Abstract
For reasons largely unknown, Caucasian women are at a significantly higher risk of developing breast cancer than Asian women. Over a decade ago, mutations in BRCA1/2 were identified as genetic risk factors; however, the discovery of additional breast cancer genes and genes contributing to racial disparities are lacking. We report a functional germline mutation (polymorphism) in the galectin-3 gene at position 191 (rs4644) substituting proline with histidine (P64H), which results in susceptibility to matrix metalloproteinase cleavage and acquisition of resistance to drug induced apoptosis. This substitution correlates with incidence of breast cancer and racial disparity. Genotype analysis of 338 Caucasian (194 disease free and 144 breast cancer patients) and 140 Asian (79 disease free and 61 breast cancer patients) women showed that the allele homozygous for H64 exists in disease free Caucasian and Asian women at a frequency of 12% and 5% respectively versus 37% and 82% in breast cancer patients. The data indicate that H/H allele is associated with increased breast cancer risk in both races. The data implicate galectin-3 H64 in breast cancer and explain, in part, the noted racial disparity, thus providing a novel target for diagnosis and treatment.
Keywords: Galectin-3, Breast Cancer, nsSNP, racial disparity
Introduction
Breast cancer is the second leading cause of cancer deaths among women. It is estimated that in 2008 about 182,460 new cases of invasive breast cancer will be diagnosed among women in the United States (1). Although many epidemiological and etiological risk factors have been identified, the cause of any individual breast cancer is often unknown. Breast cancer incidence and mortality can vary tremendously with race. Population genetics studies revealed genetic variation within racial or ethnic subpopulations, but to what degree such genetic variability accounts for the differences in disease outcomes is still unknown. Investigation of the risk factors associated with racial and ethnic populations may highlight how they might affect prevalence, severity and responses to treatment.
To date, numerous attempts have been made at identifying potential breast cancer risk alleles, genetic signatures and their distribution across populations. However, since the identification of the two major breast cancer hereditary susceptibility genes BRCA1 and BRCA2 that are responsible for ~5% of cases, no other obvious candidate genes contributing to a significant breast cancer risk have been identified (2). Of note, within BRCA1 and BRCA2 carriers, broad variation in breast cancer risk exists with additional factors contributing to breast cancer development (3). Mutations in p53, ATM, CHEK2, PIK3CA and over-expression of genes such as HER-2, CCND1 and others by gene amplification have been identified in sub-fractions of breast cancer cases (4, 5). A number of recent reports have focused on single nucleotide polymorphisms (SNP) with the notion that such a genetic variations can/may contribute to breast cancer development as well as other cancers (3, 6). Genomic-wide association studies (GWAS) and intensive bioinformatic searches of existing databases demonstrated that non-synonymous (ns) SNPs of a few candidate genes exhibit limited association with breast cancer (3, 4, 6). While a powerful experimental tool, GWAS is not without challenges; critical to success is the development of robust study designs, sufficient sample sizes, rigorous phenotypes and comprehensive maps. Candidate gene studies conducted to date have focused mainly on established oncogenes or genes of perceived interest while many other genes of potential significance have been overlooked. For example the commercially available Affymetrix Genome-Wide Human SNP Array 6.0 features 1.8 million genetic markers, includes more than 906,600 SNPs while still missing numerous others. The Human Hap550 chip from Illumina suffers from a similar shortcoming. One such missing nsSNP that has not been evaluated to date is the rs4644 in the galectin-3 gene. Galectin-3 is a member of the galectin gene family expressing binding/specificity to β-galactoside residues through evolutionarily conserved sequence elements of a carbohydrate recognition/binding domain (CRD) and a collagen-α-like motif cleavable by matrix metalloproteases (MMPs) (7). Clinical investigations have shown a correlation between expression of galectin-3 and the malignant properties of several types of cancer (8); consequently, galectin-3 is thought to be a cancer-associated protein. An allelic variation (C to A) in the DNA sequence of galectin-3 at position 191 (rs4644), leading to the expression of either proline (P) or histidine (H) at position 64, was previously noted and regarded as a non-tumor specific variation (9). Recently we have addressed the functional significance of this variation. Amino acids Ala62-Tyr63 of galectin-3 harbor the cleavage site for MMPs (7) and substitution of the subsequent amino acid H64 with P resulted in resistance to cleavage by MMPs (10). Introducing galectin-3 H64 into galectin-3 null cells resulted in acquisition of tumorigenicity and metastatic phenotype in experimental models (11). In sharp contrast, cells transfected with galectin-3 P64 result in reduced tumorigenicity associated with abrogated angiogenesis and increased apoptosis suggesting that cleavage of galectin-3 H64 by MMPs plays a role during tumor development/progression (10). In this manuscript we have performed the genotype analysis of the nsSNP (rs4644) in cancer free and breast cancer patients and correlated it with the incidence of breast cancer in Caucasian and Asian populations.
Materials and Methods
Control and Patient Population
The racial distribution of SNP rs4644 in the normal population was retrieved from Ensembl release 50 (http://www.ensembl.org/Homo_sapiens/snpview?snp=rs4644) (50 Caucasian and 59 Asians women), blood DNA samples from randomly selected women from the Southeast Michigan Tri-county area (139 Caucasian) and an additional 25 disease free volunteers (5 Caucasian and 20 Asian). DNA samples from the breast cancer patients were isolated from the frozen tissues of 43 Caucasian and 45 Asian (Asterand, Detroit, MI) and 71 Caucasian and 1 Asian women (Southeast Michigan Tri-county area). Matched DNA (from blood, frozen normal and cancer) from 15 Asian breast cancer patients was obtained from the Medical School of Zhengzhou University, 10 Caucasian matched frozen cancer and normal breast DNA were obtained from Asterand. Matched DNA from frozen cancer, normal tissue and blood, and RNA from cancer and normal tissue from 17 Caucasian patients were obtained from the Biospecimens Core of the Mayo Breast Cancer SPORE (Rochester, MN). The mean age of the combined breast cancer group was 51.7 years. In all cases, a pathologist independently confirmed the diagnosis. DNAs were isolated using AccuPrep® Genomic DNA extraction kit (Bioneer, Alameda, CA).
PCR, Sequencing and Restriction Fragment Length Polymorphism (RFLP)
The following primer pair was designed to amplify exon 3 of human galectin-3 gene
| Sense | 5′ CTCCATGATGCGTTATCTGGGTCTGG 3′ |
| Anti-sense | 3′ CCTATGGCGCCCCTGCTGGGCCACTG 5′ |
The target sequence was amplified using Eppendorf® Thermal Cycler by QIAGEN Fast Cycling PCR. The resulting 324 bp DNA fragment was purified using QIAquick® PCR Purification Kit (Qiagen, Valencia, CA), visualized on 1% agarose gel containing Syto® 60 stain, scanned by Odyssey infrared imaging system (LI-COR Biosciences, Lincoln, NE) and sequenced using the sense primer at Applied Genomics Technology Center, Core facility at Wayne State University and Karmanos Cancer Institute. PCR-based RFLP was performed by digestion of the PCR product with NcoI (Invitrogen Corporation, Carlsbad, CA) as an additional method to confirm the genotype. Both experiments were performed in a blinded manner by two investigators independently on two different dates. To verify that the DNA samples were not cross-contaminated, the entire length of each PCR product was sequenced and distribution of a different nsSNP in exon 3 was observed, implying no cross-contamination.
Cell Lines and Antibodies
The human breast cancer cell lines SK-BR-3, MDA-MB-435, MDA-MB-231, MDA-MB-468, normal fibroblast cells IMR90 and HS68 were maintained in McCoy medium (Invitrogen Corporation, Carlsbad, CA) supplemented with 10% fetal bovine serum (Atlanta Biologicals®, Lawrenceville, GA). Normal breast cell line MCF10A was maintained in DMEM F-12 medium (Invitrogen Corporation, Carlsbad, CA) with 0.1 μg/ml cholera toxin, 10 μg/ml insulin, 0.5 μg/ml hydrocortisone, 0.02 μg/ml epidermal growth factor, 5% horse serum (Sigma-Aldrich Inc., St-Louis, MO). BT-549 and its stably transfected cell clones, BT-549 Gal-3 H64, BT-549 Gal-3 P64 were maintained in Dulbecco’s Minimal Essential Medium (Invitrogen Corporation, Carlsbad, CA) containing 10% fetal bovine serum, essential and non-essential amino acids (Invitrogen), vitamins and antibiotics (Mediatech Cellgro Inc., Herdon, VA), SUM-102, SUM-149 and SUM 1315 were maintained in Ham’s F-12 medium supplemented with 5% fetal bovine serum, 5 μg/ml insulin, 1 μg/ml hydrocortisone (Sigma-Aldrich Inc., St-Louis, MO). All cell lines were grown in 5% CO2 incubator at 37°C. Cells were grown to 80% confluence and genomic DNA was extracted using AccuPrep® Genomic DNA extraction kit (Bioneer, Alameda, CA). Anti-poly (ADP-ribose) polymerase (PARP), mouse monoclonal antibody was purchased from BioMol (Plymouth Meeting, PA). Monoclonal anti-β-actin (clone AC-15) was purchased from Sigma-Aldrich (St. Louis, MO). Hybridoma expressing galectin-3 monoclonal antibody TIB-166 was purchased from American Type Culture Collection (Manassas, VA). The polyclonal antibody against galectin-3 was prepared as described11.
Cell Viability and Apoptosis
BT-549, BT-549 Gal-3 H64 and BT-549 Gal-3 P64 cells were seeded at a density of 2500 cells per well in triplicates onto 24 well culture dishes. In one set of wells 25 μM cisplatin (Sigma-Aldrich Inc., St. Louis, MO) was added after 24 hr of seeding, the other non-treated set served as control. Cell viability was determined after 72 hr by the mitochondrial-dependent reduction of 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) (Sigma-Aldrich Inc., St. Louis, MO) to formazan.
To determine if cell death is due to apoptosis, 1×106 cells were seeded in 100 mm Petri dishes and treated similarly with cisplatin. After 24, 48, 72 hr the cells were trypsinized, lysed and equivalent number of cells (1×105) were subjected to SDS-PAGE and Western blot analysis with a 1:200 dilution of PARP mouse monoclonal antibody (BioMol, Plymouth Meeting, PA). Blots were also immuno-reacted with a 1:5000 dilution of anti-β-actin mouse monoclonal antibody to normalize for variation in protein loading. Following washes the blots were reacted with secondary antibody mix containing 1:5000 dilutions of IR Dye 680 or IR Dye 800 conjugated corresponding antibody (Molecular Probes, Eugene, OR) and scanned by Odyssey infrared imaging system to identify the respective proteins.
Immunohistochemical Analysis
A breast cancer progression tissue array (BR480) containing 48 tissue cores of 2 mm size was purchased from U.S. Biomax (Rockville, MD). Four μm serial sections were deparafinized, rehydrated, blocked and incubated with primary antibodies (anti-galectin-3 monoclonal, anti-galectin-3 polyclonal) at 4°C overnight at the suitable dilution and linked with the appropriate host secondary antibodies (Vector Laboratories, Burlingame, CA) tagged with Avidin biotinylated horseradish peroxidase, colorized with 3′-3′-diaminobenzidine and counterstained with hematoxylin. Visualization and documentation were accomplished with an OLYMPUS BX40 microscope supporting a Sony DXC-979MD 3CCCD video camera and stored with the M5+ micro-computer imaging device (Interfocus, Cambridge, UK).
Statistical analysis
The assumption of Hardy-Weinberg equilibrium in the control groups was evaluated using Fisher exact tests. Odds ratios (ORs) were used to estimate the magnitude of the association of breast cancer with the alleles HH and HP. Cornfield’s method was used to estimate 95% confidence intervals (CIs). If there was no evidence of heterogeneity in the ORs based on Tarone’s test, the Mantel-Haenszel combined OR was calculated to estimate the overall OR across ethnic groups. Two-sided p-values are reported. Calculations were made with Stata SE 10.2.
The assumption of Hardy-Weinberg equilibrium was found to be tenable in both the Asian control group (p=0.12) and the Caucasian control group (p=0.44). The Biostatistics Core of the Karmanos Cancer Institute performed statistical analysis. Microsoft Excel software was used for calculating statistical significance of apoptosis assay by 2 sample t test: using unequal variance. P values of <0.05 were considered statistically significant.
Results and Discussion
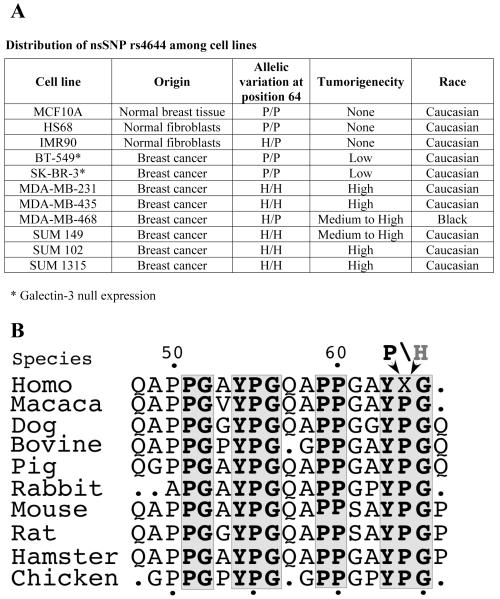
To examine the possibility that the P64H substitution contributes to breast cancer, we initially screened a panel of commonly used human breast cancer cell lines, analyzed their galectin-3 genomic sequence and compared it with their reported experimental tumorigenicity in nude mice. The normal human breast epithelial cell line (MCF10A) was found to be homozygous for the P allele similar to normal human fibroblast cells (HS68 and IMR90). Two breast carcinoma cell lines BT-549 and SK-BR-3 (11) that are galectin-3 null and exhibited none to very low tumorigenicity in experimental models were homozygous for P allele at DNA level, while the tumorigenic human breast carcinoma cell lines, MDA-MB-231, MDA-MB-435, SUM 149, SUM 1315 and SUM 102 were homozygous for the H allele and the MDA-MB-468 (12, 13) was heterozygous carrying both alleles (Fig. 1A). Thus, we hypothesized that in these cell lines galectin-3 P64 and galectin-3 H64 could be associated with normal and tumor phenotypes respectively. Next we questioned whether this genotype variation is human specific or is evolutionarily conserved. Homology search of galectin-3 from chicken to mammals revealed that appearance of H64 variant is relatively late occurrence in evolution and found only in humans (Fig. 1B).
Figure 1.
A. Distribution of nsSNP rs4644 among cell lines and their tumorigenicity.
B. Multiple sequence alignment for galectin-3 of chicken and a few mammalian species. The dots in alignment represent gaps. Grey boxes show the identity between the proteins. X represents either H or P. The numbers represent the position of amino acid in human galectin-3.
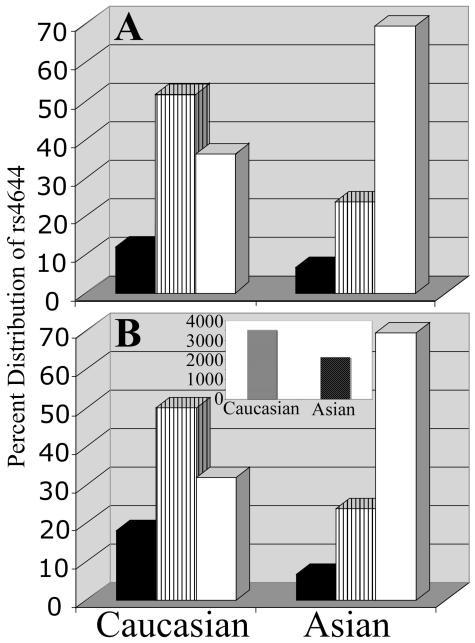
Genomic DNAs from healthy women were isolated from two control studies: one, random digit dialing, the other, volunteers. The data independently obtained from sequencing and RFLP analysis revealed that 64% of the cancer-free Caucasian women were either homozygous (12%) or heterozygous (52%) for H64 and the remaining 36% were homozygous for P64. In contrast, 70% of the cancer-free control Asian women were homozygous for P64, the remaining 30% carried at least one H64 allele, 5% of which were homozygous and 25% were heterozygous (Fig 2A). This racial difference of genotype distribution is in accordance with the data retrieved from available databases (Ensembl release 50) (Fig. 2B).
Figure 2.
Genotype distribution of nsSNP rs4644 in normal population by race. A. Genotype distribution of rs4644 from 144 cancer-free Caucasian and 20 Asian volunteer women.
B. Data from 50 Caucasian and 59 Asian unrelated women were retrieved from Ensembl release 46 (from the database we have excluded family related members). (http://www.ensembl.org/Homo_sapiens/snpview?snp=rs4644).
Black bar: H- homozygous alleles; lined bar: H/P- heterozygous alleles; Open bar: P- homozygous alleles.
Inset: Breast cancer incidence by race. Rates are expressed as cases per 100,000. Statistics were generated from malignant cases only. Surveillance, Epidemiology, and End Results (SEER) Program (www.seer.cancer.gov) SEER Stat Database: Incidence - SEER 17 Regs Limited-Use, Nov 2006 Sub (1995-2004), National Cancer Institute, DCCPS, Surveillance Research Program, Cancer Statistics Branch, released April 2007, based on the November 2006 submission.
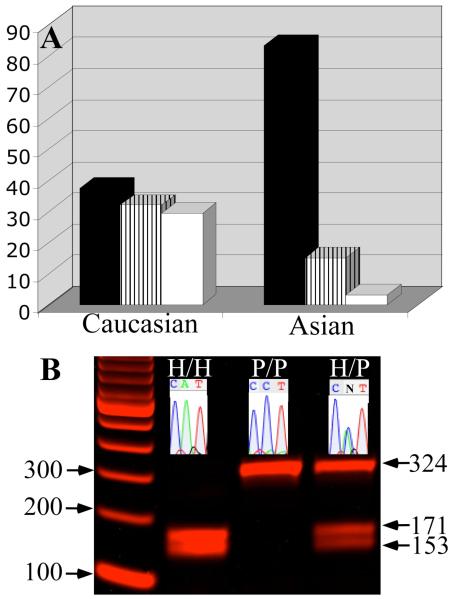
The National Cancer Institute SEER data indicated that Caucasian women have ~1.5 times higher propensity to develop breast cancer as compared to Asian women (Fig. 2B inset). Genotype distribution of rs4644 in Caucasian and Asian breast cancer patients showed that 70% and 97% of the tumor samples from Caucasians and Asians, respectively, harbor the H64 allele, of which 37% of the Caucasians and 82% of the Asians were homozygous (Fig. 3A) (Supplementary data Table 1s).
Figure 3.
Genotype distribution of nsSNP rs4644 in breast cancer patients by race. A. Genotype distribution of rs4644 from 141 Caucasian and 61 Asian breast cancer patients was determined using independent sequencing and RFLP analyses.
B. RFLP and sequence analysis of the C191A site of the galectin-3 gene. 324-bp amplified DNA was digested with NcoI and the resulting fragments were separated by electrophoresis on a 2% agarose gel at 100 V for 30 min. PCR product containing the H allele can be digested by NcoI creating two fragments of 171 and 153 bp length (lanes H/H and H/P). Sequencing of the galectin-3 gene around the C191A site was performed in Applied Genomics Technology Center using the sense primer (inset: DNA sequence chromatograms indicating allelic variation).
The odds ratio of breast cancer for the H/H genotype compared to the P/P genotype for Caucasian and Asian patients were 2.7; 95% CI- 1.4-4.9, P=0.001 and 94.6; 95% CI, 10.0-892.4, P<0.001 respectively.
Kienan et al have recently reported that Asian and European ancestors shared the same population bottleneck expanding out of Africa, but that both also experienced a more recent genetic drift, which was greater in Asians. We presume that this genetic drift led in part to an enrichment in the Asian population of P genotype due to evolutionary selection (14). Breast cancer is a complex disease, consisting of diverse structures, genetic, epigenetic variations and clinical outcomes. Genetic linkage studies have not identified a significant number of breast cancer-causing genes other than BRCA1/2 (15). This lack of identifiable susceptibility genes suggests that breast cancer is most probably conferred by a large number of loci, each contributing in a small additive manner to the overall breast cancer risk (4). In addition, variation in risk for breast cancer has been attributed in part to diet, local environmental factors, and/or yet to be identified genetic predisposing factors. Thus, to date the majority of clinical cases lack identifiable risk factor(s) other than gender, age, race, and family history. The differences in the propensity to develop breast cancer are likely attributable to multiple genes/factors and the data presented here suggest that the reported H64 and P64 allelic variation may contribute, in part, to the racial disparity observed between Caucasian and Asian women in the incidence of breast cancer. There are statistically significant differences in the odds of breast cancer for the H/H genotype compared to the P/P genotype for Caucasian and Asian patients (odds ratio, 2.7; 95% CI- 1.4-4.9, P=0.001 and odds ratio, 94.6; 95% CI, 10.0-892.4, P<0.001 respectively).
To establish whether this tumor-specific genotype reflects a somatic or germline change and/or loss of heterozygosity, genotyping was performed in RNA and/or DNA isolated from cancer tissue, surrounding normal tissue and blood of Caucasian and Asian breast cancer patients (Supplementary data Table 2s). The genotype distribution of rs4644 from these samples was analyzed using two independent methods: DNA sequencing and PCR-based RFLP (Fig. 3B). The data illustrated that the genotype of the normal DNA as well as RNA when available (both from blood and normal tissue) matched the genotype in tumor tissue, implying that this genetic variation was germline.
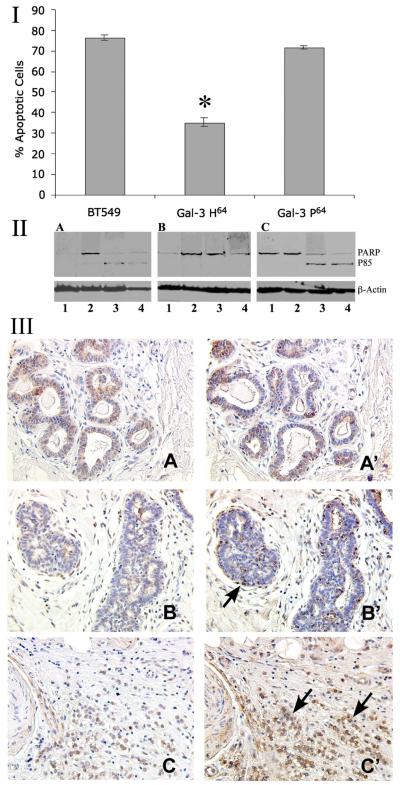
Galectin-3 has been implicated with the progression and metastasis of several types of cancers by affecting cell growth, differentiation, transformation, angiogenesis, immune response and apoptosis (16). While most of these functions are positively influenced by galectin-3, it may act as a double-edged sword either protecting against or stimulating cell death. Over-expression of the galectin-3 H64 variant resulted in resistance to cisplatin-induced apoptosis in transfected BT-549 breast cancer cells (17). Of note, the secreted form could lead to apoptosis of tumor infiltrating T cells leading to cancer cells’ evasion from the host immune system, which is regulated in part by a functional crosstalk between intra-cellular and extra-cellular galectins (18). To investigate the possible functional significance of this allelic variation, we used BT-549, a galectin-3 null cell line to generate cell clones stably over-expressing either galectin-3 P64 or H64 variant allele (10). BT-549 cell clones harboring either H64 or P64 allele were treated with cisplatin. A dose and time dependent decrease in the number of viable cells was observed. Cells expressing the H64 galectin-3 variant were resistant to apoptosis with only 36.2±2.2% apoptotic cells, whereas the parental and P64 cell clones were highly susceptible to drug induced apoptosis with 76.5±1.34% and 71.5±3.02% respectively (Fig. 4I). That the cell death was due to apoptosis was confirmed by the cleavage of PARP. Fig. 3II indicates that parental and galectin-3 P64 expressing cells showed the 85 kDa cleaved polypeptide of PARP, while galectin-3 H64 cells were refractive to PARP cleavage.
Figure 4.
Cisplatin induced apoptosis of BT-549 cells and cloned variants.
I: Percent apoptosis in cells treated with 25uM cisplatin for 72 hr at 37°C and analyzed following MTT assay. ODs in the untreated control cells were given a value of 100 and relative percent apoptosis was calculated as 100%, and the values are the average of triplicate experiments. Bars represent ±SD. * P>0.001
II: Western blot analysis of the expression of PARP in cells treated with 50 μM cisplatin after 24, 48, and 72 hr. A: BT-549; B: BT-549 Gal-3 H64; C: BT-549 Gal-3 P64. β-Actin was used as loading control. 1- Untreated; 2- 24 hr; 3- 48 hr; 4- 72 hr. Data shown is representative of three clones for each variant.
III: Immunohistochemical analysis of intact and cleaved galectin-3 in breast cancer progression. A-C: Intact protein was visualized using monoclonal anti-galectin-3 antibody TIB166; A’-C’: full length plus cleaved galectin-3 using polyclonal galectin-3 antibody hL31. For detailed description see(10). A-A’: Normal breast tissue; B-B’: Ductal hyperplasia; C-C’: Infiltrating lobular carcinoma. Arrows in B’ and C’ indicate the cleaved protein. Representative pictures are presented from a study of 5 normal breast tissues, 10 lobular hyperplasia and 8 infiltrating lobular carcinoma cases. x200
The biological functions regulated by galectin-3 include mRNA splicing, cell growth, cell cycle and apoptosis resistance, while secreted galectin-3 modulates cellular adhesion and signaling, immune response, angiogenesis and tumorigenesis by binding to cell surface glycoconjugates such as laminin, fibronectin, and collagen IV(16). The functions regulated by secreted galectin-3 such as angiogenesis and tumor growth were inhibited only in BT-549 cell clones harboring the MMP cleavage resistant galectin-3 P64 variant (10).
An important role for galectin-3 in breast cancer progression has been suggested since it was found to be significantly elevated in the sera of breast cancer patients as compared with healthy controls (19). Furthermore, galectin-3 localizes in cells proximal to the stroma in comedo-ductal carcinoma in situ (DCIS), which could lead to increased invasive potential by inducing interactions with the stromal counterparts (20). Secreted galectin-3 H64 binds to the endothelial cells of the micro vessels and induces migration, morphogenesis and angiogenesis (21). The cleaved galectin-3 H64 bound to the endothelial cell surface with higher affinity (~20 times) than the full-length protein in a carbohydrate-dependent manner, and the MMPs cleavage resistant galectin-3 P64 didn’t mimic galectin-3 H64 functions (10). Of note, in vivo cleavage of galectin-3 was found to be a surrogate marker for the activity of MMPs in the growing breast cancer (10). Thus, we examined here, whether and at what progressive stage this cleavage of galectin-3 could be detected in breast cancer. Immunohistochemical analysis of a breast cancer progression tissue array with specific antibodies to detect intact (Fig. 4III A-C) and cleaved galectin-3 (Fig. 4III A’-C’) revealed that in the normal breast tissue (A-A’), no cleavage of galectin-3 could be detected. In ductal hyperplasia the intact protein could be detected on the tubular side of the ducts, whereas the cleaved protein could be detected adjacent to the stroma (B-B’). In the infiltrating lobular carcinoma most of the invasive cell clusters were positive for the cleaved product (C-C’). These results emphasize the significance of P64H mutation in breast cancer progression and highlight the role of MMPs in this process.
Since galectin-3 is not a classical oncogene but rather a cancer helper gene product, we suggest that it most probably exerts its function during the progression stage, i.e., following cancer initiation and promotion by other genes/factors. Future larger scale studies will be required to determine which allele is dominant in an H/P heterozygote and whether this affects breast cancer development.
In conclusion, we provide novel data implicating a galectin-3 germline non-synonymous polymorphism in breast cancer and note that variation in allelic frequencies for this nsSNP between Caucasian and Asian women in the general population may explain, in part, the evolutionary differential incidence of breast cancer observed in the two races.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank Drs. PVM Shekhar and George Brush for critical evaluation of the manuscript, Nancy K. Levin for management and maintenance cancer samples database and all the volunteers who participated in this study. The samples were obtained with approval of IRBs HIC#: 017208M1X and HIC#: 10350MP4E
Financial support: The financial support by National Institute of Health (R37CA46120-19 to A.R. and R01CA87895 to A.G.S.), National Institutes of Health Mayo Breast Cancer SPORE Grant (CA116201 to F.J.C. and W.L.L.) and the Karmanos Cancer Institute’s Strategic Research Grant to A.R. and P.N.M. is thankfully acknowledged.
REFERENCES
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.King MC, Marks JH, Mandell JB. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science. 2003;302:643–6. doi: 10.1126/science.1088759. [DOI] [PubMed] [Google Scholar]
- 3.Begg CB, Haile RW, Borg A, et al. Variation of breast cancer risk among BRCA1/2 carriers. JAMA. 2008;299:194–201. doi: 10.1001/jama.2007.55-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Savas S, Schmidt S, Jarjanazi H, Ozcelik H. Functional nsSNPs from carcinogenesis-related genes expressed in breast tissue: potential breast cancer risk alleles and their distribution across human populations. Human Genomics. 2006;2:287–96. doi: 10.1186/1479-7364-2-5-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holst F, Stahl PR, Ruiz C, et al. Estrogen receptor alpha (ESR1) gene amplification is frequent in breast cancer. Nat Genet. 2007;39:655–60. doi: 10.1038/ng2006. [DOI] [PubMed] [Google Scholar]
- 6.Gonzalez-Neira A, Rosa-Rosa JM, Osorio A, et al. Genomewide high-density SNP linkage analysis of non-BRCA1/2 breast cancer families identifies various candidate regions and has greater power than microsatellite studies. BMC Genomics. 2007;8:299. doi: 10.1186/1471-2164-8-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ochieng J, Fridman R, Nangia-Makker P, et al. Galectin-3 is a novel substrate for human matrix metalloproteinases-2 and -9. Biochemistry. 1994;33:14109–14. doi: 10.1021/bi00251a020. [DOI] [PubMed] [Google Scholar]
- 8.Califice S, Castronovo V, Van Den Brule F. Galectin-3 and cancer. Int J Oncol. 2004;25:983–92. [PubMed] [Google Scholar]
- 9.Lotz MM, Andrews CW, Jr., Korzelius CA, et al. Decreased expression of Mac-2 (carbohydrate binding protein 35) and loss of its nuclear localization are associated with the neoplastic progression of colon carcinoma. Proc Natl Acad Sci U S A. 1993;90:3466–70. doi: 10.1073/pnas.90.8.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nangia-Makker P, Raz T, Tait L, Hogan V, Fridman R, Raz A. Galectin-3 cleavage: a novel surrogate marker for matrix metalloproteinase activity in growing breast cancers. Cancer Res. 2007;67:11760–8. doi: 10.1158/0008-5472.CAN-07-3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nangia-Makker P, Thompson E, Hogan V, Ochieng J, Raz A. Induction of tumorigenicity by galectin-3 in a non-tumorigenic human breast carcinoma cell line. Int J Oncol. 1995;7:1079–87. doi: 10.3892/ijo.7.5.1079. [DOI] [PubMed] [Google Scholar]
- 12.Zhang RD, Fidler IJ, Price JE. Relative malignant potential of human breast carcinoma cell lines established from pleural effusions and a brain metastasis. Invasion Metastasis. 1991;11:204–15. [PubMed] [Google Scholar]
- 13.Hoffmeyer MR, Wall KM, Dharmawardhane SF. In vitro analysis of the invasive phenotype of SUM 149, an inflammatory breast cancer cell line. Cancer Cell Int. 2005;5:11. doi: 10.1186/1475-2867-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keinan A, Mullikin JC, Patterson N, Reich D. Measurement of the human allele frequency spectrum demonstrates greater genetic drift in East Asians than in Europeans. Nat Genet. 2007;39:1251–5. doi: 10.1038/ng2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith P, McGuffog L, Easton DF, et al. A genome wide linkage search for breast cancer susceptibility genes. Genes Chromosomes Cancer. 2006;45:646–55. doi: 10.1002/gcc.20330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dumic J, Dabelic S, Flogel M. Galectin-3: an open-ended story. Biochim Biophys Acta. 2006;1760:616–35. doi: 10.1016/j.bbagen.2005.12.020. [DOI] [PubMed] [Google Scholar]
- 17.Yoshii T, Fukumori T, Honjo Y, Inohara H, Kim HR, Raz A. Galectin-3 phosphorylation is required for its anti-apoptotic function and cell cycle arrest. J Biol Chem. 2002;277:6852–7. doi: 10.1074/jbc.M107668200. [DOI] [PubMed] [Google Scholar]
- 18.Fukumori T, Takenaka Y, Oka N, et al. Endogenous galectin-3 determines the routing of CD95 apoptotic signaling pathways. Cancer Res. 2004;64:3376–9. doi: 10.1158/0008-5472.CAN-04-0336. [DOI] [PubMed] [Google Scholar]
- 19.Iurisci I, Tinari N, Natoli C, Angelucci D, Cianchetti E, Iacobelli S. Concentrations of galectin-3 in the sera of normal controls and cancer patients. Clin Cancer Res. 2000;6:1389–93. [PubMed] [Google Scholar]
- 20.Shekhar MP, Nangia-Makker P, Tait L, Miller F, Raz A. Alterations in galectin-3 expression and distribution correlate with breast cancer progression: functional analysis of galectin-3 in breast epithelial-endothelial interactions. Am J Pathol. 2004;165:1931–41. doi: 10.1016/S0002-9440(10)63245-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nangia-Makker P, Honjo Y, Sarvis R, et al. Galectin-3 induces endothelial cell morphogenesis and angiogenesis. Am J Pathol. 2000;156:899–909. doi: 10.1016/S0002-9440(10)64959-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.