Abstract
Ischemia exists in many diseased tissues including arthritic joints, atherosclerotic plaques and malignant tumors. Macrophages accumulate in these sites and upregulate hypoxia-inducible transcription factors (HIFs) 1 and 2 in response to the hypoxia present. Here we show that the gene expression profile in primary human and murine macrophages changes markedly when they are exposed to hypoxia for 18h. For example, they were seen to upregulate the cell surface receptors, CXCR4 and GLUT1, and the potent, tumor-promoting cytokines, VEGFA, interleukins 1β and 8, adrenomedullin, CXCR4 and angiopoietin-2. Hypoxia also stimulated their expression and/or phosphorylation of various proteins in the NF-κB signalling pathway. We then used both genetic and pharmacological methods to manipulate the levels of HIFs 1α and 2α or NF-κB in primary macrophages in order to elucidate their role in the hypoxic induction of many of these key genes. These studies showed that both HIFs 1 and 2, but not NF-κB, are important transcriptional effectors regulating the responses of macrophages to such a period of hypoxia. Further studies using experimental mouse models are now warranted to investigate the role of such macrophage responses in the progression of various diseased tissues like malignant tumors.
Keywords: macrophage, hypoxia, HIF, NF-κB
INTRODUCTION
Cells experience sustained periods of hypoxia in such diseased tissues as malignant tumors, atherosclerotic plaques and arthritic joints.1–3 The predominant transcription factors mediating the effects of hypoxia on gene expression are hypoxia-inducible factors (HIFs) 1 and 2.4,5 These consist of distinct, hypoxia-responsive α subunits and an identical, constitutively expressed β subunit. In the presence of oxygen, the α subunits are hydroxylated by oxygen-sensitive enzymes called prolyl hydroxylases (PDHs), which targets them for degradation by a ubiquitin-proteasomal pathway.4 In hypoxia, HIFα subunits accumulate and translocate to the nucleus, couple with the HIF-1β subunit and bind to hypoxic response elements (HREs) in the promoters of various genes, activating their transcription.4,5
Macrophages accumulate in most ischemic diseased sites including tumors,6–9 where they accumulate both HIF 1α and 2α,10,11 and upregulate HIF target genes like the potent proangiogenic growth factor, VEGFA.12 There are conflicting views of the relative contribution of each HIF to the regulation of hypoxic gene expression in these cells. Some studies suggest that the main form of HIF upregulated by TAMs is HIF-2,11,13 and over-expression of HIF-2α in normoxic human macrophages upregulates various pro-angiogenic genes.14 However, human macrophages also markedly upregulate HIF-1α when exposed to hypoxia in vitro and in tumors,10 and HIF-1α-deficient murine macrophages express lower levels of such HIF-regulated genes as VEGF and the glucose receptor, GLUT1 in hypoxia than their wild type counterparts.15
Interestingly, the exact contribution of HIFs 1 and 2 to the regulation of hypoxic gene expression appears to vary between different cell types. HIF-1, for example, mediates the induction of virtually all hypoxia-activated genes in mouse embryonic fibroblasts and human breast tumor cells,16,17 whereas HIF-2 performs this function in renal tumor cells.17 This depends partly on the cell-type specific expression of other transcription factors like Elk-1 which bind to the promoters of some genes conferring HIF-2 target specificity on them.18,19
Hypoxia may also employ another transcription factor, NF-κB, as two major components of canonical NF-κB signaling, κB kinase β (IKKβ) and p65 (RelA) are activated when murine macrophages experience short-term (≤4h) hypoxia. This then upregulates their expression of both HIF-1α and various HIF target genes.20–22
In the present study, we show that exposure to hypoxia for 18h markedly upregulates a broad array of tumor-promoting genes in primary macrophages, and then investigated the role of HIFs 1 and 2 and NF-κB in this phenomenon.
MATERIALS & METHODS
Cells
Two forms of primary macrophages were used in this study: macrophages differentiated in vitro from human peripheral blood (monocyte-derived macrophages or ‘MDMs’) or bone marrow-derived macrophages (‘BMDMs’) derived from bone marrow progenitors isolated from wt mice or mice bearing deletions in the HIF-1α or HIF-2 α genes.
Isolation and culture of human MDMs
Monocytes were isolated from Buffy coats (National Blood Service, Sheffield, UK) as previously described.10 50×106 mononuclear cells was seeded in Iscove’s Modified Dulbecco’s Media (BioWhittaker UK Ltd, Wokingham, UK) with 5% human AB serum (neat AB serum contains ~1 ng/ml human CSF-1) and 2mM L-Glutamine (All from Sigma, Poole, UK) and incubated at 37°C, 5% CO2. After 2h, adherent cells were washed and cultured for 7 days to allow differentiation into MDMs.
Isolation and culture of murine BMDMs
As previously described, 22 BMDMs were isolated from the bones of wild type mice or mice bearing a targeted deletion of (i) the HIF-1α gene in myeloid cells (2loxP/1loxP, LysM Cre/+ mice15) or (ii) the HIF-2α gene in myeloid cells (2loxP/1loxP, LysM Cre/+ mice; HongxiaZ, Simon CS submitted).
Bone marrow aspirates were washed and resuspended in medium with 10% heat-inactivated FCS (BioWhittaker UK Ltd, Wokingham, UK), 2mM L-Glutamine (Sigma), 100IU/ml penicillin and 100μg/ml streptomycin (BioWhittaker UK Ltd, Wokingham, UK), murine macrophage colony stimulating factor (M-CSF) (PeproTech Ltd, London, UK) and cultured at 37°C, 5% CO2 for 7 days to allow macrophage differentiation. Their purity was assessed after 7 days using an F4/80 antibody. Only BMDMS cultures of >90% purity were used in subsequent experiments.
Successful deletion of HIFs 1 or 2α has been demonstrated previously using Southern and/or immunoblotting assays of extracts from hypoxic BMDMs from the HIF-1α LysM-Cre mice23 and HIF-2α LysM-Cre (H.Z. Imtiyaz & M.C. Simon, submitted) mice used in this study.
Normoxic and hypoxic cell cultures
Human MDMs or murine BMDMs were subjected to severe hypoxia (< 0.5% O2) or normoxia (20.9% O2) in 5% CO2 humidified multi-gas incubators (Heto, Camberly, UK) for 18h.
siRNA treatment of human MDMs in vitro
siRNA duplexes for HIF-1α or HIF-2α were synthesized by Eurogentec laboratories. A randomly scrambled duplex was synthesized as a negative control. The HIF-1α siRNA duplex sequences were comprised of sense 5-CUGAUGACCAGCAACUUGAdTdT-3 and antisense 5-UCAAGUUGCUGGUCAUCAGdTdT-3. The HIF-2α siRNA duplex sequences were sense 5-CAGCAUCUUUGAUAGCAGUdTdT-3 and antisense 5-ACUGCUAUCAAAGAUGCUGdTdT-3. The scrambled non-specific duplex sequences were sense 5-AGUUCAACGACCAGUAGUCdTdT-3 and antisense 5-GACUACUGGUCGUUGAdTdT-3. Transient siRNA transfections were carried out using RNAifect as described by the manufacturer’s instructions (Qiagen, Crawley, West Sussex, UK). Five-day human MDMs were washed and incubated in 100μl, siRNA complex for 48h. Cells were then washed, fresh media added and cells incubated in normoxia or hypoxia for 18h as described earlier.
RNA and protein extraction from human MDMs
Total RNA was prepared using RNeasy kit (Qiagen) according to the manufacturer’s instructions and stored at −80°C. For protein extraction, cells were lysed with lysis buffer (50 mM pH 8.0 Tris-HCl, 150 mM NaCl, 1% Triton-X-100 and 1 protease inhibitor tablet (Roche, Mannheim, Germany). Protein levels were measured using the BCA protein assay (Sigma Aldrich Inc, Poole, Dorset, UK).
RNA and protein extraction from murine BMDMs
Total RNA and protein isolation was prepared using NucleoSpin RNA/Protein kit (Macherey-Nagel, Duren, Germany) and stored at −80°C for RNA and −20°C for protein. For HIF-2α −/− BMDMs, whole cell extracts were prepared using RIPA lysis buffer (50mM Tris pH 8.0, 150 mM NaCl, 1% NP40, 0.1% SDS, 0.25% deoxycholate, 1mM EDTA) containing phosphotase inhibitors (sodium fluoride 0.1 mM, sodium orthovanadate 1 mM, sodium pyrophosphate 2 mM and β-glycerophosphate 10mM). Again, protein extracts were stored at −20°C until used for immunoblotting.
Transcriptional profile analysis
Human Genome U133A plus 2.0 gene chip arrays (Affymetrix UK, UK) that detect 47,000 transcripts were used. Total RNA was reverse transcribed to generate cDNA libraries using oligo dT and superscript II (Invitrogen, Paisley, UK). cDNA was amplified using MEGscript T7 kit and cleaned using Gene Chip Cleanup (both Affymetrix, High Wycombe, UK). Labelled cRNA was synthesized using Gene Chip IVT kit and then hybridized to the arrays following the manufacturer’s instructions (Affymtreix, High Wycombe, UK). Gene chips were processed using an Affymetrix GeneChip scanner 3000.
To verify the results obtained by using Affymetrix arrays, total RNA was extracted from 2 separate experiments, reverse transcribed, amplified and hybridized to Sentrix HumanRef-8_V2 Bead Chip from Illumina (San Diego, CA, USA) according to the manufacturer’s protocols. After washing and drying, the Beadarray was scanned using an Illumina Bead Station 500X which employs SentrixScan Application V2.7.2 software. Illumina Bead Studio software was used for quality control assessment and normalization of data using the LOESS normalization method from BioConductor R packages.
Genes that were upregulated in both arrays by > 1.5-fold or downregulated by <0.67-fold in hypoxia relative to normoxia were considered differentially expressed. One Affymetrix and an Illumina microarray were conducted on RNA isolated from separate experiments. Their combined use was considered to be the first level of screening for the most robust hypoxia robust genes in human macrophages. Only mRNA species regulated by hypoxia on all arrays were considered to be reproducibly regulated by hypoxia and worthy of further study. Using this criterion, 148 genes were upregulated and 60 genes downregulated by hypoxia. A panel of selected genes were then further analysed using real-time PCR.
Real-time-PCR
cDNAs was prepared from 1μg total RNA using SuperScript Synthesis kit (Invitrogen, Paisley, UK) and amplified with TaqMan gene expression master mix and pre-designed gene probes using a ABI 7900HT Sequence Detection System (Applied Biosystems, Warrington, UK). The human TaqMan gene expression assay probes used were VEGF, IL-1α, IL-1β, IL-6, CXCL8, CXCR4 (chemokine C-X-C receptor 4), adrenomedullin (ADM), STAT4, adenosine receptor 2A (ADORA2A) intercellular adhesion molecule 1 (ICAM1), heme oxygenase 1 (HMOX1), Prolyl Hydroxylase 2 (PHD2), CITED2, Heat shock 70kDa protein 1B (HSPA1B) ADAM metallopeptidase domain 8 (ADAM8) ERO1-like (ERO1L) matrix metalloproteinase 7 (MMP7), glucose transporter 1 (GLUT-1) and β-2-microglobulin as the endogenous control (Applied Biosystems, Warrington, UK). The murine TaqMan probes used for murine homologs of these were also supplied by Applied Biosystems, Warrington, UK. Real-time PCR cycling conditions for both human and murine samples were 2 min at 50°C then 95°C for 10 min followed by 40 cycles of 15 seconds at 95°C followed by 1 minute at 60°C. In addition, the human NF-κB signalling genes were analysed using SyBr green real-time PCR. The primer sequences used were NFKBIA fwd- TCGCAGTGGACCTGCAAAAT rev-TGAGCTGGTAGGGAGAATAGC, IKKα fwd- CACCATCCACACCTACCCTG rev-CTTATCGGGGATCAACGCCAG, IKKγ fwd-CGTACTGGGCGAAGAGTCTC rev-GGCTGGCTTGGAAATGCAG, NFKB1 (p50) fwd-TGCCAACAGATGGCCCATAC rev-TGTTCTTTTCACTAGAGGCACCA, and Rel A fwd-TTGAGGTGTATTTCACGGGACC rev-GCACATCAGCTTGCGAAAAGG. Real-time PCR was done using SyBr Green PCR Master Mix, detected by ABI-Prism 5700 Sequence Detector and data processed using Gene Amp software (Applied Biosystems, Warrington, UK) The murine TaqMan probes used for murine homologs of Rel A and IKKβ were also supplied by Applied Biosystems, UK. The threshold cycle (Ct) of all human and murine data was normalised against their respective endogenous controls (unaltered by hypoxia). Real-Time PCR were analysed in RNA extracts generated in 3–5 independent experiments and then fold changes in expression relative to normoxic cells calculated with ΔCt values of the sample and reference gene using the formula 2−ΔΔCt.
Immunoblotting studies
Immunoblotting for human HIFs 1α and 2α were conducted as described previously10,11 using 1:1000 anti-human HIF-1α monoclonal antibody supplied by BD Biosciences, Oxford, UK or 1:1000 anti-human HIF-2α monoclonal antibody from Novus, Soham, UK. Both blots were incubated with HRP-conjugated anti-mouse antibody (Dako, Copenhagen, Denmark) and protein bands visualized using an enhanced chemilluminescence detection system (ECL) (Amersham Biosciences, Buckinghamshire, UK). In all cases expression of β-actin was used as a loading control. For NF-κB immunoblotting assays, an anti-human Phospho-NFκB p65, total NF-κB p65, Phospho-IKKα/IKKβ or total IKKα/β (Cell Signalling Technology, Danvers, MA) was used at a dilution of 1:500 or 1:1000 and incubated overnight at 4°C.
Cytokine release assay
Cell supernatants were centrifuged for 5 min at 400 g and filtered to eliminate cell debris and then stored at −20°C. The levels of VEGF, IL8 and IL-1β in these supernatants were measured using a BD FACS Array bioanalyzer (BD).
Role of NF-κB in hypoxic gene regulation in primary macrophages
This was investigated in two ways. First, human MDMs were exposed to a specific NF-κB inhibitor, 4-Methyl-Nl-(3-phenyl-propyl)-benzene-1,2-diamine (JSH-23) (Merck Chemicals, Nottingham, UK) which blocks translocation of phophorylated NF-κB (p65) to the nucleus of cells and its subsequent activation of NF-κB gene targets.24 MDMs were exposed to medium alone or medium containing 40μM JSH-23 (or the equivalent amount of the vehicle for JSH-23, DMSO) for 1.5h, washed and incubated in normoxia or hypoxia for 18h. Normoxic MDMs were also exposed to 10ng/ml rec. human TNF-α (PeproTech, London, UK) for 18h as a positive control for NF-κB activation. RNA and nuclear proteins were then extracted from parallel cultures of MDMS after these treatments for real-time RT-PCR and immunoblot analysis respectively. Some cells were also fixed in 3% formaldehyde in PBS for 15 min, washed and permeabilized with ice cold 100% methanol for 10 min and blocked with 5% goat serum in 0.3% Triton x-100/PBS solution for 1 h. NF-κB p65 was detected using a rabbit anti-mouse antibody (1:25, Cell Signaling Technology, Danvers, USA) followed by addition of goat anti-rabbit Alexa-488 secondary antibody (Invitrogen, Paisley, UK) (1:250 dilution). Cells were counter-stained with 300nM DAPI (Molecular Probes Inc.,) and then photgraphed on a confocal fluorescent microscope (at x400 magnification). Twelve areas of cells were photographed for each treatment group and the degree of nuclear p65 immunofluorescence (ie. Alexa-488-labeled nuclei) in each DAPI-stained nuclei quantified using Analysis D software (Olympus). The proportion of green fluorescence per nuclei was then calculated for all nuclei in 5 fields of view/treatment. The number of all MDMs in each field of view containing Alexa-488-labeled (p65+) nuclei was also counted. To confirm JSH-23 inhibition of NF-κB activity in hypoxic MDMs, EMS As for NF-κB binding to an NF-κB DNA consensus site were conducted as described previously by us25 on lysates from MDMs exposed to nomoxia, hypoxia or hypoxia plus JSH-23 (all in the presence of DMSO as the vehicle for JSH-23). Protein extracts from parallel cultures of MDMs were also immunoblotted for HIFs 1 and 2α (as described above).
The second approach was to infect MDMs with an adenovirus expressing a dominant negative inhibitor of IKKβ to block phosphorylation/activation of p65/RelA. After 4 days in culture, MDMs were exposed to 50ng/ml rec. human M-CSF for 24 h to stimulate upregulation of integrin αvβ5 (required for adenovirus infection of macrophages26). The adv-IKKβDN and control adv (Adv-GFP) (a gift from Dr Thorsten Hagemann, London) were E1/E3-deleted, of the Ad5 serotype, and used to transfect MDMS as described previously.27 MDMS were infected for 2h with 100 multiplicity of infection (MOI) of either adenovirus in serum-free medium. The adenovirus was then removed and fresh medium containing 2% AB serum added. MDMS were maintained for a further 2 days in culture and then exposed to hypoxia or normoxia for 18h. This infection protocol markedly reduces the activity of p65/RelA in human MDMS46 and human endothelial cells.28
Immunofluorescent labelling of IL-1β expressed by TAMs in hypoxic areas of murine 4T1 mammary tumors
Frozen sections of 4T1 murine mammary tumors were generated in a previous study.29 These had been grown in the mammary fat pads of female BALB/c mice, and removed and snap frozen 2h after injection of mice with the hypoxic cell marker, pimonidazole.29 Sections (7 μM) were blocked with FcR Blocking Reagent (Miltenyi Biotec, Surrey, UK) in TBS-0.05% Tween 20 (TBST) for 30 min at room temperature and then incubated with rat anti-F4/80-Alexa 488 (1 μg/mL, clone CL:A3–1; AbD Serotec, Oxford, UK), goat anti-mouse IL-1β (15 μg/ml; R&D Systems, Abingdon, UK) and rabbit anti-PIMO (1:4000, a gift from James Raleigh) for 30 min at room temperature. Negative controls included substitution of primary antibodies with species-matched, non-specific antibodies. Sections were then washed twice and incubated in Donkey anti-goat-Alexa 568 (8 μg/mL; Molecular Probes, Eugene, OR, USA) or Alexa 647-conjugated goat anti-rabbit (8 μg/mL; Molecular Probes) secondary antibodies for 30 min at room temperature in the dark and 30nM DAPI (Molecular Probes) for 2 min.
Gene Set Enrichment Analysis
Gene set enrichment analysis (GSEA) was performed as described previously30 on gene lists ranked by level of hypoxic gene induction (hypoxia/normoxia fold induction) separately for both the Affymetrix and Illumina gene expression data sets. Correlations to the predefined Curated and TFT: transcription factor targets gene set collections were analyzed with the GSEA Pre-ranked tool using 1000 permutations. Further information regarding the gene sets used in these analyses is available in the Molecular Signatures Database (MSigDB) (www.broad.mit.edu/gsea/msigdb).
Statistics
All experiments were repeated 3–6 times. Statistical analyses were performed using the one or two-tailed Student’s t test to determine statistical significance after checking the data for normality (as appropriate). P values of <0.05 were considered statistically significant. All data are expressed as means ± SEMs.
RESULTS
Evidence of distinct transcription signaling in primary human macrophages experiencing hypoxia
Hypoxic MDMs upregulated both HIF-1α and HIF-2α and this was markedly inhibited by prior treatment with siRNA to either HIFα (Fig 1A). As in previous publications,31,32 genes were defined as being differentially regulated in hypoxia if they exhibited >1.5 fold increase in gene expression (Table 1) or downregulated if they showed <0.67 fold change (Table 2) compared to normoxic cultures. A comparison of our human MDM microarray results (Tables 1 & 2) with those obtained previously for related human myeloid cell types exposed to hypoxia (monocytes and monocyte-derived dendritic cells31,32) shows that some genes were seen to be regulated by all three cell types (upregulated: VEGFA, CXCR4, TNFα, TIMP1, PHD3, Aldolases A & C, Enolase 2, TREM1, NCF1; downregulated: Cathepsin C). However, some genes regulated by hypoxia in MDMs are not similarly regulated by hypoxia in these other two cell types such as IL-1β, IL-12p40, Ang-2, endothelin 1, STATS 4 & 6, CCLs 3 & 5, CCR7, HMOX1 & hsp70 (upregulated) and CD36, PECAM1 (CD31), HIF-2α & MHCII DMβ) (downregulated) (Tables 1 and 2). A number of key genes were selected and their upregulation confirmed using qRT-PCR (Table 1). Macrophages were also shown to express abundant IL-1β protein in pimonidazole-stained (hypoxic) areas of murine 4T1 mammary tumors (Fig. 2C).
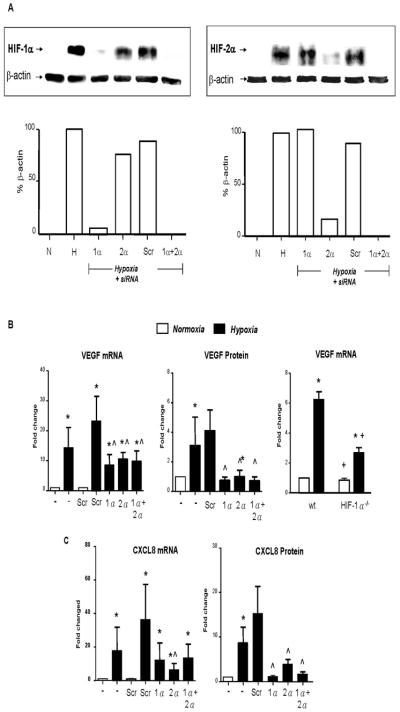
Figure 1. Role of HIFs 1α and 2α in the hypoxic induction of VEGF and CXCL8: insights from siRNA knockdown studies and use of macrophages bearing a deletion in the HIF-1α gene.
A: Immunoblots of HIF-1α or HIF-2 α in MDM lysates following their exposure to normoxia (20.9% O2; ‘N’) or hypoxia (0.1% O2; ‘H’) for 18h, or hypoxia for 18h following exposure to siRNA for HIF-1α (‘1 α’), HIF-2 (‘2α’), both HIFs 1α and 2α together (‘1 α +2 α’ or a scrambled control (‘Scr’). Loading controls were β-actin. Below each gel picture is the densitometric analysis of HIF expression relative to its β-actin loading control. B & C: Effects of HIF-1α and 2α knockdown on the hypoxic induction of VEGF (B) and CXCL8 (IL-8; C) mRNA and protein. In the case of VEGF, gene expression was also assayed in normoxic and hypoxic BMDMs from conditional knockout HIF-1 α−/− mice in vitro by qRT-PCR (right hand panel in B). It was not possible to do this for CXCL8 as this gene is not expressed in mice. Pooled data from 6 replicate experiments are shown. * p<0.05 compared to corresponding normoxic group; ^ P<0.05 compared to the scr siRNA/hypoxia group; + P<0.05 compared to macrophages from wt mice exposed to hypoxia.
Table 1.
Selected genes upregulated by hypoxia in human monocyte-derived macrophages in vitro.
| Hypoxic Induction (fold change) | |||||
|---|---|---|---|---|---|
| Gene Symbol | Full Name | Accession No. | Main Functions of Gene Product | ||
| Array 1 (Affymetrix) | Array 2 (Illumina) | ||||
| Cytokines, & their receptors | |||||
| IL-6* | Interleukin 6 (interferon, beta 2) | NM_000600 | Regulates host defence, acute phase reactions, immune responses, inflammation, cell proliferation, hematopoiesis, angiogenesis and metastasis | 62 | 7 |
| IL-23A* | Interleukin 23, alpha (subunit p19) | NM_016584 | Subunit of IL-23. Regulates T cells & promotes angiogenesis | 31 | 23 |
| IL-1α* | Interleukin 1 alpha | NM_000575 | Multifunctional, pro-inflammatory, proangiogenic cytokine. | 26 | 12 |
| VEGFA*^ | Vascular endothelial growth factor A | NM_003376 | Stimulates angiogenesis & chemotactic for monocytes & other myeloid cells | 14 | 14 |
| TNFA* | Tumor necrosis factor alpha (TNF superfamily, member 2; cachexin) | NM_000594 | Regulates immunity to pathogen, inflammation, apoptosis & proliferation, differentiation, angiogenesis | 11 | 32 |
| WNT5A | Wingless-type MMTV integration site family, member 5A | NM_003392 | Binds to receptor, frizzled-5, and regulates tumor cell migration/invasion | 11 | 10 |
| IL-12B* | Interleukin 12B (p40 subunit, natural killer cell stimulatory factor 2, cytotoxic lymphocyte maturation factor 2) | NM_002187 | A subunit of the cytokine, IL-12, which activates T cells. Also anti-angiogenic factor. | 9 | 10 |
| IL-1β* | Interleukin 1 beta | NM_000576 | Multifuctional, pro-inflammatory, proangiogenic cytokine. | 9 | 8 |
| ADM* | Adrenomedullin | NM_001124 | Vasodilator. Also regulates cell responses to oxidative stress & hypoxic injury. Pro-angiogenic cytokine. | 8 | 17 |
| TNFAIP6 | Tumor necrosis factor, alpha-induced protein 6 | NM_007115 | Hyaluronan-binding protein involved in extracellular matrix stability and cell migration | 5 | 17 |
| MIF* | Macrophage migration inhibitory factor | NM_002415 | Pleoitrophic cytokine with multiple effects on inflammation including immobilising macrophages | 4 | 4 |
| IL-18* | Interleukin 18 (interferon-gamma-inducing factor) | NM_001562 | Pro-inflammatory cytokine that stimulates T and NK cells to secrete interferon-γ (IFN-γ). | 3 | 2 |
| IL-8* | Interleukin 8 | NM_000584 | Pro-angiogenic & chemoattractant for neutrophils | 2 | 43 |
| EDN1* | Endothelin 1 | NM_001955 | Vasoconstrictor. Regulates vascular homeostasis. Chemotactic for monocytes | 2 | 4 |
| CLCF1 | Cardiotrophin-like cytokine factor 1 (Neurotrophin-1) | NM_013246 | IL-6 family protein. Stimulates IL-1 via IL-6R and STAT3. Also stimulates B-cells functions. | 2 | 5 |
| CSF2* | Colony stimulating factor 2 (GM-CSF) | NM_000758 | Stimulates stem cells in the bone marrow to produce granulocytes and monocytes. | 10 | 9 |
| ANGPT2* | Angiopoietin 2 (Ang-2) | NM_001147 | Pro-angiogenic cytokine. Destabilises blood vessels. Chemoattractant for endothelial cells and Tie2+ monocytes. | 2 | 3 |
| Chemokines & their receptors | |||||
| CCL20* | Chemokine (C-C motif) ligand 20 (MIP-3) | NM_004591 | Chemotactic for lymphocytes and neutrophils | 34 | 18 |
| CXCL2* | Chemokine (C-X-C motif) 2 (MIP-2α) | NM_002089 | Chemotactic for neutrophils and hematopoietic stem cells | 23 | 4 |
| CXCL1* | Chemokine (C-X-C motif) 1 (MSGA-α) | NM_001511 | Chemotactic for neutrophils | 20 | 27 |
| CCR7* | Chemokine (C-C motif) receptor 7 | NM_001838 | Receptor for chemokines, CCL19 and CCL21 | 10 | 52 |
| CCL5* | Chemokine (C-C motif) 5 (Rantes) | NM_002985 | Chemotactic for T cells, eosinophils, and basophils; recruits leukocytes to inflammatory sites | 9 | 10 |
| CCL3* | Chemokine (C-C motif) 3 (MIP-1a) | NM_002983 | Recruitment and activation of neutrophils | 3 | 6 |
| CXCR4* | Chemokine (C-X-C motif) receptor 4 | NM_003467 | Receptor for SDF-1 (CXCL12) – which regulates hematpoietic stemm and myeloid cell recruitment by tissues. Involved in proliferation and metatasis of tumor cells. Co-receptor for entry of HIV into T cells. | 2 | 6 |
| Intracellular enzymes & metabolism | |||||
| EGLN3 | HIF prolyl hydroxylase 3 (PHD3) | NM_022073 | One of 3 PHD enzymes that hydroxylate HIFs, resulting in their binding to VHL and degradation. Regulates HIF-2α more than HIF-1α. | 54 | 12 |
| CA12 | Carbonic anhydrase XII | NM_001218 | Enzyme that catalyzes the reversible hydration of carbon dioxide. Acidifies extracellular milieu o tumour cells, stimulating their growth/invasion. | 32 | 81 |
| ALDOC | Aldolase C, fructose-bisphosphate | NM_005165 | Glycolytic enzyme. Catalyzes the breakdown of fructose 1,6-bisphosphate. | 23 | 23 |
| SLC2A5* | Solute carrier family 2 (facilitated glucose/fructose transporter), members 5 (GLUT-5) | NM_003039 | Transports fructose and glucose into the cell. | 12 | 11 |
| NCF1 | Neutrophil cytosolic factor 1 | NM_000265 | Superoxide production | 8 | 6 |
| SLC2A1 | Solute carrier family 2 (facilitated glucose transporter), member 1 (GLUT-1) | NM_006516 | Transports glucose into the cell. | 7 | 23 |
| HMOX1* | Heme oxygenase (decycling) 1 | NM_002133 | Essential enzyme in heme catabolism -cleaves heme to form biliverdin | 7 | 4 |
| SLC2A6 | Solute carrier family 2 (facilitated glucose transporter), member 6 (GLUT-6) | NM_017585 | Transports glucose into the cell. | 6 | 3 |
| SLC2A3 | Solute carrier family 2 (facilitated glucose transporter), member 3 (GLUT-3) | NM_006931 | Transports glucose into the cell. | 5 | 14 |
| PFKFB3 | 6-phosphofructo-2-kinase/fructose-2, 6-biphosphatase 3 | NM_004566 | Synthesis and degradation of fructose 2, 6-bisphosphate | 4 | 7 |
| SLC7A5 | Solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 | NM_003486 | Involved in cellular amino acid uptake. | 3 | 3 |
| PFKP | Phosphofructokinase | NM_002627 | Glycolytic enzyme | 2 | 3 |
| HK1 | Hexokinase 1 | NM_000188 | Commits glucose to the glycolytic pathway | 2 | 2 |
| ALDOA | Aldolase A | NM_000034 | Glycolytic enzyme. Catalyzes the breakdown of fructose 1, 6-bisphosphate | 2 | 2 |
| EGLN1* | HIF prolyl hydroxylase 2 (PHD2) | NM_022051 | One of 3 PHD enzymes that hydroxylate HIFs, resulting in their binding to VHL and degradation. Regulates HIF-1α more than HIF-2α. | 2 | 5 |
| Extracellular enzymes/molecules | |||||
| SERPINE1 | Serpin peptidase inhibitor | NM_000602 | Regulation of fibrinolysis | 7 | 5 |
| ADAM 8 | ADAM metallopeptidase domain 8 | NM_001109 | Membrane-anchored protein involved cell-cell and cell-matrix interactions | 6 | 5 |
| CFB | Complement factor B | NM_001710 | A component of the alternative pathway of complement activation. | 6 | 5 |
| F3* | Coagulation factor III (Thromboplastin, tissue factor; CD 142) | NM_001993 | Cell surface glycoprotein that cleaves prothrombin to thrombin, promoting coagulation. | 3 | 7 |
| TIMP1 | TIMP metallopeptidase inhibitor 1 | NM_003254 | Inhibits activity of most known MMPs. Stimulates proliferation in a wide range of cell types. Also anti-apoptotic. | 2 | 3 |
| MMP7 | Matrix metallopeptidase 7 (matrilysin, uterine) | NM_002423 | Enzyme with broad substrate specificity in ECM. Promotes wound healing, angiogenesis, tumour invasion and metastasis. | 2 | 2 |
| Cell viability | |||||
| SERPINB2* | Serpin peptidase inhibitor, clade B (ovalbumin), member 2 (PAI-2) | NM_002575 | Inhibits serine protease, tissue-type-and urokinase-type plasminogen activator; tPA, uPA. Also regulates gene expression, cell proliferation, differentiation, and apoptosis. | 102 | 4 |
| PTGS2 (COX2)* | Prostaglandin-endoperoxide synthase 2 (cyclo-oxygenase-2) | NM_000963 | Pro-inflammatory - stimulates expression of prostanoids | 27 | 8 |
| EN02 | Enolase 2 | NM_001975 | Glycolytic enzyme | 8 | 10 |
| IGFBP6 | Insulin-like growth factor binding protein 6 | NM_002178 | Binds and prolong the half-life of the IGFs. | 4 | 2 |
| BNIP3L | BCL2/adenovirus E1B 19kDa interacting protein 3-like | NM_004331 | Pro-apoptotic Bcl family protein. | 3 | 5 |
| IER3* | Immediate early response 3 (IEX-2orDIF-2) | NM_003897 | Regulates growth and apoptosis (inhibits NF-kB induced apoptosis) | 2 | 13 |
| NRG1* | Neuroregulin 1 | NM_006096 | Growth factor which regulates cell apoptosis and proliferation (protects cells in ischemia). | 2 | 4 |
| Receptors & cell adhesion/signalling | |||||
| DDIT4 | DNA-damage-inducible transcript 4 (DIG2 or REDD-1) | NM_019058 | A stress response gene, an essential regulator of the checkpoint kinase, mTOR. | 103 | 54 |
| HIG2 | Hypoxia-inducible protein 2 | NM_001098786 | Growth factor that stimulates tumour cell growth. | 12 | 21 |
| NFKB2* | Nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) | NM_002502 | Central mediator of non-canonical NFkB signalling pathway in cells | 11 | 3 |
| TRAF1* | TNF receptor-associated factor 1 | NM_005658 | One of the TNFR members - forms a complex with TRAF2 to activate MAPK8/JNK and NF-kappaB. Mediates the anti-apoptotic signals from TNF receptors | 9 | 8 |
| ADORA2A* | Adenosine A2a receptor | NM_000675 | Receptor for adenosine. | 8 | 33 |
| TREM1* | Triggering receptor expressed on myeloid cells 1 | NM_018643 | Receptor of lg superfamily expressed on human myeloid cells. Regulates their inflammatory functions. | 5 | 6 |
| MAP3K8 | Mitogen-activated protein kinase kinase kinase 8 | NM_005204 | Cell signalling & cycling | 4 | 6 |
| TNS1 | Tensin 1 | NM_022648 | Involved in cell migration and links signal transduction pathways to the cytoskeleton. | 3 | 7 |
| TNIP2 | TNFAIP3 interacting protein 2 (ABIN-2) | NM_024309 | Inhibits NF-kappa-B activation. | 3 | 4 |
| RelA (p65) | Nuclear factor of kappa light polypeptide gene enhancer in B-cells 3(p65/RelA) | NM_021975 | Central mediator of canonical NFkB signalling pathway in cells | 3 | 3 |
| TNFRSF1B | Tumor necrosis factor receptor superfamily, member 1B (TNF-R2 or CD120P) | NM_001066 | One of 2 main TNFRs (I and II) mediating the effects of TNFalpha on cells. | 3 | 2 |
| TNFAIP3* | Tumor necrosis factor, alpha-induced protein 3 | NM_006290 | Interacts with NAF1 & inhibits TNFalpha-induced NF-kappa-B-dependent gene expression | 4 | 5 |
| ANPEP | Aminopeptidase M or N (CD13 or APN) | NM_001150 | Zinc-binding metalloprotease. | 3 | 2 |
| STAT4 | signal transducer and activator of transcription 4 | NM_003151 | Signal transduction and activation of transcription. Involved in IL12 signalling | 4 | 4 |
| STAT6 | signal transducer and activator of transcription 6 | NM_003153 | Plays a central role in regulating the alternative (M2) activation of macrophages in response to interleukin 4 | 4 | 2 |
| ETS2 | v-ets erythroblastosis virus E26 oncogene homolog 2 (ETS2) | NM_005239 | Transcription factor that regulates MMP-9 expression in macrophages and drives mammary tumor progression | 3 | 3 |
| NFKBIE* | Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon | NM_004556 | Inhibits DNA-binding of NF-kappa-B p50-p65 & p50-c-Rel complexes. | 2 | 2 |
| NFKBIA* | Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha | NM_020529 | Inhibits the activity of dim eric NF-kappa-B/REL complexes by trapping REL dimers in the cytoplasm | 2 | 3 |
| IRAK3 | Interleukin-1 receptor-associated kinase 3 | NM 007199 | Inhibits dissociation of IRAK1 & IRAK4 from the Toll-like receptor signalling complex | 2 | 4 |
| RIPK2 | Receptor-interacting serine-threonine kinase 2 | NM_003821 | Apoptosis & activates NFkB. | 2 | 4 |
| Transcription, translation & RNA processing | |||||
| ERO1L | Endoplasmic reticulum oxidoreductin-1 (Ero1)-L alpha | NM_014584 | Endoplasmic reticular enzyme that helps to maintain VEGF secretion under hypoxia. | 3 | 6 |
| HSPA1B | Heat shock 70kDa protein 1B | NM_005346 | Intracellular enzyme. Stabilizes existing proteins against aggregation & mediates the folding of newly translated proteins. | 3 | 3 |
| BTG1 | B-cell translocation gene 1 | NM_001731 | Anti-proliferative protein | 3 | 2 |
| ELL2 | Elongation factor, RNA polymerase II, 2 | NM_012081 | Increase the catalytic rate of RNA polymerase II transcription | 2 | 2 |
Genes shown previously to be regulated by NF-κB (or where the gene promoters contain NF-κB binding sites).
It is currently debatable as to whether the human VEGFA gene is an NF-kB target gene as there is not a well characterized kB site in its promoter. In mice, there is a putative kB site but it is not clear if it is functional. Gene names that are underlined are ones that were also shown in the present study to be upregulated by hypoxia using qRT-PCR.
Table 2.
Selected genes downregulated by hypoxia in human monocyte-derived macrophages in vitro.
| Main Function of Gene Product | Hypoxic Downregulation (fold change) | ||||
|---|---|---|---|---|---|
| Gene Symbol | Full Name | Accession No. | |||
| Array 1 (Affymetrix) | Array 2 (lllumina) | ||||
| Cell adhesion and cell junction molecules | |||||
| CD36 | CD36 molecule (thrombospondin receptor) | NM_000072 | A multi-functional class B scavenger receptor - binds thrombospondin, apoptotic cells and LDLs | 0.16 | 0.17 |
| VCL | Vinculin | NM_014000 | A cytoskeletal protein associated with cell-cell and cell-matrix junctions. Involved in cell adhesion, cell morphology and locomotion | 0.42 | 0.65 |
| PECAM1 | Platelet/endothelial cell adhesion molecule (CD31 antigen) | NM_000442 | Surface receptor expressed by endothelial cells, platelets and various other cells. Helps macrophages to remove aged neutrophils. | 0.62 | 0.52 |
| Cell Metabolism | |||||
| ACAT1 | Acetyl-Coenzyme A acetyltransferase 1 | NM_000019 | Plays a role in lipoprotein assembly and dietary cholesterol absorption | 0.25 | 0.38 |
| NME1 | Non-metastatic cells 1 protein (NM23A) | NM_000269 | A nucleoside diphosphate kinase linked to metastasis suppression in some cell types. | 0.29 | 0.58 |
| PDHB | Pyruvate dehydrogenase (lipoamide) beta | NM_000925 | Catalyzes the overall conversion of pyruvate to acetyl-CoA and CO(2) | 0.37 | 0.49 |
| ST3GAL5 | ST3 beta-galactoside alpha-2,3-sialyltransferase 5 | NM_003896 | Promotes cell differentiation, modulation of cell proliferation, signal transduction, and integrin-mediated cell adhesion. | 0.40 | 0.54 |
| LYPLA3 | lysophospholipase 3 (lysosomal phospholipase A2) | NM_012320 | Regulates the multifunctional lysophospholipids in cell membranes | 0.49 | 0.48 |
| Intracellular transport | |||||
| TOMM22 | Translocase of outer mitochondrial membrane 22 homolog (yeast) | NM_020243 | Mitochondrial membrane protein. Imports cytosolic preproteins into the mitochondrion. | 0.26 | 0.53 |
| HLA-DMB | Major histocompatibility complex, class II, DM beta | NM_002118 | Plays a central role in the peptide loading of MHC class II molecules. | 0.29 | 0.60 |
| SLC17A5 | Solute carrier family 17 (anion/sugar transporter), member 5 | NM_012434 | Primary solute translocator for anionic substances | 0.34 | 0.57 |
| ST6GAL1* | ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 | NM_003032 | Transfers sialic acid from the donor of substrate CMP-sialic acid to galactose containing acceptor substrates | 0.42 | 0.31 |
| MRPL3 | Mitochondrial ribosomal protein L3 | NM_007208 | Helps with protein synthesis within the mitochondrion | 0.47 | 0.50 |
| Receptors & cell signalling | |||||
| TNFSF13B* | Tumor necrosis factor (ligand) superfamily, member 13b | NM_006573 | Promotes cell proliferation | 0.39 | 0.39 |
| RASGRP3 | RAS guanyl releasing protein 3 (calcium and DAG-regulated) | NM_015376 | Guanine nucleotide exchange factor (GEF) for Ras and Rap1 | 0.13 | 0.38 |
| TFRC | Transferrin receptor (p90, CD71) | NM_003234 | Regulates iellular uptake of iron and iron metabolism | 0.25 | 0.19 |
| HIF-1A | Hypoxia-inducible factor, alpha subunit (HIF-2α) | NM_001530 | Regulates response of cells to hypoxia | 0.36 | 0.21 |
| EPAS1 | Endothelial PAS domain protein 1 (HIF-2α) | NM_001430 | Regulates response of cells to hypoxia | 0.33 | 0.21 |
| TLR4 | Toll-like receptor 4 | NM_003266 | Cell surface receptor that binds many ligands including bacterial LPS and fibrinogen | 0.26 | 0.63 |
| MAPRE2 | Micro tubule-associated protein, RP/EB family, member 2 | NM_014268 | Involved in microtubule polymerization, cell migration | 0.35 | 0.52 |
| CTSC | Cathepsin C | NM_001814 | A lysosomal cysteine proteinase that activates many serine proteinases in immune/inflammatory cells | 0.41 | 0.48 |
| PRCP | Prolylcarboxypeptidase (angiotensinase C) | NM_005040 | A lysosomal prolylcarboxypeptidase, which cleaves C-terminal amino acids linked to proline in peptides | 0.43 | 0.48 |
| SPARC | Secreted protein, acidic, | NM_003118 | A calcium binding glycoprotein that also | 0.46 | 0.26 |
genes shown previously to be regulated by NF-κB (or where the gene promoters contain NF-κB binding sites).
Figure 2. Hypoxic upregulation of IL-1β by human MDMs in vitro and by TAMs in hypoxic areas of murine mammary tumors: role of HIFs 1 and 2.
A: IL-1β mRNA levels and protein release by human MDMs following their exposure to normoxia (20.9% O2; ‘N’) or hypoxia (0.1% O2; ‘H’) for 18h, or hypoxia for 18h following exposure to siRNA for HIF-1 α (‘1 α’), HIF-2α (‘2 α’), both HIFs 1 α and 2 α together (‘1 α +2 α’) or a scrambled control (‘Scr’). B: Hypoxic induction of IL-1β mRNA by bone marrow-derived macrophages derived from wt or HIF-1α−/− mice. C: Upregulated expression of IL-1β by F4/80+ macrophages in pimonodazole-stained (ie. hypoxic; ‘H’) compared to pimonodazole-unstained (ie. normoxic; ‘N’) areas of murine mammary (4T1) tumours (see yellow arrows on the merged ‘H’ image). Pooled data from 3 replicate experiments are shown. * p<0.05 compared to corresponding normoxic group; ^ P<0.05 compared to the Scr siRNA/hypoxia group; + P<0.05 compared to macrophages from wt mice exposed to hypoxia.
Genetic manipulation of HIFs 1 and 2α demonstrates the co-regulation of genes in primary human macrophages experiencing hypoxia
The hypoxic accumulation of both HIFs 1 and 2α was ablated following transfection with siRNA for either α subunits. Both VEGFA mRNA and protein were markedly increased by hypoxia and this was significantly inhibited by siRNA for either HIFα subunit (Fig. 1B, left and middle panels). It may appear that the hypoxic induction of VEGF mRNA is higher in hypoxic macrophages treated with the scrambled control siRNA than in the ‘no siRNA’ group. However, this failed to reach statistical significance. This was also the case for these 2 groups in panels E, G, I and K of Fig. 3.
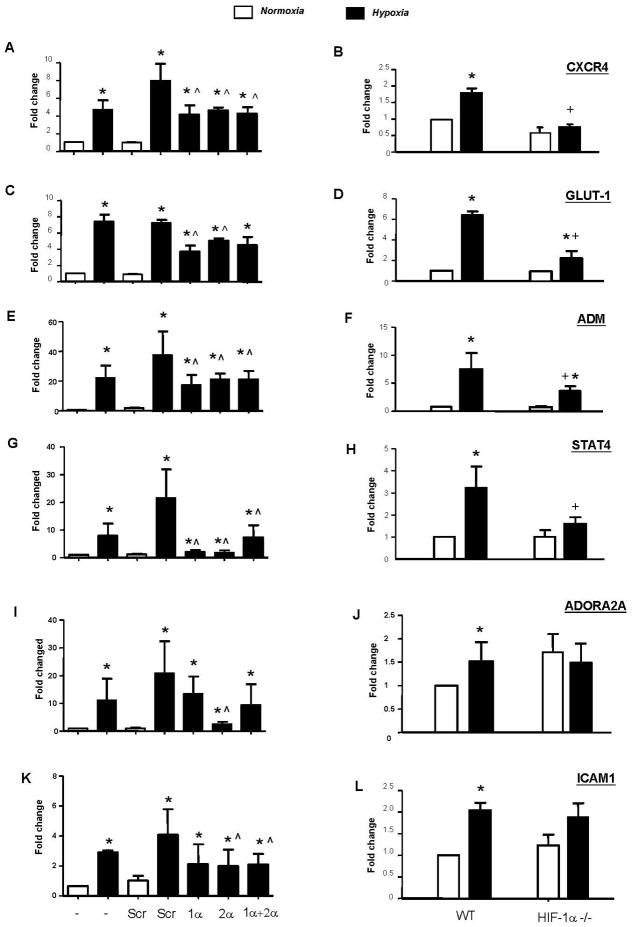
Figure 3. Role of HIFs 1α and 2α in the hypoxic induction of other key genes by macrophages.
Panels A, C, E, G, I & K: hypoxic induction of mRNA for CXCR4, GLUT-1, ADM, STAT4, ADORA2A and ICAM1 (as measured by qRT-PCR) following exposure of primary human macrophages to normoxia (20.9% O2; ‘N’) or hypoxia (0.1% O2; ‘H’) for 18h, or hypoxia following exposure to siRNA for HIF-1 α (‘1 α’), HIF-2 α (‘2 α’), both HIFs 1 α and 2 α together (‘1 α +2 α’) or a scrambled control (‘Scr’). Panels B, D, F, H, J & L: hypoxic induction of the same genes in murine BMDMs from wt and HIF-1α−/− mice. Pooled data from 3 replicate experiments are shown. *p<0.05 compared to corresponding normoxic group; ^P<0.05 compared to the Scr siRNA/hypoxia group; +P<0.05 compared to macrophages from wt mice exposed to hypoxia.
CXCL8 mRNA and protein release were also upregulated in hypoxic MDMS (Fig. 1C) and while both HIFα siRNA treatments reduced hypoxia-induced CXCL8 mRNA, only the effect of HIF-2α siRNA reached significance. However, both HIFα siRNA species significantly reduced CXCL8 protein release (Fig. 1C). The inhibitory effect of HIF siRNA on the hypoxic induction of both VEGF and CXCL8 appeared to be slightly greater at the protein than the mRNA level.
Hypoxia also upregulated IL-1β mRNA and protein and this was significantly inhibited by exposure to siRNA for either HIF α subunit (Fig 2A). We then investigated the role of HIFs-1 and 2 in the hypoxic regulation of several other genes listed in Table 1. The hypoxic upregulation of mRNA for CXCR4, GLUT1, adrenomedulin (ADM) and STAT-4 was significantly (p<0.05) reduced by HIF-1α or 2α siRNA (Fig. 3A, C, E & G). In contrast to the other genes investigated, the hypoxic induction of adenosine A2a receptor (ADORA2A) and ICAM1 mRNA was significantly (p<0.05) inhibited only by HIF-2α siRNA (Fig. 3I & K).
Transcriptional signaling in primary human MDMs experiencing hypoxia for 18h is independent of NF-κB
(i) Gene Set Enrichment Analysis
To assess the likelihood of NF-κB playing a role in hypoxic signal transduction in human macrophages we first searched our data for correlations with several published gene sets relating to hypoxia-regulated regulated genes in other cell types (eg. the ‘HYPOXIA_REVIEW’ gene set33 - Fig. 4, upper panels). This highlighted a significant degree of enrichment of known hypoxia-regulated genes in our array data, showing that hypoxia induced gene expression changes in MDMs follow a consensus hypoxia gene expression profile (Fig. 4 upper panels). This was evident for both the Affymetrix array data (Normalized Enrichment Score (NES): 2.2; False Discovery Rate (FDR) q<0.001) and the Illumina data (NES: 2.24; q<0.001). Table 1 shows that many genes upregulated by hypoxic MDMs have previously been identified as NF-κB target genes. In the GSEA analysis, the hypoxic MDMS array data also correlated significantly with several NFκB-related gene sets (eg. the V$NFKAPPAB_01 geneset34 - Fig. 4, lower panels). Again, this was evident for both the Affymetrix data (NES: 1.69, q=0.02) and the Illumina data (NES: 1.67; q=0.12).
Figure 4. Gene enrichment analysis to compare key genes upregulated by hypoxia in human MDMs and known NF-κB -regulated genes.
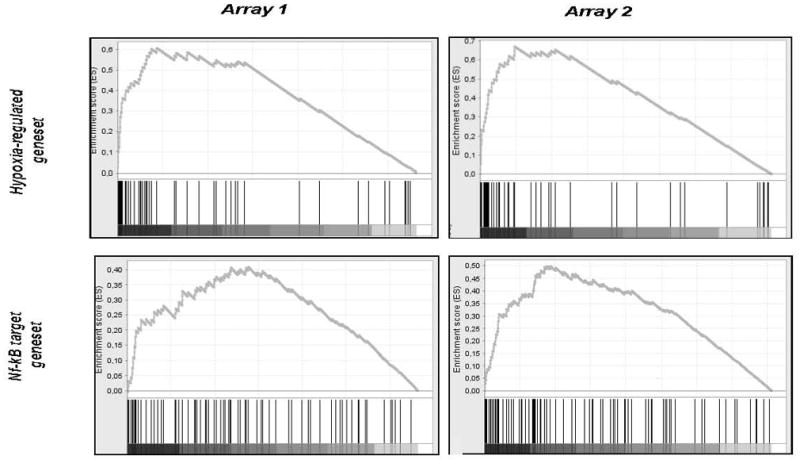
Hypoxia upregulated genes identified in two separate macrophage cultures using Affymetrix or Illumina microarrays were ranked by level of hypoxia-mediated induction. The ranked gene lists were then compared to both a previously published geneset for hypoxia-regulated genes in tumour cells (the ‘hypoxia regulated’ gene set - top row) or genes previously shown to be NFκB target genes (the ‘NFκB target’ geneset – bottom row) by geneset enrichment analysis. The ‘hypoxia regulated’ gene set (top row) was significantly enriched in the hypoxic macrophage geneset identified on both the Affymetrix (‘Array 1’; NES=2.2, q<0.001) and the Illumina (‘Array 2’; NES=2.24, q<0.001) arrays. The NFκB target gene set (lower row) was also enriched in the hypoxic macrophage geneset on both Affymetrix (NES=1.69, q=0.02) and Illumina (NES=1.67, q=0.12) arrays.
(ii) Hypoxic upregulation of NFκB signaling in human macrophages: role of HIFs 1 and 2
The effect of exposure of human MDMs to hypoxia for 18h on NF-κB signalling was then assessed. IKKβ and γ, IκBα, NF-κB1 (p50) and p65/RelA mRNA levels were upregulated (and IKKα mRNA slightly downregulated) in MDMs exposed to 0.1 %O2 for 18h. This hypoxic regulation (with the exception of IKKs α and γ) was inhibited using siRNA to knock down either HIF-1 or 2α (Fig. 5A). Fig. 5C shows that, while there was a small hypoxic induction of total IKKβ protein, the hypoxic upregulation of p65/RelA mRNA was not mirrored by a similar upregulation of total p65/RelA protein, suggesting a differential effect of hypoxia on mRNA versus protein expression for p65/RelA. By contrast, the phosphorylation of both IKKα/β and p65/RelA was upregulated in hypoxic human MDMs (Fig. 5C).
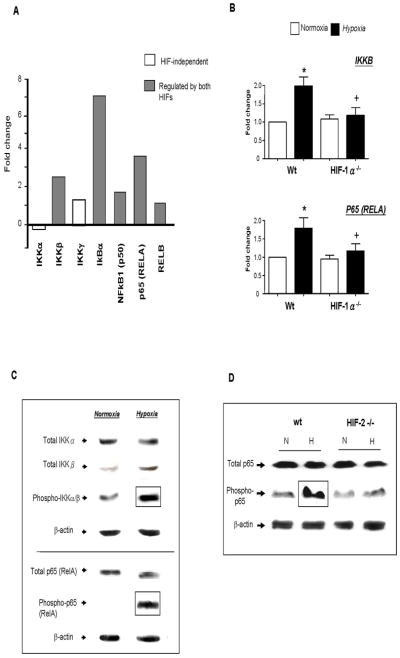
Figure 5. Effect of hypoxia on the expression and/or phosphorylation of components of the canonical NF-κB signaling pathway in MDMs: regulation by HIFs 1α and 2α.
Panel A: Fold induction (hypoxia (0.1% O2)/normoxia (20.9% O2) of mRNA levels for individual NF-κB signaling proteins in primary human MDMs. The contribution of both HIFs 1 and 2 to the regulation of many of these genes was also assessed using siRNA to knock down the expression of each α subunit in MDMs. Panel B: effect of normoxia (N) or hypoxia (0.1% O2; H) for 18h on the expression of mRNA for IKKβ and p65 in murine bone marrow-derived macrophages from wt or HIF-1 α−/− mice. *p<0.05 compared to corresponding normoxic group; +P<0.05 compared to macrophages from wt mice exposed to hypoxia. Panel C: immunoblots showing the effect of exposure to normoxia or hypoxia (0.1% O2) for 18h on the levels of total and phosphorylated IKKβ and p65/RelA in primary human MDMs. Panel D: effects of normoxic (N) or hypoxic (0.5% O2; ‘H’) culture on the level of total or phosphorylated p65 protein in murine bone marrow-derived macrophages from wt or HIF-2α−/− mice. Similar results were obtained using BMDMs from wt and HIF-1−/− mice (data not shown).
(iii) Role of NF-κB in the transcriptional responses of human macrophages to hypoxia
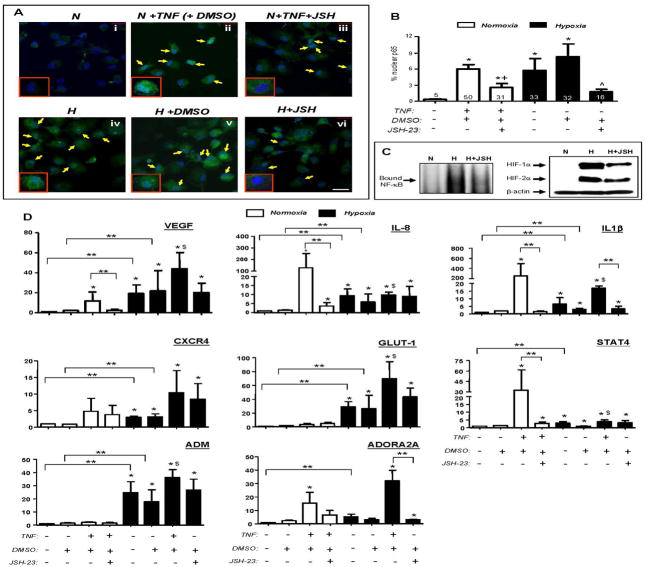
Fig. 6(A and B) illustrates the effects of the NF-kB inhibitor, JSH-23 on the hypoxic induction of various genes in human MDMs. This shows that immunoreactive p65 was cytoplasmic in normoxic MDMs but transported to the nucleus upon exposure to TNFα or 18h hypoxia. In both cases, this was significantly (P<0.05) inhibited by prior exposure to JSH-23. EMSA assays confirmed the induction of NF-κB DNA binding in hypoxic MDMs, and the inhibition of this by JSH-23 (Fig. 6C). JSH-23-treated cells also exhibited slightly lower levels of HIFs 1 and 2α (particularly HIF-1α) than MDMs exposed to hypoxia alone (Fig. 6C). We then investigated the effect of JSH-23 inhibition of NF-κB activity on the induction of 8 hypoxia-regulated genes listed in Table 1. Exposure to TNFα for 18h significantly (P<0.05) increased the expression of VEGF, CXCL8, G1UT1, STAT4 and ADORA2A in a manner that was inhibited by JSH-23 (Fig. 6). Hypoxia significantly (P<0.05) upregulated all eight genes studied (Fig. 6D) in a manner that was not reduced by prior exposure of cells to JSH-23.
Figure 6. Inhibition of nuclear translocation of p65 has no effect on the hypoxic induction of various genes in human MDMs in vitro.
Panels A & B: Effect of the p65 inhibitor, JSH-23, (or its vehicle, DMSO) on the nuclear translocation of p65 induced by TNFα or hypoxia by human MDMs. ‘N’ = normoxia; ‘H’ = hypoxia (0.1% O2). The ‘% nuclear p65 immunofluorescence’ = the % of the total, DAPI-stained (blue) area of MDM nuclei that was GFP+ (green). The figures at the base or just above each bar represents the average % of all MDM nuclei immunofluorescent for p65 (panel B). * P<0.05 w.r.t. ‘normoxia alone’ group; + P<0.05 w.r.t TNFα + DMSO group. ^ P<0.05 w.r.t. hypoxia + DMSO group. Panel C: effect of JSH-23 on NF-κB binding and accumulation of HIFs 1 and 2α in hypoxic MDMs (N = normoxia; H = hypoxia, H + JSH-23 = hypoxia following JSH.23 treatment. All three groups received the vehicle for JSH-23, DMSO); (i) left panel: electromobility shift assay showing NF-κB binding to a DNA consensus sequence, and (ii) right hand panel: immunoblots for HIFs 1 and 2α. Panel D: Effect of JSH-23 blockade of p65 function on the fold induction of VEGF, CXCL8, IL-1β, CXCR4, GLUT-1, STAT4, ADM and ADORA2A by TNFα or hypoxia. *P<0.05 w.r.t. ‘normoxia with DMSO’ alone; ** P<0.05 w.r.t. group indicated; $ P<0.05 w.r.t. TNF + DMSO group. Pooled data from 3 replicate experiments are shown.
Infection of MDMs with adv-IKKβDN significantly (P<0.05) inhibited their TNFα–induced expression of CXCL8 mRNA (Fig. 7A) as well as the nuclear accumulation of phospho-p65/RelA by MDMS after 18h of exposure to hypoxia (Fig. 7B). The control adv vector had no such effect. However, adv-IKKβDN blockade of hypoxia-induced phospho-p65/RelA failed to reduce the hypoxic induction of VEGF, CXCL8, GLUT-1, CXCR4or ADM mRNA.
Figure 7. IKKβ inhibition has no effect on the hypoxic induction of various genes in human MDMs in vitro: use of a recombinant adenovirus expressing a dominant negative inhibitor of IKKβ (adv-IKKβDN).
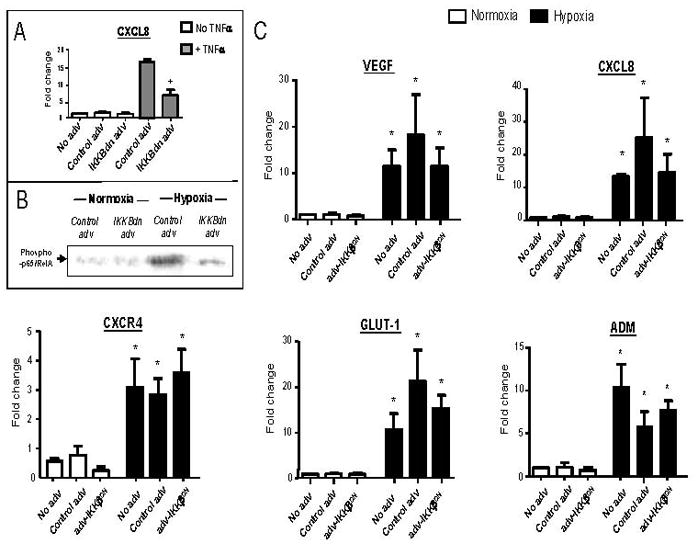
MDM infection with adv-IKKβDN (but not control adv) significantly inhibited both TNFα-induced gene expression of CXCL8 (Panel A) and hypoxia-induced nuclear accumulation of phospho-p65/RelA (Panel B) by human MDM. Panel C: Hypoxia significantly increased the expression of VEGF, CXCL8, CXCR4, GLUT-1 and ADM mRNA in untreated and adenovirally-infected MDM compared to respective normoxic MDM controls. However, there was no significant difference in the expression of these genes between hypoxic MDM infected with adv-IKKβDN or the control adenovirus. N=3. * P<0.05 w.r.t. respective normoxic group, + P<0.05 w.r.t TNFα + control adenovirus group.
Hypoxic regulation of genes in primary murine macrophages: role of HIFs 1 and 2
BMDMs from HIF-1α−/− mice were only able to mount partial VEGFA and IL-1β responses to hypoxia (Fig 1B, right panel and Fig. 2B). The hypoxic upregulation of CXCR4 and STAT4 was lost in HIF-1α null BMDMs (Fig. 3B & H). This contrasts with our aforementioned human MDMs data showing that these were regulated by both HIFs 1 and 2 (Fig. 3A & G). The fact that GLUT1 and ADM was reduced but not ablated in HIF-1α null BMDMs (Fig. 3D & F) agrees with our finding that these 2 genes are co-regulated by HIFs 1 and 2 in human MDMs (Fig. 3C & E). Also in agreement with the human MDMs data (Fig. 3I & K), the hypoxic upregulation of neither the ADORA2A nor ICAM1 genes was inhibited in hypoxic HIF-1α null BMDMs (Fig. 3J & L).
Fig. 5B shows that hypoxic upregulation of IKKβ and p65/RelA mRNA levels was lost in HIF-1α−/− BMDMs. Moreover, the hypoxic induction of phosphorylated p65/RelA was lost in HIF-2a−/− BMDMs (Fig. 5D). Similar results were seen for HIF-1α −/− BMDMs (data not shown).
DISCUSSION
Our data show that exposure to hypoxia activates a distinct transcriptional profile in primary human macrophages, including the upregulation of VEGFA, ILs-1α and β, IL-8, STAT4, ADM; the receptors, glucose transporter, GLUT1, CXCR4 and the adenosine receptor 2 A (ADORA2). Some were seen to also be regulated by hypoxia in monocytes and immature dendritic cells (VEGFA, GLUT1 and CXCR4).31,32 However, others like IL-1β, ADORA2 A and STAT4 were only altered in hypoxic macrophages. These differences could be due to variations in the severity and/or duration of hypoxia applied to cells,31,32 and/or may reflect differences in the transcription factors employed by these 3 cell types in hypoxia. For example, hypoxic human monocytes exposed to a similar level and duration of hypoxia as in the current study failed to upregulate HIFs 1 and 2α but rather other transcription factors like ATF-4 and Egr-1.35 Moreover, the ability to regulate hypoxic gene expression via HIFs is maturation-linked in macrophages.36 Although dendritic cells accumulate HIF-1α in hypoxia,37 immature forms of this cell type upregulate other hypoxia-responsive genes like CCL20 via upregulated p50/p50 NFκB homodimers rather than HIFs.38 A study of the responses of such related myeloid cell types to identical hypoxic conditions would be interesting but beyond the remit of this study.
As macrophages are known to express receptors for both VEGF39 and IL-140, it is possible that that during such exposure to hypoxia, their hypoxia-induced release might have then stimulated the expression of other genes in macrophages (making it look as if they are also directly upregulated by hypoxia when, in fact, the effect is indirect). However, hypoxic gene expression by human MDMs is not reduced in the presence of either a neutralising VEGF antibody or an IL-1 receptor antagonist (Fang H-Y, Murdoch C, Hughes R and Lewis CE, unpublished observations).
We also show for the first time that genes encoding the two transcription factors, STATs (signal transducers and activators of transcription) 4 and 6, are upregulated by hypoxia in macrophages. STAT 4 and 6 are known to mediate the marked effects of two central immunomodulatory cytokines, IL-12 and IL-4, respectively.41,42 It remains to be seen whether their hypoxic induction could ‘prime’ macrophages to the effects of these cytokines.
Our HIF siRNA studies showed that both HIFs play a part in regulating the hypoxic induction of the known HIF target genes, VEGF, GLUT1, CXCR4, IL-8 and ADM by MDMs. Furthermore, hypoxic induction of these genes was reduced but not lost in murine macrophages from HIF-1α−/− mice. Similar results were obtained for the hypoxic upregulation of VEGF and ADM in murine HIF-2α−/− BMDMs (H.Z. Imtiyaz, M.C. Simon & colleagues, submitted).
The pluripotent cytokine, IL-1β, stimulates many steps in tumor progression43 and was upregulated by hypoxic MDMs. We show that TAMs express abundant IL-1β in hypoxic areas of murine mammary (4T1) tumors. The IL-1β gene promoter bears multiple HREs and is transactivated by HIF-144,45 Our HIF siRNA knock down studies show that HIFs 1 and 2 co-regulate the hypoxic induction of IL-1β in macrophages – a finding confirmed by the hypoxic upregulation of IL-1β being only partially diminished in murine BMDMs from HIF-1α−/− (Fig. 1) and HIF-2α−/− mice (H.Z. Imtiyaz, M.C. Simon and colleagues, submitted).
It remains to be seen whether HIFs 1 and 2 bind to different HREs on the promoters of the above co-regulated genes or whether other, unknown mechanisms underpin the phenomenon of dual HIF responsiveness. Furthermore, as mentioned previously, this may vary between cell types as HIF-1 has been shown to be the primary regulator of various genes in some cell types,16,46 while other cells employ HIF-2 or both HIFs in their regulation.18,19,47 Interestingly, when just one HIF was inhibited using siRNA the other did not appear to compensate for its loss and maintain maximal hypoxic induction. It is known that many HIF-target genes have multiple HREs. If, once HIFs 1 and 2 have bound to different HREs in a given promoter they then co-operate, both might be required for maximal gene transcription.. This may be similar to the molecular ‘co-operation’ that takes place between HIF-2 and Elk-1 on some gene promoters.18,19
The complete knockdown of both HIFs 1α and 2α in MDMs failed to completely block the hypoxic upregulation of most of the HIF-target genes discussed above. This suggests that other transcription factors may also be involved in regulating their hypoxic induction. The transcription factor, NF-κB, may be one such factor. This has been shown recently to be activated in macrophages by short-term (2–4h) exposure to hypoxia, with the expression and/or phosphorylation of IKKβ, IKBα and p65/RelA in macrophages, as well as the nuclear translocation and DNA-binding activity of p65 being upregulated.20–22 There also appears to be a close interplay between NF-κB and HIF-1 as p65/p50 heterodimers bind to the HIF-1α gene promoter and drives its expression under hypoxia. Interestingly, HIF-2α is not upregulated by NF-κB in murine macrophages during short-term hypoxia (4h).22 The present report shows that p65 protein is phosphorylated and binds DNA in the nuclei of MDMs in hypoxia. Furthermore we show that both HIFs 1 and 2 contribute to the maintenance of high levels of p65 expression and phosphorylation in such cells.
As many of the genes we found to be markedly upregulated in human macrophages by hypoxia had previously been identified as potential NF-κB target genes (Table 1 and Fig. 5), we examined the role of p65 in the hypoxic upregulation of the most highly ones. Studies using the synthetic inhibitor of nuclear translocation of p65, JSH-23,24 or an adenoviral inhibitor of IKKβ showed that NF-κB is not essential for their induction during an 18h exposure to hypoxia. The upregulation of HIF-1α in macrophages exposed to short-term hypoxia (4h) is partially dependent on NF-kB22 so the fact that both HIFs 1 and 2α continued to be upregulated in JSH-treated MDMs following hypoxia in our study suggests that, either p65 inhibition was incomplete or that the accumulation of these sub units during a more sustained period (18h) of hypoxia is independent of NF-κB.
While both forms of NF-κB inhibition resulted in the marked inhibition of TNFα-induced CXCL8 (as well as other genes examined with JSH-23), it had no detectable effect on the hypoxic expression of any of the genes examined. These data are supported by the recent finding that that the hypoxic induction of several such NF-κB target genes in murine BMDMs does not involve activation of NF-κB (H.Z. Imtiyaz & M.C. Simon, submitted).
Our data indicate that NF-κB signaling may not contribute to the induction of these genes by macrophages in response to an 18h exposure to hypoxia. At first glance, this appears to contrast with the finding that hypoxic induction of HIF-1α and various HIF target genes was diminished in BMDMs from IKKβ−/− mice following exposure to short-term hypoxia (ie. 4h).22 However, it may be that there is a switch from acute, NF-κB-dependent hypoxic responses in macrophages that are critical for innate immunity (eg. bacterial infection) to an NF-κB-independent, HIF-driven response to the chronic hypoxia present at sites like tumors. Clearly, a detailed investigation of the role of NF-κB in the temporal and gene-specific responses of macrophages to hypoxia is now warranted.
Taken together, our data show that when macrophages experience hypoxia for 18h it elicits a profound change in their expression of various tumor-promoting genes. While this study provides invaluable insights into the basic repertoire of such macrophage responses, it should be remembered that macrophages in hypoxic areas of complex tissues like tumors are a heterogenous mix of cells including immature, monocyte-like cells and mature macrophages.6 Moreover, the responses of these cells to hypoxia will also be influenced by a host of tumor-derived signals like cytokines and enzymes. Further in vivo studies are now warranted to investigate the role of hypoxic macrophage responses within the complex milieu of tumors.
Acknowledgments
The authors gratefully acknowledge grant support from the Yorkshire Cancer Research, UK and Breast Cancer Campaign, UK for different parts of this study. The authors also thank Drs Bosco & Varesio (Italy) and Michael Karin (USA) for their helpful comments in the preparation of this manuscript. They also thank Dr Thorsten Hagemann (London) for providing the control and IKKBdn adenoviruses and Dr Sheila Francis (Sheffield) for her help with the adenoviral infections.
Footnotes
Author contribution: Hsin-Yu Fang designed & performed research. Russell Hughes - designed & performed research. Craig Murdoch - designed & performed research. Seth Coffelt designed & performed research. Subhra K. Biswas - designed & performed research. Adrian L. Harris contributed vital new reagents or analytical tools, analyzed data and wrote the paper. Randall S. Johnson contributed vital new reagents or analytical tools and analyzed data. Hongxia Z. Imityaz performed research. M. Celeste Simon contributed vital new reagents and analytical tools, analyzed data. Erik Fredlund designed & performed research. Florian Greten designed & performed research. Jordi Rius designed & performed research. Claire E. Lewis contributed vital reagents and analytical tools, analyzed data, wrote the paper.
References
- 1.Vaupel P, Kelleher DK, Hockel M. Oxygen status of malignant tumors: pathogenesis of hypoxia and significance for tumor therapy. Semin Oncol. 2001;28:29–35. doi: 10.1016/s0093-7754(01)90210-6. [DOI] [PubMed] [Google Scholar]
- 2.Taylor PC, Sivakumar B. Hypoxia and angiogenesis in rheumatoid arthritis. Curr Opin Rheumatol. 2005;17:293–8. doi: 10.1097/01.bor.0000155361.83990.5b. [DOI] [PubMed] [Google Scholar]
- 3.Björnheden T, Levin M, Evaldsson M, Wiklund O. Evidence of hypoxic areas within the arterial wall in vivo. Arterioscler Thromb Vase Biol. 1999;19:870–6. doi: 10.1161/01.atv.19.4.870. [DOI] [PubMed] [Google Scholar]
- 4.Bertout JA, Patel SA, Simon MC. The impact of O2 availability on human cancer. Nat Rev Cancer. 2008;8:967–75. doi: 10.1038/nrc2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Semenza GL. Hypoxia-inducible factor 1: oxygen homeostasis and disease pathophysiology. Trends Mol Med. 2001;7:345–350. doi: 10.1016/s1471-4914(01)02090-1. [DOI] [PubMed] [Google Scholar]
- 6.Lewis CE, Pollard JW. Distinct role of macrophages in different tumor microenvironments. Cancer Res. 2006;66:605–12. doi: 10.1158/0008-5472.CAN-05-4005. [DOI] [PubMed] [Google Scholar]
- 7.Murdoch C, Giannoudis A, Lewis CE. Mechanisms regulating the recruitment of macrophages into hypoxic areas of tumors and other ischemic tissues. Blood. 2004;104:2224–34. doi: 10.1182/blood-2004-03-1109. [DOI] [PubMed] [Google Scholar]
- 8.Lewis C, Murdoch C. Macrophage responses to hypoxia: implications for tumor progression and anti-cancer therapies. Am J Pathol. 2005;167:627–35. doi: 10.1016/S0002-9440(10)62038-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murdoch C, Lewis CE. Macrophage migration and gene expression in response to tumor hypoxia. Int J Cancer. 2005;117:701–8. doi: 10.1002/ijc.21422. [DOI] [PubMed] [Google Scholar]
- 10.Burke B, Tang N, Corke KP, Tazzyman D, Ameri K, Wells M, Lewis CE. Expression of HIF-1 alpha by human macrophages: implications for the use of macrophages in hypoxia-regulated cancer gene therapy. J Pathol. 2002;196:204–212. doi: 10.1002/path.1029. [DOI] [PubMed] [Google Scholar]
- 11.Talks KL, Turley H, Gatter KC, Maxwell PH, Pugh CW, Ratcliffe PJ, Harris AL. The expression and distribution of the hypoxia-inducible factors HIF-1 alpha and HIF-2 alpha in normal human tissues, cancers, and tumor-associated macrophages. Am J Pathol. 2000;157:411–421. doi: 10.1016/s0002-9440(10)64554-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis JS, Landers RJ, Underwood JC, Harris AL, Lewis CE. Expression of vascular endothelial growth factor by macrophages is up-regulated in poorly vascularized areas of breast carcinomas. J Pathol. 2000;192:150–8. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH687>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 13.Leek RD, Talks KL, Pezzella F, Turley H, Campo L, Brown NS, Bicknell R, Taylor M, Gatter KC, Harris AL. Relation of hypoxia-inducible factor-2 alpha (HIF-2 alpha) expression in tumor-infiltrative macrophages to tumor angiogenesis and the oxidative thymidine phosphorylase pathway in Human breast cancer. Cancer Res. 2002;62:1326–9. [PubMed] [Google Scholar]
- 14.White JR, Harris RA, Lee SR, et al. Genetic amplification of the transcriptional response to hypoxia as a novel means of identifying regulators of angiogenesis. Genomics. 2004;83:1–8. doi: 10.1016/s0888-7543(03)00215-5. [DOI] [PubMed] [Google Scholar]
- 15.Cramer T, Yamanishi Y, Clausen BE, Förster I, Pawlinski R, Mackman N, Haase VH, Jaenisch R, Corr M, Nizet V, Firestein GS, Gerber HP, Ferrara N, Johnson RS. HIF-1 alpha is essential for myeloid cell-mediated inflammation. Cell. 2003;112:645–57. doi: 10.1016/s0092-8674(03)00154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park SK, Dadak AM, Haase VH, Fontana L, Giaccia AJ, Johnson RS. Hypoxia-induced gene expression occurs solely through the action of hypoxia-inducible factor 1 alpha (HIF-1 alpha): role of cytoplasmic trapping of HIF-2alpha. Mol Cell Biol. 2003;23:4959–4971. doi: 10.1128/MCB.23.14.4959-4971.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sowter HM, Raval RR, Moore JW, Ratcliffe PJ, Harris AL. Predominant role of hypoxia-inducible transcription factor (Hif)-1alpha versus Hif-2alpha in regulation of the transcriptional response to hypoxia. Cancer Res. 2003;63:6130–6134. [PubMed] [Google Scholar]
- 18.Hu CJ, Sataur A, Wang L, Chen H, Simon MC. The N-terminal transactivation domain confers target gene specificity of hypoxia-inducible factors HIF-1alpha and HIF-2alpha. Mol Biol Cell. 2007;18(11):4528–42. doi: 10.1091/mbc.E06-05-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elvert G, Kappel A, Heidenreich R, Englmeier U, Lanz S, Acker T, Rauter M, Plate K, Sieweke M, Breier G, Flamme I. Cooperative interaction of hypoxia-inducible factor-2alpha (HIF-2 alpha) and Ets-1 in the transcriptional activation of vascular endothelial growth factor receptor-2 (Flk-1) J Biol Chem. 2003;278:7520–30. doi: 10.1074/jbc.M211298200. [DOI] [PubMed] [Google Scholar]
- 20.Van Uden P, Kenneth NS, Rocha S. Regulation of hypoxia-inducible factor-1 alpha by NF-kappa B. Biochem J. 2008;412:477–84. doi: 10.1042/BJ20080476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cummins EP, Berra E, Comerford KM, Ginouves A, Fitzgerald KT, Seeballuck F, Godson C, Nielsen JE, Moynagh P, Pouyssegur J, Taylor CT. Prolyl hydroxylase-1 negatively regulates IkappaB kinase-beta, giving insight into hypoxia-induced NFkappaB activity. Proc Natl Acad Sci USA. 2006;103(48):18154–9. doi: 10.1073/pnas.0602235103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rius J, Guma M, Schachtrup C, Akassoglou K, Zinkernagel AS, Nizet V, Johnson RS, Haddad GG, Karin M. NF-kappaB links innate immunity to the hypoxic response through transcriptional regulation of HIF-1alpha. Nature. 2008;453:807–11.22. doi: 10.1038/nature06905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peyssonnaux C, Datta V, Cramer T, Doedens A, Theodorakis EA, Gallo RL, Hurtado-Ziola N, Nizet V, Johnson RS. HIF-1alpha expression regulates the bactericidal capacity of phagocytes. J Clin Invest. 2005;115:1806–15. doi: 10.1172/JCI23865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shin HM, Kim MH, Kim BH, Jung SH, Kim YS, Park HJ, Hong JT, Min KR, Kim Y. Inhibitory action of novel aromatic diamine compound on lipopolysaccharide-induced nuclear translocation of NF-kappaB without affecting IkappaB degradation. FEBS Lett. 2004;571:50–4. doi: 10.1016/j.febslet.2004.06.056. [DOI] [PubMed] [Google Scholar]
- 25.Greten FR, Arkan MC, Bollrath J, Hsu LC, Goode J, Miething C, Göktuna SI, Neuenhahn M, Fierer J, Paxian S, Van Rooijen N, Xu Y, O’Cain T, Jaffee BB, Busch DH, Duyster J, Schmid RM, Eckmann L, Karin M. NF-kappaB is a negative regulator of IL-1beta secretion as revealed by genetic and pharmacological inhibition of IKKbeta. Cell. 2007;130(5):918–31. doi: 10.1016/j.cell.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bondeson J, Browne KA, Brennan FM, Foxwell BM, Feldmann M. Selective regulation of cytokine induction by adenoviral gene transfer of IkappaBalpha into human macrophages: lipopolysaccharide-induced, but not zymosan-induced, proinflammatory cytokines are inhibited, but IL-10 is nuclear factor-kappaB independent. J Immunol. 1999;162:2939–2945. [PubMed] [Google Scholar]
- 27.Hagemann T, Lawrence T, McNeish I, et al. “Re-educating” tumor-associated macrophages by targeting NF-kappaB. J Exp Med. 2008;205:1261–1268. doi: 10.1084/jem.20080108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oitzinger W, Hofer-Warbinek R, Schmid JA, Koshelnick Y, Binder BR, de Martin R. Adenovirus-mediated expression of a mutant IkappaB kinase 2 inhibits the response of endothelial cells to inflammatory stimuli. Blood. 2001;97:1611–7. doi: 10.1182/blood.v97.6.1611. [DOI] [PubMed] [Google Scholar]
- 29.Ryan RM, Green J, Williams PJ, Tazzyman S, Hunt S, Harmey JH, Kehoe SC, Lewis CE. Bacterial delivery of a novel cytolysin to hypoxic areas of solid tumors. Gene Therapy. 2009 doi: 10.1038/gt.2008.188. (in press) [DOI] [PubMed] [Google Scholar]
- 30.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bosco MC, Puppo M, Santangelo C, Anfosso L, Pfeffer U, Fardin P, Battaglia F, Varesio L. Hypoxia modifies the transcriptome of primary human monocytes: modulation of novel immune-related genes and identification of CC-chemokine ligand 20 as a new hypoxia-inducible gene. J Immunol. 2006;177:1941–55. doi: 10.4049/jimmunol.177.3.1941. [DOI] [PubMed] [Google Scholar]
- 32.Ricciardi A, Elia AR, Cappello P, Puppo M, Vanni C, Fardin P, Eva A, Munroe D, Wu X, Giovarelli M, Varesio L. Transcriptome of hypoxic immature dendritic cells: modulation of chemokine/receptor expression. Mol Cancer Res. 2008;6:175–85. doi: 10.1158/1541-7786.MCR-07-0391. [DOI] [PubMed] [Google Scholar]
- 33.Harris AL. Hypoxia - a key regulatory factor in tumour growth. Nat Rev Cancer. 2002;2(l):38–47. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- 34.Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434(7031):338–45. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elbarghati L, Murdoch C, Lewis CE. Effects of hypoxia on transcription factor expression in human monocytes and macrophages. Immunobiology. 2008;213:899–908. doi: 10.1016/j.imbio.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 36.Oda T, Hirota K, Nishi K, Takabuchi S, Oda S, Yamada H, Arai T, Fukuda K, Kita T, Adachi T, Semenza GL, Nohara R. Activation of hypoxia-inducible factor 1 during macrophage differentiation. Am J Physiol Cell Physiol. 2006;291(1):C104–13. doi: 10.1152/ajpcell.00614.2005. [DOI] [PubMed] [Google Scholar]
- 37.Elia AR, Cappello P, Puppo M, Fraone T, Vanni C, Eva A, Musso T, Novelli F, Varesio L, Giovarelli M, Varesio L, Giovarelli M. Human dendritic cells differentiated in hypoxia down-modulate antigen uptake and change their chemokine expression profile. J Leukoc Biol. 2008;84(6):1472–82. doi: 10.1189/jlb.0208082. [DOI] [PubMed] [Google Scholar]
- 38.Battaglia F, Delfino S, Merello E, Puppo M, Piva R, Varesio L, Bosco MC. Hypoxia transcriptionally induces macrophage-inflammatory protein-3alpha/CCL-20 in primary human mononuclear phagocytes through nuclear factor (NF)-kappaB. J Leukoc Biol. 2008;83:648–62. doi: 10.1189/jlb.0607349. [DOI] [PubMed] [Google Scholar]
- 39.Lee MC, Wei SC, Tsai-Wu JJ, Wu CH, Tsao PN. Novel PKC signaling is required for LPS-induced soluble Flt-1 expression in macrophages. J Leukoc Biol. 2008;84:835–41. doi: 10.1189/jlb.1007691. [DOI] [PubMed] [Google Scholar]
- 40.Diekensheets HL, Donnelly RP. I FN-gamma and IL-10 inhibit induction of IL-1 receptor type I and type II gene expression by IL-4 and IL-13 in human monocytes. J Immunol. 1997;159:6226–33. [PubMed] [Google Scholar]
- 42.Cho SS, Bacon CM, Sudarshan C, Rees RC, Finbloom D, Pine R, O’Shea JJ. Activation of STAT4 by IL-12 and IFN-alpha: evidence for the involvement of ligand-induced tyrosine and serine phosphorylation. J Immunol. 1996;157(11):4781–9. [PubMed] [Google Scholar]
- 42.Hou J, Schindler U, Henzel WJ, Ho TC, Brasseur M, McKnight SL. An interleukin-4-induced transcription factor: IL-4 Stat. Science. 1994;265 (5179):1701–6. doi: 10.1126/science.8085155. [DOI] [PubMed] [Google Scholar]
- 43.Apte RN, Dotan S, Elkabets M, White MR, Reich E, Carmi Y, Song X, Dvozkin T, Krelin Y, Voronov E. The involvement of IL-1 in tumorigenesis, tumor invasiveness, metastasis and tumor-host interactions. Cancer Metastasis Rev. 2006;25(3):387–408. doi: 10.1007/s10555-006-9004-4. [DOI] [PubMed] [Google Scholar]
- 44.Hirani N, Antonicelli F, Strieter RM, Wiesener MS, Ratcliffe PJ, Haslett C, Donnelly SC. The regulation of interleukin-8 by hypoxia in human macrophages - a potential role in the pathogenesis of the acute respiratory distress syndrome (ARDS) Mol Med. 2001;7(10):685–97. [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang W, Petrovic JM, Callaghan D, Jones A, Cui H, Howlett C, Stanimirovic D. Evidence that hypoxia-inducible factor-1 (HIF-1) mediates transcriptional activation of interleukin-1beta (IL-1beta) in astrocyte cultures. J Neuroimmunol. 2006;174(1–2):63–73. doi: 10.1016/j.jneuroim.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 46.Carroll VA, Asheroft M. Role of hypoxia-inducible factor (HIF)-1 alpha versus HIF-2alpha in the regulation of HIF target genes in response to hypoxia, insulin-like growth factor-I, or loss of von Hippel-Lindau function: implications for targeting the HIF pathway. Cancer Res. 2006;66(12):6264–70. doi: 10.1158/0008-5472.CAN-05-2519. [DOI] [PubMed] [Google Scholar]
- 47.Meade ES, Ma YY, Guller S. Role of hypoxia-inducible transcription factors 1alpha and 2alpha in the regulation of plasminogen activator inhibitor-1 expression in a human trophoblast cell line. Placenta. 2007;28:1012–9. doi: 10.1016/j.placenta.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]