
Figure 4.
Part of the MD-simulated TS3 structure for the cocaine hydrolysis catalyzed by A199S/A328W/Y332G BChE. The geometry was energy-minimized molecular mechanically starting from the MD-simulated average structure. Highlighted as balls are the H atoms (grey balls) and O atoms (red balls) involved in the key hydrogen bonds with the oxyanion hole.
Conclusion
A series of molecular dynamics (MD) simulations and quantum mechanical/molecular mechanical (QM/MM) calculations consistently reveal a unique role of the oxyanion hole (consisting of G116, G117, and A199) in the process of human butyrylcholinesterase (BChE)-catalyzed hydrolysis of (−)-cocaine, as compared to that in mouse acetylcholinesterase (AChE)-catalyzed hydrolysis of acetylcholine (ACh). The initial step of the acylation in AChE-catalyzed hydrolysis of ACh was known to involve three hydrogen bonds of ACh carbonyl oxygen with the NH groups of G121, G122, and A204; two hydrogen bonds with the NH groups of G121 and G122 in the Michaelis-Menten complex and three hydrogen bonds with the NH groups of G121, G122, and A204 in the transition state and intermediate. However, for BChE-catalyzed hydrolysis of cocaine, only G117 has a hydrogen bond with the carbonyl oxygen (O31) of the cocaine benzoyl ester in the prereactive BChE-cocaine complex. In going from the prereactive complex to the transition states and intermediates, another hydrogen bond forms between O31 of cocaine and NH hydrogen of A199 while the hydrogen bond with G117 is kept, but G116 is not available to form a hydrogen bond with O31 of cocaine in the acylation because an unexpected hydrogen bond exists between the NH hydrogen of G116 and the carboxyl group of E197 side chain. The NH hydrogen of G116 is partially available to form a weak hydrogen bond with O31 of cocaine only in some structures involved in the deacylation.
Based on the MD simulations and the hydrogen bonding energy estimation, the change of the estimated hydrogen bonding energy between the oxyanion hole and O31 of cocaine during the reaction process demonstrates how the protein environment can affect the energy barrier for each step of BChE-catalyzed hydrolysis of cocaine. The results demonstrate that the hydrogen bonding effects on the energy barriers for the (less-important) second and fourth reaction steps are insignificant. However, the hydrogen bonding energy estimated based on the MD simulations increases from ES to TS1 and from INT2 to TS3, demonstrating that the enhanced hydrogen bonding between O31 of cocaine and the oxyanion hole of BChE in the transition states should lower the energy barriers for the first and third reaction steps of the catalytic hydrolysis (i.e. the initial step of the acylation and the initial step of deacylation). Future rational design of the BChE mutants with a potentially higher catalytic activity for the cocaine hydrolysis may focus on how to enhance the hydrogen bonding of cocaine O31 with the oxyanion hole of the BChE mutants in the transition states, particularly for the rate-determining step. The computational results also predict that mutation A199S/A328W/Y332G could significantly decrease the energy barrier for the third reaction step.
Supplementary Material
Table S1. Summary of the MD-simulated and optimized key distances (Å) and the root-mean-square deviation (RMSD) of the simulated structures from the initial structure.
Figure S1. Plots of the key internuclear distances (in Å) versus the simulation time in the simulated the prereactive BChE-cocaine complex (ES), transition states, intermediates, and product for BChEcatalyzed hydrolysis of (-)-cocaine. Traces D1, D2, and D3 refer to the distances between the carbonyl oxygen of cocaine benzoyl ester and the NH hydrogen of G116, G117, and A199, respectively. RMSD represents the root-mean-square deviation (in Å) of the simulated positions of BChE backbone atoms from those in the initial structure.
Acknowledgments
The research was supported by NIH/NIDA (grant R01DA013930 to C.-G. Zhan). The authors also acknowledge the Center for Computational Sciences (CCS) at University of Kentucky for supercomputing time on Superdome (a shared-memory supercomputer, with 4 nodes and 256 processors).
References
- 1(a).Gawin FH, Ellinwood EH., Jr. Cocaine and other stimulants - actions, abuse, and treatment. N Eng J Med. 1988;318:1173–1182. doi: 10.1056/NEJM198805053181806. [DOI] [PubMed] [Google Scholar]; (b) Landry DW. Immunotherapy for cocaine addiction. Scientific American. 1997;276:42–45. doi: 10.1038/scientificamerican0297-42. [DOI] [PubMed] [Google Scholar]
- 2.Singh S. Chemistry, design, and structure-activity relationship of cocaine antagonists. Chem Rev. 2000;100:925–1024. doi: 10.1021/cr9700538. [DOI] [PubMed] [Google Scholar]
- 3.Sparenborg S, Vocci F, Zukin S. Peripheral cocaine-blocking agents: new medications for cocaine dependence - An introduction to immunological and enzymatic approaches to treating cocaine dependence reported by Fox, Gorelick and Cohen in the immediately succeeding articles (see pages 153-174) Drug Alcohol Depend. 1997;48:149–151. doi: 10.1016/s0376-8716(97)00072-0. [DOI] [PubMed] [Google Scholar]
- 4.Gorelick DA. Enhancing cocaine metabolism with butyrylcholinesterase as a treatment strategy. Drug Alcohol Depen. 1997;48:159–165. doi: 10.1016/s0376-8716(97)00119-1. [DOI] [PubMed] [Google Scholar]
- 5.Kamendulis LM, Brzezinski MR, Pindel EV, Bosron WF, Dean RA. Metabolism of cocaine and heroin is catalyzed by the same human liver carboxylesterases. J Pharmacol Exp Ther. 1996;279:713–717. [PubMed] [Google Scholar]
- 6.Poet TS, McQueen CA, Halpert JR. Participation of cytochromes P4502B and P4503A in cocaine toxicity in rat hepatocytes. Drug Metab Dispos. 1996;24:74–80. [PubMed] [Google Scholar]
- 7.Pan W-J, Hedaya MA. Cocaine and alcohol interactions in the rat: Contribution of cocaine metabolites to the pharmacological effects. J Pharm Sci. 1999;88:468–476. doi: 10.1021/js980283h. [DOI] [PubMed] [Google Scholar]
- 8.Sukbuntherng J, Martin DK, Pak Y, Mayersohn M. Characterization of the properties of cocaine in blood: Blood clearance, blood to plasma ratio, and plasma protein binding. J Pharm Sci. 1996;85:567–571. doi: 10.1021/js960026h. [DOI] [PubMed] [Google Scholar]
- 9.Browne SP, Slaughter EA, Couch RA, Rudnic EM, McLean AM. The influence of plasma butyrylcholinesterase concentration on the in vitro hydrolysis of cocaine in human plasma. Biopharmaceutics & Drug Disposition. 1998;19:309–314. doi: 10.1002/(sici)1099-081x(199807)19:5<309::aid-bdd108>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 10(a).Carmona GN, Baum I, Schindler CW, Goldberg SR, Jufer R. Plasma butyrylcholinesterase activity and cocaine half-life differ significantly in rhesus and squirrel monkeys. Life Sci. 1996;59:939–943. doi: 10.1016/0024-3205(96)00392-x. [DOI] [PubMed] [Google Scholar]; (b) Lynch TJ, Mattes CE, Singh A, Bradley RM, Brady RO, Dretchen KL. Cocaine detoxification by human plasma butyrylcholinesterase. Toxicol. Appl Pharmacol. 1997;145:363–371. doi: 10.1006/taap.1997.8187. [DOI] [PubMed] [Google Scholar]
- 11.Mattes CE, Lynch TJ, Singh A, Bradley RM, Kellaris PA, Brady RO, Dretchen KL. Therapeutic use of butyrylcholinesterase for cocaine intoxication. Toxicol Appl Pharmacol. 1997;145:372–380. doi: 10.1006/taap.1997.8188. [DOI] [PubMed] [Google Scholar]
- 12.Mattes CE, Belendiuk GW, Lynch TJ, Brady RO, Dretchen KL. Butyrylcholinesterase: an enzyme antidote for cocaine intoxication. Addict Biol. 1998;3:171–188. doi: 10.1080/13556219872236. [DOI] [PubMed] [Google Scholar]
- 13.Gateley SJ. Activities of the enantiomers of cocaine and some related compounds as substrates and inhibitors of plasma butyrylcholinesterase. Biochem Pharmacol. 1991;41:1249–1254. doi: 10.1016/0006-2952(91)90665-r. [DOI] [PubMed] [Google Scholar]
- 14.Gatley SJ, MacGregor RR, Fowler JS, Wolf AP, Dewey SL, Schlyer DJ. Rapid stereoselective hydrolysis of (+)-cocaine in baboon plasma prevents its uptake in the brain - implications for behavioral-studies. J Neurochem. 1990;54:720–723. doi: 10.1111/j.1471-4159.1990.tb01933.x. [DOI] [PubMed] [Google Scholar]
- 15(a).Masson P, Legrand P, Bartels CF, Froment M-T, Schopfer LM, Lockridge O. Role of aspartate 70 and tryptophan 82 in binding of succinyldithiocholine to human butyrylcholinesterase. Biochemistry. 1997;36:2266–2277. doi: 10.1021/bi962484a. [DOI] [PubMed] [Google Scholar]; (b) Masson P, Xie W, Froment MT, Levitsky V, Fortier PL, Albaret C, Lockridge O. Interaction between the peripheral site residues of human butyrylcholinesterase, D70 and Y332, in binding and hydrolysis of substrates. Biochim Biophys Acta. 1999;1433:281–293. doi: 10.1016/s0167-4838(99)00115-6. [DOI] [PubMed] [Google Scholar]; (c) Xie W, Altamirano CV, Bartels CF, Speirs RJ, Cashman JR, Lockridge O. An improved cocaine hydrolase: The A328Y mutant of human butyrylcholinesterase is 4-fold more efficient. Mol Pharmacol. 1999;55:83–91. doi: 10.1124/mol.55.1.83. [DOI] [PubMed] [Google Scholar]; (d) Duysen EG, Bartels CF, Lockridge O. Wild-type and A328W mutant human butyrylcholinesterase tetramers expressed in Chinese hamster ovary cells have a 16-hour half-life in the circulation and protect mice from cocaine toxicity. J Pharmacol Exp Ther. 2002;302:751–758. doi: 10.1124/jpet.102.033746. [DOI] [PubMed] [Google Scholar]; (e) Nachon F, Nicolet Y, Viguie N, Masson P, Fontecilla-Camps JC, Lockridge O. Engineering of a monomeric and low-glycosylated form of human butyrylcholinesterase - Expression, purification, characterization and crystallization. Eur J Biochem. 2002;269:630–637. doi: 10.1046/j.0014-2956.2001.02692.x. [DOI] [PubMed] [Google Scholar]; (f) Zhan CG, Landry DW. Theoretical studies of competing reaction pathways and energy barriers for alkaline ester hydrolysis of cocaine. J Phys Chem A. 2001;105:1296–1301. [Google Scholar]
- 16.Berkman CE, Underiner GE, Cashman JR. Stereoselective inhibition of human butyrylcholinesterase by phosphonothiolate analogs of (+)- and (−)-cocaine. Biochem Pharmcol. 1997;54:1261–1266. doi: 10.1016/s0006-2952(97)00403-6. [DOI] [PubMed] [Google Scholar]
- 17(a).Sun H, El Yazal J, Lockridge O, Schopfer LM, Brimijoin S, Pang YP. Predicted Michaelis-Menten complexes of cocaine-butyrylcholinesterase - Engineering effective butyrylcholinesterase mutants for cocaine detoxication. J Biol Chem. 2001;276:9330–9336. doi: 10.1074/jbc.M006676200. [DOI] [PubMed] [Google Scholar]; (b) Sun H, Shen ML, Pang YP, Lockridge O, Brimijoin S. Cocaine metabolism accelerated by a re-engineered human butyrylcholinesterase. J Pharmacol Exp Ther. 2002;302:710–716. doi: 10.1124/jpet.302.2.710. [DOI] [PubMed] [Google Scholar]; (c) Sun H, Pang YP, Lockridge O, Brimijoin S. Re-engineering butyrylcholinesterase as a cocaine hydrolase. Mol Pharmacol. 2002;62:220–224. doi: 10.1124/mol.62.2.220. [DOI] [PubMed] [Google Scholar]
- 18.Zhan CG, Zheng F, Landry DW. Fundamental reaction mechanism for cocaine hydrolysis in human butyrylcholinesterase. J Am Chem Soc. 2003;125:2462–2474. doi: 10.1021/ja020850+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamza A, Cho H, Tai H-H, Zhan CG. Molecular dynamics simulation of cocaine binding with human butyrylcholinesterase and its mutants. J Phys Chem B. 2005;109:4776–4782. doi: 10.1021/jp0447136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, Kua J, McCammon JA. Role of the catalytic triad and oxyanion hole in acetylcholinesterase catalysis: An ab initio QM/MM study. J Am Chem Soc. 2002;124:10572–10577. doi: 10.1021/ja020243m. [DOI] [PubMed] [Google Scholar]
- 21.Case DA, Pearlman DA, Caldwell JW, Cheatham TE, III, Wang J, Ross WS, Simmerling CL, Darden TA, Merz KM, Stanton RV, Cheng AL, Vincent JJ, Crowley M, Tsui V, Gohlke H, Radmer RJ, Duan Y, Pitera J, Massova I, Seibel GL, Singh UC, Weiner PK, Kollman PA. AMBER 7. University of California; San Francisco: 2002. [Google Scholar]
- 22.Nicolet Y, Lockridge O, Masson P, Fontecilla-Camps JC, Nachon F. Crystal structure of human butyrylcholinesterase and of its complexes with substrate and products. J Biol Chem. 2003;278:41141–41147. doi: 10.1074/jbc.M210241200. [DOI] [PubMed] [Google Scholar]
- 23(a).Bernstein FC, Koetzle TF, Williams GJ, Meyer EF, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M. Protein data bank - computer-based archival file for macromolecular structures. J Mol Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]; (b) http://www.rcsb.org/pdb/
- 24.Sali A, Blundell TL. Definition of general topological equivalence in protein structures - a procedure involving comparison of properties and relationships through simulated annealing and dynamic-programming. J Mol Biol. 1990;212:403–428. doi: 10.1016/0022-2836(90)90134-8. [DOI] [PubMed] [Google Scholar]
- 25.Sali A, Blundell TL. Comparative protein modeling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 26.Jorgensen WL, Chandrasekhar J, Madura JD, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 27(a).Zhan CG, de Souza O Norberto, Rittenhouse R, Ornstein RL. Determination of two structural forms of catalytic bridging ligand in zinc-phosphotriesterase by molecular dynamics simulation and quantum chemical calculation. J Am Chem Soc. 1999;121:7279–7282. [Google Scholar]; (b) Koca J, Zhan CG, Rittenhouse R, Ornstein RL. Mobility of the active site bound paraoxon and sarin in zinc-phosphotriesterase by molecular dynamics simulation and quantum chemical calculation. J Am Chem Soc. 2001;123:817–826. doi: 10.1021/ja000439r. [DOI] [PubMed] [Google Scholar]; (c) Koca J, Zhan CG, Rittenhouse RC, Ornstein RL. Coordination number of zinc ions in the phosphotriesterase active site by molecular dynamics and quantum mechanics. J Comput Chem. 2003;24:368–378. doi: 10.1002/jcc.10217. [DOI] [PubMed] [Google Scholar]
- 28.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. Molecular-dynamics with coupling to an external bath. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- 29.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical-integration of cartesian equations of motion of a system with constraints - molecular-dynamics of n-alkanes. J Comp Phys. 1977;23:327–341. [Google Scholar]
- 30.Darden TA, Lee H, Pedersen LG. Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 31.Dapprich S, Komaromi I, Byun KS, Morokuma K, Frisch MJ. A new ONIOM implementation in Gaussian98. Part I. The calculation of energies, gradients, vibrational frequencies and electric field derivatives. J Mol Struct (Theochem) 1999;461:1–21. [Google Scholar]
- 32.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JA, Jr., Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA. Gaussian 03, Revision A.1. Gaussian, Inc.; Pittsburgh, PA: 2003. [Google Scholar]
- 33.Vreven T, Morokuma K. The ONIOM (our own N-layered integrated molecular orbital + molecular mechanics) method for the first singlet excited (S1) state photoisomerization path of a retinal protonated Schiff base. J. Chem. Phys. 2000;113:2969–2975. [Google Scholar]
- 34.Frisch M, Vreven T, Schlegel HB, Morokuma K. Efficient and reliable geometry optimization for QM/MM methods. J. Comput. Chem. 2003;24:760–769. doi: 10.1002/jcc.10156. [DOI] [PubMed] [Google Scholar]
- 35.By using the geometries optimized at the HF/3-21G:Amber level with all of the transition bonds fixed, the total energies of the high-layer obtained from the single-point energy calculations at the B3LYP/6-31+G*:Amber level roughly suggest that the energy barriers associated with TS1, TS2, TS3, and TS4 are 2.8, 14.2, 6.6, and 20.4 kcal/mol, respectively. We emphasize that the energy barriers estimated without fully optimizing the geometries of the transition states are not reliable.
- 36(a).Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem. 1998;19:1639–1662. (b)Based on the general HBE equation, we have HBE(r) ≈ 5εr012/r12 – 6εr010/r10, in which r is the H…O distance in the considered hydrogen bond and r0 is the minimum value of the H…O distance for which the HBE equation can be used. We used r0 = 1.49 Å, because it is the shortest H…O distance found in all of our MD simulations and geometry optimizations. The ε value was determined by using the condition that HBE(r) = −5.0 kcal/mol when r = 1.90 Å.
- 37.Tai K, Shen T, Börjesson U, Philippopoulos M, McCammon JA. Analysis of a 10-ns molecular dynamics simulation of mouse acetylcholinesterase. Biophys J. 2001;81:715–724. doi: 10.1016/S0006-3495(01)75736-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kua J, Zhang Y, Eslami AC, Butler JR, McCammon JA. Studying the roles of W86, E202, and Y337 in binding of acetylcholine to acetylcholinesterase using a combined molecular dynamics and multiple docking approach. Protein Science. 2003;12:2675–2684. doi: 10.1110/ps.03318603. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Summary of the MD-simulated and optimized key distances (Å) and the root-mean-square deviation (RMSD) of the simulated structures from the initial structure.
Figure S1. Plots of the key internuclear distances (in Å) versus the simulation time in the simulated the prereactive BChE-cocaine complex (ES), transition states, intermediates, and product for BChEcatalyzed hydrolysis of (-)-cocaine. Traces D1, D2, and D3 refer to the distances between the carbonyl oxygen of cocaine benzoyl ester and the NH hydrogen of G116, G117, and A199, respectively. RMSD represents the root-mean-square deviation (in Å) of the simulated positions of BChE backbone atoms from those in the initial structure.