Abstract
Background
We examined the association between twelve single nucleotide polymorphisms (SNPs) in the alpha-adducin (ADD1) and guanine nucleotide binding protein (G protein) beta polypeptide 3 (GNB3) genes and systolic (SBP), diastolic (DBP), and mean arterial (MAP) pressure responses to salt-intake.
Methods
A 7-day low-sodium (51.3 mmol sodium/day) followed by a 7-day high-sodium intervention (307.8 mmol sodium/day) was conducted among 1,906 Han participants from rural north China. BP measurements were obtained at baseline and the end of each intervention period using a random-zero sphygmomanometer.
Results
We identified a significant association between a rare ADD1 variant rs17833172 and SBP, DBP, and MAP responses to high-sodium (p-values<0.0001) and DBP response to low-sodium (p-value=0.002). Participants homozygous for the variant A allele of this marker had SBP, DBP, and MAP responses (95% confidence interval) to high-salt of 1.6 (−1.8, 4.9), −0.8 (−5.6, 4.0), and −0.1 (−4.0, 3.9) mmHg, respectively, versus corresponding responses of 4.6 (2.5, 6.6), 1.7 (−0.2, 3.6), and 2.7 (0.9, 4.4) mmHg, respectively, for those who were heterozygous or homozygous for the G allele. In addition, participants with at least one copy of the A allele of SNP rs1129649 of the GNB3 gene had significantly decreased MAP response to low-salt compared to homozygotes for the C allele (p-value=0.004) with responses of −3.4 (−3.8, −3.0) versus −4.2 (−4.6, −3.8) mmHg, respectively.
Conclusions
These data support a role for the ADD1 and GNB3 genes in BP salt-sensitivity. Future studies aimed at replicating these novel findings are warranted.
Keywords: blood pressure, genetics, polymorphism, dietary sodium, salt sensitivity, ADD1, GNB3
INTRODUCTION
Individuals with salt-sensitivity of blood pressure (BP) are usually unable to excrete excess sodium without experiencing a concurrent increase in arterial pressure 1, 2. These individuals are at increased risk for essential hypertension, cardiovascular disease (CVD), and premature death 3, 4. There is strong evidence that genetic mechanisms may underlie the inter-individual variation in BP response to salt intake 5–7. Genes encoding intracellular messenger proteins that influence renal sodium handling are biologically plausible candidate genes that could be involved with BP salt-sensitivity. Both the guanine nucleotide binding protein (G protein) beta polypeptide 3 (GNB3) and alpha-adducin (ADD1) genes have been implicated in BP salt-sensitivity due to their potential biological effects on sodium homeostasis via sodium-proton exchanger activity and renal tubular sodium reabsorption, respectively 7. Several reports have investigated the association between the GNB3 C825T and ADD1 Gly460Trp polymorphisms and BP responses to sodium 7. However, there have been no studies aimed at characterizing other variants in these genes that could play an important role in BP salt-sensitivity.
The current study was conducted in a large, homogeneous sample of Han Chinese participants, who took part in the Genetic Epidemiology Network of Salt-Sensitivity (GenSalt). The objective of our study was to examine the association between twelve single nucleotide polymorphisms (SNPs), which provided full coverage of the GNB3 and ADD1 genes in the Chinese population, and systolic (SBP), diastolic (DBP), and mean arterial (MAP) pressure responses to a dietary sodium intervention.
METHODS
Study Population
The GenSalt study was conducted in a Han Chinese population from rural areas of northern China where habitual salt-intake is high. A community-based BP screening was conducted among persons aged 18 to 60 years in the study villages to identify potential probands and their families. Those with a mean SBP between 130–160 mmHg and/or a DBP between 85–100 mmHg and no use of antihypertensive medications and their spouses, siblings and offspring were recruited for the dietary intervention study. Detailed eligibility criteria for the probands and spouses/siblings/offspring have been presented elsewhere 8. Individuals who had stage-2 hypertension, secondary hypertension, clinical cardiovascular disease, diabetes, chronic kidney disease, current use of antihypertensive or antidiabetic medications or insulin, pregnancy, heavy alcohol consumption, or current use of a low-sodium diet were excluded from the study. Among the 1,906 eligible participants from 637 families, 1,871 (98.2%) and 1,860 (97.6%) completed the low- and high-sodium interventions, respectively, and were included in the current analysis.
Institutional Review Boards at all of the participating institutions approved the GenSalt study. Written informed consents for the baseline observation and intervention program were obtained from each participant.
Dietary Intervention
The study participants received a 7-day low-sodium diet (3 grams of sodium chloride or 51.3 mmol of sodium per day) followed by a 7-day high-sodium diet (18 grams of sodium chloride or 307.8 mmol of sodium per day). During the period of sodium intervention, other dietary nutrient intake remained unchanged. Total energy intake was varied according to each participant’s baseline energy intake. All study foods were cooked without salt, and pre-packaged salt was added to the individual study participant’s meal when it was served by the study staff. To ensure study participants’ compliance to the intervention program, they were required to have their breakfast, lunch and dinner at the study kitchen under supervision of the study staff during the entire study period. The study participants were instructed to avoid consuming any foods that were not provided by study personnel. Three timed urinary specimens (one 24–hour and two overnight) were collected at baseline and at the end of each phase of intervention (days 5, 6, and 7) to monitor each participants’ compliance with their dietary sodium intervention. The timed overnight urinary excretions of sodium and potassium were converted to 24-hour values based on a formula developed from data in this study. The results from the 24-hour urinary excretions of sodium showed excellent compliance with the study diet: the mean (standard deviation) 24-hour urinary excretions of sodium and potassium were 242.4 (66.7) mmol and 36.9 (9.6) mmol at baseline, 47.5 (16.0) and 31.4 (7.7) at the end of the low-sodium intervention period, and 244.3 (37.7) and 35.7 (7.5) at the end of the high-sodium intervention period, respectively.
Phenotype Measurement
A standard questionnaire was administered by trained staff at the baseline examination to collect information on family structure, demographic characteristics, personal and family medical history, and lifestyle risk factors. Three morning BP measurements were obtained according to a standard protocol during each of the 3-days of baseline observation and on days 5, 6 and 7 of each intervention period by trained and certified observers using a random–zero sphygmomanometer 9. BP was measured with the participant in the sitting position after 5 minutes of rest. In addition, participants were advised to avoid alcohol, cigarette smoking, coffee/tea, and exercise for at least 30 minutes prior to their BP measurements. All BP observers were blinded to the participant’s dietary intervention. Body weight and height were measured twice in light indoor clothing without shoes during the baseline examination. Body mass index (BMI) was calculated as kilograms per meters squared (kg/m2).
Mean SBP, DBP and MAP responses to low-sodium were calculated as the mean of 9 measurements on days 5, 6 and 7 during the low-sodium intervention minus the mean of 9 measurements at baseline. Similarly, responses to high-sodium were calculated as the mean of 9 measurements on days 5, 6 and 7 during the high-sodium intervention minus the mean of 9 measurements on days 5, 6 and 7 during the low-sodium intervention.
Candidate Gene and SNP Selection and Genotyping
Of 30 candidate genes selected from various biological pathways involved in BP regulation and genotyped as part of the GenSalt study, the current analysis focused on two candidate genes in the intracellular messenger pathway, and included the ADD1 and GNB3 genes. Tagger software was used to identify tagSNPs in these candidate genes based on the empirical patterns of linkage disequilibrium (LD) structure in the Chinese population, as determined by the Chinese HapMap 10, 11. In addition, functional SNPs were chosen based on previously reported associations with BP. SNP genotyping was performed using SNPlex assays (Applied Biosystems) based on the oligonucleotide ligation assay (OLA) for capillary electrophoresis on an automated DNA Sequencer (ABI 3700 DNA Analyzer).
Of the fifteen genotyped SNPs, two monomorphic SNPs and one with a low genotyping call rate (54.2%) were excluded from the analysis. The Supplemental Table lists the chromosomal and physical locations, major and minor alleles, genotyping call rate, minor allele frequency (MAF), and Hardy-Weinberg (HW) p-value for the remaining twelve SNPs examined in this analysis. Likely due to its low MAF, rs17833172 deviated significantly from HW-equilibrium, with 197 expected compared to 185 observed heterozygotes (p=0.003).
Statistical Analysis
ASPEX and GRR were used to check for potential misreported relationships in the GenSalt family pedigrees 12, 13. The Mendelian consistency of the SNP genotype data was assessed by PLINK and PedCheck 14, 15, and HW-equilibrium for each SNP was examined using the PEDSTATS procedure, as implemented in Merlin 16. The genotyping call rate and MAF were also assessed for each SNP.
The mean or percentage of each baseline characteristic and BP response variable was calculated for all study participants. The association between SNP genotypes and BP responses to low- and high-sodium interventions were examined in codominant, additive, dominant and recessive genetic models using a mixed linear model. A sandwich estimator was used to account for the non-independence of family members. Age, gender, BP measurement room temperature, and study site were adjusted in multivariable analyses. To adjust for multiple comparisons, the false discovery rate (FDR) q-value was calculated for all SNPs 17. For those SNPs with a q-value<0.05 for at least one BP response phenotype, we used a mixed linear model to estimate the mean effect size and 95% confidence interval (CI) of each genotype using the appropriate genetic model. These analyses were conducted using SAS statistical software (version 9.1; SAS Institute Inc).
RESULTS
On average, participants were 38.7 years of age, had a BMI of 23.3 kg/m2 and mean SBP, DBP and MAP of 116.9, 73.7, and 88.1 mmHg, respectively, at the baseline examination (Table 1). Approximately 53% of the study population was male. SBP, DBP and MAP responses were normally distributed with average responses of −5.5, −2.8, and −3.7 mmHg, respectively, for the low-sodium intervention and 4.9, 1.9, and 2.9 mmHg, respectively, for the high-sodium intervention.
Table 1.
Characteristics of 1,906 study participants.
| Variable | Mean ± SD or Percentage |
Range |
|---|---|---|
| Age, yrs | 38.7 ± 9.6 | (15.0, 62.0) |
| Male, % | 53.0 | … |
| BMI, kg/m2 | 23.3 ± 3.2 | (14.5, 37.8) |
| Baseline BP, mm Hg | ||
| Systolic | 116.9 ± 14.2 | (80.9, 176.9) |
| Diastolic | 73.7 ± 10.3 | (38.9, 109.3) |
| Mean arterial pressure | 88.1 ± 10.9 | (58.7, 126.4) |
| BP response to low salt, mm Hg | ||
| Systolic | −5.5 ± 7.0 | (−45.8, 24.2) |
| Diastolic | −2.8 ± 5.5 | (−23.1, 18.2) |
| Mean arterial pressure | −3.7 ± 5.3 | (−28.8, 15.2) |
| BP response to high salt, mm Hg | ||
| Systolic | 4.9 ± 6.0 | (−11.3, 36.2) |
| Diastolic | 1.9 ± 5.4 | (−20.0, 27.1) |
| Mean arterial pressure | 2.9 ± 5.0 | (−14.4, 30.1) |
BMI = body mass index; BP = blood pressure; SD = standard deviation
P-value associations between each SNP and SBP, DBP and MAP responses to the low-sodium intervention are presented in Table 2. Markers rs17833172 and rs3775067 of the ADD1 gene and markers rs4963516, rs1129649, and rs3213431 of the GNB3 gene were significantly associated with SBP, DBP or MAP responses to low-sodium in at least one of the genetic models tested. After adjustment for multiple comparisons, marker rs17833172 remained significantly associated with DBP response in both the codominant and recessive genetic models (q-values=0.04 and 0.02, respectively). In addition, rs1129649 remained significantly associated with MAP responses in the dominant genetic model (q-value=0.05).
Table 2.
The association between SNPs and blood pressure response to low-sodium intervention.
| Gene | SNP | Codominant | Additive | Dominant | Recessive |
|---|---|---|---|---|---|
| P-Value | P-Value | P-Value | P-Value | ||
| Systolic Blood Pressure | |||||
| ADD1 | rs4690002 | 0.68 | 0.96 | 0.62 | 0.61 |
| rs12503220 | 0.24 | 0.97 | 0.69 | 0.13 | |
| rs1263359 | 0.45 | 0.21 | 0.25 | 0.43 | |
| rs17833172 | 0.81 | 0.78 | 0.81 | 0.51 | |
| rs3775067 | 0.03 | 0.04 | 0.008 | 0.88 | |
| rs4961 | 0.47 | 0.26 | 0.23 | 0.56 | |
| rs4963 | 0.44 | 0.22 | 0.22 | 0.44 | |
| GNB3 | rs4963516 | 0.21 | 0.09 | 0.21 | 0.13 |
| rs1129649 | 0.09 | 0.05 | 0.03 | 0.42 | |
| rs3213431 | 0.03 | 0.67 | 0.26 | 0.02 | |
| rs2301339 | 0.16 | 0.06 | 0.07 | 0.29 | |
| rs5446 | 0.90 | 0.98 | 0.89 | 0.70 | |
| Diastolic Blood Pressure | |||||
| ADD1 | rs4690002 | 0.94 | 0.76 | 0.89 | 0.72 |
| rs12503220 | 0.15 | 0.21 | 0.10 | 0.54 | |
| rs1263359 | 0.41 | 0.18 | 0.22 | 0.37 | |
| rs17833172 | 0.003† | 0.34 | 0.29 | 0.002† | |
| rs3775067 | 0.08 | 0.05 | 0.02 | 0.58 | |
| rs4961 | 0.21 | 0.08 | 0.16 | 0.14 | |
| rs4963 | 0.21 | 0.08 | 0.13 | 0.18 | |
| GNB3 | rs4963516 | 0.06 | 0.02 | 0.08 | 0.04 |
| rs1129649 | 0.02 | 0.009 | 0.004 | 0.38 | |
| rs3213431 | 0.26 | 0.41 | 0.58 | 0.11 | |
| rs2301339 | 0.22 | 0.08 | 0.18 | 0.14 | |
| rs5446 | 0.48 | 0.24 | 0.23 | 0.68 | |
| Mean Arterial Pressure | |||||
| ADD1 | rs4690002 | 0.86 | 0.84 | 0.91 | 0.64 |
| rs12503220 | 0.17 | 0.38 | 0.20 | 0.31 | |
| rs1263359 | 0.33 | 0.14 | 0.17 | 0.35 | |
| rs17833172 | 0.19 | 0.60 | 0.54 | 0.12 | |
| rs3775067 | 0.03 | 0.03 | 0.007 | 0.67 | |
| rs4961 | 0.23 | 0.09 | 0.14 | 0.22 | |
| rs4963 | 0.21 | 0.08 | 0.11 | 0.22 | |
| GNB3 | rs4963516 | 0.06 | 0.02 | 0.08 | 0.04 |
| rs1129649 | 0.02 | 0.008 | 0.004† | 0.34 | |
| rs3213431 | 0.12 | 0.68 | 0.96 | 0.04 | |
| rs2301339 | 0.15 | 0.05 | 0.09 | 0.15 | |
| rs5446 | 0.68 | 0.44 | 0.38 | 0.91 | |
ADD1 = Alpha-adducin; GNB3 = Guanine nucleotide binding protein beta polypeptide 3
False discovery rate q-value < 0.05
Table 3 presents p-value results from the high-sodium intervention. In the ADD1 gene, rs17833172 was significantly associated with SBP, DBP and MAP responses to high-sodium in the codominant and recessive genetic models, even after adjustment for multiple comparisons (all q-values<0.0001). Additionally, markers rs3213431 and rs2301339 of the GNB3 gene were significantly associated with SBP responses to high-sodium, but did not retain significance after adjusting for multiple testing.
Table 3.
The association between SNPs and blood pressure response to high-sodium intervention.
| Gene | SNP | Codominant | Additive | Dominant | Recessive |
|---|---|---|---|---|---|
| P-Value | P-Value | P-Value | P-Value | ||
| Systolic Blood Pressure | |||||
| ADD1 | rs4690002 | 0.16 | 0.25 | 0.06 | 0.94 |
| rs12503220 | 0.33 | 0.79 | 0.56 | 0.20 | |
| rs1263359 | 0.95 | 0.92 | 0.81 | 0.91 | |
| rs17833172 | 1.33E-13† | 0.13 | 0.19 | 1.77E-25† | |
| rs3775067 | 0.51 | 0.35 | 0.25 | 0.87 | |
| rs4961 | 0.79 | 0.59 | 0.82 | 0.49 | |
| rs4963 | 0.71 | 0.55 | 0.86 | 0.41 | |
| GNB3 | rs4963516 | 0.46 | 0.26 | 0.48 | 0.24 |
| rs1129649 | 0.37 | 0.16 | 0.24 | 0.28 | |
| rs3213431 | 0.04 | 0.07 | 0.02 | 0.26 | |
| rs2301339 | 0.06 | 0.03 | 0.02 | 0.24 | |
| rs5446 | 0.87 | 0.62 | 0.68 | 0.66 | |
| Diastolic Blood Pressure | |||||
| ADD1 | rs4690002 | 0.75 | 0.57 | 0.44 | 0.94 |
| rs12503220 | 0.14 | 0.32 | 0.15 | 0.23 | |
| rs1263359 | 0.72 | 0.42 | 0.46 | 0.59 | |
| rs17833172 | 1.70E-07† | 0.82 | 0.95 | 2.11E-10† | |
| rs3775067 | 0.31 | 0.12 | 0.18 | 0.30 | |
| rs4961 | 0.81 | 0.52 | 0.63 | 0.58 | |
| rs4963 | 0.78 | 0.51 | 0.50 | 0.70 | |
| GNB3 | rs4963516 | 0.52 | 0.77 | 0.78 | 0.31 |
| rs1129649 | 0.22 | 0.30 | 0.13 | 0.70 | |
| rs3213431 | 0.86 | 0.80 | 0.90 | 0.59 | |
| rs2301339 | 0.97 | 0.82 | 0.87 | 0.82 | |
| rs5446 | 0.52 | 0.25 | 0.30 | 0.43 | |
| Mean Arterial Pressure | |||||
| ADD1 | rs4690002 | 0.40 | 0.36 | 0.18 | 0.98 |
| rs12503220 | 0.16 | 0.42 | 0.21 | 0.19 | |
| rs1263359 | 0.80 | 0.53 | 0.52 | 0.73 | |
| rs17833172 | 1.82E-10† | 0.45 | 0.56 | 1.16E-16† | |
| rs3775067 | 0.31 | 0.13 | 0.14 | 0.44 | |
| rs4961 | 0.97 | 0.81 | 0.80 | 0.90 | |
| rs4963 | 0.89 | 0.81 | 0.66 | 0.96 | |
| GNB3 | rs4963516 | 0.46 | 0.49 | 0.93 | 0.22 |
| rs1129649 | 0.29 | 0.19 | 0.12 | 0.88 | |
| rs3213431 | 0.45 | 0.55 | 0.39 | 0.41 | |
| rs2301339 | 0.57 | 0.31 | 0.31 | 0.54 | |
| rs5446 | 0.62 | 0.33 | 0.38 | 0.47 | |
ADD1 = Alpha-adducin; GNB3 = Guanine nucleotide binding protein beta polypeptide 3
False discovery rate q-value < 0.05
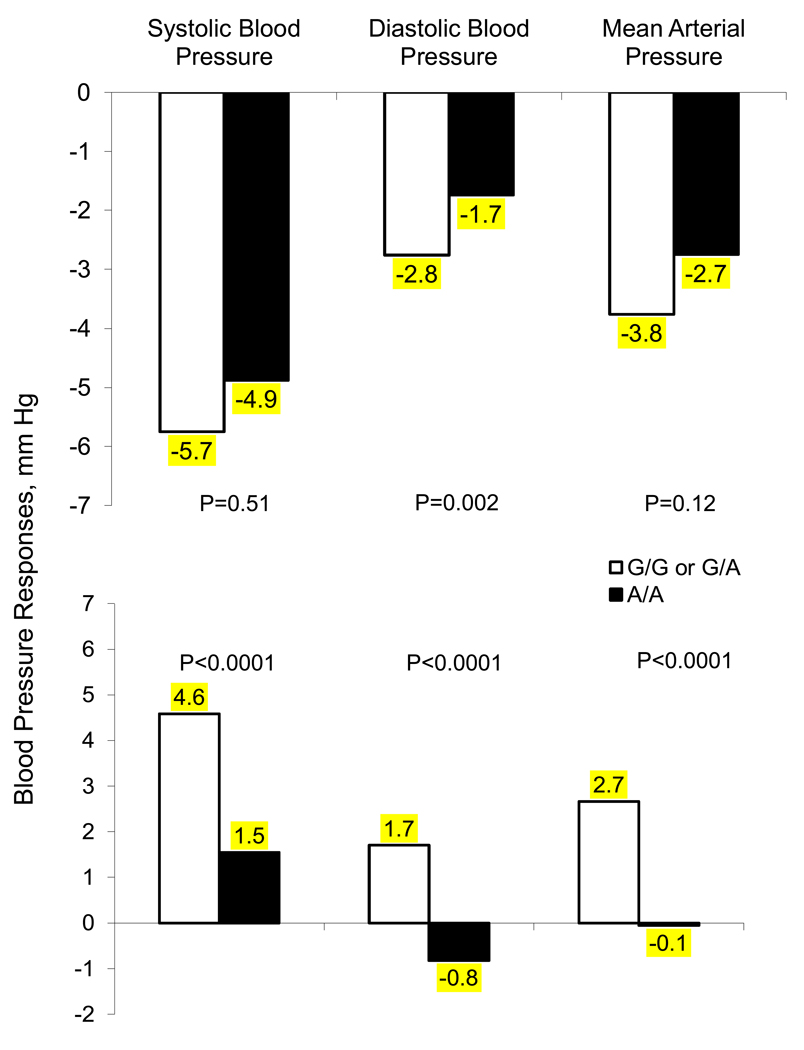
The SBP, DBP and MAP responses to low- and high-sodium intervention for participants homozygous for the A allele of the rs17833172 marker of the ADD1 gene compared to those with one or less copies are presented in Figure 1. While both the codominant and recessive models were significant for this variant, assessment of the p-values for these models and mean effect sizes for each genotype indicated a potentially better fit of the recessive model. Despite the low MAF of the A allele (4%), participants that were homozygous for this variant had a decreased response to the low- and high-sodium interventions compared to those with who were heterozygous or homozygous for the common G allele. Responses (95% CI) to the low-sodium intervention for those with G/G or G/A genotypes compared to A/A genotype were −5.8 (−8.3, −3.2) vs. −4.9 (−26.2, 16.5) for SBP, −2.8 (−4.9, −0.6) vs. −1.7 (−9.5, 6.0) for DBP, and −3.8 (−5.8, −1.7) vs. −2.8 (−15.2, 9.7) for MAP; and for the high-sodium intervention were 4.6 (2.5, 6.6) vs. 1.6 (−1.9, 4.9) for SBP, 1.7 (−0.2, 3.6) vs. −0.8 (−5.6, 4.0) for DBP, and 2.7 (0.9, 4.4) vs. −0.1 (−4.0, 3.9) for MAP.
Figure 1.
Absolute change (mm Hg) in systolic (SBP), diastolic (DBP), and mean arterial pressure (MAP) from the baseline to low (upper) and low to high (lower) sodium intervention according to SNP rs17833172 genotypes of the ADD1 gene.
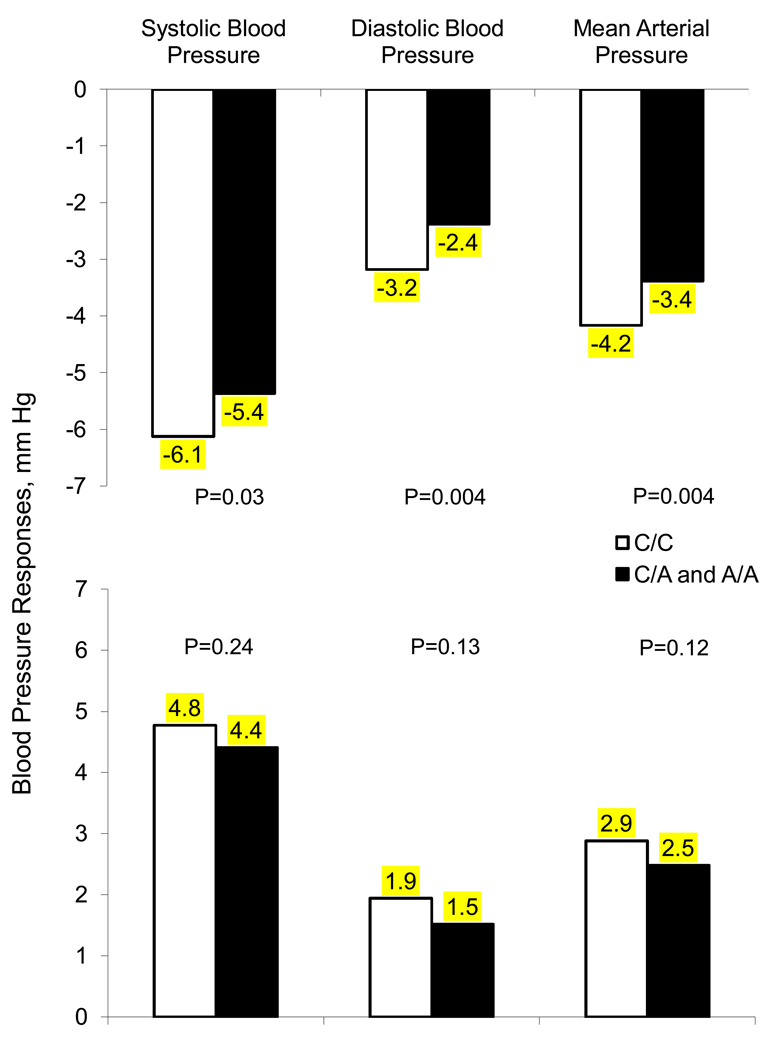
Figure 2 presents mean SBP, DBP and MAP responses to low- and high-sodium intervention among participants who had 0 compared to 1 or more copies of the minor A allele of the GNB3 marker rs1129649. Those with 1 or more copies of the A allele had a decreased response to sodium intervention. Mean SBP responses to low-sodium were −5.4 (−5.9, −4.9) vs. −6.1 (−6.7, −5.6), DBP −2.4 (−2.8, −1.9) vs. −3.2 (−3.6, −2.8), and MAP −3.4 (−3.8, −3.0) vs. −4.2 (−4.6, −3.8), respectively, for those with the C/A or A/A genotypes compared to those with C/C genotype. Similarly, for the high-sodium intervention, those who had the C/A or A/A genotypes compared to the C/C genotypes had responses of 4.4 (4.0, 4.9) vs. 4.8 (4.3, 5.2) for SBP, 1.5 (1.1, 1.9) vs. 1.9 (1.5, 2.3) for DBP, and 2.5 (2.1, 2.9) vs. 2.9 (2.5, 3.3) for MAP.
Figure 2.
Absolute change (mm Hg) in systolic (SBP), diastolic (DBP), and mean arterial pressure (MAP) from the baseline to low (upper) and low to high (lower) sodium intervention according to SNP rs1129649 genotypes of the GNB3 gene.
DISCUSSION
We identified novel genetic variants in the ADD1 and GNB3 genes that may be important predictors of salt-sensitivity of BP. ADD1 marker rs17833172 was strongly associated with SBP, DBP and MAP responses to a high dietary sodium intervention, and was also associated with DBP response to low-sodium intervention. Participants homozygous for the minor A allele appeared to have a significantly decreased response to salt-intake compared to those who were heterozygous or homozygous for the common G allele. Moreover, SNP rs1129649 of the GNB3 gene was significantly predictive of MAP response to low-sodium, with the minor A allele significantly decreasing MAP response to sodium intervention. The positive findings reported here could have important clinical and public health implications and warrant the need for further investigation of these variants.
Our study has several important strengths. To date, this is largest dietary intervention study to examine the association between genetic variants in the ADD1 and GNB3 genes and BP response to dietary sodium intervention. This is also the first study to examine salt-sensitivity in relation to multiple tag-SNPs and previously reported SNPs that provide full coverage of the ADD1 and GNB3 genes. Study attributes, including the recruitment of a homogenous sample of Han Chinese participants, makes our analysis robust to population stratification. Additionally, study participants were similar with respect to lifestyle and environmental risk factors, including diet and physical activity. Thus, confounding due to these exposures should be minimized. Moreover, we controlled for the fixed effects of age, gender, field center and room temperature in our analysis. An additional adjustment for BMI was conducted, and the results were similar to those presented in the current study. Participation in the dietary interventions was high (98.2% and 97.6%, respectively, for the low- and high-sodium interventions), and compliance with the study interventions, as assessed by urinary excretion of sodium during each intervention period, was excellent. Finally, stringent quality control procedures were employed during measurement of BP and the other study covariables, conduct of the dietary interventions, genotyping, and marker data cleaning. The limitations of our study include the relatively short duration of dietary sodium interventions. However, in a recent study among 36 Olivetti Heart Study participants an increased incidence of hypertension during 15-years of follow-up was noted among individuals with high salt-sensitivity of BP as defined by their response to a 3-day period of reduced salt intake18.
ADD1 has been a target for candidate gene studies of BP-related phenotypes because of its role in modulating the reabsorption of sodium in renal tubular cells 19. In the current study, we observed an association between the rs17833172 variant of the ADD1 gene and SBP, DBP, and MAP responses to a high-sodium intervention and DBP response to a low-sodium intervention. Two copies of the A allele of rs17833172 significantly decreased response to salt-intake. Despite the potential importance of the rs17833172 variant on BP salt-sensitivity, a word of caution regarding our result is warranted. This study represents the first examination of rs17833172, a variant that had a low MAF in the Chinese population. We highlight the need for replication of this result in other study populations. In addition, future studies of unique ADD1 variants may provide important information in relation to BP salt-sensitivity.
Many studies have examined the effects of the ADD1 Gly460Trp polymorphism (rs4961) on BP salt-sensitivity 1, 7, 20–25, BP responses to thiazide diuretics 20, 26–29, and essential hypertension 20, 21, 30, 31. While several studies have reported increased salt-sensitivity of BP among participants with the 460Tryp variant 1, 20, 23–25, the current study did not find such an association. Our results are similar to those of two separate reports of negative findings 21, 22. These differences may be explained by phenotypic discrepancies between studies. In our study and the study by Castejon and colleagues 21, salt-sensitivity was determined by BP response to a dietary sodium intervention. In contrast, most previous studies have used an acute salt-loading and depletion protocol to measure salt-sensitivity 20, 23–25. The discrepant findings reported here may also reflect population heterogeneity, as the LD structure of the Han Chinese population examined here may be different to that of the Caucasion populations examined in the majority of previous studies 1, 20, 23–25.
In this study, participants with at least one A allele of the GNB3 marker rs1129649 had a significantly decreased MAP response to low-sodium intervention. The novel finding reported here could have important implications and warrants replication. Marker rs1129649 lies in the 5’ region of the GNB3 gene. Additionally, it has been reported as both a nonsense and missense mutation of exon 15 in the leprecan-like 2 (LEPREL2) gene 32, 33. It should be noted that the GNB3 and LEPREL2 genes overlap on chromosome 12 32, 33. Functional studies will be important to determine whether this SNP influences the transcription and protein levels of the GNB3 gene or the structure of the LEPREL2 protein, and whether these changes may be associated with BP-related phenotypes.
Although the novel finding reported here has not been identified previously, many studies have examined the association between another GNB3 polymorphism, the C825T variant and BP-related phenotypes. Studies have indicated a significant association between this SNP and both essential hypertension and response to thiazide diuretics 34, 35. However, past examinations of salt-sensitivity phenotypes have not supported such an association 7, 36–38. The current study did not examine C825T specifically, but did investigate the rs2301339 variant of GNB3. Marker rs2301339 is in perfect linkage disequilibrium with the C825T polymorphism (D’ and r2 = 1.0) in the HapMap Chinese (CHB) and Caucasian (CEU) populations. Interestingly, this SNP was significantly associated with SBP response to high-sodium, and was marginally associated with SBP, DBP, and MAP responses to low-sodium in our analysis. Although these findings did not retain significance after adjustment for multiple testing, our results combined with positive findings of an association between C825T and response to thiazide diuretics warrants the need for more research of the effect of this variant on BP response to salt-intake.
In conclusion, we identified an association between two variants of the ADD1 and GNB3 genes and BP response to a sodium intervention that remained significant after adjustment for multiple comparisons. Our results are in support of a role for the ADD1 and GNB3 genes in BP salt-sensitivity and could have important clinical and public health implications. Future studies aimed at replicating these novel findings in other populations are warranted.
Supplementary Material
Acknowledgments
The Genetic Epidemiology Network of Salt Sensitivity (GenSalt) is supported by a cooperative agreement project grant (U01HL072507) from the National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, Maryland.
Footnotes
Disclosure
We declare that we have no conflict of interest to disclose.
Supplementary information is available at http://www.nature.com/ajh
REFERENCES
- 1.Grant FD, Romero JR, Jeunemaitre X, Hunt SC, Hopkins PN, Hollenberg NH, Williams GH. Low-renin hypertension, altered sodium homeostasis, and an alpha-adducin polymorphism. Hypertension. 2002;39(2):191–196. doi: 10.1161/hy0202.104273. [DOI] [PubMed] [Google Scholar]
- 2.Cowley AW., Jr Genetic and nongenetic determinants of salt sensitivity and blood pressure. Am J Clin Nutr. 1997;65(2 Suppl) doi: 10.1093/ajcn/65.2.587S. [DOI] [PubMed] [Google Scholar]
- 3.Morimoto A, Uzu T, Fujii T, Nishimura M, Kuroda S, Nakamura S, Inenaga T, Kimura G. Sodium sensitivity and cardiovascular events in patients with essential hypertension. Lancet. 1997;350(9093):1734–1737. doi: 10.1016/S0140-6736(97)05189-1. [DOI] [PubMed] [Google Scholar]
- 4.Weinberger MH, Fineberg NS, Fineberg SE, Weinberger M. Salt sensitivity, pulse pressure, and death in normal and hypertensive humans. Hypertension. 2001;37(2 Part 2):429–432. doi: 10.1161/01.hyp.37.2.429. [DOI] [PubMed] [Google Scholar]
- 5.Svetkey LP, McKeown SP, Wilson AF. Heritability of salt sensitivity in black Americans. Hypertension. 1996;28(5):854–858. doi: 10.1161/01.hyp.28.5.854. [DOI] [PubMed] [Google Scholar]
- 6.Miller JZ, Weinberger MH, Christian JC, Daugherty SA. Familial resemblance in the blood pressure response to sodium restriction. Am J Epidemiol. 1987;126(5):822–830. doi: 10.1093/oxfordjournals.aje.a114719. [DOI] [PubMed] [Google Scholar]
- 7.Beeks E, Kessels AG, Kroon AA, van der Klauw MM, de Leeuw PW. Genetic predisposition to salt-sensitivity: a systematic review. J Hyperten. 2004;22(7):1243–1249. doi: 10.1097/01.hjh.0000125443.28861.0d. [DOI] [PubMed] [Google Scholar]
- 8.GenSalt Collaborative Research G. GenSalt: rationale, design, methods and baseline characteristics of study participants. J Hum Hyperten. 2007;21(8):639–646. doi: 10.1038/sj.jhh.1002207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perloff D, Grim C, Flack J, Frohlich ED, Hill M, McDonald M, Morgenstern BZ. Human blood pressure determination by sphygmomanometry. Circulation. 1993;88(5 Pt 1):2460–2470. doi: 10.1161/01.cir.88.5.2460. [DOI] [PubMed] [Google Scholar]
- 10.The International HapMap C. The International HapMap Project. Nature. 2003;426(6968):789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 11.de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37(11):1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 12.Hinds D, Risch M. [Accessed May 20, 2008];The ASPEX package: affected sib-pair exclusion mapping v1.88. Available at: ftp://lahmed.stanford.edu/pub/aspex/doc/usage.html.
- 13.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. GRR: graphical representation of relationship errors. Bioinformatics. 2001;17(8):742–743. doi: 10.1093/bioinformatics/17.8.742. [DOI] [PubMed] [Google Scholar]
- 14.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63(1):259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30(1):97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 17.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J Roy Stat Soc. 1995;57(1):289–300. [Google Scholar]
- 18.Barba G, Galletti F, Cappuccio FP, Siani A, Venezia A, Versiero M, Della Valle E, Sorrentino P, Tarantino G, Farinaro E, Strazzullo P. Incidence of hypertension in individuals with different blood pressure salt-sensitivity: results of a 15-year follow-up study. J Hypertens. 2007;25(7):1465–1471. doi: 10.1097/HJH.0b013e3281139ebd. [DOI] [PubMed] [Google Scholar]
- 19.Manunta P, Citterio L, Lanzani C, Ferrandi M. Adducin polymorphisms and the treatment of hypertension. Pharmacogenomics. 2007;8(5):465–472. doi: 10.2217/14622416.8.5.465. [DOI] [PubMed] [Google Scholar]
- 20.Cusi D, Barlassina C, Azzani T, Casari G, Citterio L, Devoto M, Glorioso N, Lanzani C, Manunta P, Righetti M, Rivera R, Stella P, Troffa C, Zagato L, Bianchi G. Polymorphisms of alpha-adducin and salt sensitivity in patients with essential hypertension. Lancet. 1997;349(9062):1353–1357. doi: 10.1016/S0140-6736(97)01029-5. [DOI] [PubMed] [Google Scholar]
- 21.Castejon AM, Alfieri AB, Hoffmann IS, Rathinavelu A, Cubeddu LX. Alpha-adducin polymorphism, salt sensitivity, nitric oxide excretion, and cardiovascular risk factors in normotensive Hispanics. Am J Hypertens. 2003;16(12):1018–1024. doi: 10.1016/j.amjhyper.2003.07.022. [DOI] [PubMed] [Google Scholar]
- 22.Ciechanowicz A, Widecka K, Drozd R, Adler G, Cyrylowski L, Czekalski S. Lack of association between Gly460Trp polymorphism of alpha-adducin gene and salt sensitivity of blood pressure in Polish hypertensives. Kidney Blood Press Res. 2001;24(3):201–206. doi: 10.1159/000054228. [DOI] [PubMed] [Google Scholar]
- 23.Barlassina C, Schork NJ, Manunta P, Citterio L, Sciarrone M, Lanella G, Bianchi G, Cusi D. Synergistic effect of alpha-adducin and ACE genes causes blood pressure changes with body sodium and volume expansion. Kidney Int. 2000;57(3):1083–1090. doi: 10.1046/j.1523-1755.2000.00935.x. [DOI] [PubMed] [Google Scholar]
- 24.Manunta P, Cusi D, Barlassina C, Righetti M, Lanzani C, D'Amico M, Buzzi L, Citterio L, Stella P, Rivera R, Bianchi G. Alpha-adducin polymorphisms and renal sodium handling in essential hypertensive patients. Kidney Int. 1998;53:1471–1478. doi: 10.1046/j.1523-1755.1998.00931.x. [DOI] [PubMed] [Google Scholar]
- 25.Manunta P, Maillard M, Tantardini C, Simonini M, Lanzani C, Citterio L, Stella P, Casamassima N, Burnier M, Hamlyn JM, Bianchi G. Relationships among endogenous ouabain, alpha-adducin polymorphisms and renal sodium handling in primary hypertension. J Hypertens. 2008;26(5):914–920. doi: 10.1097/HJH.0b013e3282f5315f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matayoshi T, Kamide K, Takiuchi S, Yoshii M, Miwa Y, Takami Y, Tanaka C, Banno M, Horio T, Nakamura S, Nakahama H, Yoshihara F, Inenaga T, Miyata T, Kawano Y. The thiazide-sensitive Na(+)-Cl(−) cotransporter gene, C1784T, and adrenergic receptor-beta3 gene, T727C, may be gene polymorphisms susceptible to the antihypertensive effect of thiazide diuretics. Hypertens Res Clin Exper. 2004;27(11):821–833. doi: 10.1291/hypres.27.821. [DOI] [PubMed] [Google Scholar]
- 27.Glorioso N, Manunta P, Filigheddu F, Troffa C, Stella P, Barlassina C, Lombardi C, Soro A, Dettori F, Parpaglia PP, Alibrandi MT, Cusi D, Bianchi G. The role of alpha-adducin polymorphism in blood pressure and sodium handling regulation may not be excluded by a negative association study. Hypertension. 1999;34(4 Pt 1):649–654. doi: 10.1161/01.hyp.34.4.649. [DOI] [PubMed] [Google Scholar]
- 28.Sciarrone MT, Stella P, Barlassina C, Manunta P, Lanzani C, Bianchi G, Cusi D. ACE and alpha-adducin polymorphism as markers of individual response to diuretic therapy. Hypertension. 2003;41(3):398–403. doi: 10.1161/01.HYP.0000057010.27011.2C. [DOI] [PubMed] [Google Scholar]
- 29.Bianchi G, Ferrari P, Staessen JA. Adducin polymorphism: detection and impact on hypertension and related disorders. Hypertension. 2005;45(3):331–340. doi: 10.1161/01.HYP.0000156497.39375.37. [DOI] [PubMed] [Google Scholar]
- 30.Beeks E, Janssen RG, Kroon AA, Keulen ET, Geurts JM, de Leeuw PW, de Bruin TW. Association between the alpha-adducin Gly460Trp polymorphism and systolic blood pressure in familial combined hyperlipidemia. Am J Hypertens. 2001;14(12):1185–1190. doi: 10.1016/s0895-7061(01)02216-6. [DOI] [PubMed] [Google Scholar]
- 31.Shioji K, Kokubo Y, Mannami T, Inamoto N, Morisaki H, Mino Y, Tagoi N, Yasui N, Iwaii N. Association between hypertension and the alpha-adducin, beta1-adrenoreceptor, and G-protein beta3 subunit genes in the Japanese population; the Suita study. Hypertens Res Clin Exper. 2004;27(1):31–37. doi: 10.1291/hypres.27.31. [DOI] [PubMed] [Google Scholar]
- 32.EntrezGene. [Accessed October 15, 2008];National Center for Biotechnology Information. http://www.ncbi.nlm.nih.gov.
- 33.EntrezSNP. [Accessed October, 2008];National Center for Biotechnology Information.
- 34.Bagos PG, Elefsinioti AL, Nikolopoulos GK, Hamodrakas SJ. The GNB3 C825T polymorphism and essential hypertension: a meta-analysis of 34 studies including 14,094 cases and 17,760 controls. J Hypertens. 2007;25(3):487–500. doi: 10.1097/HJH.0b013e328011db24. [DOI] [PubMed] [Google Scholar]
- 35.Turner ST, Schwartz GL, Chapman AB, Boerwinkle E. C825T polymorphism of the G protein beta(3)-subunit and antihypertensive response to a thiazide diuretic. Hypertension. 2001;37(2 Part 2):739–743. doi: 10.1161/01.hyp.37.2.739. [DOI] [PubMed] [Google Scholar]
- 36.Schorr U, Blaschke K, Beige J, Distler A, Sharma AM. G-protein beta3 subunit 825T allele and response to dietary salt in normotensive men. J Hypertens. 2000;18(7):855–859. doi: 10.1097/00004872-200018070-00006. [DOI] [PubMed] [Google Scholar]
- 37.Gonzalez-Nunez D, Giner V, Bragulat E, Coca A, de la Sierra A, Poch E. [Absence of an association between the C825T polymorphism of the G-protein beta 3 subunit and salt-sensitivity in essential arterial hypertension] Nefrologia. 2001;21(4):355–361. [PubMed] [Google Scholar]
- 38.Pamies-Andreu E, Ramirez-Lorca R, Stiefel Garcia-Junco P, Muniz-Grijalbo O, Vallejo-Maroto I, Garcia Morillo S, Miranda-Guisado ML, Ortiz JV, Carneado de la Fuente J. Renin-angiotensin-aldosterone system and G-protein beta-3 subunit gene polymorphisms in salt-sensitive essential hypertension. J Hum Hypertens. 2003;17(3):187–191. doi: 10.1038/sj.jhh.1001534. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.