Abstract
Background
Melastatin (TRPM1), a.k.a. transient receptor potential cation channel, subfamily M, member 1 (TRPM-1) regulates melanocyte differentiation and proliferation. TRPM1 is transcriptionally regulated by the essential melanocyte transcription factor MITF (microphthalmia-associated transcription factor). For the most part, MITF expression is preserved during melanoma progression, while TRPM1 mRNA expression decreases or is completely lost. The loss of TRPM1 is associated with melanomas that are more aggressive.
Objective
To assess the relationship between TRPM1 mRNA expression and the expression of MITF and nine other markers of melanocytes and melanin-related proteins by immunohistochemistry in normal skin, scars, hair follicles and ordinary melanocytic nevi.
Methods
Samples of normal skin (n = 102; from tumor excisions and plastic procedures), scars (n = 5; from re-excision specimens) and compound melanocytic nevi (n = 4) were evaluated for the presence of TRPM1 mRNA transcripts as detected by chromogenic in situ hybridization (CISH). Immunohistochemical techniques were used to detect melanin-related proteins including: MITF, S100 protein, Mart-1, tyrosinase, Mel5, HMB45, tyrosinase-related protein-1 (TRP1), TRP2 and α-melanocyte stimulating hormone (αMSH). The labeling index (LI) was defined as the number of intraepidermal cells expressing mRNA or protein per one hundred basal keratinocytes.
Results
A wide range of LI was found for all markers (0–33 positive cells/100 keratinocytes). When these LI were compared, no significant differences in the expression of MITF, S100, Mart1, tyrosinase proteins and TRPM1 mRNA were identified. The LI for TRPM1 mRNA expression ranged from 74% of that for MITF to 86% for tyrosinase. The LI for TRP-1, TRP-2 and Mel5 was similar to that of TRPM1, while HMB-45 had a significantly lower LI than all other markers. TRPM1 mRNA correlated most tightly with MITF and tyrosinase expression (r = 0.81 and 0.68, respectively, both p = 0.0001). Likewise, the strongest correlation among all the melanin-related proteins existed between tyrosinase and MITF (r = 0.79, p = 0.0001). There was variable expression of melanin-related proteins when LI were analyzed by anatomic site, patient age, extent of sun-damage and proximity to a melanocytic tumor. Anogenital skin showed the highest and acral skin the lowest LI for TRPM1, MITF, S100 protein, Tyrosinase, Mel5 and HMB45. Advanced age (>60 years) was associated with decreased TRPM1 expression. Sun-damaged skin exhibited significantly increased LI as measured by MITF, S100 protein, Mart1, tyrosinase and HMB-45, but no differences for TRPM1. However, the MITF-TRPM1 differential (i.e. MITF LI-TRPM1 LI = MITF+TRPM1 – melanocytes) was significantly increased in site-matched skin (4.6 ± 4.4 vs. 1.5 ± 2.5, p = 0.01). There was a suggestion of reduced LI in normal skin in the proximity of melanoma (from melanoma re-excision specimens) for S100, HMB45 and TRPM1 mRNA. TRPM1 LI was significantly decreased in scars compared to normal skin (5.6 ± 1.4 vs. 9.7 ± 4.3, p = 0.02), this was reflected in an increase in the MITF-TRPM1 differential (9.6 ± 7.5 vs. 3.2 ± 3.1, p = 0.0001). MITF LI were consistently higher than MSLN LI at all levels of the hair follicle; notably, MITF was expressed by isthmic-bulge cells. In ordinary melanocytic nevi, MITF and TRPM1 expression decreased with melanocyte descent: there was more signal for both markers in superficial epithelioid type A melanocytes than deeper type C melanocytes.
Conclusions
By CISH, TRPM1 mRNA expression is specific for melanocytes and strongly associated with MITF and tyrosinase expression, the latter implicating a mature melanocyte phenotype. However, in normal skin, TRPM1 mRNA expression appears to be dynamic, labeling most but not all melanocytes, with variable expression ostensibly related to local environmental factors.
In addition to introducing fundamental concepts pertinent to the relationship of morphology to clinical behavior of malignant melanoma, Dr Mihm also pioneered the field of translationally relevant biomarkers. One recently described biomarker, melastatin, (TRPM1) was first identified through differential cDNA display of F1 and F10 murine melanoma cell lines that differed in metastatic capacity. This gene was selected for investigation because it was absent from highly metastatic melanoma cell lines, implying a tumor-suppressor gene function.1 TRPM1, a.k.a. transient receptor potential cation channel, subfamily M, member 1 (TRPM-1), is a melanocyte-specific gene localized to human chromosome 15.2,3 TRPM1 is the founding member of one of 7 families of TRP ion channels, which comprise more than 50 cation-permeable channels that are grouped according to structural homology: TRPM (melastatin), TRPC (canonical), TRPV (vanilloid), TRPP (polycystin), TRPML (mucolipin), TRPA (ankyrin) and TRPN (NO mechanopotential).4 TRPM channels exhibit highly variable permeability to Ca2+ and Mg2+, ranging from impermeable to Ca2+ to highly permeable to both Ca2+ and Mg2+; various gating mechanisms such as hydrogen peroxide and heat; and assorted activating mechanisms such as cooling, cooling agents and cell swelling.4 To date, the functional characteristics of TRPM1 have not been reported; however, it is suspected that TRPM1 plays a role in controlling intracellular Ca2+ concentration,5 by which loss of TRPM1 expression alters bipolar cell signaling and melanocyte function.6 Furthermore, TRPM1 is one of several TRP related to cancer (others include TRPV5, TRPV6 and TRPM8), in which altered expression of Ca2+-entry channels is suspected to play a role in tumor cell proliferation and differentiation.4 Indeed, utilizing radioactive in situ hybridization (RISH) methods, TRPM1 mRNA was expressed in normal melanocytes and nevomelanocytes, whereas some primary melanomas and all melanoma metastases showed some loss of TRPM1 mRNA.1,7,8 Fang et al.9 confirmed TRPM1’s role in differentiation by demonstrating that the differentiation inducer hexamethylene bisacetamide (HMBA) upregulates TRPM1 gene expression in pigmented melanoma cell lines. The expression of TRPM1 itself is tightly controlled by the essential melanocyte transcription factor MITF (microphthalmia-associated transcription factor).10,11
King, Mihm and colleagues12,13 demonstrated that the antibody to MITF protein is a sensitive and relatively specific marker of melanocytes and melanoma. MITF is a member of the basic helix-loop-helix leucine-zipper transcription family and has been shown to be a critical regulator of melanocyte development and survival.14 Indeed, MITF has been termed a master regulator of melanocyte development and a melanoma oncogene,15 as it governs melanoblast survival and melanocyte lineage commitment;15 modulates melanocyte differentiation gene and cell-cycle gene expression;11 is amplified in a fraction of melanomas;16 and is expressed by most melanomas.12,13 In vitro experiments have demonstrated that MITF plays both pro-and anti-tumor roles in melanoma depending on the expression level and cellular context. For example, its expression is essential for melanoma cell proliferation and survival.15 However, it can also inhibit proliferation and cellular viability as well as stimulate differentiation,17 as MITF is expressed at lower levels in melanoma than in melanocytes, where increased levels of MITF reduce melanoma proliferation even in the presence of oncogenic BRAF,18 and high levels of MITF reduce melanoma tumorigenicity. Thus, high levels of MITF predispose to cell-cycle arrest and differentiation; critically low levels lead to cell-cycle arrest and apoptosis; and only at intermediate levels is melanoma proliferation favored.17 The finding that increasing loss of TRPM1 correlates with melanoma progression and aggressive disease supports this molecular pathway of melanoma7,8,20 and points to a tightly coupled relationship between MITF and TRPM1.
In addition to TRPM1, MITF regulates the transcription of multiple gene products involved in melanogenesis, including tyrosinase, tyrosinase-related protein-1 (TRP1) (Mel5; gene – TRP1), TRP2 (dopachrome tautomerase; DCT), HMB45 (gp100, PMEL17; SILV), Mart1 (melanoma antigen recognized by T cells) (Melan A; MLANA) and melanocortin 1 receptor (MC1R), whose ligand is α-melanocyte stimulating hormone (αMSH).11,21 The specific pattern of expression of some of these markers of melanogenesis or melanocyte proteins (melanogenesis-related proteins) correlates with the specific stage of melanocyte development.22 For example, adult melanocyte stem cells express TRP2 and MITF, whereas acquisition of tyrosinase and TRP1 expression is a sign of a terminally differentiated melanocyte. In addition, a gradient of decreasing or loss of expression of specific melanin-related proteins has been described for MITF12 and Mart123 as well as for TRPM1 by RISH7 in melanocytic nevi showing zonal maturation.
Here, utilizing CISH methods,20 we sought to evaluate the extent of TRPM1 mRNA expression in melanocytes found in normal epidermis and regenerating epidermis-overlying scars. To determine whether TRPM1 mRNA correlates with a specific stage of melanocyte differentiation, its expression was compared to the immunohistochemical detection of melanogenesis-related proteins in matched skin samples. In addition, the expression of TRPM1 was examined in hair follicles and ordinary compound melanocytic nevi and compared to MITF expression.
Materials and methods
Samples
To examine the expression of melanin-related proteins and TRPM1 mRNA by epidermal melanocytes, 102 histologically normal skin samples were chosen and paraffin blocks were retrieved from cosmetic and excision specimens from the surgical pathology files at Albany Medical College (AMC) covering a 3-year period. These cases were selected to obtain a relatively even distribution of normal skin from all ages, both sexes, all anatomical sites and varied associated pathologic processes (e.g. benign or malignant skin tumors) (see Table 1 for clinical details of cohort). Analysis of hair follicle distribution of MITF protein and TRPM1 mRNA bearing cells was performed on these samples (longitudinal follicle cross-sections). Cases designated to contain regenerating epidermis included five re-excision specimens with acute scars 4–8 weeks old. None contained residual tumor; mean patient age was 39 (range 20–71 years), and sites included trunk (n = 3) and face (n = 2). These cases were excisions of dysplastic melanocytic nevus (n = 3) and skin cancer (n = 2). Cases of common melanocytic nevi included four melanocytic nevi with type A, B and C melanocytes (all cases showed partial neuritization) from the trunk (n = 2) and extremity (n = 2) of males 1–14 years old and females 3–10, 16 and 41 years old. Hematoxylin and eosin-stained sections from all material were reviewed to confirm classification and exclude any other pathologic process. The Institutional Review Board of AMC approved this study.
Table 1.
Demographics and characteristics of normal skin samples
| Clinicopathologic features | Normal skin |
|---|---|
| Samples | n = 102 |
| Age* | 38 ± 25 years 0–88 |
| ≤20 years | 32 (32%) |
| 21 – 40 years | 22 (22%) |
| 41 – 60 years | 23 (23%) |
| >60 years | 24 (24%) |
| Male: Female* | 42: 58 |
| Site | |
| Acral | 14 (14%) |
| Anogenital and areolar | 19 (19%) |
| Extremity | 15 (15%) |
| Head and neck | 21 (21%) |
| Trunk | 33 (32%) |
| Reason for excision | |
| Cosmetic or debridement | 42 (42%) |
| Benign tumor | 36 (36%) |
| Malignant tumor | 24 (24%) |
| Associated skin findings | |
| Solar elastosis | 16 (16%) |
| Adjacent melanocytic nevus | 18 (18%) |
| Adjacent melanoma | 6 (6%) |
| Adjacent carcinoma | 16 (16%) |
Age and sex information were not available for 1 and 2 patients, respectively.
Chromogenic in situ hybridization (CISH) assay
Details of the CISH assay can be found in Table 2 and have been previously published.20 Briefly, formalin-fixed, paraffin-embedded 5-μm tissue sections were cut and placed on plus slides in an RNase-free environment. The slides were then deparaffinized, digested with an appropriate concentration of proteinase K and dehydrated with ethanol. The sections were then hybridized with a colorimetric melastatin mRNA detection probe overnight at 37°C. Following a post-hybridization wash, the sections were incubated with anti-digoxigenin-AP for detection and covered with BCIP/NBT for signal generation. Sections were counterstained with Nuclear Fast Red. Bluish-purple melanocytic nuclei were scored as having retained melastatin expression, while pink melanocytic nuclei (Nuclear Fast Red counterstain) were categorized as having loss of melastatin expression. The expression of TRPM1 mRNA by melanocyte nuclei was measured per one hundred basal keratinocytes (labeling index or LI) along the dermal–epidermal junction in normal skin and in the epidermis-overlying scars. For longitudinal sections of hair follicles, the number of TRPM1 CISH+ cells in the upper third (follicular infundibulum), middle third (isthmus and bulge region) and lower third (follicular bulb and outer root sheath) of anagen follicles were counted. For melanocytic nevi, the presence, degree and pattern of TRPM1 mRNA nuclear expression were assessed.
Table 2.
Materials and methods for in situ hybridization and immunohistochemical studies
| Marker | Function | Manufacturer (provider) | Clone | Retrieval | Dilution | Blocking | Positive control |
|---|---|---|---|---|---|---|---|
| HMB45 (gp100/PMEL17/Silv) | DHICA polymerization/stablin | Cell Marque | Mouse monoclonal | Cell Conditioner 1 CC1 (Ventana) | 1: 50 | None | Melanoma |
| Mart1 | Melanosomal protein | Ventana | A103 | Citra (60 min steam) | Prediluted | None | Melanoma |
| Mel5 | TRP-1 DHICA oxidase, tyrosinase stabilization | Signet | Ta99 | Protease I (4 min) | 1: 40 | None | Normal skin |
| Mitf | Transcription regulator | Leica/Novocastra | 34CA5 | Citra (60 min steam) | 1: 10 | None | Melanoma |
| TRPM1 | Unknown, ion channel protein (Ca2+) | TriPath Oncology™, Inc., Durham, NC | NA | proteinase K (15 min at 37°C) | None | Serum-free protein block | Normal skin |
| S100 protein | Multiple, including Ca2+ homeostasis and regulation of enzyme activities | Ventana | Rabbit polyclonal | None | Prediluted | Avidin-biotin | Melanoma |
| TRP1 | DHICA oxidase, Tyro stabilization | Gift from V. Hearing68 | Rabbit polyclonal | Citra (60 min steam) | 1: 250 (32 min at 37°C) | Endogenous biotin | Melanocytic nevus, melanoma |
| TRP2 (dtc) | Dopachrome tautomerase, Tyro stabilization | Gift from V. Hearing69 | Rabbit polyclonal | Citra (60 min steam) | 1: 500 (32 min at 37°C) | Endogenous biotin | Melanocytic nevus, melanoma |
| Tyrosinase (Tyro) | Oxidation of tyrosine, DOPA, DHI | Gift from V. Hearing70 | Rabbit polyclonal | Citra (60 min steam) | 1: 500 (32 min at 37°C) | Endogenous biotin | Melanocytic nevus, melanoma |
| α-MSH | Peptide hormone | Chemicon | Polyclonal | None | 1: 600 (32 min at 37°C) | Endogenous biotin | Normal skin |
α-MSH, alpha melanocyte stimulating hormone; DHI, dihydroxyindole; DHICA, 5,6-dihydroxyindole-2-carboxylic acid; Dtc, dopachrome tautomerase; gp100, glycoprotein 100; HMB, human melanoma black; MART1, melanocyte antigen reactive to T cells; Mel-5, pigmentation-associated antigen; Mitf, microphthalmia-associated transcription factor; TRPM1, melastatin (transient receptor protein melanocyte); PMEL17, potentially malignant epithelial lesion protein 17; TRP-1, tyrosinase-related protein-1; TRP-2, tyrosinase-related protein 2.
Immunohistochemistry
The characteristics of the nine antibodies to melanin-related proteins used in this study can be found in Table 2. Briefly, the Ventana ES automated DAB immunohistochemical system (Ventana Medical Systems, Tucson, AZ, USA) was used to perform the immunohistochemistry (IHC) assays. All sections were counterstained with hematoxylin. Partial or diffuse nuclear and/or cytoplasmic labeling was counted as positive. The LIs of these IHC stains along the dermal-epidermal junction were recorded. For longitudinal sections of hair follicles, the number of MITF+ cells in the upper third (follicular infundibulum), middle third (isthmus and bulge region) and lower third (follicular bulb and outer root sheath) of anagen follicles were counted. For melanocytic nevi, the presence, degree and pattern of MITF expression were assessed.
Statistical analysis
The statistical software package STATA (College Station, TX, USA) was used for statistical analyses. To compare mean and median values of paired samples, paired student t-test and Wilcoxon signed-rank tests were employed. To compare the equality of mean values among categories, the student t-test and analysis of variance (ANOVA) were utilized, comparing two groups or more groups, respectively. For comparing the equality of populations (median), the Mann-Whitney and Kruskal-Wallis tests were employed to compare two groups or more groups, respectively. Pairwise correlations (covariances) were also assessed. A p-value ≤0.05 was considered statistically significant.
Results
Expression of TRPM1 and melanin-related proteins in normal skin
In vertical sections of normal skin, the ratio of normal melanocytes to keratinocytes has been reported to average 1: 1024,25 and range from 1: 4 on the cheek to 1: 11 on the limbs.26,27 Moreover, depending on the means of detection utilized, the number of melanocytes counted is deceptively low, because not all melanocytes actively produce melanin.28,29 CISH showed that TRPM1 mRNA was expressed only by epidermal and follicular melanocytes. For normal melanocytes of the epidermis, TRPM1 mRNA was expressed in a relatively predictable pattern, averaging 1 TRPM1+ melanocyte to 10 basal keratinocytes (95% confidence interval or CI: 1: 7 to 1: 16). However, it was evident that not all melanocytes expressed TRPM1 mRNA, as scattered non-expressing melanocytes were identified (Fig. 1). The higher average LI for MITF, S100, Mart1 and tyrosinase verified the presence of melanocytes without detectable TRPM1 mRNA. Table 3 lists the LI results for all nine melanogenesis-related proteins and TRPM1 in normal skin and overlying scars. Overall, MITF detected the largest number of melanocytes (highest average LI) along the basal layer of normal epidermis, which was significantly greater than the next two greatest LI, S100 protein and Mart1. However, comparing medians (equality of populations), MITF was not significantly greater than S100 or Mart1. MITF mean and median LI were significantly greater than tyrosinase and TRPM1 (p = 0.0001). By MITF, the melanocyte: keratinocyte ratio averaged 1: 7 (95% CI: 1: 6 to 1: 12). In decreasing order, in comparison to the mean LI for MITF, the mean LI of S100 was 95%, Mart1 92%, tyrosinase 87%, TRPM1 75%, Mel5 72%, TRP2 69%, TRP1 60% and HMB45 35%. Examining pairwise co-expression of all these melanogenesis-related proteins revealed that the strongest correlations existed between MITF and TRPM1 (r = 0.81, p = 0.0001) and between MITF and tyrosinase (r = 0.79, p = 0.0001). Both MITF and TRPM1 showed strong (>0.5) correlation coefficients for all melanin-related proteins except HMB45, and in the case of TRPM1, TRP1. Table 4 lists the correlation coefficients among all eight melanin-related proteins and TRPM1. These data reveal the existence of two or more melanocyte phenotypes in normal skin, the majority of which show a melanogenic phenotype (MITF+/TRPM1+/Tyrosinase+/TRP1 (Mel5)+/TRP2+).
Fig. 1.
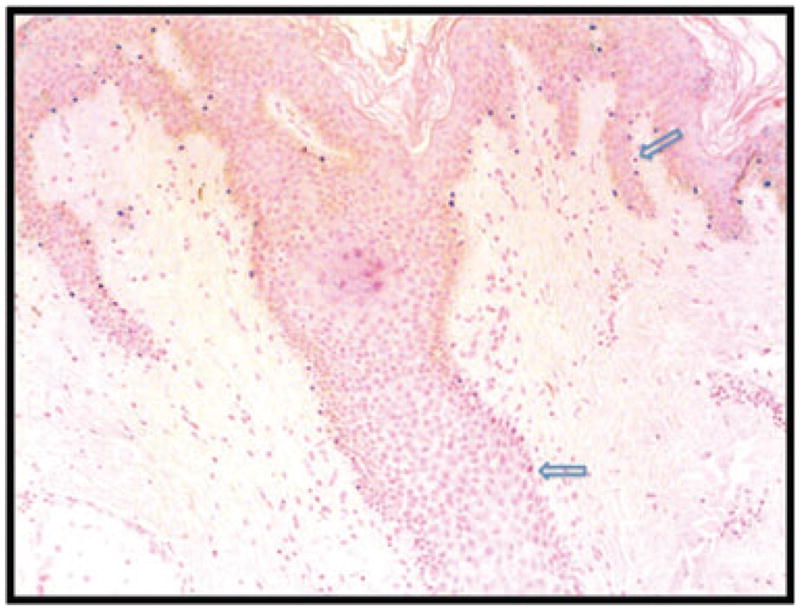
TRPM1 mRNA expression in normal skin. Most melanocytic nuclei in the epidermis and follicular infundibulum express TRPM1 mRNA in a predictable manner along the basement membrane zone at a ratio of 1: 10 melanocytes to basal keratinocytes. However, some intraepidermal melanocytes (arrows) do not express TRPM1 mRNA; these cells may be dividing/migrating melanocytes and/or reserve melanocytes.
Table 3.
Expression of TRPM1 and melanin-related proteins expression in normal skin and scars
| Normal skin |
Scar |
||||
|---|---|---|---|---|---|
| Marker | Median | Mean LI ± SD range | Median | Mean LI ± SD range | p-Value* |
| MITF | 13 | 13.0 ± 5.4 4–28 |
14 | 15.2 ± 6.9 8–26 |
NS |
| S100 protein | 12 | 12.4 ± 5.2 3–28 |
11 | 11.8 ± 5.3 6–20 |
NS |
| Mart1 | 11 | 12.0 ± 5.4 3–27 |
11 | 14.6 ± 7.4 7–24 |
NS |
| Tyro | 11 | 11.3 ± 5.1 3–33 |
9 | 10.3 ± 3.2 7–15 |
NS |
| TRPM1 mRNA | 9 | 9.7 ± 4.3 2–21 |
6 | 5.6 ± 1.4 4–7 |
0.01/0.02 |
| Mel5 | 8 | 9.3 ± 5.1 2–25 |
9 | 8.4 ± 2.1 5–10 |
NS |
| TRP2 | 8 | 9.0 ± 4.7 0–23 |
9 | 10.4 ± 3.4 7–14 |
NS |
| TRP1 | 7 | 7.8 ± 3.8 1–17 |
10 | 11.2 ± 3.9 8–18 |
0.07/0.03 |
| HMB45 | 3 | 4.5 ± 3.9 0–18 |
3 | 4.6 ± 4.7 0–12 |
NS |
| αMSH† | 62% | Weak, regional keratinocytes | 100% | Strong, diffuse keratinocytes | 0.0001 |
LI, labeling index (number of positive cells per 100 basal keratinocytes); NS, not significant at p < 0.05.
Medians/means compared by Kruskal-Wallis and t-test methods, respectively. Italicized p-value indicates a trend (p > 0.05 and ≤ 0.10).
Rare melanocytes expressed αMSH in 2/102 cases (2%); however, spinous keratinocytes expressed αMSH.
Table 4.
Pairwise covariance among melanocytic and melanogenesis-related protein markers in normal skin melanocytes
| TRPM1 | Mitf | S100 | Mart1 | Tyrosinase | Mel 5 | TRP2 | TRP1 | HMB45 | |
|---|---|---|---|---|---|---|---|---|---|
| TRPM1 | 1 | – | – | – | – | – | – | – | – |
| MITF | 0.81 | 1 | – | – | – | – | – | – | – |
| S100protein | 0.67 | 0.72 | 1 | – | – | – | – | – | – |
| Mart1 | 0.64 | 0.75 | 0.73 | 1 | – | – | – | – | – |
| Tyrosinase | 0.68 | 0.79 | 0.74 | 0.78 | 1 | – | – | – | – |
| Mel 5 | 0.58 | 0.68 | 0.68 | 0.74 | 0.65 | 1 | – | – | – |
| TRP2 | 0.56 | 0.66 | 0.66 | 0.68 | 0.68 | 0.51 | 1 | – | – |
| TRP1 | 0.38 | 0.59 | 0.59 | 0.65 | 0.62 | 0.67 | 0.62 | 1 | – |
| HMB45 | 0.27 | 0.20* | 0.20* | 0.38 | 0.43 | 0.38 | 0.24 | 0.41 | 1 |
No significant pairwise correlation was found at p ≤ 0.05.
Expression of TRPM1 and melanin-related proteins overlying scars
In the regenerating epidermis-overlying scars, melanocytes migrate from the wound edges to populate the epidermis.30 Clinically, scars are paler than the surrounding epidermis, a trait that is not attributable to loss of melanocytes or a change in melanocyte function.27,31 Rarely, scars can be pigmented.32 These two disparate phenotypes may be secondary to stromal-melanocyte interactions.33 In this study, examination of melanin-related proteins showed preservation of melanocyte numbers; however, the numbers of melanocytes expressing TRPM1 mRNA and TRP1 were significantly different. TRPM1 LI was significantly lower overlying scars than normal skin; however, TRP1 was significantly higher (Table 3).
αMSH expression in normal skin and scars
αMSH labeled very few melanocytes; 98% of samples were negative. However, in 62% of normal skin samples, keratinocytes (mostly spinous) expressed αMSH regionally, but with weak intensity. Two normal samples showed melanocytes with weak cytoplasmic expression of αMSH associated with diffuse and weak spinous keratinocyte expression of αMSH. No correlations with clinicopathologic characteristics were identified among normal samples with αMSH expression. In contrast, the epidermis overlying a scar showed diffuse, moderately intense spinous expression that was greater than basal keratinocyte expression of αMSH, and no melanocytic expression at all (Table 3).
Melanocyte distribution related to clinical and pathologic features
It has been reported that melanocyte numbers do not differ between sexes,24 but melanocyte counts/densities do change with age,24,27,34,35 anatomic site,24,32,36,37 sun exposure25,34,35,38 – 40 and history and/or presence of an adjacent melanoma or melanocytic nevus.25,41,42 Melanocyte LI for TRPM1 and melanogenesis-related proteins were compared according to the above traits.
Controlling for site, no differences were identified comparing male and female skin melanocyte counts with respect to melanogenesis-related proteins and TRPM1 mRNA. However, specific anatomic sites showed significant differences in melanocyte populations and mean LI for TRPM1 and most melanogenesis-related proteins. Anogenital and head and neck skin had the highest LI for MITF, S100, tyrosinase, TRPM1, Mel5 and HMB45; acral sites had the lowest LI see Fig. 2.
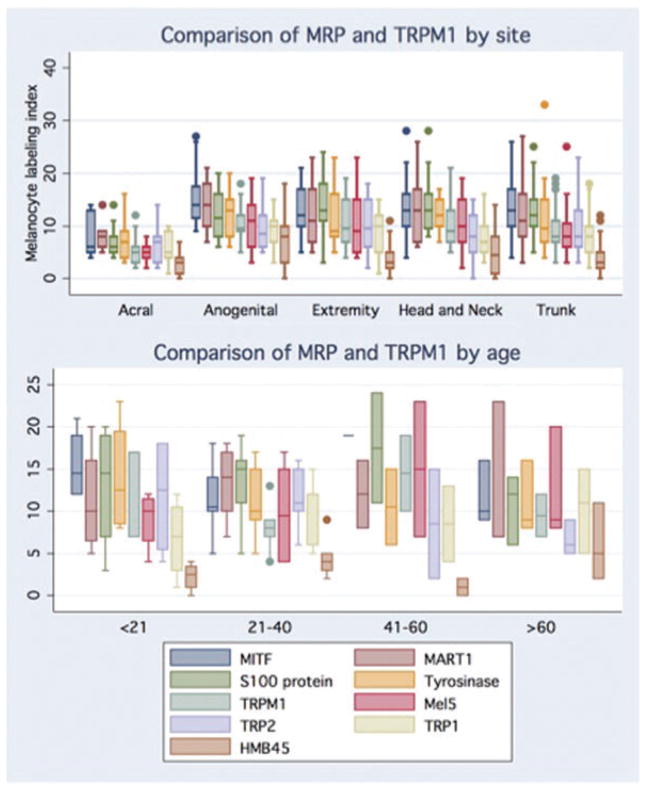
Fig. 2.
A graphic comparison of melanogenesis-related proteins (MRP) and TRPM1 mRNA labeling indices (LI) by site and age. By site, significantly higher median and mean LI were identified for Mitf, S100 protein, tyrosinase (median LI only), TRPM1, Mel5 and HMB45 (all p < 0.5). By age, significantly different median LI between age groups was identified for Mart1 and both median and mean LI for TRPM1 (p ≤ 0.05). [Box and whisker plots of LI: line in the middle of the box represents the median; the box extends from the 25th percentile to the 75th percentile (interquartile range); the whiskers mark upper and lower adjacent values; circles represent outliers.]
To identify any age-related differences in melanocyte LI, trunk samples were grouped into 20-year intervals. Samples were restricted to trunk skin because of the regional variation in melanocyte LIs, low incidence of solar elastosis and because trunk biopsies had the largest number of samples (≥4) per age group (n = 10, ≤20 years; 6, 21–40 years; 13, 41–60 years; and 4, >60 years). Based on this grouping, the following trends were identified. Young adults had larger melanocyte LI than teenagers, children or middle-aged adults; there were differences for TRPM1 LI and Mart1, and a trend toward significance for MITF (Fig. 3). Notably, older individuals had higher LI than middle-aged adults, but two of these four specimens were sun-damaged skin (none of the other trunk specimens had solar elastosis). Comparing these four specimens according to presence of solar elastosis demonstrated that the LIs were lower for MITF and Mart1 without solar elastosis (12.5/7.5 vs. 19.5/20.5 LI, respectively), but higher for TRPM1 (8.5 vs. 7 LI) (see Fig. 2).
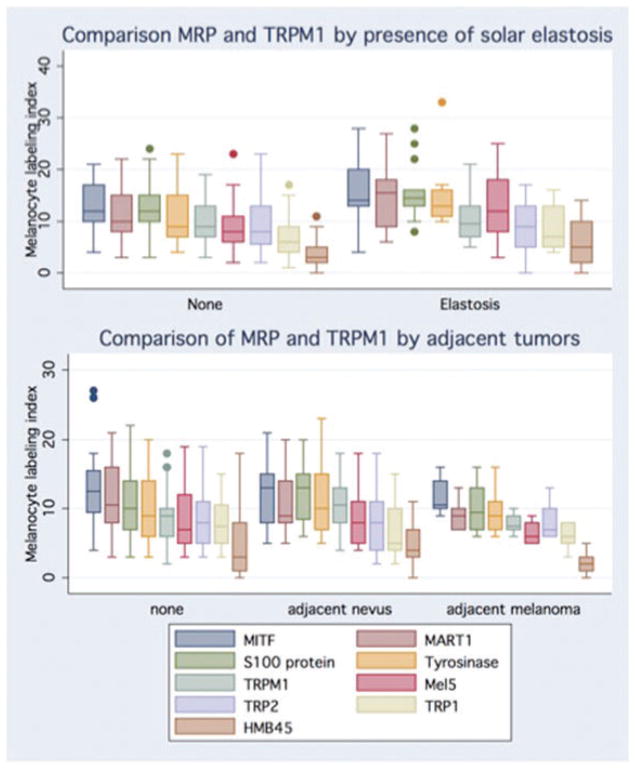
Fig. 3.
Graphic comparison of melanogenesis-related proteins (MRP) and TRPM1 labeling indices (LI) in skin with solar elastosis and skin adjacent to nevi and melanoma. Chronically sun-exposed skin had significantly higher mean LI for Mitf, S100 protein, tyrosinase, Mel5 and HMB45 compared to non-sun-damaged skin. A significantly higher TRPM1 and HMB45 LI were identified for normal skin samples adjacent to nevi compared to normal samples without or with adjacent melanoma. [Box and whisker plots of LI: line in the middle of the box represents the median; the box extends from the 25th percentile to the 75th percentile (interquartile range); the whiskers mark upper and lower adjacent values; circles represent outliers.]
Significant increases in melanocyte LI were identified in samples with solar elastosis from the extremities, trunk, and head and neck for MITF, S100, Mart1, tyrosinase, Mel5 and HMB45. However, no difference was identified for TRPM1 mRNA, TRP2 or TRP1 LIs (Figs 3 and 4). These data reproduce previous findings of melanocyte hyperplasia in sun-exposed skin25,34,35,38,39 and indicate that a larger proportion of these melanocytes are less differentiated, non-melanogenic (MSLN−/TRP1−/TRP2−) melanocytes.
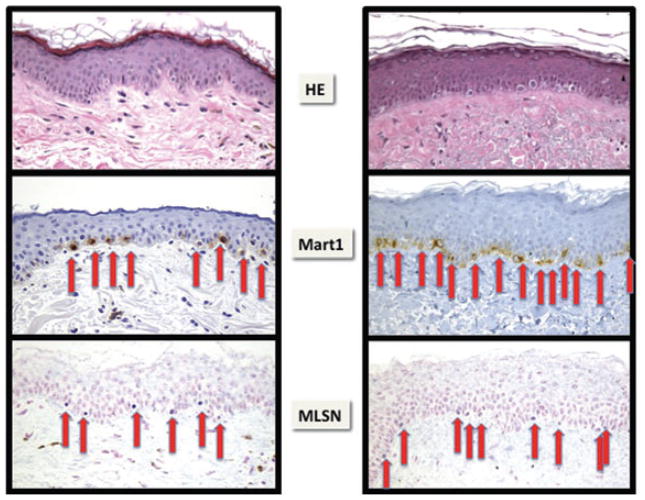
Fig. 4.
Melanocytic hyperplasia in chronically sun-exposed and damaged skin, witnessed by solar elastosis. As assessed by CISH and IHC, melanocyte density in skin showed a high degree of variability that was associated with site, age, presence of solar elastosis or presence of an adjacent nevus or melanoma. On average, MSLN identified fewer melanocytes than MITF, S100 protein or Mart1. In these examples of head and neck skin, the left panels of skin without solar elastosis have a ratio of 6 MSLN CISH-labeled melanocytes to eight Mart1-labeled melanocytes (75%). In sun-damaged skin (solar elastosis present), a marked increase in melanocytes (melanocyte hyperplasia) is evident, with a smaller ratio between TRPM1 and Mart1 expressing melanocyts of 9: 14 (64%).
Controlling for site, comparing melanocyte LI based on presence or absence of adjacent neoplasm (whether benign or malignant) or between benign and malignant tumors did not reveal any differences. However, melanocyte LI of normal skin adjacent to melanocytic nevi was higher than for skin adjacent to melanoma. This difference was significant for HMB45 and showed a similar trend for S100, tyrosinase and MSLN (Fig. 3). Fallowfield et al.25 identified a similar trend comparing patients with melanoma and atypical nevi; both counts were significantly greater than those for normal controls.
Differential expression of MITF and TRPM1 (MITF LI−TRPM1 LI=Mitf+TRPM1−melanocytes)
A comparison of the difference between the LI for MITF and TRPM1 suggests that a subpopulation of MITF+TRPM1– melanocytes exists in skin and shows variable density, based on the presence of an underlying scar, age, site, presence of solar elastosis and/or proximity to melanocyte proliferation. Significantly elevated populations of MITF+MLSN– melanocytes were found repopulating the epidermis overlying a scar, in anogenital skin, and overlying solar elastosis. Also, a trend was seen toward increased numbers of MITF+TRPM1– melanocytes in proximity to a melanoma (Table 5 and Fig. 5).
Table 5.
Differential expression of MITF compared to TRPM1 mRNA expression (number of Mitf+TRPM1– melanocytes)
| Mitf LI-TRPM1 LI differential* | |||
|---|---|---|---|
| All normal samples | (2) 3.2 ± 3.1 [0–16] | ||
| Scar | (10) 9.6 ± 7.5 [1–21] | ||
| p-value (median/mean)† | 0.03/0.0001 | ||
| Acral | (1) 2.2 ± 2.0 [0–16] | ||
| Anogenital | (5) 5.1 ± 2.8 [0–31] | ||
| Extremity | (2) 2.2 ± 1.7 [0–11] | ||
| Head and neck | (3) 3.8 ± 4.2 [1–11] | ||
| Trunk | (2) 2.8 ± 2.9 [0–7] | ||
| p-value (median/mean)‡ | 0.03/0.07 | ||
| ≤20 years (n = 10)§ | (1.5) 3.1 ± 3.4 [1–10] | ||
| 21–40 years (n = 6) | (1.5) 1.5 ± 0.6 [1–2] | ||
| 41–60 years (n = 13) | (2) 2.5 ± 3.1 [0–8] | ||
| >60 years (n = 4) | (6) 5 ± 2.6 [2–7] | ||
| p-value (median/mean)‡ | NS | ||
| Solar elastosis (n = 16)¶ | (4.5) 4.6 ± 4.4 [0–14] | ||
| No elastosis (n = 53) | (2) 1.5 ± 2.5 [0–10] | ||
| p-value (median/mean)† | 0.07/0.01 | ||
| No adjacent tumor (n = 29) | (2) 2.4 ± 1.6 [1–5] | ||
| Adjacent nevus (n = 18) | (2) 2.1 ± 2.8 [0–7] | ||
| Adjacent melanoma (n = 6) | (2.5) 3.8 ± 2.6 [2–6] | ||
| p-value (median/mean)‡ | NS/0.06 |
NS, not significant by p > 0.05.
Italicized p-value indicates a trend (p < 0.1 and >0.05).
(Median) Mean ± SD [Range].
Population median and means compared by rank sum (Mann-Whitney) test and Student’s t-test, respectively.
Differences between medians and means compared by Kruskal-Wallis and one-way analysis of variance (ANOVA), respectively.
Only trunk skin samples evaluated.
Head and neck, trunk and extremity skin examined.
Fig. 5.
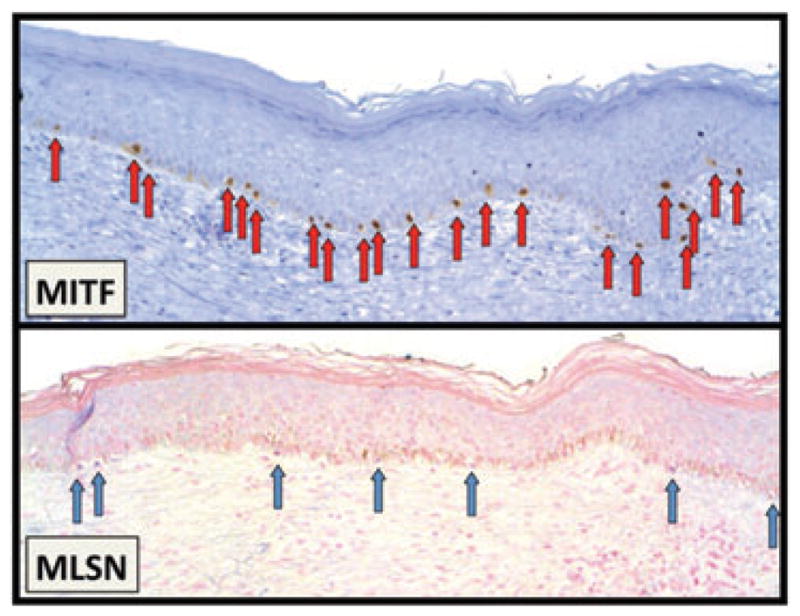
MITF+TRPM1 – cells in regenerating skin overlying a scar. In normal skin, there were median and mean differentials of 2 and 3.2 more MITF+ than TRPM1+ melanocytes. In regenerating epidermis overlying a scar, a significantly increased differential was apparent: 10 and 9.6, respectively. In this example, the differential is 14 (21 MITF+ melanocytes − 7 MSLN+ melanocytes).
Hair follicle expression of TRPM1 mRNA and MITF
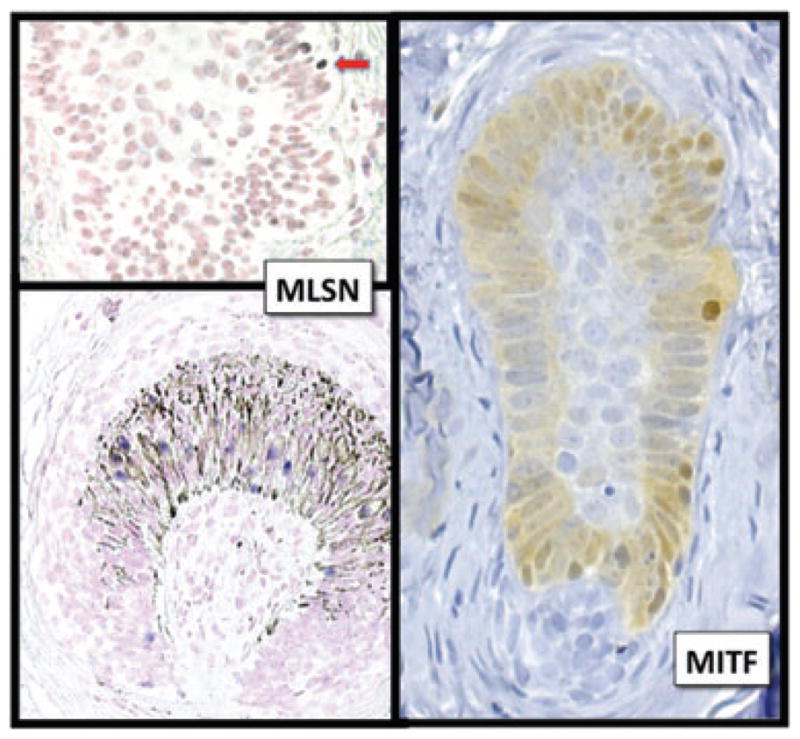
In reviewing normal head and neck samples, we analyzed the expression of TRPM1 in 22 and MITF in 38 hair follicles where more than half of the follicular unit was present. Most of these follicles were in anagen, while a minority were in the late catagen or telogen stage. MITF was expressed at significantly higher levels than TRPM1 in all regions of the hair follicle (Table 6 and Figs 6 and 7). The largest differential in expression was seen in the isthmic region, where not only did MITF label more melanocytes, but epithelioid cells also expressed MITF in the bulge, showing both cytoplasmic and nuclear localization. The finding of bulge expression of MITF indicates that stem cells for both keratinocytic and melanocytic lineages reside in this niche, a finding confirmed in fetal skin by the Murphy group43 and predicted by others.44 – 46 Lastly, rare TRPM1+ (differentiated) melanocytes were found in the isthmic region (Figs 6 and 7). These results correlate with other reports of ectopic melanocyte pigmentation/differentiation in the bulge region, which is suspected to represent premature differentiation or activation of a senescence program associated with hair graying and loss of melanocyte stem cells.46
Table 6.
Expression of Mitf and TRPM1 in hair follicles from normal skin samples
| Infundibula (upper third) | Isthmus/bulge (middle third) | Follicular bulb (lower third) | |
|---|---|---|---|
| MITF (n = 38) | (9) 8.8 ± 3.7 3–18 |
(3.5) 3.9 ± 2.6 0–9 |
(7) 9 ± 6.1 0–28 |
| TRPM1 (n = 22) | (5.5) 7.1 ± 4.6 2–24 |
(1) 1.1 ± 1.0 0–3 |
(5) 6.1 ± 4.3 1–20 |
| p-value* | (0.008) 0.02 | (0.001) 0.001 | (0.08) NS |
(Median) and mean labeling indices compared by Kruskal-Wallis and t-test methods, respectively. Italicized p-value indicates a trend (p > 0.05 and ≤ 0.10).
Fig. 6.
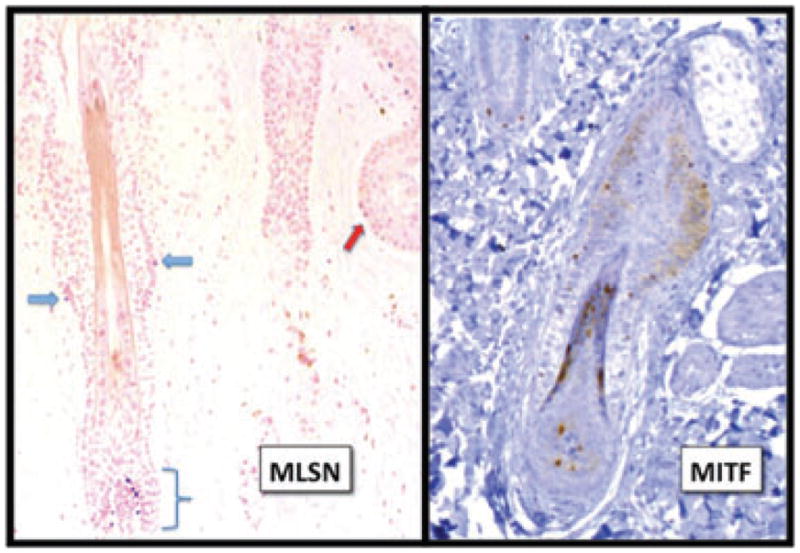
MITF and TRPM1 expression in hair follicles. Like the epidermis, MITF labeled consistently more melanocytes than TRPM1 in all segments of the hair follicle. In addition, weak but diffuse nuclear and cytoplasmic MITF expression was evident in the epithelioid cells of the bulge area (putative stem cells43). Scattered melanocytes expressed TRPM1 mRNA in the follicular bulb (bracket), isthmus (blue arrows) and infundibulum (red arrow). The finding of differentiated melanocytes in the isthmus has been described in middle-aged human hair follicles and was associated with loss of melanocyte stem cells and hair graying.46
Fig. 7.
TRPM1 marks melanocytes of the follicular bulb and isthmus (red arrow). In contrast, MITF detects a greater number of melanocytes including those with epithelioid cytology in this photo of the isthmus and early anagen bulb (right).
TRPM1 mRNA and MITF expression in ordinary compound melanocytic nevi
By RISH, Deeds et al.7 reported that 3 of 14 ordinary melanocytic nevi showed a gradient of TRPM1 mRNA expression in which higher expression was apparent in the superficial aspects of the nevus and reduced expression in the deeper dermis. This gradient expression has also been described for several melanogenesis-related proteins including MITF12 and Mart1.23 By CISH, all four compound melanocytic nevi showed similar zonal gradient patterns of TRPM1 and MITF expression: superficial, type A (pagetoid) melanocytes exhibited strong signals, and dermal type B (nevoid) and type C (lymphocyte-like and neurotized) melanocytes showed progressively weaker signals for MITF or TRPM1 mRNA with increasing dermal depth. Some melanocytes did not express either marker (Fig. 8).
Fig. 8.
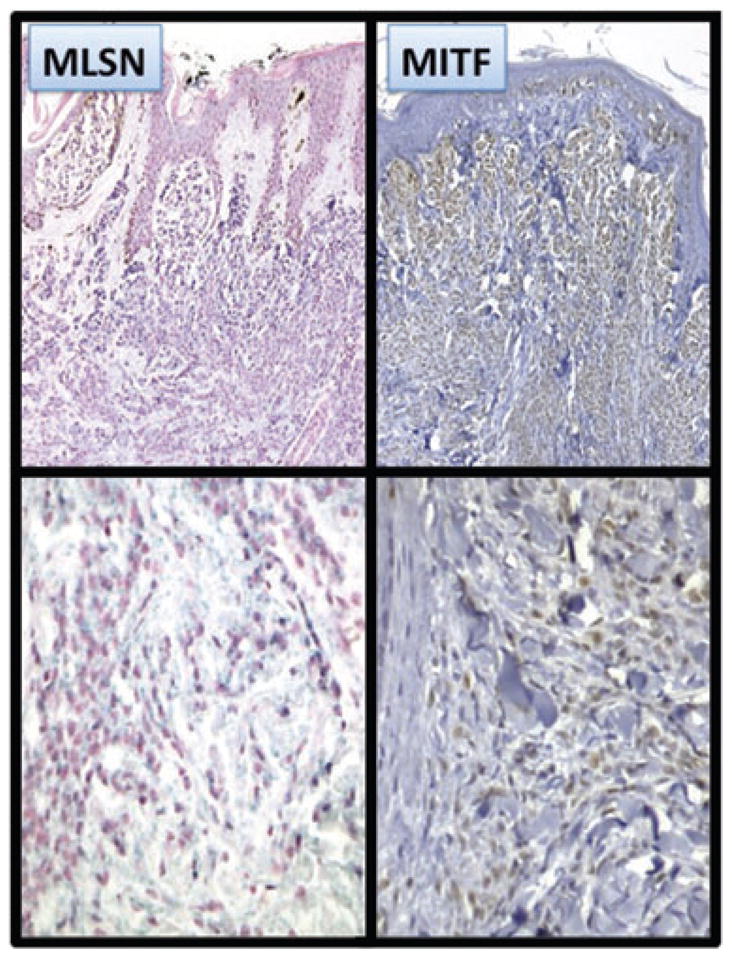
Vertically decreased expression of TRPM1 and MITF. Multiple studies have demonstrated a vertically oriented, decreasing gradient of melanin-related proteins expression in melanocytic nevi showing zonal maturation moving from junctional, type A (pagetoid) melanocytes to superficial dermal, type B (nevoid) melanocytes, to deep dermal, type C (spindle/neurotized) melanocytes for MITF12 and Mart1.23 Similarly, TRPM1 mRNA shows strong diffuse expression in papillary dermal type A melanocytes that decreases to little or no expression in deep dermal type C melanocytes, which is synchronous with MITF expression.
Discussion
This study, utilizing CISH and IHC methods, has reproduced many of the results obtained in older studies, which used the DOPA reaction or counted melanocytes on hematoxylin and eosin-stained sections.24,25,27,28,34,36,38,47 – 49 Utilizing multiple melanin-related proteins, we have also demonstrated that intraepidermal melanocyte populations are phenotypically heterogeneous and dynamic based on age, site and environmental exposures (e.g. anogenital or sun-exposed skin). Moreover, these results show that MITF is a robust, lineage-specific marker of all skin melanocytes and putative stem cells. Furthermore, TRPM1 mRNA expression, which strongly correlates with tyrosinase, Mel5 and TRP2 expression, ostensibly identifies terminally differentiated (pigmented) melanocytes. Indeed, the average ratio of melanocytes to keratinocytes for TRPM1 mRNA is equivalent to the average of 1: 10 found for the DOPA reaction,27 underscoring this observation.
The production of melanin and the capability of transferring pigment granules to surrounding keratinocytes are functions specific to the finally differentiated melanocyte. In vivo production of melanin requires expression of tyrosinase, synthesis and assembly of melanosomes, transport of tyrosinase as well as of its substrate L-tyrosine into the melanosome and generation of an intramelanosomal environment that allows for hydroxylation of tyrosine and further metabolism (melanogenesis) to melanin pigment.21 In this context, it is not surprising that the highly differentiated phenotype correlates with tyrosinase expression and to different degrees with other melanogenesis-related proteins, including structural proteins of melanosomes (HMB45 and/or Mart1) or proteins with enzymatic activity (TRP1 and TRP2). The rate-limiting enzyme of melano-genesis is tyrosinase, whose activation requires the proper intramelanosomal environment.21,44 TRP1 and TRP2 regulate the velocity of the melanogenic pathway, but they are not necessary for melanogenesis to proceed. Therefore, TRP1 and TRP2 are expressed in melanocytes at different levels of differentiation and are co-expressed with MITF, a master regulator of the differentiation program, as reported here. The high degree of correlation is significant, and again, emphasizes an important role for TRPM1 in the regulation of fully differentiated melanocyte phenotype and proliferation [reviewed in (1,7,8,50,51)].
Loss of TRPM1 mRNA is not only a signal of a less-differentiated melanocyte phenotype; it is also a sign of nevo-melanocytic maturation toward a peripheral nerve sheath cell phenotype, referred to as ‘neurotized melanocytes’. The gradient phenomenon of strong and diffuse to decreased-to-absent expression of TRPM1, MITF,12 and other melanogenesis-related proteins23 in the deep dermal aspects of common melanocytic nevi highlights the neural-crest origin of melanocytes and their re-programming to exhibit a neural phenotype.22
During childhood, as the skin surface expands, throughout adulthood to maintain melanocyte numbers, and in response to exposure to sunlight or skin wounding, melanocytes are stimulated to proliferate at a low rate. Melanocyte proliferation entails uncoupling from keratinocytes, loss of dendrites, cell division, migration along the basement membrane (a melanoblast-like phenotype), then recoupling with keratinocytes to form the epidermal melanin unit. Keratinocytes regulate melanocyte growth and expression of melanocyte cell surface receptors via cell adhesion and growth factors, which include E-cadherin, P-cadherin and desmoglein, which are regulated through growth factors such as hepatocyte growth factor (HGF), stem cell factor (SCF), platelet-derived growth factor (PDGF) and endothelin-1 (produced by fibroblasts or keratinocytes).52,53 In addition to the above, decreased expression of TRPM1 mRNA overlying scars or in sun-damaged skin appears to be another phenotypic change in regenerating melanocytes, reflecting loss of terminal differentiation and initiation of melanocyte proliferation. In addition, the finding of increased expression of TRP1 overlying scars may be the consequence of signaling through the KIT receptor (activated by PDGF and SCF) and its upregulation.11,54
Loss of keratinocyte regulation characterizes the development of melanoma and is seen in the downregulation of E and P-cadherins, upregulation of melanocyte-melanocyte and melanocyte-fibroblast adhesion molecules such as Mel-CAM and N-cadherin, expression of cell-matrix adhesion molecules such as αvβ3 integrins, and increased elaboration of metallo-proteinases.52,53 The importance of growth factor signaling in producing the malignant phenotype has been demonstrated in experimental models, where increased expression of basic fibroblastic growth factor (bFGF), HGF, SCF and endothelin-3 coupled with UV radiation produced invasive and in situ-like tumors.55,56 The melanocytic hyperplasia found in sun-damaged skin, characterized by an increased population of MITF+TRPM1 – melanocytes, may actually reflect two cell populations: regenerating melanocytes and early precursors to melanoma in situ. North et al.57 recently reported a population of histologically normal melanocytes with genetic amplifications (e.g. 11q13, Cyclin D1 locus) in unaffected skin adjacent to acral melanomas. Their study supports the concept of melanocytic ‘field cancerization’,58,59 a hypothesis that explains why melanoma can recur (locally persist) for years at, or in the proximity of, excision sites, despite wide safety margins and evidence of complete excision. Another sign of melanocyte field cancerization may be the increased numbers of MITF+TRPM1-melanocytes in the vicinity of melanomas we found here.
HMB45 is a product of the SILV gene locus and membrane-bound melanosomal protein that acts as a regulator of melanogenesis, involved in the transition from stage I to stage II melanosome maturation and stabilization of melanin intermediates.60 A minority of normal melanocytes, subsets of melanocytic nevi such as blue nevi, dysplastic nevi and some Spitz nevi and most metastatic melanomas typically express HMB45.50,61 HMB45 has been considered a marker of melanocyte activation (not proliferation62), because it is frequently expressed overlying scars, in areas of inflammation, and in regions exposed to ultraviolet radiation.33,63 – 67 In the current study, significantly increased levels of HMB45+ melanocytes were found in anogenital and head and neck skin, in skin with solar elastosis, and in the vicinity of melanocytic nevi; changes that were weakly correlated with TRPM1 mRNA expression (r = 0.27). The finding of increased HMB45 in these settings may well be related to local environmental influences including melanocyte-epidermal and melanocyte-stromal interactions.21,33,67 In the case of increased HMB45 melanocytes in the vicinity of melanocytic nevi, the existence of ‘field cells’ is also possible.
Conclusion
By CISH, TRPM1 mRNA expression has been shown to be specific for melanocytes and strongly associated with MITF and tyrosinase expression, the latter implicating a mature melanocyte phenotype. However, in normal skin, TRPM1 mRNA expression is dynamic, labels only a fraction of melanocytes and changes according to its local environment. Loss of TRPM1 mRNA expression relative to MITF was associated with regenerative (melanoblast) phenotype overlying scars. This immunophenotype could potentially represent a premalignant clone(s) overlying solar elastosis and the proximity of melanoma – so called field cells. The elevated TRPM1 LI found in anogenital skin and skin near melanocytic nevi may be related to melanocyte activation, particularly as these settings were associated with significantly greater HMB45. Overall, these results support the contention that TRPM1 mRNA expression is a marker of a fully differentiated melanocyte.
Acknowledgments
We would like to acknowledge Talat Nazir, DO for his able assistance in retrieving much of the pathologic material used in this study. The study was partially supported by NIH grant no. AR052190.
Footnotes
This work was presented in part at the 95th and 98th Annual Meetings of United States and Canada Academy of Pathology, February 2006, Atlanta, Georgia and March 2009, Boston, Massachusetts, respectively; the 28th Symposium of International Society of Dermatopathology in Paris, France, November 2007; and at the 15th PASPCR Meeting in Memphis, Tennessee, September 2009.
Conflicts of interest
None.
References
- 1.Duncan LM, Deeds J, Hunter J, et al. Down-regulation of the novel gene melastatin correlates with potential for melanoma metastasis. Cancer Res. 1998;58:1515. [PubMed] [Google Scholar]
- 2.Entrez gene: TRPM1 transient receptor potential cation channel, subfamily M, member 1 [database on the Internet] [accessed April 7, 2009]; http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=4308.
- 3.Hunter JJ, Shao J, Smutko JS, et al. Chromosomal localization and genomic characterization of the mouse melastatin gene (Mlsn1) Genomics. 1998;54:116. doi: 10.1006/geno.1998.5549. [DOI] [PubMed] [Google Scholar]
- 4.Nilius B, Owsianik G, Voets T, Peters JA. Transient receptor potential cation channels in disease. Physiol Rev. 2007;87:165. doi: 10.1152/physrev.00021.2006. [DOI] [PubMed] [Google Scholar]
- 5.Xu XZ, Moebius F, Gill DL, Montell C. Regulation of melastatin, a TRP-related protein, through interaction with a cytoplasmic isoform. Proc Natl Acad Sci USA. 2001;98:10692. doi: 10.1073/pnas.191360198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bellone RR, Brooks SA, Sandmeyer L, et al. Differential gene expression of TRPM1, the potential cause of congenital stationary night blindness and coat spotting patterns (LP) in the Appaloosa horse (Equus caballus) Genetics. 2008;179:1861. doi: 10.1534/genetics.108.088807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deeds J, Cronin F, Duncan LM. Patterns of melastatin mRNA expression in melanocytic tumors. Hum Pathol. 2000;31:1346. [PubMed] [Google Scholar]
- 8.Duncan LM, Deeds J, Cronin FE, et al. Melastatin expression and prognosis in cutaneous malignant melanoma. J Clin Oncol. 2001;19:568. doi: 10.1200/JCO.2001.19.2.568. [DOI] [PubMed] [Google Scholar]
- 9.Fang D, Setaluri V. Expression and up-regulation of alternatively spliced transcripts of melastatin, a melanoma metastasis-related gene, in human melanoma cells. Biochem Biophys Res Commun. 2000;279:53. doi: 10.1006/bbrc.2000.3894. [DOI] [PubMed] [Google Scholar]
- 10.Miller AJ, Du J, Rowan S, Hershey CL, Widlund HR, Fisher DE. Transcriptional regulation of the melanoma prognostic marker melastatin (TRPM1) by MITF in melanocytes and melanoma. Cancer Res. 2004;64:509. doi: 10.1158/0008-5472.can-03-2440. [DOI] [PubMed] [Google Scholar]
- 11.Hoek KS, Schlegel NC, Eichhoff OM, et al. Novel MITF targets identified using a two-step DNA microarray strategy. Pigment Cell Melanoma Res. 2008;21:665. doi: 10.1111/j.1755-148X.2008.00505.x. [DOI] [PubMed] [Google Scholar]
- 12.King R, Googe PB, Weilbaecher KN, Mihm MC, Jr, Fisher DE. Microphthalmia transcription factor expression in cutaneous benign, malignant melanocytic, and nonmelanocytic tumors. Am J Surg Pathol. 2001;25:51. doi: 10.1097/00000478-200101000-00005. [DOI] [PubMed] [Google Scholar]
- 13.King R, Weilbaecher KN, McGill G, Cooley E, Mihm M, Fisher DE. Microphthalmia transcription factor. A sensitive and specific melanocyte marker for Melanoma Diagnosis. Am J Pathol. 1999;155:731. doi: 10.1016/S0002-9440(10)65172-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steingrimsson E, Copeland NG, Jenkins NA. Melanocytes and the microphthalmia transcription factor network. Annu Rev Genet. 2004;38:365. doi: 10.1146/annurev.genet.38.072902.092717. [DOI] [PubMed] [Google Scholar]
- 15.Levy C, Khaled M, Fisher DE. MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol Med. 2006;12:406. doi: 10.1016/j.molmed.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 16.Garraway LA, Widlund HR, Rubin MA, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 17.Gray-Schopfer V, Wellbrock C, Marais R. Melanoma biology and new targeted therapy. Nature. 2007;445:851. doi: 10.1038/nature05661. [DOI] [PubMed] [Google Scholar]
- 18.Wellbrock C, Marais R. Elevated expression of MITF counteracts B-RAF-stimulated melanocyte and melanoma cell proliferation. J Cell Biol. 2005;170:703. doi: 10.1083/jcb.200505059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Selzer E, Wacheck V, Lucas T, et al. The melanocyte-specific isoform of the microphthalmia transcription factor affects the phenotype of human melanoma. Cancer Res. 2002;62:2098. [PubMed] [Google Scholar]
- 20.Hammock L, Cohen C, Carlson G, et al. Chromogenic in situ hybridization analysis of melastatin mRNA expression in melanomas from American Joint Committee on Cancer stage I and II patients with recurrent melanoma. J Cutan Pathol. 2006;33:599. doi: 10.1111/j.1600-0560.2006.00501.x. [DOI] [PubMed] [Google Scholar]
- 21.Slominski A, Tobin DJ, Shibahara S, Wortsman J. Melanin pigmentation in mammalian skin and its hormonal regulation. Physiol Rev. 2004;84:1155. doi: 10.1152/physrev.00044.2003. [DOI] [PubMed] [Google Scholar]
- 22.White RM, Zon LI. Melanocytes in development, regeneration, and cancer. Cell Stem Cell. 2008;3:242. doi: 10.1016/j.stem.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 23.Busam KJ, Chen YT, Old LJ, et al. Expression of melan-A (MART1) in benign melanocytic nevi and primary cutaneous malignant melanoma. Am J Surg Pathol. 1998;22:976. doi: 10.1097/00000478-199808000-00007. [DOI] [PubMed] [Google Scholar]
- 24.Cochran AJ. The incidence of melanocytes in normal human skin. J Invest Dermatol. 1970;55:65. doi: 10.1111/1523-1747.ep12290550. [DOI] [PubMed] [Google Scholar]
- 25.Fallowfield ME, Curley RK, Cook MG. Melanocytic lesions and melanocyte populations in human epidermis. Br J Dermatol. 1991;124:130. doi: 10.1111/j.1365-2133.1991.tb00421.x. [DOI] [PubMed] [Google Scholar]
- 26.Breathnach A. An atlas of of the ultrastructure of human skin. London: Churchill; 1971. [Google Scholar]
- 27.Szabo G. Quantitative histological investigation on the melanocyte system in the human epidermis. In: Gordon M, editor. Pigment cell biology. New York: Academic Press; 1959. p. 99. [Google Scholar]
- 28.Mishima Y, Sheldon W. Enzymically active and inactive melanocyte populations and ultraviolet irradiation: combined dopa-premelanin reaction and electron microscopy. J Invest Dermatol. 1967;49:273. [Google Scholar]
- 29.Dean NR, Brennan J, Haynes J, Goddard C, Cooter RD. Immunohistochemical labeling of normal melanocytes. Appl Immunohistochem Mol Morphol. 2002;10:199. doi: 10.1097/00129039-200209000-00002. [DOI] [PubMed] [Google Scholar]
- 30.Breathnach A. Melanocytes in early regenerated human epidermis. J Invest Dermatol. 1960;35:245. [Google Scholar]
- 31.Velangi SS, Rees JL. Why are scars pale? An immunohistochemical study indicating preservation of melanocyte number and function in surgical scars. Acta Derm Venereol. 2001;81:326. doi: 10.1080/000155501317140016. [DOI] [PubMed] [Google Scholar]
- 32.Botella-Estrada R, Sanmartin O, Sevila A, Escudero A, Guillen C. Melanotic pigmentation in excision scars of melanocytic and non-melanocytic skin tumors. J Cutan Pathol. 1999;26:137. doi: 10.1111/j.1600-0560.1999.tb01818.x. [DOI] [PubMed] [Google Scholar]
- 33.Carlson JA, Mu XC, Slominski A, et al. Melanocytic proliferations associated with lichen sclerosus. Arch Dermatol. 2002;138:77. doi: 10.1001/archderm.138.1.77. [DOI] [PubMed] [Google Scholar]
- 34.Gilchrest BA, Blog FB, Szabo G. Effects of aging and chronic sun exposure on melanocytes in human skin. J Invest Dermatol. 1979;73:141. doi: 10.1111/1523-1747.ep12581580. [DOI] [PubMed] [Google Scholar]
- 35.Scheibner A, McCarthy WH, Milton GW, Nordlund JJ. Langerhans cell and melanocyte distribution in “normal” human epidermis. Preliminary report. Australas J Dermatol. 1983;24:9. doi: 10.1111/j.1440-0960.1983.tb00241.x. [DOI] [PubMed] [Google Scholar]
- 36.Staricco RJ, Pinkus H. Quantitative and qualitative data on the pigment cells of adult human epidermis. J Invest Dermatol. 1957;28:33. doi: 10.1038/jid.1957.4. [DOI] [PubMed] [Google Scholar]
- 37.Rosdahl I, Rorsman H. An estimate of the melanocyte mass in humans. J Invest Dermatol. 1983;81:278. doi: 10.1111/1523-1747.ep12518318. [DOI] [PubMed] [Google Scholar]
- 38.Stierner U, Rosdahl I, Augustsson A, Kagedal B. UVB irradiation induces melanocyte increase in both exposed and shielded human skin. J Invest Dermatol. 1989;92:561. doi: 10.1111/1523-1747.ep12709572. [DOI] [PubMed] [Google Scholar]
- 39.Hendi A, Brodland DG, Zitelli JA. Melanocytes in longstanding sun-exposed skin: quantitative analysis using the MART-1 immunostain. Arch Dermatol. 2006;142:871. doi: 10.1001/archderm.142.7.871. [DOI] [PubMed] [Google Scholar]
- 40.Acker SM, Nicholson JH, Rust PF, Maize JC. Morphometric discrimination of melanoma in situ of sun-damaged skin from chronically sun-damaged skin. J Am Acad Dermatol. 1998;39:239. doi: 10.1016/s0190-9622(98)70082-9. [DOI] [PubMed] [Google Scholar]
- 41.Barlow JO, Maize J, Sr, Lang PG. The density and distribution of melanocytes adjacent to melanoma and nonmelanoma skin cancers. Dermatol Surg. 2007;33:199. doi: 10.1111/j.1524-4725.2006.33039.x. [DOI] [PubMed] [Google Scholar]
- 42.Wong CK. A study of melanocytes in the normal skin surrounding malignant melanomata. Dermatologica. 1970;141:215. doi: 10.1159/000252469. [DOI] [PubMed] [Google Scholar]
- 43.Gleason BC, Crum CP, Murphy GF. Expression patterns of MITF during human cutaneous embryogenesis: evidence for bulge epithelial expression and persistence of dermal melanoblasts. J Cutan Pathol. 2008;35:615. doi: 10.1111/j.1600-0560.2007.00881.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Slominski A, Wortsman J, Plonka PM, Schallreuter KU, Paus R, Tobin DJ. Hair follicle pigmentation. J Invest Dermatol. 2005;124:13. doi: 10.1111/j.0022-202X.2004.23528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Slominski A, Paus R, Plonka P, et al. Pharmacological disruption of hair follicle pigmentation by cyclophosphamide as a model for studying the melanocyte response to and recovery from cytotoxic drug damage in situ. J Invest Dermatol. 1996;106:1203. doi: 10.1111/1523-1747.ep12348479. [DOI] [PubMed] [Google Scholar]
- 46.Nishimura EK, Granter SR, Fisher DE. Mechanisms of hair graying: incomplete melanocyte stem cell maintenance in the niche. Science. 2005;307:720. doi: 10.1126/science.1099593. [DOI] [PubMed] [Google Scholar]
- 47.Rosdahl I. The epidermal melanocyte population and its reaction to ultraviolet light. Acta Derm Venereol Suppl (Stockh) 1979;88:1. [PubMed] [Google Scholar]
- 48.Montagna W, Chase HB. Histology and cytochemistry of human skin. X. X-irradiation of the scalp. Am J Anat. 1956;99:415. doi: 10.1002/aja.1000990304. [DOI] [PubMed] [Google Scholar]
- 49.Staricco RG. Amelanotic melanocytes in the outer sheath of the human hair follicle and their role in the repigmentation of regenerated epidermis. Ann NY Acad Sci. 1963;100:239. doi: 10.1111/j.1749-6632.1963.tb57123.x. [DOI] [PubMed] [Google Scholar]
- 50.Carlson JA, Ross JS, Slominski AJ. New techniques in dermatopathology that help to diagnose and prognosticate melanoma. Clin Dermatol. 2009;27:75. doi: 10.1016/j.clindermatol.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 51.Slominski A. Cooling skin cancer: menthol inhibits melanoma growth. Focus on “TRPM8 activation suppresses cellular viability in human melanoma”. Am J Physiol. 2008;295:C293. doi: 10.1152/ajpcell.00312.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haass NK, Herlyn M. Normal human melanocyte homeostasis as a paradigm for understanding melanoma. J Investig Dermatol Symp Proc. 2005;10:153. doi: 10.1111/j.1087-0024.2005.200407.x. [DOI] [PubMed] [Google Scholar]
- 53.Carlson JA, Linette GP, Aplin A, Ng B, Slominski A. Dermatol Clin. Vol. 25. 2007. Melanocyte receptors: clinical implications and therapeutic relevance; p. 541.p. viii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grichnik JM, Burch JA, Burchette J, Shea CR. The SCF/KIT pathway plays a critical role in the control of normal human melanocyte homeostasis. J Invest Dermatol. 1998;111:233. doi: 10.1046/j.1523-1747.1998.00272.x. [DOI] [PubMed] [Google Scholar]
- 55.Berking C, Takemoto R, Satyamoorthy K, et al. Induction of melanoma phenotypes in human skin by growth factors and ultraviolet B. Cancer Res. 2004;64:807. doi: 10.1158/0008-5472.can-03-3438. [DOI] [PubMed] [Google Scholar]
- 56.Noonan FP, Dudek J, Merlino G, De Fabo EC. Animal models of melanoma: an HGF/SF transgenic mouse model may facilitate experimental access to UV initiating events. Pigment Cell Res. 2003;16:16. doi: 10.1034/j.1600-0749.2003.00014.x. [DOI] [PubMed] [Google Scholar]
- 57.North JP, Kageshita T, Pinkel D, Leboit PE, Bastian BC. Distribution and significance of occult intraepidermal tumor cells surrounding primary melanoma. J Invest Dermatol. 2008;128:2024. doi: 10.1038/jid.2008.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Slaughter DP, Southwick HW, Smejkal W. Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin. Cancer. 1953;6:963. doi: 10.1002/1097-0142(195309)6:5<963::aid-cncr2820060515>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 59.Carlson JA, Scott D, Wharton J, Sell S. Incidental histopathologic patterns: possible evidence of ‘field cancerization’ surrounding skin tumors. Am J Dermatopathol. 2001;23:494. doi: 10.1097/00000372-200110000-00020. [DOI] [PubMed] [Google Scholar]
- 60.Slominski A. Coming of age of melanogenesis-related proteins. Arch Pathol Lab Med. 2002;126:775. doi: 10.5858/2002-126-0775-COAOMR. [DOI] [PubMed] [Google Scholar]
- 61.Busam KJ. The use and application of special techniques in assessing melanocytic tumours. Pathology. 2004;36:462. doi: 10.1080/00313020412331283824. [DOI] [PubMed] [Google Scholar]
- 62.Smoller BR, Hsu A, Krueger J. HMB-45 monoclonal antibody recognizes an inducible and reversible melanocyte cytoplasmic protein. J Cutan Pathol. 1991;18:315. doi: 10.1111/j.1600-0560.1991.tb01542.x. [DOI] [PubMed] [Google Scholar]
- 63.Abdel-Naser MB, Krasagakis K, Garbe C, Eberle J. Direct effects on proliferation, antigen expression and melanin synthesis of cultured normal human melanocytes in response to UVB and UVA light. Photodermatol Photoimmunol Photomed. 2003;19:122. doi: 10.1034/j.1600-0781.2003.00034.x. [DOI] [PubMed] [Google Scholar]
- 64.Skelton HG, 3rd, Smith KJ, Barrett TL, Lupton GP, Graham JH. HMB-45 staining in benign and malignant melanocytic lesions. A reflection of cellular activation. Am J Dermatopathol. 1991;13:543. doi: 10.1097/00000372-199113060-00004. [DOI] [PubMed] [Google Scholar]
- 65.Smoller BR, McNutt NS, Hsu A. HMB-45 recognizes stimulated melanocytes. J Cutan Pathol. 1989;16:49. doi: 10.1111/j.1600-0560.1989.tb00010.x. [DOI] [PubMed] [Google Scholar]
- 66.Lommel K, Tronnier M, Wolff HH. Activation of epidermal melanocytes is independent of epithelial proliferation and vascularization. Immunohistologic study using HMB-45 antibody with review of the literature. Hautarzt. 1996;47:616. doi: 10.1007/s001050050478. [DOI] [PubMed] [Google Scholar]
- 67.Carlson JA, Grabowski R, Mu XC, Del Rosario A, Malfetano J, Slominski A. Possible mechanisms of hypopigmentation in lichen sclerosus. Am J Dermatopathol. 2002;24:97. doi: 10.1097/00000372-200204000-00001. [DOI] [PubMed] [Google Scholar]
- 68.Boissy RE, Zhao H, Oetting WS, et al. Mutation in and lack of expression of tyrosinase-related protein-1 (TRP-1) in melanocytes from an individual with brown oculocutaneous albinism: a new subtype of albinism classified as “OCA3”. Am J Hum Genet. 1996;58:1145. [PMC free article] [PubMed] [Google Scholar]
- 69.Tsukamoto K, Jackson IJ, Urabe K, Montague PM, Hearing VJ. A second tyrosinase-related protein, TRP-2, is a melanogenic enzyme termed DOPAchrome tautomerase. EMBO J. 1992;11:519. doi: 10.1002/j.1460-2075.1992.tb05082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Virador V, Matsunaga N, Matsunaga J, et al. Production of melanocyte-specific antibodies to human melanosomal proteins: expression patterns in normal human skin and in cutaneous pigmented lesions. Pigment Cell Res. 2001;14:289. doi: 10.1034/j.1600-0749.2001.140410.x. [DOI] [PubMed] [Google Scholar]