Summary
In the last decade, the planarian has become an increasingly tractable invertebrate model for the investigation of regeneration and stem cell biology. Application of a variety of techniques and development of genomic reagents in this system have enabled exploration of the molecular mechanisms by which pluripotent somatic stem cells called neoblasts replenish, repair, and regenerate planarian tissues and organs. Recent investigations have implicated evolutionarily conserved signaling pathways in the re-establishment of anterior-posterior, dorsal-ventral, and medial-lateral polarity after injury. These studies have significantly advanced our understanding of early events during planarian regeneration, and have raised new questions about the mechanisms of stem cell-based tissue repair and renewal.
Introduction
Freshwater planarians are non-parasitic flatworms (phylum Platyhelminthes). They are able to repair and replace tissue that is lost or damaged after injury, as part of normal cellular turnover, or during growth and degrowth in response to changing nutritional availability [1–4]. These impressive restorative abilities are conferred, in part, by a population of pluripotent somatic stem cells, called neoblasts, that are distributed throughout the mesenchyme of the planarian body (Fig. 1A,B), and that differentiate into tissues and organs lost to injury. The molecular mechanisms underlying planarian regeneration and neoblast dynamics have until recently been poorly understood. However, the application of molecular and functional genomics technologies has proceeded rapidly in planarians (Box 1), allowing increasingly systematic analyses of a growing array of developmental [1,4–10] and physiological processes [11,12], as well as other problems [13,14].
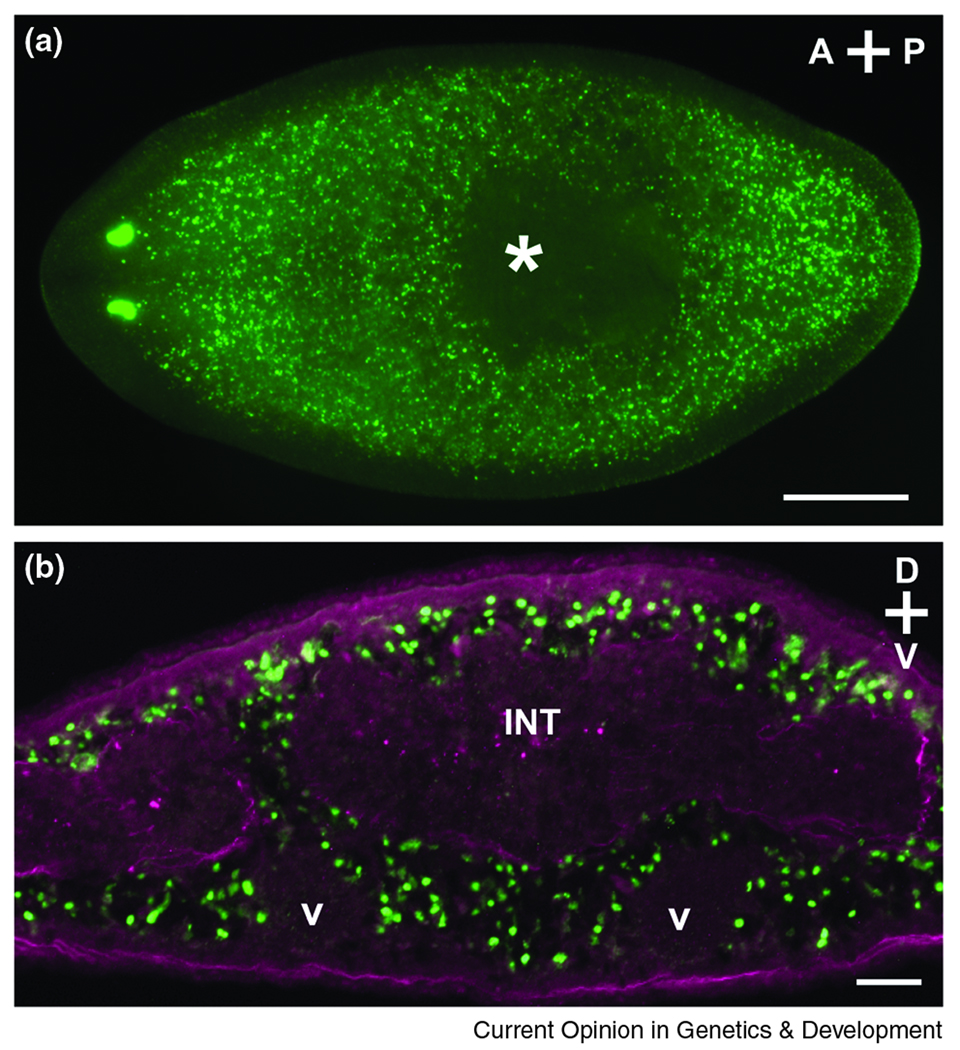
Figure 1. Planarian neoblasts.
(A) Distribution of neoblasts in an intact animal 24h after BrdU incorporation (green). S-phase cells do not reside anterior to the photoreceptors or within the pharynx (asterisk), and fragments amputated from these regions do not regenerate [1,25]. Dorsal view; anterior is to the left. Scale bar, 0.5 mm. (B) In cross section (anterior to the pharynx), neoblasts (green) are distributed mesenchymally around differentiated tissues such as the intestine (INT) and ventral nerve cords (V). Enteric and outer body wall muscles are labeled in magenta. Scale bar, 0.05 mm.
Recent technical developments have made planarians experimentally tractable
Planarians possess a rich history as an experimental subject, with rigorous studies dating back to the late 1800s [1–3,7]. Starting in the 1990s, the application of molecular cloning, immunohistochemistry, and other modern techniques initiated the re-examination of long-standing questions derived from nearly a century of surgical, physiological, histological, and ultrastructural investigations [1,10,15,16].
Experimental tractability of planarians has vastly increased in the past ten years due to a number of achievements. The selection of a European species, Schmidtea mediterranea [17], as a model planarian was made because of its small genome, diploid complement of chromosomes (four), and extant asexual and sexual strains [1]. Transcriptome characterization has been fruitful in S. mediterranea as well as the Japanese species Dugesia japonica: ~78,000 S. mediterranea expressed sequence tags (ESTs) and ~7500 D. japonica ESTs have been deposited in NCBI, the products of both published and unpublished efforts [18–20]. S. mediterranea is also the first planarian species to have its genome sequenced (Washington University Genome Sequencing Center, St. Louis, MO); annotated genomic, transcriptomic, and phenotypic data are available in a publicly accessible database [21,22]. Furthermore, gene expression can be assayed in situ [18,20,23,24], neoblasts can be labeled, isolated, and analyzed using a variety of methods [25–29], and assessment of gene function by RNA interference can be accomplished by injecting, soaking, or feeding [30–33]. These developments represent a powerful toolkit that has enabled the identification and functional analysis of genes that regulate axial patterning and neoblast dynamics during regeneration.
In order for injured planarians to regenerate in response to almost any amputation [3], robust mechanisms must exist to re-establish anterior-posterior (A–P), dorsal-ventral (D–V), and medial-lateral (M-L) axes after wounding. In the last two years, a number of investigations have revealed roles for evolutionarily conserved signaling pathways in these critical early events. Here, we survey these studies and discuss some of the unresolved questions that have emerged.
Planarians maintain and regenerate anterior-posterior polarity
As the only mitotically active somatic cells, neoblasts proliferate at sites of injury to generate the blastema, a mass of cells that differentiates into the tissues and organs lost by amputation [1]. In order for these newly generated cells to be patterned correctly, mechanisms must exist to reset polar axes after wounding; for example, if a tail is amputated, the positional identity of “posterior-most” must be re-established in order for a new tail to regenerate properly. This fact was appreciated by T. H. Morgan, whose hypotheses regarding morphogenetic gradients and polarity were formulated in part to explain the appearance of two-headed planarian regenerates: extremely thin transverse fragments would sometimes re-grow heads at both cephalic and caudal amputation sites [1,3,34–37] (Fig. 2A).
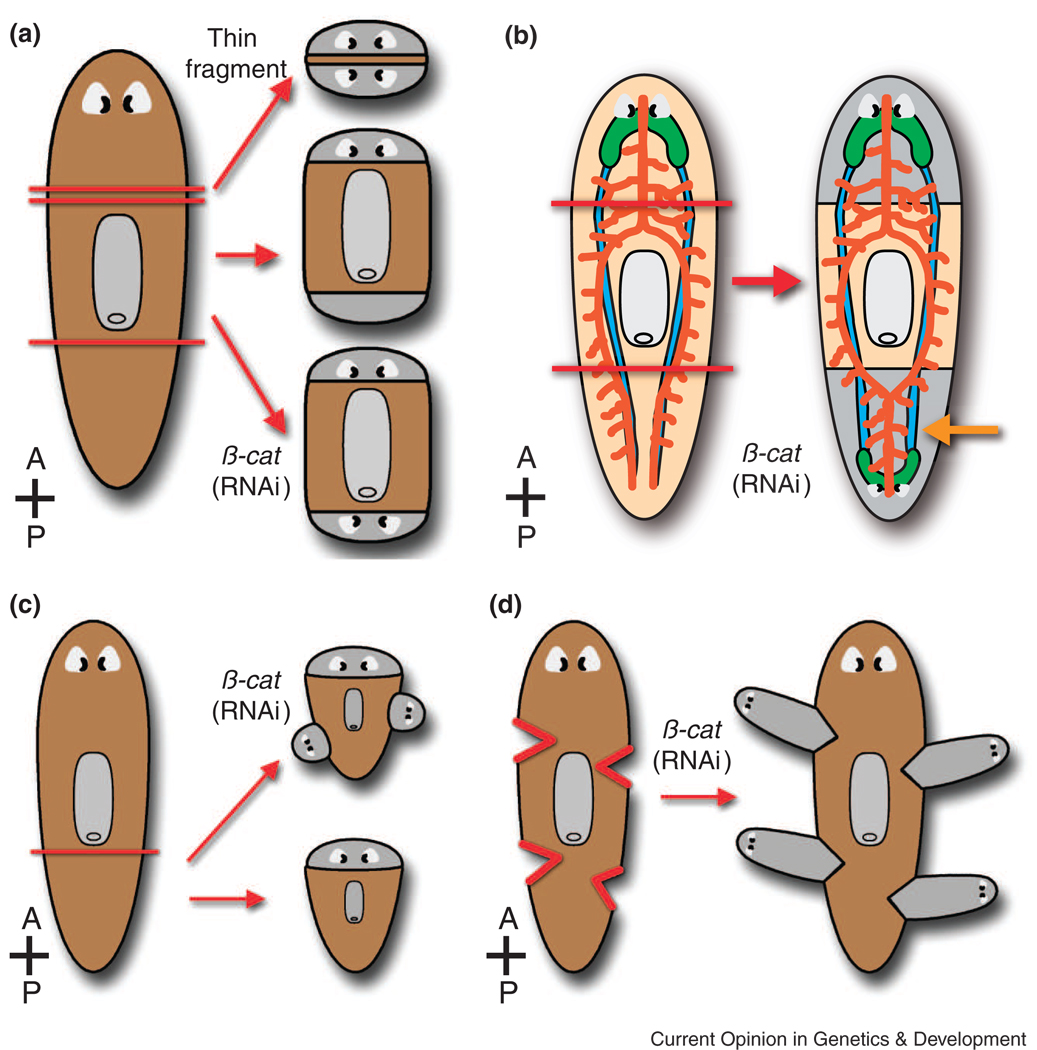
Figure 2. Patterning defects caused by β-catenin RNAi.
(A) A transverse slice from a planarian normally regenerates an anterior head and a posterior tail (middle right). Morgan observed that a very thin transverse slice (top right) sometimes regenerates a head both anteriorly and posteriorly. Knockdown of β-catenin and other Wnt pathway genes also results in regeneration of a posterior head in trunk fragments (bottom right). (B) In β-catenin(RNAi) trunk regenerates, cephalic ganglia (green) and eyes are duplicated posteriorly, while intestinal morphology (orange) is anteriorized (orange arrow). (C) and (D) β-catenin knockdown also causes growth of ectopic heads along the flanks of regenerating tail pieces (C) or at lateral incisions (D). Gray, regenerated tissue (A–D). Red lines = amputation sites (A–D).
In metazoans, Wnt proteins signal through their receptors via β-catenin-dependent (“canonical”) and -independent (“non-canonical”) pathways to regulate embryonic A–P axis formation as well as a variety of other processes [38–41]. Recently, three groups reported that in planarians, RNAi-mediated knockdown of Smed-β-catenin-1 causes the regeneration of a “posterior head” after tail amputation, replicating the two-headed phenotype that Morgan observed [34,35,42] (Fig. 2A). These animals regenerate posterior cephalic ganglia (the brain) and eyes [34,35,42], and even intestinal branches regenerate to form a single primary branch, characteristic of anterior, not posterior, patterning [34,42] (Fig. 2B). Ectopic anterior structures also appear in the uninjured regions of regenerating tail pieces [34] (Fig. 2C), in response to small lateral incisions [35] (Fig. 2D), or even in the lateral flanks of uninjured animals [34,35,42], indicating a role for β-catenin not only during regeneration, but also during normal homeostatic tissue maintenance. Furthermore, Gurley et al. [34] indirectly demonstrated that upregulation of β-catenin may be sufficient to promote posterior identity, since knockdown of Smed-APC-1 (Adenomatous Polyposis Coli, a β-catenin inhibitor) results in a reciprocal phenotype: the regeneration of posterior tissues after head amputation (an “anterior tail”).
Candidate upstream regulators of β-catenin were identified by Petersen and Reddien [35], who reported that the expression of several Wnt genes is re-established in small clusters of posterior cells within 4–5 days after amputation. In a subsequent study, Adell and colleagues reported that Smed-wntP-1, Smed-wnt11-2, and Smed-evi/wntless (necessary for Wnt secretion) are required to varying degrees for proper specification of posterior fate [43]. Additionally, in both D. japonica and S. mediterranea, knockdown of wntA results in posterior expansion of the brain [43,44].
Wnt signaling pathways have been implicated during early stages of limb and tail regeneration in zebrafish, Xenopus, and axolotl [45–48], during which their disruption inhibits the initiation of regeneration, blastema growth, and proliferation of progenitor cells. By contrast, in planarians blastema formation and growth appear to be normal in wnt and β-catenin knockdown animals, suggesting that the primary role of these genes during planarian regeneration is to regulate repatterning [34,35,42,43,49]. Taken together, the evidence suggests that Wnt signaling (through β-catenin-dependent mechanisms) promotes posterior specification and/or suppresses anterior identity in both intact and injured planarians. Since Wnt/β-catenin-mediated regulation of A–P axis formation has been studied primarily in the context of embryogenesis, observations in planarians (and other regenerating invertebrates such as Hydra) support suggestions that such a role is ancient and broadly conserved [34,35,42,43,49–53].
Questions pertaining to the role of Wnt/β-catenin signaling in planarians remain. For example, both β-catenin and APC are transcribed rather ubiquitously [34,35,42] and are upregulated in both anterior and posterior blastemas within 24 hours after amputation [34]. Since activation of these genes might occur as a nonspecific response to injury, assessment of the spatial and temporal regulation of APC, Wnt, and β-catenin proteins (for example, in which cells β-catenin translocates to the nucleus) ultimately will be needed to understand the mechanistic details of A–P axis re-establishment [50]. Additionally, other factors may regulate A–P polarity. For example, treatment with a pharmacological inhibitor of gap junctions can, like β-catenin knockdown, cause regeneration of posterior heads [54]. Likewise, a number of planarian Hox genes are expressed differentially along the A–P axis, although functional roles for these genes have not yet been reported [55–57]. Thus, although β-catenin is required to confer posterior identity, multiple pathways may interact to regulate A–P axis formation and patterning [34].
Bone Morphogenetic Protein regulates dorsal-ventral patterning
Dorsal-ventral (D–V) patterning is also important for the organization of the planarian body plan. Cephalic ganglia, nerve cords, the majority of ciliated epithelial cells, and the mouth opening are ventral, whereas eyes, testes, as well as specific populations of pigment and secretory cells are located in stereotypical positions along the D–V axis.
Bone morphogenetic protein (BMP) signaling regulates D–V polarity in metazoans, promoting ventralization in vertebrates, and dorsal identity in invertebrates [58]. In planarians, the first bmp homolog was isolated from D. japonica, where its expression in a dorsal stripe of midline cells suggested a potential role in D–V or midline patterning [59]. Three recent studies conducted in D. japonica [60] and S. mediterranea [61,62] assessed the roles of several BMP pathway genes. These included a bmp2/4/decapentaplegic homolog [60–62]; Smed-smad1 and Smed-smad4-1, members of a family of transcription factors that transduce BMP and Transforming Growth Factor β (TGF-β) receptor signaling [61,62]; and smedolloid-1, the S. mediterranea homolog of tolloid, an extracellular metalloprotease that potentiates BMP signaling by inactivating Chordin/Short Gastrulation [61]. These groups reported ventralization phenotypes such as the disappearance of dorsal markers accompanied by the ectopic dorsal expression of ventral markers [61,62], dorsal duplication of cephalic ganglia and ventral nerve cords [60–62], as well as the duplication of lateral body margins (the “D–V boundary”) [60,62] (Fig. 3A). Perhaps most dramatic was the propensity of animals to swim on their (formerly) dorsal surfaces, accompanied by a nearly complete conversion of the dorsal epithelium to a ventral-like, ciliated epidermis [61]. As with disruption of A–P patterning (above), many phenotypes were observed in both regenerating and uninjured animals, revealing a role for this signaling pathway in the maintenance of D–V patterning during normal tissue homeostasis.
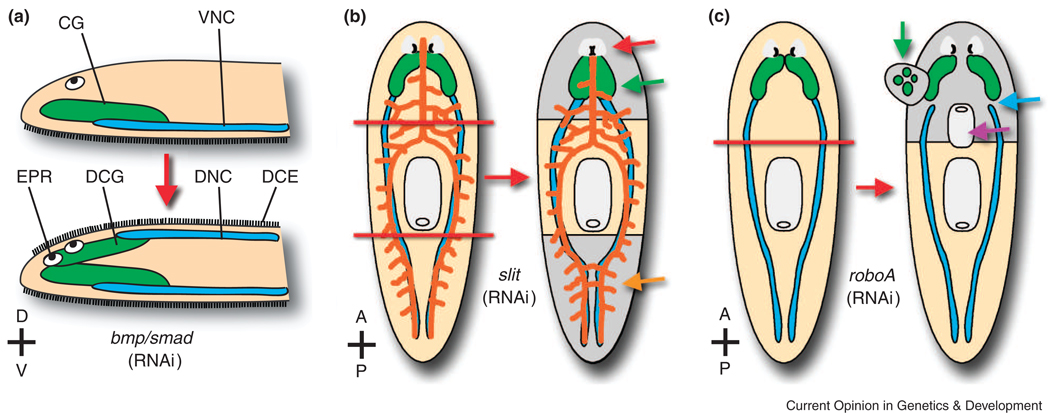
Figure 3. Patterning defects resulting from knockdown of BMP pathway, slit, and robo genes.
(A) When BMP signaling is disrupted by RNAi, normally ventral tissues develop/regenerate dorsally. EPR, ectopic photoreceptor. CG, cephalic ganglia (green). DCG, dorsal cephalic ganglia. VNC, ventral nerve cord (blue). DNC, dorsal nerve cord. DCE, dorsal ciliated epithelial cells. (B) In trunk pieces (i.e. after amputation of both head and tail), Smed-slit knockdown results in the regeneration of cephalic neural tissue that is collapsed at the midline (green arrow), fused photoreceptors (red arrow), and posterior collapse of ventral cords (blue) and intestinal branches (orange and orange arrow). (C) After head amputation, Smed-robo knockdown results in regeneration of pharynges with reversed A–P polarity (purple arrow) and ectopic dorsal cephalic outgrowths (green arrow), correlated with lack of VNC/CG connectivity (blue arrow). Cephalic ganglia are also displaced laterally. Gray, regenerated tissue (B–C). Red lines = amputation sites (B–C).
Other phenotypes suggest that BMP signaling may play roles beyond the regulation of D–V patterning. A role in medial-lateral (M-L) patterning is supported by the fact that normally, bmp expression shifts toward a lateral plane of injury, suggesting an early role in resetting the position of the midline [59,61]. Consistent with this idea, bmp, smad, and smedolloid knockdown animals fail to re-express lateral body margin markers after longitudinal amputations [61], and lateral blastemas in these animals are severely reduced or absent [61,62]. Furthermore, in animals that are cut transversely (i.e. in which heads or tails are amputated), disruption of BMP signaling causes the production of indented anterior and posterior blastemas [61,62]; in these misshapen blastemas, visual axon projections across the midline are often absent or otherwise aberrant [61]. Even more dramatically, in long-term bmp and smad RNAi experiments in uninjured animals, photoreceptors and their projections to the brain are often inappropriately duplicated on either side of the midline [60–62]. Consistent with these phenotypes, BMP signaling has been shown to regulate midline patterning during development in other organisms, for example in the mouse telencephalon [63], in the zebra fish heart and viscera [64], and during axon guidance [65].
Although indented anterior/posterior blastemas and reduced/absent lateral blastemas may be the output of disrupted early midline repatterning [61,62], these phenotypes also suggest that BMP signaling might regulate neoblast proliferation. Smed-smad4-1 knockdown does not affect neoblast proliferation 24 hours after transverse amputation [32]; the effects of Smed-bmp4-1 knockdown on early proliferation have not been reported [32,61]. However, knockdown of either of two planarian msx (muscle segment homeobox) transcription factors, Djmsh1 or Djmsh2, delays the formation of the head blastema and interferes with the normal dynamics of neoblast proliferation after injury [66]. Vertebrate MSX proteins are often coexpressed with BMPs during development and regeneration, mediating some of their activities [67–70]. Furthermore, recent evidence suggests that BMPs may function mitogenically during vertebrate limb, tail, fin, and retina regeneration [69,71,72]. Thus, the possibility that BMP signaling regulates proliferation in planarians warrants further investigation.
An alternative explanation for some phenotypes is that they occur secondarily to D–V axis disruption [61,62]. For example, Molina et al. [62] have proposed that the inappropriate differentiation of cells with ventral identity in dorsal regions may lead to ectopic D–V interactions, causing some phenotypes such as dorsal outgrowths and abnormal blastema formation. This explanation is consistent with studies in which experimental juxtaposition of dorsal and ventral tissues induces proliferation, blastema formation, and secondary axes [3,73–75]. Further complicating interpretation, variability among bmp, smad, and smedolloid phenotypes was reported. This may reflect the overlapping and non-overlapping roles of BMP pathway components [60–62], some of which (for example, members of the noggin family of BMP pathway inhibitors) have been identified only recently [76]. Given that cross-talk between signaling systems regulating A–P, D–V, and M-L polarity is likely [48,50,58,77,78], further analysis will be required to understand the multiple roles BMP signaling may play during regeneration.
On an interesting side note, Reddien and colleagues also reported that in thin lateral regenerates, bmp-expressing cells appear even in irradiated animals [61]. Stem cell-independent induction of gene expression has also been reported for a noggin-like gene in D. japonica [79]. Additionally, irradiation of either host or donor tissue does not abrogate the production of ectopic outgrowths from D–V reversed transplants [75]. These results all suggest that differentiated cells are capable of initiating or altering the expression of positional cues without input from the neoblasts, a capability that may allow rapid repatterning in response to injury [61].
Medial-lateral patterning and the central nervous system
Some planarian organs, most notably the central nervous system (CNS) and the gonads, are duplicated laterally on either side of the midline. However, the mechanisms by which bilateral symmetry is maintained and regenerated are poorly understood. In the cephalic ganglia of D. japonica, homologs of orthodenticle and orthopedia homeobox-containing genes are expressed in non-overlapping medial-lateral (M-L) domains, although the functional significance of these expression domains is unknown [23,80].
Cebrià and colleagues recently identified two planarian genes whose function is critical for proper patterning and regeneration at the midline [81,82]. Slit is a secreted extracellular ligand for the Roundabout (Robo) family of receptors, functioning in vertebrates and invertebrates as a midline repulsive cue for axons and dendrites [83]. Knockdown of Smed-slit causes regeneration of cephalic ganglia and photoreceptors that are collapsed at the midline, as well as inappropriate fusion of posterior intestinal branches (Fig. 3B) [81]. Ectopic neural tissue also appears over time at the midline in uninjured Smed-slit(RNAi) animals, suggesting a role for Slit in the normal homeostatic maintenance of CNS organization [81]. Smed-slit is expressed in both dorsal and ventral midline cells, whereas roboA is expressed in the brain, ventral cords, and early in the brain “primordia,” cells within the blastema thought to be among the first neurons to differentiate after amputation [5,6,81,82]. Thus, the regeneration of collapsed neural tissue (and the appearance of ectopic midline neuronal tissue in intact animals) may reflect the inappropriate migration or differentiation of neurons that normally respond to the midline as a repulsive boundary. Intriguingly, however, Smed-roboA knockdown results in the growth of supernumerary pharynges with reversed A–P orientation, dorsal cephalic outgrowths, and a reduced (or absent) anterior commissure connecting the two cephalic ganglia (Fig. 3C) [82]. This production of ectopic tissue (pharynges and cephalic outgrowths) in roboA(RNAi) animals correlates with the failure of the regenerated cephalic ganglia to re-establish proper connectivity with the pre-existing ventral cords [82]. In other regenerating organisms, deviation of nervous tissue can cause the growth of ectopic structures, and in urodele amphibians, regeneration is dependent on the nervous system [5,82,84]. Smed-roboA(RNAi) phenotypes support the possibility of a similar role for the planarian CNS in the general coordination of patterning [5,82]. The identification of additional ligands and/or receptors will be required in order to understand why slit and roboA phenotypes are not identical, and why roboA knockdown causes reduction of the anterior commissure.
How are neoblasts instructed by patterning cues?
Roles for Wnt/β-catenin, BMP, and Slit-Robo signaling in A–P, D–V, and M-L patterning, respectively, have been established by the studies outlined above. When axial patterning or repatterning is disrupted, the most common result is production of ectopic differentiated tissues -- anteriorized central nervous and digestive systems (wnt/β-catenin knockdown), dorsal neurons and ectopic photoreceptors (BMP pathway disruption), and supernumerary pharynges and dorsal outgrowths (roboA(RNAi)). These phenotypes imply that neoblasts are instructed to differentiate inappropriately, raising a number of questions. How do neoblasts integrate positional information? Do they respond directly to positional cues, for example to a gradient of secreted Wnt and/or BMP proteins? Or do the neoblasts respond indirectly to the axial identity of differentiated tissues, for example to signals secreted by severed nerve cords? In an uninjured animal, is neoblast fate restricted regionally? That is, do neoblasts themselves have positional identity, or do extracellular cues act only during or after lineage restriction? Many of the genes involved in re-establishment of polarity in planarians (β-catenin, APC, bmp2/4, slit, roboA) are upregulated in blastemas [34,35,42,43,62,81,82], in which early markers of neoblast differentiation are also expressed [85]. At least one of the 10 planarian Wnt receptor genes (Smed-frizzled-4) is expressed in the blastema [34]; the identity and expression pattern(s) of BMP/TGFβ receptors has not been reported. Identification of cells that co-express these genes may provide clues to the genetic programs that incorporate positional information with the control of neoblast dynamics.
Upregulation of patterning genes in the blastema also suggests that upstream components of these signaling pathways, that is, those that link wound healing with blastema growth and patterning, remain to be identified [3]. In planarians, knockdown of Djmsh1 or Djmsh2 (mentioned above) causes an overall reduction in the level of Djbmp mRNA [66], although a specific role in the blastema has not been demonstrated. Similarly, in fetal mouse digits, both Msx1 and Msx2 homeobox transcription factors are required for Bmp4 expression [70]. Also, transgenic expression of noggin in the regenerating Xenopus tadpole tail prevents upregulation of wnt3a and wnt5a, suggesting the possibility that BMP signaling may regulate Wnt expression [48]. In planarians, the drastically different phenotypes in wnt(RNAi) and bmp(RNAi) regenerates would seem to argue against such a hierarchy, but the regulatory relationships between these pathways have not yet been investigated.
Conclusions
The potential to inform our understanding of both stem cell and cancer biology has led to renewed interest in the molecular basis of regeneration in a variety of organisms [1,3,4,13,84,86–90]. Recent studies have revealed that planarians maintain and regenerate polar axes by employing evolutionarily conserved patterning mechanisms. Deeper understanding of the utilization of positional information in regenerating organisms may provide additional insights into the functions of secreted factors in adult stem cell niches [91,92], and may inform efforts to direct the differentiation of embryonic or induced pluripotent stem cells in vitro. In addition to determining the extent to which genetic programs of regeneration recapitulate those of metazoan embryogenesis [45,67,93], a second major goal is the identification of novel regulators [87]. Recently, larger scale approaches have identified hundreds of planarian genes that are expressed in dividing and differentiating neoblasts [85,94], and that are required for blastema growth, proliferation, and patterning during planarian regeneration [32]. Although many questions remain, future investigations of this intriguing invertebrate will continue to provide new perspectives on the initiation and maintenance of polarity, and the coordination of somatic stem cells by positional cues in both regenerating and fully developed tissues.
Acknowledgments
We are grateful to Tracy Chong, Jim Collins, Ryan King, Joel Stary, Jason Wever and other Newmark laboratory members for insightful comments and discussion, and to Bill Brieher and Brian Freeman for helpful critiques of this review. We apologize to colleagues whose work we were unable to discuss due to space limitations. DJF is supported by a Ruth L. Kirschstein National Research Service Award from the National Institutes of Health (F32-DK077469). Research in PAN’s laboratory is supported by the National Institutes of Health (R01-HD043403) and the National Science Foundation (IOS-0774689). PAN is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Newmark PA, Sánchez Alvarado A. Not your father's planarian: a classic model enters the era of functional genomics. Nat Rev Genet. 2002;3:210–219. doi: 10.1038/nrg759. [DOI] [PubMed] [Google Scholar]
- 2.Brøndsted HV. In: Planarian Regeneration. Kerkut GA, editor. Oxford: Pergamon Press; 1969. [Google Scholar]
- 3.Reddien PW, Sánchez Alvarado A. Fundamentals of planarian regeneration. Annu Rev Cell Dev Biol. 2004;20:725–757. doi: 10.1146/annurev.cellbio.20.010403.095114. [DOI] [PubMed] [Google Scholar]
- 4.Pellettieri J, Sánchez Alvarado A. Cell turnover and adult tissue homeostasis: from humans to planarians. Annu Rev Genet. 2007;41:83–105. doi: 10.1146/annurev.genet.41.110306.130244. [DOI] [PubMed] [Google Scholar]
- 5.Cebrià F. Regenerating the central nervous system: how easy for planarians! Dev Genes Evol. 2007;217:733–748. doi: 10.1007/s00427-007-0188-6. [DOI] [PubMed] [Google Scholar]
- 6.Agata K, Umesono Y. Brain regeneration from pluripotent stem cells in planarian. Philos Trans R Soc Lond B Biol Sci. 2008;363:2071–2078. doi: 10.1098/rstb.2008.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Newmark PA, Wang Y, Chong T. Germ Cell Specification and Regeneration in Planarians. Cold Spring Harb Symp Quant Biol. 2008 doi: 10.1101/sqb.2008.73.022. [DOI] [PubMed] [Google Scholar]
- 8.Sánchez Alvarado A. Planarian regeneration: its end is its beginning. Cell. 2006;124:241–245. doi: 10.1016/j.cell.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 9.Rossi L, Salvetti A, Batistoni R, Deri P, Gremigni V. Planarians, a tale of stem cells. Cell Mol Life Sci. 2008;65:16–23. doi: 10.1007/s00018-007-7426-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salò E, Abril JF, Adell T, Cebrià F, Eckelt K, Fernandez-Taboada E, Handberg-Thorsager M, Iglesias M, Molina MD, Rodríguez-Esteban G. Planarian regeneration: achievements and future directions after 20 years of research. Int J Dev Biol. 2008 doi: 10.1387/ijdb.072414es. [DOI] [PubMed] [Google Scholar]
- 11.Raffa RB, Rawls SM. Planaria: A model for drug action and abuse. Austin: Landes Bioscience; 2008. [Google Scholar]
- 12.González-Estévez C. Autophagy in freshwater planarians. Methods Enzymol. 2008;451:439–465. doi: 10.1016/S0076-6879(08)03227-8. [DOI] [PubMed] [Google Scholar]
- 13.Pearson BJ, Sánchez Alvarado A. Regeneration, Stem Cells, and the Evolution of Tumor Suppression. Cold Spring Harb Symp Quant Biol. 2009 doi: 10.1101/sqb.2008.73.045. [DOI] [PubMed] [Google Scholar]
- 14.Oviedo NJ, Beane WS. Regeneration: The origin of cancer or a possible cure? Semin Cell Dev Biol. 2008 doi: 10.1016/j.semcdb.2009.04.005. doi:10.1016/j.semcdb.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baguñà J, Saló E, Collet J, Auladell C, Ribas M. Cellular, molecular and genetic approaches to regeneration and pattern formation in planarians. Fortschr Zool. 1988;36:65–78. [Google Scholar]
- 16.Salò E, Baguñà J. Regeneration in planarians and other worms: New findings, new tools, and new perspectives. J Exp Zool. 2002;292:528–539. doi: 10.1002/jez.90001. [DOI] [PubMed] [Google Scholar]
- 17.Benazzi M, Banguñà J, Ballester R, Puccinelli I, Del Papa R. Further contribution to the taxonomy of the Dugesia lugubris-polychroa group with description of Dugesia mediterranea n.sp. (Tricladida, Paludicola) Boll Zool. 1975;42:81–89. [Google Scholar]
- 18.Sánchez Alvarado A, Newmark PA, Robb SM, Juste R. The Schmidtea mediterranea database as a molecular resource for studying platyhelminthes, stem cells and regeneration. Development. 2002;129:5659–5665. doi: 10.1242/dev.00167. [DOI] [PubMed] [Google Scholar]
- 19.Mineta K, Nakazawa M, Cebria F, Ikeo K, Agata K, Gojobori T. Origin and evolutionary process of the CNS elucidated by comparative genomics analysis of planarian ESTs. Proc Natl Acad Sci U S A. 2003;100:7666–7671. doi: 10.1073/pnas.1332513100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zayas RM, Hernandez A, Habermann B, Wang Y, Stary JM, Newmark PA. The planarian Schmidtea mediterranea as a model for epigenetic germ cell specification: analysis of ESTs from the hermaphroditic strain. Proc Natl Acad Sci U S A. 2005;102:18491–18496. doi: 10.1073/pnas.0509507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cantarel BL, Korf I, Robb SM, Parra G, Ross E, Moore B, Holt C, Sánchez Alvarado A, Yandell M. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2008;18:188–196. doi: 10.1101/gr.6743907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robb SM, Ross E, Sánchez Alvarado A. SmedGD: the Schmidtea mediterranea genome database. Nucleic Acids Res. 2008;36:D599–D606. doi: 10.1093/nar/gkm684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Umesono Y, Watanabe K, Agata K. A planarian orthopedia homolog is specifically expressed in the branch region of both the mature and regenerating brain. Dev Growth Differ. 1997;39:723–727. doi: 10.1046/j.1440-169x.1997.t01-5-00008.x. [DOI] [PubMed] [Google Scholar]
- 24.Pearson BJ, Eisenhoffer GT, Gurley KA, Rink JC, Miller DE, Sánchez Alvarado A. Formaldehyde-based whole-mount in situ hybridization method for planarians. Dev Dyn. 2009;238:443–450. doi: 10.1002/dvdy.21849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newmark PA, Sánchez Alvarado A. Bromodeoxyuridine specifically labels the regenerative stem cells of planarians. Dev Biol. 2000;220:142–153. doi: 10.1006/dbio.2000.9645. [DOI] [PubMed] [Google Scholar]
- 26. Hayashi T, Asami M, Higuchi S, Shibata N, Agata K. Isolation of planarian X-ray-sensitive stem cells by fluorescence-activated cell sorting. Dev Growth Differ. 2006;48:371–380. doi: 10.1111/j.1440-169X.2006.00876.x. For many years, semi-pure enriched fractions of neoblasts from dissociated planarians could only be obtained by density gradient centrifugation or filtration through small-pore nylon meshes. In an important technical achievement, Hayashi and colleagues developed a fluorescence activated cell sorting (FACS) protocol to yield highly pure fractions of living stem cells by comparing single cell suspensions from irradiated and unirradiated animals. This methodology has quickly become a standard analytical tool.
- 27.Kang H, Alvarado AS. Flow cytometry methods for the study of cell-cycle parameters of planarian stem cells. Dev Dyn. 2009 doi: 10.1002/dvdy.21928. [DOI] [PubMed] [Google Scholar]
- 28.Sánchez Alvarado A, Kang H. Multicellularity, stem cells, and the neoblasts of the planarian Schmidtea mediterranea. Exp Cell Res. 2005;306:299–308. doi: 10.1016/j.yexcr.2005.03.020. [DOI] [PubMed] [Google Scholar]
- 29.Sánchez Alvarado A. Stem cells and the Planarian Schmidtea mediterranea. C R Biol. 2007;330:498–503. doi: 10.1016/j.crvi.2007.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sánchez Alvarado A, Newmark PA. Double-stranded RNA specifically disrupts gene expression during planarian regeneration. Proc Natl Acad Sci U S A. 1999;96:5049–5054. doi: 10.1073/pnas.96.9.5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Newmark PA, Reddien PW, Cebrià F, Sánchez Alvarado A. Ingestion of bacterially expressed double-stranded RNA inhibits gene expression in planarians. Proc Natl Acad Sci U S A. 2003 100 Suppl 1:11861–11865. doi: 10.1073/pnas.1834205100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reddien PW, Bermange AL, Murfitt KJ, Jennings JR, Sánchez Alvarado A. Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria. Dev Cell. 2005;8:635–649. doi: 10.1016/j.devcel.2005.02.014. The first large-scale RNAi screen in planarians. 240 genes from a variety of functional categories (e.g. translational regulators, transcriptional regulators, signal transduction pathway components, etc.) yielded phenotypes indicating a role in regeneration, patterning, neoblast proliferation, and/or homeostatic tissue renewal.
- 33.Orii H, Mochii M, Watanabe K. A simple "soaking method" for RNA interference in the planarian Dugesia japonica. Dev Genes Evol. 2003;213:138–141. doi: 10.1007/s00427-003-0310-3. [DOI] [PubMed] [Google Scholar]
- 34. Gurley KA, Rink JC, Sánchez Alvarado A. Beta-catenin defines head versus tail identity during planarian regeneration and homeostasis. Science. 2008;319:323–327. doi: 10.1126/science.1150029. The authors identified Smed-β-catenin-1 as essential for proper regeneration of posterior structures. β-catenin(RNAi) and dishevelled(RNAi) animals regenerate caudal heads, rather than tails. Smed-APC(RNAi) animals display a reciprocal phenotype, regenerating anterior tails, demonstrating that upregulation of β-catenin may be sufficient for posteriorization. The authors also analyzed the expression of over 30 planarian Wnt pathway genes.
- 35. Petersen CP, Reddien PW. Smed-betacatenin-1 is required for anteroposterior blastema polarity in planarian regeneration. Science. 2008;319:327–330. doi: 10.1126/science.1149943. The authors showed that Smed-β-catenin-1 is required for posteriorization in intact and regenerating animals; in β-catenin(RNAi) animals, ectopic heads form even in response to small lateral incisions. The authors also identified five Wnt genes expressed in specific regions along the anteroposterior axis.
- 36.Morgan TH. Proceedings of the American Society of Zoologists: Polarity and axial heteromorphosis. Am. Nat. 1904;38:502–505. [Google Scholar]
- 37.Morgan TH. "Polarity" considered as a phenomenon of gradation of materials. J. Exp. Zool. 1905;2:495–506. [Google Scholar]
- 38.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 39.Schier AF, Talbot WS. Molecular genetics of axis formation in zebrafish. Annu Rev Genet. 2005;39:561–613. doi: 10.1146/annurev.genet.37.110801.143752. [DOI] [PubMed] [Google Scholar]
- 40.Marikawa Y. Wnt/beta-catenin signaling and body plan formation in mouse embryos. Semin Cell Dev Biol. 2006;17:175–184. doi: 10.1016/j.semcdb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 41.Hardin J, King RS. The long and the short of Wnt signaling in C. elegans. Curr Opin Genet Dev. 2008;18:362–367. doi: 10.1016/j.gde.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Iglesias M, Gomez-Skarmeta JL, Saló E, Adell T. Silencing of Smed-betacatenin1 generates radial-like hypercephalized planarians. Development. 2008;135:1215–1221. doi: 10.1242/dev.020289. With [34,35], a third example of β-catenin’s posteriorizing role. The authors found that over a longer time course, some β-catenin(RNAi) regenerates become “hypercephalized,” a phenotype that the authors interpret as “radial-like.”
- 43. Adell T, Saló E, Boutros M, Bartscherer K. Smed-Evi/Wntless is required for {beta}-catenin-dependent and -independent processes during planarian regeneration. Development. 2009;136:905–910. doi: 10.1242/dev.033761. These authors observed anteriorization simlar to the β-catenin(RNAi) phenotype by knocking down the gene encoding SMED-EVI/WNTLESS, which is required for Wnt secretion in other organisms. WntP-1 and Wnt11-2 were identified as putative posterior organizers.
- 44.Kobayashi C, Saito Y, Ogawa K, Agata K. Wnt signaling is required for antero-posterior patterning of the planarian brain. Dev Biol. 2007;306:714–724. doi: 10.1016/j.ydbio.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 45.Kawakami Y, Rodriguez Esteban C, Raya M, Kawakami H, Marti M, Dubova I, Izpisua Belmonte JC. Wnt/beta-catenin signaling regulates vertebrate limb regeneration. Genes Dev. 2006;20:3232–3237. doi: 10.1101/gad.1475106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stoick-Cooper CL, Weidinger G, Riehle KJ, Hubbert C, Major MB, Fausto N, Moon RT. Distinct Wnt signaling pathways have opposing roles in appendage regeneration. Development. 2007;134:479–489. doi: 10.1242/dev.001123. [DOI] [PubMed] [Google Scholar]
- 47.Yokoyama H, Ogino H, Stoick-Cooper CL, Grainger RM, Moon RT. Wnt/beta-catenin signaling has an essential role in the initiation of limb regeneration. Dev Biol. 2007;306:170–178. doi: 10.1016/j.ydbio.2007.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin G, Slack JM. Requirement for Wnt and FGF signaling in Xenopus tadpole tail regeneration. Dev Biol. 2008;316:323–335. doi: 10.1016/j.ydbio.2008.01.032. [DOI] [PubMed] [Google Scholar]
- 49.Tanaka EM, Weidinger G. Heads or tails: can Wnt tell which one is up? Nat Cell Biol. 2008;10:122–124. doi: 10.1038/ncb0208-122. [DOI] [PubMed] [Google Scholar]
- 50.Meinhardt H. Beta-catenin and axis formation in planarians. Bioessays. 2009;31:5–9. doi: 10.1002/bies.080193. [DOI] [PubMed] [Google Scholar]
- 51.Holstein TW. Wnt signaling in cnidarians. Methods Mol Biol. 2008;469:47–54. doi: 10.1007/978-1-60327-469-5. [DOI] [PubMed] [Google Scholar]
- 52.Lengfeld T, Watanabe H, Simakov O, Lindgens D, Gee L, Law L, Schmidt HA, Ozbek S, Bode H, Holstein TW. Multiple Wnts are involved in Hydra organizer formation and regeneration. Dev Biol. 2009 doi: 10.1016/j.ydbio.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 53.Martindale MQ. The evolution of metazoan axial properties. Nat Rev Genet. 2005;6:917–927. doi: 10.1038/nrg1725. [DOI] [PubMed] [Google Scholar]
- 54.Nogi T, Levin M. Characterization of innexin gene expression and functional roles of gap-junctional communication in planarian regeneration. Dev Biol. 2005;287:314–335. doi: 10.1016/j.ydbio.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 55.Bayascas JR, Castillo E, Saló E. Platyhelminthes have a hox code differentially activated during regeneration, with genes closely related to those of spiralian protostomes. Dev Genes Evol. 1998;208:467–473. doi: 10.1007/s004270050204. [DOI] [PubMed] [Google Scholar]
- 56.Orii H, Kato K, Umesono Y, Sakurai T, Agata K, Watanabe K. The planarian HOM/HOX homeobox genes (Plox) expressed along the anteroposterior axis. Dev Biol. 1999;210:456–468. doi: 10.1006/dbio.1999.9275. [DOI] [PubMed] [Google Scholar]
- 57.Nogi T, Watanabe K. Position-specific and non-colinear expression of the planarian posterior (Abdominal-B-like) gene. Dev Growth Differ. 2001;43:177–184. doi: 10.1046/j.1440-169x.2001.00564.x. [DOI] [PubMed] [Google Scholar]
- 58.De Robertis EM, Kuroda H. Dorsal-ventral patterning and neural induction in Xenopus embryos. Annu Rev Cell Dev Biol. 2004;20:285–308. doi: 10.1146/annurev.cellbio.20.011403.154124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Orii H, Kato K, Agata K, Watanabe K. Molecular cloning of Bone Morphogenetic Protein (BMP) gene from the planarian Dugesia japonica. Zoolog Sci. 1998;15:871–877. [Google Scholar]
- 60.Orii H, Watanabe K. Bone morphogenetic protein is required for dorso-ventral patterning in the planarian Dugesia japonica. Dev Growth Differ. 2007;49:345–349. doi: 10.1111/j.1440-169X.2007.00931.x. [DOI] [PubMed] [Google Scholar]
- 61. Reddien PW, Bermange AL, Kicza AM, Sánchez Alvarado A. BMP signaling regulates the dorsal planarian midline and is needed for asymmetric regeneration. Development. 2007;134:4043–4051. doi: 10.1242/dev.007138. The authors analyzed the function of Smed-bmp4-1, Smed-smad4-1, and smedolloid-1, reporting both ventralization and midline patterning phenotypes. These included the appearance of a ciliated dorsal epidermis accompanied by dorsal duplication of ventral nerve cords; some animals even swim on their formerly dorsal surfaces (functional ventralization). A role for BMP signaling in medial-lateral patterning was indicated by duplication of photoreceptors and defects in visual axon regeneration at the midline, the inability of lateral regenerates to produce blastemas or re-express lateral margin markers, and indented or absent anterior and posterior blastemas.
- 62. Molina MD, Salò E, Cebrià F. The BMP pathway is essential for re-specification and maintenance of the dorsoventral axis in regenerating and intact planarians. Dev Biol. 2007;311:79–94. doi: 10.1016/j.ydbio.2007.08.019. Analyzing the roles of Smed-BMP and Smed-Smad1, the authors reported disruption of dorsal-ventral patterning (dorsal duplication of cephalic ganglia and nerve cords, ectopic expression of ventral markers dorsally), midline patterning (expansion of the dorsal midline, duplication of photoreceptors), and defects in blastema growth.
- 63.Chizhikov VV, Millen KJ. Roof plate-dependent patterning of the vertebrate dorsal central nervous system. Dev Biol. 2005;277:287–295. doi: 10.1016/j.ydbio.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 64.Monteiro R, van Dinther M, Bakkers J, Wilkinson R, Patient R, ten Dijke P, Mummery C. Two novel type II receptors mediate BMP signalling and are required to establish left-right asymmetry in zebrafish. Dev Biol. 2008;315:55–71. doi: 10.1016/j.ydbio.2007.11.038. [DOI] [PubMed] [Google Scholar]
- 65.Charron F, Tessier-Lavigne M. Novel brain wiring functions for classical morphogens: a role as graded positional cues in axon guidance. Development. 2005;132:2251–2262. doi: 10.1242/dev.01830. [DOI] [PubMed] [Google Scholar]
- 66.Mannini L, Deri P, Gremigni V, Rossi L, Salvetti A, Batistoni R. Two msh/msx-related genes, Djmsh1 and Djmsh2, contribute to the early blastema growth during planarian head regeneration. Int J Dev Biol. 2008;52:943–952. doi: 10.1387/ijdb.072476lm. [DOI] [PubMed] [Google Scholar]
- 67.Yokoyama H. Initiation of limb regeneration: the critical steps for regenerative capacity. Dev Growth Differ. 2008;50:13–22. doi: 10.1111/j.1440-169X.2007.00973.x. [DOI] [PubMed] [Google Scholar]
- 68.Beck CW, Christen B, Slack JM. Molecular pathways needed for regeneration of spinal cord and muscle in a vertebrate. Dev Cell. 2003;5:429–439. doi: 10.1016/s1534-5807(03)00233-8. [DOI] [PubMed] [Google Scholar]
- 69.Beck CW, Christen B, Barker D, Slack JM. Temporal requirement for bone morphogenetic proteins in regeneration of the tail and limb of Xenopus tadpoles. Mech Dev. 2006;123:674–688. doi: 10.1016/j.mod.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 70.Han M, Yang X, Farrington JE, Muneoka K. Digit regeneration is regulated by Msx1 and BMP4 in fetal mice. Development. 2003;130:5123–5132. doi: 10.1242/dev.00710. [DOI] [PubMed] [Google Scholar]
- 71.Haynes T, Gutierrez C, Aycinena JC, Tsonis PA, Del Rio-Tsonis K. BMP signaling mediates stem/progenitor cell-induced retina regeneration. Proc Natl Acad Sci U S A. 2007;104:20380–20385. doi: 10.1073/pnas.0707202104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smith A, Avaron F, Guay D, Padhi BK, Akimenko MA. Inhibition of BMP signaling during zebrafish fin regeneration disrupts fin growth and scleroblasts differentiation and function. Dev Biol. 2006;299:438–454. doi: 10.1016/j.ydbio.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 73.Chandebois R. The dynamics of wound closure and its role in the programming of planarian regeneration. I. Blastema emergence. Dev Growth Differ. 1979;21:195–204. doi: 10.1111/j.1440-169X.1979.00195.x. [DOI] [PubMed] [Google Scholar]
- 74.Kato K, Orii H, Watanabe K, Agata K. The role of dorsoventral interaction in the onset of planarian regeneration. Development. 1999;126:1031–1040. doi: 10.1242/dev.126.5.1031. [DOI] [PubMed] [Google Scholar]
- 75.Kato K, Orii H, Watanabe K, Agata K. Dorsal and ventral positional cues required for the onset of planarian regeneration may reside in differentiated cells. Dev Biol. 2001;233:109–121. doi: 10.1006/dbio.2001.0226. [DOI] [PubMed] [Google Scholar]
- 76.Dolores Molina MA, Salò E, Cebrià F. Expression pattern of the expanded noggin gene family in the planarian Schmidtea mediterranea. Gene Expr Patterns. 2009;9:246–253. doi: 10.1016/j.gep.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 77.Meinhardt H. Models of biological pattern formation: from elementary steps to the organization of embryonic axes. Curr Top Dev Biol. 2008;81:1–63. doi: 10.1016/S0070-2153(07)81001-5. [DOI] [PubMed] [Google Scholar]
- 78.Guo X, Wang XF. Signaling cross-talk between TGF-beta/BMP and other pathways. Cell Res. 2009;19:71–88. doi: 10.1038/cr.2008.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ogawa K, Ishihara S, Saito Y, Mineta K, Nakazawa M, Ikeo K, Gojobori T, Watanabe K, Agata K. Induction of a noggin-like gene by ectopic DV interaction during planarian regeneration. Dev Biol. 2002;250:59–70. doi: 10.1006/dbio.2002.0790. [DOI] [PubMed] [Google Scholar]
- 80.Umesono Y, Watanabe K, Agata K. Distinct structural domains in the planarian brain defined by the expression of evolutionarily conserved homeobox genes. Dev Genes Evol. 1999;209:31–39. doi: 10.1007/s004270050224. [DOI] [PubMed] [Google Scholar]
- 81. Cebrià F, Guo T, Jopek J, Newmark PA. Regeneration and maintenance of the planarian midline is regulated by a slit orthologue. Dev Biol. 2007;307:394–406. doi: 10.1016/j.ydbio.2007.05.006. Smed-slit(RNAi) causes the regeneration of neural tissue (cephalic ganglia, ventral nerve cords, and photoreceptors) that is collapsed at the midline. Ectopic neural tissue also appears at the midline even in uninjured animals, suggesting a role for slit during homeostatic maintenance of CNS organization.
- 82. Cebrià F, Newmark PA. Morphogenesis defects are associated with abnormal nervous system regeneration following roboA RNAi in planarians. Development. 2007;134:833–837. doi: 10.1242/dev.02794. Knockdown of Smed-roboA causes the production of ectopic dorsal cephalic outgrowths and supernumerary pharynges, phenotypes that are correlated with a lack of connectivity between the newly regenerated brain lobes and the pre-existing ventral nerve cords. These data suggest that the planarian CNS may play a central role during regeneration, as in annelids and amphibians.
- 83.Dickson BJ, Gilestro GF. Regulation of commissural axon pathfinding by slit and its Robo receptors. Annu Rev Cell Dev Biol. 2006;22:651–675. doi: 10.1146/annurev.cellbio.21.090704.151234. [DOI] [PubMed] [Google Scholar]
- 84.Brockes JP, Kumar A. Comparative aspects of animal regeneration. Annu Rev Cell Dev Biol. 2008;24:525–549. doi: 10.1146/annurev.cellbio.24.110707.175336. [DOI] [PubMed] [Google Scholar]
- 85. Eisenhoffer GT, Kang H, Sánchez Alvarado A. Molecular analysis of stem cells and their descendants during cell turnover and regeneration in the planarian Schmidtea mediterranea. Cell Stem Cell. 2008;3:327–339. doi: 10.1016/j.stem.2008.07.002. Microarray-based transcriptome analysis revealed 259 genes whose expression is downregulated in planarians one and seven days after lethal irradiation. Genes downregulated early are expressed in mitotically active neoblasts, while those downregulated later are expressed primarily in postmitotic progeny. BrdU pulse-chase experiments demonstrated that cells expressing ‘late’ genes are the differentiating progeny of those expressing ‘early‘ genes. Analysis using a subset of these markers to label single sorted cells revealed considerable heterogeneity among neoblasts and their immediate progeny.
- 86.Poss KD. Getting to the heart of regeneration in zebrafish. Semin Cell Dev Biol. 2007;18:36–45. doi: 10.1016/j.semcdb.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 87.Yin VP, Poss KD. New regulators of vertebrate appendage regeneration. Curr Opin Genet Dev. 2008;18:381–386. doi: 10.1016/j.gde.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Slack JM, Lin G, Chen Y. The Xenopus tadpole: a new model for regeneration research. Cell Mol Life Sci. 2008;65:54–63. doi: 10.1007/s00018-007-7431-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sánchez Alvarado A, Tsonis PA. Bridging the regeneration gap: genetic insights from diverse animal models. Nat Rev Genet. 2006;7:873–884. doi: 10.1038/nrg1923. [DOI] [PubMed] [Google Scholar]
- 90.Stoick-Cooper CL, Moon RT, Weidinger G. Advances in signaling in vertebrate regeneration as a prelude to regenerative medicine. Genes Dev. 2007;21:1292–1315. doi: 10.1101/gad.1540507. [DOI] [PubMed] [Google Scholar]
- 91.Li L, Xie T. Stem cell niche: structure and function. Annu Rev Cell Dev Biol. 2005;21:605–631. doi: 10.1146/annurev.cellbio.21.012704.131525. [DOI] [PubMed] [Google Scholar]
- 92.Morrison SJ, Spradling AC. Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell. 2008;132:598–611. doi: 10.1016/j.cell.2008.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bryant SV, Endo T, Gardiner DM. Vertebrate limb regeneration and the origin of limb stem cells. Int J Dev Biol. 2002;46:887–896. [PubMed] [Google Scholar]
- 94. Rossi L, Salvetti A, Marincola FM, Lena A, Deri P, Mannini L, Batistoni R, Wang E, Gremigni V. Deciphering the molecular machinery of stem cells: a look at the neoblast gene expression profile. Genome Biol. 2007;8:R62. doi: 10.1186/gb-2007-8-4-r62. Microarray-based analysis of gene expression in D. japonica after lethal and sublethal irradiation identified 60 downregulated and 63 upregulated genes, respectively. Expression of a subset of the downregulated genes in neoblasts was verified by in situ hybridization; not all genes are coexpressed, indicating heterogeneity of the stem cell population.