Abstract
Arbuscular mycorrhizal (AM) symbioses are mutualistic associations between soil fungi and most vascular plants. The symbiosis significantly affects the host physiology in terms of nutrition and stress resistance. Despite the lack of host range specificity of the interaction, functional diversity between AM fungal species exists. The interaction is finely regulated according to plant and fungal characters, and plant hormones are believed to orchestrate the modifications in the host plant. Using tomato as a model, an integrative analysis of the host response to different mycorrhizal fungi was performed combining multiple hormone determination and transcriptional profiling. Analysis of ethylene-, abscisic acid-, salicylic acid-, and jasmonate-related compounds evidenced common and divergent responses of tomato roots to Glomus mosseae and Glomus intraradices, two fungi differing in their colonization abilities and impact on the host. Both hormonal and transcriptional analyses revealed, among others, regulation of the oxylipin pathway during the AM symbiosis and point to a key regulatory role for jasmonates. In addition, the results suggest that specific responses to particular fungi underlie the differential impact of individual AM fungi on plant physiology, and particularly on its ability to cope with biotic stresses.
Keywords: Arbuscular mycorrhiza, hormones, jasmonates, LC-MS/MS analysis, microarrays, oxylipins
Introduction
About 80% of all terrestrial plants, including most agricultural and horticultural crop species, are able to establish mutualistic associations with soil fungi from the phylum Glomeromycota (Smith and Read, 2008). The resulting symbiosis is known as arbuscular mycorrhiza (AM) and is widely distributed throughout the world. This association is considered to be older than 400 million years and a key step in the evolution of terrestrial plants (Smith and Read, 2008). Arbuscular mycorrhizal fungi (AMF) are obligate biotrophs that colonize the root cortex of the host plant inter- and intracellularly forming specialized and highly branched structures called arbuscules (Parniske, 2008). The fungus obtains carbohydrates from the host plant and, in return, they assist the plant in the acquisition of mineral nutrients (mainly phosphorus) and water. AM symbiosis is maintained throughout the life of the plant, affecting its physiology significantly (Parniske, 2008). Accordingly, the symbiosis not only influences plant nutrition, but also impacts the plant's ability to overcome biotic and abiotic stresses (Pozo and Azcón-Aguilar, 2007). While AMF are considered non-specific with respect to host range, there are differences in their growth patterns within the roots, functionality, and with regards to their effects on plant nutrition and resistance to stress (Cavagnaro et al., 2001; Pozo et al., 2002; Smith et al., 2004).
AM establishment and functioning require a high degree of coordination between the two partners based on a finely regulated molecular dialogue that orchestrates complex symbiotic programmes (Paszkowski, 2006; Hause et al., 2007; Requena et al., 2007). Upon recognition of the fungal partner, the plant actively accommodates the fungus in the root tissue (Genre et al., 2008) and controls its proliferation, which implies an important transcriptional reprogramming in the plant. Understanding the molecular basis of the AM symbiosis is an ongoing challenge, and, so far, some plant genes specifically associated with the establishment and development of the symbiosis have been identified in different plant species, mostly in legumes (Grunwald et al., 2004; Hohnjec et al., 2005; Liu et al., 2007; Siciliano et al., 2007; Guether et al., 2009). Some of the changes in the host are related to modifications in the relative abundance of plant hormones, most of which are thought to play a role in the symbiosis (Hause et al., 2007). Among plant hormones, ethylene (ET), salicylic acid (SA), abscisic acid (ABA), and jasmonic acid (JA) are known to be key elements in fine-tuning the plant defence response during interaction with other organisms (Pieterse et al., 2009). In the case of the interaction with AMF, it is accepted that there is an inverse correlation between root colonization and the levels of ET and SA (Blilou et al., 1999; Herrera-Medina et al., 2003, 2007; Riedel et al., 2008). Conversely, a positive correlation for ABA and mycorrhizal establishment has been evidenced (Herrera-Medina et al., 2007). JA and its derivatives, known as jasmonates, have received special attention since they are believed to play a major role in the AM symbiosis. However, experimental data are highly controversial (Gutjahr and Paszkowski, 2009; Hause and Schaarschmidt, 2009). Increased JA levels in mycorrhizal roots compared with non-mycorrhizal controls have been described in Medicago truncatula (Hause et al., 2002; Meixner et al., 2005), while they remained unaltered in Nicotiana attenuata (Riedel et al., 2008). In addition, studies using reverse genetics approaches with plant mutants affected in JA biosynthesis or signalling have shown positive and negative regulatory roles of the JA pathway in the symbiosis (Isayenkov et al., 2005; Herrera-Medina et al., 2008; Tejeda-Sartorius et al., 2008). Besides the use of different plant and experimental systems, these controversies might be partly due to the overlapping yet distinct signalling activities of its precursor oxo-phytodienoic acid (OPDA) and jasmonate derivatives such as the isoleucine conjugate JA–Ile (Stintzi et al., 2001; Taki et al., 2005; Wang et al., 2008). Moreover, jasmonates belong to a diverse class of lipid metabolites known as oxylipins that include other biologically active molecules (Wasternack, 2007; Mosblech et al., 2009).
In the present study, the agriculturally and economical important crop tomato has been used as a model system to carry out integrative analysis of the transcriptional and metabolic changes that take place during AM symbiosis. The plant response to two related AMF, Glomus mosseae and Glomus intraradices, that showed in previous studies different colonization patterns and functionality, was compared in an attempt to provide insights into the common and differential host responses to AMF. The effects of AMF colonization on the content of ABA, SA, ET, and different JA-related compounds in the host plant were assessed, and correlated with the modifications in their transcriptional profiles. The results provide original insights into our understanding of the AM symbiosis and its impact on plant physiology, and they pave the way for further analyses of the regulatory network controlling this association.
Materials and methods
Plant growth, AM inoculation, and chemical treatments
The AMF G. mosseae (BEG 12) and G. intraradices (BEG 121) were maintained as a soil–sand-based inoculum. Tomato seeds (Solanum lycopersicum L. cv. MoneyMaker) were surface sterilized in 4% sodium hypochlorite containing 0.02% (v/v) Tween-20, rinsed thoroughly with sterile water and germinated for 3 d in a container with sterile vermiculite at 25 °C in darkness. Subsequently, individual seedlings were transferred to 0.25 l pots with a sterile sand:soil (4:1) mixture. Pots were inoculated by adding 10% (v:v) G. mosseae or G. intraradices inoculum. The same amount of soil:sand mix but free from AMF was added to control plants. All plants received an aliquot of a filtrate (<20 μm) of both AM inocula to homogenize the microbial populations. For each treatment, a total of nine plants were used. Plants were randomly distributed and grown in a greenhouse at 24/16 °C with a 16/8 h photoperiod and 70% humidity, and watered three times a week with Long Ashton nutrient solution (Hewitt, 1966) containing 25% of the standard phosphorus concentration. Plants were harvested after 9 weeks of growth, and the fresh weight of shoots and roots was determined. An aliquot of each individual root system was reserved for mycorrhizal quantification. For microarray and hormone analyses, three pools each consisting of roots from three independent plants were used.
For methyl jasmonate (MeJA) treatment tomato plants were grown hydroponically in 3.0 l plastic containers with Long Ashton nutrient solution containing 25% of the standard phosphorus concentration and with constant aeration. The nutrient solution was replaced once a week. Four-week-old plants were individually transferred to 50 ml plastic tubes filled with nutrient solution with or without 50 μM MeJA (Sigma-Aldrich) and maintained for 24 h. Then, the roots were rinsed with sterilized deionized water and stored at –80 °C until use.
Mycorrhizal colonization determination
Roots were stained with trypan blue (Phillips and Hayman, 1970) and examined using a Nikon Eclipse 50i microscope and bright-field conditions. The percentage of total root colonization and frequency of intraradical fungal structures, arbuscules, and vesicles was determined by the gridline intersection method (Giovannetti and Mosse, 1980).
Phosphorus content
The total phosphorus content of the leaves was measured at the CEBAS-CSIC (Spain). Shoots were briefly rinsed with deionized water and mature leaves were oven-dried at 60 °C for 72 h, weighed, and ground to a fine powder. Then, samples were extracted with deionized water. Tissue phosphorus concentrations were determined by inductively coupled plasma optical emission spectrometry (ICP-OES) (Iris Intrepid II, Thermo Electron Corporation) after acid digestion. Three biological replicates each consisting of a pool of leaves from three independent plants were measured for each treatment.
Hormone quantification
OPDA, JA, JA–Ile, ABA, and SA were analysed by ultraperformance liquid chromatography coupled to mass spectrometry (UPLC-MS) as described by Flors et al. (2008). A 50 mg aliquot of dry tissue was used per sample. A mixture of internal standards containing 100 ng of [2H6]ABA, 100 ng of dihydrojasmonic acid, 100 ng of prostaglandin B1, and 100 ng of [2H5]SA was added to each sample prior to extraction. Individual calibration curves for each tested compound and internal standard were performed before the analysis. The tissue was immediately homogenized in 2.5 ml of ultra pure water and centrifuged at 5000 g for 40 min. Then, the supernatant was acidified and partitioned against diethyl-ether, dried, and resuspended in 1 ml of water/methanol (90:10, v/v). A 20 μl aliquot of this solution was injected into a Waters Acquity UPLC system (Waters). The UPLC was interfaced into a triple quadrupole tandem mass spectrometer (TQD, Waters). LC separation was performed using an Acquity UPLC BEH C18 analytical column (Waters) at a flow rate of 300 μl min−1. Quantifications were carried out with MassLynx 4.1 software (Waters) using the internal standards as a reference for extraction recovery and the standard curves as quantifiers.
Ethylene release was determined by gas chromatography–mass spectrometry (GC-MS). Excised roots were placed on wet filter paper in a 90 mm diameter Petri dish, sealed with sticky tape and a rubber stopper on the top, and incubated for 1 h at room temperature. A 1 ml aliquot of headspace gas per plate was sampled with a syringe, and ET was measured in a Hewlett Packard 5890 gas chromatograph fitted with a flame ionization detector (FID). Analyses were carried out at 65 °C with the injector and FID held at 120 °C and 105 °C, respectively. Five independent replicates per treatment were measured.
RNA isolation
Total RNA was extracted using Tri-Reagent (Sigma-Aldrich) according to the manufacturer's instructions. The RNA was treated with RQ1 DNase (Promega), purified through a silica column using the NucleoSpin RNA Clean-up kit (Macherey-Nagel), and stored at –80 °C until use.
Microarray hybridization and data analysis
The Affymetrix GeneChip Tomato Genome Array (Affymetrix) was used. A 5 μg aliquot of total RNA was used as starting material. cDNA synthesis, cRNA production, and fragmentation were carried out as described in the Expression Analysis Technical Manual (Affymetrix). The GeneChip Arrays were hybridized, stained, washed, and screened according to the manufacturer's protocol at the Unidad de Genómica of the Universidad Complutense de Madrid (http://www.ucm.es/info/gyp/genomica/) (Madrid, Spain). Three biological replicates, each consisting of pools of three independent plants, were used for microarray analysis of roots colonized or not by G. mosseae or G. intraradices. For the MeJA experiment, two biological replicates from MeJA- or mock-treated roots were used. Probe signal summarization, normalization, and background subtraction were performed using the multichip analysis RMA algorithm (Irizarry et al., 2003) in the ‘affy’ package with default parameters. The statistical test for differentially expressed genes was performed using the software ‘Cyber-T’ (Baldi and Long, 2001), which allows a better variance estimation by calculating the moderated t-statistic using empirical Bayesian techniques. Genes were considered as differentially regulated if P <0.01 and the ratio compared with the controls was ≥1.7 or ≤0.6. For the MeJA treatment, genes were considered as differentially regulated if P <0.01 and the ratio compared with the controls was ≥2 or ≤0.5. Updated annotation of the differentially regulated genes was obtained by tblastx against the NCBI nr-database. An E-value <10−15 was required to take into account the blast result.
Microarray data will be deposited in the Tomato Functional Genomics database (http://ted.bti.cornell.edu/cgi-bin/TFGD/array/home.cgi).
Gene expression analysis by real-time quantitative RT-PCR (qPCR)
Real-time qPCR was performed using the iCycler iQ5 system (Bio-Rad) and gene-specific primers (Supplementary Table S1 available at JXB online). The first-strand cDNA was synthesized with 1 μg of purified total RNA using the iScript cDNA Synthesis kit (Bio-Rad) according to the manufacturer's instructions. Three independent biological replicates were analysed per treatment. Relative quantification of specific mRNA levels was performed using the comparative 2–Δ(ΔCt) method (Livak and Schmittgen, 2001). Expression values were normalized using the housekeeping gene SlEF, which encodes for the tomato elongation factor-1α.
Statistical analysis
Data for hormone and phosphorus content and mycorrhization levels of tomato roots were subjected to one-way analysis of variance (ANOVA) using the software SPSS Statistics v. 14.1 for Windows. When appropriate, Fisher's LSD test was applied.
Results
Root colonization by G. mosseae and G. intraradices, and physiological status of the plant
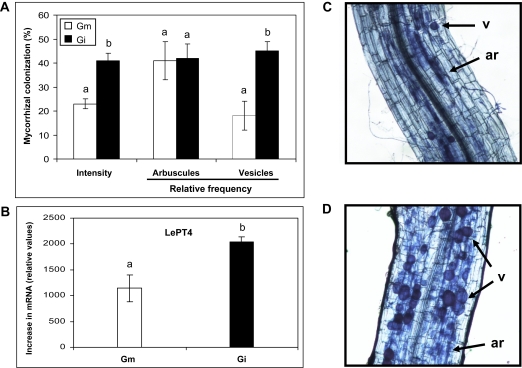
In order to identify plant responses directly related to the AM symbiosis and not to potential nutritional effects, an experimental system that allowed a robust mycorrhizal colonization without significant changes in plant growth and nutrition was used. Nine weeks after inoculation the shoot and root mass and phosphorus (Pi) content of the tomato plants colonized by either G. mosseae or G. intraradices were not significantly different from those in the non-mycorrhizal controls (Supplementary Table S2 at JXB online). The colonization intensities of G. mosseae (23%) and G. intraradices (41%) differed significantly (P <0.01). Moreover, both AMF showed different colonization patterns. They both developed intraradical hyphae, arbuscules, and vesicles, but with different frequencies. Although the relative abundance of arbuscules was similar in both interactions, the proportion of vesicles, fungal reservoir structures, was higher in G. intraradices- but scarce in G. mosseae-colonized roots (Fig. 1A, C, D). Besides the presence of fungal structures, the functionality of the symbiosis was also checked by molecular methods. The tomato gene LePT4 encodes a phosphate transporter specific for the AM symbiosis which is expressed in arbusculated cells and considered a marker for a functional symbiosis (Balestrini et al., 2007). A high LePT4 expression was detected in all mycorrhizal roots, although the levels were almost 2-fold higher in G. intraradices- than in G. mosseae-colonized roots (Fig. 1B), in agreement with the differences observed in root colonization (absolute arbuscule abundance).
Fig. 1.
Mycorrhizal colonization and expression analysis of the marker gene LePT4 of tomato roots inoculated with G. mosseae or G. intraradices. (A) Intensity of mycorrhizal colonization by G. mosseae (Gm) or G. intraradices (Gi), and relative frequency of arbuscules and vesicles. (B) Gene expression analysis by real-time qPCR for the mycorrhizal marker gene LePT4. Data points represent the means of five (A) or three (B) replicates (±SE). Data not sharing a letter in common differ significantly (P <0.01) according to Fisher's LSD test. The right-hand panels show photographs of root samples after trypan blue staining. (C) Glomus mosseae colonizes the root cortex to a lower extent, forming a large number of arbuscules but a limited number of vesicles. (D) Glomus intraradices extensively colonize the root cortex forming arbuscules and a large number of vesicles. Arrows indicate arbuscules (ar) and vesicles (v).
Impact of root colonization by G. mosseae or G. intraradices on defence-related hormones
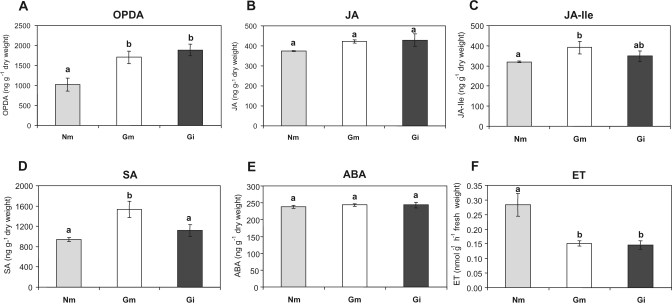
To investigate the responses of tomato plants to AMF and evaluate the impact of the interaction on hormone homeostasis, the levels of JA, ABA, SA, and ET in tomato roots colonized by the two fungi were determined. Analysis by UPLC-coupled tandem mass spectrometry (LC-MS/MS) allowed the simultaneous quantification of free JA, the JA precursor OPDA, JA–Ile, ABA, and SA from each sample. The levels of OPDA were significantly (P <0.05) higher in roots colonized by both AMF (Fig. 2A). In contrast, JA levels were not altered in mycorrhizal roots (Fig. 2B). Unlike free JA, the levels of its bioactive derivative JA–Ile were higher in G. mosseae-colonized roots compared with non-mycorrhizal controls, whereas no significant differences were observed upon G. intraradices colonization (Fig. 2C). In spite of this, differences between roots colonized by both fungi were not statistically significant. It was noteworthy that a clear increase on SA content was only detected in G. mosseae-colonized roots (Fig. 2D). Conversely, ABA content did not change upon mycorrhizal colonization by either of the AMF studied (Fig. 2E), but a clear reduction of ET levels in roots colonized by both AMF compared with non-mycorrhizal plants was observed by GC-MS (Fig. 2F).
Fig. 2.
Hormonal content in non-mycorrhizal (Nm) and G. mosseae- (Gm) and G. intraradices- (Gi) colonized roots. Levels of (A) oxo-phytodienoic acid (OPDA), (B) free jasmonic acid (JA), (C) jasmonic acid isoleucine (JA–Ile), (D) salicylic acid (SA), (E) abscisic acid (ABA), and (F) ethylene (ET). Data points represent the means of five replicates (±SE). Data not sharing a letter in common differ significantly (P <0.05) according to Fisher's LSD test.
Changes in plant gene expression during interaction with G. mosseae or G. intraradices
To gain further insight into the plant changes related to the symbiosis, a global gene expression profiling of the roots from plants colonized by G. mosseae or G. intraradices as compared with non-mycorrhizal plants was performed. The Affymetrix Tomato Genome Genechip Array containing >9200 tomato genes was used. In agreement with the similarity in the Pi content of the plants, no differences were observed in the expression of Pi nutrition marker genes present in the array such as the Pi transporter LePT1, acid and purple phosphatases and kinases, and other marker genes for Pi starvation such as the iron deficiency-specific-4 (IDS4) and the tomato Pi starvation-induced (TPSI1). Thus, the transcriptional changes reported in the present study are not expected to be related to differences in Pi nutrition.
As a first approach, the aim was to determine whether the hormonal changes observed in mycorrhizal plants (Fig. 2) correlated with transcriptional regulation of their metabolic genes. For that, the expression of tomato genes present in the array involved in the metabolism of the hormones analysed was scrutinized (Supplementary Table S3 at JXB online). Consistent with the results from the hormonal analysis, only an induction of the genes encoding enzymes related to the metabolism and regulation of jasmonates, and more generally to oxylipins—LOXA, AOS1, AOS3, JAME, and the jasmonate ZIM domain 2 (JAZ2)—was observed in mycorrhizal plants. However, no changes for the genes related to ABA, SA, and ET were observed, except for the gene encoding a 1-aminocyclopropane-1-carboxylic acid oxidase (ACO1) (Supplementary Table S3).
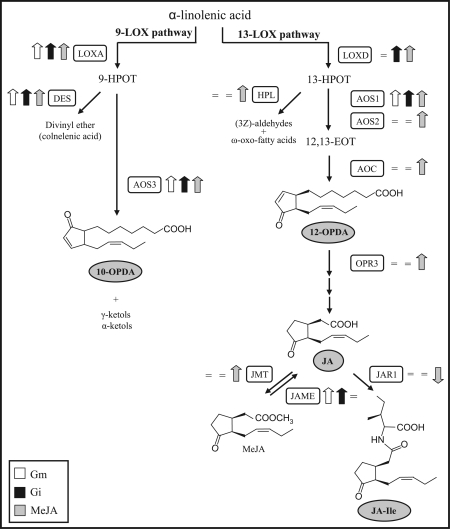
Comparing the global transcriptional profiles, 162 genes were differentially regulated (P <0.01) in the roots upon G. mosseae colonization. Of these, 101 genes (62%) were up-regulated. In G. intraradices-colonized roots the expression of 173 genes differed significantly from non-mycorrhizal roots, of which 103 genes (60%) were up-regulated. When comparing the expression profiles in the two mycorrhizal systems, 59 genes were co-regulated during the interaction with both AMF (Table 1). The overlap was considerably higher for up-regulated genes. Among the co-induced genes, previously described markers of the AM symbiosis were found such as those encoding a chitinase, glutathione S-transferase, β-1,3-glucanase, patatin, β-D-xylosidase, a pathogenesis PR10-like protein, and DXS-2, a key enzyme of the mevalonate-independent pathway of carotenoid biosynthesis (Hohnjec et al., 2005; Liu et al., 2007; Siciliano et al., 2007; Walter et al., 2007; Floss et al., 2008), suggesting a role in the AM interaction conserved across plant species. Besides these markers, the microarray analysis revealed the common induction in mycorrhizal roots of key genes in the biosynthesis of oxylipins. In plants there are two main branches of the oxylipin pathway, determined by two different types of lipoxygenases (LOXs), 9-LOX and 13-LOXs (Fig. 3). The 13-LOX pathway leads to the biosynthesis of JA and derivatives, and, as described above, some genes in this pathway were up-regulated during AM interaction (Fig. 3). In addition, the microarray analysis revealed the induction of genes encoding key enzymes of the 9-LOX branch, LOXA and AOS3. LOXA is involved in the production of lipid 9-hydroperoxides by adding molecular oxygen to either linolenic or linoleic acid at the C-9 position (Ferrie et al., 1994). AOS3 encodes a root-specific and jasmonate-regulated allene oxide synthase that catalyses the biosynthesis of γ- and α-ketols, and 10-OPDA, an isomer of the JA precursor 12-OPDA (Fig. 3) (Itoh et al., 2002; Grechkin et al., 2008). Furthermore, the gene encoding a divinyl ether synthase (DES), also related to the 9-LOX branch and involved in the formation of the divinyl ether fatty acids colnelenic and colnelic (Itoh and Howe, 2001), was also induced by the two AMF (Table 1 and Fig. 3).
Table 1.
Genes regulated in roots colonized by both G. mosseae and G. intraradices compared with non-mycorrhizal roots, and their changes in expression after MeJA treatment
| ID | Annotation | Ratio Gm | Ratio Gi | MeJA |
| BT014524 | Serine protease/subtilisin-like | 28.06 | 36.26 | 0.72 |
| X72729 | Ripening-related protein ERT1b | 25.96 | 38.42 | 1.25 |
| M69248 | Pathogenesis protein PR1b1 | 19.71 | 16.74 | 2.56a |
| AW622368 | Esterase/lipase/thioesterase | 17.59 | 42.64 | 0.11a |
| AI897365 | Putative proteinase inhibitor | 17.34 | 7.42 | 543.59a |
| AI487223 | Anthocyanin acyltransferase | 8.63 | 3.40 | 112.45a |
| AF454634 | Allene oxide synthase 3 (AOS3) | 8.49 | 3.42 | 236.13a |
| AB015675 | Copalyl diphosphate synthase | 7.69 | 10.41 | 1.25 |
| U09026 | Lipoxygenase A (LOXA) | 6.59 | 2.78 | 13.95a |
| AB010991 | 3b-hydroxylase (Le3OH-1) | 5.13 | 9.60 | 0.94 |
| BG631079 | β-1,3-Glucanase | 5.07 | 5.35 | 0.72 |
| BI933750 | 1-Deoxy-D-xylulose 5-phosphate synthase 2 (DXS-2) | 5.00 | 9.92 | 2.65a |
| AB041811 | β-D-Xylosidase (LEXYL1) | 4.72 | 4.49 | 1.00 |
| BG125734 | Calcium/lipid-binding protein | 4.49 | 15.13 | 0.91 |
| BT013355 | Pathogenesis protein PR-P2 | 4.45 | 6.57 | 9.79a |
| BG626023 | Electron carrier (ACD1-like) | 4.12 | 6.88 | 1.10 |
| BI423134 | Germin-like protein | 3.92 | 5.78 | 1.20 |
| BI423255 | Germin-like protein (GLP6) | 3.44 | 4.67 | 1.18 |
| X94946 | Proteinase inhibitor II (Cevi57) | 3.42 | 2.12 | 4.18a |
| BM412305 | EF-hand-containing protein | 3.24 | 4.56 | 0.81 |
| BF114155 | Glutathione S-transferase | 3.20 | 2.76 | 9.19a |
| AF090115 | Heat shock protein HSP17.4 | 3.20 | 7.48 | 1.58 |
| CK720570 | Patatin-like protein | 3.19 | 6.04 | 2.89a |
| AF515615 | Lysine-rich protein (TSB) | 3.18 | 3.67 | 0.47a |
| AI895164 | Fatty acid desaturase (FAD) | 3.05 | 3.12 | 0.46a |
| AJ785041 | Cytochrome P450 CYP81C6v2 | 2.89 | 2.65 | 360.27a |
| BT014484 | Glucosyltransferase | 2.83 | 3.91 | 6.26a |
| BG630947 | β-Galactosidase (TBG5) | 2.79 | 5.27 | 0.55 |
| BT014016 | Cysteine synthase (cs1) | 2.76 | 5.94 | 0.99 |
| BI923212 | Germin-like protein (GLP9) | 2.71 | 3.04 | 0.75 |
| CK716273 | Miraculin-like protein | 2.69 | 2.48 | 2.27a |
| AF049898 | Gibberellin 20-oxidase-1 (20ox-1) | 2.67 | 3.36 | 0.61 |
| AW220405 | Germin-like protein (ger2a) | 2.56 | 2.86 | 0.73 |
| AW626187 | Unkonwn | 2.48 | 2.83 | 0.74 |
| AW034398 | Subtilisin-like protease (sbt4a) | 2.47 | 3.19 | 1.70 |
| U30465 | Class II chitinase (Chi2;1) | 2.41 | 3.01 | 6.34a |
| CN384809 | 1-Aminocyclopropane-1-carboxylic acid oxidase (ACO) | 2.35 | 2.27 | 12.03a |
| AF317515 | Divinyl ether synthase (DES) | 2.29 | 1.96 | 3.55a |
| X79337 | Ribonuclease le | 2.24 | 1.88 | 2.44a |
| BG628191 | Jasmonate ZIM domain 2 (JAZ2) | 2.23 | 2.49 | 14.78a |
| L77963 | Metallothionein II-like protein (MTA) | 2.23 | 2.06 | 0.31a |
| AY455313 | Methylesterase/methyl jasmonate esterase (JAME) | 2.16 | 2.82 | 1.16 |
| AJ271093 | Allene oxide synthase 1 (AOS1) | 2.14 | 2.52 | 23.83a |
| AA824679 | Dihydrolipoamide S-acetyltransferase (LTA2) | 2.00 | 1.96 | 0.51 |
| Y15846 | Pathogenesis protein PR10-like | 1.93 | 2.72 | 0.52 |
| AI899627 | Unknown | 1.92 | 2.47 | 3.89a |
| BT013881 | Cytochrome P450 (CYP721A7v1) | 1.86 | 2.45 | 1.42 |
| BG625959 | Enoyl-[acyl-carrier-protein] reductase | 1.81 | 2.24 | 0.23a |
| AI780669 | Glutathione S-transferase | 0.49 | 0.50 | 7.29a |
| BM411685 | Zinc finger (B-box type) | 0.48 | 0.49 | 1.80 |
| AW649455 | GDSL-motif lipase/hydrolase | 0.48 | 0.54 | 0.33a |
| AF437878 | bHLH transcriptional regulator | 0.44 | 0.32 | 2.29a |
| AW218614 | UDP-glycosyltransferase | 0.43 | 0.36 | 4.85a |
| AW442015 | Cysteine-type endopeptidase | 0.40 | 0.60 | 0.67 |
| BI207994 | Cytochrome P450 CYP72A15 | 0.36 | 0.54 | 4.17a |
| M21775 | Metallocarboxypeptidase inhibitor | 0.36 | 0.35 | 0.82 |
| AI771889 | Cytochrome P450 CYP72A57 | 0.27 | 0.44 | 5.12a |
| BI205190 | UDP-glycosyltransferase | 0.25 | 0.39 | 21.66a |
| BM411019 | Unknown | 0.16 | 0.39 | 1.96 |
Genes significantly up- or down-regulated in roots colonized by G. mosseae (Gm) and G. intraradices (Gi) compared with non-mycorrhizal roots are sorted according to the fold change in expression in G. mosseae-colonized roots. MeJA shows the changes in expression levels of mycorrhiza-regulated genes upon treatment with 50 μM methyl jasmonate compared with mock-treated roots.
Significant (P <0.01) changes in MeJA-treated roots.
Fig. 3.
Induction of the oxylipin biosynthethic pathway in arbuscular mycorrhizal symbiosis. Metabolic scheme of the oxylipin pathway including the 9-LOX and 13-LOX branches (modified after Wasternack, 2007). Shaded boxes show the metabolites analysed by LC-MS/MS in the present study. Thick arrows indicate the direction of the changes in expression levels compared with non-mycorrhizal control roots (up- or down-regulated) of the genes coding for the enzyme cited, as determined by the transcriptomic analysis; = indicates no changes in gene expression. LOX, lipoxygenase; AOS, allene oxide synthase; AOC, allene oxide cyclase; OPR3, oxo-phytodienoic acid reductase; JMT, jasmonic acid carboxyl methyltransferase; JAME, methyl jasmonate esterase; JAR1, jasmonate-amino synthetase; HPL, hydroperoxide lyase; and DES, divinyl ether synthase.
Besides the common genes, there were specific sets of genes regulated only by either G. mosseae or G. intraradices. Among those significantly (P <0.01) induced exclusively in G. intraradices-colonized roots, a group of genes related to the biosynthesis of carotenoids, namely DXR, PSD, ZDS, and CtrL-b, stood out (Supplementary Table S4 and Fig. S1 at JXB online). The production of the carotenoid cleavage products mycorradicin and cyclohexenone derivatives is associated with mycorrhization. Indeed, mycorradicin—known as the ‘yellow pigment’—is responsible for the typical yellow coloration of some mycorrhizal roots and has been correlated with a functional symbiosis (Walter et al., 2007). The first step in this biosynthetic pathway is catalysed by DXS-2, induced transcriptionally during interaction with both fungi but to a higher extent in G. intraradices- than in G. mosseae-colonized roots (Table 1). Although not significant under the present selection criteria, a moderate increase in the expression of DXR, PSD, ZDS, and CtrL-b was also observed in G. mosseae-colonized roots. Thus, the induction of the carotenoid pathway is not exclusive to G. intraradices-colonized roots; more probably it correlates with the root colonization level.
With regards to G. mosseae, a major part of the differentially regulated genes upon mycorrhization were related to defence and wounding responses associated with jasmonates (Supplementary Table S5 at JXB online), including the typical JA marker genes encoding proteinase inhibitors I and II (PinI and II), multicystatin (MC), polyphenol oxidase, and threonine deaminase (Wasternack et al., 2006). Remarkably, despite the strong induction of these genes in G. mosseae-mycorrhizal roots, they remained unaltered in G. intraradices-colonized roots.
To confirm the regulation of the different signalling pathways, the expression of the hormone biosynthetic genes and several response marker genes was analysed by real-time qPCR. In addition, potential markers of AM symbiosis, orthologues of genes previously described as AM induced in other plant species, such as DXS-2, PR-10, and the chitinase Chi2;1, were also checked (Table 2). There was a very good correlation between the expression data obtained by qPCR and microarray analysis, confirming the global transcript profiling analysis. The analysis highlights the induction of the key biosynthetic genes from the 9-LOX branch (LOXA, AOS3, and DES) in mycorrhizal roots, the induction of LOXA and AOS3 being markedly higher in G. mosseae-colonized roots (Table 2), and the striking differential induction in those roots of the typical JA-regulated, wound-related genes such as PinII and MC.
Table 2.
Expression analyses of marker genes for AM symbiosis and hormone pathways by quantitative real time RT-PCR (qPCR) in roots colonized by G. mosseae (Gm) or G. intraradices (Gi)
| ID | Annotation | Pathway/marker | Microarray |
qPCR |
||
| Gm | Gi | Gm | Gi | |||
| BI933750 | 1-Deoxy-D-xylulose 5-phosphate synthase 2 (DXS-2) | AM | 5.00a | 9.92a | 8.59a | 20.08a |
| U30465 | Class II chitinase (Chi2;1) | AM | 2.41a | 3.01a | 2.70a | 2.23a |
| Y15846 | Pathogenesis protein PR10-like | AM | 1.93a | 2.72a | 4.82a | 5.81a |
| AY885651 | Phosphate transporter LePT4 | AM | – | – | 1141.69a | 2046.48a |
| U09026 | Lipoxygenase A (LOXA) | Oxylipins | 6.59a | 2.78a | 7.46a | 3.38a |
| AF454634 | Allene oxide synthase 3 (AOS3) | Oxylipins | 8.49a | 3.42a | 16.60a | 6.28a |
| AF317515 | Divinyl ether synthase (DES) | Oxylipins | 2.29a | 1.96a | 3.28a | 2.94a |
| U37840 | Lipoxygenase D (LOXD) | JA | 1.27 | 2.17a | 0.79 | 2.35a |
| AJ271093 | Allene oxide synthase 1 (AOS1) | JA | 2.14a | 2.52a | 2.51a | 3.32a |
| AF230371 | Allene oxide synthase 2 (AOS2) | JA | 0.97 | 0.89 | 0.98 | 0.84 |
| AF384374 | Allene oxide cyclase (AOC) | JA | 1.34 | 1.29 | 1.32 | 1.41 |
| AJ278332 | 12-Oxophytodienoate 3 reductase (OPR3) | JA | 0.94 | 1.05 | 1.09 | 1.15 |
| AF083253 | Multicystatin (MC) | JA | 9.27a | 1.64 | 8.35a | 0.76 |
| K03291 | Proteinase inhibitor II (PinII) | JA | 5.92a | 0.94 | 7.39a | 1.06 |
| CN384809 | 1-Aminocyclopropane-1-carboxylic acid oxidase (ACO1) | ET | 2.35a | 2.27a | 0.79 | 1.76 |
| X58885 | Ethylene-forming enzyme (EFE) | ET | 0.69 | 1.13 | 0.61 | 1.54 |
| AY394002 | CTR1-like protein kinase (CTR4) | ET | 1.11 | 1.13 | 0.71 | 0.83 |
| Z97215 | 9-cis-Epoxycarotenoid (NCED1) | ABA | 0.96 | 0.69 | 0.68 | 0.57 |
| X51904 | Le4 | ABA | – | – | 0.82 | 0.48 |
| M69247 | Pathogenesis-related protein PR1a | SA | ND | ND | Ct 30.67 | ND |
Numbers indicate fold change in expression levels compared with non-mycorrhizal controls.
Ct, the threshold cycle; ND. non-detected;– not present in the tomato array.
Significant (P <0.01) changes.
According to the ABA content, the levels of NCED—encoding a 9-cis-epoxycarotenoid enzyme—involved in ABA biosynthesis, and the ABA-inducible gene Le4 (Kahn et al., 1993) remained unaltered in the mycorrhizal plants. The qPCR analysis showed unaltered expression of all the ET-related genes tested, not validating the up-regulation of ACO1 observed in the microarray analysis. In relation to SA, the gene coding for PR1a, a common marker of SA-regulated responses (Uknes et al., 1993), was only detected in G. mosseae-colonized roots (Table 2), supporting the increased SA levels detected previously by LC-MS/MS.
The mycorrhiza-related transcriptome is partially mimicked by MeJA treatment
The induction of genes associated with jasmonates in mycorrhizal roots suggested a possible role for these hormones in the plant response to AMF. To assess whether this is the case, a microarray analysis was carried out to identify JA-regulated genes, and the transcriptional profile was compared with those obtained from mycorrhizal roots. Root treatment with MeJA resulted in 1398 differentially regulated genes (P <0.01), most of them being up-regulated (∼60%) (Supplementary Table S6 at JXB online). Even though two different systems were used for growing the plants, 66% of the genes co-regulated upon G. mosseae and G. intraradices colonization were also regulated by MeJA (Table 1). Among them, the jasmonate biosynthetic and regulatory genes derived from the 13-LOX branch and induced by mycorrhiza—AOS1 and JAZ2—were also significantly (P <0.01) induced by MeJA. Other JA metabolic genes such as AOC, OPR3, and the jasmonic acid carboxyl methyl transferase (JMT), not altered by mycorrhiza, were also up-regulated by MeJA, supporting the reported positive feedback in jasmonate biosynthesis (Fig. 3 and Supplementary Table S6). In addition, the genes involved in defence responses to wounding and herbivory, known to be regulated by JA in aerial parts of the plant (Wasternack et al., 2006), were all up-regulated by MeJA in the roots. Remarkably, as stated before, most of these genes were induced in G. mosseae-mycorrhizal roots (Supplementary Table S5), but not in those colonized by G. intraradices. The genes LOXA, AOS3, and DES, induced by both AMF and acting on the 9-LOX branch, were also induced by MeJA, illustrating a positive regulation of this branch by JA (Table 1 and Supplementary Table S6).
Discussion
It is widely accepted that AM establishment induces transcriptional changes in the host plant (Grunwald et al., 2004; Hohnjec et al., 2005; Liu et al., 2007; Fiorilli et al., 2009), and the involvement of a number of plant hormones in mycorrhiza formation and functioning has been proposed (Hause et al., 2007; Herrera-Medina et al., 2007; Riedel et al., 2008; Fiorilli et al., 2009; Grunwald et al., 2009). However, the precise mechanisms underlying plant–AMF interactions are still unknown. Here, the plant response to the colonization by the two AMF G. mosseae and G. intraradices was analysed using tomato as the model plant. A different level and structure of mycorrhizal colonization were observed when comparing both fungi, G. intraradices being the most effective colonizer. This higher colonization level intensity was accompanied by a larger induction of host genes coding for symbiosis-related elements such as the mycorrhiza-specific phosphate transporter LePT4 and those involved in the biosynthesis of mycorrhiza-related carotenoids (Walter et al., 2007). Differences in the colonization by G. intraradices and G. mosseae isolates have been previously shown in several plant species (Pozo et al., 2002; Feddermann et al., 2008), supporting that at least part of the morphological features in the AM colonization, and probably of the hormonal and transcriptional changes in the host, are related to the AMF genotype.
Simultaneous quantification of several hormones by LC-MS/MS and GC-MS revealed significant changes in the levels of defence-related hormones in mycorrhizal tomato roots, depending on the AMF involved. While ABA and free JA levels remained unaltered in mycorrhizal plants compared with controls, ET and OPDA levels were significantly down- and up- regulated, respectively, during the interaction with both fungi. In contrast, the levels of the JA derivative JA–Ile and SA were elevated exclusively in roots colonized by G. mosseae, the AMF with a lower colonization level. The significant reduction in ET production in roots colonized by both AMF is in agreement with studies in other mycorrhizal plants and during plant interaction with other beneficial endophytic fungi (Barazani et al., 2007; Riedel et al., 2008). This reduction, together with data from pharmacological and genetic approaches to analyse the role of ET in the symbiosis, indicates that a precise regulation of ET levels is required for AM establishment (Azcón-Aguilar et al., 1981; Zsogon et al., 2008).
As mentioned above, OPDA levels were elevated in roots colonized by both AMF. Surprisingly, the higher content of OPDA was not accompanied by increased levels of free JA. Free JA increased up to several fold in roots of barley, cucumber, M. truncatula, and soybean upon mycorrhization (Hause et al., 2002; Vierheilig and Piche, 2002; Meixner et al., 2005; Stumpe et al., 2005). Accordingly, a conserved role for JA in the establishment and functionality of the AM symbiosis was proposed. However, analysis of the JA content in N. attenuata showed unaltered levels in mycorrhizal roots (Riedel et al., 2008). It is also noteworthy that no changes in OPDA content were found in mycorrhizal barley or Medicago roots (Hause et al., 2002; Stumpe et al., 2005). Therefore, it is possible that OPDA and other oxylipins, but not free JA, are the main players in orchestrating the plant response to mycorrhizal fungi in the Solanaceae. In this regard, organ- and plant species-specific patterns of accumulation of different JA-related compounds have been described, tomato being among the plants with higher levels of OPDA relative to the JA concentration before and after wounding (Miersch et al., 2008) and in response to pathogen attack (Vicedo et al., 2009).
At the transcriptional level, some of the genes related to the biosynthesis and metabolism of jasmonates showed a moderate increase in their expression in mycorrhizal roots. LOXD and AOS1 are involved in the early steps of the pathway, so their higher expression levels might support the increase in the JA precursor 12-OPDA. Induction of the JA biosynthetic genes AOS and AOC was previously shown in arbusculated cells in barley and M. truncatula roots (Hause et al., 2002). However, changes in the expression of AOC and later biosynthetic genes were not detected in the tomato array. The picture is more complex because, besides JA, other oxylipins play important roles in biological processes such as plant defence and development (Wasternack, 2007; Mosblech et al., 2009). Interestingly, the microarray analysis revealed a strong induction of the genes coding for LOXA and AOS3, key enzymes in the 9-LOX branch of the oxylipin pathway which give rise to the formation of ketols and 10-OPDA. 10-OPDA is a structural isomer of the JA precursor 12-OPDA (Itoh et al., 2002; Grechkin et al., 2008). Because of the transcriptional activation of the 9- and 13-LOX branches in mycorrhizal roots and the inability of LC-MS/MS to discriminate between the two isomers, it is likely that the increased OPDA levels observed correspond to a mixture of both isomers. In addition to its function as a JA precursor, 12-OPDA per se plays a role in plant defence signalling, regulating the expression of a specific subset of genes (Stintzi et al., 2001; Taki et al., 2005). Despite the lack of information about 10-OPDA in vivo, a similar role for this OPDA isomer in plant defence has been postulated (Itoh et al., 2002). Further supporting the activation of the 9-LOX branch, up-regulation of DES in mycorrhizal tomato roots was also found. The encoded enzyme catalyses the biosynthesis of colnelenic and colneleic acids, for which a role in defence against plant pathogens has been proposed in tobacco and potato (Mosblech et al., 2009). It is noteworthy that the 9-LOX branch is largely root specific. Indeed, AOS3 is exclusively expressed in the roots (Itoh et al., 2002), and LOXA and DES show only very low basal expression levels in shoots (Ferrie et al., 1994; Itoh and Howe, 2001). Recently, the relevance of the 9-LOX pathway in plant interactions with nematodes and pathogens has been demonstrated (Vellosillo et al., 2007; Gao et al., 2008). Thus, it is plausible that the activation of the 9-LOX pathway is part of the strategy of the plant to control AMF development within the roots. Supporting this hypothesis, increased mycorrhization levels were found in the tomato mutant jai1 (Herrera-Medina et al., 2008; JAL-R et al., unpublished data), in which AOS3 expression is undetectable (Itoh et al., 2002). Moreover, although LOXA and AOS3 were significantly induced in roots colonized by both AMF, a higher up-regulation was found in the interaction with the lower colonization rate, tomato–G. mosseae. As a side effect, higher levels of 9-LOX-derived products may be responsible for the enhanced resistance to root pathogens in mycorrhizal plants. We previously compared G. mosseae and G. intraradices in terms of their ability to protect tomato plants against Phytophthora parasitica var. nicotianae, and only G. mosseae efficiently induced resistance (Pozo et al., 2002). More recently, resistance to this pathogen has been demonstrated to depend on the 9-LOX branch in tobacco roots (Fammartino et al., 2007). Accordingly, the stronger activation of this branch of the oxylipin pathway in G. mosseae-colonized roots may contribute to the enhanced resistance of these roots against P. parasitica.
In addition to the higher up-regulation of 9-LOX biosynthetic genes, G. mosseae differentially triggered other metabolic changes related to defence in tomato roots. Only roots colonized by this AMF showed a moderate but significant increase in the levels of JA–Ile, one of the most active forms of JA with specific biological roles (Staswick and Tiryaki, 2004; Fonseca et al., 2009). JA–Ile is formed by the action of JAR1, which catalyses the conjugation of JA to isoleucine synthesized from threonine by a threonine deaminase (TD). Remarkably, a gene encoding a TD was up-regulated >6-fold in roots colonized by G. mosseae, but not by G. intraradices, which is in agreement with the differences in JA–Ile content between roots colonized by the two AMF. In N. atenuatta the defensive role of this enzyme is linked to its mediation of JA–Ile signalling, leading to the accumulation of direct defences such as protease inhibitors (Kang et al., 2006). In agreement with the elevated levels of JA–Ile only in G. mosseae-colonized roots, all wound-inducible marker genes (coding for proteinase inhibitors I and II, polyphenol oxydase, arginase 2, multicystatin, etc.) were up-regulated exclusively in those roots. It is noteworthy that only JA–Ile, and not JA or OPDA, is able to promote interaction of the SCFcoi1 ubiquitin ligase complex and JAZ proteins in Arabidopsis. This interaction liberates MYC2, a positive regulator of JA (reviewed in Memelink, 2009) which is essential in rhizobacteria-induced systemic resistance (Pozo et al., 2008). Thus, it is tempting to speculate that elevated JA–Ile levels and related transcripts in G. mosseae-colonized plants may be related to its ability to induce mycorrhiza-induced resistance.
As for JA–Ile, SA content and expression of its marker gene PR1a were significantly elevated only in G. mosseae-colonized roots. SA is a key phytohormone in the regulation of plant defence responses, especially in interactions with biotrophic pathogens. In agreement with the biotrophic character of AMF, a negative regulatory role of SA in the AM symbiosis has been proposed (Gutjahr and Paszkowski, 2009). Indeed, an inverse correlation between SA levels and AM colonization was found in pea and tobacco (Blilou et al., 1999; Herrera-Medina et al., 2003). Accumulation of SA in G. mosseae-colonized barley roots has also been described (Khaosaad et al., 2007). Therefore, the enhanced SA levels could modulate the plant control of AMF proliferation within the roots. Additionally, because of the role of SA in regulating systemic defence responses and induced resistance (Vlot et al., 2009), the increase in SA levels might be key in the induction of resistance by mycorrhiza.
Taken together, the results show common and divergent responses of the plant to mycorrhizal colonization by different AMF at the hormonal and transcriptional levels. Remarkably, the differences in the response are very significant: only ∼35% of the genes regulated during the interaction with G. mosseae and G. intraradices overlap. A similar overlap (∼30%) was found in the response of M. truncatula to the same fungi (Hohnjec et al., 2005). The differential responses found in tomato plants interacting with G. mosseae, namely stronger induction of the oxylipin 9-LOX branch, increased SA and JA–Ile contents, and the associated induction of Pr1a and jasmonate-related defence genes points to a more exhaustive control of the fungal partner by the plant that may explain the reduced colonization level of G. mosseae when compared with G. intraradices. Moreover, it might be that these changes in the host also contribute to the bioprotection ability of this AMF.
In conclusion, it is shown here that the maintenance of AM symbiosis implies changes in the content of several phytohormones, which correlate with changes in the expression of genes involved in their biosynthesis and the responses they regulate. A crucial role for oxylipins in this mutualistic symbiosis has been proposed for the first time, and indications of their regulation by jasmonates suggested. Additionally, a different plant response to the colonization by particular AMF is demonstrated, which may underlie the differential impact of individual AMF on plant physiology and, particularly, on its ability to cope with biotic stresses. Further research is required to elucidate the role of the 9-LOX-derived oxylipins, as well as the metabolic and transcriptional changes needed for the long-term maintenance of the AM symbiosis and its benefits to the host.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Scheme of the biosynthesis of the apocarotenoids cyclohexenone and mycorradicin in mycorrhizal roots (adapted from Walter et al., 2007). Thick arrows indicate significant up-regulation of the corresponding genes as determined by the transcriptomic analysis of mycorrhizal roots (G. mosseae or G. intraradices colonized) compared with non-mycorrhizal controls; = indicates no changes in gene expression. DXS-2, 1-deoxy-D-xylulose 5-phosphate synthase 2; DXR, 1-deoxy-D-xylulose reductase; PSY, phytoene synthase; PDS, phytoene desaturase; ZDS, zeta-carotene desaturase; CrtL-b, lycopene cyclase b; and CCD1, carotenoid cleavage dioxygenase 1.
Table S1. Primer sequences used in the real-time qPCR analysis.
Table S2. Mycorrhization, root and shoot fresh weights, and phosphorus content in tomato plants colonized by G. mosseae (Gm) or G. intraradices (Gi). Nm indicates control non-mycorrhizal plants.
Table S3. Expression level of genes involved in hormone metabolism in roots colonized by G. mosseae (Gm) or G. intraradices (Gi).
Table S4. Genes specifically induced or repressed in tomato roots colonized by G. intraradices (Gi). Fold change in their expression level compared with non-mycorrhizal plants or upon methyl jasmonate (MeJA) treatment.
Table S5. Genes specifically induced or repressed in tomato roots colonized by G. mosseae (Gm). Fold change in their expression level compared with non-mycorrhizal plants or upon methyl jasmonate (MeJA) treatment.
Table S6. Genes showing significant differential expression in tomato roots upon treatment with 50 μM MeJA.
Supplementary Material
Acknowledgments
This work was supported by grants AGL2006-08029 from the National R&D Plan of the MINCIN and PERG-02-2007-224751 from the Marie Curie programme from the European Commission. JAL-R is supported by a postdoctoral contract (JAE-Doc) from the CSIC, and IF by a PhD fellowship from the MINCIN. We thank Jesús García-Cantalejo, Unidad de Genómica del Parque Científico-Universidad Complutense, Madrid for the microarray analyses, the SCIC Universitat Jaume I for technical assistance in hormone determinations, and the CEBAS-CSIC for phosphorus measurements. We acknowledge Ted Farmer from the University of Lausanne and B Mauch-Mani from the University of Neuchatel for providing the JA–Ile standard.
References
- Azcón-Aguilar C, Rodríguez-Navarro DN, Barea JM. Effects of ethrel on the formation and responses to VA mycorrhiza in Medicago and Triticum. Plant and Soil. 1981;60:461–468. [Google Scholar]
- Baldi P, Long AD. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics. 2001;17:509–519. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- Balestrini R, Gomez-Ariza J, Lanfranco L, Bonfante P. Laser microdissection reveals that transcripts for five plant and one fungal phosphate transporter genes are contemporaneously present in arbusculated cells. Molecular Plant-Microbe Interactions. 2007;20:1055–1062. doi: 10.1094/MPMI-20-9-1055. [DOI] [PubMed] [Google Scholar]
- Barazani O, Von Dahl CC, Baldwin IT. Sebacina vermifera promotes the growth and fitness of Nicotiana attenuata by inhibiting ethylene signaling. Plant Physiology. 2007;144:1223–1232. doi: 10.1104/pp.107.097543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blilou I, Ocampo JA, Garcia-Garrido JM. Resistance of pea roots to endomycorrhizal fungus or Rhizobium correlates with enhanced levels of endogenous salicylic acid. Journal of Experimental Botany. 1999;50:1663–1668. [Google Scholar]
- Cavagnaro TR, Gao LL, Smith FA, Smith SE. Morphology of arbuscular mycorrhizas is influenced by fungal identity. New Phytologist. 2001;151:469–475. [Google Scholar]
- Fammartino A, Cardinale F, Gobel C, Mene-Saffrane L, Fournier J, Feussner I, Esquerre-Tugaye MT. Characterization of a divinyl ether biosynthetic pathway specifically associated with pathogenesis in tobacco. Plant Physiology. 2007;143:378–388. doi: 10.1104/pp.106.087304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feddermann N, Boller T, Salzer P, Elfstrand S, Wiemken A, Elfstrand M. Medicago truncatula shows distinct patterns of mycorrhiza-related gene expression after inoculation with three different arbuscular mycorrhizal fungi. Planta. 2008;227:671–680. doi: 10.1007/s00425-007-0649-1. [DOI] [PubMed] [Google Scholar]
- Ferrie BJ, Beaudoin N, Burkhart W, Bowsher CG, Rothstein SJ. The cloning of two tomato lipoxygenase genes and their differential expression during fruit ripening. Plant Physiology. 1994;106:109–118. doi: 10.1104/pp.106.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorilli V, Catoni M, Miozzi L, Novero M, Accotto GP, Lanfranco L. Global and cell-type gene expression profiles in tomato plants colonized by an arbuscular mycorrhizal fungus. New Phytologist. 2009;184:975–987. doi: 10.1111/j.1469-8137.2009.03031.x. [DOI] [PubMed] [Google Scholar]
- Flors V, Ton J, van Doorn R, Jakab G, García-Agustín P, Mauch-Mani B. Interplay between JA, SA and ABA signalling during basal and induced resistance against Pseudomonas syringae and Alternaria brassicicola. The Plant Journal. 2008;54:81–92. doi: 10.1111/j.1365-313X.2007.03397.x. [DOI] [PubMed] [Google Scholar]
- Floss DS, Hause B, Lange PR, Kuster H, Strack D, Walter MH. Knock-down of the MEP pathway isogene 1-deoxy-d-xylulose 5-phosphate synthase 2 inhibits formation of arbuscular mycorrhiza-induced apocarotenoids, and abolishes normal expression of mycorrhiza-specific plant marker genes. The Plant Journal. 2008;56:86–100. doi: 10.1111/j.1365-313X.2008.03575.x. [DOI] [PubMed] [Google Scholar]
- Fonseca S, Chico JM, Solano R. The jasmonate pathway: the ligand, the receptor and the core signalling module. Current Opinion in Plant Biology. 2009;12:539–547. doi: 10.1016/j.pbi.2009.07.013. [DOI] [PubMed] [Google Scholar]
- Gao XQ, Starr J, Gobel C, Engelberth J, Feussner I, Tumlinson J, Kolomiets M. Maize 9-lipoxygenase ZmLOX3 controls development, root-specific expression of defense genes, and resistance to root-knot nematodes. Molecular Plant-Microbe Interactions. 2008;21:98–109. doi: 10.1094/MPMI-21-1-0098. [DOI] [PubMed] [Google Scholar]
- Genre A, Chabaud M, Faccio A, Barker DG, Bonfante P. Prepenetration apparatus assembly precedes and predicts the colonization patterns of arbuscular mycorrhizal fungi within the root cortex of both Medicago truncatula and Daucus carota. The Plant Cell. 2008;20:1407–1420. doi: 10.1105/tpc.108.059014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannetti M, Mosse B. Evaluation of techniques for measuring vesicular arbuscular mycorrhizal infection in roots. New Phytologist. 1980;84:489–500. [Google Scholar]
- Grechkin AN, Mukhtarova LS, Latypova LR, Gogolev Y, Toporkova YY, Hamberg M. Tomato CYP74C3 is a multifunctional enzyme not only synthesizing allene oxide but also catalyzing its hydrolysis and cyclization. Chembiochem. 2008;9:2498–2505. doi: 10.1002/cbic.200800331. [DOI] [PubMed] [Google Scholar]
- Grunwald U, Guo WB, Fischer K, Isayenkov S, Ludwig-Muller J, Hause B, Yan XL, Kuster H, Franken P. Overlapping expression patterns and differential transcript levels of phosphate transporter genes in arbuscular mycorrhizal, Pi-fertilised and phytohormone-treated Medicago truncatula roots. Planta. 2009;229:1023–1034. doi: 10.1007/s00425-008-0877-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunwald U, Nyamsuren O, Tarnasloukht M, Lapopin L, Becker A, Mann P, Gianinazzi-Pearson V, Krajinski F, Franken P. Identification of mycorrhiza-regulated genes with arbuscule development-related expression profile. Plant Molecular Biology. 2004;55:553–566. doi: 10.1007/s11103-004-1303-y. [DOI] [PubMed] [Google Scholar]
- Guether M, Balestrini R, Hannah M, He J, Udvardi MK, Bonfante P. Genome-wide reprogramming of regulatory networks, transport, cell wall and membrane biogenesis during arbuscular mycorrhizal symbiosis in Lotus japonicus. New Phytologist. 2009;182:200–212. doi: 10.1111/j.1469-8137.2008.02725.x. [DOI] [PubMed] [Google Scholar]
- Gutjahr C, Paszkowski U. Weights in the balance: jasmonic acid and salicylic acid signaling in root–biotroph interactions. Molecular Plant-Microbe Interactions. 2009;22:763–772. doi: 10.1094/MPMI-22-7-0763. [DOI] [PubMed] [Google Scholar]
- Hause B, Maier W, Miersch O, Kramell R, Strack D. Induction of jasmonate biosynthesis in arbuscular mycorrhizal barley roots. Plant Physiology. 2002;130:1213–1220. doi: 10.1104/pp.006007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hause B, Mrosk C, Isayenkov S, Strack D. Jasmonates in arbuscular mycorrhizal interactions. Phytochemistry. 2007;68:101–110. doi: 10.1016/j.phytochem.2006.09.025. [DOI] [PubMed] [Google Scholar]
- Hause B, Schaarschmidt S. The role of jasmonates in mutualistic symbioses between plants and soil-born microoganisms. Phytochemistry. 2009;70:1589–1599. doi: 10.1016/j.phytochem.2009.07.003. [DOI] [PubMed] [Google Scholar]
- Herrera-Medina MJ, Gagnon H, Piche Y, Ocampo JA, Garcia-Garrido JM, Vierheilig H. Root colonization by arbuscular mycorrhizal fungi is affected by the salicylic acid content of the plant. Plant Sciences. 2003;164:993–998. [Google Scholar]
- Herrera-Medina MJ, Steinkellner S, Vierheilig H, Ocampo JA, Garcia-Garrido JM. Abscisic acid determines arbuscule development and functionality in the tomato arbuscular mycorrhiza. New Phytologist. 2007;175:554–564. doi: 10.1111/j.1469-8137.2007.02107.x. [DOI] [PubMed] [Google Scholar]
- Herrera-Medina MJ, Tamayo MI, Vierheilig H, Ocampo JA, Garcia-Garrido JM. The jasmonic acid signalling pathway restricts the development of the arbuscular mycorrhizal association in tomato. Journal of Plant Growth Regulation. 2008;27:221–230. [Google Scholar]
- Hewitt EJ. Sand and water culture methods used in the study of plant nutrition. Technical communication no. 22. London, UK: Commonwealth Agriculture Bureau; 1966. [Google Scholar]
- Hohnjec N, Vieweg ME, Puhler A, Becker A, Kuster H. Overlaps in the transcriptional profiles of Medicago truncatula roots inoculated with two different Glomus fungi provide insights into the genetic program activated during arbuscular mycorrhiza. Plant Physiology. 2005;137:1283–1301. doi: 10.1104/pp.104.056572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Isayenkov S, Mrosk C, Stenzel I, Strack D, Hause B. Suppression of allene oxide cyclase in hairy roots of Medicago truncatula reduces jasmonate levels and the degree of mycorrhization with Glomus intraradices. Plant Physiology. 2005;139:1401–1410. doi: 10.1104/pp.105.069054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh A, Howe GA. Molecular cloning of a divinyl ether synthase. Identification as a CYP74 cytochrome P-450. Journal of Biological Chemistry. 2001;276:3620–3627. doi: 10.1074/jbc.M008964200. [DOI] [PubMed] [Google Scholar]
- Itoh A, Schilmiller AL, McCaig BC, Howe GA. Identification of a jasmonate-regulated allene oxide synthase that metabolizes 9-hydroperoxides of linoleic and linolenic acids. Journal of Biological Chemistry. 2002;277:46051–46058. doi: 10.1074/jbc.M207234200. [DOI] [PubMed] [Google Scholar]
- Kahn TL, Fender SE, Bray EA, Oconnell MA. Characterization of expression of drought and abscisic acid-regulated tomato genes in the drought-resistant species Lycopersicon pennellii. Plant Physiology. 1993;103:597–605. doi: 10.1104/pp.103.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JH, Wang L, Giri A, Baldwin IT. Silencing threonine deaminase and JAR4 in Nicotiana attenuata impairs jasmonic acid–isoleucine-mediated defenses against Manduca sexta. The Plant Cell. 2006;18:3303–3320. doi: 10.1105/tpc.106.041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaosaad T, Garcia-Garrido JM, Steinkellner S, Vierheilig H. Take-all disease is systemically reduced in roots of mycorrhizal barley plants. Soil Biology and Biochemistry. 2007;39:727–734. [Google Scholar]
- Liu J, Maldonado-Mendoza I, Lopez-Meyer M, Cheung F, Town CD, Harrison MJ. Arbuscular mycorrhizal symbiosis is accompanied by local and systemic alterations in gene expression and an increase in disease resistance in the shoots. The Plant Journal. 2007;50:529–544. doi: 10.1111/j.1365-313X.2007.03069.x. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2_DDCt method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Meixner C, Ludwig-Muller J, Miersch O, Gresshoff P, Staehelin C, Vierheilig H. Lack of mycorrhizal autoregulation and phytohormonal changes in the supernodulating soybean mutant nts1007. Planta. 2005;222:709–715. doi: 10.1007/s00425-005-0003-4. [DOI] [PubMed] [Google Scholar]
- Memelink J. Regulation of gene expression by jasmonate hormones. Phytochemistry. 2009;70:1560–1570. doi: 10.1016/j.phytochem.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Miersch O, Neumerkel J, Dippe M, Stenzel I, Wasternack C. Hydroxylated jasmonates are commonly occurring metabolites of jasmonic acid and contribute to a partial switch-off in jasmonate signaling. New Phytologist. 2008;177:114–127. doi: 10.1111/j.1469-8137.2007.02252.x. [DOI] [PubMed] [Google Scholar]
- Mosblech A, Feussner I, Heilmann I. Oxylipins: structurally diverse metabolites from fatty acid oxidation. Plant Physiology and Biochemistry. 2009;47:511–517. doi: 10.1016/j.plaphy.2008.12.011. [DOI] [PubMed] [Google Scholar]
- Parniske M. Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nature Reviews Microbiology. 2008;6:763–775. doi: 10.1038/nrmicro1987. [DOI] [PubMed] [Google Scholar]
- Paszkowski U. A journey through signaling in arbuscular mycorrhizal symbioses. New Phytologist. 2006;172:35–46. doi: 10.1111/j.1469-8137.2006.01840.x. [DOI] [PubMed] [Google Scholar]
- Phillips JM, Hayman DS. Improved procedures for clearing roots and staining parasitic and vesicular–arbuscular mycorrhizal fungi for rapid assessment of infection. Transactions of the British Mycology Society. 1970;55:158–161. [Google Scholar]
- Pieterse CMJ, Leon-Reyes A, Van der Ent S, Van Wees SCM. Networking by small-molecule hormones in plant immunity. Nature Chemical Biology. 2009;5:308–316. doi: 10.1038/nchembio.164. [DOI] [PubMed] [Google Scholar]
- Pozo MJ, Azcón-Aguilar C. Unravelling mycorrhiza-induced resistance. Current Opinion in Plant Biology. 2007;10:393–398. doi: 10.1016/j.pbi.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Pozo MJ, Cordier C, Dumas-Gaudot E, Gianinazzi S, Barea JM, Azcón-Aguilar C. Localized versus systemic effect of arbuscular mycorrhizal fungi on defence responses to Phytophthora infection in tomato plants. Journal of Experimental Botany. 2002;53:525–534. doi: 10.1093/jexbot/53.368.525. [DOI] [PubMed] [Google Scholar]
- Pozo MJ, Van Der Ent S, Van Loon LC, Pieterse CMJ. Transcription factor MYC2 is involved in priming for enhanced defense during rhizobacteria-induced systemic resistance in Arabidopsis thaliana. New Phytologist. 2008;180:511–523. doi: 10.1111/j.1469-8137.2008.02578.x. [DOI] [PubMed] [Google Scholar]
- Requena N, Serrano E, Ocon A, Breuninger M. Plant signals and fungal perception during arbuscular mycorrhiza establishment. Phytochemistry. 2007;68:33–40. doi: 10.1016/j.phytochem.2006.09.036. [DOI] [PubMed] [Google Scholar]
- Riedel T, Groten K, Baldwin IT. Symbiosis between Nicotiana attenuata and Glomus intraradices: ethylene plays a role, jasmonic acid does not. Plant, Cell and Environment. 2008;31:1203–1213. doi: 10.1111/j.1365-3040.2008.01827.x. [DOI] [PubMed] [Google Scholar]
- Siciliano V, Genre A, Balestrini R, Cappellazzo G, deWit PJGM, Bonfante P. Transcriptome analysis of arbuscular mycorrhizal roots during development of the prepenetration apparatus. Plant Physiology. 2007;144:1455–1466. doi: 10.1104/pp.107.097980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SE, Read DJ. Mycorrhizal symbiosis. 3rd edn. London: Academic Press; 2008. [Google Scholar]
- Smith SE, Smith FA, Jakobsen I. Functional diversity in arbuscular mycorrhizal (AM) symbioses: the contribution of the mycorrhizal P uptake pathway is not correlated with mycorrhizal responses in growth or total P uptake. New Phytologist. 2004;162:511–524. [Google Scholar]
- Staswick PE, Tiryaki I. The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. The Plant Cell. 2004;16:2117–2127. doi: 10.1105/tpc.104.023549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi A, Weber H, Reymond P, Browse J, Farmer EE. Plant defense in the absence of jasmonic acid: the role of cyclopentenones. Proceeding of the National Academy of Sciences, USA. 2001;98:12837–12842. doi: 10.1073/pnas.211311098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpe M, Carsjens JG, Stenzel I, Gobel C, Lang I, Pawlowski K, Hause B, Feussner I. Lipid metabolism in arbuscular mycorrhizal roots of Medicago truncatula. Phytochemistry. 2005;66:781–791. doi: 10.1016/j.phytochem.2005.01.020. [DOI] [PubMed] [Google Scholar]
- Taki N, Sasaki-Sekimoto Y, Obayashi T, et al. 12-Oxo-phytodienoic acid triggers expression of a distinct set of genes and plays a role in wound-induced gene expression in Arabidopsis. Plant Physiology. 2005;139:1268–1283. doi: 10.1104/pp.105.067058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tejeda-Sartorius M, de la Vega OM, Delano-Frier JP. Jasmonic acid influences mycorrhizal colonization in tomato plants by modifying the expression of genes involved in carbohydrate partitioning. Physiologia Plantarum. 2008;133:339–353. doi: 10.1111/j.1399-3054.2008.01081.x. [DOI] [PubMed] [Google Scholar]
- Uknes S, Dincher S, Friedrich L, Negrotto D, Williams S, Thompson TH, Potter S, Ward E, Ryals J. Regulation of pathogenesis-related protein-1a gene expression in tobacco. The Plant Cell. 1993;5:159–169. doi: 10.1105/tpc.5.2.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellosillo T, Martinez M, Lopez MA, Vicente J, Cascon T, Dolan L, Hamberg M, Castresana C. Oxylipins produced by the 9-lipoxygenase pathway in Arabidopsis regulate lateral root development and defense responses through a specific signaling cascade. The Plant Cell. 2007;19:831–846. doi: 10.1105/tpc.106.046052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicedo B, Flors V, Leyva MD, Finiti I, Kravchuk Z, Real MD, Garcia-Agustin P, Gonzalez-Bosch C. Hexanoic acid-induced resistance against Botrytis cinerea in tomato plants. Molecular Plant-Microbe Interactions. 2009;22:1455–1465. doi: 10.1094/MPMI-22-11-1455. [DOI] [PubMed] [Google Scholar]
- Vierheilig H, Piche Y. Signalling in arbuscular mycorrhiza: facts and hypotheses. In: Buslig B, Manthey J, editors. Flavonoids in cell function. New York: Kluwer Academic/Plenum Publishers; 2002. pp. 23–39. [DOI] [PubMed] [Google Scholar]
- Vlot AC, Dempsey DA, Klessig DF. Salicylic acid, a multifaceted hormone to combat disease. Annual Reviews of Phytopathology. 2009;47:177–206. doi: 10.1146/annurev.phyto.050908.135202. [DOI] [PubMed] [Google Scholar]
- Walter MH, Floss DS, Hans J, Fester T, Strack D. Apocarotenold biosynthesis in arbuscular mycorrhizal roots: contributions from methylerythritol phosphate pathway isogenes and tools for its manipulation. Phytochemistry. 2007;68:130–138. doi: 10.1016/j.phytochem.2006.09.032. [DOI] [PubMed] [Google Scholar]
- Wang L, Allmann S, Wu JS, Baldwin IT. Comparisons of lipoxygenase3- and jasmonate-resistant4/6-silenced plants reveal that jasmonic acid and jasmonic acid–amino acid conjugates play different roles in herbivore resistance of Nicotiana attenuata. Plant Physiology. 2008;146:904–915. doi: 10.1104/pp.107.109264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C. Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Annals of Botany. 2007;100:681–697. doi: 10.1093/aob/mcm079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C, Stenzel I, Hause B, Hause G, Kutter C, Maucher H, Neumerkel J, Feussner I, Miersch O. The wound response in tomato—role of jasmonic acid. Journal of Plant Physiology. 2006;163:297–306. doi: 10.1016/j.jplph.2005.10.014. [DOI] [PubMed] [Google Scholar]
- Zsogon A, Lambais MR, Benedito VA, Figueira AVO, Peres LEP. Reduced arbuscular mycorrhizal colonization in tomato ethylene mutants. Scientia Agricola. 2008;65:259–267. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.