SUMMARY
The Gram-negative oral anaerobe Prevotella intermedia forms an iron(III) protoporphyrin IX pigment from haemoglobin. The microorganism expresses a 90 kDa cysteine protease, Interpain A (InpA), a homologue of Streptococcus pyogenes streptopain (SpeB). The role of InpA in haemoglobin breakdown and haem release was investigated. At pH 7.5, InpA mediated oxidation of oxyhaemoglobin to hydroxymethaemoglobin (in which the haem iron is oxidised to the Fe(III) state and which carries OH− as the sixth co-ordinate ligand) by limited proteolysis of globin chains as indicated by SDS-PAGE and MALDI-TOF analysis. Prolonged incubation at pH 7.5, did not result in further haemoglobin protein breakdown, but in the formation of a haemoglobin haemichrome (where the haem Fe atom is co-ordinated by another amino acid ligand in addition to the proximal histidine) stable to degradation by InpA. InpA-mediated haem release from hydroxymethaemoglobin-agarose was minimal compared with trypsin at pH 7.5. At pH 6.0, InpA increased oxidation at a rate greater than auto-oxidation, producing aquomethaemoglobin (with H2O as sixth co-ordinate ligand), and resulted in its complete breakdown and haem loss. Aquo-methaemoglobin proteolysis and haem release was prevented by blocking haem dissociation by ligation with azide, whilst InpA proteolysis of haem-free globin was rapid even at pH 7.5. Both oxidation of oxyhaemoglobin and breakdown of methaemoglobin by InpA were inhibited by the cysteine-protease inhibitor E64. In summary we conclude that InpA may play a central role in haem acquisition by mediating oxyhaemoglobin oxidation, and by degrading aquomethaemoglobin in which haem-globin affinity is weakened under acidic conditions.
Keywords: Prevotella intermedia, interpain A, protease, haemoglobin, methaemoglobin, haem
INTRODUCTION
The genera Prevotella and Porphyromonas belong to a group of Gram-negative, black-pigmenting oral anaerobes which are associated with periodontal diseases in humans and other animals [1]. The black pigments, which develop upon prolonged incubation on blood-containing media, are composed of iron (III) protoporphyrin IX, Fe(III)PPIX, and are derived from the breakdown of haemoglobin. The haem pigment produced by Porphyromonas gingivalisis is composed of iron(III) protoporphyrin IX in the µ-oxo bishaem or dimeric form, (Fe(III)PPIX)2O [2], whilst that from both Prevotella intermedia and Prevotella nigrescens is in the form of monomeric haematin, Fe(III)PPIX.OH [3]. These pigments serve a defensive role as they make use of the intrinsic ferrihaem catalase activity to destroy hydrogen peroxide [4]. In addition, the formation of the µ-oxo dimer by P. gingivalis from ferrous haemoglobin is a chemical mechanism which can tie up oxygen and thus promote anaerobiosis [2, 5].
The mechanism of protease-mediated haem release from haemoglobin by P. gingivalis involves the concerted sequential action of both the arginine- and lysine-specific cysteine protease gingipains [5, 6, 7]. Haemoglobin breakdown has also been demonstrated for Prevotella species [8, 9, 10] but the way in which haem is released from haemoglobin by these bacteria is unclear. Guan et al. [10] have shown that the culture supernatant of P. intermedia could degrade haemoglobin over a wide pH range, but with optimal activity at around pH 5. Whilst di - and tripeptidyl peptidases and general thiol-dependent proteolytic activity have been partially characterised from P. intermedia and Prevotella nigrescens [11, 12], little is known regarding the individual endopeptidases of these organisms. The only characterised endopeptidase from Prevotella spp. is interpain A (InpA) [13], a cysteine protease which is an orthologue of SpeB of Streptococcus pyogenes and periodontain of P. gingivalis (PrtP) [14]. Several other InpA orthologue genes are apparently present in the genomes of the Bacteroidetes [15]. However, in strict contrast to SpeB, gingipains and many other bacterially-derived cysteine proteases [16], little is known about the role of interpain-like enzymes in bacterial pathogenicity and/or haem acquisition.
The gingipains of P. gingivalis have been shown to play an essential function in formation of haem pigmentation of P. gingivalis, where, at slightly alkaline pH, they work sequentially to firstly oxidise oxyhaemoglobin to methaemoglobin (where the haem iron is in the Fe(III) state and OH− is the sixth co-ordinate ligand),and, finally, to degrade this species to release Fe(III) iron protoporphyrin IX, which becomes dimerised and incorporated into the pigment [6, 7, 17]. To elucidate the mechanism of haem acquisition by P. intermedia, we have examined the role of interpain A in haemoglobin breakdown. We demonstrate here that at pH 7.5, InpA mediates hydroxymethaemoglobin formation from oxyhaemoglobin and that under acidic conditions which may be generated as a result of their saccharolytic metabolism, the aquo- (or acid) form of methaemoglobin is rendered susceptible to InpA enzymic attack and haem release.
EXPERIMENTAL
Interpain isolation and purification
InpA was expressed as a recombinant protein in Escherichia coli and purified by affinity chromatography on Fast Flow Ni-NTA Sepharose (Qiagen) followed by anion exchange chromatography (MonoQ, GE Healthcare) as described previously [13]. The amount of active enzyme in wild-type InpA preparations was determined by active site titration using inhibitor E-64 (Sigma). Briefly, recombinant protein was activated at 37°C for 15 min in 0.1M Tris-HCl, 5mM EDTA, pH 7.5, freshly supplemented with 2mM DTT and then pre-incubated with increasing concentrations of E-64 for 30 min at room temperature. Residual enzyme activity was determined by measurement of fluorescence (λex = 380 nm and λem = 460 nm) of AMC released from Boc-Val-Leu-Lys-AMC (PeptaNova) added to the reaction mixture at 250µM final concentration and using the microplate spectrofluorimeter SpectraMax Gemini EM (Molecular Devices). The concentration of active InpA was determined by active site titration using an appropriate dilution of a standardized 1 mM aqueous solution of the inhibitor E-64 (Sigma Chemicals Ltd.) needed for total inactivation of the proteinase. Before use, InpA was pre-activated by incubation for 15 min in 0.1M NaCl, 0.1M Tris-HCl, pH 7.5, supplemented with 2mM DTT. For use in haemoglobin degradation assays, this was replaced with the above buffer minus DTT by ultrafiltration using 10 kDa cut-off Microcons (Amico Ltd.).
Haemoglobin preparations
Oxyhaemoglobin was prepared from fresh horse erythrocytes as previously described [5] and stored as a concentrated solution (approximately 1mM) at −80°C in 0.14M NaCl, 0.1M Tris-HCl, pH 7.5, until required. Stock preparations of methaemoglobin were prepared from the oxygenated protein by treatment with NaNO2 as previously described [7]. Azido-methaemoglobin was formed from methaemoglobin by incubation with 0.4mM NaN3 in 0.2M phosphate buffer, pH 6.0, for 18 h at room temperature [7]. Bovine haemoglobin-agarose was obtained from Sigma Chemical Company (product number H8756). In assays to determine haem release from haemoglobin subunits, haemoglobin-agarose beads were firstly washed in 0.14M NaCl buffered with 0.1M Tris-HCl buffer at pH 7.5, to remove any un-conjugated haemoglobin. The beads were then incubated with InpA and at various time periods samples of these were removed, pelleted by centrifugation at 5,000 × g for 1 min and the supernatant solutions carefully removed. Iron protoporphyrin IX released from the protein was assayed using the pyridine haemochromogen method as described by Gallagher and Elliot [18], after firstly reducing the sample with 10mM sodium dithionite, followed by the addition of 0.1M pyridine.
Haem-free globin
Haem-free globin was prepared as described by Ascoli et al. [19]. Briefly, a 3% w/v solution of freshly prepared horse oxyhaemoglobin was added slowly to 30-fold volume of vigorously stirred cold (−20°C) acetone containing 5mM HCl. Precipitated globin was collected by low speed centrifugation (5,000 × g,10 min, 0°C), treated again with cold acid-acetone until no red colour remained, re-suspended in a minimum volume of water, dialysed against sodium bicarbonate (0.1g l−1), and then against 0.01M phosphate buffer, pH 7.2. The denatured globin precipitating during this step was removed by centrifugation (10,000g 15 min at 5°C), and the soluble globin fraction recovered by freeze-drying after dialysis against water at 4°C.
SDS-PAGE and tetramethylbenzidine staining for haem-associated peroxidase
SDS-PAGE was carried out as previously described on 15% polyacrylamide gels [20]. For haem-associated peroxidase staining in the gels, samples were prepared in non-reducing sample buffer by incubation at 37°C for 1h. For some experiments, samples were solubilised by heating at 100°C for 5 min as specified in the text.
Densitometry
Densitometry was carried out on coomassie blue-stained bands using UVIband gel analysis software (UVItech Ltd., Cambridge, UK) after digital image capture using a UMax Powerlook 1000 flatbed transmission scanner.
Measurement of methaemoglobin formation
The change in the concentration of methaemoglobin formed as a result of oxyhaemoglobin oxidation was estimated by difference spectroscopy as a function of the change in area of the Soret band [21, 22], and as a function of the Q band absorbances as previously described [17]. The rate of haemoglobin (HbO2) oxidation was also followed quantitatively using plots of –ln([HbO2]t / [HbO2]0) versus time t, where the ratio of HbO2 concentration after time t to that at time t=0 was monitored by the absorbance changes of the alpha band (576nm) of oxyhaemoglobin [23].
MALDI-TOF mass spectrometry
Samples were analysed using a Micromass M@LDI mass spectrometer, using an α-cyano-4-hydroxy-cinnamic acid matrix (Sigma Chemicals). The spectra were recorded in the positive ion mode and the mass range scanned was 800 to 4000 Daltons. The spectrometer was calibrated using a mixture of authentic peptide samples. The observed peptide masses were compared to those predicted by theoretical cleavage using the PeptideMass program (ExPASy).
RESULTS AND DISCUSSION
For the arguments presented previously [7], P. gingivalis and other black-pigmenting species inhabiting the gingival sulcus or periodontal pocket, may be exposed, from time-to-time, to oxygenated haemoglobin. However, the refractory nature of the oxyhaemoglobin molecule to proteolysis and haem release is overcome by P. gingivalis by firstly promoting its oxidation to methaemoglobin through the proteolytic action of R-gingipain [5,6,7]. In methaemoglobin, the affinity of globin for haem in which the iron is in the Fe(III) state is greatly relaxed, and the resultant haem-free protein is rendered susceptible to proteolysis by the lysine-specific gingipain [7]. K-gingipain can also attack hydroxymethaemoglobin produced non-proteolytically by treatment of haemoglobin with sodium nitrite [17].
Formation of methaemoglobin by InpA
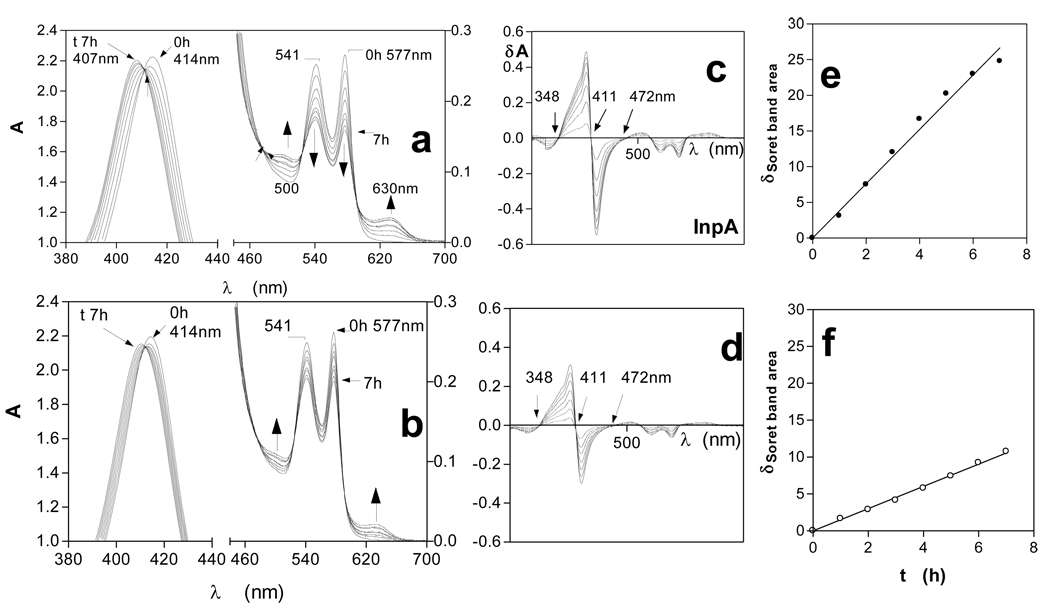
In this study, incubation of horse oxyhaemoglobin at pH 7.5 over a seven hour period in the presence of InpA (Fig 1a) resulted in spectral changes indicative of oxidation to hydroxymethaemoglobin i.e., increases in extinction at 500 and 630nm, and decreases at 541 and 577nm. These were accompanied by an increase in absorbance and blue-shift in λmax of the Soret band to approximately 406nm. The same spectral changes were observed during incubation of oxyhaemoglobin in the absence of the protease (Fig 1b). Isosbestic points were observed in both sets of spectra (at 348 and 472nm). This indicated that only two absorbing species were present and showed that InpA had mediated the direct conversion of oxyhaemoglobin into the methaemoglobin form. When 10mM Na2S2O4 was added to the haemoglobin samples after 7h incubation, to chemically reduce the haem iron and to deplete O2 in the buffer, both were converted to deoxyhaemoglobin (Soret λmax at 429nm and 555nm Q band), confirming that methaemoglobin had originally been produced (data not shown).
Figure 1.
UV-visible spectra of horse oxyhaemoglobin during oxidation by interpain A.
Oxyhaemoglobin (4µM with respect to tetramer) was incubated with 2µM interpain A (a). Oxyhaemoglobin auto-oxidation is shown in (b). Difference spectra derived from data in a) and b), showing the changes in the Soret band region of oxyhaemoglobin during oxidation mediated by InpA (c), and during auto-oxidation (d). (e) and (f) show the show the linear increases in change in Soret band region area indicative of methaemoglobin formation for InpA-mediated oxidation and auto-oxidation respectively. Buffer was 0.1M Tris-HCl, 0.14M NaCl, pH 7.5, and incubations were carried out at 37°C.
By subtracting the initial time zero spectrum from those at subsequent time periods shown in Figs 1a and b, a series of difference spectra were obtained with peak maxima at 402nm and minima at 420nm, with a zero cross-over points at 411nm (Figs 1 c and d), which is also characteristic of methaemoglobin formation [21, 22]. Summation of the integrated peak areas between 348 and 411nm, and trough areas between 411nm and 472nm, giving the total area change of the Soret band during the change from the oxy- to the met- state, allowed assessment of the extent of methaemoglobin formation [22]. This showed that InpA had mediated a 3-fold increase in oxidation rate compared to auto-oxidation (Fig 1 e and f). Incubation of oxyhaemoglobin with InpA in the presence of the specific cysteine protease inhibitor E-64 (500µM) had the effect of reducing the oxidation rate by approximately 80% (data not shown), thus demonstrating that the ability to mediate oxyhaemoglobin oxidation was a result of its enzyme activity.
Mass spectrometry analysis of haemoglobin breakdown during InpA-mediated oxidation
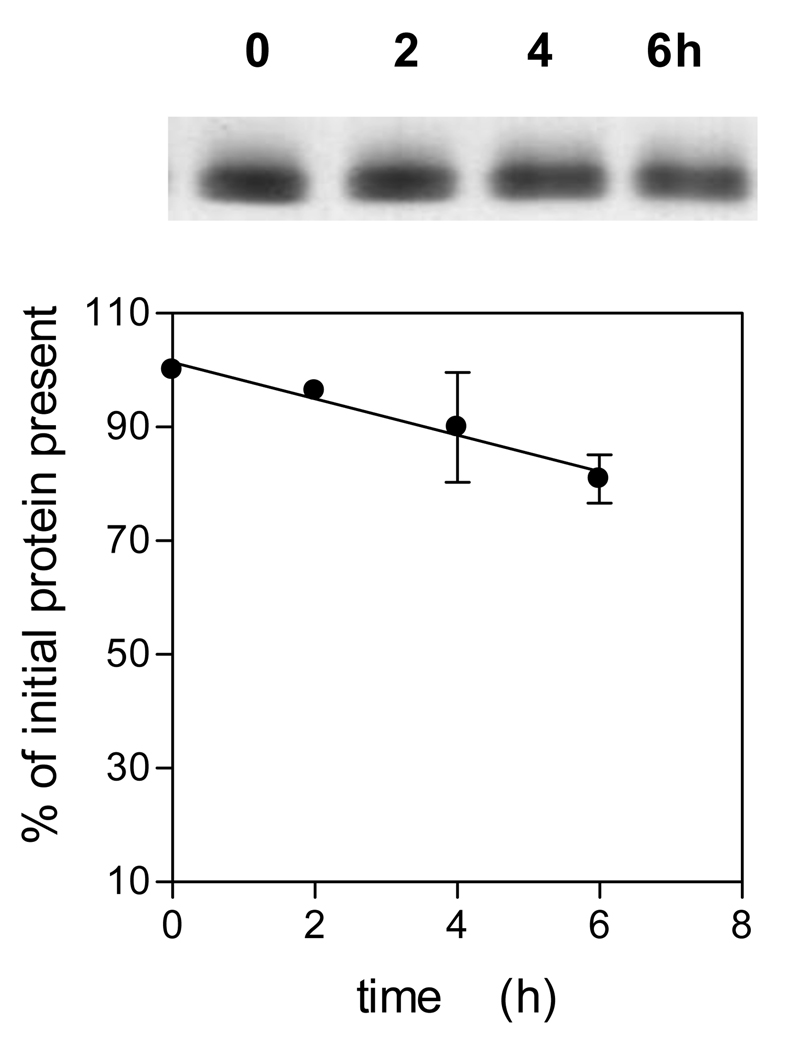
To gain some insight into which residues were attacked during InpA-mediated oxidation of oxyhaemoglobin at pH 7.5, the enzyme was incubated with oxyhaemoglobin at 37°C and samples periodically removed and the protease inactivated by incubation for 30 min with E-64 (as above) before subjection to MALDI-TOF mass spectrometric analysis. Table 1 shows the major digestion fragments identified as arising from alpha and beta globin chains during InpA-mediated oxidation of oxyhaemoglobin. These indicated cleavages at the C-terminal sides of lysine or arginine and alanine or phenylalanine which are in keeping with the specificities for hydrolysis of chromogenic p-nitrolanilide-derivatized peptides and processing sites of N- and C-terminal profragments cleaved during the autoproteolytic maturation of prointerpain, respectively (Potempa et al., unpublished findings). Noteworthy, was the detection of peptides of masses 1356.92 and 2227.66, which indicate cleavage of residues around the distal region of the haem pocket of the alpha and beta chains. Importantly, SDS-PAGE analysis of the Hb samples incubated with InpA and solubilised by heating at 100°C clearly showed a limited degree of protein breakdown (circa 20%) had occurred over the same time period (Fig 3), suggesting that although the globin structure has been proteolytically cleaved giving rise to oxidation, its gross structure had remained stable. Indeed, the complex structure of haemoglobin is maintained by numerous interactions between individual residues within the globin chain, and with the haem moiety, as well as interfacial interactions between the globin chains of haemoglobin tetramers and dimers. These interactions not only enable haemoglobin to alter shape in response to oxygen loading, but may also prevent complete unfolding of globin chains during incisions made by InpA. This would also explain why peptides are only released from the protein under the extreme conditions of denaturation during the MALDI process or boiling.
Table 1. Digestion fragments identified as arising from proteolysis of alpha and beta oxyhaemoglobin chains during InpA-mediated oxidation.
Oxyhaemoglobin (4 µM) was incubated at 37°C with 2µM InpA and samples removed periodically, incubated for 30 min with E-64 (0.5mM) to inhibit protease activity, and subjected to MALDI-TOF mass spectrometry (see text for details).
| fragment size (Da) and time of appearance |
corresponding predicted fragment size (Da) |
Chain designation and residue number |
amino acid sequence |
|---|---|---|---|
| 1126.75 (5h) |
1126.56 | β-96 to 104 | K…96LHVDPENFR104… |
| 1356.92 (4h) |
1356.65 | β-46 to 59 | F…46GDLSNPGAVMGNPK59… |
| 1417.92 (2h) |
1417.72 | α-111 to 123 | A…111VHLPNDFTPAVHA123… |
| 1902.36 (8h) |
1902.94 | α-8 to 26 | K…8TNVKAAWSKVGGHAGEYGA26… |
| 2227.66 (4h) |
2227.08 | β-41 to 61 | R…41FFDSFGDLSNPGAVMGNPKVK61… |
| 2227.10 | α-41 to 60 | K…41TYFPHFDLSHGSAQVKAHGK60… | |
| 2037.65 (2h) |
2037.97 | β-86 to 103 | F…86AALSELHCDKLHVDPENF103… |
Figure 3.
SDS-PAGE of oxyhaemoglobin during incubation with interpain A.
Incubation conditions and enzyme and substrate conditions were as described for Fig 1. The gel was stained with coomassie blue and densitometrically scanned to quantify the loss of protein during incubation, and expressed as % of initial protein present. Samples were solubilised by heating at 100°C for 5 min. The data points represent the mean and standard deviation of three separate experiments.
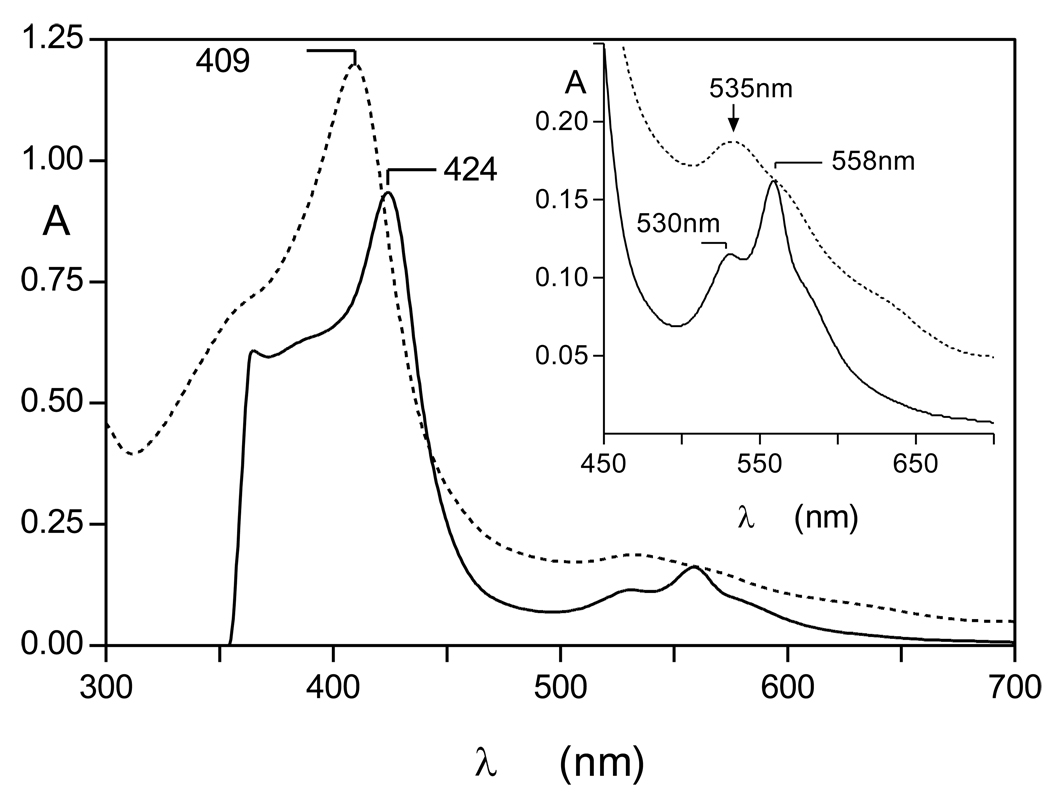
Longer term incubation of InpA with oxyhaemoglobin at pH 7.5 produces a haemoglobin haemichrome
Although short term incubation of oxyhaemoglobin with InpA resulted in hydroxymethaemoglobin formation, interaction for periods of 24h or greater resulted in a characteristic haemichrome spectrum [24] with Soret λmax 409 nm and a broad Q band at 535nm (Fig 2). The haemichrome nature of this product was confirmed by reduction to a haemochrome with sodium dithionite (424nm Soret and 530 and 558nm Q bands; [24, 25]) (Fig 2, solid lines). Whilst UV-visible spectroscopy appeared to show that there had been no detectable InpA-mediated breakdown of haemoglobin over the initial six hours, SDS-PAGE (where samples had been solubilised by heating at 100°C) and densitometry showed that approximately 20% of the globin chains were degraded after 6h of incubation (Fig 3). It is noteworthy that like the Arginine-specific gingipains HRgpA [17] and RgpB (Smalley et al., unpublished observations), InpA also promoted haemoglobin oxidation at pH 7.5. This is a pH at which the natural oxidation rate of oxyhaemoglobin is lowest [26]. However, the hydroxy-methaemoglobin product formed by InpA at this pH was refractory to further breakdown by InpA and further incubation resulted in conversion to a haemichrome. This is puzzling given the numerous K and R residues present in α and β globin chains, the broad pH optima of InpA proteolytic activity in the range from pH 5.5 to 8.0, and the preference of InpA for arginine, lysine, alanine and phenylalanine residues at the C-terminal side of hydrolyzed peptide bonds (Potempa et al., unpublished findings).
Figure 2.
UV-visible spectrum of the haemoglobin haemichrome formed after longer incubation of oxyhaemoglobin with interpain A at pH 7.5. The haemichrome produced after 24h incubation (dotted line) gave rise to a haemochrome spectrum upon addition of 10mM Na2S2O4 (solid line). Incubation conditions and enzyme and haemoglobin concentrations were as for Figure1. Inset shows the Q-band region.
Relative susceptibilities of methaemoglobin and aquomethaemoglobin to degradation by InpA
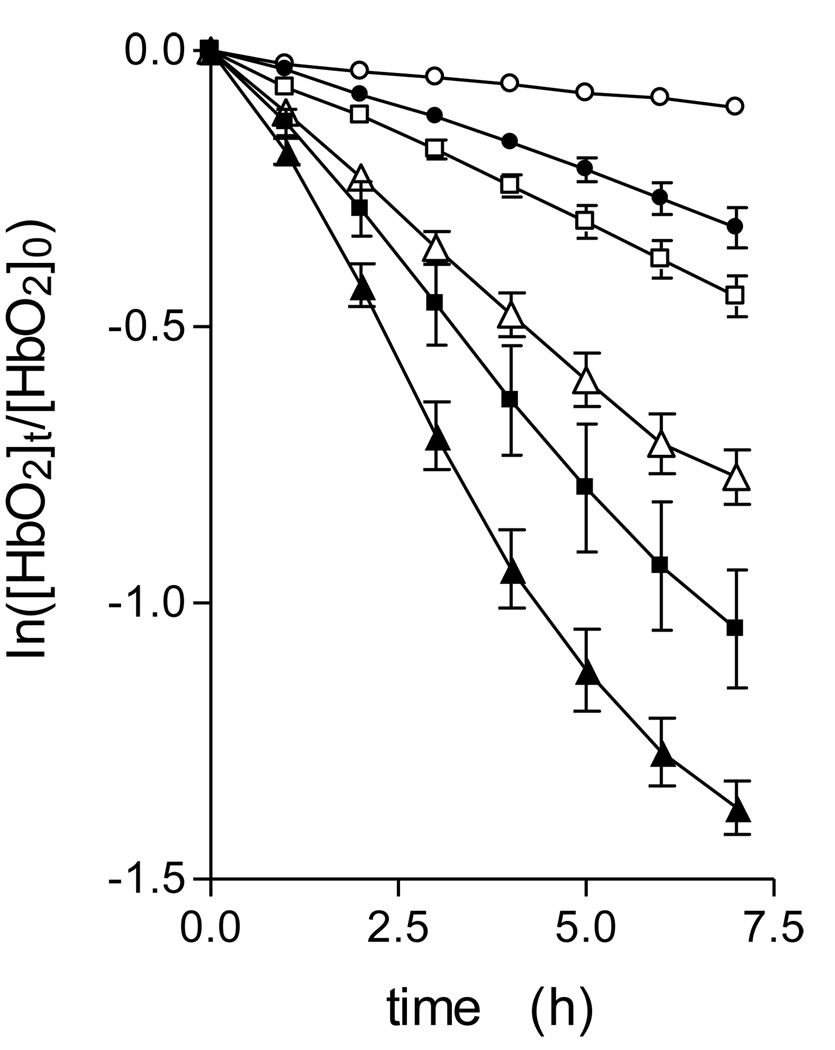
Prevotella species display acidic terminal growth pHs [27, 28, 29]. Both binding to the outer membrane and degradation of haemoglobin mediated by enzymes in the growth supernatant of P. intermedia is enhanced at acid pHs [10, 30]. In addition, pigment production during growth on blood agar is associated with a drop in pH to around 5.8 [3]. Accordingly, we investigated the pH-dependency of oxyhaemoglobin oxidation and breakdown by InpA. To measure the rate of haemoglobin oxidation whilst obviating any complication relating to changes in Soret band area due to degradation and loss of haem from the protein, the rate of haemoglobin oxidation was quantified by following the change in A576nm versus time [23]. This gave linear plots of the initial oxidation rate over the 6 hour incubation period (Fig 4), which clearly demonstrated that InpA increased oxidation above that of the auto-oxidation rate at each pH examined. In this context, it is noteworthy that whilst InpA increased the oxidation rate at pH 7.5 (a pH at which the auto-oxidation rate of both the alpha and beta haemoglobin chains is naturally lowest [26]), it was also able to increase the oxidation rate above the auto-oxidation rate at the acid pHs, conditions where the natural oxidation rate of oxyhaemoglobin is high [26]. This is relevant to the biology of the sub-gingival environment since Prevotella species produce a potent haemolysin [31, 32, 33], and any haemoglobin released through the haemolysis of extra-vasated erythrocytes will be readily oxidised in the absence of intra-erthrocytic reductants and the enzyme methaemoglobin reductase.
Figure 4.
Oxyhaemoglobin oxidation rates in the presence of interpain A.
Rates were measured as a function of the change in A577nm in the presence of interpain A at pH 7.5(●), 6.5 (■), and 6.0 (▲). Open symbols depict the auto-oxidation rate at each pH in the absence of enzyme. Incubations were carried out at 37°C with 4µM oxyhaemoglobin (as tetramer) and 2µM InpA, in 0.14M NaCl, buffered with 0.1M Tris-HCl (pH 7.5) or 0.2M phosphate (pH 6.5 and 6.0). The data points represent the mean and standard deviation of four separate determinations, except for those at pH 7.5, where n=7.
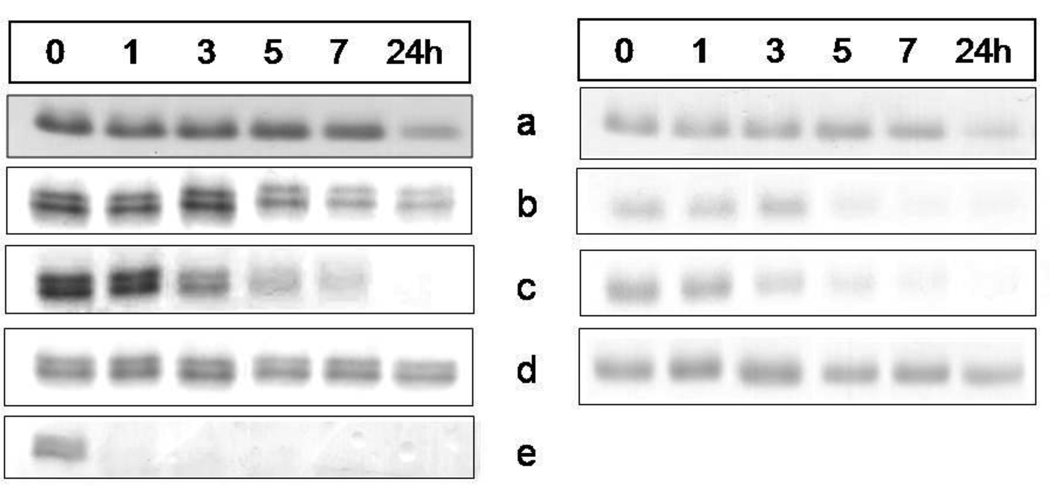
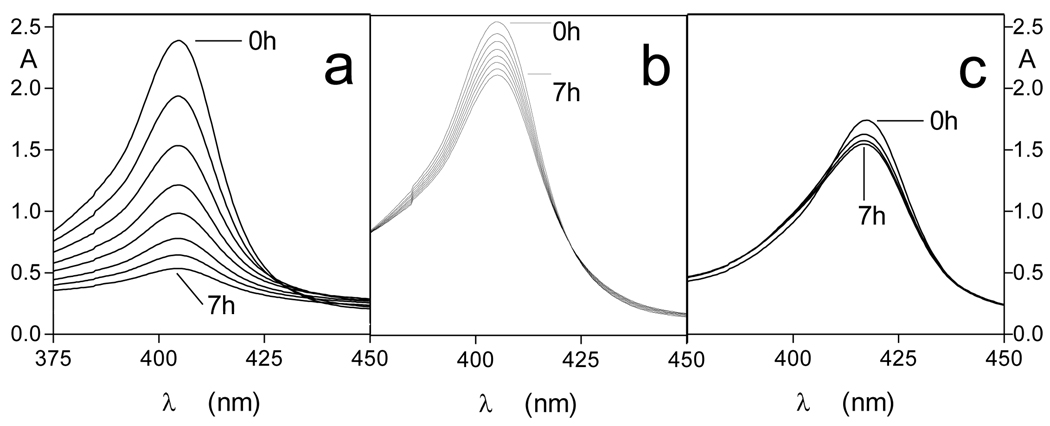
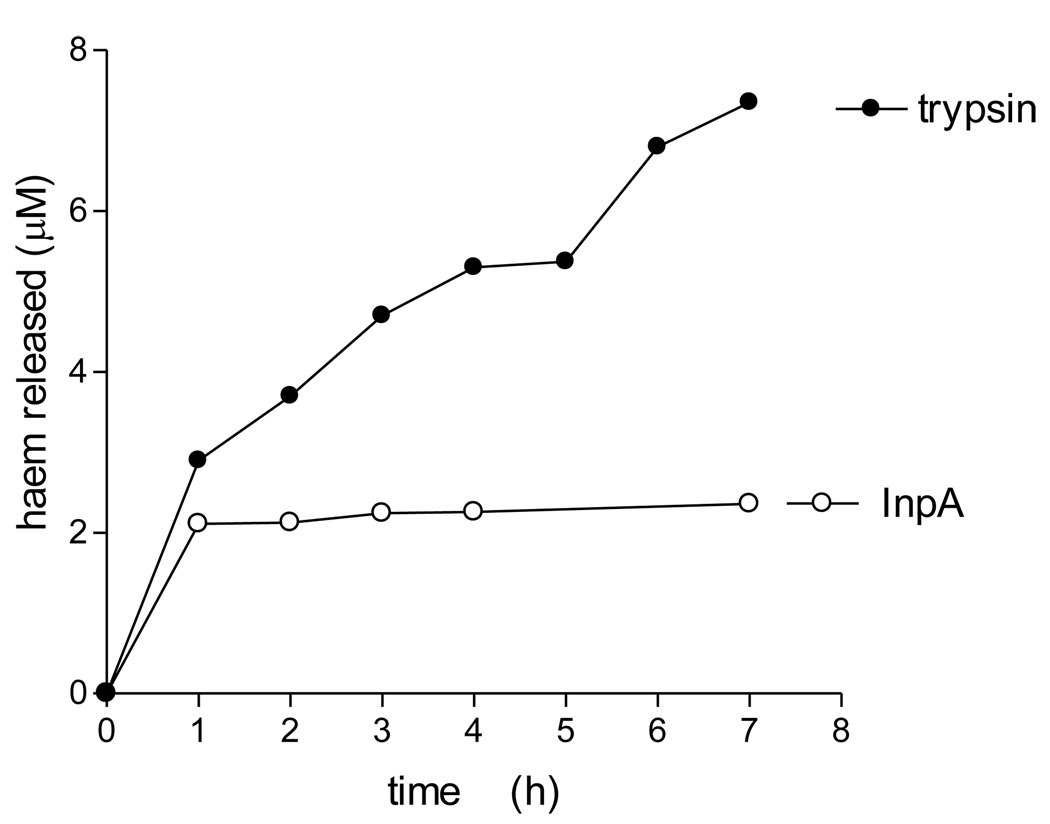
We have previously shown that whilst oxyhaemoglobin is not degraded by the lysine-specific gingipain, it is susceptible to breakdown by this protease when converted to the hydroxy-methaemoglobin form, either through oxidation mediated by NaNO2 or via pre-treatment with the arginine-specific gingipain HRgpA [7, 17]. To test whether hydroxymet- and aquomethaemoglobin species were susceptible to breakdown by InpA, oxyhaemoglobin was firstly auto-oxidised by incubation for 24h at 37°C at pH 7.5, 6.0 and 5.5. Under these conditions, approximately 95% of the oxyhaemoglobin was converted into the aquomet - and hydroxy-methaemoglobin forms. The methaemoglobin preparations (4µM as tetramer) were then exposed at 37°C to InpA (2µM) and sampled periodically for SDS-PAGE. Gels were firstly stained with TMB-H2O2 to reveal haemoglobin-associated haem peroxidase activity, and then counterstained for protein with coomassie blue (Fig 5). Densitometry revealed that during incubation with InpA at pH 7.5, the globin chains were still intact (Fig 5, panel A, gel a). Moreover, TMB staining (Fig 5, panel B, gel a) showed that little or no haem had been lost from hydroxymethaemoglobin. In contrast however, the aquomethaemoglobin preparations were degraded to a much greater degree, especially at pH 5.5 (Fig 5, panel A, gels b and c), and proteolysis of the haemoglobin chains was also accompanied by a greater haem loss as shown by the reduced level of TMB staining (Fig 5, panel B, gels b and c). The susceptibility of aquomethaemoglobin and hydroxymethaemoglobin to InpA was also examined spectroscopically. These substrates (~ 4µM on a tetramer basis) were prepared by oxidation of oxyhaemoglobin with NaNO2 in either pH 5.5 or pH 7.5 buffers, to give the aquo- or hydrox- forms, respectively, and were then exposed to 2µM InpA. As can be seen in Fig 6, InpA effected an almost complete breakdown aquomethaemoglobin (panel a) compared to the hydroxymet- form (panel b) over 7h as evidenced by the collapse of the Soret band. In addition, spectrophotometric analysis also confirmed that azidomethaemoglobin, in which the haem was stabilised by N3− ligation, was degraded minimally by InpA (Fig 6 c). However, trypsin (2µM) brought about complete breakdown of the hydroxymet-haemoglobin even at pH 7.5 (Fig 7). The trypsin-mediated breakdown of the hydroxymethaemoglobin protein chains was also reflected in the loss of Soret band intensity (Fig 7). The reduced ability of InpA to degrade hydroxy-methaemoglobin and to release haem at pH 7.5 was confirmed in a separate experiment in which hydroxy-methaemoglobin-agarose was incubated with InpA or trypsin. Haem released from the immobilised haemoglobin was detected as the pyridine haemochromogen after reduction with sodium dithionite, followed by reaction with pyridine. This showed that InpA was only able to release approximately 20% of the total haemoglobin haem liberated by trypsin over a period of 7h (Fig 8). Significantly, this observation also correlates perfectly with the 20% degradation of oxyhaemoglobin by InpA over the same time period (Fig 3).
Figure 5.
SDS-PAGE showing the effect of interpain A on methaemoglobin species and haem-free globin chains
Hydroxy-methaemoglobin, pH 7.5, (a); aquomethaemoglobin, pH 6.0, (b), and pH 5.5 (c); azido-methaemoglobin, pH 6.0, (d); haem-free globin, pH 7.5, (e). Haemoglobin substrates at 4µM (with respect to tetramer) were incubated with InpA (2µM) at 37°C and aliquots withdrawn at indicated time points were subjected to the SDS-PAGE analysis. The hydroxy-methaemoglobin and aquomethaemoglobin species were formed by auto-oxidation of oxyhaemoglobin at 37°C for 24h at pHs 7.5, 6.0 and 5.5, and constituted 95% of the total haemoglobin present. Panel A, gels stained with coomassie blue; panel B, TMB-stained for haem-associated peroxidise activity. Azido-methaemoglobin was prepared by incubating aquomethaemoglobin in the presence of 400µM NaN3.
Figure 6.
Soret band regions of (a) aquomethaemoglobin, (b) hydroxymethaemoglobin, and (c) azidomethaemoglobin during incubation with InpA. Substrate and enzyme concentrations were 4µM (as tetramer) and 2µM, respectively. Incubations were carried out at 37°C.
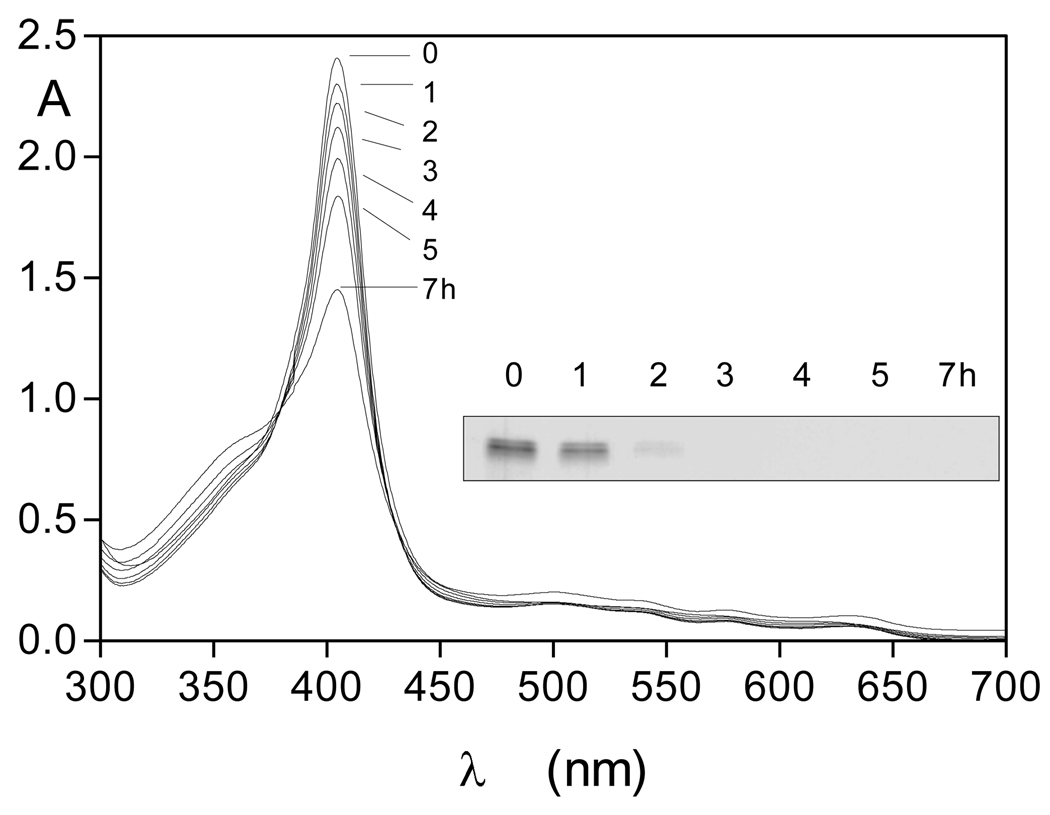
Figure 7.
Trypsin-mediated breakdown of hydroxy-methaemoglobin as shown by loss of Soret band intensity and by SDS-PAGE.
The gel was stained with coomassie blue. Haemoglobin was at 4µM and trypsin at 2µM. Incubation was carried out at 37°C in 0.1M Tris-HCl, 0.14M NaCl, pH 7.5.
Figure 8.
Proteolytic release of haem from bovine hydroxy-methaemoglobin-agarose mediated by trypsin and interpain A
Suspensions of methaemoglobin-agarose beads (4 µM with respect to haemoglobin tetramer) were incubated at 37°C with trypsin or InpA (2 µM) in 0.1M Tris-HCl, 0.14M NaCl, pH 7.5. Beads were pelleted by centrifugation and the supernatant buffer assayed at intervals for released free haem using the pyridine haemochrome assay.
Azide ligation prevents InpA breakdown of aquomethaemoglobin at pH 6.0
Aquomethaemoglobin was efficiently degraded by InpA. However, when the haem iron of aquomethaemoglobin was ligated with azide, the protein was resistant to breakdown by InpA at pH 6.0. Neither globin chain proteolysis nor haem loss was observed (Figs 5, panels A and B, gel d,). In stark contrast, haem-free globin (at the same concentration as the methaemoglobin substrates) was completely degraded within a few minutes by InpA, even at pH 7.5 (Fig 5A, gel e).
The difference in susceptibility of the aquo-met- versus the hydroxymethaemoglobin substrate is, at first sight, puzzling, as these two forms are structurally indistinguishable. Although InpA displays a broad pH activity profile centred on pH 8 versus the synthetic chromogenic peptide substrates bearing P1 Lys or Arg residues, it also has an acid pH optimum (~ pH 6) towards protein substrates azocoll, azoalbumin and azocasein (Potempa et al., unpublished data), which would explain the efficient proteolysis at acid pH. However, if haemoglobin breakdown was simply related to a pH effect on the enzyme, then the extent of proteolysis of azide-liganded aquomethaemoglobin would be commensurate with that observed for the non-azide bound substrate at pH 6, but this was not the case. The mass spectrometric detection of peptide fragments arising from both alpha and beta globin chains during incubation of oxyhaemoglobin with InpA demonstrates that proteolytic attack occurs during oxidation. The reason that gross breakdown of either oxyhaemoglobin or hydroxymethaemoglobin does not occur is that proteolytic modification resulting in oxidation at pH 7.5, brought about the production of a stable haemichrome, largely resistant to further breakdown by the enzyme. Rather, the underlying reason for proteolysis of aquomethaemoglobin by InpA resides in the fact that haem dissociation is greatly increased when haemoglobin is in the aquomet- compared to the hydroxymet- form, which in turn facilitates the proteolysis of the newly formed haem-free globin.
Mechanism of InpA-mediated oxidation
As a functional oxygen carrier, the oxyhaemoglobin haem iron exists in the Fe2+ form. However, the ferrous iron can spontaneously oxidize (auto-oxidation) into the ferric state (methaemoglobin) with the production of superoxide anion, O2− [23, 24], and in this form is incapable of binding oxygen. Because O2 is a poor one-electron acceptor, the Fe-O2 bond is relatively stable which poses a considerable thermodynamic barrier for electron transfer. For this reason, auto-oxidation via spontaneous superoxide dissociation is considered unlikely [34]. Rather, the accepted mechanism is the entry of OH− or H2O molecules into the haem pocket from the solvent, to effect a stoichiometric nucleophilic displacement of O2− [23, 26]. These nucleophiles remain bound to the iron at the sixth co-ordinate position, forming either aquomethaemoglobin (or acid form), for bound H2O, or hydroxymethaemoglobin, for bound OH−. The formation of these species is indicative of the oxidation process. During InpA-mediated oxyhaemoglobin oxidation, we detected several peptides by mass spectrometry, indicative of scission at sites around the haem pocket. Such cleavage events, whilst not leading to extensive proteolysis, may instead lead to structural changes which facilitate the displacement of O2− as evidenced by format ion of the hydroxymet- and aquomethaemoglobin species at pH 7.5 and 6.0, respectively, and in keeping with the accepted paradigm for the oxidation process [23, 34, 35].
Even minor structural changes to the haem pocket of haemoglobin disturb its function. For example, 46phe→val mutation in myoglobin markedly increases the rate of oxidation by facilitating access of water molecules [36]. This is because phenylalanine46 stabilises the distal histidine64, and removal of this contact through mutation to valine opens the distal pocket. In this context, it is noteworthy that a prominent peptide of molecular mass1356.92 (β chain, 45F…46GDLSNPGAVMGNPK59) was observed early in the time course of haemoglobin digestion by InpA, and is evidence that InpA disrupts the distal region of the haem pocket. Given that phe45 of the β-chain shares a similar orientation to the distal histidine as phe46 of myoglobin, and the structural similarities between these proteins, it is possible to speculate that this residue plays a similar role in dictating the degree of auto-oxidation, and that cleavage by InpA at this site may account for the increased oxidation rate of oxyhaemoglobin. It is also noteworthy that the abnormal M haemoglobins which have a high propensity to auto-oxidise have amino acid substitutions in or near the haem pocket [37].
The possibility that oxidation was a result of the presence of contaminant metal ions which are well know to bring about oxidation was also tested by conducting incubations of InpA with oxyhaemoglobin in the presence of EDTA (10µM). Under these conditions the rates of methaemoglobin formation brought about by InpA (determined by plots of δA576nm as above) measured over a 5 hour period were the same in both the presence and absence of EDTA (data not presented). It was concluded from this that the InpA-mediated oxidation was unrelated to the presence of metal ions.
Although oxidation of oxyhaemoglobin weakens the haem iron-proximal histidine bond, permitting haem dissociation [38, 39], breakdown of haemoglobin by InpA at acidic pH may also be aided, in addition to the proton-assisted oxidation [26], by the low pH which per se enhances haem mobility and dissociation from the protein, especially from the β chain [39]. We have shown that P. intermedia and P. nigrescens engender a low pericellular pH during pigment formation [3]. This would promote dissociation of haemoglobin tetramers into αβ dimers and enhance haem loss from the latter [40]. It should also be noted that αβ dimers auto-oxidise more rapidly than tetramers [41].
This is the first paper to describe a specific role of a P. intermedia protease in both haemoglobin oxidation and breakdown and subsequent haem release. Interpain A may thus play a major role in pigment formation by this black-pigmenting organism. Together with our previous work [7, 17] these present findings have revealed a new paradigm for haem acquisition from haemoglobin by black - pigmenting species, which is dependent upon initial haemoglobin oxidation which facilitates haem dissociation and globin breakdown.
ACKNOWLEDGEMENTS
JWS is grateful to the School of Dental Science, University of Liverpool, for financial support for DB. This study was also supported in part by grants from the National Institutes of Health, USA [DE 09761], and the Department of Scientific Research, Polish Ministry of Science and Higher Education [1642/B/P01/2008/35]. We thank Mark Prescott and Dr Deborah Ward of the Proteomics Laboratory, University of Liverpool, School of Biological Sciences, who carried out the mass spectrometry.
Abbreviations
- E64
trans-Epoxysuccinyl-L-leucylamido (4-guanidino) butane
- InpA
Interpain A
REFERENCES
- 1.Holt SC, Ebersole JL. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the red complex, a prototype polybacterial pathogenic consortium in periodontitis. Periodontology 2000. 2005;38:72–122. doi: 10.1111/j.1600-0757.2005.00113.x. [DOI] [PubMed] [Google Scholar]
- 2.Smalley JW, Silver J, Marsh PJ, Birss AJ. The periodontopathogen Porphyromonas gingivalis binds iron protoporphyrin IX in the µ-oxo dimeric form: an oxidative buffer and possible pathogenic mechanism. Biochem. J. 1998;331:681–677. doi: 10.1042/bj3310681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smalley JW, Silver J, Birss AJ, Withnall R, Titler PJ. The haem pigment of the oral anaerobes Prevotella nigrescens and Prevotella intermedia is composed of iron(III) protoporphyrin IX in the monomeric form. Microbiology. 2003;49:1711–1718. doi: 10.1099/mic.0.26258-0. [DOI] [PubMed] [Google Scholar]
- 4.Smalley JW, Birss AJ, Silver J. The periodontal pathogen Porphyromonas gingivalis harnesses the chemistry of the µ-oxo bishaem of iron protoporphyrin IX to protect against hydrogen peroxide. FEMS Microbiol. Lett. 2000;183:159–164. doi: 10.1111/j.1574-6968.2000.tb08951.x. [DOI] [PubMed] [Google Scholar]
- 5.Smalley JW, Birss AJ, Withnall R, Silver J. Interactions of Porphyromonas gingivalis with oxyhaemoglobin and deoxyhaemoglobin. Biochem. J. 2002;362:239–245. doi: 10.1042/0264-6021:3620239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smalley JW, Thomas MF, Birss AJ, Withnall R, Silver J. A combination of both arginine- and lysine-specific gingipain activity of Porphyromonas gingivalis is necessary for the generation of the µ-oxo bishaem-containing pigment from haemoglobin. Biochem. J. 2004;379:833–840. doi: 10.1042/BJ20031221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smalley JW, Birss AJ, Szmigielski B, Potempa J. Mechanism of methaemoglobin breakdown by the lysine-specific gingipain of the periodontal pathogen Porphyromonas gingivalis. Biol. Chem. 2008;389:1235–1238. doi: 10.1515/BC.2008.XXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zwickel J, Weiss EI, Schejter A. Degradation of native hemoglobin following hemolysis by Prevotella loescheii. Infect. Immun. 1992;60:1721–1723. doi: 10.1128/iai.60.4.1721-1723.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leung KP, Subramaniam PS, Okamoto M, Fukushima H, Lai CH. The binding and utilization of hemoglobin by Prevotella intermedia. FEMS Microbiol. Lett. 1998;162:227–233. doi: 10.1111/j.1574-6968.1998.tb13003.x. [DOI] [PubMed] [Google Scholar]
- 10.Guan SM, Nagata H, Shizukuishi S, Wu JZ. Degradation of human hemoglobin by Prevotella intermedia. Anaerobe. 2006;12:279–282. doi: 10.1016/j.anaerobe.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 11.Fujimura S, Ueda O, Shibata Y, Hirai K. Isolation and properties of a tripeptidyl peptidase from a periodontal pathogen Prevotella nigrescens. FEMS Microbiol. Lett. 2003;219:305–309. doi: 10.1016/S0378-1097(03)00048-X. [DOI] [PubMed] [Google Scholar]
- 12.Shibata Y, Miwa Y, Hirai K, Fujimura S. Purification and partial characterization of a dipeptidyl peptidase from Prevotella intermedia. Oral Microbiol. Immunol. 2003;18:196–198. doi: 10.1034/j.1399-302x.2003.00057.x. [DOI] [PubMed] [Google Scholar]
- 13.Mallorquí-Fernández N, Manandhar SP, Mallorquí-Fernández G, Usón I, Wawrzonek K, Kantyka T, Solà M, Thøgersen IB, Enghild JJ, Potempa J, Gomis-Rüth FX. A new autocatalytic activation mechanism for cysteine proteases revealed by Prevotella intermedia interpain A. J. Biol. Chem. 2008;283:2871–2882. doi: 10.1074/jbc.M708481200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nelson D, Potempa J, Kordula T, Travis J. Purification and characterization of a novel cysteine proteinase (periodontain) from Porphyromonas gingivalis. Evidence for a role in the inactivation of human alpha1-proteinase inhibitor. J. Biol. Chem. 1999;274:12245–12251. doi: 10.1074/jbc.274.18.12245. [DOI] [PubMed] [Google Scholar]
- 15.Potempa J, Golonka E, Filipek R, Shaw LN. Fighting an enemy within: cytoplasmic inhibitors of bacterial cysteine proteases. Mol. Microbiol. 2005;57:605–610. doi: 10.1111/j.1365-2958.2005.04714.x. [DOI] [PubMed] [Google Scholar]
- 16.Potempa J, Pike RN. Corruption of innate immunity by bacterial proteases. Innate Immun. 2009;1:70–87. doi: 10.1159/000181144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smalley JW, Birss AJ, Szmigielski B, Potempa J. Sequential action of R- and K-specific gingipains of Porphyromonas gingivalis in the generation of the haem-containing pigment from oxyhaemoglobin. Arch. Biochem. Biophys. 2007;465:44–49. doi: 10.1016/j.abb.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 18.Gallagher WA, Elliot WB. The formation of pyridine haemochromogen. Biochem. J. 1965;97:187–193. doi: 10.1042/bj0970187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ascoli F, Fanelli MR, Antonini E. Preparation and properties of apohaemoglobin and reconstituted haemoglobins. Meth. Enzymol. 1981;76:72–87. doi: 10.1016/0076-6879(81)76115-9. [DOI] [PubMed] [Google Scholar]
- 20.Charalabous P, Risk JM, Jenkins R, Birss AJ, Hart CA, Smalley JW. Characterisation of a bi-functional catalase-peroxidase of Burkholderia cenocepacia. FEMS Immunol. Med. Microbiol. 2007;50:37–44. doi: 10.1111/j.1574-695X.2007.00224.x. [DOI] [PubMed] [Google Scholar]
- 21.Kelm M, Dahmann R, Wink D, Feelisch M. The nitric oxide / superoxide assay. J. Biol. Chem. 1997;272:9922–9932. doi: 10.1074/jbc.272.15.9922. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y, Samson FE, Nelson SR, Pazdernik TL. Nitric oxide detection with intracerebral microdialysis: Important considerations in the application of the hemoglobin-trapping technique. J. Neurosci. Meth. 1996;68 doi: 10.1016/0165-0270(96)00058-1. l65-I73. [DOI] [PubMed] [Google Scholar]
- 23.Tsuruga M, Shikama K. Biphasic nature in the oxidation reaction of human oxyhemoglobin. Biochim. Biophys. Acta. 1997;133:96–104. doi: 10.1016/s0167-4838(96)00156-2. [DOI] [PubMed] [Google Scholar]
- 24.Rifkind JM, Abugo O, Levy A, Heim J. Detection, formation and relevance of hemichromes and hemochromes. Meth. Enzymol. 1994;231:449–480. doi: 10.1016/0076-6879(94)31030-0. [DOI] [PubMed] [Google Scholar]
- 25.Rachmilewitz EA. Formation of hemichromes from oxidised subunits. Ann. New York Acad. Sci. 1969;165:171–184. doi: 10.1111/j.1749-6632.1969.tb27787.x. [DOI] [PubMed] [Google Scholar]
- 26.Shikama K. The molecular mechanisms of autoxidation for myoglobin and haemoglobin: a venerable puzzle. Chem. Rev. 1998;98:1357–1372. doi: 10.1021/cr970042e. [DOI] [PubMed] [Google Scholar]
- 27.Shah HN, Williams RAD, Bowden GH, Hardie JM. Comparison of the properties of Bacteroides melaninogenicus from human dental plaque and other sites. J. Appl. Bacteriol. 1976;41:473–492. doi: 10.1111/j.1365-2672.1976.tb00660.x. [DOI] [PubMed] [Google Scholar]
- 28.Takahashi N, Saito K, Schachtele CF, Yamada T. Acid tolerance and acid-neutralizing activity of Porphyromonas gingivalis, Prevotella intermedia and Fusobacterium nucleatum. Oral Microbiol. Immunol. 1997;12:323–328. doi: 10.1111/j.1399-302x.1997.tb00733.x. [DOI] [PubMed] [Google Scholar]
- 29.Takahashi N, Yamada T. Glucose metabolism by Prevotella intermedia and Prevotella nigrescens. Oral Microbiol. Immunol. 2000;15:188–195. doi: 10.1034/j.1399-302x.2000.150307.x. [DOI] [PubMed] [Google Scholar]
- 30.Guan SM, Nagata H, Maeda K, Kuboniwa M, Minamino N, Shizukuishi S. Purification and characterization of a hemoglobin-binding outer membrane protein of Prevotella intermedia. FEMS Microbiol. Lett. 2004;235:333–339. doi: 10.1016/j.femsle.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 31.Beem JE, Nesbitt WE, Leung KP. Identification of hemolytic activity in Prevotella intermedia. Oral Microbiol.Immunol. 1998;13:97–105. doi: 10.1111/j.1399-302x.1998.tb00719.x. [DOI] [PubMed] [Google Scholar]
- 32.Okamoto M, Meada N, Kondo K, Leung KP. Hemolytic and hemagglutinating activities of Prevotella intermedia and Prevotella nigrescens. FEMS Microbiol. Lett. 1999;178:299–304. doi: 10.1111/j.1574-6968.1999.tb08691.x. [DOI] [PubMed] [Google Scholar]
- 33.Silva TA, Rodrigues PH, Ribeiro RN, Noronha FSM, Farias LMF, Carvalho MAR. Hemolytic activity of Prevotella intermedia and Prevotella nigrescens stains: influence of abiotic factors in solid and liquid assays. Res. Microbiol. 2003;154:29–35. doi: 10.1016/s0923-2508(02)00003-7. [DOI] [PubMed] [Google Scholar]
- 34.Shikama K. Nature of the FeO2 bonding in myoglobin: an overview from physical to clinical biochemistry. Experentia. 1985;41:701–706. doi: 10.1007/BF02012563. [DOI] [PubMed] [Google Scholar]
- 35.Shikama K. Autoxidation of oxymyoglobin: a meeting point of the stabilization and the activation of molecular oxygen. Biol. Rev. Camb. Philos. Soc. 1990;65:517–527. doi: 10.1111/j.1469-185x.1990.tb01236.x. [DOI] [PubMed] [Google Scholar]
- 36.Lai HH, Li T, Lyons DS, Phillips GN, Olson JS, Gibson QH. Phe-46(CD4) orients the distal histidine for hydrogen bonding to bound ligands in sperm whale myoglobin. Proteins. 1995;22:322–339. doi: 10.1002/prot.340220404. [DOI] [PubMed] [Google Scholar]
- 37.Percy MJ, McFerran NV, Lappin TRJ. Disorders of oxidised haemoglobin. Blood Rev. 2005;19:61–68. doi: 10.1016/j.blre.2004.02.001. [DOI] [PubMed] [Google Scholar]; 37 Brantley RE, Smerdon SJ, Wilkinson AJ, Singleton EW, Olson JS. The mechanism of auto-oxidation of myoglobin. J. Biol. Chem. 1993;268:6995–7010. [PubMed] [Google Scholar]
- 38.Hargrove MS, Singleton EW, Quillin ML, Ortiz LA, Phillips GN, Olson JS. His64(E7)>Tyr apomyoglobin as a reagent for measuring rates of hemin dissociation. J. Biol. Chem. 1994;269:4207–4214. doi: 10.2210/pdb1mgn/pdb. [DOI] [PubMed] [Google Scholar]
- 39.Katz S, Beall JA, Crissman JK. Structure-volume relationship. Dilatometric study of the acid-base reaction involving human oxy- and methemoglobins in water and denaturing media. Biochem. 1973;12:4180–4185. doi: 10.1021/bi00745a022. [DOI] [PubMed] [Google Scholar]
- 40.Zhang L, Levy A, Rifkind JM. Autoxidation of hemoglobin enhanced by dissociation into dimers. J. Biol. Chem. 1991;266:24698–24701. [PubMed] [Google Scholar]