Abstract
Sessile inhabitants of marine intertidal environments commonly face heat stress, an important component of summer mortality syndrome in the Pacific oyster Crassostrea gigas. Marker-aided selection programs would be useful for developing oyster strains that resist summer mortality; however, there is currently a need to identify candidate genes associated with stress tolerance and to develop molecular markers associated with those genes. To identify candidate genes for further study, we used cDNA microarrays to test the hypothesis that oyster families that had high (>64%) or low (<29%) survival of heat shock (43°C, 1 h) differ in their transcriptional responses to stress. Based upon data generated by the microarray and by real-time quantitative PCR, we found that transcription after heat shock increased for genes putatively encoding heat shock proteins and genes for proteins that synthesize lipids, protect against bacterial infection, and regulate spawning, whereas transcription decreased for genes for proteins that mobilize lipids and detoxify reactive oxygen species. RNAs putatively identified as heat shock protein 27, collagen, peroxinectin, S-crystallin, and two genes with no match in Genbank had higher transcript concentrations in low-surviving families than in high-surviving families, whereas concentration of putative cystatin B mRNA was greater in high-surviving families. These ESTs should be studied further for use in marker-aided selection programs. Low survival of heat shock could result from a complex interaction of cell damage, opportunistic infection, and metabolic exhaustion.
Keywords: Oyster, Bivalve, Microarray, Gene expression, Heat shock, Crassostrea gigas, Crassostrea virginica, Summer mortality
Introduction
Summer mortality syndrome causes mass mortalities of Pacific oysters Crassostrea gigas during warm summer months and has been problematic for oyster growers in the USA, France, and Japan since the 1960s (reviewed by Cheney et al. 2000 and Samain et al. 2007). Summer mortality kills both juvenile and adult oysters and results from complex interactions between environmental factors such as temperature, dissolved oxygen, primary productivity, and sediment characteristics (Soletchnik et al. 1999; Cheney et al. 2000; Soletchnik et al. 2005, 2007; Li et al. 2009), physiological condition including reproductive status and glycogen metabolism (Soletchnik et al. 1999; Berthelin et al. 2000), and pathogens (Lacoste et al. 2001; Friedman et al. 2005; Garnier et al. 2007). Resistance to summer mortality has a genetic basis in at least some populations (Dégremont et al. 2007), and selective breeding for oyster stocks that resist summer mortality is a promising way to reduce losses. However, the complex dynamics of summer mortality in USA waters precludes selection of broodstock solely upon the basis of field trials because mass mortalities are unpredictable in both time and space. Marker-aided selection could be used to reliably identify stocks or families that have increased resistance to stress (Cnaani 2006; Rothschild and Ruvinsky 2007), but there are presently needs to identify genes and markers, and to understand the physiological basis of stress resistance in oysters.
Heat stress is an inevitable consequence of life in the marine intertidal zone; and during periods coinciding with summer mortality in the Pacific Northwest region of the USA, ambient temperature during tidal emersion can exceed 40°C (Cheney et al. 2000; Hamdoun et al. 2003). In marine bivalves, heat shock results in oxidative stress (Abele et al. 2002; Verlecar et al. 2007), impairs immune defenses through reduced hemocyte phagocytosis and killing capacity (Hégaret et al. 2003, 2004; Chen et al. 2007), and may lead to metabolic exhaustion and susceptibility to opportunistic infection, particularly when coupled with high energetic investment in reproduction (Li et al. 2007, 2009). All of these factors contribute to summer mortality syndrome, but heat shock under laboratory conditions is a convenient means to elicit the transcription of genes with potential as genetic markers for superior survival under summer mortality-inducing conditions.
Transcriptome profiling is a useful first step in studies of the response of oysters to stress. Recently, an oyster cDNA microarray consisting of expressed sequence tag (EST) libraries from several different tissues of C. gigas and the Atlantic oyster Crassostrea virginica exposed to hypoxia, pesticides, bacterial and protozoan infection, hydrocarbons, hyperthermia, and summer mortality conditions became available (Jenny et al. 2007). The microarray contains 4,460 elements from C. virginica and 2,320 elements from C. gigas, and 16 non-oyster DNAs that can be used as negative and positive controls. We used this tool and real-time quantitative polymerase chain reaction (RT-QPCR) to test the hypothesis that oyster families that differ in survival after heat shock also differ in their transcriptional responses to this stress.
Materials and Methods
Screening of Families
To identify oyster families that differed in their tolerance of heat shock, during Summer 2003, we heat-shocked 100 juvenile oysters (1–2 cm) from each of 44 families from a selectively bred cohort of oyster families produced according to Langdon et al. (2003). The heat shock treatment consisted of exposure for 1 h to sand-filtered, UV-sterilized water heated to 43°C (Clegg et al. 1998; Hamdoun et al. 2003) followed by recovery in troughs supplied with sand-filtered seawater at ambient temperature (~14°C) without the addition of algal ration. These heat shock conditions were based upon preliminary experiments showing a mortality of approximately 50% for groups of juvenile oysters (spat) sampled from all families (data not shown). Families were tentatively classified as low-surviving or high-surviving when their survival after heat shock as juveniles was <29% and >64%, respectively.
During Fall 2003, we deployed 150 unstressed juveniles from each of four low- and four high-surviving families in Yaquina Bay estuary, Newport, Oregon, USA at an intertidal site located approximately 15 km from the mouth of the bay (44.6° N, 124.1° W). Spat were deployed within three individual family-specific rectangular mesh bags (0.53×0.81 m, 7-mm mesh) at a density of 50 animals per bag, and each of the three family-specific bags was planted at the same tidal height (+0 MLLW; mean lower low water). During Summer 2005, we recovered the families, and allowed 1 month for acclimation in on-land troughs supplied with sand-filtered seawater at ambient temperature (~14°C). Animals were fed a mixture of Isochrysis galbana and Chaetoceros calcitrans at a concentration of approximately 50,000–80,000 cells/ml. We heat-shocked (43°C, 1 h) 20 individuals from each of the eight families and monitored their survival as before. Four of the eight families retained their classification as low- or high-surviving (two families of each type) and were used for the microarray experiment.
Tissue Collection
We chose to study gill tissue because it is easy to collect without contamination from other organs and because it is a large organ with high surface area that has been shown to be perturbed by thermal stress (Meistertzheim et al. 2007). Gill of 45 oysters from each of four families (two high- and two low-surviving; n=180 animals in total) were sampled in this study. We sampled these oysters across three independent heat shock experiments (Fig. 1a). In each of these experiments, we first collected pre-stress (control) samples by excising ~50 mg gill of the second lamellibranch from three individuals per family, pooling the three gill samples (families were kept separate), and discarding the remaining carcasses. Thereafter, we simultaneously exposed 12 previously unstressed animals per family for 1 h to UV-filtered water heated to 40°C (previously determined to be non-lethal) in flow-through systems, transported them to a different flow-through system supplied with sand-filtered water at ambient temperature (~14°C), and we collected pools of gill of three individuals per family, as described above, after 1-h, 3-h, 6-h, and 24-h recovery. Thus, in total we prepared 60 samples representing three biological replicates of gill from two high- and two low-surviving families at five points in time (Fig. 1a). In addition to the 180 animals that were sampled, we treated five animals per family exactly as their siblings and monitored them for 6 days to ensure that heat shock was not lethal, and we also monitored five animals alongside their heat-shocked siblings to ensure that general handling and storage conditions were not lethal.
Fig. 1.
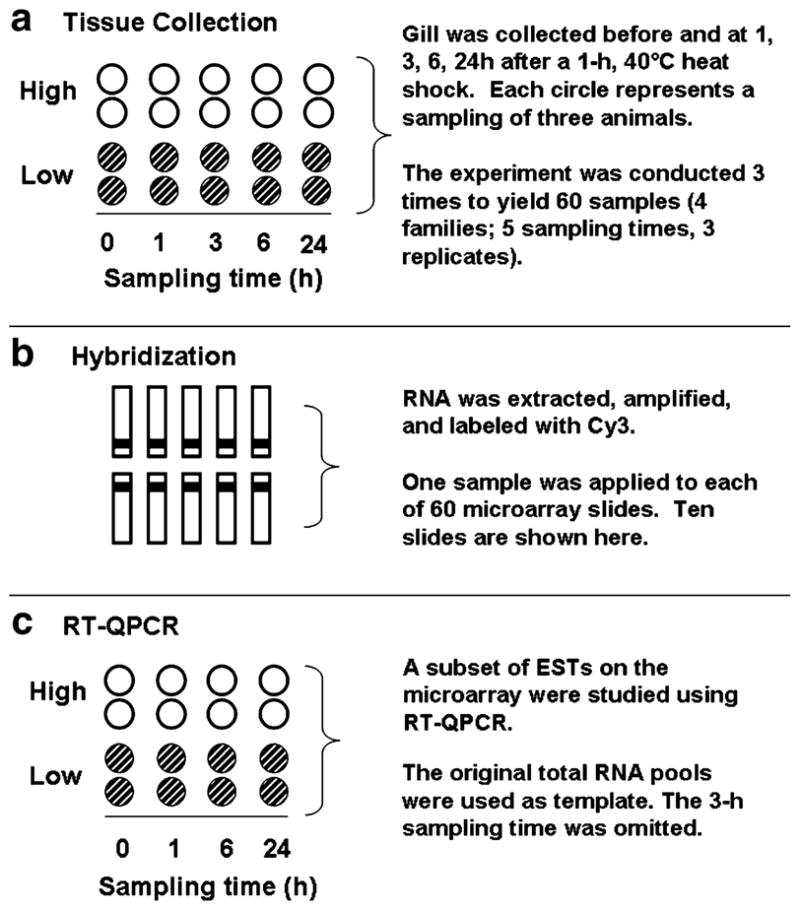
Summary of experimental design. a Gills from two oyster families characterized by high (white circles) or low survival (hatched circles) of a 1 h, 43°C heat shock were collected before and at 1, 3, 6, and 24 h after a non-lethal heat shock (40°C, 1 h) in an experiment that was repeated three times to yield 60 samples. Each circle represents a sample that contained gill of three oysters. b One sample of cy3-labeled antisense RNA was hybridized to a single microarray slide (eg, a single-color microarray experiment). Slides were hybridized in batches of ten, as shown. c A subset of ESTs was studied using real-time quantitative polymerase chain reaction (RT-QPCR). The original samples of total RNA were used as template. Samples collected after 3 h recovery of heat shock were omitted
We homogenized the gill tissue by placing the three pieces (representing three animals) in a homogenizer tube containing refrigerated buffer RLT (Qiagen, Valencia, CA, USA), and disrupted the pieces using a Teflon©-coated homogenizer fitted to a hand-held drill. Pooling individuals at this stage precluded estimating the variance among individuals within families, and thus reduced the power of statistical tests. However, this study was intended primarily to identify genes for further study in experiments designed for estimating variance in a larger number of families, potentially without a temporal component.
RNA preparation and amplification
We extracted total RNA from gill suspensions using Qiagen® RNeasy kits (Qiagen, Valencia, CA, USA), including the on-column DNAse treatment, according to the manufacturer’s instructions. We quantified total RNA using a SPECTRAmax PLUS spectrophotometer (Molecular Devices, Sunnyvale, CA, USA) and produced amplified amino allyl RNA (aRNA) for hybridization to microarrays using 1 μg of total oyster RNA as template for the Amino Allyl MessageAmp™ Kit (Ambion, Austin, TX, USA), according to the manufacturer’s instructions. In each amplification reaction, we included a mixture of RNA standards produced from plasmid DNA stocks of Karenia brevis genes according to Jenny et al. (2007). This mixture contained 100 ng chlorophyll A/B binding protein (Genbank #CO059871) RNA, 10 ng photosystem core protein (Genbank #CO062297) RNA, 1 ng flavodoxin (Genbank #CO065421) RNA, and 0.1 ng photolyase (Genbank #CO064781) RNA. Therefore, each reaction contained 1.1111 μg of template.
Microarray Hybridization
We used a single-dye approach, in which RNA pools representing one oyster family, sampling time, and biological replicate were prepared in 90-μl mixtures and hybridized to single arrays (Fig. 1b). Slides were hybridized over an 8-day period in batches of ten samples per run, and samples were randomly selected for inclusion in any run. Each 90-μl mixture contained 20 μg of Cy3-coupled aRNA added to a non-commercial hybridization buffer [50% formamide, 2.4% sodium dodecyl sulfate (SDS), 4× standard saline phosphate EDTA (SSPE), 2.5× Denhardt’s solution, plus 1 μg Cot-DNA and 1 μg polydATP; Jenny et al. 2007]. We boiled the mixtures for 1 min and incubated them for 1 h in a covered thermocycler heated to 50°C, during which time we prepared the microarrays for hybridization by soaking in 0.2% SDS solution for 2 min, boiling in water for 2 min, and drying by brief pulsed centrifugation. To reduce non-specific background, dried slides were pre-hybridized with a non-commercial blocking solution (1.6× Denhardt’s solution, 33.3% formamide, 1.6% SDS, 2.6× SSPE, and 0.1 μM salmon sperm DNA; Jenny et al. 2007) for 1 h at 50°C in a humidified hybridization oven (Boekel Inslide-Out™, Boekel, Festerville, PA, USA). After this preparation, we added the aRNA mixtures to the slides, covered them with coverslips (Lifterslips, Erie Scientific, Portsmouth, NH, USA) and stored them overnight (12 h) at 50°C in the hybridization oven. We then washed away unbound aRNAs by quickly dipping the slides in 2× saline sodium citrate (SSC), 0.1% SDS solution to remove cover-slips and soaking them for 15 min each in 0.2× SSC, 0.1% SDS, followed by 0.2× SSC, and 0.1× SSC. Slides were dried by pulsed centrifugation and scanned within 15 min.
Microarray Data Acquisition and Analysis
We used a ScanArray Express (Perkin Elmer, Boston, MA, USA) microarray scanner to acquire images of each slide using 70% PMT gain and 90% laser power. The QuantArray software package (Perkin Elmer, Boston, MA, USA) was used to acquire raw fluorescence data, background, and spot quality information from the scanned images using the included histogram spot segmentation method. Each spot image was visually inspected for overall quality, and damaged spots were excluded prior to statistical analysis. The hybridization data and the MIAME protocols are currently available at the Marine Genomics website (http://www.marinegenomics.org), and also at the Gene Expression Omnibus website (http://www.ncbi.nlm.nih.gov/projects/geo; Series GSE12070, Accn #’s GSM304764 through GSM304823).
We accounted for mean-versus-variance dependence of the raw intensity data by transforming the entire dataset using a robust generalized log transformation, variance stabilized normalization (vsn) method (Huber et al. 2002) as implemented in the Bioconductor suite for the R software environment (Ihaka and Gentleman 1996; Gentleman et al. 2004). All spots were included in the transformation. Only spots with an average vsn-adjusted signal of 6 for at least one of 5 times points were included in our analysis; this intensity cut-off corresponds to the average vsn-transformed signal generated by the K. brevis RNA gene for photolyase for all slides (average signal± SEM=5.8±0.03).
Spots were printed in duplicate (Jenny et al. 2007) and we averaged the two replicate spots after vsn-transformation but before statistical analysis. We used analysis of variance to compare the means of vsn-adjusted signals for all spots among sampling times, between low- and high-surviving family types, between individual families within family types, and to test for interactions between time and family type using the GLM procedure in SAS (SAS Institute Inc. 1999). The statistical model was: Signal = Time + Family Type + Family ID (Family Type) + Family Type × Time, where “Time” refers to sampling time, “Family Type” refers to low-surviving versus high-surviving families, “Family ID (Family Type)” refers to individual families within either family type, and “Family Type × Time” is the interaction of time and family type. To avoid issues with multiple hypothesis tests, we calculated Q-values for each EST using the software Q-value (Storey 2002; Storey and Tibshirani 2003). The use of Q-values is based on false discovery rate approaches that estimate the proportion of false positives among large numbers of tests (Benjamini and Hochberg 1995). We adopted a false discovery rate of 5% and effects were considered to be significant when Q<0.05.
The identities of the ESTs printed on the microarray were determined using the algorithms BLASTX and BLASTN which are available at the National Center for Biotechnology Information. An EST was considered to have significant database match when the expected value (E) was less than 1.E-05. We aligned ESTs with matching sequences using CLUSTALX, and excluded the shorter EST of any pairs with identical nucleotide sequences.
To visually summarize temporal transcription for the ESTs that differed among sampling times, we grouped them into clusters using the software SSClust (Ma et al. 2006). This program plots each cluster as a mean times series with 95% confidence intervals for ESTs with similar profiles, and should be regarded as a guide to overall change over time rather than an absolute representation of the transcription profile of each EST contained within that cluster. Data for each EST were converted to a common scale by dividing mean vsn-adjusted signal for each sampling time by the average of all values for that EST. Raw clustering output is included as supplementary material.
Real-Time Quantitative Polymerase Chain Reaction
We used real-time quantitative PCR to verify the microarray data of 15 ESTs for the 0, 1, 6, and 24-h sampling times (Fig. 1c). We assayed all ESTs that were significant for the terms Family Type and Family Type × Time, and for a subset of ESTs that were significantly different among sampling times, were considered to be relevant to heat shock based upon literature searches, and for which reliable primers could be designed (described below). Complementary DNA (cDNA) was reverse-transcribed from the original pools of total RNA using the ABI® High Capacity cDNA Synthesis kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions and using the included random primers. All RT-QPCR reactions were conducted using an ABI7500 quantitative PCR thermocycler (Applied Biosystems, Foster City, CA, USA), had concentrations of 50 nM forward and reverse primers, and each reaction contained 3 ng of cDNA, 12.5 μl of 2× SYBR Green master mix (Applied Biosystems, Foster City, CA, USA) and water resulting in a total reaction volume of 25 μl. We used the thermocycling protocol: (1) 50°C, 2 min, (2) 95°C, 10 min for transcription enzyme activation, and (3) 40 or 45 cycles of (a) 95°C for 15 seconds and (b) 58°C or 59°C for 1 min (Table 1).
Table 1.
Accession number, identity, forward primer sequence, reverse primer sequence, annealing temperature (Ta), and number of cycles (C) for quantitative RT-QPCR assays
| EST Putative identity |
Forward primer Reverse primer |
Ta | C | Efficiency |
|---|---|---|---|---|
| AB122066 | F: GGAAGCTGCTGAGATGGGAA | 1.98±0.004 | ||
| Elongation factor 1α | R: TCCAACACCCAGGCGTATTT | |||
| AJ305315 | F: AGGGCATTTCATGTCCGAAG | 58 | 40 | 2.01±0.002 |
| Heat shock protein 70 | R: TGCTCAGACATCCAAGGAAGG | |||
| AJ565456 | F: ACCACAAAAGCAGTTGCCGT | 58 | 40 | 2.08±0.001 |
| S-crystallin | R: TGAGGGATAAGGCGACCATC | |||
| AJ565627 | F: GTACTTCAATGAGATGCCGTGG | 59 | 40 | 1.84±0.153 |
| Nucleoredoxin | R: AGGTCACGTTCACTGAAGGGA | |||
| AJ565582 | F: CCTTCGGATCGTCTGGAGAA | 59 | 40 | 1.98±0.004 |
| Delta-9 desaturase | R: CCGAAGTGTAAAAGAGCCATCAG | |||
| AM237676 | F: GCCAAACCTCGCCTACCTTC | 58 | 40 | 2.00±0.004 |
| Peroxinectin | R: GTGGAGTTGACGCGTGACATA | |||
| AM237729 | F: GAGATCGTGGAGGACAAGAAAGA | 58 | 45 | 1.97±0.005 |
| No match | R: CTGCTCCGATCTCGTCAGC | |||
| AM237796 | F: GCTGTGGAGTGGTTACCAGGA | 59 | 40 | 2.01±0.004 |
| Galectin | R: GAGAGTAACCCAGCTCCCCG | |||
| BQ426456 | F: AGGCCATTGAAGCCACAAAT | 58 | 40 | 2.00±0.005 |
| Neuropeptide Y | R: TCCACAAAGGACTCTTGCCAT | |||
| BQ426550 | F: TGACGCAATGGATTTTCTGC | 58 | 40 | 1.86±0.005 |
| Heat shock protein 27 | R: GCCTGGTATCCAGGTCGGTAG | |||
| BQ426623 | F: GCGCGACATTTTCGATTTTC | 59 | 40 | 1.88±0.007 |
| No match | R: CCATTGGTGATGACGTAACGC | |||
| BQ426658 | F: AGAAGTGTGGAGAAATTTTGTCCAG | 59 | 40 | 1.92±0.005 |
| No match | R: TGGGATCAGATCCCATGCTC | |||
| BQ426884 | F: ACGATGTCAGGGACAACTTTCTGT | 58 | 45 | 1.96±0.002 |
| No match | R: CGCAGAGGTAATCTTTTAAACGTCA | |||
| BQ426927 | F: GACATGGTCGTCCAGATCGG | 59 | 40 | 2.02±0.005 |
| SOCS2 | R: CCACCCTGATTGACTTAACGTGA | |||
| CX069163 | F: TATGAAGTAGCAGCATGATGACTCC | 58 | 40 | 2.02±0.004 |
| Collagen | R: TCGTGGCTACCTAATGGCG | |||
| CX069133 | F: GTTCCTGCCACGACCTGAGT | 58 | 40 | 2.01±0.004 |
| Cystatin B | R: GCTGGACACTTACACCCCAGTC |
Mean efficiency (Er) ± SEM was calculated by using the software LinReg PCR (Ramakers et al. 2003). Target concentration was normalized to that of the endogenous control ef1 (AB122066)
Taris et al. (2008) found that RT-QPCR analysis of pooled oyster RNA samples can lead to misleading inferences for gene transcription among oyster families due to polymorphisms among individuals within such pools. Because each of the samples used in this work contained RNAs from three individual oysters, we designed RT-QPCR assays by testing multiple primer pairs for the four individual families by preparing pools of cDNA that each contained all replications of an individual family at a single sampling time (4 times×4 families=16 pools in total). Assays were deemed suitable for use when multiple primer pairs produced similar amplification profiles among the pools and also produced a single PCR product, as determined by examining post-reaction dissociation plots. Preference for individual primer pairs that met these criteria was based on high amplification efficiencies which were estimated as described below.
Raw RT-PCR data were acquired using the SDS 1.4 software (Applied Biosystems, Foster City, CA, USA) and expressed as the relative concentration of target to an internal endogenous control gene, elongation factor 1 (ef1; Genbank #AB122066). We used the formula , where Er is the reaction-specific amplification efficiency estimated using LinRegPCR software (Ramakers et al. 2003), cT is the cycle number at which amplification reached a detection threshold, target is the EST under consideration, and ef1 is the endogenous control gene. We chose ef1 because it was stably expressed in C. gigas exposed to a variety of conditions in other studies (Huvet et al. 2004; Taris et al. 2008). Additionally, analysis of variance revealed no significant difference in average cT of ef1 among sampling times (P=0.3114), between low- and high-surviving families (P=0.9209), and the interaction of family type and time (P=0.9399) in the samples used for this study.
We used analysis of variance to test for differences among RT-QPCR data using the same statistical model as used for the microarray data. Additionally, we performed linear contrasts using the CONTRAST statement within the GLM procedure of SAS. These contrasts tested for differences in mean relative transcript concentrations between adjacent sampling times and for differences in mean relative transcript concentrations between low- and high-surviving families at each time point. We did not include linear contrasts for the microarray data to avoid issues with multiple testing.
Results and Discussion
Microarray
In total, 1,675 averaged spot pairs for individual ESTs were included in our analysis after quality control, of which ~66% originated from C. gigas libraries and ~34% originated from C. virginica libraries (Table 2). This represented ~25% of the features included on the microarray. The mean vsn-adjusted signals of 110 ESTs (~7% of all ESTs included in the analysis) differed significantly among sampling times, of which the majority originated from C. gigas (Table 2). Based on the microarray data, mean vsn-adjusted signals of four ESTs differed significantly between family types, and one significant interaction of time and family type was detected (Table 2). We classified the 110 ESTs that differed among sampling times into ten functional categories based on searches of published literature (Table 3). Over half (54%) of the ESTs had no match in Genbank. The identities and accession numbers of all ESTs for which significant effects were detected are reported in Table 4. These identities are based upon database searches and represent current knowledge of the poorly characterized genomes of C. gigas and C. virginica. Many of the genes may belong to diverse protein families whose members have discrete, non-overlapping biological functions. We therefore stress that further study is needed to characterize the genes discussed here, that many of the identities are tentative, and that interpretations of the data may change as more genetic information becomes available.
Table 2.
Summary of results for microarray experiment: source of ESTs included in the analysis and the percentage that originated from either Crassostrea gigas and percentage of ESTs included in the analysis from either species that differed among sampling times; number and percentage of ESTs included in the analysis from either species that differed between family types or had significant interactions of time and family type
| Source | Included in analysis | Different among times | Different among family types | Interaction of time and family type |
|---|---|---|---|---|
| C. gigas | 1,102 (66%) | 90 (8.1%) | 4 (0.004%) | 1 (<0.001%) |
| C. virginica | 573 (34%) | 21 (3.6%) | 0 (0%) | 0 (0%) |
| Total | 1,675 (100%) | 111 (6.5%) | 4 (0.02%) | 1 (<0.001%) |
Table 3.
Number and percentage of ESTs that differed among sampling times from 10 functional categories, and the number and percentage of those that originated from C. gigas or C. virginica EST libraries
| Functional category | Number | No. from C. gigas | No. from C. virginica |
|---|---|---|---|
| Molecular chaperones | 10 (9.0%) | 8 (80%) | 2 (20%) |
| Protein degradation | 4 (3.6%) | 3 (75%) | 1 (25%) |
| Growth/reproduction | 3 (2.7%) | 3 (100%) | 0 (0%) |
| Antioxidant/detoxification | 2 (1.8%) | 2 (100%) | 0 (0%) |
| Lipid metabolism | 5 (4.5%) | 5 (100%) | 0 (0%) |
| Cellular immunity | 5 (4.5%) | 5 (100%) | 0 (0%) |
| Transcription/Translation | 5 (4.5%) | 1 (20%) | 4 (80%) |
| Cytoskeletal organization | 7 (6.3%) | 7 (100%) | 0 (0%) |
| Other processes | 10 (9.0%) | 8 (80%) | 2 (20%) |
| No match in Genbank | 59 (53.6%) | 47 (80%) | 12 (20%) |
| Total | 110 (100.0%) | 50 (81%) | 21 (19%) |
Table 4.
Accession number, cluster (“Cl”; refers to Fig. 1), Q-value, identity, species match, and accession number, and E-scores of ESTs for which vsn-adjusted signals in gill of oysters differed significantly (Q<0.05)
| Accession | Cl. | Q | Identity | Species | Accession | E |
|---|---|---|---|---|---|---|
| Among sampling times | ||||||
| Molecular chaperones | ||||||
| AJ305315 | E | 1.E-10 | Heat shock cognate 70 | Crassostrea gigas | AJ305315 | 0 |
| AJ565518 | E | 1.E-03 | EKN1 | Strongylocentrotus purpuratus | XP_787717 | 4.E-47 |
| AM237785 | A | 6.E-14 | Heat shock protein 27 | Carassius auratus | AAV97950 | 8.E-07 |
| BQ426550 | A | 1.E-04 | Heat shock protein 27 | Homo sapiens | AAA62175 | 8.E-07 |
| BQ426606 | E | 3.E-08 | Heat shock protein 90 | Chlamys farreri | AAR11781 | 6.E-112 |
| BQ426928 | E | 3.E-09 | Heat shock protein 70 | Crassostrea gigas | AF144646 | 1.E-114 |
| CB617443 | D | 1.E-09 | Heat shock protein 90 | Crassostrea gigas | ABS18268 | 2.E-110 |
| CB617445 | NA | 4.E-02 | Chaperonin subunit 6 | Nasonia vitripennis | XP_001606486 | 3.E-27 |
| CD648461 | NA | 1.E-05 | Small heat shock protein | Uncultured cnidarian | ABA42878 | 3.E-10 |
| CV089199 | D | 6.E-06 | Heat shock cognate 72 | Crassostrea gigas | AAD31042 | 6.E-133 |
| Protein degradation | ||||||
| BQ427148 | NA | 4.E-02 | Siah-interacting protein | Xenopus laevis | NP_000180214 | 8.E-18 |
| CB617402 | C | 2.E-06 | UBE 1 | Xenopus laevis | AAH47256 | 2.E-52 |
| CB617456 | C | 3.E-06 | Valosin-containing protein | Homo sapiens | AAH07562 | 4.E-84 |
| CV132727 | A | 2.E-02 | Ribosomal Protein L40a | Lycosa singoriensis | ABX75386 | 3.E-35 |
| Growth and reproduction | ||||||
| BQ426456 | NA | 2.E-03 | Neuropeptide Y | Haliotis discus hannai | ABH10673 | 7.E-12 |
| BQ426927 | E | 8.E-03 | SOCS2 | Apis mellifera | XP_394764 | 4.E-13 |
| CK172319 | C | 4.E-07 | Temptin | Aplysia brasiliana | AAS92605 | 6.E-27 |
| Antioxidant/detoxification | ||||||
| AJ565456 | B | 1.E-03 | S-crystallin | Strongylocentrotus purpuratus | XP_791441 | 9.E-33 |
| CX069146 | B | 3.E-03 | Glutathione peroxidase | Hymeniacidon perlevis | ABB91779 | 4.E-53 |
| Lipid metabolism | ||||||
| AJ565582 | NA | 8.E-14 | Delta-9-desaturase | Danio rerio | NP_942110 | 8.E-08 |
| BQ426641 | B | 9.E-03 | Patatin-like phospholipase | Strongylocentrotus purpuratus | XP_001185333 | 5.E-47 |
| BQ426935 | C | 6.E-03 | SREBP-1 | Gallus gallus | NP_989457 | 2.E-41 |
| CB617498 | C | 9.E-12 | Delta-9-desaturase | Ctenopharyngodon idella | CAB53008 | 4.E-69 |
| CK172312 | B | 4.E-03 | Fatty acid binding protein | Crassostrea gigas | AAT44355 | 5.E-08 |
| Cellular immunity | ||||||
| AJ565627 | E | 6.E-14 | Nucleoredoxin | Strongylocentrotus purpuratus | XP_781174 | 3.E-30 |
| AM237796 | C | 2.E-02 | Galectin 4 | Crassostrea gigas | CAD79473 | 4.E-88 |
| BQ427041 | B | 2.E-03 | 5-lipoxygenase | Strongylocentrotus purpuratus | XP_001198535 | 3.E-24 |
| CB617516 | B | 3.E-05 | Protein,COMM domain | Xenopus laevis | NP_001087091 | 8.E-44 |
| CX069133 | C | 5.E-08 | Cystatin B | Gallus gallus | XP_416492 | 3.E-18 |
| RNA transcription/protein translation | ||||||
| CD648403 | D | 2.E-08 | FUSE binding protein 1 | Xenopus tropicalis | NP_989293 | 6.E-08 |
| CD649011 | B | 5.E-02 | Y-box factor protein | Lymnaea stagnalis | AAT97092 | 6.E-31 |
| CV087081 | B | 6.E-03 | RPS27-1 | Crassostrea gigas | CAD91436 | 0 |
| CV087137 | A | 1.E-02 | 18S rRNA gene | Crassostrea gigas | AM182263 | 0 |
| CX069191 | NA | 3.E-02 | Ribosomal protein P1 | Biomphalaria glabrata | AAZ39530 | 4.E-24 |
| Cytoskeletal organization | ||||||
| AJ565479 | NA | 2.E-03 | Fascin homolog 2 | Xenopus tropicalis | NP_001093724 | 1.E-60 |
| BQ426318 | C | 5.E-02 | Ankyrin w/MYND | Bos taurus | NP_00168852 | 1.E-52 |
| BQ426459 | A | 6.E-03 | Ankyrin 2,3/unc44 | Strongylocentrotus purpuratus | XP_001179527 | 3.E-20 |
| BQ426555 | B | 5.E-02 | Arp2/3 16 kd subunit | Aedes aegypti | EAT39104 | 2.E-10 |
| BQ426750 | E | 9.E-04 | Transgelin | Bombyx mori | NP_001040372 | 7.E-46 |
| BQ426862 | E | 5.E-05 | Profilin | Entamoeba histolytica | CAA62418 | 5.E-08 |
| CB617442 | NA | 1.E-03 | Tubulin β5 | Crassostrea gigas | AAU93877 | 1.E-65 |
| Other processes | ||||||
| AM237712 | NA | 6.E-12 | Inhibitor of apoptosis | Ochlerotatus triseriatus | AAL46972 | 2.E-12 |
| BQ426894 | NA | 9.E-03 | Hypothet. dehydrogenase | Gallus gallus | XP_426310 | 2.E-16 |
| BQ426907 | C | 1.E-02 | Arginase | Equus caballus | XP_001503335 | 8.E-58 |
| BQ426918 | B | 3.E-02 | Aldehyde dehydrogenase | Mus musculis | NP_082546 | 3.E-42 |
| BQ426932 | B | 1.E-04 | Diaphorase 1 | Xenopus laevis | NP_001080477 | 2.E-84 |
| BQ427069 | NA | 2.E-03 | Alcohol dehydrogenase | Lysiphlebus testaceipes | AAY63990 | 6.E-104 |
| CD526814 | F | 8.E-15 | Senescence associated | Brugia malayi | EDP31077 | 2.E-55 |
| CD526816 | F | 6.E-14 | Tetraspanin 66E | Drosophila melanogaster | NP_523985 | 2.E-07 |
| CD647828 | NA | 3.E-02 | Bm1_17870 | Brugia malayi | EDP36122 | 6.E-22 |
| CF369127 | B | 1.E-02 | LOC496342 protein | Strongylocentrotus purpuratus | XP_792838 | 3.E-46 |
| AJ565488 | E | 4.E-02 | Not known | |||
| ESTs with no match in Genbank | ||||||
| AJ565533 | A | 1.E-06 | Not known | |||
| AJ565662 | E | 7.E-03 | Not known | |||
| AJ565673 | B | 1.E-03 | Not known | |||
| AJ565697 | A | 5.E-24 | HSP24 | Branchiostoma lan | CAE83570 | 8.E-02 |
| AJ565827 | NA | 2.E-20 | Not known | |||
| AJ565831 | A | 9.E-10 | Not known | |||
| AJ565846 | NA | 3.E-07 | Not known | |||
| AM237667 | A | 1.E-04 | Not known | |||
| AM237694 | NA | 2.E-09 | Not known | |||
| AM237730 | A | 5.E-10 | Not known | |||
| AM237767 | A | 5.E-14 | Not known | |||
| AM237771 | NA | 2.E-12 | Not known | |||
| BG624437 | B | 8.E-03 | Not known | |||
| BG624595 | A | 5.E-10 | Not known | |||
| BG624747 | A | 1.E-03 | Not known | |||
| BQ426330 | C | 4.E-02 | Not known | |||
| BQ426395 | NA | 3.E-04 | tRNA transferase | Nematostella vectensis | XP_001626290 | 4.E-04 |
| BQ426399 | NA | 2.E-05 | Not known | |||
| BQ426402 | C | 6.E-03 | Not known | |||
| BQ426405 | C | 7.E-04 | Not known | |||
| BQ426413 | A | 2.E-17 | Not known | |||
| BQ426454 | NA | 2.E-02 | Not known | |||
| BQ426620 | A | 1.E-07 | Not known | |||
| ESTs with no match in Genbank, continued | ||||||
| BQ426623 | B | 2.E-02 | Not known | |||
| BQ426654 | C | 9.E-08 | Not known | |||
| BQ426851 | NA | 2.E-14 | Transcription factor | Ciona intestinalis | BAE06318 | 5.E-04 |
| BQ426884 | NA | 5.E-09 | Not known | |||
| BQ426924 | NA | 9.E-04 | Not known | |||
| BQ426995 | NA | 1.E-06 | Not known | |||
| BQ427058 | NA | 3.E-18 | Not known | |||
| BQ427084 | NA | 1.E-02 | Not known | |||
| BQ427134 | NA | 2.E-03 | Not known | |||
| BQ427151 | A | 9.E-09 | Uroplakin 1 | Heliocidaris tuberculata | ABE27955 | 1.E-04 |
| BQ427169 | NA | 9.E-06 | Not known | |||
| CB617327 | NA | 5.E-03 | Not known | |||
| CB617408 | D | 7.E-08 | Not known | |||
| CB617424 | NA | 3.E-04 | Not known | |||
| CB617426 | D | 5.E-14 | Not known | |||
| CB617465 | C | 2.E-04 | Not known | |||
| CB617473 | NA | 1.E-05 | Not known | |||
| CB617482 | NA | 1.E-06 | Not known | |||
| CB617484 | NA | 4.E-04 | Not known | |||
| CB617540 | B | 1.E-04 | Not known | |||
| CD526789 | F | 9.E-09 | Not known | |||
| CD526790 | F | 4.E-08 | Not known | |||
| CD526794 | A | 3.E-04 | Not known | |||
| CD526795 | NA | 8.E-06 | Not known | |||
| CD526862 | F | 9.E-14 | Not known | |||
| CD526863 | F | 3.E-20 | Not known | |||
| CD526872 | F | 1.E-06 | Not known | |||
| CD648405 | E | 3.E-02 | Not known | |||
| CD648964 | NA | 4.E-02 | Not known | |||
| CK172325 | NA | 9.E-15 | Not known | |||
| CK172347 | NA | 1.E-02 | Not known | |||
| CK240435 | NA | 4.E-03 | Not known | |||
| CV132419 | A | 2.E-05 | Not known | |||
| CV132796 | NA | 8.E-03 | Not known | |||
| CV132891 | C | 5.E-03 | Not known | |||
| Between family types or the interaction of time and family type | ||||||
| ESTs with significant effect of family type | ||||||
| AM237676 | NA | 3.E-02 | Peroxinectin | Pacifastacus leniusculus | CAA62752 | 6.E-29 |
| AM237729 | NA | 3.E-02 | Not known | |||
| BQ426658 | NA | 3.E-03 | Not known | |||
| CX069163 | NA | 6.E-03 | Collagen-like protein | Suberites domuncula | CAC81019 | 7.E-09 |
| ESTs with significant interaction of time and family type | ||||||
| BQ426623 | NA | 2.E-02 | Not known | |||
Each EST has been grouped according to the functional categories listed in Table 3
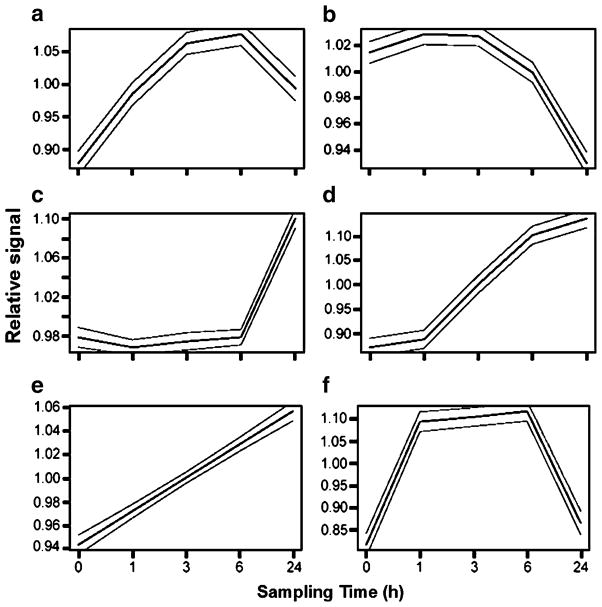
We grouped 73 of the 110 ESTs into six descriptive clusters (Fig. 2). The cluster designation for each EST is listed alongside its accession number in Table 4. Clusters a and f contained ESTs in which transcript concentration increased and then decreased within the 24-h sampling period. Clusters d and e contain ESTs in which transcription increased monotonically. Cluster c contains ESTs in which transcription remained low until after the 6-h sampling period and then increased sharply. Cluster b contains ESTs in which transcription was suppressed after heat shock. Not all ESTs could be unambiguously included in any particular cluster, and we identified these ESTs by visually inspecting the raw output and excluding ESTs whose expression profile was markedly dissimilar to any of the clusters.
Fig. 2.
Time-course clusters (95% confidence intervals) for ESTs in which transcription level differed among sampling times in gill of heat-shocked (40°C, 1 h) oysters before and at 1, 3, 6, and 24 h after heat shock. Data were clustered using the software SSClust (Ma et al. 2006). The vsn-transformed data for each sampling time are presented relative to the average for all sampling times for a given EST
RT-QPCR
We selected a subset of ESTs from six of the functional categories listed in Table 3 for verification using RT-QPCR based upon their potential relevance to heat shock and reports in other studies of summer mortality in bivalves. These data are reported in Figs. 3, 4, and 5; in each figure, microarray data are presented on the left and RT-QPCR data on the right. The six categories from which we selected ESTs included: molecular chaperones and co-chaperones (Fig. 3), growth and reproduction (Fig. 3), antioxidant and detoxification enzymes (Fig. 4), lipid metabolism (Fig. 4), cellular immunity (Figs. 4 and 5), and ESTs with no match in Genbank (Fig. 5). In general, patterns of change in transcript concentration over time were similar between the microarray and RT-QPCR methodologies (discussed below) and we therefore consider those microarray data to be reliable. The transcription profiles for ESTs discussed below that were not verified using RT-QPCR are available as supplementary material.
Fig. 3.
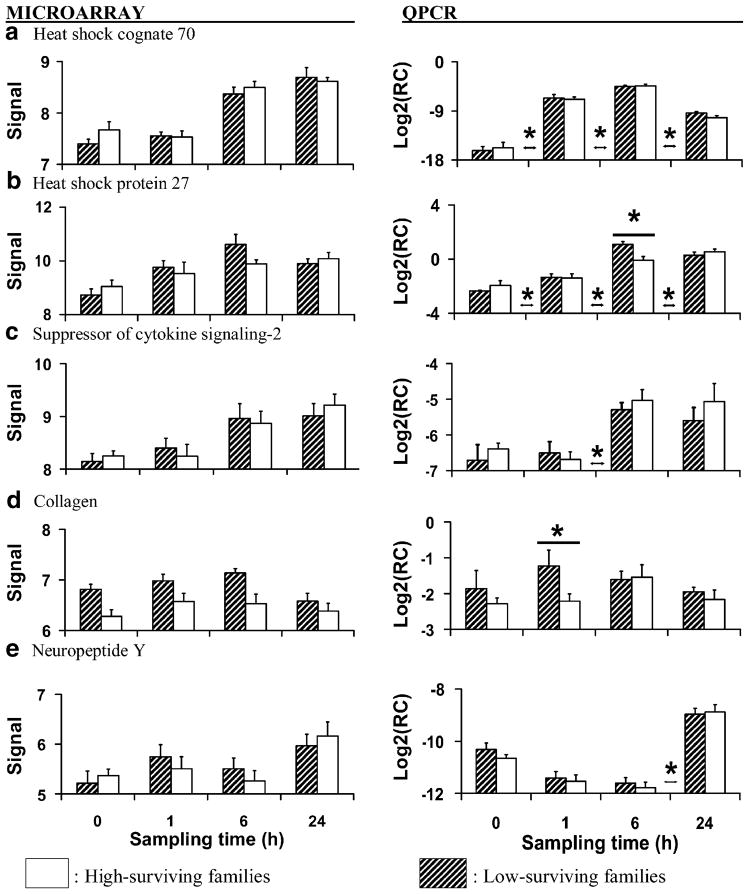
Average (±SEM) microarray-generated vsn-adjusted signal data and RT-QPCR-generated relative concentration data (target concentration relative to elongation factor 1; Log2RC) for heat shock protein genes and genes putatively related to growth and reproduction measured in gill before and at 1, 6, and 24 h after heat shock (40°C, 1 h). Each bar represents three replicates of two families (six samples total) with either low survival (hatched bars) or high survival (white bars) after heat shock. Asterisks above or between individual sampling times indicate significant (P<0.05) differences between family types at that time or between sampling times, respectively
Fig. 4.
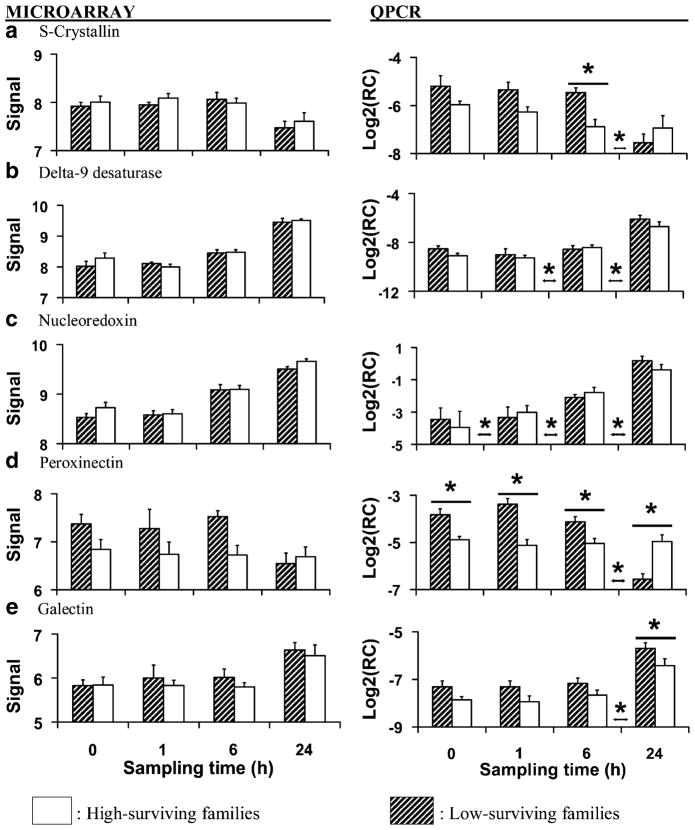
Average (±SEM) microarray-generated vsn-adjusted signal and RT-QPCR-generated relative concentration data (target concentration relative to elongation factor 1; Log2RC) for genes putatively encoding antioxidant enzymes, genes involved in lipid synthesis and genes involved in cellular immunity measured in gill before and at 1, 6, and 24 h after heat shock (40°C, 1 h). Each bar represents three replicates of two families (six samples total) with either low survival (hatched bars) or high survival (white bars) after heat shock. Asterisks above or between individual sampling times indicate significant (P<0.05) differences between family types at that time or between sampling times, respectively
Fig. 5.
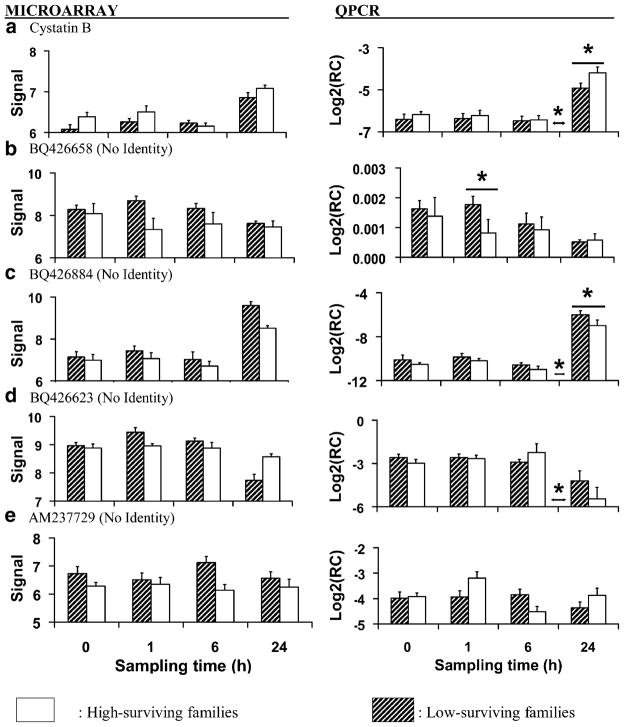
Average (±SEM) microarray-generated vsn-adjusted signal data and RT-QPCR-generated relative concentration data (target concentration relative to elongation factor 1; RC or Log2RC) for genes involved in cellular immunity measured in gill before and at 1, 6, and 24 h after heat shock (40°C, 1 h). Each bar represents three replicates of two families (six samples total) with either low survival (hatched bars) or high survival (white bars) after heat shock. Asterisks above or between individual sampling times indicate significant (P<0.05) differences between family types at that time or between sampling times, respectively
The outcomes of the statistical tests for both the microarray data and the RT-QPCR data are reported in Table 5. In contrast to the profiles of change over time, microarray data and RT-QPCR data were not always similar for ESTs whose concentrations differed between family types, and only agreed in general for collagen (Genbank # CX069163) and peroxinectin (Genbank #AM237676). The RT-QPCR data did not support the difference between family types for Genbank # AM237729 or the significant interaction (Genbank # BQ426623) indicated by the microarray data; these are discussed below in the section “ESTs with no match in Genbank.” In the RT-QPCR data only, we detected an overall effect of family type for mRNA concentrations of galectin and an EST with no match in Genbank (BQ426884), and a significant interaction between time and family type for heat shock protein 27. Linear contrasts revealed significant differences between the family types at individual sampling times for S-crystallin and cystatin B.
Table 5.
Statistical results for ESTs studied in gill of families with high (>64%) or low (<29%) survival (“FT”) after heat shock (43°C, 1 h) in which transcription was measured using a cDNA microarray and RT-QPCR
| EST | Microarray |
RT-QPCR |
||||
|---|---|---|---|---|---|---|
| Time | FT | Time × FT | Time | FT | Time × FT | |
| Heat shock cognate 72 | <0.0001 | 0.9935 | 0.9842 | <0.0001 | 0.7884 | 0.6863 |
| Heat shock protein 27 | <0.0001 | 0.9881 | 0.8032 | <0.0001 | 0.4360 | 0.0150 |
| SOCS2 | 0.0078 | 0.9877 | 0.9881 | <0.0001 | 0.3032 | 0.7171 |
| Collagen | 0.7152 | 0.0065 | 0.9823 | 0.1621 | 0.0417 | 0.2461 |
| Neuropeptide Y | 0.0023 | 0.9865 | 0.9619 | <0.0001 | 0.2264 | 0.4574 |
| S-Crystallin | 0.0013 | 0.8785 | 0.9865 | 0.0041 | 0.0610 | 0.1578 |
| Delta 9-desaturase | <0.0001 | 0.9596 | 0.9823 | <0.0001 | 0.1580 | 0.5841 |
| Nucleoredoxin | <0.0001 | 0.4558 | 0.9842 | <0.0001 | 0.4798 | 0.0709 |
| Peroxinectin | 0.5497 | 0.0254 | 0.7992 | 0.0228 | <0.0001 | <0.0001 |
| Galectin | 0.0207 | 0.8660 | 0.9894 | <0.0001 | 0.0021 | 0.9720 |
| Cystatin B | <0.0001 | 0.4962 | 0.8309 | <0.0001 | 0.0661 | 0.3960 |
| BQ426658 | 0.4715 | 0.0027 | 0.5672 | 0.0115 | 0.1054 | 0.3353 |
| BQ426884 | <0.0001 | 0.1474 | 0.9164 | <0.0001 | 0.0041 | 0.5580 |
| BQ426623 | <0.0001 | 0.9557 | 0.0171 | <0.0001 | 0.4356 | 0.2682 |
| AM237729 | 0.9842 | 0.0329 | 0.8127 | 0.1843 | 0.4669 | 0.1160 |
Differences were considered significant at Q<0.05 or P<0.05
SOCS2 suppressor of cytokine signaling 2
At least two phenomena may have contributed to the discordance between the microarray data and the RT-QPCR data. First, the microarray contained targets derived from cDNA libraries and these targets ranged from ~100 bp to several kilobases in length (Jenny et al. 2007). Therefore, it is possible that mRNA of multiple genes bound to conserved regions contained within these targets, as discussed below for both heat shock protein 70 and S-crystallin. In contrast, RT-QPCR assays designed to further explore expression of those targets amplified, to the best of our knowledge, only single gene products.
Second, sequence polymorphisms in C. gigas coding regions are thought to occur on average every 60 base pairs (Sauvage et al. 2007). Family-specific sequence mismatches could have influenced transcript binding to microarray probes, which would reduce the correspondence between microarray signal and RT-QPCR data. Studies that employ RT-QPCR to compare gene transcription among C. gigas individuals and families are known to be influenced by polymorphisms among individuals that can lead to misleading inferences (Taris et al. 2008). We tested each EST using multiple primers to increase our confidence that inferences were not influenced by polymorphisms of individuals within pools, but we cannot completely exclude the possibility of bias due to RT-QPR artifacts and caution should still be exercised in their interpretation.
Molecular Chaperones
Based on the microarray data, concentrations of mRNAs encoding heat shock protein 70, heat shock protein 90, and small stress proteins increased progressively following heat shock in both low-and high-surviving families (microarray data only; Table 4; Fig. 2a, d, e). This is not surprising because rapid synthesis of heat shock proteins is a nearly universal response to heat shock and other stressors (Lindquist 1986; Hochacka and Somero 2002). Heat shock proteins refold denatured proteins and fold newly synthesized proteins into functional conformations (Parsell and Lindquist 1993; Zhao and Houry 2007) and small stress proteins participate in protein folding, block apoptosis and protein translation during heat shock, and bind lipid membranes to ensure structural integrity (Bruey et al. 2000; Cuesta et al. 2000; Wang and Spector 2000; Tsvetkova et al. 2002; Doerwald et al. 2006). The presence of increased levels of heat shock proteins after thermal stress confers the ability to withstand subsequent exposure to otherwise lethal temperatures in marine bivalves and other animals and is an important aspect of bivalve adaptation to their environment (Clegg et al. 1998; Hamdoun et al. 2003).
We verified the mRNA concentrations of heat shock cognate 70 (Genbank #AJ305315) and small heat shock protein 27 (Genbank #BQ426550) using RT-QPCR (microarray and RT-QPCR data; Table 5; Fig. 3a, b). The microarray probe for the heat shock cognate 70 gene (Boutet et al. 2003) is a full-length mRNA sequence that contains six exons and encodes a highly conserved protein with homology to several other heat shock proteins. Therefore, the microarray signal for this particular probe may be a measure of heat shock protein 70 family gene transcription rather than that of an individual gene. We designed primers to amplify only the single gene product of heat shock cognate 70 and found that its relative mRNA concentration increased after heat shock between the pre-stress and 1-h sampling times and then decreased after 6 h, whereas transcription of heat shock protein 70 family genes appeared to increase between the 1-h and 6-h sampling times and remained elevated thereafter (Fig. 3a). Although we detected no family-level differences in the transcription of this heat shock protein gene, heat shock protein 70 protein concentrations in gill were reported to be greater in C. gigas families susceptible to summer mortality than in resistant families both before and during experimental hypoxia (Samain et al. 2007).
Based on the RT-QPCR data, there was a significant interaction between time and family type for the relative concentration of heat shock protein 27 (Table 5). The significant interaction resulted from the significantly greater concentration of transcript in low-surviving families at 6 h after heat shock (Fig. 3b). Taken together with the findings of Samain et al. (2007), it is possible that a correlate of high sensitivity to stress is an exaggerated requirement for heat shock protein (both transcription and translation) to repair damage caused by heat shock and environmental stress, and that this may also impose considerable metabolic costs. Li et al. (2007) found that heat shock and heat shock protein 70 production reduced tissue energy levels (adenylate energy charge and mantle glycogen levels) in reproductively mature C. gigas, and that the combination of reproductive activity and mounting a heat shock response led to metabolic exhaustion. Therefore, we speculate that high production of heat shock protein gene transcripts and proteins could be deleterious under certain conditions due to trade-offs between the need to salvage damaged proteins and the energetic costs this imposes on other biological functions.
Growth and Reproduction
The relative mRNA concentration of suppressor of cytokine signaling-2 (Genbank #BQ426927; microarray and RT-QPCR data; Table 5; Fig. 3c) increased between 1 and 6 h after heat shock and remained elevated thereafter. Suppressor of cytokine signaling proteins disrupt signaling cascades by targeting cytokines for proteasomal degradation (Alexander 2002; Yoshimura et al. 2007). In mammals, suppressor of cytokine signaling-2 negatively regulates growth by inhibiting signal transduction by insulin-like growth factor-1 and growth hormone (Greenhalgh and Alexander 2004; Rincón et al. 2007). In C. gigas, insulin-related receptors have been identified, and recombinant human insulin-like growth factor-1 has been shown to stimulate tissue growth (Griscourt et al. 2003). Thus, it is possible that suppressor of cytokine signaling-2 expression after heat shock resulted in suppressed growth which would be advantageous for reallocating energy to meet the needs of mounting a stress response.
The mRNA concentration of collagen (Genbank #CX069163; microarray and RT-QPCR data; Table 5; Fig. 3d) was greater overall in low-surviving than in high-surviving families based on both the microarray data and RT-QPCR data, but in the RT-QPCR dataset the significant family effect may have resulted mainly from the significantly greater concentration in low-surviving families at 1 h after heat shock. Collagen is an abundant protein that is part of the extracellular matrix that is deposited or broken down as part of the process of tissue growth and repair (Woessner 1998; Ziegler et al. 2002; Montagnani et al. 2005). We speculate that its increased transcription may reflect differences in tissue growth and remodeling between the family types. Reduced growth in the high-surviving families could have consequences for stress tolerance because more energy would be available to fuel stress responses.
The mRNA concentrations of temptin (Genbank #CK172319; microarray data only; Table 4; Fig. 2c) and of neuropeptide Y (Genbank #BQ426456; microarray and RT-QPCR data; Table 5; Fig. 3e) increased after 6 h. Exposure to high temperature is a common method of inducing spawning in oysters in hatcheries (e.g. Tibile and Singh 2003), and the transcription data for temptin and neuropeptide Y suggest that spawning could be influenced by simultaneous positive and negative regulation. Temptin is a component of the protein pheromone complex used by the mollusc Aplysia to attract mates and stimulate spawning behavior (Cummins et al. 2004) and may play a role in promoting C. gigas spawning. Neuropeptide Y secretion is associated with increased hunger and decreased sexual activity in mammals (Kalra and Kalra 2004).
Receptors for neuropeptide Y have been identified in molluscs (Tensen et al. 1998) and its expression could suppress spawning, which would be advantageous under certain conditions. For example, in France oyster families that are susceptible to summer mortality invest more energy into gametogenesis and spawn repeatedly, whereas resistant families invest less energy into gametogenesis and fully spawn once per reproductive season (Samain et al. 2007). Tissue energy reserves in oysters are lowest during summer months (Berthelin et al. 2000) and the combination of reproductive activity and low energy reserves can lead to metabolic exhaustion and death by opportunistic infection (Li et al. 2007). Although we detected no differences at the family level in gills of heat-shocked oysters, others have suggested that neuropeptide Y could be responsible for differing patterns of spawning in summer mortality susceptible and resistant families and may be of interest for future research (Samain et al. 2007).
Antioxidant and Detoxification Enzymes
The concentration of glutathione peroxidase (Genbank #CX069146; micro-array data only; Table 4, Fig. 2b) mRNA decreased after heat shock. Heat shock causes increased production of reactive oxygen radicals that form long-lived toxic byproducts that damage cells and membranes (Flanagan et al. 1998; Arnaud et al. 2002; Bruskov et al. 2002). Antioxidant enzymes such as superoxide dismutase, catalase, and glutathione peroxidase reduce oxidative damage by detoxifying free oxygen radicals and their byproducts (Foreman and Fisher 1981; Storey 1996; Young and Woodside 2001). Although we detected no family-level differences in the microarray data, suppressed expression of glutathione peroxidase in gill after heat shock could be a potentially deleterious consequence of heat shock.
The concentration of S-crystallin (Genbank #AJ565456; microarray and RT-QPCR data; Table 5; Fig. 4a) mRNA decreased after 6 h, and based on the RT-QPCR data was greater in low-surviving families at 6 h after stress. The protein S-crystallin is derived from glutathione S-transferase and is a major protein of vertebrate and invertebrate eye lenses that appears to lack enzymatic function (Chiou et al. 1995; Tomarev and Piatigorsky 1996; Chuang et al. 1999; Blanchette et al. 2007). As with most of the probes included on the microarray, it is possible that the probe for this S-crystallin EST hybridized with mRNA from multiple genes including glutathione S-transferase, leading to the discrepancy between the microarray data and the RT-PCR data. We have included the putative S-crystallin under the heading of “detoxification” because of its similarity to glutatione S-transferase, although as is the case for each of the ESTs discussed in this report, further characterization is needed to determine whether it truly encodes S-crystallin, its function, and the significance of enhanced transcription in low-surviving families.
Lipid Metabolism
The transcription of delta-9-desaturase (Genbank #AJ565582; microarray and RT-QPCR data; Table 5; Fig. 4b) and of sterol regulatory element binding protein transcription factor 1 (Genbank #BQ426935; microarray data only; Table 4; Fig. 2c) increased after the 6-h sampling time. Heat shock damages lipid membranes through changes to the physical state of the membrane and through oxidative stress (Storey 1996; Hochacka and Somero 2002). Delta 9-desaturase is required for synthesis of unsaturated fatty acids and sterol regulatory element binding protein transcription factor 1 activates genes involved in cholesterol synthesis (Ntambi 1999; Martin et al. 2006; Bengoechea-Alsono and Ericcson 2007). Therefore, the increased transcription of these genes could reflect repair and synthesis of lipid membranes that were damaged by heat shock. We verified the concentrations of delta-9-desaturase using RT-QPCR because in C. gigas its transcription is altered by exposure to chronic hypoxia (David et al. 2005), chronic elevated temperature (25°C, 3 d; Meistertzheim et al. 2007), and hydrocarbon exposure (Boutet et al. 2004), and because polymorphisms in this gene have been linked with summer mortality syndrome (David et al. 2007). However, we detected no family-level differences in its expression in heat-shocked gill.
The transcription of patatin-like phospholipase (#BQ426641; microarray data only; Table 4) and of fatty acid binding protein (Genbank #CK172312; microarray data only; Table 4) decreased after heat shock. Patatin-like phospholipase metabolizes fatty tissue in mammals (Schweiger et al. 2006), and fatty acid binding protein binds and transport lipids (Storch and Thumser 2000). These data suggest that mobilization of lipid reserves was suppressed by heat shock, which would be deleterious to mounting stress responses and gametogenesis during seasonal periods when tissue reserves of glycogen are low (Berthelin et al. 2000). Disruption of lipid deposition into gonads during spring is thought to contribute to summer mortality in C. gigas reared in Marennes–Oléron Bay, France (Soletchnik et al. 2006), and C. gigas families that were susceptible to summer mortality syndrome had lower transcription levels of fatty acid binding protein than resistant families following injection with pathogenic Vibrios (Huvet et al. 2004).
Immune and Inflammatory Responses
The relative concentration of nucleoredoxin (Genbank #AJ565627; microarray and RT-QPCR data; Table 5; Fig. 4c) mRNA increased after each of the sampling times. This suggests a rapid and intense inflammatory response to heat shock. Nucleoredoxin resides in the nucleus and participates in the regulation of transcription factors including the rapid-acting nuclear factor κB (NF-κB) that enables cells to quickly react to harmful stimuli and to mount immune responses (Hirota et al. 2000).
Based on the RT-QPCR data, there was a significant interaction between time and family type for the relative mRNA concentration of peroxinectin (Genbank #AM237676; microarray and RT-QPCR data; Table 5; Fig. 4d). In the low-surviving families, relative mRNA concentration was greater before and at 1 and 6 h after heat shock and then decreased after 6 h, whereas transcript concentration did not change in the high-surviving families (Fig. 4d). Peroxinectin is a cell-adhesion protein with peroxidase activity identified in crustaceans including freshwater crayfish Pacifastacus leniusculus (Johansson et al. 1995), white shrimp Litopenaeus vannamei (Liu et al. 2005), and prawn Machrobrachium rosenbergii (Liu et al. 2007). It is synthesized and stored in hemocytes and when released mediates hemocyte adhesion to foreign particles (Liu et al. 2007). Peroxinectin has not been previously studied in C. gigas but it may perform adhesive and defensive functions in oysters. The potential significance of the pattern of peroxinectin transcription in low-surviving families is discussed below.
The relative mRNA concentration of another cell adhesion protein, galectin (Genbank #AM237796; microarray and RT-QPCR data; Table 5, Fig. 4e), increased after 6 h and based on the RT-QPCR data was greater overall in low- than in high-surviving families. In vertebrates, galectins facilitate cell adhesion and participate in various aspects of cellular immunity (Perillo et al. 1998; Rabinovich et al. 2002; Levroney et al. 2005). In oysters, galectins serve a defensive and digestive role by binding to pathogens and algae and promoting phagocytosis of these particles by macrophages (Tasumi and Vasta 2007; Yamaura et al. 2008).
Oysters are constantly exposed to bacteria and pathogens as they filter-feed. One interpretation of higher transcription of adhesive proteins in stress-sensitive oyster families is that they are more susceptible to opportunistic infection before and after stress, and that this susceptibility is due in part to decreased ability of hemocytes to bind to and phagocytose bacteria; greater transcription of adhesive proteins compensate partially for lower levels of innate immunity. Hemocyte adhesion in marine bivalves is reduced by some bacteria including Vibrio aestuarianus and Vibrio tapetis (Choquet et al. 2003; Labreuche et al. 2006), and substrate adhesion by hemocytes from summer mortality-susceptible oysters reared at the Rivière d’ Auray, France, was compromised by Vibrio sp. strain S322 to a greater extent than in resistant families during summer months (Lambert et al. 2007). However, substrate adhesion was greater in hemocytes of the same susceptible families than in resistant families before the seasonal period of mortality (Samain et al. 2007), and therefore the linkage between expression of adhesion proteins and family-specific resistance to infection requires further clarification. We did not measure bacterial loads of dead and surviving oysters in this study and therefore do not know if infections lead to mortality. However, Samain et al. (2007) found that C. gigas families that were susceptible to summer mortality syndrome had higher quantities of Vibrio aestuarianus after heat shock than did resistant families, and it is reasonable to speculate that opportunistic infection contributed to morality in the low-surviving families used in the present study.
The relative concentration of cystatin B (Genbank #CX069133; microarray and RT-QPCR data; Table 5, Fig. 5a) mRNA increased after 6 h and based upon the RT-QPCR data was greater in high-surviving than in low-surviving families at 24 h after heat shock (Fig. 5a). Cystatins bind to and inhibit proteolytic cathepsins that are secreted by lysosomes as part of routine protein turnover and by pathogens as a means of acquiring nutrition from host cells (Ulrich 1995; Rinne et al. 2002; Vergote et al. 2005).
Protein turnover (the net result of synthesis and degradation) accounts for a large portion of maintenance metabolism energy expenditure in marine bivalves (Hawkins 1991; Hawkins and Day 1996) and lower rates of protein turnover in mussels Mytilus edulis afforded energy savings that consequently lead to enhanced survival under stressful conditions (Hawkins et al. 1987, 1989). One potential advantage of higher levels of cystatin B in the high-surviving oyster families could have been reduced energy loss due to protein turnover resulting in more energy for mounting stress responses.
Pathogens of oysters secrete proteases that damage host cells and protease inhibitors such as cystatin B may protect against these offensive processes during infection (Romestead et al. 2002; Xue et al. 2006) or may regulate host cathepsin activity. Protease inhibition in C. virginica families bred to resist Perkinsus marinus infection was negatively correlated with parasite loads (Oliver et al. 2000). Cystatin B has been explicitly associated with immune defense of arthropods (Kanost 1999), snails (Guillou et al. 2007), and marine invertebrates (Guegan et al. 2003; Kang et al. 2006). Snail Biomphalaria glabrata strains that were resistant to infection by Echinostoma caproni had higher induced levels of a type-2 cystatin protein after infection that is thought to inhibit the parasite-derived cathepsins used to digest host cells (Vergote et al. 2005). Therefore, in addition to retarding protein turnover, higher levels of cystatin B in the oysters studied here could have augmented cellular defense following heat shock.
ESTs without Identity
In this study, transcript concentrations of dozens of unidentified ESTs were altered by heat shock, and some appeared to differ between family types. Transcription of Genbank EST #BQ426658 was greater overall in low-surviving families in the microarray dataset, but RT-QPCR data indicated a significant difference between low- and high-surviving families only at 1 h after stress (microarray and RT-QPCR data; Table 5; Fig. 5b). Another unidentified EST (Genbank #BQ426884) was strongly upregulated after 6 h, and was more abundant in low-surviving families than in high-surviving families at this time (microarray and RT-QPCR data; Table 5; Fig. 5c). The significant differences in the microarray dataset for the interaction of time and family type observed for Genbank #BQ426623 and between family types for Genbank #AM237729 were not supported by RT-QPCR (microarray and RT-QPCR data; Table 5; Fig. 5d, e). The field of genomics in marine bivalves is still in its infancy, and the results obtained from each of these unidentified ESTs are difficult to interpret. Recently, Tanguy et al. (2008) sequenced over 10,000 ESTs and found that no more than 27% could be assigned to a functional category. As our knowledge of genes and the functions of their products expands in C. gigas and other molluscs, we expect that this dataset will continue to yield insights into the pathways that are perturbed by heat shock.
Conclusions
The response of oyster gills to heat shock included immediately increased transcription of genes that encode heat shock proteins, and increased transcription after 6 h of genes whose products synthesize lipids, participate in cellular immunity, and influence reproductive activity. Potentially deleterious consequences of heat shock included suppressed mobilization of stored lipid reserves and decreased transcription (relative to pre-stress levels) of antioxidant genes. Overall, a clear picture of gene expression over time was presented by the data, but differences between the high- and low-surviving families require further validation because of some discordance between the microarray and RT-QPCR data. The tentative identification of the ESTs requires further characterization before definitive statements on biological functions can be made. Furthermore, because only two high- and low-surviving families were studied here, we cannot exclude the possibility that some of the correlations between gene expression and family type were fortuitous, and future studies should evaluate these genes in a larger group of families.
With the exception of one EST that appears to encode cystatin B, transcript concentrations of ESTs that differed between family types were greater in low-surviving families than in high-surviving families. It seems counterintuitive that low-surviving families expressed potentially stress-relevant genes at higher levels than did high-surviving families. It is possible that low-surviving families required higher transcript levels to supply proteins (e.g. heat shock proteins and lectins) that mitigate cellular damage and opportunistic infection. However, the increased production of both transcripts and proteins could come at an energetic cost above that of maintenance metabolism, and the resulting increase in protein turnover could have lead to metabolic exhaustion (Hawkins et al. 1987, 1989; Li et al. 2007). A complex interaction of cell damage, opportunistic infection, and exhaustion is a plausible explanation for mortality at 43°C when considering that tissue energy stores are low when oysters are in reproductive condition (Berthelin et al. 2000) and lipid mobilization may be suppressed by heat shock.
We identified a number of ESTs whose transcription differed between oyster families with high or low survival after heat shock. With further characterization, these ESTs could provide the basis for broodstock selection for increased thermal tolerance and potentially also for disease resistance.
Acknowledgments
Funding for this research was provided by the Oyster Disease Research Program, NOAA agreement no. NA16RG1039, Sea Grant Project SAQ-08-NSI. Additional funding was provided by the Lylian Brucefield Scholarship and the Mamie Markham Award. We thank A. Barton, F. Evans, D. Mosher, and D. Stick for rearing the oyster families used during this experiment and for assistance during the heat-shocking experiments. We thank Oregon Oyster Farms, Newport, Oregon for the generous use of their facilities. We thank R. Chapman, Y. A. Chen, M. Cook, P. Cupit, P. Gross, M. Lundqvist, D. MacKillan, A. Mancia, N. Taris, H. Trent, and G. Warr for technical assistance and support during the hybridization of the microarrays and during data analysis. We thank W. Huber and W. Zhong for their advice during data analysis using vsn and SSClust, respectively. We thank A. Huvet, N. Taris, and two anonymous reviewers for thoughtful review of this manuscript. Any use of trade, firm, or corporation names in this publication is for the information and convenience of the reader. Such use does not constitute an official endorsement or approval by the United States Department of Agriculture or the Agricultural Research Service of any product or service to the exclusion of others that may be suitable.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10126-009-9181-6) contains supplementary material, which is available to authorized users.
Contributor Information
R. Paul Lang, Email: rlang@agcenter.lsu.edu, Department of Fisheries and Wildlife, Coastal Oregon Marine Experiment Station, Hatfield Marine Science Center, Oregon State University, Newport, OR 97365, USA. Department of Entomology, Louisiana State University Agricultural Center, 404 Life Sciences Building, Baton Rouge, LA 70803, USA.
Christopher J. Bayne, Department of Zoology, Oregon State University, Corvallis, OR 97331, USA
Mark D. Camara, United States Department of Agriculture, Agricultural Research Service, Hatfield Marine Science Center, Newport, OR 97365, USA
Charles Cunningham, Department of Biology, University of New Mexico, Albuquerque, NM 87131, USA.
Matthew J. Jenny, Department of Biology, Woods Hole Oceanographic Institution, Woods Hole, MA 02543, USA
Christopher J. Langdon, Department of Fisheries and Wildlife, Coastal Oregon Marine Experiment Station, Hatfield Marine Science Center, Oregon State University, Newport, OR 97365, USA
References
- Abele D, Heise K, Pörtner HO, Puntarulo S. Temperature-dependence of mitochondrial function and production of reactive oxygen species in the intertidal mud clam Mya arenaria. J Exp Biol. 2002;205:1831–1841. doi: 10.1242/jeb.205.13.1831. [DOI] [PubMed] [Google Scholar]
- Alexander WS. Suppressors of cytokine signaling (SOCS) in the immune system. Nat Rev Immunol. 2002;6:410–416. doi: 10.1038/nri818. [DOI] [PubMed] [Google Scholar]
- Arnaud C, Joyeux M, Garrel C, Godin-Ribout D, Demenge P, Ribuot C. Free-radical production triggered by hyperthermia contributes to heat stress-induced cardioprotection in isolated rat hearts. Brit J Pharm. 2002;135:1776–1782. doi: 10.1038/sj.bjp.0704619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57:289–300. [Google Scholar]
- Bengoechea-Alsono M, Ericcson J. SREBP in signal transduction: cholesterol metabolism and beyond. Curr Op Cell Biol. 2007;19:215–222. doi: 10.1016/j.ceb.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Berthelin C, Kellner K, Mathieu M. Storage metabolism in the Pacific oyster (Crassostrea gigas) in relationship to summer mortalities and reproductive cycle (West Coast of France) Comp Bio Phys B. 2000;125:359–369. doi: 10.1016/s0305-0491(99)00187-x. [DOI] [PubMed] [Google Scholar]
- Blanchette B, Feng X, Singh BR. Marine glutathione S-transferases. Mar Biotech. 2007;9:513–542. doi: 10.1007/s10126-007-9034-0. [DOI] [PubMed] [Google Scholar]
- Boutet I, Tanguy A, Rousseau S, Auffret M, Moraga D. Molecular identification and transcription of heat shock cognate 70 (hsc70) and heat shock protein 70 (hsp70) genes in the Pacific oyster Crassostrea gigas. Cell Stress Chap. 2003;8:76–85. doi: 10.1379/1466-1268(2003)8<76:miaeoh>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutet I, Tanguy A, Moraga D. Response of the Pacific oyster Crassostrea gigas to hydrocarbon contamination under experimental conditions. Gene. 2004;329:147–157. doi: 10.1016/j.gene.2003.12.027. [DOI] [PubMed] [Google Scholar]
- Bruey J, Ducasse C, Bonniaud P, Ravagnan L, Susin S, Diaz-Latoud C, Gurbuxani S, Arrigo AP, Kroemer G, Solary E, Garrido C. Hsp27 negatively regulates cell death by interacting with cytochrome c. Nat Cell Biol. 2000;2:645–652. doi: 10.1038/35023595. [DOI] [PubMed] [Google Scholar]
- Bruskov V, Malakhova L, Masalimov Z, Chernikov A. Heat-induced formation of reactive oxygen species and 8-oxoguanine, a biomarker of damage to DNA. Nuc Ac Res. 2002;30:1354–1363. doi: 10.1093/nar/30.6.1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Yang H, Delaporte M, Zhao S. Immune condition of Chlamys farreri in response to acute temperature challenge. Aquaculture. 2007;271:479–487. [Google Scholar]
- Cheney D, MacDonald B, Elston R. Summer mortality of Pacific oysters, Crassostrea gigas (Thunberg): Initial findings on multiple environmental stressors in Puget Sound, Washington, 1998. J Shellfish Res. 2000;19:353–359. [Google Scholar]
- Chiou SH, Yu CW, Lin CW, Pan FM, Lu SF, Lee HJ, Change GG. Octopus S-crystallins with endogenous glutathione S-transferase: cDNA sequence determination, transcription and comparison of structure and kinetic mechanism with authentic enzymes. Biochem J. 1995;309:793–800. doi: 10.1042/bj3090793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet G, Soudant P, Lambert C, Nicolas J-L, Paillard C. Reduction of adhesion properties of Ruditapes philippinarum hemocytes exposed to Vibrio tapetis. Dis Aquat Org. 2003;57:109–116. doi: 10.3354/dao057109. [DOI] [PubMed] [Google Scholar]
- Chuang C-C, Wu S-H, Chiou S-H, Chang G-G. Homology modeling of cephalopod lens S-crystallin: a natural mutant of sigma-class glutathione transferase with diminished endogenous activity. Biophys J. 1999;76:679–690. doi: 10.1016/S0006-3495(99)77235-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegg JS, Uhlinger K, Jackson S, Cherr G, Rifkin E, Friedman C. Induced termotolerance and the heat-shock protein 70 family in the Pacific oyster Crassostrea gigas. Mol Mar Biol Biotechnol. 1998;7:21–30. [Google Scholar]
- Cnaani A. Genetic perspective on stress response and disease resistance in aquaculture. Israeli J Aquaculture. 2006;58:375–383. [Google Scholar]
- Cuesta R, Laroia G, Schneider R. Chaperone Hsp27 inhibits translation during heat shock by binding eIF4G and facilitating dissociation of cap-initiation complexes. Genes Dev. 2000;14:1460–1470. [PMC free article] [PubMed] [Google Scholar]
- Cummins S, Nichols A, Amare A, Hummons A, Sweedler J, Nagle G. Characterization of Aplysia rtifact and temptin, two novel water-borne protein pheromones that act in concert with attractin to stimulate mate attraction. J Biol Chem. 2004;279:25614–25622. doi: 10.1074/jbc.M313585200. [DOI] [PubMed] [Google Scholar]
- David E, Tanguy A, Pichavant K, Moraga D. Response of the Pacific oysterCrassostrea gigas to hypoxia exposure under experimental conditions. FEBS J. 2005;272:5635–5652. doi: 10.1111/j.1742-4658.2005.04960.x. [DOI] [PubMed] [Google Scholar]
- David E, Boudry P, Degremont L, Tanguy A, Quere N, Samain J-F, Moraga D. Genetic polymorphism of glutamine synthetase and delta-9 desaturase in families of Pacific oyster Crassostrea gigas and susceptibility to summer mortality. J Exp Marine Biol Ecol. 2007;349:272–283. [Google Scholar]
- Dégremont L, Ernaude B, Bédier E, Boudry P. Summer mortality of hatchery-reared Pacific oyster (Crassostrea gigas). I. Estimation of genetic parameters for survival and growth. Aquaculture. 2007;262:41–53. [Google Scholar]
- Doerwald L, van Genesena S, Onnekinka C, Marín-Vinadera L, de Lange F, deonga W, Lubsen N. The effect of αB-crystallin and Hsp27 on the availability of translation initiation factors in heat-shocked cells. Cell Mol Life Sci. 2006;63:735–743. doi: 10.1007/s00018-005-5582-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan S, Moseley P, Buettner G. Increased flux of free radicals in cells subjected to hyperthermia: detection by electron paramagnetic resonance spin trapping. FEBS Lett. 1998;431:285–286. doi: 10.1016/s0014-5793(98)00779-0. [DOI] [PubMed] [Google Scholar]
- Foreman H, Fischer AB. Antioxidant defenses. In: Gilbert D, editor. Oxygen and living processes. Springer; New York: 1981. pp. 235–250. [Google Scholar]
- Friedman CS, Estes R, Stokes NA, Burge C, Hargove J, Barber B, Elston R, Burreson E, Reece K. Herpes virus in juvenile Pacific oysters Crassostrea gigas from Tomales Bay, California, coincides with summer mortality episodes. Dis Aquat Org. 2005;63:33–41. doi: 10.3354/dao063033. [DOI] [PubMed] [Google Scholar]
- Garnier M, Labreuche Y, Garcia C, Robert M, Nicolas J-L. Evidence for the involvement of pathogenic bacteria in summer mortalities of the Pacific oyster Crassostrea gigas. Micro Ecol. 2007;53:187–196. doi: 10.1007/s00248-006-9061-9. [DOI] [PubMed] [Google Scholar]
- Gentleman R, Carey V, Bates D, Bolstad B, Dettling M, Dudoi S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Torsten H, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini A, Sawitzki G, Smith C, Smyth G, Tierny L, Yang J, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Bio. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenhalgh C, Alexander W. Suppressors of cytokine signaling and regulation of growth hormone action. Growth Horm IGF Res. 2004;3:200–206. doi: 10.1016/j.ghir.2003.12.011. [DOI] [PubMed] [Google Scholar]
- Griscourt L, Bonnec G, Boujard D, Mathieu M, Kellner K. Insulin-like system and growth regulation in the Pacific oyster Crassostrea gigas: hrIGF-1 effect on protein synthesis of mantle edge cells and transcription of an homologous insulin receptor-related receptor. Gen Comp Endo. 2003;13:44–56. doi: 10.1016/s0016-6480(03)00217-x. [DOI] [PubMed] [Google Scholar]
- Gueguen Y, Cadoret JP, Flament D, Barreau-Roumiguiere C, Girardot AL, Garnier J, Hoareau A, Bachere E, Escoubas JM. Immune gene discovery by expressed sequence tags generated from hemocytes of the bacteria-challenged oyster, Crassostrea gigas. Gene. 2003;303:139–145. doi: 10.1016/s0378-1119(02)01149-6. [DOI] [PubMed] [Google Scholar]
- Guillou F, Mitta G, Galinier R, Coustau C. Identification and transcription of gene transcripts generated during an anti-parasitic response in Biomphalaria glabrata. Dev Comp Immunol. 2007;31:657–671. doi: 10.1016/j.dci.2006.10.001. [DOI] [PubMed] [Google Scholar]
- Hamdoun AM, Cheney DP, Cherr GN. Phenotypic plasticity of HSP70 and HSP70 gene transcription in the Pacific oyster (Crassostrea gigas): implications for thermal limits and induction of thermal tolerance. Biol Bull. 2003;205:160–169. doi: 10.2307/1543236. [DOI] [PubMed] [Google Scholar]
- Hawkins AJS. Protein turnover: a functional appraisal. Funct Ecol. 1991;5:222–233. [Google Scholar]
- Hawkins AJS, Day A. The metabolic basis of genetic differences in growth efficiency among marine animals. J Exp Mar Biol Ecol. 1996;203:93–115. [Google Scholar]
- Hawkins AJS, Wilson IA, Bayne BL. Thermal responses reflect protein turnover in Mytilus edulis L. Func Ecol. 1987;1:339–351. [Google Scholar]
- Hawkins AJS, Rusin J, Bayne BL, Day A. The metabolic/physiological basis of genotype-dependent mortality during copper exposure in Mytilus edulis. Mar Env Res. 1989;28:253–257. [Google Scholar]
- Hégaret H, Wikfors G, Soudant P. Flow cytometric analysis of hemocytes from oysters, Crassostrea virginica, subjected to a sudden temperature evelation II. Hemocyte functions: aggregation, viability, phagocytosis, and respiratory burst. J Exp Mar Biol Ecol. 2003;293:249–265. [Google Scholar]
- Hégaret H, Wikfors GH, Soudant P, Delaporte M, Alix JH, Smith BC, Dixon MS, Quere C, Le Cox JR, Paillard C, Moal J, Samain J-F. Immunological competence of eastern oysters, Crassostrea virginica, fed with different microalgal diets and challenged with a temperature elevation. Aquaculture. 2004;234:541–560. [Google Scholar]
- Hirota K, Matsui M, Murata M, Takashima Y, Cheng F, Itoh T, Fukuda K, Junji Y. Nucleoredoxin, glutaredoxin, and thioredoxin differentially regulate NF-κB, AP-1, and CREB activation in HEK293 cells. Biochem Biophys Res Comm. 2000;274:177–182. doi: 10.1006/bbrc.2000.3106. [DOI] [PubMed] [Google Scholar]
- Hochacka P, Somero G. Biochemical adaptation. 2. Oxford; New York: 2002. [Google Scholar]
- Huvet A, Herpin A, Degremont L, Labreuche Y, Samain J-F, Cunningham C. The identification of genes from the oyster Crassostrea gigas that are differentially expressed in progeny exhibiting opposed susceptibility to summer mortality. Gene. 2004;343:211–220. doi: 10.1016/j.gene.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Huber W, von Heydebreck A, Sultmann H, Poustka A, Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential transcription. Bioinformatics. 2002;18(Suppl 1):S96–S104. doi: 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
- Ihaka R, Gentleman R. R: a language for data analysis and graphics. J Comp Graphical Stat. 1996;5(3):299–314. [Google Scholar]
- Jenny M, Chapman R, Mancia A, Chen Y-A, McKillen D, Trent H, Lang P, Escoubas J-M, Bachere E, Boulo V, Liu J, Cunningham C, Cupit P, Tanguy A, Guo X, Moraga D, Boutet I, Huvet A, De Guise S, Almeida J, Warr G. A cDNA Microarray for Crassostrea virginica and C. gigas. Mar Biotech. 2007;9:577–591. doi: 10.1007/s10126-007-9041-1. [DOI] [PubMed] [Google Scholar]
- Johansson M, Lind M, Holmblad T, Thörnqvist P, Söderhäll K. Peroxinectin, a novel cell adhesion protein from crayfish blood. Biochem Biophys Res Com. 1995;216:1079–1087. doi: 10.1006/bbrc.1995.2731. [DOI] [PubMed] [Google Scholar]
- Kalra SP, Kalra PS. NPY an endearing journey in search of neurochemical on/off switch for appetite, sex, and reproduction. Peptides. 2004;25:465–471. doi: 10.1016/j.peptides.2004.03.001. [DOI] [PubMed] [Google Scholar]
- Kang Y-S, Kim Y-M, Park K-I, Cho S, Choi K-S, Cho M-C. Analysis of EST and lectin transcriptions in hemocytes of Manila clams (Ruditapes philippinarum) (Bivalvia: Mollusca) infected with Perkinsus oleseni. Dev Comp Immunol. 2006;30:1119–1131. doi: 10.1016/j.dci.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Kanost M. Serine proteinase inhibitors in arthropod immunity. Dev Comp Immunol. 1999;23:291–301. doi: 10.1016/s0145-305x(99)00012-9. [DOI] [PubMed] [Google Scholar]
- Labrueche Y, Lambert C, Soudant P, Boulo V, Huvet A, Nicolas J-L. Cellular and molecular hemocyte reponses to the Pacific oyster, Crassostrea gigas, following bacterial infection with Vibrio aestuarianus strain 01/32. Microbes Infect. 2006;8:2715–2724. doi: 10.1016/j.micinf.2006.07.020. [DOI] [PubMed] [Google Scholar]
- Lacoste A, Jalabert F, Malham S, Cueff A, Gélébeart F, Cordevant C, Lange M, Poulet S. A Vibrio splendidus strain is associated with summer mortality of juvenile oysters Crassostrea gigas in the bay of Morlaix (North Brittany, France) Dis Aquat Org. 2001;46:139–145. doi: 10.3354/dao046139. [DOI] [PubMed] [Google Scholar]
- Lambert C, Soudant P, Dégremont L, Delaporte M, Moal J, Boudry P, Jean F, Huvet A, Samain JF. Hemocyte characteristics in families of oysters, Crassotrea gigas, selected for differential survival during summer mortality and reared in three sites. Aquaculture. 2007;270:276–288. [Google Scholar]
- Langdon C, Evans F, Jacobson D, Blouin M. Yields of cultured Pacific oysters Crassostrea gigas Thunberg improved after one generation of selection. Aquaculture. 2003;220:227–244. [Google Scholar]
- Levroney E, Aguilar H, Fulcher J, Kohatsu L, Pace K, Pang M, Gurney K, Baum L, Benhur L. Novel innate immune functions for galectin-1: Galectin-1 inhibits cell fusion by Nipah virus envelope glycoproteins and augments dendritic cell secretion of proinflammatory cytokines. J Immunol. 2005;175:413–420. doi: 10.4049/jimmunol.175.1.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist S. The heat shock response. Ann Rev Biochem. 1986;55:1151–1191. doi: 10.1146/annurev.bi.55.070186.005443. [DOI] [PubMed] [Google Scholar]
- Li Y, Qin J, Abbott C, Li X, Benkendorff Synergistic impacts of heat shock and spawning on the physiology and immune health of Crassostrea gigas: an explanation for summer mortality in Pacific oysters. Am J Physiol Regul Integr Comp Physiol. 2007;293:2353–2362. doi: 10.1152/ajpregu.00463.2007. [DOI] [PubMed] [Google Scholar]
- Li Y, Qin J, Abbott C, Li X, Benkendorff Spawning-dependent stress response to food deprivation in Pacific oyster Crassostrea gigas. Aquaculture. 2009;286:309–317. [Google Scholar]
- Liu CH, Cheng W, Chen JC. The peroxinectin of white shrimp Litopenaeus vannamei is synthesized in the semi-granular and granular cell, and its transcription is upregulated with Vibrio alginolyticus infection. Fish Shell Immunol. 2005;18:431–444. doi: 10.1016/j.fsi.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Liu CH, Yeh SP, Hsu PY, Cheng W. Peroxinectin gene transcription of the giant freshwater prawn Machobranchium rosenbergii under intrinsic, immunostimulant, and chemotherapeutant influences. Fish Shell Immunol. 2007;22:408–417. doi: 10.1016/j.fsi.2006.06.015. [DOI] [PubMed] [Google Scholar]
- Ma P, Castillo-David C, Zhong W, Liu J. A data-driven clustering method for time course gene transcription data. Nuc Ac Res. 2006;34:1261–1269. doi: 10.1093/nar/gkl013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Oh C-S, Jiang Y. Regulation of long chain fatty acid synthesis in yeast. Biochim Biophys Acta. 2006;1771:271–285. doi: 10.1016/j.bbalip.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Meistertzheim A-L, Tanguy A, Moraga D, Thebault M-T. Identification of differentially expressed genes of the Pacific oyster Crassostrea gigas exposed to prolonged thermal stress. FEBS J. 2007;24:6392–6402. doi: 10.1111/j.1742-4658.2007.06156.x. [DOI] [PubMed] [Google Scholar]
- Montagnani C, Tirape A, Boulo V, Escobas JM. The two Cg-timp mRNAs expressed in oyster hemocytes are generated by two gene families and differentially expressed during ontogenesis. Dev Comp Immunol. 2005;29:831–839. doi: 10.1016/j.dci.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Ntambi J. Regulation of stearoyl-CoA desaturase by polyun-saturated fatty acids and cholesterol. J Lipid Res. 1999;40:1549–1558. [PubMed] [Google Scholar]
- Oliver JL, Gaffney PM, Allen SK, Faisal M, Kaattari SL. Protease inhibitory activity in selectively bred families of Eastern oysters. J Aquat Anim Health. 2000;12:136–145. [Google Scholar]
- Parsell DA, Lindquist S. The function of heat-shock proteins in stress tolerance: degradation and reactivation of damaged proteins. Annu Rev Genet. 1993;27:437–496. doi: 10.1146/annurev.ge.27.120193.002253. [DOI] [PubMed] [Google Scholar]
- Perillo N, Marcus M, Baum L. Galectins: versatile modulators of cell adhesion, cell proliferation, and cell death. J Mol Med. 1998;76:402–412. doi: 10.1007/s001090050232. [DOI] [PubMed] [Google Scholar]
- Rabinovich G, Rubinstein N, Fainboim Unlocking the secrets of galectins: a challenge at the frontier of glyco-immunology. J Leukoc Bio. 2002;71:741–752. [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RH, Moorman AF. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett. 2003;339:62–66. doi: 10.1016/s0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- Rincón G, Young AE, Bannasch DL, Medrano JF. Charaterization of variation in the canine suppressor of cytokine signaling-2 (SOCS-2) gene. Genet Mol Res. 2007;6:144–151. [PubMed] [Google Scholar]
- Rinne R, Saukko P, Järvinen M, Lehesjoki A-E. Reduced cystatin B activity correlates with enhanced cathepsin activity in progressive myoclonus epilepsy. Ann Med. 2002;34:380–385. doi: 10.1080/078538902320772124. [DOI] [PubMed] [Google Scholar]
- Romestead B, Corbier F, Roch P. Protease inhibitors and haemagglutins associated with resistance to the protozoan parasite, Perkinsus marinus, in the Pacific oyster, Crassostrea gigas. Parasitol. 2002;125:323–329. doi: 10.1017/s0031182002002135. [DOI] [PubMed] [Google Scholar]
- Rothschild M, Ruvinsky A. Marker-assisted selection for aquaculture species. In: Liu Z, editor. Aquaculture genome technologies. Blackwell; Ames: 2007. pp. 199–215. [Google Scholar]
- SAS Institute Inc. SAS/STAT User’s Guide. Version 8. SAS Institute; Cary: 1999. [Google Scholar]
- Samain JF, Dégremont L, Soletchnik P, Haure J, Bédier E, Ropert M, Moal J, Huvet A, Bacca H, Van Wormhoudt A, Delaporte M, Costil K, Pouvreau S, Lambert C, Boulo V, Soudant P, Nicolas JL, Le Roux F, Renault T, Gagnaire B, Geret F, Boutet I, Burgeot T, Boudry P. Genetically based resistance to summer mortality in the Pacific oyster (Crassostrea gigas) and its relationship with physiological, immunological characteristics and infection processes. Aquaculture. 2007;268:227–243. [Google Scholar]
- Sauvage C, Bierne N, Boudry P. Single nucleotide polymorphisms and their relationship to codon usage bias in the Pacific oyster Crassostrea gigas. Gene. 2007;406:13–22. doi: 10.1016/j.gene.2007.05.011. [DOI] [PubMed] [Google Scholar]
- Schweiger M, Schreiber R, Haemmerle G, Lass A, Fledlius C, Jacobsen P, Zechner R, Zimmermann R. Adipose triglyceride lipase and hormone-sensitive lipase are the major enzymes in adipose tissue triacylglycerol catabolism. J Biol Chem. 2006;281:40236–40241. doi: 10.1074/jbc.M608048200. [DOI] [PubMed] [Google Scholar]
- Soletchnik P, Le Moine O, Faury N, Razet D, Geairon P, Goulletquer P. Summer mortality of the oyster in the Bay Marennes-Oleron: spatial variability of environment and biology using a geographical information system (GIS) Aquat Living Resou. 1999;12:131–144. [Google Scholar]
- Soletchnik P, Lambert C, Costil K. Summer mortality of Crassostrea gigas (Thunberg) in relation to rearing environment conditions. J Shellfish Res. 2005;24:197–207. [Google Scholar]
- Soletchnik P, Faury N, Goulletquer P. Seasonal changes in carbohydrate metabolism and its relationship with summer mortality of Pacific oyster Crassostrea gigas (Thunberg) in Marennes-Oléron bay (France) Aquaculture. 2006;252:328–338. [Google Scholar]
- Soletchnik P, Ropert M, Mazuie J, Fleury P, Le Coz F. Relationships between oyster mortality patterns and environmental data from monitoring databases along the coasts of France. Aquaculture. 2007;271:384–400. [Google Scholar]
- Storch J, Thumser A. The fatty acid transport and function of fatty acid-binding proteins. Biochim Biophys Act. 2000;1486:28–44. doi: 10.1016/s1388-1981(00)00046-9. [DOI] [PubMed] [Google Scholar]
- Storey K. Oxidative stress: animal adaptations in nature. Braz J Med Biol Res. 1996;29:1715–1733. [PubMed] [Google Scholar]
- Storey JD. A direct approach to false discovery rate. J R Statist Soc B. 2002;64:479–498. [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genome-wide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanguy A, Bierne N, Saaverdra C, Pina B, Bachere E, Kube M, Bazin E, Bonhomme F, Boudry P, Boulo V, Boutet I, Cancela L, Doussat C, Favrel P, Huvet A, Jarque S, Jollivet D, Klages S, Lapegue S, Leite R, Moal J, Moraga D, Reinhardt R, Samain J-F, Zouros E, Canario A. Increasing genomic information in bivalves through new EST collections in four species: development of new genetic markers for environmental studies and genome evolution. Gene. 2008;408:27–36. doi: 10.1016/j.gene.2007.10.021. [DOI] [PubMed] [Google Scholar]
- Taris N, Lang P, Camara M. Sequence polymorphism can produce serious artifacts in real-time PCR assays: hard lessons from Pacific oysters. BMC Genomcs. 2008;9:234. doi: 10.1186/1471-2164-9-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasumi S, Vasta G. A galectin of unique domain organization from hemocytes of the Eastern oyster (Crassostrea virginica) is a receptor for the protistan parasite Perkinsus marinus. J Immunol. 2007;179:3086–3098. doi: 10.4049/jimmunol.179.5.3086. [DOI] [PubMed] [Google Scholar]
- Tensen CP, Cox KJ, Burke JF, Leurs R, van der Schors RC, Geraerts WP, Vreuhdenhil E, Heerikhuizen H. Molecular cloning and characterization of an invertebrate homologue of a neuropeptide Y receptor. Eur J Neurosci. 1998;10:3409–3416. doi: 10.1046/j.1460-9568.1998.00350.x. [DOI] [PubMed] [Google Scholar]
- Tibile R, Singh H. Larval rearing and spat production of edible oyster Crassostrea gryphoides. Aquaculture Res. 2003;34:785–792. [Google Scholar]
- Tomarev S, Piatgorrsky J. Lens crystallins of invertebrates. Diversity and recruitment from detoxification enzymes and novel proteins. Eur J Biochem. 1996;235:449–465. doi: 10.1111/j.1432-1033.1996.00449.x. [DOI] [PubMed] [Google Scholar]
- Tsvetkova N, Horváth I, Török Z, Wolkers Wm Balogi Z, Shigapova N, Crowe L, Tablin F, Vierling E, Crowe J, Vigh L. Small heat-shock proteins regulate lipid polymorphism. Proc Natl Acad Sci USA. 2002;99:13504–13509. doi: 10.1073/pnas.192468399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich K. Comparitive animal biochemistry. Springer; New York: 1995. p. 248. [Google Scholar]
- Vergote D, Bouchut A, Sautiere PE, Roger E, Galinier R, Rognon A, Coustau C, Slazet M, Mitta G. Characterization of proteins differentially present in the plasma of Biomphalaria glabrata susceptible or resistant to Echinostoma caproni. Internat Jour Parasit. 2005;32:215–224. doi: 10.1016/j.ijpara.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Verlecar XN, Jena KB, Chainy GBN. Biochemical markers of oxidative stress in Perna viridis exposed to mercury and temperature. Chemico-Biological Interact. 2007;167:219–226. doi: 10.1016/j.cbi.2007.01.018. [DOI] [PubMed] [Google Scholar]
- Wang K, Spector A. α-Crystallin prevents irreversible protein denaturation and acts cooperatively with other heat-shock proteins to renature the stabilized partially denatured protein in an ATP-dependent manner. Eur J Biochem. 2000;267:4705–4712. doi: 10.1046/j.1432-1327.2000.01521.x. [DOI] [PubMed] [Google Scholar]
- Woessner J. The matrix metalloproteinase family. In: Parks WC, Mecham RP, editors. Matrix metalloproteinases. Academic; San Diego: 1998. pp. 1–14. [Google Scholar]
- Xue QG, Waldrop GL, Chey KL, Itoh N, Ogawa M, Cooper R, Losso J, La Peyre JF. A novel slow-tight binding serine protease inhibitor from eastern oyster (Crassostrea virginica) plasma inhibits perkinsin, the major extracellular protease of the oyster protozoan parasite Perkinsus marinus. Comp Biochem Physiol B. 2006;145:16–26. doi: 10.1016/j.cbpb.2006.05.010. [DOI] [PubMed] [Google Scholar]
- Yamaura K, Takahashi K, Suzuki T. Identification and tissue transcription analysis of C-type lectin and galectin in Pacific oyster, Crassostrea gigas. Comp Biochem Phys B. 2008;149:168–175. doi: 10.1016/j.cbpb.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Yoshimura A, Naka T, Kubo M. SOCS proteins, cytokine signaling and immune regulation. Nat Rev Immu. 2007;7:454–465. doi: 10.1038/nri2093. [DOI] [PubMed] [Google Scholar]
- Young IS, Woodside JV. Antioxidants in health and disease. J Clin Pathol. 2001;54:176–186. doi: 10.1136/jcp.54.3.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler G, Paynter K, Fisher D. Matrix metalloproteinase-like activity from hemocytes of the eastern oyster, Crassostrea virginica. Comp Biochem Physiol B. 2002;131:361–370. doi: 10.1016/s1095-6433(01)00518-9. [DOI] [PubMed] [Google Scholar]
- Zhao R, Houry W. Molecular interaction network of the Hsp90 chaperone system. In: Csermely P, Vigh L, editors. Molecular aspects of the stress response: Chaperones, membranes, and networks (Advances in experimental medicine and biology 594) Springer; New York: 2007. pp. 27–36. [DOI] [PubMed] [Google Scholar]