Abstract
The p53 tumor suppressor is a multifaceted transcription factor that responds to a diverse array of stresses that include DNA damage and aberrant oncogene signaling. On activation, p53 prevents the emergence of cancer cells by initiating cell cycle arrest, senescence (terminal cell cycle arrest), or apoptosis. Although its role in assuring longevity by suppressing cancer is well established, recent studies obtained largely from genetically engineered mouse models suggest that p53 may regulate longevity and aging. In some contexts, it appears that altered p53 activity may enhance longevity, and in others, it appears to suppress longevity and accelerate aging phenotypes. Here, we discuss how genetically engineered mouse models have been used to explore antiproliferative functions of p53 in cancer suppression and how mouse models with altered aging phenotypes have shed light on how p53 might influence the aging process.
Studies of mice with mutations in the p53 pathway reveal that it regulates aging in different ways. This may reflect a role preventing the emergence of cancer stem cells.
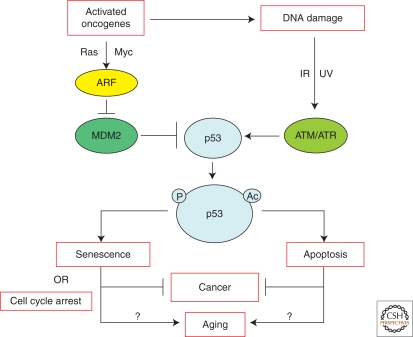
Aging is a fundamental biological process and a very complex phenomenon affecting virtually all systems and biological functions in different ways in different individuals of a species. It has been defined by Arking as “the time-independent series of cumulative, progressive, intrinsic, and deleterious functional and structural changes that usually begin to manifest themselves at reproductive maturity and eventually culminate in death” (Arking 1998). Many age-associated pathologies have been identified, cancer being one of the more prominent (Campisi 2000; Balducci and Beghe 2001; Balducci and Ershler 2005). In human populations, cancer incidence increases geometrically after the fifth decade of life. Many of these cancers arise primarily through accumulation of genetic lesions in genes that protect genomic stability or fidelity of cell division (Kinzler and Vogelstein 1996; Kinzler and Vogelstein 1997; Hanahan and Weinberg 2000). Such protective genes can be considered longevity assurance genes as they prevent early death through cancer. The subset of longevity assurance genes that prevent or retard cancers are often tumor suppressor genes. Tumor suppressor genes suppress cancers by multiple pathways and mechanisms, but serve to prevent the emergence or progression of a cancer cell (Lowe et al. 2004; Campisi 2005; Sherr 2004). The prototypical tumor suppressor and longevity assurance gene is p53, illustrated by studies showing that when both alleles of p53 are deleted in the mouse germline, the median lifespan is reduced to 4.5 months compared with 30 months for wild-type p53 mice of the same genetic background (Donehower 1996; Venkatachalam et al. 2001). The dramatic lifespan reduction in p53 null mice is solely a result of early tumors. p53 is a multifunctional transcriptional factor that can suppress cancer through both transcriptional and nontranscriptional mechanisms (Levine 1997; Vousden 2000; Moll et al. 2005; Green and Kroemer 2009). p53 has been consistently shown to be a stress response protein that invokes various antiproliferative programs in response to cellular damage and other stresses (Levine 1997; Ljungman 2000; Vousden and Lu 2002; Rodier et al. 2007) (Fig. 1). These responses include transient or terminal cell cycle arrest (senescence) or apoptosis, which prevent damaged cells from further division.
Figure 1.
p53 responds to DNA damage and activated oncogenes by distinct pathways to prevent genomic instability and cancer. Note that aberrant oncogene signaling may activate a DNA damage response through induction of replicative stress. DNA damage results in activation of p53 through ATM/ATR kinases phosphorylating p53, whereas aberrantly activated oncogenes induce ARF, which sequesters the E3 ubiquitinase MDM2, allowing p53 stabilization. Activated and stabilized p53 induces either apoptosis or transient or permanent cell cycle arrest (senescence). The activated p53 responses prevent the progression (senescence) or promote the elimination (apoptosis) of a nascent cancer cell. It has been hypothesized that these antiproliferative effects of p53 could promote aging through effects on stem and progenitor cell functionality.
Do these antiproliferative functions, that are critical for suppression of nascent cancer cells, have any implications for organismal aging or longevity? It is well established that mammalian cells incur various types of damage with age, and the damage itself or the cellular damage responses could both contribute to aging phenotypes (Garinis et al. 2008; Maslov and Vijg 2009). Many have hypothesized that tissue atrophy and functional decline are a result not only of increased cellular damage and genomic instability in mature somatic cells, but of failures in regenerative function in stem and progenitor cell compartments (Van Zant and Liang 2003; Chen 2004; Dumble et al. 2004; Pelicci 2004; Sharpless and DePinho 2004; Campisi 2005; Rando 2006; Rodier et al. 2007; Sharpless and DePinho 2007; Rossi et al. 2008; Ruzankina et al. 2008). Thus, one possibility is that declines in stem/progenitor cell self-renewal and differentiation are critical components of aging and that age-associated changes in p53 antiproliferative function could have effects on stem/progenitor function. In the last decade, evidence has gradually accumulated that p53 may directly impact on aging or aging-associated pathologies. The first clues were provided by genetically engineered mice with p53 germline alterations or mutations in pathways that impacted p53 signaling (Rodier et al. 2007; Serrano and Blasco 2007). Related studies in mouse models with mutations in the p53 family member p63 and other animal models (worms and flies) supported the idea that the p53 family members influence longevity and may be responsible for some age-associated pathologies (Bauer et al. 2005; Keyes et al. 2005; Bauer et al. 2007; Ventura et al. 2009). In addition, two large scale epidemiological studies in human populations have suggested that inherited polymorphisms in the p53 gene could affect not only relative cancer risk but overall longevity (Van Heemst et al. 2005; Orsted et al. 2007). In this article, representative studies that mechanistically link p53 not only to cancer suppression, but also to aging and longevity, are discussed. Heavy emphasis is placed on studies of genetically engineered mouse cancer models, in part because of the author's familiarity with such approaches and in part because so many key insights into the role of p53 in the cancer/aging interface have been obtained from these tools in the last two decades.
THE ANTICANCER FUNCTIONS OF p53
Upstream Regulation of p53: DNA Damage
The p53 protein acts as a stress responder to an array of both intrinsic and extrinsic insults (Ljungman 2000; Vousden 2002). Because the various components of the p53 signaling pathway are covered in great detail in other articles on this topic, only some of the salient features relevant to aging will be discussed here. These stress signals include many types of DNA damage, hypoxia, alterations in ribonucleoside triphosphate pools and ribosomal biogenesis, heat and cold shock, mitotic spindle poisons, improper protein folding in the cell, shortened telomeres, and aberrantly activated oncogenes (Ljungman 2000; Vousden 2002). The impact of damage caused by DNA strand breaks has been particularly well studied and involves the activation of key phosphoinositide 3-kinase-related kinase (PIKK) sensor kinase family members such as ataxia telangiectasia mutated (ATM), ATR, and DNA-PK (Abraham 2001; Shiloh 2003). These kinases in turn initiate a kinase cascade that results in phosphorylation of numerous downstream targets important in DNA repair and cell cycle checkpoint engagement. Elledge and colleagues have identified at least 700 potential targets of the ATM/ATR kinases (Matsuoka et al. 2007). One of the critical targets of the PIKKs is p53, which is phosphorylated directly at serines 15 and 37 (Canman et al. 1998; Tibbetts et al. 1999). In addition, ATR and ATM activate the kinases Chk1 and Chk2, which phosphorylate p53 at serine 20 (Hirao et al. 2000; Abraham 2001; Shiloh 2003). Such phosphorylation events are important for p53 functional activation, as defects in phosphorylation at these sites decrease p53 transcriptional functions (Appella and Anderson 2001; Sluss et al. 2004). Phosphorylation at serine 20 in particular has been shown to be important in preventing binding to p53 by the E3 ubiquitinase Mdm2 and facilitating p53 proteolytic degradation (Chehab et al. 1999). Under normal conditions, the p53 protein is kept at low concentrations and low activity levels by its major negative regulator, Mdm2, which confers a very short half life (6–40 min) on the p53 protein (Chene 2003; Kruse and Gu 2009). Thus, p53 phosphorylation results in p53 stabilization and facilitates its nuclear localization, where it acts as a potent transcriptional activator of a number of genes involved in an array of cellular functions, including DNA repair, apoptosis, and cell cycle regulation (Levine 1997; Vousden 2002; Lavin and Gueven 2006). It should be noted that p53 is phosphorylated at many sites and by many other kinases, and that it is posttranslationally modified through acetylation, methylation, neddylation, and ubiquitination (Appella and Anderson 2001; Lavin and Gueven 2006). Moreover, its stability is regulated by other molecules, such as Mdmx, Pirh2, Cop-1, HAUSP, ARF-BP1, and Pin1 (Brooks and Gu 2006; Kruse and Gu 2009). However, those modifications that impact the Mdm2-p53 interaction appear to play the single largest role in p53 regulation and stability.
Recent studies indicate that the DNA damage response may be directly involved in preventing progression of precancerous cells. The Halazonetis and Bartek laboratories have shown that some early precancerous lesions display activation of the DNA damage response pathway (Bartkova et al. 2005; Gorgoulis et al. 2005; Bartkova et al. 2006; Halazonetis et al. 2008). High levels of phosphorylation of ATM, Chk2, and p53, indicating strong activation of the DNA damage response, were observed in precancerous lesions, but not normal tissues or fully malignant tumors (Bartkova et al. 2005; Gorgoulis et al. 2005). Moreover, the DNA damage activation is closely accompanied by elevated senescence or apoptotic responses, depending on the precancerous cell type (Bartkova et al. 2006; Halazonetis et al. 2008). These strong antiproliferative responses act as a barrier to prevent progression of the precancerous cell. The initiating events for the DNA damage response are likely to be a result of the aberrant activation of a cellular oncogene, which is believed to deregulate the cell cycle so that DNA replication is compromised, resulting in a state of replication stress that is accompanied by increased fork stalling and DNA double-strand breaks (Halazonetis et al. 2008). It is believed that the increased DNA double-strand breaks directly activate the DNA damage response through ATM, ATR, and other damage sensors. As indicated previously, tumors that progress beyond the precancerous stages into fully malignant tumors lose the protective DNA damage response. In many cases, this appears to be a result of p53 mutation or deletion. Loss of activated wild-type p53, with its strong induction of apoptosis or senescence, would likely be a critical event in malignant progression of the precancerous lesion (Halazonetis et al. 2008). The Bartek-Halazonetis oncogene-induced DNA damage model for cancer development and progression is an appealing one that may explain many aspects of early cellular anticancer responses. In particular, it nicely highlights how DNA damage activation of p53 and its attendant antiproliferative functions may play a central role in the barrier to tumor progression.
Upstream Regulation of p53: Oncogenic Stress
Oncogenic activation of the p53 antiproliferative response is also mediated by the ARF protein encoded by the CDKN2A locus, which also encodes the p16INK4a tumor suppressor protein through an alternative reading frame (Sherr 2001; Kim and Sharpless 2006). ARF is generally not expressed at detectable levels in unstressed cells, but overexpression or mutational activation of prototypical oncogenes such as Myc, Ras, and E2F up-regulates ARF protein expression (Kim and Sharpless 2006). In turn, ARF antagonizes Mdm2 E3 ubiquitin ligase activity and thus facilitates the stabilization and activation of p53. Once activated, p53 will then perform its transcriptional functions and fulfill its antiproliferative roles. ARF generally triggers cell-cycle arrest, but oncogene-induced signals conveyed through collateral pathways can alter the response from growth arrest to apoptosis (Sherr 2006). ARF expression is not believed to be initiated by DNA damage, but rather by aberrant oncogene signaling. In experiments performed in my laboratory, mice that completely lack Arf and p53 genes have a tumor incidence similar to that of p53-null mice, suggesting that tumor suppression by ARF proceeds completely through p53 (Moore et al. 2003). In addition, ARF−/− mice do not develop tumors as quickly as p53−/− mice, indicating that p53 is not solely dependent on ARF activity for its tumor suppressor functions (Moore et al. 2003). Presumably, the DNA damage induction of p53 through ATM/ATR and other PIKKs accounts for some of the ARF-independent p53 tumor suppressor function. Thus, p53 clearly can integrate multiple upstream signals to enhance its suppression of oncogenically activated precancerous cells. Both DNA damage and ARF-mediated pathways are likely to play a role in mediating p53 effects on mammalian longevity.
Finally, another activator of p53 function resulting from oncogenic stress may be the Seladin-1 (DHCR24) protein. Previously known as a neuroprotective gene (Peri and Serio 2008), Seladin-1 was isolated in a screen for inducers of Ras-induced senescence in rodent fibroblasts (Wu et al. 2004). Following Ras-induced oncogenic or oxidative stress, Seladin-1 binds to the p53 amino terminus and displaces Mdm2 from p53, resulting in p53 stabilization. Knockdown of Seladin-1 results in abrogation of Ras-induced senescence in rodent and human fibroblasts, resulting in Ras-mediated transformation (Wu et al. 2004). This transducer of oncogenic stress is interesting from an aging perspective because it is specifically activated by reactive oxygen species (and not other types of mutagens), which some theories of aging postulate to be a critical mediator of intrinsic aging (Sohal and Weindruch 1996).
Anticancer Outcomes Regulated by p53: Apoptosis
If the primary role of p53 is to suppress cancer formation, how does it do it? What are the anticancer or antiproliferative functions of p53? Once p53 becomes activated by DNA damage, it may induce cell cycle arrest, accompanied by DNA repair. If the DNA damage is transient or limited, it is generally repaired and p53 then returns to prestress levels of activity and the cell can continue normal division. In other cases, where DNA damage is high or is continuous (as in organisms with inherited deficiencies in DNA repair or DNA damage responses), p53 activation may be extended and induce more permanent cellular outcomes, such as senescence or apoptosis. Senescence is generally considered a permanent form of G1 cell cycle arrest. In some contexts, rather than cell cycle arrest or senescence, p53 may induce apoptosis, a form of genetically programmed cell death (Vousden and Lu 2002; Rodier et al. 2007). Which outcome is chosen may be dependent on a variety of factors, including environmental growth factor levels, p53 levels, the intensity or nature of the mutagenic or oncogenic inducer, or the cell type (Vousden and Lu 2002; Aylon and Oren 2007). Regardless of the path taken, however, the emergence of a proliferating precancerous or cancerous cell has been prevented. That either senescence or apoptosis are key barriers to cancer in vivo has become increasingly evident from studies on human tissues or in genetically altered mice (Rodier et al. 2007; Serrano and Blasco 2007). Histopathological examination of tissues with preneoplastic lesions often shows an increase in fractions of apoptotic or senescent cells accompanying an elevated proliferation rate (Bartkova et al. 2005; Gorgoulis et al. 2005; Bartkova et al. 2006; Halazonetis et al. 2008). In mice expressing an activated oncogene in specific tissues, fractions of apoptotic or senescent cells in the affected tissues are dramatically increased (Collado et al. 2005; Calcagno et al. 2008; Murphy et al. 2008; Efeyan et al. 2009).
The induction of apoptosis by p53 can occur by both transcriptional and nontranscriptional mechanisms. Activated p53 induces increased transcription of a variety of proapoptotic factors such as bax, puma, perp, noxa, and apaf-1, which lead to the intrinsic mitochondrial release of cytochrome c, formation of the the apoptosome, caspase activation, and the resulting execution of cell death functions (Vousden 2000; Vousden and Lu 2002). In addition to its transcriptional activation activities, p53 acts as a transcriptional repressor to inhibit expression of antiapoptotic genes such as Bcl-2, PTEN, IGF-BP3, and survivin (Vousden and Lu 2002). Finally, p53 facilitates apoptosis directly and by nontranscriptional mechanisms through interaction with the multidomain members of the Bcl-2 family to induce mitochondrial outer membrane permeabilization and release of cytochrome c (Moll et al. 2005; Green and Kroemer 2009). Robust p53-mediated apoptosis depends on both transcriptional and nontranscriptional p53 functions.
The importance of p53-mediated apoptosis as a potent tumor suppressor mechanism has been indicated by studies of a number of tumor susceptible mouse models with activated oncogenes. In the first of such studies, mouse models with inactivated Rb in the eye and brain resulted in hyperproliferative lesions or weak tumors with high levels of apoptosis (Howes et al. 1994; Pan and Griep 1994; Symonds et al. 1994). When p53 was removed through viral oncoprotein inactivation or crossing into a p53 null background, tumor proliferation was greatly enhanced and apoptotic cells virtually eliminated. Similarly, lymphoma prone Eμ-myc transgenic mice develop lymphomas with a median incidence of 5–6 months (Adams et al. 1985). Examination of lymphoma tissue sections revealed high levels of apoptosis, but if null alleles for p53 or p19ARF, the upstream regulator of p53, were introduced into the Eμ myc line, tumor incidence was greatly accelerated and evidence of apoptosis in the tumors was greatly diminished (Eischen et al. 1999; Schmitt et al. 1999). Thus, the induction of apoptosis through p53 signaling pathway clearly delays tumor progression in multiple contexts, at least in part through apoptotic cell death.
Recently, development of conditionally activated wild-type p53 models was also used by the Evan laboratory to show that restoration of p53 function can cause dramatic tumor regression (Christophorou et al. 2005; Martins et al. 2006). Lymphomas that arise in the absence of p53 maintain an intact p53 signaling pathway that can be completely restored when functional wild-type p53 is reintroduced (Martins et al. 2006). The Trp53ER(TAM) inducible p53 model was used to activate p53 in the Eμ-myc lymphoma model by tamoxifen injection to regress naturally arising lymphomas (Christophorou et al. 2005). The lymphoma regression was accompanied by massive tumor cell apoptosis (Martins et al. 2006). Thus, the in vivo evidence for the role of p53-dependent apoptosis raises the possibility that restoration of p53 function may have dramatic therapeutic effects, although one caveat is that the model showed occasional tumor relapses after p53 induction because of loss of ARF (Cdkn2aARF) expression or the inducible Trp53 allele itself (Martins et al. 2006).
In a related study with the Trp53ER(TAM) inducible p53 model, ionizing radiation (IR) treatment of these mice was shown to induce early lymphomas in the absence of tamoxifen (no p53). Interestingly, when mice were briefly treated with tamoxifen at the same time as IR treatment, high levels of apoptosis were observed in the irradiated tissues, a result expected when wild-type p53 was restored (Christophorou et al. 2006). Surprisingly, no delay in subsequent IR-induced lymphomagenesis was observed when p53 was briefly activated concurrently with IR, despite the abundant p53-induced apoptosis. However, when p53 was briefly restored eight days post IR treatment, there was a significant delay in lymphoma incidence (Christophorou et al. 2006). These unexpected results indicated that the robust p53-mediated apoptotic response immediately after irradiation was actually irrelevant for tumor suppression. Moreover, the lymphoma resistance accompanying delayed p53 activation was completely dependent on p19ARF. Thus, at least in this context of DNA damage, the primary mode of p53 tumor suppression is not through immediate apoptotic elimination of cells with damaged DNA, but rather through 19ARF-mediated suppression of rare oncogenically activated cell clones.
Recent experiments by my laboratory tested the role of the p53 tumor suppressor response to DNA damage in a different manner than the Evan laboratory. We crossed floxed p53 mice to mice with a Cre recombinase gene fused to a mutant tamoxifen-responsive form of the estrogen receptor (CreERT2) knocked into the ubiquitously expressing Rosa26 locus (Hinkal et al. 2009b). With these bi-allelic mice, we could somatically delete p53 at any time in virtually all tissues by tamoxifen injection. So, in contrast to the experiment described previously in which p53 was activated during or before IR, p53 was deleted either before, during, or up to 4 weeks after radiation. Surprisingly, we found that regardless of when p53 was deleted (2 weeks before, at the same time, 2 weeks after, or 4 weeks after IR), lymphomas developed with the same median latency (Hinkal et al. 2009b). These results appeared to confirm the Evan result that the immediate p53-mediated apoptotic response to DNA damage does not protect against subsequent tumor development. However, in contrast to the earlier result showing that induction of wild-type p53 8–14 days after IR delayed tumors, our data indicated that even maintenance of wild-type p53 up to 4 weeks post-IR provided little or no protective effect from subsequent lymphomas. Thus, a prolonged and sustained wild-type p53 response is critical for protection from the oncogenic effects of DNA damage.
Anticancer Outcomes Regulated by p53: Senescence
The alternative mechanism of p53 anticancer function is through induction of a permanent cell cycle arrest called senescence. The senescence response causes a number of alterations in cellular phenotypes. These alterations include a permanent proliferative arrest while maintaining stable metabolic functions, a resistance to apoptosis, and altered gene expression patterns (Campisi 2001; Rodier et al. 2007). Senescence-associated markers such as senescence-associated β-galactosidase, p16, and senescence-associated heterochromatin foci (SAHFs), and secretion of a number of cytokines are often observed in these cells.
One of the primary mediators of this permanent arrest is the cyclin-dependent kinase (CDK) inhibitor p21, also a key inducer of the transient cell cycle arrest (el-Deiry et al. 1993; Harper et al. 1993; Noda et al. 1994). Activated p53 transcriptionally up-regulates p21, which then specifically binds to and inhibits the G1 cyclin-CDK complexes, resulting in block in entry of the cell into S phase. This CDK suppression impacts on the retinoblastoma (Rb) gene pathway by maintaining Rb protein hypophosphorylation, which prevents E2F family members from initiating an S phase transcription program (Classon and Harlow 2002; Campisi 2005). The critical importance of p53 and p21 in senescence initiation is illustrated by experiments in which their levels are reduced in senescent cells. In this situation, the senescence response can actually be reversed, and cells can re-enter the cell cycle (Brown et al. 1997; Wei et al. 2001; Rodier et al. 2007).
Oncogenic stress has been shown to be an inducer of p53-mediated senescence from in vitro studies (Serrano et al. 1997; Lin et al. 1998; Zhu et al. 1998). Overexpression of Ras in primary human or rodent cells results in senescence accompanied by p53 and p16INK4a activation (Serrano et al. 1997). Inactivation of p53 can reverse the senescence effect. In vivo, the function of p53-induced senescence as a potent anticancer barrier has been implicated in the correlative studies of Halazonetis and Bartek, who showed that some precancerous lesions show not only indicators of an enhanced DNA damage response, but also evidence of increased senescence markers (Bartkova et al. 2005; Gorgoulis et al. 2005; Bartkova et al. 2006). In fully malignant tumors, many of the same DNA damage and senescence markers that were activated in premalignant lesions were greatly reduced, indicating that circumvention of the senescence and DNA damage responses were a critical component of tumor progression. The major culprit in this progression process was believed to be loss or mutation of p53, although other genetic lesions in the DNA damage response pathway might also facilitate progression (Halazonetis et al. 2008). In studies of a similar nature in mice, in a model in which oncogenic Ras could be selectively activated in the lung and pancreas, early stage premalignant lung adenomas and pancreatic intraductal neoplasias showed high levels of several senescence markers, whereas malignant lung adenocarcinoma and pancreatic ductal adenocarcinomas displayed a comparative absence of such markers (Collado et al. 2005).
Other genetically engineered mouse cancer models have provided additional mechanistic insights into this issue. For example, Schmitt et al. (2002) showed that senescence is a chemotherapy-drug-responsive program in vivo that improves treatment outcome when apoptosis is blocked by Bcl-2 overexpression (Schmitt et al. 2002). Tumors deficient for p53 showed impaired treatment responses and reduced senescence. Subsequent studies by the Jacks and Lowe laboratories with inducible wild-type p53 mouse models showed that restoration of p53 function can cause dramatic tumor regression in part through induction of senescence (Ventura et al. 2007; Xue et al. 2007). The Jacks laboratory showed that induction of wild-type p53 led to regression of naturally arising lymphomas and sarcomas (Ventura et al. 2007). The Lowe laboratory used a different shRNA-based approach to activate wild-type p53 in a mouse liver carcinoma model (Xue et al. 2007). Both sarcomas and liver tumors arising in these models tended to show high levels of cellular senescence after wild-type p53 activation. In the liver carcinoma model, p53-induced senescent cells produced inflammatory cytokines and an innate immune response that resulted in tumor cell clearance (Xue et al. 2007). In these models, senescence did not act as a block to progression of a premalignant lesion, but rather prevented further division of already malignant cells. Thus, restoration of p53 activity and p53-induced senescence may be a viable therapeutic strategy in suppressing both early and late stages of tumor progression.
The studies previously mentioned suggest that the induction of senescence, either through p53 or through other mechanisms, may serve only as an antioncogenic mechanism. However, as the Campisi laboratory has shown, senescent cells may have pro-oncogenic effects under some circumstances. They showed that senescent cells may affect their immediate microenvironment through altered secretory processes (Krtolica et al. 2001; Campisi 2005). Senescent human fibroblasts secrete epithelial growth factors, inflammatory cytokines, and a number of matrix metalloproteinases (Krtolica et al. 2001). In transplant experiments, it was shown that senescent fibroblasts could stimulate premalignant and malignant epithelial cells to proliferate in culture and form tumors in mice. Moreover, senescent cells could disrupt normal epithelial cell functions and differentiation (Parrinello et al. 2005). These studies are highly intriguing and provide one potential mechanism for the dramatic increases in cancers observed in aged individuals.
Attenuation of p53 Anticancer Function
Cancer rates increase dramatically with age in both humans and mice (Balducci and Beghe 2001; Campisi 2003). Generally, this increase has been attributed in part to accumulation of mutations in cancer-associated genes that occasionally result in an emergent cancer cell (Kinzler and Vogelstein 1996; Kinzler and Vogelstein 1997; Hanahan and Weinberg 2000). Certainly, epigenetic, microenvironment, and systemic alterations may also contribute to tumor initiation, but genetic and epigenetic alterations have been given primacy, because virtually all cancers have multiple mutations and altered gene expression that are often consistent in each cell of the tumor. Thus, the fact that half of all human cancers display loss-of-function mutations or deletions in the p53 gene is a strong argument for the importance of p53 anticancer function (Soussi and Lozano 2005). But is mutation the only way in which p53 function can be circumvented in the evolution of a tumor cell? We now know that even when the p53 gene is not mutated, p53 signaling can be rendered dysfunctional or minimally functional by alterations in upstream regulatory genes such as Mdm2, Mdm4, ARF, and Wip1 (Momand et al. 1998; Marine et al. 2007; Lu et al. 2008; Matheu et al. 2008). For example, in a comprehensive genomics survey of 91 human glioblastomas, it was shown that although only 35% of the tumors had inactivating mutations in the p53 gene, amplification of Mdm2 or Mdm4 genes (potent suppressors of p53) or deletion of the Cdkn2aARF gene was observed so frequently in the remaining tumors that 87% of all glioblastomas had some key alteration in the p53 signaling pathway (The Cancer Genome Atlas Research Network 2008).
A recent study by the Levine laboratory has revealed yet another potential mechanism for circumvention of p53 anticancer activity, through an age-associated decline in overall function. Feng et al. found that p53 activity was significantly decreased in old tissues compared with young tissues (Feng et al. 2007). Treatment of young and old mice with ionizing radiation, followed by harvesting of the tissues after six hours and examination of the tissues for p53 protein and various markers of p53 function was performed. The p53 protein levels were increased as expected in irradiated young mouse spleens. However, spleens from old mice showed significantly less p53 protein levels than their young mouse counterparts. To assess p53 function, two assays were described. First, because p53 is a transcription factor that activates known target genes, RNA levels of six such p53 target genes were examined before and after irradiation. Tissues from old mice showed a less effective p53 target induction than that seen in tissues from young mice. In the second assay, apoptotic cell numbers in irradiated young and old spleens were measured, and old spleens showed less efficient apoptosis than young spleens, another indicator of reduced p53 function with age (Feng et al. 2007).
Why wild-type p53 functions decline with age in mice is unclear. However, one clue arose from the authors' discovery that stress-activated ATM (ataxia telangiectasia mutated) protein levels and activation markers decreased with age along with decreasing p53 activity (Feng et al. 2007). This is important because ATM is a pivotal DNA damage sensor kinase that phosphorylates p53 and facilitates p53 activation (Abraham 2001; Shiloh 2003). The age-associated declines in ATM and p53 signaling functions in mice provide a potential mechanism for the age-associated increases in cancer rates. In those tumor types where p53 mutations and signaling inactivation is rare, perhaps a decline in p53 functionality with age could nevertheless play a role in cancer incidence. An obvious question is whether the age-associated decline in p53 function is specific to mice or can it be extrapolated to humans.
In a different experimental approach to assess the role of p53 tumor suppressor function with aging, my laboratory globally deleted p53 in the mouse at different ages to determine age-associated effects on tumor progression in this context (Hinkal et al. 2009b). Floxed p53 mice containing a tamoxifen-inducible Cre recombinase gene enabled tamoxifen-initiated somatic deletion of both p53 alleles in most tissues at 3, 6, and 12 months of age. The resulting tumors in these mice were virtually identical in type to those observed in germline p53−/− mice but arose later depending on the age of somatic p53 deletion. However, somatic global p53 deletion resulted in age-specific differences in post-deletion tumor latencies. Deletion of p53 at 12 months led to a significantly more rapid tumor incidence than at 3 months, which is consistent with a model in which tissues accumulate oncogenically activated cells with age and these are held in check by wild-type p53. Although these experiments did not directly address whether p53 function declines with age, they did emphasize that maintenance of intact p53 function does become progressively more important in preventing cancer as the organism ages.
THE REGULATION OF AGING AND LONGEVITY BY p53 IN GENETICALLY ENGINEERED MICE
Prematurely Aging Mice with Activated p53
As indicated previously, p53 can suppress cancer formation by different mechanisms. In addition, p53 has a profound impact on organismal longevity. Mice missing p53 have a median lifespan of 4.5 months and maximal lifespan of 10 months compared with 27 months and 38 months, respectively, for normal mice of the same background (Venkatachalam et al. 2001). Of course, longevity assurance on the part of p53 is primarily a result of its prevention of early cancers. Recent evidence suggests that p53 may regulate aging and longevity aside from its specific anticancer functions. The first clue was provided by reports describing telomerase-deficient (Terc−/−) mice and Ku80-deficient (Ku80−/−) mice. Terc−/− mice intercrossed for four to six generations had shortened telomeres that were associated with reduced longevity and accelerated aging phenotypes (Rudolph et al. 1999). Fibroblasts and testicular cells derived from these mice contained significantly elevated levels of activated p53. Crossing of these mice into a p53 null background resulted in a loss of the aging phenotypes associated with telomerase deficiency (Chin et al. 1999). Ku80−/− mice, which are defective for DNA double-strand break repair show a number of early aging phenotypes including skin atrophy, osteopenia, hepatocellular degeneration, and reduced longevity (Vogel et al. 1999). Moreover, fibroblasts derived from these mice underwent premature senescence in culture. When Ku80−/− fibroblasts were made deficient for p53, the premature senescence phenotypes were rescued, suggesting that p53 might play a role in the organismal aging phenotypes as well (Lim et al. 2000).
Other mutant mouse models that display early senescent or progeroid phenotypes with a p53 connection include the Brca1Δ11/Δ11 hypomorph mutant mice and the Zmpste24−/− mice. The Brca1Δ11/Δ11 mice die during embryogenesis but Brca1Δ11/Δ11 p53+/− females survive and develop mammary tumors, whereas the males showed evidence of accelerated aging phenotypes (Cao et al. 2003). Because Brca1 is an important mediator of DNA break repair and recombination, the additional damage from Brca1 deficiency may elevate p53 activity and this could lead to some of the aging phenotypes. The Zmpste24−/− mouse was characterized as a mouse model of human progeria, a syndrome that is accompanied by a number of early accelerated aging pathologies, including cardiovascular defects and death by the teenage years (Martin 2005). Zmpste24 is a gene responsible for processing of lamin A (which is mutated in the germlines of human progeria patients). The Zmpste24−/− mice had a very short life span and phenocopied some of the human progeroid syndrome symptoms (Pendas et al. 2002). Interestingly, the Zmpste24−/− mice had a robust up-regulation of numerous p53 target genes, indicating an enhanced p53 response (Varela et al. 2005). In addition, the accelerated aging phenotypes and reduced life span of the Zmpste24 null mice were partially rescued by the absence of p53 (Varela et al. 2005), indicating that activated p53 was likely responsible for some of the progeroid phenotypes.
p53 Mutant Mice with Altered Aging and Longevity Phenotypes
A potential direct linkage of p53 with organismal aging was reported in 2002 by my laboratory (Tyner et al. 2002). We had serendipitously generated a mutant p53 allele that could express only a truncated portion of p53 derived from its carboxyl terminus. Instead of the targeted allele behaving as a loss-of-function p53 allele, it actually behaved as a hypermorphic p53 allele. A caveat for this model is that it is haploinsufficient for 24 genes upstream of p53, so haploinsufficiency effects cannot be ruled out as contributors to the resulting phenotypes. This model, called the p53+/m mouse, is resistant to spontaneous cancers, displaying a 6% lifetime incidence versus 45% for p53+/+ mice of similar genetic background (Tyner et al. 2002). However, the p53+/m mice had a 23% shortened longevity compared with wild-type littermates and displayed a number of accelerated aging phenotypes, including organ atrophy, osteoporosis, skin atrophy, and reduced tolerance to stresses (Tyner et al. 2002). The early aging phenotypes were dependent on the presence of wild-type p53, as the presence of the “m” truncated p53 allele along with a p53 null allele developed tumors at the same rates as p53−/− mice. We hypothesized that the truncated “m” form of p53 interacted with wild-type p53 to increase wild-type p53 activity (Tyner et al. 2002). In fact, subsequent cell culture experiments showed that coexpression of p53 m protein with wild-type p53 drove p53 into the nucleus, stabilized it, and activated p53 transactivation functions (Moore et al. 2007).
A second p53 hypermorphic model has been reported by Scrable and colleagues (Maier et al. 2004) and expresses a 44 kD truncated naturally occurring isoform of p53 (Maier et al. 2004). Like the p53+/m mice, the p44 TG mouse displayed a dramatically shortened lifespan and a number of early aging phenotypes. The p44 TG showed a reduced cancer incidence, indicating an augmentation of wild-type p53 tumor suppressor activity. The mechanism of these early aging phenotypes was ascribed by the authors, at least in part, to hyperactivation of the IGF-1 signaling axis (Maier et al. 2004).
The descriptions of the previous two p53 mutant mice suggest the simple relationship that more p53 activity equals more cancer resistance and reduced longevity. However, subsequent reports by the Serrano and Perry laboratories indicate that the relationship of p53 and longevity was a bit more complex (Garcia-Cao et al. 2002; Mendrysa et al. 2006; Matheu et al. 2007). Serrano and colleagues addressed the overexpression issue in a different way by engineering mice with only one or two extra copies of genomic p53 along with flanking upstream and downstream regulatory sequences (Garcia-Cao et al. 2002). These “super p53” mice showed an enhanced p53 response to DNA damage and were more resistant to spontaneous and carcinogen-induced tumors than normal mice. Importantly, the super p53 mice had longevities similar to that of normal mice, and showed no obvious early aging phenotypes (Garcia-Cao et al. 2002). Similarly, another mouse model, generated by Perry and collaborators, was hypomorphic for Mdm2 activity (Mendrysa et al. 2006). Because Mdm2 facilitates p53 degradation, it was not surprising that the Mdm2 hypomorphs had an enhanced p53 response to DNA damage and were tumor resistant. Like the super p53 mice, longevity and aging phenotypes appeared to be normal in the Mdm2 hypomorphs (Mendrysa et al. 2006).
Although all four of the p53 hypermorph models described previously showed significantly enhanced cancer resistance, the longevity differences between the p53+/m and p44 TG models and the super p53 and hypomorphic Mdm2 models raise obvious questions. My laboratory and the Serrano laboratory have hypothesized that the super p53 mice maintain a relatively normal regulation of p53, so that increases in p53 activity only appear in times of stress (Klatt and Serrano 2003; Dumble et al. 2004). In contrast, the p53+/m and p44 TG mice may have somewhat abnormal p53 regulation in that there may be low level chronic activation of p53 that results in the early aging phenotypes. Thus, the p53+/m mice may phenocopy some of the other accelerated aging models, such as the Terc−/−, Ku80−/−, Brca1−/−, and Zmpste24−/− mice that are likely to elicit continuous low-level p53 activation caused by shortened telomeres, unresolved DNA damage, or cellular stresses. Clearly, further experiments will be required to understand the mechanisms by which p53 influences longevity and aging phenotypes.
After the original description of the super p53 mice, the Serrano laboratory has further improved on them by crossing them to other lines. Recently, they described super p53/p19ARF mice (Matheu et al. 2007). These mice, in addition to an additional p53 allele, have an additional p19ARF allele, which can augment further p53 activation in response to oncogenic stress. Amazingly, not only were these mice virtually cancer-free, but they actually showed decreased levels of age-associated damage and extended median longevities relative to normal mice and super p53 mice (Matheu et al. 2007). The authors hypothesized that their results revealed a previously unknown antiaging mechanism and provided a rationale for the coevolution of cancer resistance and longevity. This is an exciting result, but it will be interesting to see if stressors that activate the DNA damage response pathways of p53 rather than the oncogenic stress pathways that activate ARF will show the same longevity protection effects.
The super p53 mice have been further augmented by the generation of telomerase reverse transcriptase (TERT) overexpressing alleles in the background of overexpressed p53, p16INK4a, and p19ARF (Tomas-Loba et al. 2008). These mice showed improved epithelial cell fitness, delayed aging phenotypes, and an extended median lifespan. Because inbred mouse telomeres are very long and age-associated telomere shortening is unlikely to have any effects, this result indicates that telomerase my provide beneficial antiaging functions independent from its primary telomere maintenance role. The Serrano laboratory has also generated super ARF transgenic mice with one or two complete copies of the ARF gene and found that these mice, although showing male fertility defects, had reduced cancer and modestly increased median longevities (Matheu et al. 2009). Thus, most of the longevity extension effects in the super p53/ARF mice were conveyed by ARF, which the authors hypothesize facilitates longevity by preventing unnecessary proliferation.
Concluding Remarks: Insights and Hypotheses from Mouse Model Studies
The mouse models described previously, while showing divergent results with respect to longevity and aging, have nevertheless provided some useful insights into how p53 might regulate the aging process. The role of p53 in assuring longevity through prevention of cancer is well established. But how does it specifically affect aging and longevity aside from cancer prevention? One likely hypothesis is that p53 has evolved to protect organisms that maintain renewable tissues (stem and progenitor cells) during the adult reproductive phase. As cells are lost or become dysfunctional, they must be replaced in a manner that minimizes genetic changes that are associated with DNA replication and mitosis. Mutations in stem cells can be propagated to numerous progeny and have active cell division programs that can be readily subverted and deregulated to form cancer stem cells. The importance of cancer stem cells and their derivation from normal stem cells is increasingly recognized (Reya et al. 2001; Pardal et al. 2003). p53 family members are likely to play a major role in preventing the emergence of cancer stem cells. Indeed, reductions in p53 dosage have been shown to enhance stem cell division as well as other functional attributes (Palacios et al. 1996; Shounan et al. 1996; Hirabayashi et al. 2002; TeKippe et al. 2003; Dumble et al. 2007). Most recently, two studies have shown that p53 deficiency in hematopoietic multipotent progenitor cells and neural stem/progenitor cells contribute to enhanced proliferative capacity of these cells (Akala et al. 2008; Zheng et al. 2008). We have shown that two stem cell compartments in the accelerated aging p53+/m mice show reduced self-renewal and differentiation potential compared with their p53+/+ counterparts (Dumble et al. 2007; Gatza et al. 2008). In addition, neural progenitor cells from the accelerated aging p44 TG mice generated by the Scrable lab display defects in number and regenerative potential (Medrano et al. 2009). Finally, the finding by the Levine laboratory that p53 activity declines in older mice suggests that the impact of this decreased function could remove an important barrier to the emergence of cancer stem cells (Feng et al. 2007).
The hypothesized ability of p53 to inhibit the emergence of cancer stem cells has implications for a potential role in aging. It has been proposed that aging phenotypes may result in part from functional depletion of the stem and progenitor cell compartments because of age-associated reductions in self-renewal, proliferation, and differentiation (Van Zant and Liang 2003; Chen 2004; Dumble et al. 2004; Pelicci 2004; Sharpless and DePinho 2004; Campisi 2005; Rando 2006; Rodier et al. 2007; Sharpless and DePinho 2007; Rossi et al. 2008; Ruzankina et al. 2008). Recent studies showing increases of the senescence effector p16INK4A protein levels and activity correlating with reduced function in aged stem and progenitor cells support this idea (Janzen et al. 2006; Krishnamurthy et al. 2006; Molofsky et al. 2006). The antiproliferative responses of p53 may also play a role in the decline of stem cell functionality with age (Dumble et al. 2007). The enhanced p53 activity observed in some mouse accelerated aging models may affect the ability of stem cells to maintain organ homeostasis, resulting in organ atrophy and reduced overall functionality. On the other hand, the improved aging parameters in the super p53/ARF mice suggest that the possibility that stem cell functionality is somehow maintained at more optimal levels in the presence of a more robust ARF-p53 pathway (Matheu et al. 2007). Perhaps clearance of dysfunctional or senescent stem cells is more efficient in the super p53/ARF mice than the p53+/m mice where stem cells may be induced to become senescent and ineffectively removed. Consistent with this scenario, we have recently shown that p53+/m mice are defective for p53-mediated apoptosis and display an enhanced age-associated accumulation of senescent cells compared with wild-type mice (Hinkal et al. 2009a).
To conclude, p53 is a potent tumor suppressor and longevity assurance gene by virtue of its induction of cell cycle arrest, senescence, or apoptosis in response to DNA damage or oncogenic stress. Such stress response functions, although largely beneficial early in life by eliminating early cancers, may prove to be of mixed benefit later in life when the antiproliferative functions of p53 may impinge on stem and progenitor cell behavior. However, there are competing narratives regarding the role of p53 in aged organisms. The results from the two p53 mutant models with accelerated aging suggest that in aged animals, overactive p53 inhibits stem and progenitor cell function, whereas if properly regulated and boosted by increased ARF expression, it may have beneficial effects on stem cells later in life. And how does the observation that p53 activity may decline with age fit into the stem cell/aging picture, aside from a likely enhancement of cancer rates? Obviously, it will take numerous additional studies to fully unravel the complexities by which p53 influences intrinsic aging processes.
ACKNOWLEDGMENTS
I thank the Ellison Medical Foundation for support during the writing of this manuscript.
Footnotes
Editors: Arnold J. Levine and David Lane
Additional Perspectives on The p53 Family available at www.cshperspectives.org
REFERENCES
- The Cancer Genome Atlas Research Network 2008. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455:1061–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham RT 2001. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev 15:2177–2196 [DOI] [PubMed] [Google Scholar]
- Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, Palmiter RD, Brinster RL 1985. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature 318:533–538 [DOI] [PubMed] [Google Scholar]
- Akala OO, Park IK, Qian D, Pihalja M, Becker MW, Clarke MF 2008. Long-term haematopoietic reconstitution by Trp53−/−p16Ink4a−/−p19Arf−/− multipotent progenitors. Nature 453:228–232 [DOI] [PubMed] [Google Scholar]
- Appella E, Anderson CW 2001. Post-translational modifications and activation of p53 by genotoxic stresses. Eur J Biochem 268:2764–2772 [DOI] [PubMed] [Google Scholar]
- Arking R 1998. The Biology of Aging. Sinauer Associates, Inc; 570 p. [Google Scholar]
- Aylon Y, Oren M 2007. Living with p53, dying of p53. Cell 130:597–600 [DOI] [PubMed] [Google Scholar]
- Balducci L, Beghe C 2001. Cancer and age in the USA. Crit Rev Oncol Hematol 37:137–145 [DOI] [PubMed] [Google Scholar]
- Balducci L, Ershler WB 2005. Cancer and ageing: A nexus at several levels. Nat Rev Cancer 5:655–662 [DOI] [PubMed] [Google Scholar]
- Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, et al. 2005. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434:864–870 [DOI] [PubMed] [Google Scholar]
- Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, et al. 2006. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature 444:633–637 [DOI] [PubMed] [Google Scholar]
- Bauer JH, Chang C, Morris SN, Hozier S, Andersen S, Waitzman JS, Helfand SL 2007. Expression of dominant-negative Dmp53 in the adult fly brain inhibits insulin signaling. Proc Natl Acad Sci 104:13355–13360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer JH, Poon PC, Glatt-Deeley H, Abrams JM, Helfand SL 2005. Neuronal expression of p53 dominant-negative proteins in adult Drosophila melanogaster extends life span. Curr Biol 15:2063–2068 [DOI] [PubMed] [Google Scholar]
- Brooks CL, Gu W 2006. p53 ubiquitination: Mdm2 and beyond. Mol Cell 21:307–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JP, Wei W, Sedivy JM 1997. Bypass of senescence after disruption of p21CIP1/WAF1 gene in normal diploid human fibroblasts. Science 277:831–834 [DOI] [PubMed] [Google Scholar]
- Calcagno SR, Li S, Colon M, Kreinest PA, Thompson EA, Fields AP, Murray NR 2008. Oncogenic K-ras promotes early carcinogenesis in the mouse proximal colon. Int J Cancer 122:2462–2470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J 2000. Cancer, aging and cellular senescence. In Vivo 14:183–188 [PubMed] [Google Scholar]
- Campisi J 2001. Cellular senescence as a tumor-suppressor mechanism. Trends Cell Biol 11:S27–31 [DOI] [PubMed] [Google Scholar]
- Campisi J 2003. Cancer and ageing: Rival demons? Nat Rev Cancer 3:339–349 [DOI] [PubMed] [Google Scholar]
- Campisi J 2005. Senescent cells, tumor suppression, and organismal aging: Good citizens, bad neighbors. Cell 120:513–522 [DOI] [PubMed] [Google Scholar]
- Canman CE, Lim DS, Cimprich KA, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan MB, Siliciano JD 1998. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science 281:1677–1679 [DOI] [PubMed] [Google Scholar]
- Cao L, Li W, Kim S, Brodie SG, Deng CX 2003. Senescence, aging, and malignant transformation mediated by p53 in mice lacking the Brca1 full-length isoform. Genes Dev 17:201–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chehab NH, Malikzay A, Stavridi ES, Halazonetis TD 1999. Phosphorylation of Ser-20 mediates stabilization of human p53 in response to DNA damage. Proc Natl Acad Sci 96:13777–13782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J 2004. Senescence and functional failure in hematopoietic stem cells. Exp Hematol 32:1025–1032 [DOI] [PubMed] [Google Scholar]
- Chene P 2003. Inhibiting the p53-MDM2 interaction: An important target for cancer therapy. Nat Rev Cancer 3:102–109 [DOI] [PubMed] [Google Scholar]
- Chin L, Artandi SE, Shen Q, Tam A, Lee SL, Gottlieb GJ, Greider CW, DePinho RA 1999. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell 97:527–538 [DOI] [PubMed] [Google Scholar]
- Christophorou MA, Martin-Zanca D, Soucek L, Lawlor ER, Brown-Swigart L, Verschuren EW, Evan GI 2005. Temporal dissection of p53 function in vitro and in vivo. Nat Genet 37:718–726 [DOI] [PubMed] [Google Scholar]
- Christophorou MA, Ringshausen I, Finch AJ, Swigart LB, Evan GI 2006. The pathological response to DNA damage does not contribute to p53-mediated tumour suppression. Nature 443:214–217 [DOI] [PubMed] [Google Scholar]
- Classon M, Harlow E 2002. The retinoblastoma tumour suppressor in development and cancer. Nat Rev Cancer 2:910–917 [DOI] [PubMed] [Google Scholar]
- Collado M, Gil J, Efeyan A, Guerra C, Schuhmacher AJ, Barradas M, Benguria A, Zaballos A, Flores JM, Barbacid M, et al. 2005. Tumour biology: Senescence in premalignant tumours. Nature 436:642. [DOI] [PubMed] [Google Scholar]
- Donehower LA 1996. The p53-deficient mouse: A model for basic and applied cancer studies. Semin Cancer Biol 7:269–278 [DOI] [PubMed] [Google Scholar]
- Dumble M, Gatza C, Tyner S, Venkatachalam S, Donehower LA 2004. Insights into aging obtained from p53 mutant mouse models. Ann N Y Acad Sci 1019:171–177 [DOI] [PubMed] [Google Scholar]
- Dumble M, Moore L, Chambers SM, Geiger H, Van Zant G, Goodell MA, Donehower LA 2007. The impact of altered p53 dosage on hematopoietic stem cell dynamics during aging. Blood 109:1736–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efeyan A, Murga M, Martinez-Pastor B, Ortega-Molina A, Soria R, Collado M, Fernandez-Capetillo O, Serrano M 2009. Limited role of murine ATM in oncogene-induced senescence and p53-dependent tumor suppression. PLoS One 4:e5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL 1999. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev 13:2658–2669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75:817–825 [DOI] [PubMed] [Google Scholar]
- Feng Z, Hu W, Teresky AK, Hernando E, Cordon-Cardo C, Levine AJ 2007. Declining p53 function in the aging process: A possible mechanism for the increased tumor incidence in older populations. Proc Natl Acad Sci 104:16633–16638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Cao I, Garcia-Cao M, Martin-Caballero J, Criado LM, Klatt P, Flores JM, Weill JC, Blasco MA, Serrano M 2002. “Super p53” mice exhibit enhanced DNA damage response, are tumor resistant and age normally. Embo J 21:6225–6235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garinis GA, van der Horst GT, Vijg J, Hoeijmakers JH 2008. DNA damage and ageing: New-age ideas for an age-old problem. Nat Cell Biol 10:1241–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatza CE, Dumble M, Kittrell F, Edwards DG, Dearth RK, Lee AV, Xu J, Medina D, Donehower LA 2008. Altered mammary gland development in the p53+/m mouse, a model of accelerated aging. Dev Biol 313:130–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RA Jr, Kastrinakis NG, Levy B, et al. 2005. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 434:907–913 [DOI] [PubMed] [Google Scholar]
- Green DR, Kroemer G 2009. Cytoplasmic functions of the tumour suppressor p53. Nature 458:1127–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halazonetis TD, Gorgoulis VG, Bartek J 2008. An oncogene-induced DNA damage model for cancer development. Science 319:1352–1355 [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA 2000. The hallmarks of cancer. Cell 100:57–70 [DOI] [PubMed] [Google Scholar]
- Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ 1993. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75:805–816 [DOI] [PubMed] [Google Scholar]
- Hinkal GW, Gatza CE, Parikh N, Donehower LA 2009a. Altered senescence, apoptosis, and DNA damage response in a mutant p53 model of accelerated aging. Mech Ageing Dev 130:262–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinkal GW, Parikh N, Donehower LA 2009b. Timed somatic deletion of p53 in mice reveals age-associated differences in tumor progression. PLoS ONE 4:e6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi Y, Matsuda M, Aizawa S, Kodama Y, Kanno J, Inoue T 2002. Serial transplantation of p53-deficient hemopoietic progenitor cells to assess their infinite growth potential. Exp Biol Med (Maywood) 227:474–479 [DOI] [PubMed] [Google Scholar]
- Hirao A, Kong YY, Matsuoka S, Wakeham A, Ruland J, Yoshida H, Liu D, Elledge SJ, Mak TW 2000. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science 287:1824–1827 [DOI] [PubMed] [Google Scholar]
- Howes KA, Ransom N, Papermaster DS, Lasudry JG, Albert DM, Windle JJ 1994. Apoptosis or retinoblastoma: Alternative fates of photoreceptors expressing the HPV-16 E7 gene in the presence or absence of p53. Genes Dev 8:1300–1310 [DOI] [PubMed] [Google Scholar]
- Janzen V, Forkert R, Fleming HE, Saito Y, Waring MT, Dombkowski DM, Cheng T, Depinho RA, Sharpless NE, Scadden DT 2006. Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16(INK4a). Nature 443:421–426 [DOI] [PubMed] [Google Scholar]
- Keyes WM, Wu Y, Vogel H, Guo X, Lowe SW, Mills AA 2005. p63 deficiency activates a program of cellular senescence and leads to accelerated aging. Genes Dev 19:1986–1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WY, Sharpless NE 2006. The regulation of INK4/ARF in cancer and aging. Cell 127:265–275 [DOI] [PubMed] [Google Scholar]
- Kinzler KW, Vogelstein B 1996. Lessons from hereditary colorectal cancer. Cell 87:159–170 [DOI] [PubMed] [Google Scholar]
- Kinzler KW, Vogelstein B 1997. Cancer-susceptibility genes. Gatekeepers and caretakers. Nature 386:761–763 [DOI] [PubMed] [Google Scholar]
- Klatt P, Serrano M 2003. Engineering cancer resistance in mice. Carcinogenesis 24:817–826 [DOI] [PubMed] [Google Scholar]
- Krishnamurthy J, Ramsey MR, Ligon KL, Torrice C, Koh A, Bonner-Weir S, Sharpless NE 2006. p16(INK4a) induces an age-dependent decline in islet regenerative potential. Nature 443:453–457 [DOI] [PubMed] [Google Scholar]
- Krtolica A, Parrinello S, Lockett S, Desprez PY, Campisi J 2001. Senescent fibroblasts promote epithelial cell growth and tumorigenesis: A link between cancer and aging. Proc Natl Acad Sci 98:12072–12077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse JP, Gu W 2009. Modes of p53 regulation. Cell 137:609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin MF, Gueven N 2006. The complexity of p53 stabilization and activation. Cell Death Differ 13:941–950 [DOI] [PubMed] [Google Scholar]
- Levine AJ 1997. p53, the cellular gatekeeper for growth and division. Cell 88:323–331 [DOI] [PubMed] [Google Scholar]
- Lim DS, Vogel H, Willerford DM, Sands AT, Platt KA, Hasty P 2000. Analysis of ku80-mutant mice and cells with deficient levels of p53. Mol Cell Biol 20:3772–3780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin AW, Barradas M, Stone JC, van Aelst L, Serrano M, Lowe SW 1998. Premature senescence involving p53 and p16 is activated in response to constitutive MEK/MAPK mitogenic signaling. Genes Dev 12:3008–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljungman M 2000. Dial 9–1–1 for p53: Mechanisms of p53 activation by cellular stress. Neoplasia 2:208–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe SW, Cepero E, Evan G 2004. Intrinsic tumour suppression. Nature 432:307–315 [DOI] [PubMed] [Google Scholar]
- Lu X, Nguyen TA, Moon SH, Darlington Y, Sommer M, Donehower LA 2008. The type 2C phosphatase Wip1: An oncogenic regulator of tumor suppressor and DNA damage response pathways. Cancer Metastasis Rev 27:123–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier B, Gluba W, Bernier B, Turner T, Mohammad K, Guise T, Sutherland A, Thorner M, Scrable H 2004. Modulation of mammalian life span by the short isoform of p53. Genes Dev 18:306–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marine JC, Dyer MA, Jochemsen AG 2007. MDMX: From bench to bedside. J Cell Sci 120:371–378 [DOI] [PubMed] [Google Scholar]
- Martin GM 2005. Genetic modulation of senescent phenotypes in Homo sapiens. Cell 120:523–532 [DOI] [PubMed] [Google Scholar]
- Martins CP, Brown-Swigart L, Evan GI 2006. Modeling the therapeutic efficacy of p53 restoration in tumors. Cell 127:1323–1334 [DOI] [PubMed] [Google Scholar]
- Maslov AY, Vijg J 2009. Genome instability, cancer and aging. Biochim Biophys Acta; 1790:963–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheu A, Maraver A, Serrano M 2008. The Arf/p53 pathway in cancer and aging. Cancer Res 68:6031–6034 [DOI] [PubMed] [Google Scholar]
- Matheu A, Maraver A, Collado M, Garcia-Cao I, Canamero M, Borras C, Flores JM, Klatt P, Vina J, Serrano M 2009. Anti-aging activity of the Ink4/Arf locus. Aging Cell 8:152–161 [DOI] [PubMed] [Google Scholar]
- Matheu A, Maraver A, Klatt P, Flores I, Garcia-Cao I, Borras C, Flores JM, Vina J, Blasco MA, Serrano M 2007. Delayed ageing through damage protection by the Arf/p53 pathway. Nature 448:375–379 [DOI] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. 2007. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316:1160–1166 [DOI] [PubMed] [Google Scholar]
- Medrano S, Burns-Cusato M, Atienza MB, Rahimi D, Scrable H 2009. Regenerative capacity of neural precursors in the adult mammalian brain is under the control of p53. Neurobiol Aging 30:483–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendrysa SM, O'Leary K A, McElwee MK, Michalowski J, Eisenman RN, Powell DA, Perry ME 2006. Tumor suppression and normal aging in mice with constitutively high p53 activity. Genes Dev 20:16–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll UM, Wolff S, Speidel D, Deppert W 2005. Transcription-independent pro-apoptotic functions of p53. Curr Opin Cell Biol 17:631–636 [DOI] [PubMed] [Google Scholar]
- Molofsky AV, Slutsky SG, Joseph NM, He S, Pardal R, Krishnamurthy J, Sharpless NE, Morrison SJ 2006. Increasing p16(INK4a) expression decreases forebrain progenitors and neurogenesis during ageing. Nature 443:448–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momand J, Jung D, Wilczynski S, Niland J 1998. The MDM2 gene amplification database. Nucleic Acids Res 26:3453–3459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore L, Lu X, Ghebranious N, Tyner S, Donehower LA 2007. Aging-associated truncated form of p53 interacts with wild-type p53 and alters p53 stability, localization, and activity. Mech Ageing Dev 128:717–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore L, Venkatachalam S, Vogel H, Watt JC, Wu CL, Steinman H, Jones SN, Donehower LA 2003. Cooperativity of p19ARF, Mdm2, and p53 in murine tumorigenesis. Oncogene 22:7831–7837 [DOI] [PubMed] [Google Scholar]
- Murphy DJ, Junttila MR, Pouyet L, Karnezis A, Shchors K, Bui DA, Brown-Swigart L, Johnson L, Evan GI 2008. Distinct thresholds govern Myc's biological output in vivo. Cancer Cell 14:447–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijnik A, Woodbine L, Marchetti C, Dawson S, Lambe T, Liu C, Rodrigues NP, Crockford TL, Cabuy E, Vindigni A, et al. 2007. DNA repair is limiting for haematopoietic stem cells during ageing. Nature 447:686–690 [DOI] [PubMed] [Google Scholar]
- Noda A, Ning Y, Venable SF, Pereira-Smith OM, Smith JR 1994. Cloning of senescent cell-derived inhibitors of DNA synthesis using an expression screen. Exp Cell Res 211:90–98 [DOI] [PubMed] [Google Scholar]
- Orsted DD, Bojesen SE, Tybjaerg-Hansen A, Nordestgaard BG 2007. Tumor suppressor p53 Arg72Pro polymorphism and longevity, cancer survival, and risk of cancer in the general population. J Exp Med 204:1295–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios R, Bucana C, Xie X 1996. Long-term culture of lymphohematopoietic stem cells. Proc Natl Acad Sci 93:5247–5252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H, Griep AE 1994. Altered cell cycle regulation in the lens of HPV-16 E6 or E7 transgenic mice: Implications for tumor suppressor gene function in development. Genes Dev 8:1285–1299 [DOI] [PubMed] [Google Scholar]
- Pardal R, Clarke MF, Morrison SJ 2003. Applying the principles of stem-cell biology to cancer. Nat Rev Cancer 3:895–902 [DOI] [PubMed] [Google Scholar]
- Parrinello S, Coppe JP, Krtolica A, Campisi J 2005. Stromal-epithelial interactions in aging and cancer: Senescent fibroblasts alter epithelial cell differentiation. J Cell Sci 118:485–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelicci PG 2004. Do tumor-suppressive mechanisms contribute to organism aging by inducing stem cell senescence? J Clin Invest 113:4–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendas AM, Zhou Z, Cadinanos J, Freije JM, Wang J, Hultenby K, Astudillo A, Wernerson A, Rodriguez F, Tryggvason K, et al. 2002. Defective prelamin A processing and muscular and adipocyte alterations in Zmpste24 metalloproteinase-deficient mice. Nat Genet 31:94–99 [DOI] [PubMed] [Google Scholar]
- Peri A, Serio M 2008. Neuroprotective effects of the Alzheimer's disease-related gene seladin-1. J Mol Endocrinol 41:251–261 [DOI] [PubMed] [Google Scholar]
- Rando TA 2006. Stem cells, ageing and the quest for immortality. Nature 441:1080–1086 [DOI] [PubMed] [Google Scholar]
- Reya T, Morrison SJ, Clarke MF, Weissman IL 2001. Stem cells, cancer, and cancer stem cells. Nature 414:105–111 [DOI] [PubMed] [Google Scholar]
- Rodier F, Campisi J, Bhaumik D 2007. Two faces of p53: Aging and tumor suppression. Nucleic Acids Res 35:7475–7484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi DJ, Jamieson CH, Weissman IL 2008. Stems cells and the pathways to aging and cancer. Cell 132:681–696 [DOI] [PubMed] [Google Scholar]
- Rudolph KL, Chang S, Lee HW, Blasco M, Gottlieb GJ, Greider C, DePinho RA 1999. Longevity, stress response, and cancer in aging telomerase-deficient mice. Cell 96:701–712 [DOI] [PubMed] [Google Scholar]
- Ruzankina Y, Asare A, Brown EJ 2008. Replicative stress, stem cells and aging. Mech Ageing Dev 129:460–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt CA, Fridman JS, Yang M, Lee S, Baranov E, Hoffman RM, Lowe SW 2002. A senescence program controlled by p53 and p16INK4a contributes to the outcome of cancer therapy. Cell 109:335–346 [DOI] [PubMed] [Google Scholar]
- Schmitt CA, McCurrach ME, de Stanchina E, Wallace-Brodeur RR, Lowe SW 1999. INK4a/ARF mutations accelerate lymphomagenesis and promote chemoresistance by disabling p53. Genes Dev 13:2670–2677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M, Blasco MA 2007. Cancer and ageing: Convergent and divergent mechanisms. Nat Rev Mol Cell Biol 8:715–722 [DOI] [PubMed] [Google Scholar]
- Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW 1997. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88:593–602 [DOI] [PubMed] [Google Scholar]
- Sharpless NE, DePinho RA 2004. Telomeres, stem cells, senescence, and cancer. J Clin Invest 113:160–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpless NE, DePinho RA 2007. How stem cells age and why this makes us grow old. Nat Rev Mol Cell Biol 8:703–713 [DOI] [PubMed] [Google Scholar]
- Sherr CJ 2001. The INK4a/ARF network in tumour suppression. Nat Rev Mol Cell Biol 2:731–737 [DOI] [PubMed] [Google Scholar]
- Sherr CJ 2004. Principles of tumor suppression. Cell 116:235–246 [DOI] [PubMed] [Google Scholar]
- Sherr CJ 2006. Divorcing ARF and p53: An unsettled case. Nat Rev Cancer 6:663–673 [DOI] [PubMed] [Google Scholar]
- Shiloh Y 2003. ATM and related protein kinases: Safeguarding genome integrity. Nat Rev Cancer 3:155–168 [DOI] [PubMed] [Google Scholar]
- Shounan Y, Dolnikov A, MacKenzie KL, Miller M, Chan YY, Symonds G 1996. Retroviral transduction of hematopoietic progenitor cells with mutant p53 promotes survival and proliferation, modifies differentiation potential and inhibits apoptosis. Leukemia 10:1619–1628 [PubMed] [Google Scholar]
- Sluss HK, Armata H, Gallant J, Jones SN 2004. Phosphorylation of serine 18 regulates distinct p53 functions in mice. Mol Cell Biol 24:976–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohal RS, Weindruch R 1996. Oxidative stress, caloric restriction, and aging. Science 273:59–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soussi T, Lozano G 2005. p53 mutation heterogeneity in cancer. Biochem Biophys Res Commun 331:834–842 [DOI] [PubMed] [Google Scholar]
- Symonds H, Krall L, Remington L, Saenz-Robles M, Lowe S, Jacks T, Van Dyke T 1994. p53-dependent apoptosis suppresses tumor growth and progression in vivo. Cell 78:703–711 [DOI] [PubMed] [Google Scholar]
- TeKippe M, Harrison DE, Chen J 2003. Expansion of hematopoietic stem cell phenotype and activity in Trp53-null mice. Exp Hematol 31:521–527 [DOI] [PubMed] [Google Scholar]
- Tibbetts RS, Brumbaugh KM, Williams JM, Sarkaria JN, Cliby WA, Shieh SY, Taya Y, Prives C, Abraham RT 1999. A role for ATR in the DNA damage-induced phosphorylation of p53. Genes Dev 13:152–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomas-Loba A, Flores I, Fernandez-Marcos PJ, Cayuela ML, Maraver A, Tejera A, Borras C, Matheu A, Klatt P, Flores JM, et al. 2008. Telomerase reverse transcriptase delays aging in cancer-resistant mice. Cell 135:609–622 [DOI] [PubMed] [Google Scholar]
- Tyner SD, Venkatachalam S, Choi J, Jones S, Ghebranious N, Igelmann H, Lu X, Soron G, Cooper B, Brayton C, et al. 2002. p53 mutant mice that display early ageing-associated phenotypes. Nature 415:45–53 [DOI] [PubMed] [Google Scholar]
- Van Heemst D, Mooijaart SP, Beekman M, Schreuder J, de Craen AJM, Brandt BW, Slagboom PE, Westendorp RGJ, Long Life Study Group 2005. Variation in the human TP53 gene affects old age survival and cancer mortality. Exp Gerontol 40:11–15 [DOI] [PubMed] [Google Scholar]
- Van Zant G, Liang Y 2003. The role of stem cells in aging. Exp Hematol 31:659–672 [DOI] [PubMed] [Google Scholar]
- Varela I, Cadinanos J, Pendas AM, Gutierrez-Fernandez A, Folgueras AR, Sanchez LM, Zhou Z, Rodriguez FJ, Stewart CL, Vega JA, et al. 2005. Accelerated ageing in mice deficient in Zmpste24 protease is linked to p53 signalling activation. Nature 437:564–568 [DOI] [PubMed] [Google Scholar]
- Venkatachalam S, Tyner SD, Pickering CR, Boley S, Recio L, French JE, Donehower LA 2001. Is p53 haploinsufficient for tumor suppression? Implications for the p53+/− mouse model in carcinogenicity testing. Toxicol Pathol 29 Suppl:147–154 [DOI] [PubMed] [Google Scholar]
- Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T 2007. Restoration of p53 function leads to tumour regression in vivo. Nature 445:661–665 [DOI] [PubMed] [Google Scholar]
- Ventura N, Rea SL, Schiavi A, Torgovnick A, Testi R, Johnson TE 2009. p53/CEP-1 increases or decreases lifespan, depending on level of mitochondrial bioenergetic stress. Aging Cell [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel H, Lim DS, Karsenty G, Finegold M, Hasty P 1999. Deletion of Ku86 causes early onset of senescence in mice. Proc Natl Acad Sci 96:10770–10775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden KH 2000. p53: Death star. Cell 103:691–694 [DOI] [PubMed] [Google Scholar]
- Vousden KH 2002. Activation of the p53 tumor suppressor protein. Biochim Biophys Acta 1602:47–59 [DOI] [PubMed] [Google Scholar]
- Vousden KH, Lu X 2002. Live or let die: The cell's response to p53. Nat Rev Cancer 2:594–604 [DOI] [PubMed] [Google Scholar]
- Wei W, Hemmer RM, Sedivy JM 2001. Role of p14(ARF) in replicative and induced senescence of human fibroblasts. Mol Cell Biol 21:6748–6757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Miloslavskaya I, Demontis S, Maestro R, Galaktionov K 2004. Regulation of cellular response to oncogenic and oxidative stress by Seladin-1. Nature 432:640–645 [DOI] [PubMed] [Google Scholar]
- Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, Cordon-Cardo C, Lowe SW 2007. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 445:656–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Ying H, Yan H, Kimmelman AC, Hiller DJ, Chen AJ, Perry SR, Tonon G, Chu GC, Ding Z, et al. 2008. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature 455:1129–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Woods D, McMahon M, Bishop JM 1998. Senescence of human fibroblasts induced by oncogenic Raf. Genes Dev 12:2997–3007 [DOI] [PMC free article] [PubMed] [Google Scholar]