Abstract
Accurate copying of the genome by DNA polymerases is challenging due in part to the continuous damage inflicted on DNA, which results from its contact with reactive oxidative species (ROS), producing lesions such as 7,8-dihydroxy-8-oxoguanine (8-oxoG). The deleterious effects of 8-oxoG can be attributed to its dual coding potential that leads to G ➔ T transversions. The wild type (wt) pol α family DNA polymerase from bacteriophage RB69 (RB69pol) inserts dCMP in preference to dAMP when situated opposite 8-oxoG by > 2 orders of magnitude as demonstrated using pre-steady-state kinetics (kpol/Kd,app). In contrast, the Y567A mutant of RB69pol inserts both dCMP and dAMP opposite 8-oxoG rapidly and with equal efficiency. We have determined the structures of pre-insertion complexes for the Y567A mutant with dATP and dCTP opposite a templating 8-oxoG in a 13/18mer primer-template (P/T) at resolutions of 2.3 and 2.1 Å, respectively. Our structures show that the 8-oxoG residue is in the anti conformation when paired opposite dCTP, but it flips to a syn conformation forming a Hoogstein base-pair with an incoming dATP. Although the Y567A substitution does not significantly change the volume of the pocket occupied by anti-8-oxoG, it does provide residue G568 the flexibility to move deeper into the minor groove of the P/T to accommodate, and stabilize, syn-8-oxoG. These results support the hypothesis that it is the flexibility of the Nascent base-pair Binding Pocket (NBP) in the Y567A mutant that allows efficient insertion of dAMP opposite 8-oxoG.
DNA polymerases ensure accurate replication of genomic DNA, making errors only once in every 103 to 106 insertion events. When available, intrinsic exonuclease activity in the same polypeptide chain decreases the error rate by an additional 100-fold. Extrinsic exonucleases and nucleotide base excision repair, and mismatch repair systems (NER, BER, and MMR) further increase replication fidelity. Overall, the combined effects of these systems lead to only one error per 109–10 base-pairs polymerized (1, 2).
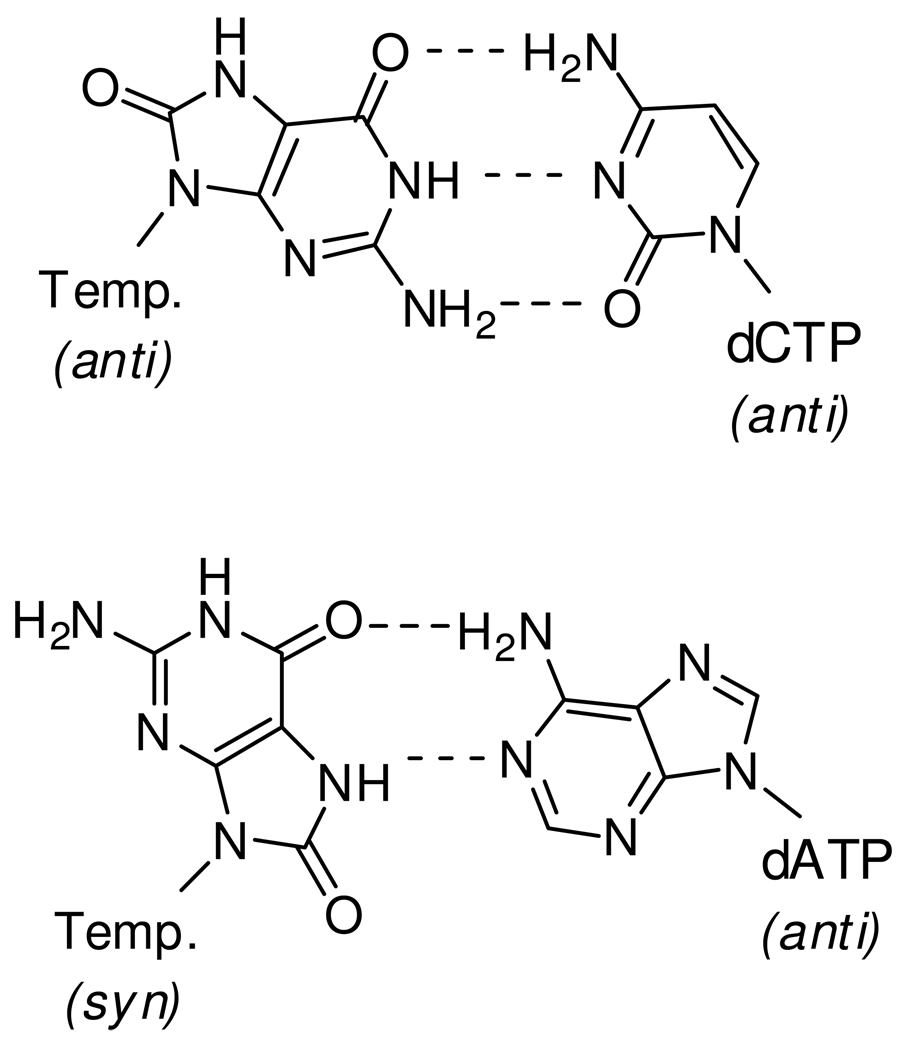
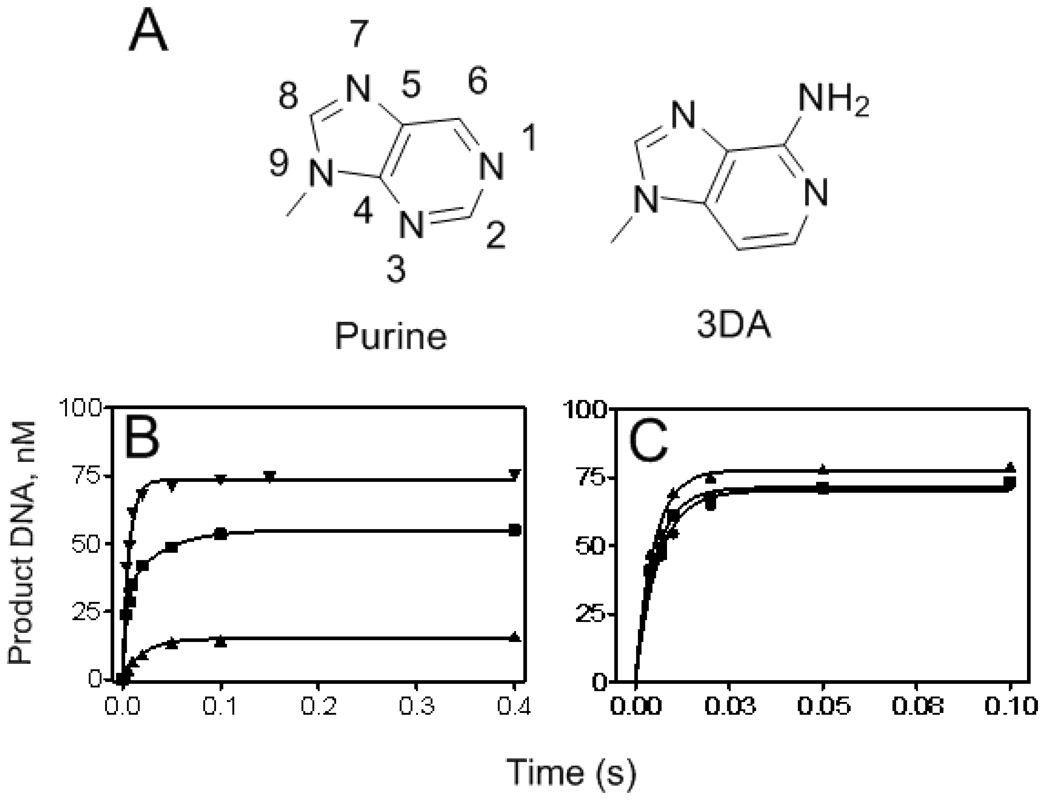
7,8-Dihydro-8-oxoguanine (8-oxoG) is a common DNA base adduct that has high mutagenic potential (3, 4). It has been estimated that approximately 100 – 400 of these adducts are formed within a mammalian cell per day (5). Reactive oxygen species (ROS), such as the hydroxyl radical •OH, directly attack the C-8 position of guanine (3). This adduct, which is simply guanine with a C-8 carbonyl oxygen (O-8), has the potential to form two hydrogen bonds with dATP when 8-oxoG flips from an anti to a syn conformation (Fig. 1), making it an acceptable template for dATP as well as dCTP, resulting in G ➔ T transversions which have been linked to cancer and other diseases (6, 7).
Figure 1.
Structures of an incoming dCTP or dATP paired opposite a templating 8-oxoG. 8-OxoG likely flips into the syn conformation to pair opposite dATP to form a base pair comparable to dATP:T, with two inter-base hydrogen bonds.
DNA polymerases vary widely in their ability to select against insertion of dAMP opposite 8-oxoG, even within the same polymerase family. It has been shown that most A family pols, such as KF, pol II, and T7 preferentially insert dCMP opposite 8-oxoG, whereas pol BF, an A family DNA pol from Bacillus stearothermophilus, prefers to insert dAMP by 10-fold over dCMP (8–11). Beese et al. (8) have shown that the ability of BF to preferentially insert dAMP relative to dCMP opposite 8-oxoG was likely due, in part, to the formation of a hydrogen bond between the O-8 of syn-8-oxoG and an amide hydrogen of Q797, in addition to the two inter-base hydrogen bonds (8). In contrast, Ellenberger et al. showed that T7 pol lowers the efficiency of dAMP insertion relative to dCMP opposite syn-8-oxoG because of an unfavorable steric and/or electrostatic interaction between N-2 of syn-8-oxoG and the protonated primary amine of K536 (11, 12). DNA polymerases from the B family, such as RB69pol, human pol α, calf thymus pol δ, and the DNA pol from bacteriophage φ29 (φ29pol) vary widely in their ability to insert dAMP relative to dCMP opposite 8-oxoG (13–16). For example, human pol α was recently shown to favor dATP, although both were inserted with very low efficiency (13), but calf thymus pol δ, in the presence of PCNA, favors dCTP by several fold (14).
RB69pol was previously found to discriminate against dAMP relative to dCMP opposite 8-oxoG (15). Modeling studies have suggested that the low efficiency of dAMP insertion opposite 8-oxoG by wt RB69pol was due to a steric clash between syn-8-oxoG and residue G568 (15), although the authors suggested that the adjacent Y567 residue was helping to create a steric environment for G568, thereby indirectly causing destabilization of syn-8-oxoG (14). Steady-state kinetic analysis of the Y390S replacement in φ29pol (the equivalent of Y567 in RB69pol) showed a decrease in discrimination for dAMP relative to dCMP insertion opposite 8-oxoG, lending support to this conclusion (16). Modeling studies have suggested that the equivalent tyrosine residue in DNA pols of other families, e.g. Y955 and Y530 of the family A DNA pols human pol γ and T7 pol, respectively, and Y271 of the family × human pol β, would also act as steric blocks for insertion of dAMP opposite 8-oxoG (11, 17–19), and it had been found that the human pol γ Y955C mutant reduced the discrimination of dAMP opposite 8-oxoG in a similar manner as the φ29pol Y390S mutant (16, 19). Interestingly, the pol γ Y955C mutant had been discovered while screening patients suffering from progressive external ophthalmoplegia (PEO), a disease that leads to terminal dementia, suggesting a link between dementia and oxidative stress (19).
To test if Y567 in RB69pol modulates the insertion of dAMP opposite 8-oxoG, we compared the pre-steady-state kinetic parameters for dAMP and dCMP insertion opposite 8-oxoG by wt RB69pol, and its Y567A and L561A mutants. We included the RB69pol L561A mutant because its side chain is close to the templating base (Fig. 2), and replacing Leu with the much smaller Ala would be expected to allow 8-oxoG more freedom to “sample” alternative conformations upon binding of an incoming dATP. We found that the Y567A mutant increased dAMP insertion opposite 8-oxoG by almost 3 orders of magnitude relative to wt RB69pol, but the L561A mutant did not. To understand why wt was so restrictive and Y567A so permissive, we determined crystal structures of the pre-insertion ternary complexes of the Y567A mutant with dCTP and dATP opposite a templating 8-oxoG (with 2.1 and 2.3 Å resolution, respectively). When we compared these structures with those of the wt:dCTP:8-oxoG and wt:dTTP:A ternary complexes obtained previously (15, 20), we were able to rationalize the kinetic behavior of the wt and the Y567A mutant in structural terms as described below.
Figure 2.
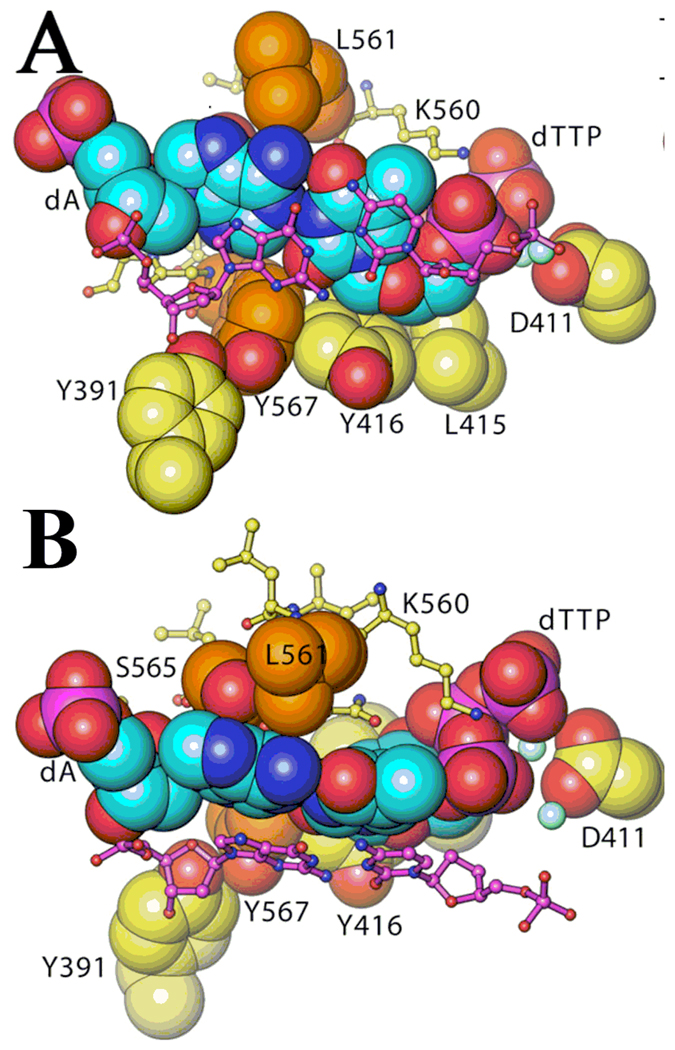
Steric relationship of some of the NBP residues surrounding an incoming dTTP paired opposite a templating A (from Franklin et al., PDB code 1IG9 (20)). A. The nascent base-pair and surrounding residues are shown from the duplex side of the primer-template in space-filling form (the terminal base-pair is shown in stick form (purple) for clarity). L561 and Y567 are shown in orange. B. Orthogonal view of (A), showing the close proximity of L561 with the templating base.
EXPERIMENTAL PROCEDURES
Materials
Materials and reagents were of the highest quality commercially available. dNTPs were purchased from Roche (Burgess Hill, UK), T4 polynucleotide kinase was purchased from New England Biolabs (Ipswich, MA), and [γ-32P]ATP was purchased from MP Biomedicals (Irvine, CA). d(3DA)TP was generously provided by Prof. Robert D. Kuchta (University of Colorado, Boulder, CO).
Enzymes
Wild-type RB69pol and the Y567A and L561A mutants in the exonuclease-deficient background (D222A and D327A), were over-expressed in Escherichia coli, purified, and stored as previously described (21).
DNA Substrates
Sequences of primer-templates (P/T) used in this study are shown in Table 1. Oligonucleotides were synthesized at the Keck facilities (Yale University). Primers were labeled on the 5’-end with 32P using T4 polynucleotide kinase and [γ-32P]ATP (except when used for crystallography) and annealed to unlabeled templates as previously described (22, 23).
Table 1.
Primer-template squences used in this study *
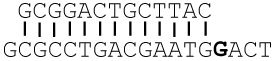 |
DG |
 |
DOG |
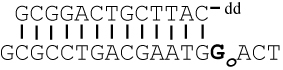 |
DOGdd |
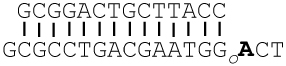 |
DC:OG |
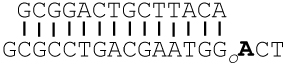 |
DA:OG |
 |
DA |
 |
D3DA |
The templating base is in bold. Go = 8-oxoG, and A3D = 3-deazaadenine (3DA). DOGdd was used to grow the crystal structures discussed.
Chemical Quench Experiments
Experiments were performed at 23°C with a buffer solution of 66 mM Tris-HCl (pH 7.4). Rapid chemical quench experiments were carried out using the KinTek rapid quench instrument (Model RQF-3; KinTek Corp., University Park, PA). For k pol and K d,app determinations, experiments were performed under single-turnover conditions, with 10-fold excess RB69pol over P/T. Although this high excess of RB69pol over P/T should ensure single-turnover, the condition for this to occur is that all of the P/T binds productively to RB69pol. This condition is not always met, as we had discovered previously with the RB69pol “triple mutant” L561A/S565G/Y567A (24). To ensure that single-turnover conditions were obeyed we added heparin (1 mg/ml final concentration) to trap any enzyme that was released from the P/T. In some instances, the heparin trap could not be used because the rate of product formation was too low to obtain accurate kinetic parameters, e.g., insertion of dAMP opposite 8-oxoG by wt. Briefly, enzyme and P/T from one syringe were rapidly mixed with Mg2+, heparin, and various [dNTP]s from the other syringe for times ranging from as short as 5 ms to as long as 1 min. For reactions that required longer times, experiments were performed manually on the bench-top. The final concentrations after mixing were 1 µM enzyme, 90 nM P/T, and 10 mM Mg2+. Reaction mixtures were quenched with 0.5 M EDTA (pH 8.0). Substrates and products were separated by PAGE (19:1% (w/v) acrylamide:bisacrylamide gels containing 8M urea), visualized using a STORM imager (Molecular Imaging), and quantified using Imagequant (GE Healthcare) and GraphPad Prizm.
Data Analysis
The amount of product formed versus time for each [dNTP] were fitted by non-linear regression to the general form of Equation 1 to obtain observed rates of product formation, k obs:
| (Eq.1) |
where Y is the concentration of the DNA product formed during the reaction, C is the offset constant, A i the observed amplitude of product formation, and ki the observed rate constant. The kinetic parameters k pol (the rate of phosphoryl transfer) and K d,app (defined as the [dNTP] at which the rate of phosphoryl transfer reaches 1/2 k pol), were obtained by fitting plots of k obs versus [dNTP] to Equation 2:
| (Eq.2) |
where k obs represents the observed rate at a given [dNTP]. Note that the K d,app values are not ground-state dissociation constants of dNTP binding. This is because the observed [dNTP]-dependence of rates of product formation are affected by “hidden” steps that occur subsequent to dNTP binding but prior to phosphoryl transfer, such as the step defining an induced-fit conformational change. Interestingly, insertion of d(hypoxanthine)TP (dITP) opposite A, and dGMP opposite dAP (2-aminopurine) (Table 2), resulted in k obs values that decreased with increasing [dITP], a similar situation to what Johnson et al. (25, 26) found with AZT and d(8-oxoG)MP insertion opposite T and C, respectively, by the human mitochondrial DNA polymerase γ. (The complex kinetics may have been caused by a slow release of pyrophosphate subsequent to chemistry, thereby allowing pyrophosphorolysis to compete with phosphodiester bond formation (26)). Values of K d,app were estimated in these cases by fitting plots of k obs versus [dNTP] to Equation 3:
| (Eq.3) |
where Δk obs is the overall change in the observed rate given the range of [dNTP].
Table 2.
Pre-steady-state kinetic parameters for insertion of dNMPs by wild type RB69pol and its Y567A mutant.
| Enzyme | dNTP | Template |
kpol (s−1) |
Kd,app (µM) |
kpol/ Kd,app (µM−1 s−1) |
|---|---|---|---|---|---|
| Wild Type | dCTP | 8-oxoG | 55 | 97 | 0.57 |
| dATP | 8-oxoG | 1.7 | 1500 | 1.1 × 10−3 | |
| L561A | dCTP | 8-oxoG | 76 | 11 | 6.9 |
| dATP | 8-oxoG | 9.5 | 980 | 9.7 × 10−3 | |
| Y567A | dCTP | 8-oxoG | 91 | 29 | 3.1 |
| dATP | 8-oxoG | 110 | 48 | 2.3 | |
| dGTP | A | 0.015 | 250 | 6.0 × 10−5a | |
| dITP | A | ~ 0.50 | ~ 1000 | 5.0 × 10−4b | |
| dGTP | dAP | ~ 0.15 | ~ 200 | 8.0 × 10−4b | |
| dITP | dAP | NDc | NDc | 3.0 × 10−5d |
Data obtained previously (33).
Values are estimates because the kobs values decreased with increasing [dNTP]. Kd,app and kpol values in these cases were obtained by fitting kobs versus [dNTP] to Equation 2.
ND = Not Determined because the value of Kd,app was too high (> 2 mM).
The kpol/Kd,app second order value was obtained in this case by calculating the slope of a linear line fitted to the data: y = mx + b, where m = slope = Δ kobs/Δ[dNTP] = kpol/Kd,app. Of the values shown, standard deviations were within 10–20% and ~30% for kpol and Kd,app values, respectively.
Crystallization of RB69pol ternary complexes
DOGdd was used for all crystallizations (Table 1). The primer was dideoxy-terminated at the 3’-end to prevent nucleotide insertion. The RB69pol Y567A mutant was mixed in an equimolar ratio with freshly annealed DOG dd. dCTP or dATP was then added to give a 2.5mM final concentration. For equilibration, using micro-batch vapor-diffusion methods for crystallography, equal volumes of a well solution containing 150 mM CaCl2, 14%(w/v) PEG 350 monomethyl ether (MME), and 100 mM Na Cacodylate (pH 6.5) was added to the RB69pol: DOG dd:dCTP mixture; and 150 mM CaCl2, 11%(w/v) PEG 350 MME, and 100 mM Na Cacodylate (pH 6.5) was added to the RB69pol:DOG dd:dATP mixture. Crystals typically grew in 2–3 days at 20°C. The square rod-shaped crystals had typical dimensions of 0.2 mm × 0.1mm × 0.1mm. Crystals were transferred from the mother liquor to a cryoprotectant/precipitant stabilization solution containing 20% (w/v) PEG 350 MME, 100 mM CaCl2, and 100 mM Na Cacodylate (pH 6.5). Crystals were then gradually transferred into a cryoprotectant/precipitant solution with the same components but with up to 30% (w/v) PEG 350 MME prior to storage in liquid nitrogen.
Data collection, structure determination, and refinement
Crystals were first characterized using home radiation sources. X-ray diffraction data were collected using synchrotron radiation sources at the beamline X29 at Brookhaven National Laboratory (NSLS) (Upton, NY) at a wavelength of 1.075 Å and at 110 K. Crystals belonged to orthorhombic space groups P212121 with different unit cell parameters (Table 3). Data were integrated and scaled using the HKL2000 program suites (27).
Table 3.
Data Collection and Refinement Statistics.
| Ternary complex with: | dATP vs d8oxoG | dCTP vs d8oxoG |
|---|---|---|
| Unit cell [a,b,c(Ǻ)] | 78.148,117.603,130.681 | 76.394,121.089,122.857 |
| Resolution range (Highest-resolution shell)(Ǻ) | 50.0–2.30 (2.38–2.30) | 50.0–2.05 (2.12–2.05) |
| No. of unique reflections | 51799(4177) | 65594(5020) |
| Redundancy | 4.2(3.0) | 3.8(2.3) |
| Completement (%) | 95.3 | 90.7 |
| Rmerge | 8.3(>100) | 7.3(96.2) |
| Signal to noise ratio (I/δI) | 15.3(0.95) | 16.5(1.03) |
| Model Contents: | ||
| no. of amino acid residues | 901 | 901 |
| no. of water molecules | 175 | 566 |
| no. of Ca2+ ions | 4 | 3 |
| no. of template nucleotides | 16 | 16 |
| no. of primer nucleotides | 13 | 13 |
| no. of dNTP molecules | 1 | 1 |
| Rf (%) | 20.40 | 20.92 |
| Rfree (%) | 26.83 | 25.26 |
| RMSD bonds(Ǻ) | 0.0092 | 0.0091 |
| RMSD angles(°) | 1.307 | 1.237 |
Structures were solved using molecular replacement methods using AMORE (28), starting with the ternary complex structure of wild type RB69pol (20), and refined using REFMAC5 (29). Using the program Coot (30), DNA duplex and dNTP were built into the electron density maps phased by the partially refined polymerase model. We observed two Ca2+ ions occupying the A and B metal ion sites, as had been observed previously (20). Structure refinement statistics are summarized in Table 3. Figures obtained from the crystal structures were made using the program Ribbons (31).
PDB accession numbers
Coordinates and structure factors for the Y567A mutant dCTP:anti-8-oxoG and dATP:syn-8-oxoG ternary complex structures have been deposited in the Protein Data Bank under accession codes 3LZJ, and 3LZI, respectively.
RESULTS
Insertion of dAMP relative to dCMP opposite 8-oxoG by wild type and mutant RB69 pols
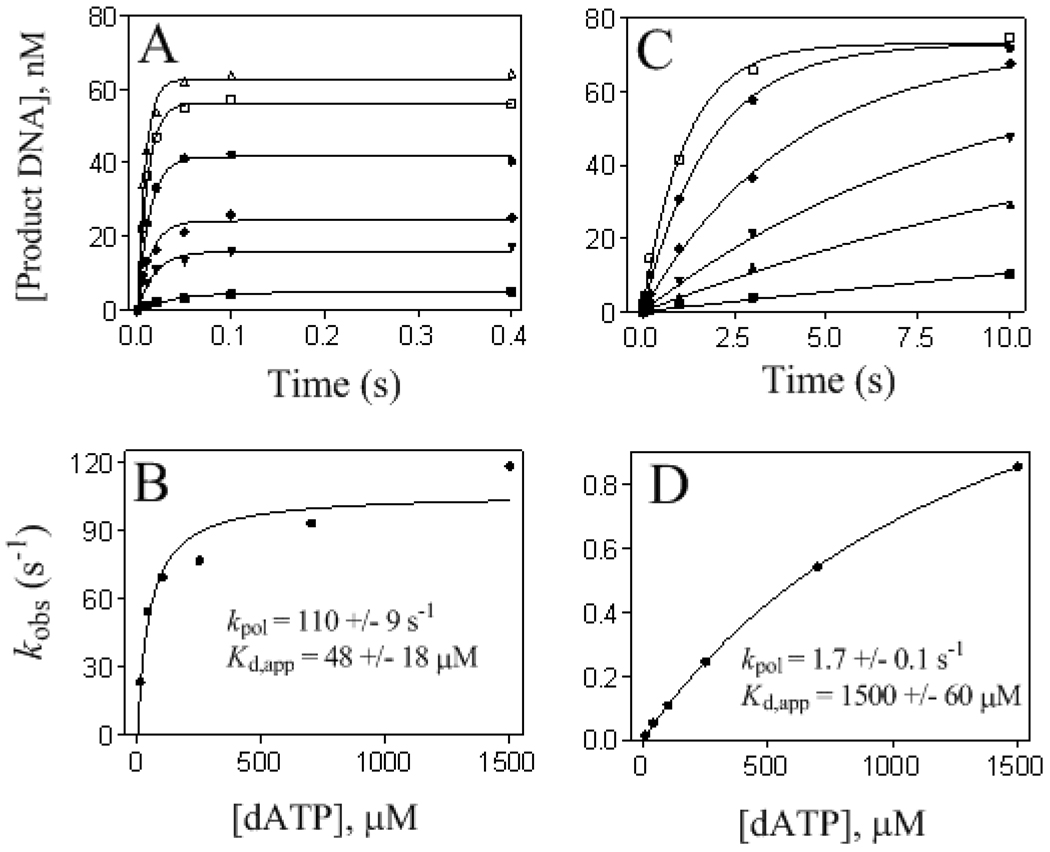
Steady-state kinetic analysis had previously shown that wt RB69pol inserted dCMP opposite 8-oxoG 20-times more efficiently than dAMP (15). We confirmed this result, but under pre-steady-state conditions, i.e., comparing k pol/K d,app specificity constants, we found a 500-fold difference between the ability of wt to insert dCMP versus dAMP opposite 8-oxoG (Table 2). In theory, k pol/K d,app should be equal to the steady-state approximation k cat/K m (32), but in our case, the pre-steady-state values differed from steady-state values. Fig. 3A,B shows a representative set of data used to obtain k pol and K d,app values when inserting a dNMP opposite 8-oxoG in the presence of a heparin trap (see Experimental Procedures), in this case dAMP opposite 8-oxoG catalyzed by the Y567A mutant. No discernable "burst" of dAMP insertion opposite 8-oxoG by the wt was observed even at 500 µM dATP (data not shown), so kinetic parameters could only be estimated from data derived from progress curves representing multiple turnovers (without a heparin trap) (Fig. 3C,D ). In this case, the specificity constants k cat/K m and k pol /K d,app are approximately equal because phosphoryl transfer was rate-limiting (33).
Figure 3.
Kinetics of dAMP insertion opposite 8-oxoG by the Y567A mutant (A and B) and the wt (C and D). A. Progress curves with various [dATP], 10, 40, 100, 250, 700, and 1500 µM (bottom to top), fit to single exponential equations. 1 mg/ml Heparin was used to prevent rebinding of the enzyme to the P/T. B. Plot of kobs versus [dATP] fit to Equation 2 to obtain kpol and Kd,app. C. Same as A but with the wt. Progress curves were obtained without a heparin trap. D. Same as B using the results of C.
We wanted to find out which NBP residues serve as a barrier to insertion of dAMP opposite 8-oxoG. The crystal structure (20) of the wt RB69pol:DA:dTTP complex showed that the O-helix residue L561, which is in close proximity to the adenine templating base (Fig. 2B ), could hinder the binding of syn-8-oxoG if it was the templating base instead of A. However, discrimination against dAMP insertion by the L561A mutant remained as high as wild type, with a 700-fold preference for incorporation of dCMP over dAMP (Table 2). We also substituted Y567 with Ala, and we found that the Y567A mutant incorporated both dAMP and dCMP opposite 8-oxoG with equal efficiency, in contrast to the results obtained with L561A. The individual kinetic parameters were within two or three fold of the values obtained for insertion of dCMP opposite 8-oxoG by wt, so the Y567A mutant caused RB69pol to lose fidelity solely by having the ability to increase the efficiency of dAMP insertion (Table 2).
Can other dNTP derivatives that form nascent Pu-Pu base-pairs, such as hypoxanthine (I):A, be utilized for primer extension?
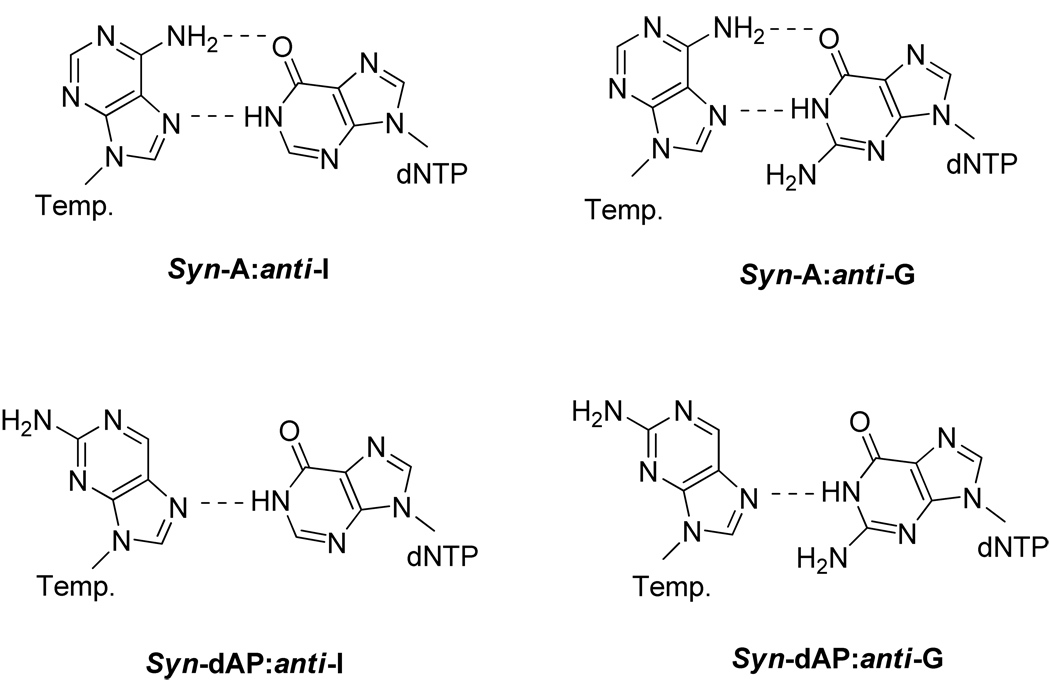
Because the Y567A mutant did not discriminate against the formation of a dATP:syn-8-oxoG base-pair, we wanted to find out if the Y567A mutant would utilize other nascent purine:purine base-pairs, e.g. a dlTP:dA base-pair, with comparable efficiency. If negative results were obtained it could be argued that the dATP:syn-8-oxoG base-pair presented a unique situation. We had previously shown that the Y567A mutant inefficiently inserted dGMP opposite dA, despite the fact that a dGTP:syn-A base-pair could form two inter-base hydrogen bonds, just like dATP:syn-8-oxoG (Fig. 4) (Table 2) (24). To test if other Pu:Pu mis-pairs could be utilized by the Y567A mutant, we investigated the ability of the Y567A mutant to insert dIMP opposite A as well as dGMP and dIMP opposite dAP (Fig. 4), and found very low levels of insertion (Table 2), suggesting that the Y567A mutant still retained some ability to discriminate against the formation of other Pu:Pu mispairs.
Figure 4.
Purine:purine nascent base pairs poorly utilized by the Y567A mutant.
Can a dTTP:3DA nascent base-pair be used for primer-extension by wt RB69pol or its Y567A mutant?
Because dATP insertion opposite 8-oxoG by the Y567A mutant was unique as being the only Pu:Pu mispair that could be efficiently utilized, there is likely to be some specific feature(s) of the templating 8-oxoG that the Y567A mutant recognizes to utilize dATP. Since 8-oxoG was the only purine base with a C-8 carbonyl oxygen that was tested, it seemed reasonable to assume that it could be a hydrogen bond acceptor of the Cα hydrogen atom (Cα-H) of residue G568 (34) which could firmly anchor the templating 8-oxoG and enhance the rate of phosphoryl transfer. By analogy, the N-3 of a templating purine in the anti-conformation may also be a H-bond acceptor of the Cα-H of G568, because of the close proximity between G568 and templating purines in the RB69pol ternary structures (15, 20). Assuming that a Cα-H•••N hydrogen bond could help to stabilize a nascent pyrimidine:purine (py:pu) base pair, removing the N-3 from a templating dA and replacing it with C–H (forming 3DA, Fig. 5A ) would be expected to cause destabilization of the nascent base-pair, leading to a less efficient insertion of the “correct” dNMP. Insertion of dTMP opposite a templating 3DA, by the wt and the Y567A mutant, resulted in progress curves with significant reduction in efficiency, relative to dTMP insertion opposite A (Fig. 5B ). In contrast, the reciprocal situation of inserting d(3DA)MP opposite of T by either wt or the Y567A mutant resulted in k pol values and amplitudes comparable to that obtained when dAMP was inserted opposite T (Fig. 5C ).
Figure 5.
Progress curves for wt RB69pol and the Y567A mutant for insertion of d(3DA)MP opposite T and dTMP opposite 3DA. A. Shown is the purine numbering scheme (left) and 3-deazaadenine (3DA) (right). B. Rate of DNA product formation for dTMP insertion opposite 3DA by wt (▲), dTMP opposite A by wt (▼), and dTMP opposite 3DA by the Y567A mutant (■). C. Rate of DNA product formation for d(3DA)TP insertion opposite T by wt (■), dATP opposite T by wt (●), and dTMP opposite 3DA by the Y567A mutant ▲.
Extention past C:8-oxoG and A:8-oxoG base-pairs
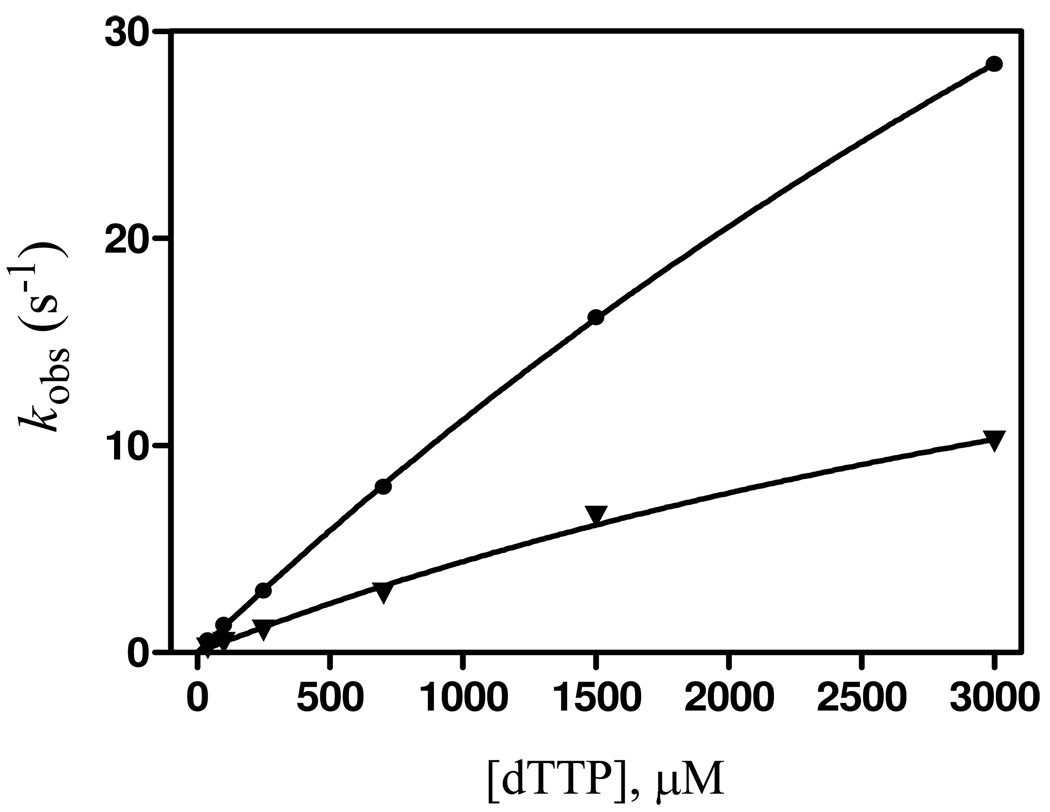
Bypass of base-pairs containing 8-oxoG as the templating base was very inefficient with wt RB69pol (Table 4). The main effect of having an 8-oxoG in the penultimate position 3’ to the templating base was to increase the K d,app for correct incoming dNTPs, by at least two orders of magnitude relative to the situation where extension occurs with a normal P/T that does not have an 8-oxoG lesion (Table 4). With the wt, the K d,app values were too high (> 2 mM) for an accurate determination of k pol, but it was apparent that extrapolation of the plots of k obs versus [dTTP] (the next correct dNTP) would give higher k pol and larger K d,app values for insertion past an A:8-oxoG base-pair, relative to insertion past a C:8-oxoG base-pair, resulting in approximately equal efficiencies for insertion of dTMP opposite A when bypassing A:8-oxoG and C:8-oxoG base-pairs (Fig. 6). This result concurs with a previous report by Kisker et al. (15). In contrast, the Y567A mutant bypassed both A:8-oxoG and C:8-oxoG pairs much more efficiently, with k pol/K d,app values 20-fold and 60-fold higher than with wt, respectively.
Table 4.
Kinetic parameters for insertion of correct dNMPs past C:8oxoG and A:8oxoG terminal base-pairs.
Figure 6.
kobs versus [dTTP] plots of wt RB69pol extending primers beyond A:8-oxoG (●) and C:8-oxoG (▼) terminal base-pairs.
Crystal structures of the Y567A mutant in complex with a dideoxy primer-template containing a templating 8-oxoG opposite dATP or dCTP
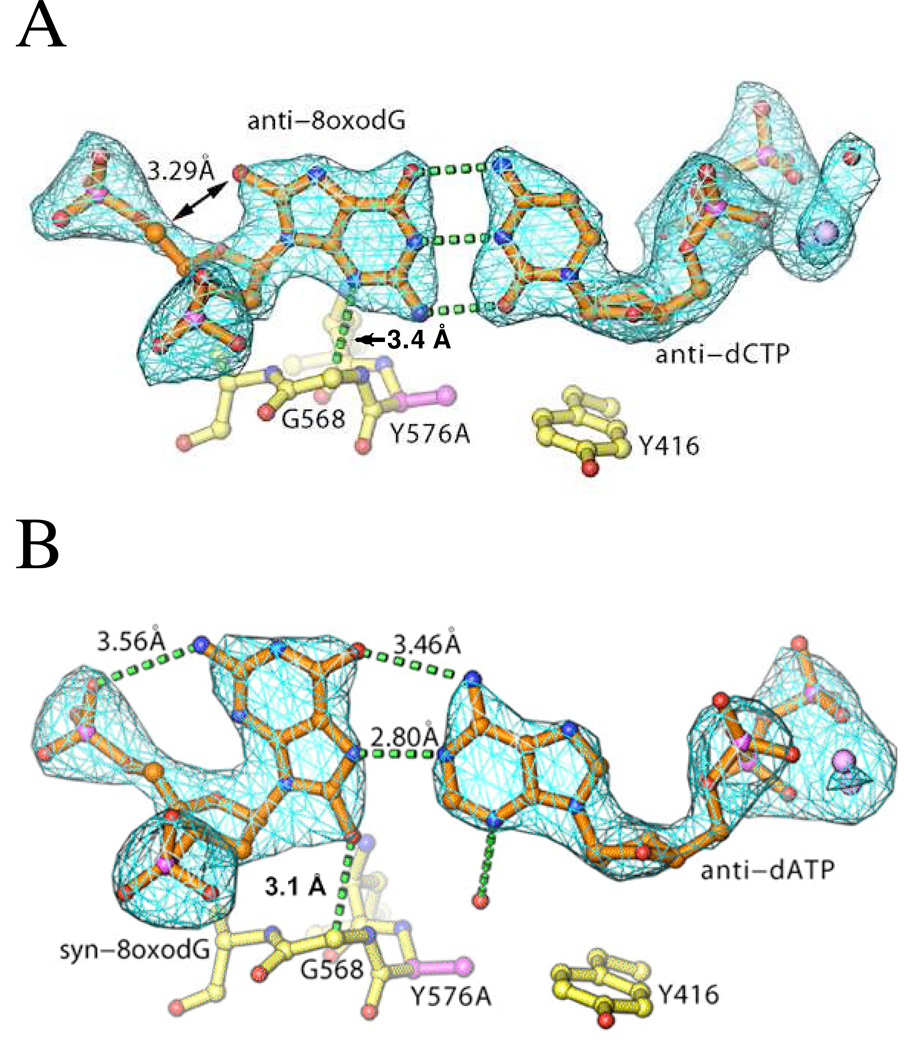
To correlate the kinetic results with the RB69pol structure it was necessary to obtain high quality crystals of the various complexes under study. Crystal structures of wt RB69pol ternary complexes containing a templating 8-oxoG and an incoming dCTP have already been reported (15), but there was no information for ternary complexes of the RB69pol Y567A mutant with P/Ts having a templating 8-oxoG opposite dCTP and dATP. To fill this gap, we were able to obtain crystal structures of the above mentioned ternary complexes, with 2.1 Å and 2.3 Å resolution, respectively. The structures were well refined, with free R-factors of ~ 25 % and 26 %, respectively (Table 3). Note that calcium ions were used as the metal ion cofactor to crystallize these complexes, instead of magnesium. Unlike magnesium, the cofactor that brings about rapid phosphoryl transfer, calcium did not lead to phosphoryl transfer, even in the presence of the 3’-OH (which was absent in these structures), so it is very likely that these structures represent inactive complexes. However, the goal here was to obtain new insights into base selectivity by determining; (i) the precise interactions between the incoming dNTP and 8-oxoG; (ii) the location and geometry of the nascent base-pair with respect to residues in the NBP, and; (iii) by comparing these structures with those from previous studies that used virtually identical conditions to crystallize RB69pol (15, 20). With the Y567A mutant:dCTP:8-oxoG structure, we observed three hydrogen bonds between the incoming dCTP and 8-oxoG, with the 8-oxoG oriented in the anti-conformation (Fig. 7A ). When we superimposed the palm domains of the wt:dCTP:anti-8-oxoG structure (15) and the Y567A mutant:dCTP:anti-8-oxoG structure, there was an ~ 0.7 Å shift of the A567 and G568 residues laterally toward the Y416 side chain, relative to the wt, while maintaining approximately the same distance between the Cα-H of G568 and the N-3 of anti-8-oxoG (Fig. 8). The shift of these residues also permitted the nascent base-pair to move in the same direction, presumably helping it to alleviate some strain between the 2’-deoxyribose 5’-carbon and O-8 of anti-8-oxoG (represented by circular arrows, Fig. 8). Such a shift would be unlikely to occur in the wt RB69pol because of the Y567 side chain, which is hydrogen bonded to the side chain of Y391 and packed against the side chain of Y416, creating considerable rigidity in this region.
Figure 7.
Crystal structures of the RB69pol Y567A mutant in complex with anti-8-oxoG:dCTP and syn-8-oxoG:dATP. A. Fo-Fc electron density map of the anti-8-oxoG:dCTP nascent base-pair. B. Fo-Fc electron density map of the syn-8-oxoG:dATP nascent base-pair.
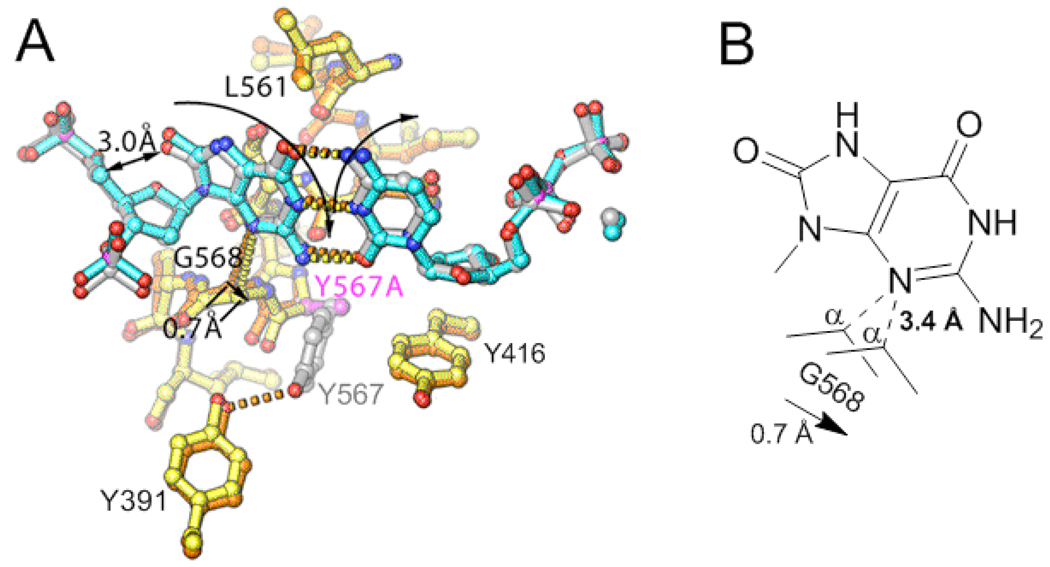
Figure 8.
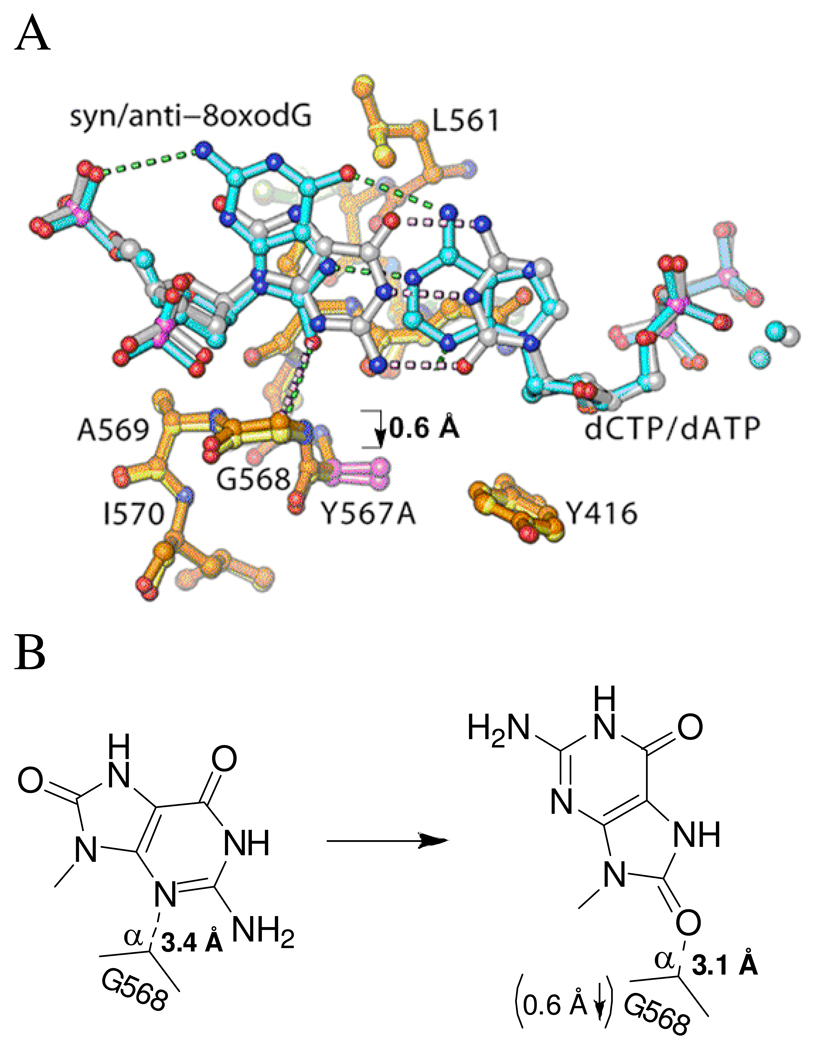
Comparison of the crystal structures of wt and the Y567A mutant in complex with anti-8-oxoG:dCTP (14). A. Superimposed palm domains showing that the Y567A substitution resulted in a shift of G568 by ~ 0.7 Å laterally toward Y416, relative to wt. Residues of the Y567A mutant and wt are shown in yellow and orange, respectively, and the individual residues Y567 and Y567A are shown in gray and magenta, respectively. The nascent base-pair in the wt and Y567A mutant structures are shown in silver and blue, respectively. B. Schematic illustration of the spatial relationship between G568 and the nascent base-pair as shown in (A).
From the omit Fobs-Fcalc electron density maps of the two Y567A mutant ternary structures, we can see that the templating 8-oxoG adopts a syn conformation when it pairs with dATP, with two inter-base hydrogen bonds helping to stabilize it in the syn orientation in a manner analogous to that found with the A family DNA pols, BF pol and T7 pol (8, 12) (Fig. 7B ). In addition, G568 was in close proximity to syn-8-oxoG so that an H-bond could form with the Cα-H of G568 and the O-8 of syn-8-oxoG (inter-atom distance of 3.1 Å) (Fig. 7B ). Superimposing the palm domains of Y567A mutant:dCTP:anti-8-oxoG with the corresponding Y567A mutant:dATP:syn-8-oxoG complex revealed that residues A567, G568, and A569 in the immediate vicinity of Y567A, shifted towards Y416. There were no other significant changes (for example, L561, I570, and Y416 remained fixed) (Fig. 9). Interestingly, the Cl’-Cl’ distance of the anti-dATP:syn-8-oxoG base-pair, often used as a measure of base-pair size, was 10.2 Å, which is less than that of the typical purine-pyrimidine base pair (10.6 Å).
Figure 9.
Comparison of the Y567A mutant anti-8-oxoG:dCTP and syn-8-oxoG:dATP ternary crystal structures. A. Superimposed palm domains showing that 8-oxoG flips from anti to a syn conformation resulting in a shift of A567 and G568 into the minor groove of the primer-template by ~ 0.6 Å. The Y567A:anti-8-oxoG:dCTP backbone is shown in orange, and the Y567A:syn-8-oxoG:dATP backbone in yellow. B. Schematic illustration of the spatial interaction between G568 and the nascent base-pair as shown in (A).
DISCUSSION
It has been estimated that reactive oxygen species (ROS), e.g. •OH, produce 100 – 400 8-oxoG residues per day per mammalian cell (5). The presence of 8-oxoG residues in DNA has been implicated in numerous diseases, including cancer (35, 36), presumably because of its dual coding potential that leads to G –> T tranversions. To help counter the dire effects of ROS mediated mutations, it is important to understand how human DNA polymerases insert dAMP opposite 8-oxoG. Because of the similarity between human DNA pols α and δ and RB69pol, the latter can serve as a good model for the human pols.
Several reports have proposed that the two hydrogen bonds formed between dATP and syn-8-oxoG (Fig. 1) are partially responsible for its efficient utilization (4, 6, 37). Crystal structures of dATP:syn-8-oxoG base-pairs within a DNA duplex show that they are compatible with B-form DNA, and their Tm values confirm that DNA duplexes containing these base-pairs are at least as stable as those that do not (38–40). However, residues within the NBP of RB69pol, such as G568, can alter the stability of this base-pair, which could affect insertion of dAMP opposite 8-oxoG (8, 12, 15). Based on a comparison of the structures of a wt RB69pol:P/T binary complex containing an abasic site, and a ternary complex containing a dCTP:anti-8-oxoG base-pair, it was argued that G568 becomes “strained” upon binding of a template base into the polymerase NBP because the templating base would have to protrude into the minor groove of the DNA substrate by 1.9 Å (15). To explain the lower efficiency of dAMP insertion opposite 8-oxoG by wt RB69pol, it was proposed that syn-8-oxoG would cause G568 to become even more “strained” by forcing it further into the minor groove of the DNA (15). Because the Y567A mutant increases misinsertion (41), Kisker et al. (15) speculated that the Y567A substitution would relieve the strain imposed on G568 by providing some “wiggle room” in the NBP. The superimposed structures of wt and the Y567A mutant complexed with dCTP:anti-8-oxoG supports this idea by showing that; (i) the Y567A substitution causes G568 to shift by ~ 0.7 Å toward the space formerly occupied by the phenyl ring of Y567 (Fig. 8) and; (ii) that the efficiency of dCMP insertion opposite 8-oxoG (k pol/K d,app) increases 5-fold compared to wt (Table 2).
Comparison of the Y567A:anti-8-oxoG:dCTP structure with the Y567A:syn-8-oxoG:dATP structure shows that G568 recedes into the DNA minor-groove by ~ 0.6 Å (Fig. 9), suggesting that the Y567A mutant provides G568 enough flexibility so that syn-8-oxoG can be easily accommodated. However, if G568 “strains” when packed against syn-8-oxoG, we would have expected the relative efficiency of dAMP versus dCMP insertion opposite 8-oxoG to decrease. Since this did not occur, G568 may not be “strained” after all. In fact, relative to the wt, the Cα-H of G568 is closer to O-8 of syn-8-oxoG (3.1 Å) (Fig. 7B ) in the Y567A mutant than to the N-3 atom of anti-8-oxoG (3.4 Å) (Fig. 7A ). These findings, and our results with nucleotide analogs (vide supra), suggests that G568 provides additional stability to a nascent base-pair, containing a templating 8-oxoG, by G568 donating its Cα-H to O-8 of syn-8-oxoG, or to N-3 of anti-8-oxoG. Previous studies with transmembrane proteins have shown that hydrogen bonds can form between Cα hydrogens of glycine and backbone carbonyl oxygens, i.e., Cα-H•••O=C, stabilizing helical structures that transverse the lipid bilayer (34). These H-bonds provide about 3 kcals/mole if they are at an inter-atom distance of 3.0 Å, which is approximately the distance between syn-8-oxoG and G568 (Fig. 7B ). This is half the strength of an N•••H-O hydrogen bond when it is in a hydrophobic environment like the NBP (42, 43). The finding that the purine analog 3DA (where N-3 is replaced with C-H, and thus cannot interact with G568) caused a significant reduction in dTMP insertion opposite 8-oxoG supports the notion that a Cα-H•••N bond helps to stabilize this Pu:Py base-pair as well (Fig. 5B ). Because this latter finding also applies to Watson-Crick base-pairs, the stability of a nascent base-pair may not be fully optimized by forming inter-base hydrogen bonds and/or by fitting snugly inside the NBP.
The finding that the Y567A substitution did not appreciably expand the NBP pocket in the vicinity of templating 8-oxoG in a manner that would relieve a steric clash until 8-oxoG flipped into a syn conformation (compare the schematics of Fig. 8B and Fig. 9B ), suggests that the Y567A mutant allows efficient dAMP insertion opposite 8-oxoG by allowing G568 to have more flexibility (Fig. 8A ). Our previous analysis of RB69pol NBP mutants demonstrated that a larger NBP volume did not affect the rates of correct dNMP insertion, but did give higher rates of misinsertion (24). The data presented here concurs with this, by demonstrating that an expansion of the NBP leads to a higher efficiency of dAMP insertion opposite 8-oxoG, without appreciably altering the kinetic parameters of dCMP insertion opposite 8-oxoG. However, because dAMP is inserted opposite 8-oxoG more efficiently by the Y567A mutant than wt, because of a more flexible NBP, we argue that misinsertion occurs not only when steric clashes are removed, but also when NBP residues fail to provide the interactions necessary to keep the pocket rigid.
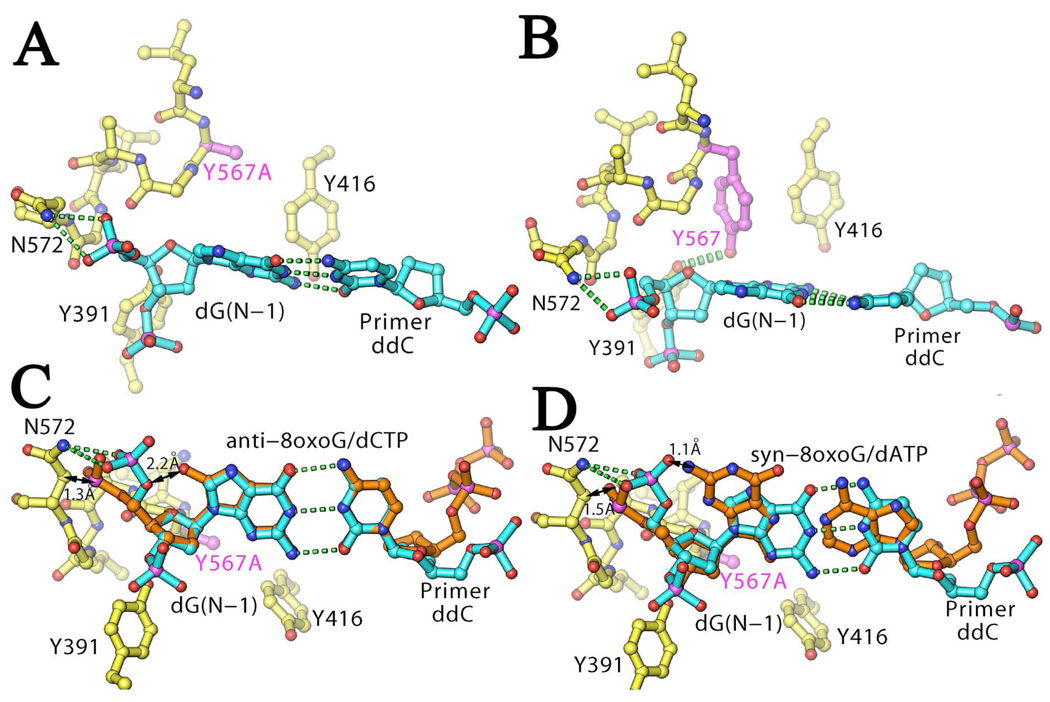
Once inserted, a terminal base-pair containing a templating 8-oxoG dramatically reduces primer extension by the next correct incoming dNTP. Interestingly, bypass of a C:8-oxoG base-pair by wt RB69pol occurs just as efficiently as the bypass of A:8-oxoG (Table 4), in contrast to what has been reported for all other DNA pols which more readily bypass a terminal A:8-oxoG pair (e.g. (9, 14, 44)) except for pol η (45). Superposition of the nascent base-pairs of our RB69pol Y567A:anti-8-oxoG:dCTP or Y567A:syn-8-oxoG:dATP structures onto the terminal dG:dC base-pair from the structure of the first RB69pol ternary complex determined (20) (Fig. 10) provides new insights into the post-chemistry translocation event and primer-extension beyond the 8-oxoG base-pair, namely that when the terminal 8-oxoG base-pair fits into the cavity of wt RB69pol (within the active site), regardless of its conformation (syn8-oxoG or anti-8-oxoG), 8-oxoG clashes into its own 5’-bridging phosphate, or this phosphate clashes into the main chain backbone (N572, which itself donates two hydrogen bonds to the phosphate backbone) to avoid a steric clash with 8-oxoG. Specifically, if the 5’-bridging phosphate assumes the canonical conformation of the templating nucleotide residue (shown in blue, Fig. 10C,D ), there would be a severe steric clash with O-8 of anti-8-oxoG or N-2 of syn-8-oxoG (Fig. 10C,D ). Thus, translocation must be coupled with a reorientation of the phosphate backbone of the templating nucleotide residues, and this explains why insertion of the next dNMP proceeds very slowly (Table 4). The modeling also suggests that the width of the binding pocket for the (N-1) base-pair is too small for the 8-oxoG base-pair to fit into the space provided by RB69pol. However, it appears that the 3’-OH of the new priming nucleotide residue is still in optimal alignment to facilitate rapid catalysis, suggesting that there are likely some differences between our modeling results and what occurs on the phosphoryl transfer pathway. We are in the process of obtaining the relevant crystal structures to identify these differences.
Figure 10.
Computer modeling of wt and the Y567A mutant ternary complex with the observed dATP:8-oxoG or dCTP:8oxoG translocating from the nascent base-pairing position to the product position. A. Structure showing the terminal template base G (dG(N-1)) opposite the dideoxy-terminated nucleotide dC in the wt RB69pol ternary complex structure with Y567A modeled into the structure (PDB entry 1IG9). The mutation site is shown in magenta. Hydrogen bonds are shown in green dotted lines. B. Corresponding wt structure showing that Y567 and Y391 can form a hydrogen bond. C. Superposition of the anti-8-oxoG:dCTP base-pair onto the dG(N-1):dC base-pair. Black arrows illustrate which interactions are unfavorable. D. Superposition of the syn-8-oxoG:dATP base-pair onto the dG(N-1):dC base-pair. Black arrows illustrate which interactions are unfavorable.
As with the wt, the precise structural features that allow the Y567A mutant to increase extension of a primer beyond a terminal 8-oxoG base-pair in the duplex cannot be identified without the relevant ternary crystal structures. However, based on what is known about the kinetic and structural consequences regarding the flexibility that occurs with G568 in the Y567A mutant, we can anticipate that a similar situation might occur with respect to the binding pocket of the terminal base-pair in the DNA duplex. Because the hydrogen bond between Y567 and Y391 in the wt likely provides rigidity for the NBP (Fig. 10B ), its absence should increase the flexibility of the local structures supporting the terminal base-pair of the P/T, thus making it possible to accommodate terminal dN:8-oxoG base-pairs. In addition, the increased flexibility due to the Y567A substitution may allow the base-pair containing 8-oxoG to recede further into the minor groove.
Despite the considerable sequence similarity between pols of the B family (46), the efficiency of dAMP and dCMP insertion opposite 8-oxoG varies considerably (13–15). However, conserved NBP residues, namely Y567 (using the RB69pol numbering scheme), would be expected to have similar roles in modulating dNMP insertion by these DNA pols, as well as by DNA pols from other families. Y567 is part of the highly conserved DNA polymerase motif B (47), and previous studies have used modeling of the pre-insertion ternary complexes of DNA polymerases which have an anti-dCTP:anti-8-oxoG nascent base-pair to show that the tyrosine residues corresponding to Y567 of RB69pol, namely Y390 of φ29pol, Y530 of T7 DNApol, Y955 of human pol γ, and Y271 of human pol β, likely “block” the stable formation of an A:8-oxoG base-pair either by preventing 8-oxoG or the incoming adenine base from flipping into the syn conformation (11, 16–19). As expected, substitution of conserved Tyr with residues having smaller side chains, i.e., Y390S in φ29pol and Y955C in human pol γ (16, 19), led to significant loss of discrimination against dAMP insertion relative to dCMP opposite 8-oxoG, but these losses were primarily due to a reduction in the insertion efficiency of dCMP, a result in contrast with that of the Y567A mutant of RB69pol (Table 2). These results suggest that RB69pol uses the conserved Y567 residue to discriminate against dAMP insertion opposite 8-oxoG in a different way than other DNA polymerases, even with DNA polymerases from the same family (e.g., φ29pol). However, we cannot rule out the possibility that the substitution of a Tyr with different residues (Ser, Cys, and Ala) would contribute significantly to these very different kinetic outcomes. Studies are underway to determine the effect of different Y567 substitutions on the fidelity of the 8-oxoG bypass.
In summary, the data presented here show that the role(s) of each conserved NBP residue in enhancing or reducing dNMP insertion efficiencies is largely influenced by nearby residues in the NBP. In addition, interactions that are not usually considered to be important, i.e., Cα-H•••O=C or Cα-H•••N hydrogen bonds, can stabilize nascent base-pairs in a fashion that leads to rapid phosphoryl transfer. This underscores the need to better understand how NBP residues interact with nascent base-pairs as well as with each other.
ABBREVIATIONS
- 8-OxoG
7,8-dihydroxy-8-oxoguanine
- pols
DNA polymerases
- wt
wild-type RB69pol
- RB69pol
bacteriophage RB69 DNA polymerase
- NBP
nascent base-pair binding pocket
- 3DA
3-deazaadenine
- dAP
2-aminopurine
- I
hypoxanthine
- Pu
purine
- Py
pyrimidine
- Cα-H
methylene hydrogen atom of Gly
- dNMP
deoxynucleoside monophosphate
- dNTP
deoxynucleoside triphosphate
- kobs
observed rate constant
- kpol
maximum rate of dNMP insertion
- Kd,app
[dNTP] that supports the half-maximal rate of dNMP insertion
- PCNA
Proliferating Cell Nuclear Antigen
- T7
DNA polymerase from T7 bacteriophage
- KF
klenow fragment of Escherichia coli DNA polymerase I
- pol II
E. coli DNA polymerase II
Footnotes
Author Contributions: J.B. determined the kinetic parameters. M.W. crystallized and determined the structures of RB69pol:8-oxoG ternary complexes. G.B. collected data at APS. J.B., J.W. and W.H.K. helped to interpret the data, designed experiments and contributed to writing the paper.
This work was supported by USPHS Grant GM063276-05.
REFERENCES
- 1.Roberts JK, Kunkel TA, editors. DNA replication in eukaryotic cells: concepts, enzymes and systems. Plainview, NY: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- 2.Kunkel TA, Erie DA. DNA mismatch repair. Annu. Rev. Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 3.Marnett LJ. Oxyradicals and DNA damage. Carcinogenesis. 2000;21:361–370. doi: 10.1093/carcin/21.3.361. [DOI] [PubMed] [Google Scholar]
- 4.Hainaut P, Hernandez T, Robinson A, Rodriguez-Tome P, Flores T, Hollstein M, Harris CC, Montesano R. IARC Database of p53 gene mutations in human tumors and cell lines: updated compilation, revised formats and new visualisation tools. Nucleic Acids Res. 1998;26:205–213. doi: 10.1093/nar/26.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 6.Shibutani S, Takeshita M, Grollman AP. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 7.Sekiguchi M, Tsuzuki T. Oxidative nucleotide damage: consequences and prevention. Oncogene. 2002;21:8895–8904. doi: 10.1038/sj.onc.1206023. [DOI] [PubMed] [Google Scholar]
- 8.Hsu GW, Ober M, Carell T, Beese LS. Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase. Nature. 2004;431:217–221. doi: 10.1038/nature02908. [DOI] [PubMed] [Google Scholar]
- 9.Lowe LG, Guengerich FP. Steady-state and pre-steady-state kinetic analysis of dNTP insertion opposite 8-oxo-7,8-dihydroguanine by Escherichia coli polymerases I exo- and II exo. Biochemistry. 1996;35:9840–9849. doi: 10.1021/bi960485x. [DOI] [PubMed] [Google Scholar]
- 10.Furge LL, Guengerich FP. Analysis of nucleotide insertion and extension at 8-oxo-7,8-dihydroguanine by replicative T7 polymerase exo- and human immunodeficiency virus-1 reverse transcriptase using steady-state and pre-steady-state kinetics. Biochemistry. 1997;36:6475–6487. doi: 10.1021/bi9627267. [DOI] [PubMed] [Google Scholar]
- 11.Brieba LG, Eichman BF, Kokoska RJ, Doublie S, Kunkel TA, Ellenberger T. Structural basis for the dual coding potential of 8-oxoguanosine by a high-fidelity DNA polymerase. EMBO J. 2004;23:3452–3461. doi: 10.1038/sj.emboj.7600354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brieba LG, Kokoska RJ, Bebenek K, Kunkel TA, Ellenberger T. A lysine residue in the fingers subdomain of T7 DNA polymerase modulates the miscoding potential of 8-oxo-7,8-dihydroguanosine. Structure. 2005;13:1653–1659. doi: 10.1016/j.str.2005.07.020. [DOI] [PubMed] [Google Scholar]
- 13.Patro JN, Urban M, Kuchta RD. Interaction of human DNA polymerase α and DNA polymerase I from Bacillus stearothermophilus with hypoxanthine and 8-oxoguanine nucleotides. Biochemistry. 2009;48:8271–8278. doi: 10.1021/bi900777s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Einolf HJ, Guengerich FP. Fidelity of nucleotide insertion at 8-oxo-7,8-dihydroguanine by mammalian DNA polymerase δ. Steady-state and pre-steady-state kinetic analysis. J. Biol. Chem. 2001;276:3764–3771. doi: 10.1074/jbc.M006696200. [DOI] [PubMed] [Google Scholar]
- 15.Freisinger E, Grollman AP, Miller H, Kisker C. Lesion (in)tolerance reveals insights into DNA replication fidelity. EMBO J. 2004;23:1494–1505. doi: 10.1038/sj.emboj.7600158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Vega M, Salas M. A highly conserved Tyrosine residue of family B DNA polymerases contributes to dictate translesion synthesis past 8-oxo-7,8-dihydro-2'-deoxyguanosine. Nucleic Acids Res. 2007;35:5096–5107. doi: 10.1093/nar/gkm545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Schlick T. Distinct energetics and closing pathways for DNA polymerase β with 8-oxoG template and different incoming nucleotides. BMC structural biology. 2007;7:7. doi: 10.1186/1472-6807-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krahn JM, Beard WA, Miller H, Grollman AP, Wilson SH. Structure of DNA polymerase β with the mutagenic DNA lesion 8-oxodeoxyguanine reveals structural insights into its coding potential. Structure. 2003;11:121–127. doi: 10.1016/s0969-2126(02)00930-9. [DOI] [PubMed] [Google Scholar]
- 19.Graziewicz MA, Bienstock RJ, Copeland WC. The DNA polymerase γ Y955C disease variant associated with PEO and parkinsonism mediates the incorporation and translesion synthesis opposite 7,8-dihydro-8-oxo-2'-deoxyguanosine. Hum. Mol. Genet. 2007;16:2729–2739. doi: 10.1093/hmg/ddm227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Franklin MC, Wang J, Steitz TA. Structure of the replicating complex of a pol α family DNA polymerase. Cell. 2001;105:657–667. doi: 10.1016/s0092-8674(01)00367-1. [DOI] [PubMed] [Google Scholar]
- 21.Zhang H, Rhee C, Bebenek A, Drake JW, Wang J, Konigsberg W. The L561A substitution in the nascent base-pair binding pocket of RB69 DNA polymerase reduces base discrimination. Biochemistry. 2006;45:2211–2220. doi: 10.1021/bi052099y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuchta RD, Mizrahi V, Benkovic PA, Johnson KA, Benkovic SJ. Kinetic mechanism of DNA polymerase I (Klenow) Biochemistry. 1987;26:8410–8417. doi: 10.1021/bi00399a057. [DOI] [PubMed] [Google Scholar]
- 23.Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 24.Zhang H, Beckman J, Wang J, Konigsberg W. RB69 DNA polymerase mutants with expanded nascent base-pair-binding pockets are highly efficient but have reduced base selectivity. Biochemistry. 2009;48:6940–6950. doi: 10.1021/bi900422b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanes JW, Thai DM, Johnson KA. Incorporation and replication of 8-oxo-deoxyguanosine by the human mitochondrial DNA polymerase. J. Biol. Chem. 2006;281:36241–36248. doi: 10.1074/jbc.M607965200. [DOI] [PubMed] [Google Scholar]
- 26.Hanes JW, Johnson KA. A novel mechanism of selectivity against AZT by the human mitochondrial DNA polymerase. Nucleic. Acids. Res. 2007;35:6973–6983. doi: 10.1093/nar/gkm695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter CW, Sweet RM, editors. Methods Enzymol. 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 28.Navaza J. AMoRE: an automated package for molecular replacement. Acta Cryst. 1994;A50:157–163. [Google Scholar]
- 29.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Cryst. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 30.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 31.Carson M. Ribbons 2.0. J. Appl. Cryst. 1991;24:958–961. [Google Scholar]
- 32.Bertram JG, Oertell K, Petruska J, Goodman MF. DNA polymerase fidelity: comparing direct competition of right and wrong dNTP substrates with steady state and pre-steady state kinetics. Biochemistry. 2010;49:20–28. doi: 10.1021/bi901653g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsai YC, Johnson KA. A new paradigm for DNA polymerase specificity. Biochemistry. 2006;45:9675–9687. doi: 10.1021/bi060993z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Senes A, Ubarretxena-Belandia I, Engelman DM. The Cα -H---O hydrogen bond: a determinant of stability and specificity in transmembrane helix interactions. Proc. Natl. Acad. Sci. U. S. A. 2001;98:9056–9061. doi: 10.1073/pnas.161280798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malins DC, Haimanot R. Major alterations in the nucleotide structure of DNA in cancer of the female breast. Cancer Res. 1991;51:5430–5432. [PubMed] [Google Scholar]
- 36.Degan P, Shigenaga MK, Park EM, Alperin PE, Ames BN. Immunoaffinity isolation of urinary 8-hydroxy-2‧-deoxyguanosine and 8-hydroxyguanine and quantitation of 8-hydroxy-2‧-deoxyguanosine in DNA by polyclonal antibodies. Carcinogenesis. 1991;12:865–871. doi: 10.1093/carcin/12.5.865. [DOI] [PubMed] [Google Scholar]
- 37.David SS, O‧Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McAuley-Hecht KE, Leonard GA, Gibson NJ, Thomson JB, Watson WP, Hunter WN, Brown T. Crystal structure of a DNA duplex containing 8-hydroxydeoxyguanine-adenine base pairs. Biochemistry. 1994;33:10266–10270. doi: 10.1021/bi00200a006. [DOI] [PubMed] [Google Scholar]
- 39.Kouchakdjian M, Bodepudi V, Shibutani S, Eisenberg M, Johnson F, Grollman AP, Patel DJ. NMR structural studies of the ionizing radiation adduct 7-hydro-8-oxodeoxyguanosine (8-oxo-7H-;dG) opposite deoxyadenosine in a DNA duplex. 8-Oxo-7H–dG(syn).dA(anti) alignment at lesion site. Biochemistry. 1991;30:1403–1412. doi: 10.1021/bi00219a034. [DOI] [PubMed] [Google Scholar]
- 40.Lipscomb LA, Peek ME, Morningstar ML, Verghis SM, Miller EM, Rich A, Essigmann JM, Williams LD. X-ray structure of a DNA decamer containing 7,8-dihydro-8-oxoguanine. Proc. Natl. Acad. Sci. U. S. A. 1995;92:719–723. doi: 10.1073/pnas.92.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang G, Wang J, Konigsberg W. Base selectivity is impaired by mutants that perturb hydrogen bonding networks in the RB69 DNA polymerase active site. Biochemistry. 2005;44:3338–3346. doi: 10.1021/bi047921x. [DOI] [PubMed] [Google Scholar]
- 42.Vargas R, Garza J, Dixon DA, Hay BP. How strong is the Cα-H---O=C Hydrogen Bond? J. Am. Chem. Soc. 2000;122:4750–4755. [Google Scholar]
- 43.Scheiner S, Kar T, Gu Y. Strength of the Cα-H---O hydrogen bond of amino acid residues. J. Biol. Chem. 2001;276:9832–9837. doi: 10.1074/jbc.M010770200. [DOI] [PubMed] [Google Scholar]
- 44.Zang H, Irimia A, Choi JY, Angel KC, Loukachevitch LV, Egli M, Guengerich FP. Efficient and high fidelity incorporation of dCTP opposite 7,8-dihydro-8-oxodeoxyguanosine by Sulfolobus solfataricus DNA polymerase Dpo4. J. Biol. Chem. 2006;281:2358–2372. doi: 10.1074/jbc.M510889200. [DOI] [PubMed] [Google Scholar]
- 45.Haracska L, Yu SL, Johnson RE, Prakash L, Prakash S. Efficient and accurate replication in the presence of 7,8-dihydro-8-oxoguanine by DNA polymerase η. Nat. Genet. 2000;25:458–461. doi: 10.1038/78169. [DOI] [PubMed] [Google Scholar]
- 46.Wang TS, Wong SW, Korn D. Human DNA polymerase α: predicted functional domains and relationships with viral DNA polymerases. FASEB J. 1989;3:14–21. doi: 10.1096/fasebj.3.1.2642867. [DOI] [PubMed] [Google Scholar]
- 47.Saturno J, Lazaro JM, Esteban FJ, Blanco L, Salas M. φ29 DNA polymerase residue Lys383, invariant at motif B of DNA-dependent polymerases, is involved in dNTP binding. J. Mol. Biol. 1997;269:313–325. doi: 10.1006/jmbi.1997.1053. [DOI] [PubMed] [Google Scholar]