Abstract
Simple visual inspection of bacteria indicated that, at least in some otherwise symmetric cells, structures such as flagella were often seen at a single pole. Because these structures are composed of proteins, it was not clear how to reconcile these observations of morphological asymmetry with the widely held view of bacteria as unstructured “bags of enzymes.” However, over the last decade, numerous GFP tagged proteins have been found at specific intracellular locations such as the poles of the cells, indicating that bacteria have a high degree of intracellular organization. Here we will explore the role of chromosomal asymmetry and the presence of “new” and “old” poles that result from the cytokinesis of rod-shaped cells in establishing bipolar and monopolar protein localization patterns. This article is intended to be illustrative, not exhaustive, so we have focused on examples drawn largely from Caulobacter crescentus and Bacillus subtilis, two bacteria that undergo dramatic morphological transformation. We will highlight how breaking monopolar symmetry is essential for the correct development of these organisms.
Bacteria have a far more complex internal organization than we thought. Polarity proteins may mark “old” and “new” poles, and each gene seems to have a defined spatial location.
Although prokaryotes with dramatic, colorful stripes such as Blake's “tygers” have not been seen, many bacteria found in nature show morphological polarity (Young 2006). This could simply be a consequence of the elaborations of bacterial cellular architecture, akin to the famous decorative but not structurally essential Spandrels in the Basilica di San Marco in Venice that are a side-effect of an adaptation, rather than a direct product of natural selection (Gould and Lewontin 1979). However, it is more likely that this polarity can be traced to a particular function in cellular physiology. An example is the ActA protein of Listeria monocytogenes that is localized at a single bacterial pole (Theriot et al. 1992; Goldberg and Theriot 1995). The interaction between ActA and the Arp2/3 complex induces actin filament formation at that pole and therefore serves to propel the bacterium (Loisel et al. 1999). In this article, we will address the mechanisms underlying such asymmetric protein distributions.
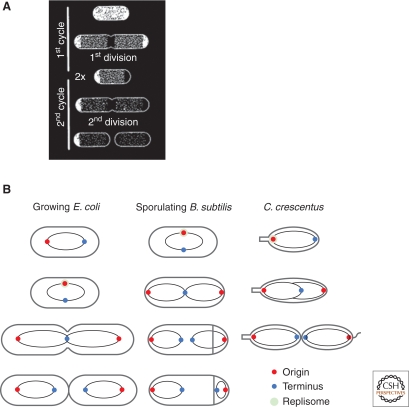
At least two aspects of prokaryotic cell physiology are intrinsically polar. First, cytokinesis in rod-shaped organisms occurs typically in the middle, so cells have an “old” pole and a “new” pole. Even in cells with a coccoid (round) morphology, the two hemispheres have different ages. At a molecular level, the poles are zones of inert peptidoglycan (Fig. 1A) resulting from the absence of new synthesis (de Pedro et al. 1997). Thus, the differential age of a pole could be reflected in its differential “inertness.” Because surface exposed proteins can become immobilized in these zones (de Pedro et al. 2004), these proteins could serve as landmarks for the establishment of morphological structures. However, few demonstrations of an interaction between peptidoglycan and a protein that result in a particular pattern of protein localization have been reported, so this mechanism remains largely hypothetical. A second basis for intrinsic polarity derives from the asymmetry of the haploid bacterial chromosome (Rocha 2008). Because bacterial chromosomes have a stereotypical layout within the bacterial cell and genes are located in either origin proximal or original distal positions, genetic loci have a defined spatial distribution (Teleman et al. 1998; Viollier et al. 2004; Berlatzky et al. 2008) that could serve as a template to direct asymmetric protein localization (Fig. 1B). Although this mechanism is appealing, not least for its simplicity, the colocalization of genes and their encoded proteins remains largely speculative (Norris et al. 2007). As discussed in more detail in the following, however, the chromosomal position of two genes necessary for cell fate determination in B. subtilis does play an important role in the activity of their respective proteins.
Figure 1.
Intrinsic polarity in bacteria. (A) The poles of rod shaped cells are zones of inert peptidoglycan. D-Cys-labeled Escherichia coli was chased in the absence of label for two mass doubling times. White indicates labeled, stable murein; black indicates unlabeled, presumably recently inserted murein. Illustration provided by Anu Janakiraman. (B) Asymmetric orientation of bacterial chromosomes. Origins of replication are red, termini of replication are blue, and the replisome is green. (Left) The haploid bacterial B. subtilis or E. coli chromosomes are orientated in slow growing cells with the origin located near one pole and the terminus located near the other pole. (Middle) During sporulation in B. subtilis, the chromosome is initially bisected by the asymmetric septum, resulting in a period of transient genetic asymmetry before completion of translocation. (Right) Chromosome replication in C. crescentus initiates at one pole, followed by transit of the newly replicated origin to the other pole.
Because most prokaryotes show at least one of these kinds of cellular polarity, the question becomes how do proteins become asymmetrically localized within the cell? Proteins can localize to one or both poles, to the mid-cell, or to helices spanning the length of the cell (Graumann 2007). Here, we will explore how some of these patterns are established and then focus on their function in cellular physiology.
HOW ARE THE CELL POLES IDENTIFIED?
Many bacteria are rod-shaped and, by first approximation, are symmetric (Janakiraman and Goldberg 2004). However, even rods that appear symmetric show deviations in this symmetry, such as slight differences in the shape of the two polar caps (Guberman et al. 2008; Itan et al. 2008). Although it is not clear how these deviations are used by the cell, the two poles are functionally distinct in that cells that inherit the “older” pole (Fig. 1A) show a diminished growth rate and an increased incidence of death (Stewart et al. 2005) perhaps because of asymmetric segregation of protein aggregates (Lindner et al. 2008). More generally, the relative age of the poles may be reflected in their molecular constituents, such as proteins and/or peptidoglycan. However, before addressing differences in the poles, it will be useful to examine how they are distinguished from the longitudinal axis of the cell.
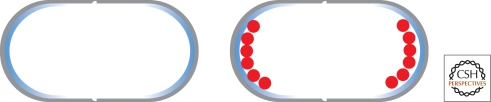
One way to distinguish the poles would be membrane curvature because the longitudinal axis of the cell is curved only in one direction (the curvature associated with the cylinder wall), whereas the cell poles are curved in two dimensions. The membrane phospholipid cardiolipin has an inherent curvature preference resulting from the energetics of the particular geometry of the intermolecular interaction between cardiolipin molecules that have a small head-to-tail ratio (McAuley et al. 1999). This characteristic results in the formation of cardiolipin domains at the cell poles via microphase separation of the membrane (Huang et al. 2006; Mukhopadhyay et al. 2008). Thus, a protein that has a preference for cardiolipin will localize to these patches at the cell pole (Fig. 2). This mechanism has been shown to underlie the polar localization of the E. coli proline transporter/osmosensor ProP (Romantsov et al. 2007) because it is found in an even cellular distribution in bacteria that are missing the cardiolipin synthase genes (Romantsov et al. 2008). Mutations of ProP that affect its osmosensing prevent polar enrichment, indicating a relationship between function and localization, although how this is achieved remains unclear.
Figure 2.
Lipid domains at cell poles. The membrane phospholipid cardiolipin favors rounded surfaces because of the energetics of intermolecular interactions between the lipid molecules. Thus, the cardiolipin (blue) forms domains at the cell poles and proteins (red) that preferentially interact with the lipid are therefore localized to the cell poles.
Another protein that preferentially binds cardiolipin in vitro is the glycosyltransferase MurG (van den Brink-van der Laan et al. 2003), a key player in peptidoglycan synthesis. In C. crescentus, MurG localization to the mid-cell is dependent, perhaps indirectly, on the cell division nucleator, the tubulin-like protein FtsZ, although the role of cardiolipin was not reported (Aaron et al. 2007). It remains unclear, therefore, whether the preferential binding of MurG that was observed in vitro has a functional role in vivo. In fact, although both curved membranes and polarity are present during B. subtilis sporulation where a large number of proteins are targeted to the spherical membrane surrounding the developing forespore, mutations that disable cardiolipin synthesis only reduce sporulation slightly (Kawai et al. 2006). Thus, although lipid–protein interactions may mediate polar targeting, few proteins likely rely exclusively on this mechanism.
Proteins themselves could have a higher affinity for cell poles. B. subtilis DivIVA is found at the cell poles and this localization is required for both the polar localization of MinC, a protein necessary for proper mid-cell division (Marston et al. 1998), as well as that of RacA, a protein necessary for polar attachment of chromosomes during sporulation (Ben-Yehuda et al. 2003). Because DivIVA polar localization does not appear to be mediated by either FtsZ or a protein involved in septal peptidoglycan synthesis (Hamoen and Errington 2003) and expression of a GFP fusion of B. subtilis DivIVA in the fission yeast Schizosaccharomyces pombe results in a polar fluorescent signal (Edwards et al. 2000), DivIVA may recognize some physical aspect of the pole, such as curvature, through a polar targeting sequence (Perry and Edwards 2004). Alternatively, the interaction of DivIVA with a chromosomal partitioning protein, which itself localizes to the origin region of the chromosome, suggests that chromosomal asymmetry may be the ultimate driving force for the polar DivIVA distribution (Perry and Edwards 2006), but this appealing possibility has not yet been examined.
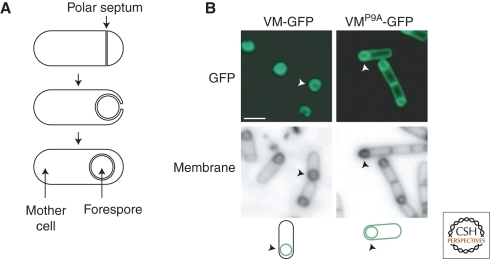
Specific hydrophobic amino acid side-chains in the amphipathic α-helix of B. subtilis SpoVM are necessary for its specificity for the forespore membrane as compared with the cytoplasmic membrane during sporulation (Ramamurthi et al. 2006). Specifically, a change from a proline to an alanine (Ramamurthi et al. 2009) causes the protein to lose this preference (Fig. 3). In vitro experiments indicate that SpoVM directly senses the curvature of the membrane and thereby discriminates between the positive curvature of the forespore membrane and the negative curvature of the cytoplasmic membrane (Ramamurthi et al. 2009). Because amphipathic α-helices are important for membrane interactions in a number of bacterial proteins such as MinD (Szeto et al. 2003; Zhou and Lutkenhaus 2003) and FtsA (Pichoff and Lutkenhaus 2005), this mechanism could underlie polar targeting more generally. Interestingly, eukaryotic proteins containing a BAR domain and an amphipathic helix bind membranes and are sensitive to curvature, so this mechanism may be phylogenetically conserved (Peter et al. 2004).
Figure 3.
Asymmetric localization of SpoVM. (A) Stages of sporulation. (Top) Division creates a mother cell and a smaller forespore. (Middle) The mother cell engulfs the forespore. (Bottom) The forespore is pinched off as a protoplast. (B) SpoVM-GFP localizes to the surface of the forespore, whereas SpoVMP9A-GFP localizes to all membranes. Arrowheads identify the cell depicted in the illustrations. Illustration provided by Kumaran Ramamurthi.
However, not all polar proteins appear to use this mechanism. During chromosome segregation in C. crescentus, the sister chromosomes become anchored at cellular opposite poles. A protein DNA-complex comprising a centromere-like sequence and a specific DNA-binding protein that recognizes this sequence is necessary for this event. PopZ, a protein with intrinsic polar affinity appears to mediate attachment of this complex to the cell poles (Bowman et al. 2008; Ebersbach et al. 2008). However, PopZ is found localized at regular intervals in the chromosome-free regions of filamentous cells generated by blocking cell division. Thus, the polar localization of PopZ is achieved independently of division and does not appear to be dictated by the curvature.
HOW ARE THE CELL POLES DISTINGUISHED?
We will now turn our attention to proteins found only at a single pole. Like L. monocytogenes ActA, Shigella flexneri IcsA is necessary for the actin polymerization that drives bacterial motility in the host cell cytoplasm. IcsA belongs to a large family of auto-transporters that, along with other members of this family, is found localized at the old cell pole (Jain et al. 2006). In fact, even in artificially generated, spherical E. coli, IcsA is found preferentially at the cell “tips” (Pradel et al. 2007). This distribution is not dependent on the Min system that is responsible for preventing polar division, the chromosomal position of the gene, or on the Sec pathway of protein secretion. Thus, although origin of the polar distribution of IcsA remains mysterious, it could remain at the poles through an interaction with stable, polar molecules such as components of the lipopolysaccharide layer (Jain et al. 2006).
Two possible mechanisms could derive information from mid-cell cytokinesis to generate polarity. First, an essential and early player in this event is the tubulin-like protein FtsZ that forms polymers at the eventual site of cell division. Because structures such as the medial FtsZ rings that appear to be symmetric with respect to the poles are composed of subunits with intrinsic asymmetry (Lowe and Amos 1998), one could imagine that this asymmetry could be used to direct proteins to one pole if there was a single polymeric ring and the targeted protein selectively bound only one face. However, the structure of this polymer in vivo is not clear (Li et al. 2007) and it may form multiple coil-like structures at mid-cell that do not have definitive orientation with respect to the poles (Michie et al. 2006). Regardless, proteins with affinity for the FtsZ ring (or, more generally, with other components of the division apparatus) would end up at the “new” cell pole of the daughter cells. A protein that might act in such a way is C. crescentus TipN that is required for the polar localization of the pilus, flagellum, and signaling apparatus. TipN could serve as a “birthscar” or a “landmark” protein that interacts with the FtsZ polymer at the septum and therefore marks the new pole (Huitema et al. 2006; Lam et al. 2006). This mechanism would require an asymmetric TipN-FtsZ interaction that has not been shown, but, if so, TipN could serve as the initial intrinsic cue of the polarization of the flagellated pole.
A second mechanism depends on later events in cytokinesis, specifically synthesis of the cell wall peptidoglycan that comprises the septum. Cell poles are largely inert with respect to peptidoglycan synthesis (de Pedro et al. 1997) because the completion of septation at mid-cell marks the end of de novo synthesis. Thus, proteins involved in this synthesis could remain associated with the peptidoglycan, and the extent of this association would be correlated with the “age” of the poles. That is, the “new” pole would have more of these associated proteins than “old” poles and these proteins could serve as landmarks for morphological asymmetries. Candidate proteins include transpeptidases and transglycosylases necessary for peptidoglycan synthesis during septation. One of these proteins, E. coli FtsI, is seen only occasionally at cell poles, well after septation, although whether this localization reflects the relative time following septation is not known (Weiss et al. 1997). The E. coli Tsr chemotaxis protein localizes to cell poles (Liberman et al. 2004) and the fluorescent signal of a Tsr-GFP fusion was correlated with the age of the pole as determined by cell lineage analysis (Ping et al. 2008). In slower growing cells, there was a strong difference between the old and the new pole Tsr-GFP signal, and this relation was also observed, albeit less robustly, in faster growing cells (Ping et al. 2008). Thus, although the mechanistic basis of these different mechanisms remains obscure, rod-shaped bacteria can distinguish their poles from the rest of the cell and further distinguish the two similar looking, but not identical, poles.
HOW DO PROTEINS GET TO A CELL POLE?
One can envision several ways to direct a protein to a pole. For example, if the macromolecular complexes responsible for protein synthesis and/or secretion were asymmetrically distributed, then this could serve as a targeting mechanism. A fluorescent protein fusion of the large ribosomal protein L1 is observed near the poles of growing B. subtilis cells (Mascarenhas et al. 2001). Although this localization could simply be a passive consequence of exclusion from the medially located nucleoid rather than a directed targeting mechanism, this distribution appeared to be dependent on the active synthesis of RNA. However, one difficulty in interpreting this result is that this fluorescent fusion reports the distribution of inactive ribosomes and proteins not associated with ribosomes in addition to the relevant active ribosomes. Interestingly, protein components of the secretion apparatus are located at the cell poles in the Gram-positive bacterium Streptococcus pyogenes (Rosch and Caparon 2004). Thus, a “landmark” membrane protein could end up at a cell pole because of localized and coupled synthesis and secretion.
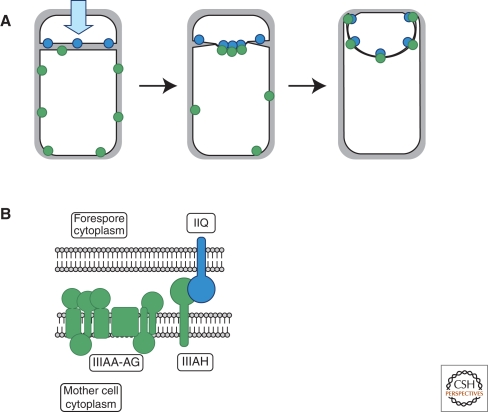
An alternative mechanism that could target proteins to the poles and does not depend on an enzymatic reaction, such as protein secretion is called “diffusion and capture,” where proteins freely diffuse through the cytoplasm or, in the case of integral membrane proteins, through the membrane, until they bind to a protein or a protein complex (Rudner et al. 2002). If these “capturing” proteins are themselves localized, then this interaction will result in the localization of the “diffusing” proteins. Such a mechanism occurs during B. subtilis sporulation in which a membrane protein is targeted to a polar septum. One might expect that this protein, SpoIIIAH, would be found distributed in the entire membrane surrounding the cellular compartment where it is expressed. However, SpoIIIAH is only observed in the portion of the membrane that abuts the membrane of the adjacent compartment (Blaylock et al. 2004). This asymmetric localization is determined by the presence of a protein in this adjacent membrane, SpoIIQ, which interacts with SpoIIIAH across the intermembrane space (Fig. 4) via the extracellular domains of both proteins and thereby restricts SpoIIIAH localization (Blaylock et al. 2004).
Figure 4.
Asymmetric localization of SpoIIQ and SpoIIIAH. (A) SpoIIQ (blue circles) is initially localized to the forespore membrane, whereas SpoIIIAH (green circles) is initially randomly distributed throughout the mother cell membrane. An interaction between SpoIIIAH and SpoIIQ across the forespore septum results in the colocalization of the two proteins at this polar structure. (B) Detailed view of the septum (as in the cell shown in A), showing the forespore membrane (FM) and the mother cell membrane (MM). SpoIIQ (“IIQ,” blue) and SpoIIIAH (“IIIAH,” green) interact in the intermembrane space mediating SpoIIIAH localization. Figure adapted from Blaylock et al. (2004) with permission.
POLARITY IN PROKARYOTIC DEVELOPMENTAL PATHWAYS
Although all rod-shaped bacteria are by definition polar, many bacteria undergo much more complex morphological differentiation. These changes are dictated in response to either extrinsic or intrinsic cues and often result in a change of daughter cell fate. They typically begin with the establishment of cellular asymmetry in the precursor cell. This process can reinforce a pre-existing polarity, as is the case with C. crescentus, or initiate a new cellular asymmetry, as is the case with B. subtilis when it undergoes sporulation.
C. crescentus
A single flagellated C. crescentus cell gives rise to unequal daughter cells. The “swarmer” cell is flagellated, whereas the “stalked” cell lacks a polar flagellum and instead has a polar stalklike appendage that facilitates adherence to surfaces. C. crescentus is intrinsically polar because the flagellum is found only at one pole, in contrast with B. subtilis undergoing sporulation where the newly defined polarity results from the asymmetric division of a previously symmetric cell. Extensive analysis has been performed on how this intrinsic polarity generates dissimilar progeny in C. crescentus (Laub et al. 2007; Brown et al. 2009), but we will focus here on a single example.
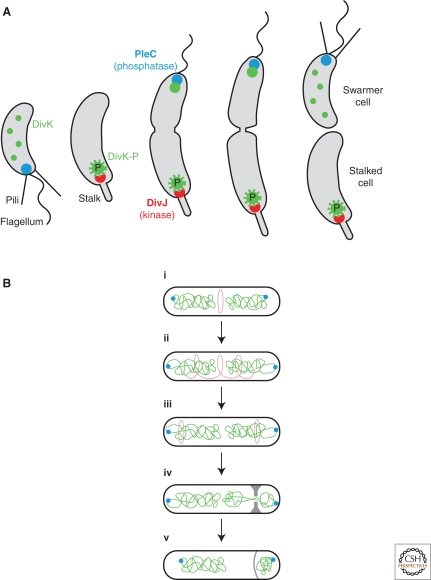
A key master regulator of differentiation is CtrA, the DNA-binding response regulator that has numerous downstream targets in the C. crescentus cell cycle, including the genes encoding components of the flagellum (Laub et al. 2002). The activity of CtrA is dependent on its phosphorylation state and this modification also has consequences for the partitioning of CtrA into the stalked cell. DivK, a response regulator present in both daughter cells immediately after cell division, controls CtrA phosphorylation and localization by participating in a multiple protein phosphorelay that mediates phosphotransfer from the DivJ kinase to CtrA (Wu et al. 1998). The ability of DivK to regulate CtrA differentially in the two daughter cells is dependent on its phosphorylation state that is itself the result of the action of both the DivJ and the PleC kinase. These two proteins localize to opposite poles of the cell at division (Wheeler and Shapiro 1999), suggesting that the phosphorylation state of DivK differs in the two daughter cells (Ryan et al. 2004). DivK thus has different activities in the two daughter cells and this ultimately results in their differential fates (Fig. 5A) (Matroule et al. 2004; Ryan et al. 2004; Biondi et al. 2006).
Figure 5.
Cell polarity in bacterial differentiation. (A) C. crescentus generates two different daughter cells during each cell cycle, the flagellated swarmer cell and the stalk cell. A key determinant of this differential cell fate is DivK, a target of the kinase DivJ and the phosphatase PleC that are themselves located at opposite poles of the dividing cell. Illustration provided by Zemer Gitai. (B) B. subtilis undergoes a transition from a symmetric division to an asymmetric division on entry into sporulation. The FtsZ ring is normally at mid-cell (i), but migrates (ii) in sporulation to polar positions (iii). One of these Z-rings becomes the site of asymmetric septum synthesis (iv). Following completion of chromosome segregation into the forespore (v), the process of engulfment starts beginning with the septum becoming curved. Adapted from Ben-Yehuda et al. 2003 with permission.
How is this protein asymmetry set up? MreB is a prokaryotic actin-like protein that forms pole-to-pole spirals in a number of bacteria, including C. crescentus. Depletion of MreB results in a disruption of chromosomal origin localization as well as inappropriate targeting of proteins such as DivK (as well as PleC, DivJ, and CckA) (Gitai et al. 2004). However, the dependence of MreB polar localization on TipN (see previous discussion) (Lam et al. 2006) suggests that this mechanism may be quite complex. In addition, it is not clear how MreB might mediate cellular polarization. Several scenarios have been proposed, largely based on the role of actin as a polarizing factor in eukaryotes. For example, the inherent polarity of MreB filaments might direct proteins to one pole, as occurs with myosin and actin during polarized growth of budding yeast (Li and Wai 2004; see Slaughter et al. 2009), although the direction of MreB polymerization appears to be independent of cellular polarity (Kim et al. 2006). Alternatively, the differential properties of the MreB polymer ends (Kim et al. 2006) could allow for asymmetric motility as is observed in vitro with actin (Li and Wai 2004). Finally, MreB may play a similar role in other bacteria because the absence of E. coli MreB achieved through either genetic or chemical means led to the loss of polar targeting of several proteins including IcsA (Nilsen et al. 2005) and the chemoreceptor Tar (Shih et al. 2005).
B. subtilis
When Bacilli are exposed to nutrient-limiting conditions, the previously symmetric cells initiate a developmental sequence that ultimately results in the establishment of a striking cellular polarity with the formation of a heat and desiccation resistant spore at one pole (Errington 2003). A key step in this pathway is the formation of a single, asymmetrically positioned septum. This event can be viewed as “symmetry-breaking” because the cell goes from being symmetric to asymmetric with respect to septum position (Fig. 5B). Although sporulating cells can (rarely) make two polar septa (Hilbert and Piggot 2004) and in fact, early in sporulation, FtsZ rings are seen near to both poles (Ben-Yehuda and Losick 2002), wild-type cells typically make a single septum. If cells were able to only construct a single septum at a time, presumably because of insufficient factors necessary for the simultaneous synthesis of two septa, then this limitation would be responsible for the pronounced polarity seen in most sporulating bacteria. Overexpression of part of the biosynthetic cascade underlying septum formation might therefore allow simultaneous formation of two symmetric and polar septa, but this possibility has not been examined.
Is this developmental asymmetry connected to (or even dependent on) the old pole/new pole asymmetry? Initial microscopic examination of sporulating cells suggested that there was a roughly equal chance of sporulation occurring at the old and new poles but necessary assumptions concerning the origins of the poles hampered a more accurate assessment. More recently, time-lapse microscopy has facilitated a careful lineage analysis that confirmed the absence of any preference in the choice of pole that will be the site of septum formation (Veening et al. 2008). Interestingly, this lack of preference is not observed in a related bacterium, Bacillus megaterium, where asymmetric septum formation at the old pole is strongly (but not absolutely) favored (Hitchins 1975).
A second example of polarity in this process occurs during the attachment of one of the chromosome origins to a cell pole. When the asymmetric sporulation septum forms over that attached chromosome, the origin-proximal one-third of the chromosome is located in the forespore, the smaller compartment, and genes at origin distal positions are located outside the forespore (Fig. 1B). So, during the time it takes for chromosome translocation to complete, origin distal genes are excluded from the forespore. This transient genetic asymmetry has important implications because, as discussed below, the differential activation of both compartment-specific transcription factors is dependent on the chromosomal position of genes encoding proteins necessary for this activation (Dworkin 2003). The DNA-binding protein RacA is essential for the proper attachment of this chromosome origin to a cell pole, a necessary step in the establishment of this genetic asymmetry (Fig. 5B). Somewhat surprisingly, however, racA mutants are only slightly defective in sporulation. An answer to this apparent contradiction emerged during careful inspection of racA mutant cells, where it became clear that even in the absence of RacA function that led to a failure of polar attachment and the formation of an empty forespore, most cells were able to successfully attach the other chromosome at the second pole (Ben-Yehuda et al. 2003). Thus, RacA allows the cell to “escape” from an initial bad morphological decision at one pole and ultimately establish cellular polarity by completing attachment at the other pole.
One way to convert chromosomal polarity to cellular polarity occurs during sporulation (Dworkin and Losick 2001) where the cell is divided into two differently sized compartments preceding the completion of chromosome segregation. As described above, this results in the transient absence of origin distal genes from the smaller of the compartments. One of these genes encodes an unstable inhibitor of a transcription factor, so during its period of exclusion from this compartment (∼20 min at 37°C), the levels of this inhibitor protein fall in the smaller compartment and thereby contribute to the activation of the transcription factor selectively in this compartment. Thus, the presence of the gene at an original distal position (genetic polarity) is converted into the activation of the transcription factor in one of two compartments (morphological polarity). A similar “position” effect is also observed in the activation of a second transcription factor that is active only in the larger compartment. The chromosomal position of the gene encoding a component of the signaling pathway necessary for activation of these transcription factors greatly affects the activity of this pathway and therefore the ability of the cells to progress in sporulation (Khvorova et al. 2000; Zupancic et al. 2001). Because the period of this transient genetic asymmetry is determined by the rate of chromosome translocation, the position of this signaling gene on the chromosome directly affects the activation kinetics of the transcription factor. In fact, manipulation of the position of this gene results in the appearance of novel developmental fates, including the formation of cells containing spores at both poles (Eldar et al. 2009).
OTHER EXAMPLES OF POLARITY
Bacterial morphologies are diverse and range from relatively subtle deviations from linearity to intricate branching patterns. C. crescentus is a curved rod and inactivation of the gene encoding a protein, crescentin, with homology with eukaryotic intermediate filament (IF) proteins is sufficient to transform the crescent-shaped rod to a straight rod. Crescentin localizes to the curved side of the cell and was proposed to interact with other cytoskeletal elements to promote this distortion (Ausmees et al. 2003). Thus, in this organism, cellular polarity is not just anterior–posterior, but dorsal–ventral. This class of proteins may play a broader role in morphology because a homolog plays a role in establishment of the tip hyphal structure in actinomycetes. In particular, atomic force microscopy has shown that the FliP homolog in Streptomyces coelicolor contributes significantly to the mechanical rigidity of the hyphal tip, consistent with the proposed role of CreS (Bagchi et al. 2008). A wide variety of bacteria, including those with rod-like shapes, such as B. subtilis, have proteins with a similar IF-like architecture (Bagchi et al. 2008), suggesting that they may play a role in a broader range of morphological types and could provide a structural basis for cellular polarity.
In some actinomycetes, hyphae that originate from the lateral walls in addition to the more commonly seen tips generate a complex pattern of branching. S. coelicolor DivIVA is necessary for the initiation of branching through the formation of DivIVA foci followed by localized peptidoglycan synthesis (Hempel et al. 2008). Interestingly, these foci formed preferentially at curved hyphal walls, similar to the polar localization observed with B. subtilis DivIVA as discussed above. Because E. coli can be induced to branch following mutation of a single penicillin-binding protein (Nelson and Young 2000), this mechanism of branching (and subsequent loss of symmetry) may be broadly conserved.
CONCLUSIONS
Although it is clear that polarity plays an important and diverse role in microbial physiology, the mechanisms responsible for establishing these morphological polarities have remained relatively obscure. However, this situation is changing as we begin to understand the relationship between physical aspects of polarity (e.g., membrane curvature) and the localization of the proteins that comprise polar complexes. The identification of bacterial proteins that sense curvature (e.g., B. subtilis SpoVM) or sense lipids such as cardiolipin whose cellular distribution is sensitive to curvature (e.g., E. coli ProP) that we have discussed in this article will facilitate more specific tests of the roles of these physical attributes in establishing cellular polarity. For example, membrane curvature could be monitored by the presence of fluorescently labeled cardiolipin-enriched domains simultaneously with protein localization to these domains. It will be informative to see how this mechanism relates to curvature-detecting proteins in eukaryotic cells by determining whether these proteins also recognize bacteria ends.
Once the poles are distinguished from the rest of the cell, the examples described in this article illustrate that this symmetry is broken by differentiating the two poles and that this process is essential for successful establishment of cell type in both B. subtilis and C. crescentus. The relationship between the chromosomal position of a gene and the eventual subcellular polarity of its encoded protein that plays a central role in cell type specification in B. subtilis sporulation may be broadly used to target proteins. In fact, the stereotyped spatial organization of the nucleoid in bacterial cells appears similar to the stereotyped organization of eukaryotic chromosomes in the nucleus where particular genetic loci are often found at similar locations within the nucleus (Fedorova and Zink 2009). Thus, eukaryotes may also use the chromosomal position of a gene as a mechanism to break morphological symmetry.
ACKNOWLEDGMENTS
I thank Anu Janakiraman, Zemer Gitai, Michael Laub, and especially Frederico Gueiros for comments on an earlier version. Anu Janakiraman, Zemer Gitai, Richard Losick, and Kumaran Ramamurthi generously provided illustrations. Work in my laboratory is supported by the National Institutes of Health (NIH) (GM081368 and AI076841) and by the Irma T. Hirschl Trust.
Footnotes
Editors: Rong Li and Bruce Bowerman
Additional Perspectives on Symmetry Breaking in Biology available at www.cshperspectives.org
REFERENCES
- Aaron M, Charbon G, Lam H, Schwarz H, Vollmer W, Jacobs-Wagner C 2007. The tubulin homologue FtsZ contributes to cell elongation by guiding cell wall precursor synthesis in Caulobacter crescentus. Mol Microbiol 64:938–952 [DOI] [PubMed] [Google Scholar]
- Ausmees N, Kuhn JR, Jacobs-Wagner C 2003. The bacterial cytoskeleton: An intermediate filament-like function in cell shape. Cell 115:705–713 [DOI] [PubMed] [Google Scholar]
- Bagchi S, Tomenius H, Belova LM, Ausmees N 2008. Intermediate filament-like proteins in bacteria and a cytoskeletal function in Streptomyces. Mol Microbiol 70:1037–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Yehuda S, Losick R 2002. Asymmetric cell division in B. subtilis involves a spiral-like intermediate of the cytokinetic protein FtsZ. Cell 109:257–266 [DOI] [PubMed] [Google Scholar]
- Ben-Yehuda S, Rudner DZ, Losick R 2003. RacA, a bacterial protein that anchors chromosomes to the cell poles. Science 299:532–536 [DOI] [PubMed] [Google Scholar]
- Berlatzky IA, Rouvinski A, Ben-Yehuda S 2008. Spatial organization of a replicating bacterial chromosome. Proc Natl Acad Sci 105:14136–14140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biondi EG, Reisinger SJ, Skerker JM, Arif M, Perchuk BS, Ryan KR, Laub MT 2006. Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature 444:899–904 [DOI] [PubMed] [Google Scholar]
- Blaylock B, Jiang X, Rubio A, Moran CP Jr, Pogliano K 2004. Zipper-like interaction between proteins in adjacent daughter cells mediates protein localization. Genes Dev 18:2916–2928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman GR, Comolli LR, Zhu J, Eckart M, Koenig M, Downing KH, Moerner WE, Earnest T, Shapiro L 2008. A polymeric protein anchors the chromosomal origin/ParB complex at a bacterial cell pole. Cell 134:945–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PJ, Hardy GG, Trimble MJ, Brun YV 2009. Complex regulatory pathways coordinate cell-cycle progression and development in Caulobacter crescentus. Adv Microb Physiol 54:1–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pedro MA, Quintela JC, Holtje JV, Schwarz H 1997. Murein segregation in Escherichia coli. J Bacteriol 179:2823–2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pedro MA, Grunfelder CG, Schwarz H 2004. Restricted Mobility of Cell Surface Proteins in the Polar Regions of Escherichia coli. J Bacteriol 186:2594–2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworkin J 2003. Transient genetic asymmetry and cell fate in a bacterium. Trends Genet 19:107–112 [DOI] [PubMed] [Google Scholar]
- Dworkin J, Losick R 2001. Differential gene expression governed by chromosomal spatial asymmetry. Cell 107:339–346 [DOI] [PubMed] [Google Scholar]
- Ebersbach G, Briegel A, Jensen GJ, Jacobs-Wagner C 2008. A self-associating protein critical for chromosome attachment, division, and polar organization in caulobacter. Cell 134:956–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards DH, Thomaides HB, Errington J 2000. Promiscuous targeting of Bacillus subtilis cell division protein DivIVA to division sites in Escherichia coli and fission yeast. EMBO J 19:2719–2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldar A, Chary VK, Xenopoulos P, Fontes ME, Losin OC, Dworkin J, Piggot PJ, Elowitz MB 2009. Partial penetrance facilitates developmental evolution in bacteria. Nature 460:510–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errington J 2003. Regulation of endospore formation in Bacillus subtilis. Nat Rev Microbiol 1:117–126 [DOI] [PubMed] [Google Scholar]
- Fedorova E, Zink D 2009. Nuclear genome organization: Common themes and individual patterns. Curr Opin Genet Dev [DOI] [PubMed] [Google Scholar]
- Gitai Z, Dye N, Shapiro L 2004. An actin-like gene can determine cell polarity in bacteria. Proc Natl Acad Sci 101:8643–8648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg MB, Theriot JA 1995. Shigella flexneri surface protein IcsA is sufficient to direct actin-based motility. Proc Natl Acad Sci 92:6572–6576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ, Lewontin RC 1979. The spandrels of San Marco and the Panglossian paradigm: A critique of the adaptationist programme. Proc R Soc Lond B Biol Sci 205:581–598 [DOI] [PubMed] [Google Scholar]
- Graumann PL 2007. Cytoskeletal elements in bacteria. Annu Rev Microbiol 61:589–618 [DOI] [PubMed] [Google Scholar]
- Guberman JM, Fay A, Dworkin J, Wingreen NS, Gitai Z 2008. PSICIC: Noise and asymmetry in bacterial division revealed by computational image analysis at sub-pixel resolution. PLoS Comput Biol 4:e1000233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamoen LW, Errington J 2003. Polar targeting of DivIVA in Bacillus subtilis is not directly dependent on FtsZ or PBP 2B. J Bacteriol 185:693–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hempel AM, Wang SB, Letek M, Gil JA, Flardh K 2008. Assemblies of DivIVA mark sites for hyphal branching and can establish new zones of cell wall growth in Streptomyces coelicolor. J Bacteriol 190:7579–7583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilbert DW, Piggot PJ 2004. Compartmentalization of gene expression during Bacillus subtilis spore formation. Microbiol Mol Biol Rev 68:234–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchins AD 1975. Polarized relationship of bacterial spore loci to the “old” and “new” ends of sporangia. J Bacteriol 121:518–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang KC, Mukhopadhyay R, Wingreen NS 2006. A curvature-mediated mechanism for localization of lipids to bacterial poles. PLoS Comput Biol 2:e151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huitema E, Pritchard S, Matteson D, Radhakrishnan SK, Viollier PH 2006. Bacterial birth scar proteins mark future flagellum assembly site. Cell 124:1025–1037 [DOI] [PubMed] [Google Scholar]
- Itan E, Carmon G, Rabinovitch A, Fishov I, Feingold M 2008. Shape of nonseptated Escherichia coli is asymmetric. Phys Rev E Stat Nonlin Soft Matter Phys 77:061902. [DOI] [PubMed] [Google Scholar]
- Jain S, van Ulsen P, Benz I, Schmidt MA, Fernandez R, Tommassen J, Goldberg MB 2006. Polar localization of the autotransporter family of large bacterial virulence proteins. J Bacteriol 188:4841–4850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janakiraman A, Goldberg MB 2004. Recent advances on the development of bacterial poles. Trends Microbiol 12:518–525 [DOI] [PubMed] [Google Scholar]
- Kawai F, Hara H, Takamatsu H, Watabe K, Matsumoto K 2006. Cardiolipin enrichment in spore membranes and its involvement in germination of Bacillus subtilis Marburg. Genes Genet Syst 81:69–76 [DOI] [PubMed] [Google Scholar]
- Khvorova A, Chary VK, Hilbert DW, Piggot PJ 2000. The chromosomal location of the Bacillus subtilis sporulation gene spoIIR is important for its function. J Bacteriol 182:4425–4429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Gitai Z, Kinkhabwala A, Shapiro L, Moerner WE 2006. Single molecules of the bacterial actin MreB undergo directed treadmilling motion in Caulobacter crescentus. Proc Natl Acad Sci 103:10929–10934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam H, Schofield WB, Jacobs-Wagner C 2006. A landmark protein essential for establishing and perpetuating the polarity of a bacterial cell. Cell 124:1011–1023 [DOI] [PubMed] [Google Scholar]
- Laub MT, Chen SL, Shapiro L, McAdams HH 2002. Genes directly controlled by CtrA, a master regulator of the Caulobacter cell cycle. Proc Natl Acad Sci 99:4632–4637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laub MT, Shapiro L, McAdams HH 2007. Systems biology of Caulobacter. Annu Rev Genet 41:429–441 [DOI] [PubMed] [Google Scholar]
- Li R, Wai SC 2004. Bacterial cell polarity: A “swarmer-stalked” tale of actin. Trends Cell Biol 14:532–536 [DOI] [PubMed] [Google Scholar]
- Li Z, Trimble MJ, Brun YV, Jensen GJ 2007. The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO J 26:4694–4708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberman L, Berg HC, Sourjik V 2004. Effect of chemoreceptor modification on assembly and activity of the receptor-kinase complex in Escherichia coli. J Bacteriol 186:6643–6646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner AB, Madden R, Demarez A, Stewart EJ, Taddei F 2008. Asymmetric segregation of protein aggregates is associated with cellular aging and rejuvenation. Proc Natl Acad Sci 105:3076–3081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loisel TP, Boujemaa R, Pantaloni D, Carlier MF 1999. Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature 401:613–616 [DOI] [PubMed] [Google Scholar]
- Lowe J, Amos LA 1998. Crystal structure of the bacterial cell-division protein FtsZ. Nature 391:203–206 [DOI] [PubMed] [Google Scholar]
- Marston AL, Thomaides HB, Edwards DH, Sharpe ME, Errington J 1998. Polar localization of the MinD protein of Bacillus subtilis and its role in selection of the mid-cell division site. Genes Dev 12:3419–3430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascarenhas J, Weber MH, Graumann PL 2001. Specific polar localization of ribosomes in Bacillus subtilis depends on active transcription. EMBO Rep 2:685–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matroule JY, Lam H, Burnette DT, Jacobs-Wagner C 2004. Cytokinesis monitoring during development; rapid pole-to-pole shuttling of a signaling protein by localized kinase and phosphatase in Caulobacter. Cell 118:579–590 [DOI] [PubMed] [Google Scholar]
- McAuley KE, Fyfe PK, Ridge JP, Isaacs NW, Cogdell RJ, Jones MR 1999. Structural details of an interaction between cardiolipin and an integral membrane protein. Proc Natl Acad Sci 96:14706–14711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michie KA, Monahan LG, Beech PL, Harry EJ 2006. Trapping of a spiral-like intermediate of the bacterial cytokinetic protein FtsZ. J Bacteriol 188:1680–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay R, Huang KC, Wingreen NS 2008. Lipid localization in bacterial cells through curvature-mediated microphase separation. Biophys J 95:1034–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DE, Young KD 2000. Penicillin binding protein 5 affects cell diameter, contour, and morphology of Escherichia coli. J Bacteriol 182:1714–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen T, Yan AW, Gale G, Goldberg MB 2005. Presence of multiple sites containing polar material in spherical Escherichia coli cells that lack MreB. J Bacteriol 187:6187–6196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris V, den Blaauwen T, Doi RH, Harshey RM, Janniere L, Jimenez-Sanchez A, Jin DJ, Levin PA, Mileykovskaya E, Minsky A, et al. 2007. Toward a hyperstructure taxonomy. Annu Rev Microbiol 61:309–329 [DOI] [PubMed] [Google Scholar]
- Perry SE, Edwards DH 2004. Identification of a polar targeting determinant for Bacillus subtilis DivIVA. Mol Microbiol 54:1237–1249 [DOI] [PubMed] [Google Scholar]
- Perry SE, Edwards DH 2006. The Bacillus subtilis DivIVA protein has a sporulation-specific proximity to Spo0J. J Bacteriol 188:6039–6043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT 2004. BAR domains as sensors of membrane curvature: The amphiphysin BAR structure. Science 303:495–499 [DOI] [PubMed] [Google Scholar]
- Pichoff S, Lutkenhaus J 2005. Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Mol Microbiol 55:1722–1734 [DOI] [PubMed] [Google Scholar]
- Ping L, Weiner B, Kleckner N 2008. Tsr-GFP accumulates linearly with time at cell poles, and can be used to differentiate ‘old’ versus ‘new’ poles, in Escherichia coli. Mol Microbiol 69:1427–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradel N, Santini CL, Bernadac A, Shih YL, Goldberg MB, Wu LF 2007. Polar positional information in Escherichia coli spherical cells. Biochem Biophys Res Commun 353:493–500 [DOI] [PubMed] [Google Scholar]
- Ramamurthi KS, Clapham KR, Losick R 2006. Peptide anchoring spore coat assembly to the outer forespore membrane in Bacillus subtilis. Mol Microbiol 62:1547–1557 [DOI] [PubMed] [Google Scholar]
- Ramamurthi KS, Lecuyer S, Stone HA, Losick R 2009. Geometric cue for protein localization in a bacterium. Science 323:1354–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha EP 2008. The organization of the bacterial genome. Annu Rev Genet 42:211–233 [DOI] [PubMed] [Google Scholar]
- Romantsov T, Helbig S, Culham DE, Gill C, Stalker L, Wood JM 2007. Cardiolipin promotes polar localization of osmosensory transporter ProP in Escherichia coli. Mol Microbiol 64:1455–1465 [DOI] [PubMed] [Google Scholar]
- Romantsov T, Stalker L, Culham DE, Wood JM 2008. Cardiolipin controls the osmotic stress response and the subcellular location of transporter ProP in Escherichia coli. J Biol Chem 283:12314–12323 [DOI] [PubMed] [Google Scholar]
- Rosch J, Caparon M 2004. A microdomain for protein secretion in Gram-positive bacteria. Science 304:1513–1515 [DOI] [PubMed] [Google Scholar]
- Rudner DZ, Pan Q, Losick RM 2002. Evidence that subcellular localization of a bacterial membrane protein is achieved by diffusion and capture. Proc Natl Acad Sci 99:8701–8706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan KR, Huntwork S, Shapiro L 2004. Recruitment of a cytoplasmic response regulator to the cell pole is linked to its cell cycle-regulated proteolysis. Proc Natl Acad Sci 101:7415–7420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih YL, Kawagishi I, Rothfield L 2005. The MreB and Min cytoskeletal-like systems play independent roles in prokaryotic polar differentiation. Mol Microbiol 58:917–928 [DOI] [PubMed] [Google Scholar]
- Slaughter BD, Smith SE, Li R 2009. Symmetry breaking in the life cycle of the budding yeast. Cold Spring Harb Perspect Biol 1:a003384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart EJ, Madden R, Paul G, Taddei F 2005. Aging and death in an organism that reproduces by morphologically symmetric division. PLoS Biol 3:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szeto TH, Rowland SL, Habrukowich CL, King GF 2003. The MinD membrane targeting sequence is a transplantable lipid-binding helix. J Biol Chem 278:40050–40056 [DOI] [PubMed] [Google Scholar]
- Teleman AA, Graumann PL, Lin DC, Grossman AD, Losick R 1998. Chromosome arrangement within a bacterium. Curr Biol 8:1102–1109 [DOI] [PubMed] [Google Scholar]
- Theriot JA, Mitchison TJ, Tilney LG, Portnoy DA 1992. The rate of actin-based motility of intracellular Listeria monocytogenes equals the rate of actin polymerization. Nature 357:257–260 [DOI] [PubMed] [Google Scholar]
- van den Brink-van der Laan E, Boots JW, Spelbrink RE, Kool GM, Breukink E, Killian JA, de Kruijff B 2003. Membrane interaction of the glycosyltransferase MurG: A special role for cardiolipin. J Bacteriol 185:3773–3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veening JW, Stewart EJ, Berngruber TW, Taddei F, Kuipers OP, Hamoen LW 2008. Bet-hedging and epigenetic inheritance in bacterial cell development. Proc Natl Acad Sci 105:4393–4398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viollier PH, Thanbichler M, McGrath PT, West L, Meewan M, McAdams HH, Shapiro L 2004. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc Natl Acad Sci 101:9257–9262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss DS, Pogliano K, Carson M, Guzman LM, Fraipont C, Nguyen-Disteche M, Losick R, Beckwith J 1997. Localization of the Escherichia coli cell division protein Ftsl (PBP3) to the division site and cell pole. Mol Microbiol 25:671–681 [DOI] [PubMed] [Google Scholar]
- Wheeler RT, Shapiro L 1999. Differential localization of two histidine kinases controlling bacterial cell differentiation. Molecular Cell 4:683–694 [DOI] [PubMed] [Google Scholar]
- Wu J, Ohta N, Newton A 1998. An essential, multicomponent signal transduction pathway required for cell cycle regulation in Caulobacter. Proc Natl Acad Sci 95:1443–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young KD 2006. The selective value of bacterial shape. Microbiol Mol Biol Rev 70:660–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Lutkenhaus J 2003. Membrane binding by MinD involves insertion of hydrophobic residues within the C-terminal amphipathic helix into the bilayer. J Bacteriol 185:4326–4335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zupancic ML, Tran H, Hofmeister AE 2001. Chromosomal organization governs the timing of cell type-specific gene expression required for spore formation in Bacillus subtilis. Mol Microbiol 39:1471–1481 [DOI] [PubMed] [Google Scholar]