Abstract
Ascidian larvae have a hollow, dorsal central nervous system that shares many morphological features with vertebrate nervous systems yet is composed of very few cells. We show here that a null mutation in the gene dmrt1 in the ascidian Ciona savignyi results in profound abnormalities in the development of the sensory vesicle (brain), as well as other anterior ectodermal derivatives, including the palps and oral siphon primordium (OSP). Although the phenotype of the mutant embryos is variable, the majority have a complete loss of the most anterior structures (palps and OSP) and extensive disruption of sensory structures, such as the light-sensitive ocellus, in the sensory vesicle. dmrt1 is expressed early in the blastula embryo in a small group of presumptive ectodermal cells as they become restricted to anterior neural, OSP and palp fates. Despite the early and restricted expression of dmrt1, we were unable, using several independent criteria, to observe a defect in the mutant embryos until the early tailbud stage. We speculate that the variability and late onset in the phenotype may be due to partially overlapping activities of other gene products.
Keywords: Ascidian, Brain, Doublesex, Nervous system
INTRODUCTION
The tunicate and vertebrate central nervous systems (CNS) share a similar but divergent anatomy and ontogeny, reflecting their common chordate ancestry. The tunicate CNS has been most extensively studied in embryonic and larval ascidians (Imai and Meinertzhagen, 2007a; Imai and Meinertzhagen, 2007b; Lemaire et al., 2002; Meinertzhagen et al., 2004). The adult tunicate CNS is less well characterized but is highly divergent from those of other adult chordates. The ascidian larval nervous system is notable for its anatomical and functional simplicity in comparison to those found in vertebrates, and contains a limited number of sensory systems and behavioral outputs. The simplicity of the ascidian nervous system is also reflected in the early lineage restriction of the various domains of the brain and rostral nerve cord (Lemaire et al., 2002).
We describe here a mutant line in the ascidian Ciona savignyi that disrupts the development of anterior neural plate derivatives. The genetic lesion responsible for this phenotype maps to a member of the doublesex/mab-3-related (dmrt) family of transcription factors. The Drosophila doublesex and the Caenorhabditis elegans mab-3 genes share a common DNA-binding motif known as the DM domain. These genes were originally characterized for their roles in sex determination in flies and worms. Subsequently, DM domain-containing genes have also been found in most vertebrate species, many of which are expressed in the gonads and play a role in sex determination. Several members of the vertebrate dmrt family are expressed outside the gonads and are involved in a wider range of developmental processes (Hong et al., 2007). The DM-containing gene terra is involved in somite formation and left-right asymmetry in both zebrafish and chickens (Meng et al., 1999; Raya and Izpisua Belmonte, 2006). In Xenopus dmrt4 is expressed in the olfactory placodes and plays a role in the neurogenesis of the olfactory epithelium (Huang et al., 2005). Dmrt3 in mouse may play a similar role (Hong et al., 2007).
In the ascidian, dmrt1 is expressed in the anterior nervous system, which is derived from the anterior animal (a-Line) cells of the eight-cell-stage embryo. An FGF signal (presumably FGF 9/16/20) from vegetal blastomeres induces neural fate in the a-Line cells. FGF, along with foxA-α, activates several neural genes including otx, nodal and dmrt1 (Hudson and Lemaire, 2001; Kim and Nishida, 2001; Imai et al., 2006). Otx plays a crucial role in maintaining anterior neural identity and suppressing epidermal fate (Wada and Saiga, 1999). Recently, knockdown experiments in ascidians have shown that dmrt1, along with otx, plays a role in promoting the expression of six 1/2, six 3/6 and meis in the developing brain as well as promoting the expression of foxC, which promotes expression of palp-specific genes (Imai et al., 2006).
MATERIALS AND METHODS
C. savignyi genetics
Culturing of C. savignyi, mutagenesis, screening for recessive mutations, and amplified fragment length polymorphism (AFLP) linkage analysis were done as described previously (Hendrickson et al., 2004). Single nucleotide polymorphism (SNP) analysis was as described previously (Chiba et al., 2009).
Primers covering the entire C. savignyi dmrt1 gene were used to PCR amplify and sequence both wild-type and mutant genomic DNA. Primers 5′-CAAAACTAAGTTACGATTTGAAC-3′ (Dmrt Gup2) and 3′-GAACGACAACGCGTCATG-5′ (Dmrt Gdn7) identified a premature stop codon in exon 1 of the mutant allele.
A 25-mer antisense morpholino oligonucleotide (MO) (Gene Tools) was designed for bases 15 to 39 bp upstream of the C. savignyi dmrt1 presumptive start site: 5′-CTGTTTGCTATAATTTCTGTAACTC-3′ (dmrt-MO). Fertilized wild-type eggs were injected the dmrt-MO or control-MO, 5′-CCTCTTACCTCAGTTACAATTTATA-3′, as described (Yamada et al., 2003).
RT-PCR
For the dmrt1 developmental timecourse, total RNAs from each stage were extracted with Trizol (Invitrogen). One μg of total RNA was used for cDNA synthesis. Reverse transcription was done by SuperScript III first-strand synthesis system for RT-PCR (Invitrogen). Two μl of each first-strand cDNA were amplified 30 cycles, with the oligonucleotides 5′-GTCGCTCTTCGTCGCCAG-3′ and 5′-GAGATTGGTGGGCTTGAG-3′ for dmrt1, and 5′-AATGGATCCGGTATGTGC-3′ and 3′-AGCTTCTCTTTGATGTCACG-5′ for cytoplasmic actin.
To assess dmrt1 expression in ENU34 mutants, nested PCR was performed on total RNA from 40 wild-type or mutant mid-tailbud embryos using the oligonucleotides 5′-CAAGCGACAGAGGAGTTGAG-3′ and 5′-TGCCTGGTCATTACACGAAG-3′ for 20 cycles, then 5′-AGCTCATCCGCCCACAGTAG-3′ and 5′-AAGTCGGTGGAACAATGGAC-3′ for 25 cycles.
For six-3, otx, foxC RT-PCR in dmrt1-MO injected embryos (and control-MO injected; see above for sequences), total RNA was extracted from single early tailbud embryos or pools of four late gastrula embryos using Nucleospin XS RNA extraction kit (Clontech). RNA was reverse transcribed using SuperScript III first-strand synthesis system for RT-PCR (Invitrogen). Four microliters of each first-strand cDNA were amplified using one round of PCR and the oligonucleotides listed above for Actin. Nested PCR was performed for six 3/6 using the oligonucleotides 5′-CTAGCTGCCCTGTCTCTTTGTC-3′ and 5′-TGGCTTGAAGCTCACTACCA-3′, then 5′-CGTGGGCTAATTCCTTCTTT-3′ and 5′-CATTGCGGCCAGTTGATAAG-3′ for 35 cycles, for OTX using the oligonucleotides 5′-GCCGGTTGCTGTAGGAGTAG-3′ and 5′-TTCATGAGGGAGGAAGTTGC-3′ followed by 5′-TTCCGGAGCTACTTCCACTG-3′ and 5′-CTGCCAGAATCCAGAGTTCA-3′ for 25 cycles, and for FoxC using the oligonucleotides 5′-TTTGGTTGGATGTGTGCTGT-3′ and 5′-CGCAATGGCAATACAAAATG-3′ followed by 5′-GGAAGTCCCCTTCATCCAAT-3′ and 5′-AGCTACCTTCGCAGACGAAA-3′ for 30 cycles.
Immunostaining
Eggs from a dmrt1+/– adult were crossed to a line carrying a stably integrated transgene composed of the regulatory region of the C. savignyi Etr1 gene fused to the cDNA for Venus fluorescent protein (Veeman et al., 2010). Venus protein was visualized by immunostaining, and actin with Bodipy-FL phallacidin, as described previously (Veeman et al., 2008). Nuclei were stained with Draq5 and counted. Immunostaining for arrestin, cellular retinaldeyhyde-binding protein, opsin and synaptotagmin was as described previously (Tsuda et al., 2003; Sakurai et al., 2004; Takimoto et al., 2006).
In situ hybridization
In situ hybridization was done as described (Satou et al., 1995). For dmrt1, a ∼400 bp fragment that was PCR amplified with the oligonucleotides 5′-TTAAATACAGCGGGGATTGG-3′ and 5′-CATGACGCGTTGTCGTTC-3′ was used for probe synthesis, whereas for islet a ∼650 bp fragment that was PCR amplified with the oligonucleotides 5′-GCGGACGACACGGATGGGAT-3′ and 5′-CAGGTGGACACGACGCTCAT-3′ was used.
RESULTS
An anterior neural mutant maps to the doublesex/mab3-related-1 gene
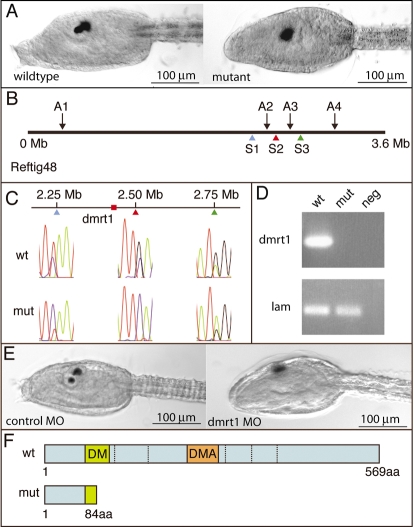
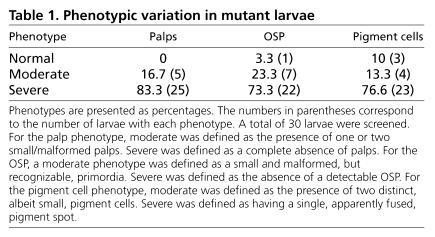
In screening ENU-mutagenized F1 C. savignyi (Moody et al., 1999) one line, ENU34, was selected for further study because of an obvious disruption in the development of the palps and the pigmented cells of the sensory vesicle (Fig. 1A). The allele responsible for the ENU34 phenotype behaved as a recessive mutation, and gave the expected 25% mutant progeny in crosses between heterozygous adults. While the mutant larvae were readily distinguishable from their wild-type siblings, there was some variation in the severity of the phenotype affecting the palps, pigments cells and oral siphon primordium (OSP) (Table 1). The OSP, also called the stomodeum, is a dorsal ectodermal derivative arising between the palps and brain (Satoh 1994; Manni et al., 2005). Despite the defects, the mutant larvae hatched at the same time as their wild-type siblings, and displayed no readily observable defects outside of the palps and brain.
Fig. 1.
Anterior neural mutant maps to dmrt1 gene. (A) Trunk region of wild-type and homozygous ENU34 mutant at larval stage. (B) Diagram of Reftig 48 showing linked AFLP (A1-A4) and SNP (S1-S3) markers. (C) Linkage analysis with SNP markers S1, S2 and S3. Representative traces are shown. (D) RT-PCR for dmrt1 and laminin α3/4/5 (lam; used as a loading control) in wild-type and homozygous ENU34 mutants. (E) A translation-blocking MO to dmrt1 phenocopies the ENU34 mutant (compare right embryo in E with right embryo in A). (F) Diagram of the predicted wild-type and ENU34 mutant C. savignyi Dmrt1 protein.
Table 1.
Phenotypic variation in mutant larvae
In order to find the genetic lesion responsible for the ENU34 phenotype, bulked segregated analysis was first used to identify amplified fragment length polymorphism (AFLP) markers linked to the mutant locus. Four linked markers (A1 to A4 in Fig. 1B) were located on a 3.6 Mb scaffold (Reftig 48) on Linkage Group 5 (Hill et al., 2008). Within Reftig 48 a region containing three clustered AFLP markers (A2 to A4) was further investigated by single nucleotide polymorphism (SNP) analysis, as we have described previously (Chiba et al., 2009). Mutant larvae from a self-fertilized heterozygous adult were collected into pools of five. Intronic DNA was amplified by PCR from the pools and sequenced to identify SNPs. DNA from phenotypically wild-type tadpoles (which would include heterozygous larvae) was also sequenced to identify SNPs. Three particularly informative SNPs (S1, S2 and S3) are shown in Fig. 1B,C. Both S1 and S3 showed weak linkage. For S1, recombination was observed in six pools of eleven tested; for S3, four of ten pools showed recombination. No recombination was observed in ten pools of five mutant tadpoles at S2. The SNP results thus define a 500 Kb region bounded by S1 and S3 that contains the genetic lesion responsible for the mutant phenotype. A candidate-gene approach was then used to identify the mutant gene.
The 500 Kb genomic region bounded by S1 and S3 is predicted to have 33 genes (http://www.ensembl.org). One gene near the center of this region, doublesex/mab3-related-1 (dmrt1), appeared to be a particularly strong candidate (Fig. 1C). Dmrt genes have been shown to be involved in neural induction and patterning in several chordates, including in C. intestinalis (Huang et al., 2005; Imai et al., 2006). When assayed by RT-PCR, we observed no detectable dmrt1 transcript in RNA samples from homozygous ENU34 tailbud embryos, whereas expression was observed in wild-type siblings (Fig. 1D). This observation is consistent with other mutants we have studied in Ciona, in which the expression of mutant genes are downregulated, presumably by a nonsense-mediated decay mechanism (Chiba et al., 2009; Jiang et al., 2005a; Jiang et al., 2005b; Veeman et al., 2008). An antisense MO was designed to block translation of C. savignyi dmrt1 transcript. Wild-type embryos injected with the dmrt1-MO appeared identical to homozygous ENU34 embryos at the larval stage (38 of 41 injected), whereas all embryos injected with a control MO developed normally (n=45) (representative larvae shown in Fig. 1E).
The C. savignyi dmrt1 gene is composed of six predicted exons that encode a 569 amino acid protein with two doublesex/mab3-related domains in exon 1 and exon 3, respectively (DM and DMA in Fig. 1F). Sequencing of genomic DNA from homozygous ENU34 larvae identified a G to A transition in the DM domain of exon 1 that would result in an early termination codon (TGG to TAG). The resulting predicted 84 amino acid protein from this transcript would lack both the DNA-binding DM domain and the DMA domain (Fig. 1F).
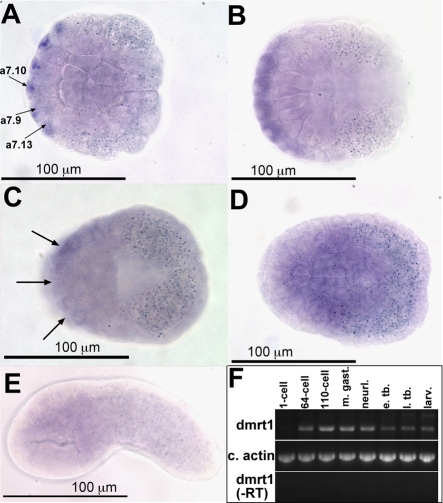
The expression of dmrt1 in the congeneric species C. intestinalis has been characterized previously by in situ hybridization (Imai et al., 2004). We found the early expression of C. savignyi dmrt1 to be indistinguishable from its ortholog in C. intestinalis. Expression was first observed at the 64-cell stage in the a7.9, a7.10 and a7.13 blastomere pairs (Fig. 2A). The a7.9 blastomeres are lineage-restricted by this stage to give rise to anterior brain and palps, whereas the a7.10 blastomeres are restricted to the OSP, brain and palps (Nishida, 1987). One daughter of the a7.13 blastomere (the a8.25 blastomere) becomes restricted to give rise to pigment and sensory vesicle cells. As in C. intestinalis, C. savignyi dmrt1 continues to be expressed in the brain, OSP and palp precursors at the 110-cell stage (Fig. 2B). Unlike in reports for C. intestinalis, faint staining was observed in the presumptive brain region of mid-gastrula [Hotta stage 12, Hotta et al. (Hotta et al., 2007)] C. savignyi embryos (arrows in Fig. 2C). Thus at the 64-, 110-cell and gastrula stages the expression pattern of dmrt1 corresponds precisely to the cell lineages disrupted in ENU34 mutant. In neither our studies of C. savignyi nor the previous studies in C. intestinalis was specific staining detectable in neurula embryos (Fig. 2D). In C. intestinalis it was observed that dmrt1 transcript, after declining to undetectable levels in gastrula and neurula stages, is then expressed in a small region of the brain at tailbud stage. We failed to detect similar expression in C. savignyi tailbud embryos (Hotta stage 20; Fig. 2E). To investigate this further we examined the timecourse of dmrt1 expression by RT-PCR (Fig. 2F). By this assay, dmrt1 expression in C. savignyi peaked at the 110-cell stage and persisted, but declined, through larval stages. Consistent with our in situ hybridization data, we did not observe a new peak of expression at the tailbud stage.
Fig. 2.
Expression of dmrt1. (A-E) Ciona savignyi dmrt1 in situ hybridization at 64-cell (A), 110-cell (B), mid-gastrula (C), neurula (D) and early tailbud (E) stages. Arrows in A indicate blastomeres with specific expression. The bilateral paired cells (a7.10, a7.0 and a7.13) are not marked. Arrows in C indicate faint staining in anterior ectoderm. (F) Timecourse of C. savignyi dmrt1 expression by RT-PCR at the indicated stages. e. tb., early tailbud; larv., larval stages; l. tb., late tailbud; m. gast., mid-gastrula; neurl., neurula.
In summary, several lines of evidence, including genetic linkage, phenocopy with MOs and expression data, allow us to conclude that the defects observed in the ENU35 mutant can be attributed to the premature stop codon found in the dmrt1 gene. Because of the severity of the truncation, as well as the absence of detectable transcript, we conclude that ENU34 has a null mutation in the dmrt1 gene. In the remainder of this manuscript the ENU34 locus will be referred to as dmrt1.
Timecourse of phenotypic onset
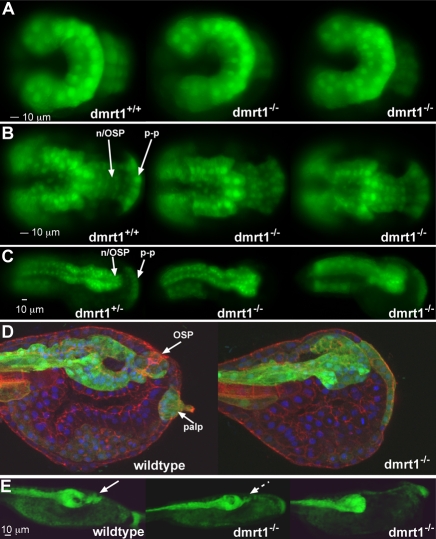
Our mutant line allowed us to examine in detail the spatial and temporal defects resulting from the complete loss of dmrt1. For this study we crossed the dmrt1 mutant line with a line carrying a stably integrated transgene composed of the regulatory region of the early pan-neural gene etr1 (Yagi and Makabe, 2001) driving expression of the fluorescent protein Venus (etr1-Venus) (Veeman et al., 2010). Expression of the transgene could be detected by antibody staining as early as the gastrula stage, whereas direct fluorescence of Venus could not be detected until the neurula stage. In examining the progeny of crossed heterozygous adults (dmrt+/–), we were unable to distinguish wild-type and mutant progeny at the late gastrula stage (Hotta stage 12-13) (Fig. 3A). However, genotyping of single transgenic embryos for the diagnostic G to A transition demonstrated that the expected fraction of embryos were homozygous for the mutation. Thus the earliest aspects of neural induction, as indicated by etr1-Venus expression, appear to occur normally in the dmrt1–/– mutants. An identical result was found at neurula stage (Hotta stage 15-16; Fig. 3B). The neural plates of dmrt1–/– embryos were indistinguishable from those of their wild-type or heterozygous siblings (either dmrt1+/+ or dmrt1+/–) both in overall morphogenesis and in etr1-Venus expression. At the neurula stage etr1 is expressed throughout the neural plate, which contains both the presumptive CNS, as well as the precursors of other anterior ectoderm derivatives such as the OSP and palps (Yagi and Makabe, 2001). At this stage, the palp precursors (`p-p' in Fig. 3B) make a characteristic fan-shaped structure at the anterior end of the neural plate. An identical structure was observed in dmrt1–/– embryos, suggesting that early morphogenesis of the palp precursors in the mutant was proceeding normally.
Fig. 3.
Brain and palp defects in dmrt1 mutants. (A-E) Central nervous system development at gastrula (A), neurula (B), early tailbud (C), late tailbud (D) and larval (E) stages. The genotypes of the embryos/larvae are indicated. n/OSP, neurohypophysis/OSP primordium; p-p, palp primordium.
At early tailbud stage, the dmrt1–/– embryos still could not be distinguished from sibling embryos by morphology. However, the etr1-Venus transgene expression pattern at this stage was consistently different in the dmrt1–/– embryos when compared with sibling embryos (Fig. 3C). For example, in the wild-type expression seen in the representative dmrt1+/– embryo, the precursors of the neurohypophysis and OSP (n/OSP in Fig. 3C) narrow and extend anteriorly, while the equivalent region of the dmrt1–/– embryos had a flat anterior boundary. We also observed by in situ hybridization that early tailbud dmrt1–/– embryos did not express the neurohypophysis/OSP marker six3/6 (n=30; data not shown), in agreement with previous reports (Imai et al., 2006). The extent of the defects in anterior neural plate derivatives caused by loss of dmrt1 is seen in confocal images of late-tailbud embryos immunostained for etr1-Venus and cytoskeletal actin. In the mid-sagittal section presented in Fig. 3D the OSP and one of the palps can be seen in the wild-type embryo. In the dmrt1–/– embryo the brain is severely shortened and is lacking the anterior protrusion making up the neurohypophysis that connects to the OSP, as well as the OSP itself (Fig. 3D). Venus protein could be detected in the anterior surface ectoderm of the dmrt1–/– tailbud embryos, indicating that the palp precursors are still present but have failed to differentiate (Fig. 3D). By the swimming larva stage (Fig. 3E) the trunk has extended and narrowed. The two dmrt1–/– larvae show the range of phenotypes observed, from having greatly diminished OSP and neurohypophysis (dashed arrow in middle larva, Fig. 3E), to having no detectable OSP and neurohypophysis (right larva). In dmrt1–/– embryos analyzed by confocal microscopy, small anterior protrusions of the brain corresponding to incomplete OSP and neurohypophyses were observed in eight of 30 larvae examined.
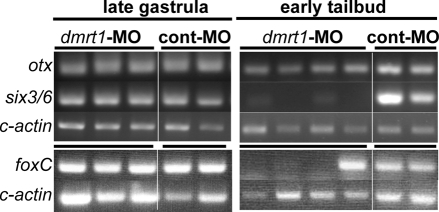
The lack of a detectable defect in etr1-Venus expression before the tailbud stage in the C. savignyi dmrt1–/– mutant (Fig. 3C) appears to differ from results in C. intestinalis in which MO knockdown of dmrt1 resulted in a significant decrease in foxC expression at gastrula stage (Imai et al., 2006). Although this difference may simply reflect the different genes examined, it may also indicate a species difference. In order to compare the two species directly, we performed a similar assay in C. savignyi in which dmrt1 was knocked down with a MO (Fig. 1E). Total RNA was prepared from single early tailbud embryos and from pools of four late gastrula embryos that had been injected with either the dmrt1-MO or control-MO (Fig. 1E). A subset of embryos from each morpholino injection session were allowed to grow to larval stage to confirm the efficacy of the knockdown. The expression levels of four genes were examined: otx; foxC; six3/6; and cytoplasmic actin (loading control). The level of expression of the four genes at late gastrula stage in the control MO and dmrt1 MO-injected embryos was indistinguishable (duplicate samples for the control-MO; and triplicate samples for the dmrt1-MO are shown; Fig. 4). However, by early tailbud stage the expression of six3/6 was greatly reduced in single embryos assayed (a representative four are shown). There also appeared to be a small decrease in the expression of otx at this stage. foxC expression was assayed in a separate MO injection series and was found to be greatly reduced in three of the four single dmrt1-MO injected embryos shown. Because we were assaying single embryos at tailbud stage, the difference in the dmrt1-MO injected embryos probably reflects embryo-to-embryo differences in success of microinjection. These RT-PCR results are consistent with the etr1-Venus results at gastrula and neurula stages (Fig. 3), and with our results for six3/6 expression in tailbud stage embryos by in situ hybridization, and suggest that dmrt1 is not essential for the initial expression of the genes assayed here, but rather for their continued expression.
Fig. 4.
Disrupted gene expression by dmrt1 knockdown. The expression of the genes otx, six3/6, c-actin and foxC were assayed by RT-PCR in single embryos at early tailbud stage, and in pools of four embryos at late gastrula stage. Embryos were injected at the one-cell stage with either a dmrt1-MO or a control-MO (cont-MO). All samples included a no-reverse transcriptase control, which showed no amplification product (data not shown).
palp defects in dmrt1–/– mutants
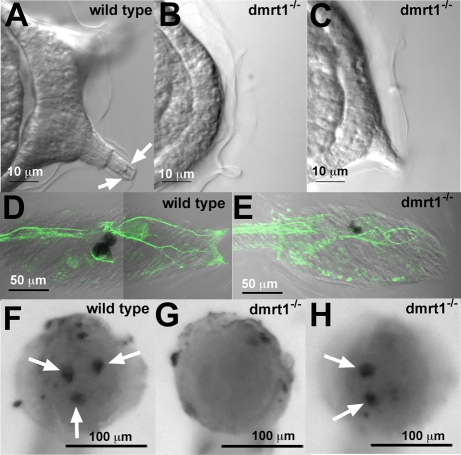
Wild-type Ciona larvae have three adhesive palps (two dorsal and one ventral) arranged in a triangular orientation on the anterior end of the larval trunk. They are composed of several distinct cell types derived from the anterior epidermis. Each of the three palps has two sensory neurons, each with a single dendrite extending anteriorly, as well as structural and secretory columnar cells (Fig. 5A). Homozygous dmrt1–/– larvae typically lack these cell types and appear to have a single layer of epidermal cells (Fig. 5B). Immunostaining with antibodies to synaptotagmin, a marker that is expressed in axons, show the palp neurons projecting to the brain in wild-type larvae (Fig. 5D). In dmrt1–/– larvae the palp neurons appear to be absent (Fig. 5E). However, axons projecting from the anterior epidermis were observed in all dmrt1–/– larvae examined (n=8), probably corresponding to projections from rostral trunk epidermal sensory neurons (Imai and Meinertzhagen, 2007b).
Fig. 5.
Palp defects in dmrt1 mutants. (A-C) Differential interference contrast images of the anterior end of wild-type (A) and dmrt–/– larvae (B,C). Arrows in A indicate endings of palp neurons. (D,E) Wild-type (D) and dmrt–/– (E) larvae stained with an antibody to synaptotagmin. (F-H) In situ hybridization for islet1 in late tailbud embryos (F, wild type; G and H, dmrt–/–). Arrows indicate specific hybridization to palp primordia.
To further characterize the development of the palps, the expression of the gene islet was examined by in situ hybridization in tailbud-stage embryos. Islet is expressed in the palp precursor cells and the sensory vesicle, beginning at the early tailbud stage. Islet production in the anterior trunk consists of two dorsal spots and a horizontal row of ventral cells that later become refined to a single spot in the middle and later tailbud stages (Giuliano et al., 1998). This triangular staining pattern corresponds to the location of the differentiated palps in the larvae (Fig. 5F). A total of 18 embryos from a self-fertilized dmrt1+/– adult were analyzed by in situ hybridization and subsequently genotyped by PCR. Fourteen embryos were either dmrt1+/– or dmrt1+/+, and showed the wild-type islet expression pattern. Of the four dmrt1–/– embryos, three showed only two spots of Islet production, and one had no production (Fig. 3G,H; and data not shown).
sensory vesicle defect in dmrt1–/– mutants
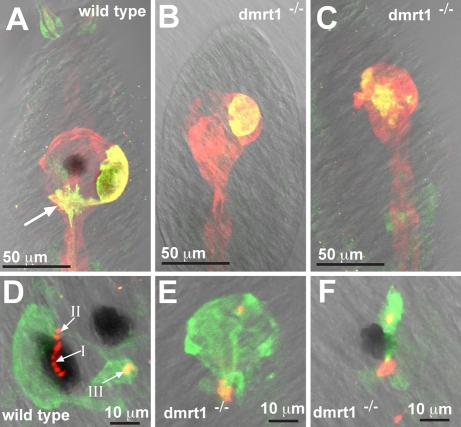
In addition to causing disruptions in the development of the palps and OSP, loss of dmrt1 also causes major disruption in the development of the sensory vesicle, which comprises the anterior brain and contains the photoreceptors (ocellus) and gravity-sensing organ (otolith). In 65% of dmrt–/– larvae, the otolith was missing (n=200), and when present, it was not properly pigmented (87%), whereas lens cells were missing or disorganized in 37% of larvae (n=200). Antibodies against Arrestin (Arr), cellular retinaldehyde-binding protein (CRALBP) and opsin were used to visualize organization of the sensory vesicle in wild-type and dmrt–/– larvae. In Ciona CRALBP is expressed broadly in the sensory vesicle and the neural tube, including the photoreceptors of the ocellus, whereas Arr is expressed exclusively in the photoreceptors. The cell bodies of the photoreceptors surround a single melanized cell on the right dorsolateral side in a hemispherical shape (Fig. 6A). Axons from these cells bundle posteriorly and then turn toward the midline near the posterior end of the sensory vesicle (arrow in Fig. 6A). Some dmrt1–/–larvae have well-organized photoreceptors on the right side of the sensory vesicle, but the entire group is often shifted anteriorly, giving the sensory vesicle an abnormal shape (Fig. 6B). More often, the photoreceptors are disorganized, often located throughout the sensory vesicle, and often decreased in number (Fig. 6C). In all dmrt1–/–larvae examined, axonal projections from the photoreceptors to the sensory vesicle were absent.
Fig. 6.
Brain defects in dmrt1 mutants. (A-C) Immunostained larvae for CRALBP (red) and Arrestin (yellow). (D-F) Immunostained larvae for Arrestin (green) and opsin-1 (red and yellow). Arrows in D indicate group I, II and III photoreceptors (Horie et al., 2008). The genotypes of all embryos are indicated.
Opsin-1 is produced in the outer segments of the photoreceptor cells. In wild-type larvae, there are three types of the photoreceptors: the group I photoreceptor cells are found within a cavity made by the pigmented ocellus cell; the group II photoreceptor cells are located outside the pigment cavity; and the group III photoreceptor cells are located near the otolith on the left ventral side of the brain vesicle (arrows Fig. 6D) (Horie et al., 2008). Photoreceptors in dmrt1–/– larvae do produce opsin-1, indicating they are properly differentiated, but there are fewer outer segments and their arrangement in the sensory vesicle is abnormal (Fig. 6E,F). Fig. S1 in the supplementary material shows opsin and arrestin staining for 14 dmrt1–/– larvae. Unlike the more anterior palps and OSP, we found no cases in which there was a complete absence of photoreceptor cells, although the number and pattern was highly variable.
DISCUSSION
Loss of dmrt1 function in C. savignyi results in profound disruptions in the development of the sensory vesicle, OSP and palps, all of which are derivatives of the anterior neural plate (Lemaire et al., 2002). The dmrt1 mutant line reported here, which is almost certainly a null based on presence of the premature stop codon early in the reading frame and the absence of detectable mutant transcript, allows for the characterization of the phenotype with a level of certainty that is not possible with MO knockdown. This is particularly important in cases such as observed here in which the phenotype is variable, and the timecourse of observed developmental defects does not correspond precisely to the timecourse of expression. We observed an apparent anterior to posterior difference in the degree of severity of the phenotype in which the most anterior of the affected organs, the palps, were in most cases completely absent. In those cases where palps could be identified, they were small and malformed. By contrast, the sensory organs and cells of the brain, including the photoreceptor and pigment cells, although malformed and reduced in number were always present.
It has been proposed that dmrt1 may function in parallel with otx (Imai et al., 2006), which may account for the late onset and variability of the phenotype. Overlapping activity from the other Ciona dmrt gene, dmrt2, is unlikely because it does not appear to be expressed during embryogenesis. Whereas ascidians have two dmrt genes, vertebrates have eight. Perhaps not surprisingly, the hermaphroditic ascidians do not appear to have a close ortholog of the vertebrate dmrt1, which is most strongly implicated in sex determination. Phylogenic analysis indicates that Ciona dmrt1 is the probable ortholog of vertebrate dmrt4 and dmrt5 (see Fig. S2 in the supplementary material). Interestingly, both vertebrate dmrt4 and dmrt5 are expressed in the brain and placodes, and MO knockdown of Xenopus dmrt4, which is expressed in the telencephalon and the olfactory placode, results in very specific defects in the olfactory placode (Huang et al., 2005). The relatively limited phenotype of dmrt4 knockdown in Xenopus compared with the ascidian phenotype described here may reflect the duplication and sub-functionalization of the dmrts following the split of the vertebrates and the tunicates.
Finally, dmrt1–/– larvae often lose the left-right asymmetry in the sensory vesicle. In wild-type larvae this asymmetry is most obvious in the two pigmented cells. These cells are specified to become pigmented at the 110-cell stage, but remain equivalent in their ability to differentiate into either the otolith or ocellus until after neural tube closure (Nishida and Satoh, 1989). During closure the cells are stochastic in their alignment along the anteroposterior axis. The anterior cell becomes the otolith and the posterior cell becomes the ocellus through a Notch/Delta-mediated mechanism (Hudson et al., 2007). By the tailbud stage, the otolith is located at the midline on the ventral floor of the sensory vesicle, whereas the ocellus is positioned posterior to the otolith on the right. In dmrt1–/– embryos the photoreceptor cells are often shifted to the anterior wall of the sensory vesicle, and the otolith is often missing or fused to the ocellus pigment cell in various locations in the sensory vesicle. This suggests that, like the terra genes in zebrafish and chick, dmrt1 has a role in establishing or maintaining left-right asymmetry. This function may be upstream or in conjunction with the Notch/Delta pathway. Future studies into the Notch/Delta and Nodal pathways (Hudson et al., 2007) in the dmrt1–/– mutant may reveal further insights into the patterning of the neural plate.
Supplementary Material
Acknowledgments
We thank Dr Di Jiang for helping with the mutagenesis screen. This work was supported by grants from the NIH (HD38701 and GM075049) to W.C.S., and from the Japanese Space Agency to M.T. Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.045302/-/DC1
References
- Chiba S., Jiang D., Satoh N., Smith W. C. (2009). Brachyury null mutant-induced defects in juvenile ascidian endodermal organs. Development 136, 35-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliano P., Marino R., Pinto M. R., De Santis R. (1998). Identification and developmental expression of Ci-isl, a homologue of vertebrate islet genes, in the ascidian Ciona intestinalis. Mech. Dev. 78, 199-202 [DOI] [PubMed] [Google Scholar]
- Hendrickson C., Christiaen L., Deschet K., Jiang D., Joly J. S., Legendre L., Nakatani Y., Tresser J., Smith W. C. (2004). Culture of adult ascidians and ascidian genetics. Methods Cell Biol. 74, 143-170 [DOI] [PubMed] [Google Scholar]
- Hill M. M., Broman K. W., Stupka E., Smith W. C., Jiang D., Sidow A. (2008). The C. savignyi genetic map and its integration with the reference sequence facilitates insights into chordate genome evolution. Genome Res. 18, 1369-1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong C., Park B., Saint-Jeannet J. (2007). The function of Dmrt genes in vertebrate development: it is not just about sex. Dev. Biol. 310, 1-9 [DOI] [PubMed] [Google Scholar]
- Horie T., Sakurai D., Ohtsuki H., Terakita A., Shichida Y., Usukura J., Kusakabe T., Tsuda M. (2008). Pigmented and nonpigmented ocelli in the brain vesicle of the ascidian larva. J. Comp. Neurol. 509, 88-102 [DOI] [PubMed] [Google Scholar]
- Hotta K., Mitsuhara K., Takahashi H., Inaba K., Oka K., Gojobori T., Ikeo K. (2007). A web-based interactive developmental table for the ascidian Ciona intestinalis, including 3D real-image embryo reconstructions: I. From fertilized egg to hatching larva. Dev. Dyn. 236, 1790-1805 [DOI] [PubMed] [Google Scholar]
- Huang X., Hong C. S., O'Donnell M., Saint-Jeannet J. P. (2005). The doublesex-related gene, XDmrt4, is required for neurogenesis in the olfactory system. Proc. Natl. Acad. Sci. USA 102, 11349-11354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson C., Lemaire P. (2001). Induction of anterior neural fates in the ascidian Ciona intestinalis. Mech. Dev. 100, 189-203 [DOI] [PubMed] [Google Scholar]
- Hudson C., Lotito S., Yasuo H. (2007). Sequential and combinatorial inputs from Nodal, Delta2/Notch and FGF/MEK/ERK signalling pathways establish a grid-like organisation of distinct cell identities in the ascidian neural plate. Development 134, 3527-3537 [DOI] [PubMed] [Google Scholar]
- Imai J. H., Meinertzhagen I. A. (2007a). Neurons of the ascidian larval nervous system in Ciona intestinalis: I. Central nervous system. J. Comp. Neurol. 501, 316-334 [DOI] [PubMed] [Google Scholar]
- Imai J. H., Meinertzhagen I. A. (2007b). Neurons of the ascidian larval nervous system in Ciona intestinalis: II. Peripheral nervous system. J. Comp. Neurol. 501, 335-352 [DOI] [PubMed] [Google Scholar]
- Imai K. S., Hino K., Yagi K., Satoh N., Satou Y. (2004). Gene expression profiles of transcription factors and signaling molecules in the ascidian embryo: towards a comprehensive understanding of gene networks. Development 131, 4047-4058 [DOI] [PubMed] [Google Scholar]
- Imai K. S., Levine M., Satoh N., Satou Y. (2006). Regulatory blueprint for a chordate embryo. Science 312, 1183-1187 [DOI] [PubMed] [Google Scholar]
- Jiang D., Munro E. M., Smith W. C. (2005a). Ascidian prickle regulates both mediolateral and anterior-posterior cell polarity of notochord cells. Curr. Biol. 15, 79-85 [DOI] [PubMed] [Google Scholar]
- Jiang D., Tresser J. W., Horie T., Tsuda M., Smith W. C. (2005b). Pigmentation in the sensory organs of the ascidian larva is essential for normal behavior. J. Exp. Biol. 208, 433-438 [DOI] [PubMed] [Google Scholar]
- Kim G. J., Nishida H. (2001). Role of the FGF and MEK signaling pathway in the ascidian embryo. Dev. Growth Differ. 43, 521-533 [DOI] [PubMed] [Google Scholar]
- Lemaire P., Bertrand V., Hudson C. (2002). Early steps in the formation of neural tissue in ascidian embryos. Dev. Biol. 252, 151-169 [DOI] [PubMed] [Google Scholar]
- Manni L., Agnoletto A., Zaniolo G., Burighel P. (2005). Stomodeal and neurohypophysial placodes in Ciona intestinalis: insights into the origin of the pituitary gland. J. Exp. Zool. B Mol. Dev. Evol. 304, 324-339 [DOI] [PubMed] [Google Scholar]
- Meinertzhagen I. A., Lemaire P., Okamura Y. (2004). The neurobiology of the ascidian tadpole larva: recent developments in an ancient chordate. Annu. Rev. Neurosci. 27, 453-485 [DOI] [PubMed] [Google Scholar]
- Meng A., Morre B., Tang H., Yuan B., Lin S. (1999). A Drosophila doublesex-related gene, terra, is involved in somitogenesis in vertebrates. Develoment 126, 1259-1268 [DOI] [PubMed] [Google Scholar]
- Moody R., Davis S. W., Cubas F., Smith W. C. (1999). Isolation of developmental mutants of the ascidian Ciona savignyi. Mol. Gen. Genet. 262, 199-206 [DOI] [PubMed] [Google Scholar]
- Nishida H. (1987). Cell lineage analysis in ascidian embryos by intracellular injection of a tracer enzyme. III. Up to the tissue restricted stage. Dev. Biol. 121, 526-541 [DOI] [PubMed] [Google Scholar]
- Nishida H., Satoh N. (1989). Determination and regualtion in the pigment cell lineage of the ascidian embryo. Dev. Biol. 132, 355-367 [DOI] [PubMed] [Google Scholar]
- Raya A., Izpisua Belmonte J. C. (2006). Left-right asymmetry in the vertebrate embryo; from early information to higher-level integration. Nat. Rev. Genet. 7, 283-293 [DOI] [PubMed] [Google Scholar]
- Sakurai D., Goda M., Kohmura Y., Horie T., Iwamoto H., Ohtsuki H., Tsuda M. (2004). The role of pigment cells in the brain of ascidian larva. J. Comp. Neurol. 475, 70-82 [DOI] [PubMed] [Google Scholar]
- Satoh N. (1994). Developmental Biology of Ascidians Cambridge: Cambridge University Press; [Google Scholar]
- Satou Y., Kusakabe T., Araki S., Satoh N. (1995). Timing of initiation of muscle-specific gene expression in the ascidian embryo precedes that of developmental fate restriction in lineage cells. Dev. Growth Differ. 37, 319-327 [DOI] [PubMed] [Google Scholar]
- Takimoto N., Kusakabe T., Horie T., Miyamoto Y., Tsuda M. (2006). Origin of the vertebrate visual cycle: III. Distinct distribution of RPE65 and beta-carotene 15,15′-monooxygenase homologues in Ciona intestinalis. Photochem. Photobiol. 82, 1468-1474 [DOI] [PubMed] [Google Scholar]
- Tsuda M., Kusakabe T., Iwamoto H., Horie T., Nakashima Y., Nakagawa M., Okunou K. (2003). Origin of the vertebrate visual cycle: II. Visual cycle proteins are localized in whole brain including photoreceptor cells of a primitive chordate. Vision Res. 43, 3045-3053 [DOI] [PubMed] [Google Scholar]
- Veeman M. T., Nakatani Y., Hendrickson C., Ericson V., Lin C., Smith W. C. (2008). Chongmague reveals an essential role for laminin-mediated boundary formation in chordate convergence and extension movements. Development 135, 33-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeman M. T., Newman-Smith E., El-Nachef D., Smith W. C. (2010). The ascidian mouth opening is derived from the anterior neuropore: Reassessing the mouth/neural tube relationship in chordate evolution. Dev. Biol. in press [DOI] [PubMed] [Google Scholar]
- Wada S., Saiga H. (1999). Vegetal cell fate specification and anterior neuroectoderm formation by Hroth, the ascidian homologue of orthodenticle/otx. Mech. Dev. 82, 67-77 [DOI] [PubMed] [Google Scholar]
- Yagi K., Makabe K. W. (2001). Isolation of an early neural maker gene abundantly expressed in the nervous system of the ascidian, Halocynthia roretzi. Dev. Genes Evol. 211, 49-53 [DOI] [PubMed] [Google Scholar]
- Yamada L., Shoguchi E., Wada S., Kobayashi K., Mochizuki Y., Satou Y., Satoh N. (2003). Morpholino-based gene knockdown screen of novel genes with developmental function in Ciona intestinalis. Development 130, 6485-6495 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.