Abstract
Biological memory can be defined as a sustained cellular response to a transient stimulus. To understand this phenomenon, we must consider how the properties of different biological systems achieve memory of a stimulus, essentially permitting a cell to produce a lasting response. One way that cells accomplish this task is through transcriptional states, which involve populations of molecules regulating gene expression. If the transcriptional response is bistable, a chemical state becomes defined as on or off and, given certain parameters, this state can be inherited through DNA replication and cell division. In this way, a cell can produce a lasting memory of a biological response.
Synthetic biologists are especially interested in transcriptional responses as a means of cellular memory because (1) much of a cell’s information processing is performed through transcription, and (2) the basic machinery for such biological behavior is well-understood. As such, transcription provides us with a set of characterized genetic units, such as promoters, activators, and repressors, that can be recombined to create novel transcriptional circuits. Furthermore, the way nature combines these biological parts to produce specific outputs, including cellular memory, has been extensively studied. Thus, we have at our fingertips the tools with which to design synthetic memory systems.
The construction of synthetic memory circuits will improve our understanding of natural networks, further aiding the creation of useful, novel biological tools. For example, a device capable of remembering a biological experience might be utilized in the long-term study of particular cells within a heterogeneous population following a defined event, or applied in industry for the sustained production of desired proteins after induction by a brief stimulus. Such bio-engineered networks exemplify a primary objective of synthetic biology: to advance simple synthetic devices into expertly constructed circuits with significant applications.
How cells make memories
More than fifty years ago, Monod and Jacob determined qualitatively how a cell might achieve biological memory through its transcriptional circuitry (Monod, et al., 1961). Only recently, however, were these circuits understood quantitatively (Alon, 2006). Synthetic biology bridges the gap between biology and mathematics, requiring our understanding of cellular memory to encompass more than half a century of scientific work. Here, we will briefly address the concepts that are fundamental to the achievement of biological memory through transcription.
The Hill function
Cells must sense and dynamically respond to both internal and external signals. This often requires the synthesis or modification of transcription factors, which form an interaction network whose design determines the speed and sustainability of protein output in response to an environmental input (Monod, et al., 1961; Alon, 2006). Depending on whether transcription factor X is an activator or repressor, the concentration of gene product Y increases or decreases from a basal level (s) as a function of the concentration of X binding to Y’s promoter (f (X)). The production of Y is further balanced by Y’s degradation and dilution rates (defined as α, with units of 1/time):
| (1) |
To more thoroughly understand how a transcriptional input produces specific gene outputs, one can use the Hill function in the above equation to define f (X) and describe the equilibrium binding of a transcription factor to its target promoter (Alon, 2006). While a transactivator Hill function (Eq. 2) varies slightly from that of a transrepressor (Eq. 3), the components of each are similar (Figure 1a):
| (2) |
| (3) |
There are three key parameters: K, β, and n. K is the activation/repression coefficient, defining the concentration of X needed to reach the threshold for activation or repression of Y; K’s value is largely related to the chemical affinity between X and its binding site on Y’s promoter (Alon, 2006). The term β defines the maximal expression level from Y’s promoter (in units of mRNAs/time), obtained when an activator is bound or a repressor is unbound. The most pertinent parameter is n, the Hill coefficient. This value governs how a network responds to transcriptional input: a smaller n (n = 1) results in a more graded response, while a larger n (n = 4) produces a bistable, switch-like response (Figure 1a). The latter behavior is essential to biological memory, because a bistable response allows a system to shift to an alternative steady state that might persist over time.
Figure 1.
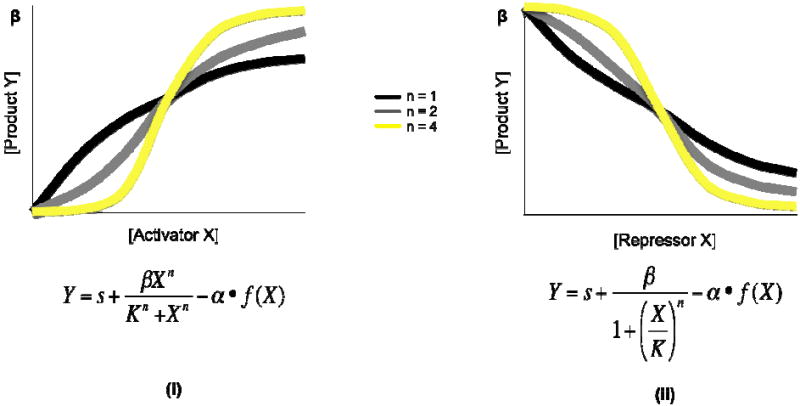
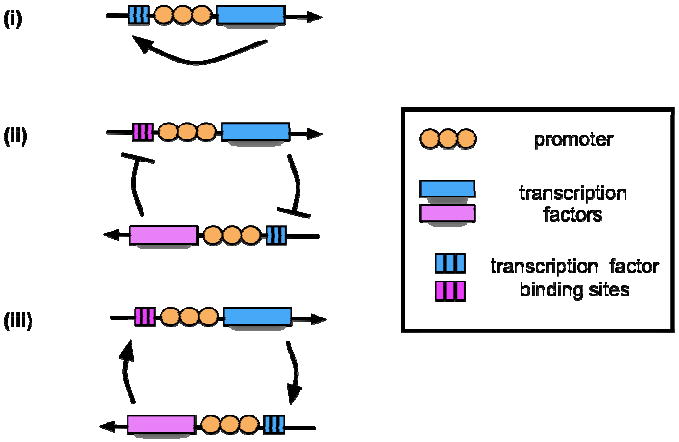
Figure 1a. Dynamics of regulated gene circuits described by Hill functions. (i) The concentration of product Y is plotted as a function of the concentration of activator X, as described by Hill functions with n = 1, 2, and 4. β is the maximal expression level from Y’s promoter when X is bound, and K defines the concentration of X needed to reach the threshold for activation of Y (ii) The concentration of product Y is plotted as a function of the concentration of repressor X, as described by Hill functions with n = 1, 2, and 4. β is the maximal expression level from Y’s promoter when X is unbound, and K defines the concentration of X need to the threshold for repression of Y.
Figure 1b. Network motifs that achieve biological memory: (i) positive feedback, (ii) double-negative feedback, and (iii) double-positive feedback.
Where does that “n” come from?
To achieve a sufficiently high Hill coefficient, biological systems employ a number of mechanisms. First, there must be non-linearity within a transcriptional circuit, meaning a functional element that guarantees a threshold-like response to a stimulus is required (Ninfa, 2004). This effect can be achieved through the affinity, cooperativity, and/or multimerization of transcription factors at their binding sites within target gene promoters. Transcription factors vary widely in their degrees of cooperativity and affinity for their binding sites, and the degree of binding can also differ depending on the number of binding sites present in a given promoter. These factors help to ensure a lasting response to a transient stimulus and are especially combative against stochastic fluctuation between steady states, an undesirable behavior in a transcriptional circuit designed to produce a lasting response.
Another requirement for attaining a large Hill coefficient is that the genetic elements we have touched upon thus far must be arranged in specific motifs that permit bistable responses (Monod, et al., 1961). To achieve this goal, nature often employs transcriptional positive feedback (Ferrell, 2002; Alon, 2006). In this network design, production of protein Y only increases once the concentration of X approaches the expression threshold for Y’s promoter, resulting in a sigmoidal response curve (Demongeot, et al., 2000). Positive feedback can be produced by either a single transcription factor self-activating in response to a stimulus (positive autofeedback), or two transcription factors regulating each other through two positive or negative interactions (double-positive / double-negative feedback) (Figure 1b). When protein concentrations reach certain thresholds, all three motifs result in a switch between two steady states. If the switch is sharp enough, a gene can become locked into an alternate steady state, even in the absence of the original inducer (Ferrell, 2002).
Finally, to achieve the desired Hill coefficient, it is important to remember that transcription-based memory is derived from proteins that are degraded at some natural rate and diluted through cell growth. If the rates of degradation and dilution are faster than protein production rates, levels of transcription factor X may not be sufficiently high to achieve or maintain the desired bistable output. Since this would result in a failure to achieve memory once the initial stimulus is absent, it is a critical parameter for the bioengineer to keep in mind.
Biological memory in nature
Many natural examples of biological memory have been discovered to possess feedback, bistability, and cooperativity. We will highlight some of this work and include examples that are not strictly transcription-based but nonetheless employ parallel mechanisms. These natural systems inform the design of synthetic memory circuits, and the diversity of mechanisms highlights the general importance of this biological phenomenon.
Phage lambda, lac operon, cell cycle
Some of the earliest work in molecular genetics elucidated the role of bistable networks in the phage lambda regulatory system, the lac operon, and the cell cycle. For example, Eisen, et al, described a multiply mutated lambda lysogen in which the toxic functions of the phage were eliminated, leaving only the regulatory essence of lambda’s two-state system (Eisen, et al., 1970); considering that this was prior to the use of restriction enzymes in genetic constructions, this work arguably represents an early example of synthetic biology limited by the technology available at that time. The phage lambda system switches between two states based on a mutual repression loop between the antagonists lambda repressor and Cro (Ptashne, 2004) (Figure 2a) and involves both positive autoregulation and double-negative feedback. Likewise, the lac operon can exist in on or off states mediated through the positive feedback loop of lactose transport (Muller-Hill, 1996). Work in Xenopus oocyte extracts has revealed that positive feedback loops govern cell cycle progression by creating a bistable system wherein levels of specific proteins alternate between two steady states, determining discrete phases and forward movement through the cycle (Novak, et al., 1993; Thron, 1996).
Figure 2. Natural mechanisms of memory.
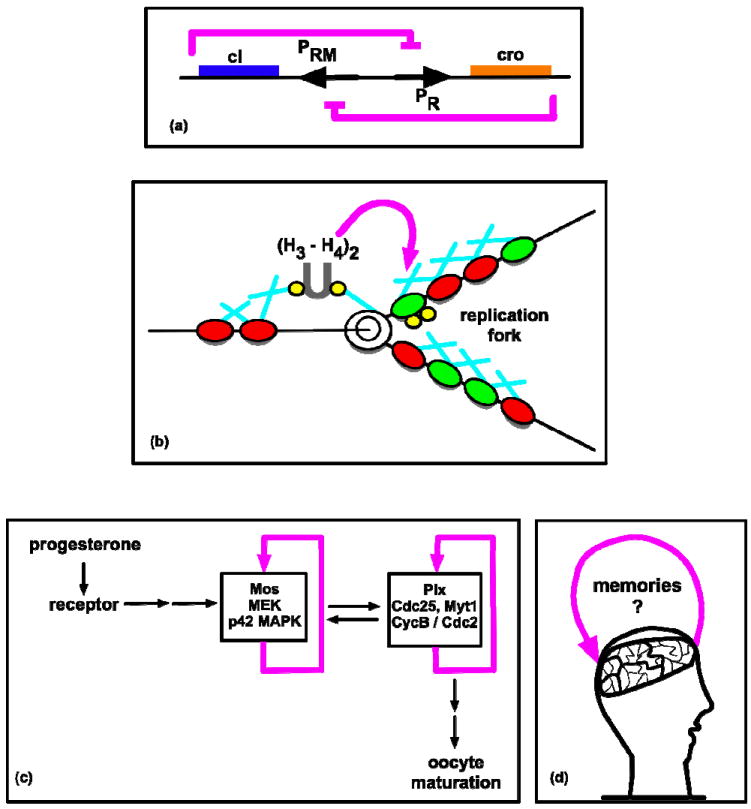
(a) The phage lambda system switches between two states based on a mutual repression loop between lambda repressor and Cro (Ptashne, 2004). (b) Replication-coupled nucleosome assembly maintains silent chromatin. Some silent nucleosomes (red) contain (H3-H4)2 tetramers (gray) with H3-K9Me methylation marks (yellow). Positive feedback can arise if newly synthesized nucleosomes (green) become methylated by histone methyltransferases bound to HP1 (blue) on adjacent nucleosomes, allowing methylated regions to persist through DNA replication and cell division (adapted from Vermaak, et al., 2003). (c) Positive-feedback-based, bistable networks govern Xenopus oocyte maturation (adpated from Xiong, et al., 2003). (d) Information storage in the brain is believed to involve bistable feedback networks (Ogasawara, et al., 2009).
Nucleosomal modification
Positive feedback is also thought to be utilized in nucleosomal modification. The mechanism is believed to allow chromosomal regions to adopt stable and heritable states, resulting in bistable gene expression that persists through DNA replication and cell division (Dodd, et al., 2007; Barrett, et al., 2008). Positive autoregulation can arise if nucleosomes carrying a specific modification recruit enzymes that catalyze similar modifications of neighboring nucleosomes, allowing a nucleosomal cluster to stably maintain itself in a particular modification state (Stefanko, et al., 2009). This phenomenon may be involved in the silencing of H3-H4 histones by H3-K9 methylation marks, to which heterochromatin-associated protein 1 (HP1) can bind (Vermaak, et al., 2003) (Figure 2b). It is hypothesized that histone methytransferases bound to HP1 can transmit methylation marks to adjacent, newly replicated nucleosomes, creating an epigenetic feedback loop of silenced chromatin.
Cell differentiation
Given the involvement of feedback motifs in epigenetic determination, it is not at all surprising to find this mechanism further used in eukaryotic cell differentiation, a process determined by epigenetic patterning. For example, transition between two Xenopus oocyte maturation stages is induced by progesterone. Once the progesterone has dissipated, commitment to maturation is maintained via positive feedback within a mitogen-activated protein (MAP) kinase cascade (Figure 2c). The role of positive feedback in binary cell-fate switches has been further observed in mouse and human embryonic stem (ES) cells. Genome-wide transcriptional studies have identified OCT4, SOX2, and NANOG as critical players in the circuitry responsible for cell differentiation (Boyer, et al., 2005; Loh, et al., 2006). Dynamic modeling has revealed that these transcription factors interact via positive feedback loops, resulting in a bistable switch that regulates when and how an ES cell differentiates (Chickarmane, et al., 2006).
The immune and nervous systems
Finally, a discussion of memory storage in the immune and nervous systems naturally arises. These systems are alike in many ways, including their capacity to manage vast quantities of information. The immune system is required to consistently recognize foreign antigens from countless sources, and each individual neuron is required to receive information from numerous synaptic connections. To file this data, each system employs molecular memory mechanisms. One tool used by the immune system is somatic V(D)J recombination, which bistably rearranges genes in response to specific foreign antigens, permitting lymphocytes to produce the necessary proteins for mounting immune responses. This mechanism results in both an immediate immune response and a population of long-lived memory cells that can mount stronger responses if the initial pathogen is ever again detected (Muotri, et al., 2006). To handle the brain’s computing load and to store information despite molecular turnover, one mechanism used by neurons might be positive feedback (Tanaka, et al., 2008; Ogasawara, et al., 2009). This activity may serve as a general method for perpetuating signal transduction networks in the brain, thus further establishing memory networks as remarkable biological tools (Figure 2d).
Synthetic memory circuits, thus far
Using natural memory circuits as guides, a variety of synthetic memory pathways have been engineered from transcription-based parts in bacterial, yeast, and mammalian cells.
Bacterial memory devices
One of the first simple bistable devices was constructed in Escherischia coli (E. coli) by Gardner, et al (Gardner, et al., 2000). The modules consist of double-negative feedback systems using well-described prokaryotic gene repressor proteins. This device demonstrates bistability: once the switch is flipped toward one steady state, it remains there in the absence of the original stimulus, until a second stimulus shifts the system to an alternate steady state. This behavior is ensured by cooperativity in the binding of repressors to DNA and by trial-and-error testing of different strength promoters (Gardner, et al., 2000; Ferrell, 2003). The Gardner circuit was the first demonstration that bistable responses can indeed be engineered into a synthetic system. Others have extended this work by building similar networks that are more robust due to further quantitative understanding of the modular components that constitute such systems (Atkinson, et al., 2003; Isaacs, et al., 2003).
These preliminary memory switches allowed for the later construction of bacterial memory networks with novel functions. This was best demonstrated when Kobayashi, et al, used a lacI/lambda repressor toggle switch as a memory circuit embedded within a larger circuit based on the E. coli SOS signaling pathway, enabling the memory circuit to sense DNA damage and retain memory of this event (Kobayashi, et al., 2004). The group further constructed a strain in which biofilm formation is induced post-DNA damage, thereby demonstrating that artificial regulatory circuits can be used to produce permanent phenotypic changes.
Yeast memory devices
The early successes in bacteria laid the groundwork for similar projects in the budding yeast Saccharomyces cerevisiae. Work on yeast memory modules largely began with Becskei and colleagues, who constructed a simple switch in which a tetracycline-dependent activator turns on its own synthesis (Becksei, et al., 2001). This design permits cells to switch between on and off states in response to increasing levels of tetracycline, largely due to the inherent cooperativity of the activator and eukaryotic transcription (Becksei, et al., 2001).
Ingolia and colleagues sought to determine how easily a monostable signaling pathway might become bistable by engineering positive feedback into the system (Ingolia, et al., 2007). To address this question, they chose the budding-yeast mating-pheromone response, a well-studied MAP kinase system stimulated by exogenous pheromone (Ingolia, et al., 2007). Using the natural phosphorylation cascade, the authors expressed a dominant active allele of a key pathway protein. Once triggered, the pathway can sustain itself because the dominant protein is able to feed back into the natural pathway, thereby producing a positive autoregulatory circuit. Furthermore, the tunability of the feedback loop was demonstrated via mutations that altered the basal and induced expression levels of the feedback promoter.
The application of quantitative approaches to reliable circuit design is well-illustrated by the work of Ajo-Franklin, et al in yeast (Ajo-Franklin, et al., 2007). Using quantitative modeling of transcription dynamics, the authors constructed a synthetic memory circuit based on transcriptional positive feedback. The device bistably responds to a pulse of galactose by producing a transcriptional activator that induces a downstream autofeedback loop. Given certain system parameters, the loop activity persists in the absence of galactose and the inducing transactivator, such that the circuit imparts memory of galactose exposure onto cells and their progeny. Computational modeling using quantitative descriptions of various tested transactivators suggested that low basal expression coupled with switch-like activation was required to maintain memory; growth rate was also found to significantly impact memory loop protein sustainability following cell division.
Mammalian memory devices
The design strategies and critical parameters discovered in nature and tested synthetically in bacteria and yeast have been further applied toward the engineering of mammalian cells. Of note is the toggle switch in Chinese hamster ovary cells, the design of which is largely based on the bacterial toggle switch (Kramer, et al., 2004). Using streptogramin and macrolide-dependent transrepressors, the system responds in a bistable manner to specific inducers. This response allows for switch-specific expression of a human glycoprotein both in culture and mice, impressively demonstrating the possibility of epigenetic transgene control through bistable circuits. Fussenegger’s group has designed a number of such modules, primarily using antibiotics to control positive or double-negative feedback networks (Kramer, et al., 2003; Weber, et al., 2007; Tigges, et al., 2009). This work lays groundwork for the further bioengineering of mammalian systems and their application in research and clinical settings.
Transcriptional memory devices vs. DNA recombinase-based circuits
The synthetic circuits discussed thus far all employ transcriptional circuitry that mimics natural networks to produce biological memory. It should be noted that memory devices have also been constructed using DNA recombinase-based systems such as Cre-Lox and FLP-FRT, which leave a permanent mark in the genome (Mortensen, et al., 2006; Friedland, et al., 2009). While this approach is useful, there are potential problems as DNA rearrangements and recombinase expression can have undesirable consequences if not properly regulated. Additionally, transgene expression via recombination is more difficult to tune or reverse due to its permanency, allowing less flexibility for a synthetic biologist interested in exploring the design capabilities of network-based memory. While both methods are entirely valid, each is appropriate in different situations and should be applied with careful consideration.
Why make memories?
Having observed the successful integration of nature’s tools into a variety of synthetic memory devices, we may now ask an important question: “What can we do with this?” The ability to construct robust, synthetic circuits enables us to engineer cells capable of recording stimulus exposure and/or maintaining desired levels of gene expression over time, in absence of inducer. Advances in synthesis and novel recombinant DNA technology allow the production of such devices from highly interchangeable units, permitting responses to an array of stimuli in a variety of cell types (Shetty, et al., 2008). A modular approach to stimulus induction of a high-fidelity memory device might allow researchers to identify cell populations responsive to specific events and track their progression through the cellular response. This would be of particular use if a device employs fluorescent markers that permit quantitative, single-cell tracking of cells within a responding population, as suggested by Ajo-Franklin (Ajo-Franklin, et al., 2007). These device characteristics may address whether response to a defined event correlates with later cell behavior. Capturing this cellular phenomenon could have great impact on the study of any disease involving the inheritance of a cellular state, such as cancer.
In addition to being read-outs of cellular experience, memory modules can potentially use their output as regulatory input to perform novel functions. Harnessing the ability to achieve long-term maintenance of desired relative protein levels, memory circuits might precisely regulate output or rapidly alternate between multiple outputs. Along these lines, one can imagine a memory module contributing to gene therapy or the synthetic differentiation of mammalian stem cells in a certain fashion after experiencing a brief stimulus.
Importantly, given the parallels to natural mechanisms of cell-based inheritance, such circuits may increase our understanding of such biological processes as cellular differentiation and tissue-formation. For example, at decreased levels of activation, a positive feedback module can spontaneously switch between steady states. This can lead to variable gene expression and possibly such undesired phenotypes as disease; in fact, unstable autocrine feedback loops have been linked to some cases of tumorigenesis (Schulze, et al., 2001). By studying the engineering of feedback loops, we may better understand how their malfunction affects biological events.
Finally, memory modules are potentially useful in industrial biotechnology. The feedback loop permits sustained induction of recombinant proteins without massive quantities of inducer. Promoters that respond to a plethora of stimuli (specific small molecules, pH, temperature, anaerobisis) already exist or can be engineered, allowing for a variety of production conditions. Of course, for these ventures to be successful, certain factors will have to be considered, including the impact of induction on protein yields, and how recombinant gene expression affects cell growth and physiology. If properly engineered, however, memory modules may help overcome high production costs associated with requiring large quantities of chemical inducer.
The precise design and implementation of systems exhibiting complex dynamic behavior remains a major goal of synthetic biology. An educated choice of network components and their fluid assembly into constructs with predictable behavior may enable synthetic circuits to increase our understanding of biology and improve our ability to engineer cells. The potential lying within memory modules may help progress the synthetic biology field toward a new phase of device production. This phase will ideally incorporate detailed quantitative and qualitative approaches into the creation of highly robust, reliable memory circuits with important, applicable functions. In sum, both nature’s wisdom and previously designed memory modules can provide bioengineering insight to supplement our movement toward devices capable of producing significant cellular memories that can last a lifetime.
Acknowledgments
Special thanks to the Silver lab, Jeffrey Way, Fred Winston, and Adrian Salic for helpful discussion. D.R.B. is supported by the NSF Synthetic Biology Engineering Research Center, and P.A.S. by grants from the National Institutes of Health.
References
- Ajo-Franklin CM, Drubin DA, Eskin JA, Gee EP, Landgraf D, Phillips I, Silver PA. Rational design of memory in eukaryotic cells. Genes Dev. 2007;21:2271–2276. doi: 10.1101/gad.1586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alon U. An introduction to systems biology: Design principles of biological circuits. Chapman & Hall/CRC Press; Boca Raton, FL: 2006. [Google Scholar]
- Atkinson MR, Savageau MA, Myers JT, Ninfa AJ. Development of genetic circuitry exhibiting toggle switch or oscillatory behavior in Escherichia coli. Cell. 2003;113:597–607. doi: 10.1016/s0092-8674(03)00346-5. [DOI] [PubMed] [Google Scholar]
- Barrett RM, Wood MA. Beyond transcription factors: the role of chromatin modifying enzymes in regulating transcription required for memory. Learning & Memory. 2008;15:460–467. doi: 10.1101/lm.917508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becksei A, Seraphin B, Serrano L. Positive feedback in eukaryotic gene networks: Cell differentiation by graded to binary response conversion. EMBO. 2001;20:2528–2535. doi: 10.1093/emboj/20.10.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chickarmane V, Troein C, Nuber UA, Sauro HM, Peterson C. Transcriptional dynamics of the embryonic stem cell switch. PLOS Comp Biol. 2006;9:1080–1092. doi: 10.1371/journal.pcbi.0020123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demongeot J, Kaufman M, Thomas R. Positive feedback circuits and memory. C R Acad Sci III. 2000;323:69–79. doi: 10.1016/s0764-4469(00)00112-8. [DOI] [PubMed] [Google Scholar]
- Dodd IB, Micheelsen MA, Sneppen K, Thon G. Theoretical analysis of epigenetic cell memory by nucleosome modification. Cell. 2007;129:813–822. doi: 10.1016/j.cell.2007.02.053. [DOI] [PubMed] [Google Scholar]
- Eisen H, Brachet P, Pereira da Silva L, Jacob F. Regulation of repressor expression in λ. PNAS. 1970;66:855–862. doi: 10.1073/pnas.66.3.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrell JE. Self-perpetuating states in signal transduction: positive feedback, double-negative feedback, and bistability. Curr Opin Chem Biol. 2002;6:140–148. doi: 10.1016/s0955-0674(02)00314-9. [DOI] [PubMed] [Google Scholar]
- Friedland AE, Lu Timothy K, Wang X, Shi D, Church G, Collins JJ. Synthetic gene networks that count. Science. 2009;324:1199–1202. doi: 10.1126/science.1172005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Murray AW. Positive-feedback loops as a flexible biological module. Curr Biol. 2007;17:668–677. doi: 10.1016/j.cub.2007.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs FJ, Hasty J, Cantor CR, Collins JJ. Prediction and measurement of an auto-regulatory genetic module. PNAS. 2003;100:7714–7719. doi: 10.1073/pnas.1332628100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Kaern M, Araki M, Chung K, Gardner TS, Cantor CR, Collins JJ. Programmable cells: Interfacing natural and engineered gene networks. PNAS. 2004;101:8414–8419. doi: 10.1073/pnas.0402940101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer BP, Weber W, Fussenegger M. Artificial regulatory networks and cascades for discrete multilevel transgene control in mammalian cells. Biotech Bioeng. 2003;83:810–820. doi: 10.1002/bit.10731. [DOI] [PubMed] [Google Scholar]
- Kramer BP, Viretta AU, Daoud-El-Baba M, Aubel D, Weber W, Fussenegger M. An engineered epigenetic transgene switch in mammalian cells. Nature Biotechnol. 2004;22:867–870. doi: 10.1038/nbt980. [DOI] [PubMed] [Google Scholar]
- Loh Y-H, Wu Q, Chew J-L, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, Wong KY, Sung KW, Lee CW, Zhao XD, Chiu KP, Lipovich L, Kuznetsov VA, Robson P, Stanton LW, Wei CL, Ruan Y, Lim B, Ng HH. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Gen. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Monod J, Jacob F. General conclusions: telenomic mechanisms in cellular metabolism, growth, and differentiation. Cold Spring Harbor Symp Quant Biol. 1961;26:389–401. doi: 10.1101/sqb.1961.026.01.048. [DOI] [PubMed] [Google Scholar]
- Mortensen R. Overview of gene targeting by homologous recombination. Curr Prot Mol Biol. 2006;23:1.1–1.12. doi: 10.1002/0471142727.mb2301s76. [DOI] [PubMed] [Google Scholar]
- Muller-Hill B. The lac operon: A short history of a genetic paradigm. Walter de Gruyter; Berlin, Germany: 1996. [Google Scholar]
- Muotri AR, Gage FH. Generation of neuronal variability and complexity. Nature. 2006;441:1087–1093. doi: 10.1038/nature04959. [DOI] [PubMed] [Google Scholar]
- Ninfa AJ, Mayo AE. Hysteresis vs. graded responses: the connections make all the difference. Sci STKE. 2004;232:e20. doi: 10.1126/stke.2322004pe20. [DOI] [PubMed] [Google Scholar]
- Novak B, Tyson JJ. Numerical analysis of a comprehensive model of M-phase control in Xenopus oocyte extracts and intact embryos. J Cell Sci. 1993;106:1153–1168. doi: 10.1242/jcs.106.4.1153. [DOI] [PubMed] [Google Scholar]
- Ogasawara H, Kawato M. Bistable switches for synaptic plasticity. Sci Signal. 2009;35:e7. doi: 10.1126/scisignal.256pe7. [DOI] [PubMed] [Google Scholar]
- Ptashne M. A genetic switch: Phage lambda revisited. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2004. [Google Scholar]
- Schulze A, Lehmann K, Jefferies HBJ, McMahon M, Downward J. Analysis of the transcriptional program induced by Raf in epithelial cells. Genes Dev. 2001;15:981–994. doi: 10.1101/gad.191101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty RP, Endy D, Knight TF., Jr Engineering BioBrick vectors from BioBrick parts. J Biol Eng. 2008;2:1–12. doi: 10.1186/1754-1611-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanko DP, Barrett RM, Ly AR, Reolon GK, Wood MA. Modulation of long-term memory for object recognition via HDAC inhibition. PNAS. 2009;106:9447–9452. doi: 10.1073/pnas.0903964106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Augustine GJ. A positive feedback signal transduction loop determines timing of cerebellar long-term depression. Neuron. 2008;59:608–620. doi: 10.1016/j.neuron.2008.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thron CD. A model for a bistable biochemical trigger of mitosis. Biophys Chem. 1996;57:239–251. doi: 10.1016/0301-4622(95)00075-5. [DOI] [PubMed] [Google Scholar]
- Tigges M, Marquez-Lago TT, Stelling J, Fussenegger M. A tunable synthetic mammalian oscillator. Nature. 2009;457:309–312. doi: 10.1038/nature07616. [DOI] [PubMed] [Google Scholar]
- Vermaak D, Ahmad K, Henikoff S. Maintenance of chromatin states: an open-and-shut case. Curr Opin Cell Biol. 2003;15:266–274. doi: 10.1016/s0955-0674(03)00043-7. [DOI] [PubMed] [Google Scholar]
- Weber W, Kramer BP, Fussenegger M. A genetic time-delay circuitry in mammalian cells. Biotech Bioeng. 2007;98:894–902. doi: 10.1002/bit.21463. [DOI] [PubMed] [Google Scholar]
- Xiong W, Ferrell JE., Jr A positive-feedback-based bistable ‘memory module’ that governs a cell fate decision. Nature. 2003;426:460–465. doi: 10.1038/nature02089. [DOI] [PubMed] [Google Scholar]


